Abstract
Natural chromosomal transformation is one of the primary driving forces of bacterial evolution. This reaction involves the recombination of the internalized linear single-stranded (ss) DNA with the homologous resident duplex via RecA-mediated integration in concert with SsbA and DprA or RecO. We show that sequence divergence prevents Bacillus subtilis chromosomal transformation in a log-linear fashion, but it exerts a minor effect when the divergence is localized at a discrete end. In the nucleotide bound form, RecA shows no apparent preference to initiate recombination at the 3′- or 5′-complementary end of the linear duplex with circular ssDNA, but nucleotide hydrolysis is required when heterology is present at both ends. RecA·dATP initiates pairing of the linear 5′ and 3′ complementary ends, but only initiation at the 5′-end remains stably paired in the absence of SsbA. Our results suggest that during gene transfer RecA·ATP, in concert with SsbA and DprA or RecO, shows a moderate preference for the 3′-end of the duplex. We show that RecA-mediated recombination initiated at the 3′- or 5′-complementary end might have significant implication on the ecological diversification of bacterial species with natural transformation.
INTRODUCTION
In eukaryotes genetic recombination during meiosis involves pairing of the homologous chromosomes that can lead, through sexual reproduction, to a novel set of genetic information that vertically pass on to the offspring. These novel alleles can then be subjected to evolutionary selection, so meiosis and meiotic genetic recombination are important players in the evolution of eukaryotic genomes (1). Bacteria, in contrast, do not have sexual reproduction, but they exchange information by horizontal gene transfer (HGT). Bacterial genomes are remarkably stable, but a substantial part of them is of foreign origin and reshaped by genetic recombination. HGT, which is considered as the major force in evolution, gene loss and gene duplication, can restore genes that are deleted or inactivated by mutations and thereby prevent the stochastic and irreversible deterioration of genomes in an asexual population, known as ‘Muller's ratchet’ (2–4). Most HGT events might be neutral or deleterious and rapidly lost, only an advantageous transfer should lead to increase genomic diversification and speciation. This advantage enables bacteria to reverse Muller’s ratchet by reducing the build-up of deleterious mutations (5), or to gain genome plasticity and genetic diversity in an evolutionary time scale that cannot be obtained through mutational processes alone (6,7).
Bacterial HGT occurs via three basic mechanisms: conjugation, transduction and natural transformation. Conjugation and transduction involve the transfer of DNA segments by bacterial parasites (conjugative plasmids or transposons, infectious viruses or virus-like particles as pathogenicity islands, gene transfer agents, etc.) (2–4). In contrast, natural transformation is a bacterial programmed mechanism of efficient uptake and incorporation of long single-stranded (ss) DNA onto the genome of the recipient bacteria (8). Natural genetic transformation, which is one of the main avenues of HGT for moving chromosomal DNA segments among bacteria, promotes the spread of antibiotic resistance and virulence gene, facilitates antigen variation to escape vaccine action, leads to non-clonal population structure, etc. (2–4). Competence in Bacillus subtilis, which occurs only in a fraction of cells and may be driven by noise, is a transient stress response that allows cells to take up DNA from the environment (8). Unlike other natural competent bacteria, during B. subtilis natural competence development, DNA replication and cell division are halted, leaving haploid cells (8). The expression of RecA, DprA, SsbB and SsbA recombination protein, among many other proteins, are induced, and the membrane specific DNA uptake machinery is built at one of the cell poles (3,4,9). The pole-localized DNA uptake machinery, which can take double-stranded (ds) DNA of any source from the environment, actively processes exogenous dsDNA and internalizes ssDNA with a mean integration length of ≈25 000 nucleotides (nt) (3,4,9). The internalized ssDNA recombine with the recipient duplex, but the molecular mechanism(s) that control this three-strand exchange reaction is poorly understood. Note that, unless stated otherwise, the indicated genes and products are of B. subtilis origin. The nomenclature used to denote the origin of proteins from other bacteria is based on the bacterial genus and species (e.g., Escherichia coli RecA is referred to as RecAEco).
Except SsbA (counterpart of SSBEco), which plays an essential role in DNA replication, the other recombination proteins are dispensable for growth. The RecA, DprA and SsbB factors, which interact with each other and with RecA in vivo, transiently localize to the cell pole, and co-localize with the DNA uptake machinery (10). The localization of SsbA remains elusive. Upon addition of DNA, the RecO mediator also co-localizes with the DNA uptake machinery (11). The absence of RecA reduced chromosomal transformation 104-fold, and the absence of both RecO and DprA lead to a 103-fold reduction of this process (12,13), suggesting that both RecO and DprA play a crucial role in RecA-mediated natural chromosomal transformation.
In E. coli cells, ≈99% of the recombination events occurring at DNA double strand breaks (DSBs) and ≈80% of the recombination events during F’ conjugation require the RecBCD enzyme (functional counterpart of Firmicutes AddAB or Actinobacteria AdnAB) for the generation of a 3′-tailed duplex (Supplementary Figure S1A). The 3′-end dsDNA indirectly determines the direction of filament extension and the polarity of the substrate for RecA-mediated recombination (Supplementary Figure S1A) (14–16). Indeed, RecA from non-natural competent bacteria (e.g. RecAEco) promotes DNA strand exchange in a 5′ → 3′ direction in the presence of ATP (17–20). Conversely, during natural transformation the DNA uptake machinery internalizes linear ssDNA with no need for recombination functions involved in end processing (Supplementary Figure S1B). Indeed, the absence of functions involved in long-range end resection (e.g., AddAB or RecJ) does not significantly affect chromosomal transformation (21).
In Streptococcus pneumoniae competent cells, RecASpn-mediated DNA exchanges occurring from 5′ → 3′, relative to the donor strand, are more efficient than in the opposite direction, thus markers located 5′ to the heterologous region are recovered with ≈4-fold preponderance (22), suggesting that RecA from competent cells might lack intrinsic bias for the 3′-end in vivo. In B. subtilis competent cells, it was shown that ssDNA internalizes in a non-polar fashion (23), and that homologous recombination (HR) is sensitive to heterologies (24–26). Alternatively, RecA might initiate pairing at any DNA region, and once a stable joint is formed, branch migration might proceed through mismatched region and one end is sufficient for efficient RecA-mediated recombination.
In vitro, RecA in the ATP·Mg2+ bound form (denoted as RecA·ATP) cannot catalyse DNA strand exchange in the absence of accessory factors. Similar results were observed when RecA·ATP from other natural competent bacteria (e.g. RecASpn or Deinococcus radiodurans [RecADra]) were analysed (27–31). In the absence of accessory factors, RecA·ATP only promotes DNA pairing and exhibits only trace DNA strand exchange activity (12,13,29). However, the presence of any of the two-component mediator (SsbA and DprA or SsbA and RecO) was necessary and sufficient to activate RecA·ATP to catalyse DNA strand exchange in vitro (32,33). Conversely, the RecA·dATP nucleoprotein filaments (NPFs) efficiently catalyses DNA strand exchange in the absence of accessory factors (12,13,29), as does RecAEco·ATP (17–20).
To gain insight into the molecular basis of chromosomal transformation, which is a three-stranded recombination reaction, we combined genetic and biochemical approaches to examine the ability of RecA to catalyse DNA strand exchange. We show that the chromosomal transformation frequency decreases linearly with increased sequence divergence. A decrease of the minimal efficient processing segment (MEPS, >40 bp) at the position of the selectable marker (+1 position) at either the 3′- or 5′-end did not decrease chromosomal transformation, suggesting that RecA forms stable joints at any DNA region and it processes those intermediates with significant high efficiency. In vitro, using the three-stranded recombination reaction and the two different two-component mediators, SsbA and RecO or DprA, we show that nucleotide-bound RecA catalyses DNA strand exchange if the linear duplex (lds) has either 3′- or 5′-end complementarity to the circular ssDNA (css). The accumulation of final recombinant products (prd, nicked circular [nc] with a duplex tail) was slightly higher when complementarity was at the 3′-end. Similar results were observed when a non-hydrolysable ATP analogue (ATPγS) was provided in place of ATP. DNA substrates with heterology at both ends required nucleotide cofactor hydrolysis for efficient strand exchange. RecA in the dATP bound form (RecA·dATP) recombined DNA even in the absence of accessory factors (28,29). Under this condition, RecA·dATP preferentially recombined a substrate with a 5′-complementary end. Here, the presence of SsbA was sufficient to overcome this bias. We proposed that RecA·ATP initiates DNA pairing with a marginal preferences for 3′-ends over 5′-ends in the presence of any of the two-component mediators. Then, joint molecule (jm) followed by translocation of the donor-recipient heteroduplex and processing of the D-loop intermediate, should increase the diversity of recombination products during HGT. In contrast, during DSB repair, RecA does not have to discriminate between 3′- or 5′- ends, because the end-processing machineries (AddAB or RecJ-RecQ[RecS]-SsbA) generate a 3′-tailed substrate for RecA·ATP to mediate recombination (see Supplementary Figure S1A) (15,16,34,35).
MATERIALS AND METHODS
Strains and plasmids
E. coli BL21(DE3)[pLysS] cells were used for SsbA, DprA and RecO protein over-expression. Plasmids pCB722-borne ssbA, pCB888-borne dprA and pCB669-borne recO gene, under the control of a phage T7 promoter, were used to overexpress SsbA, DprA and RecO proteins, respectively, as described (12,28,36). B. subtilis BG214 (also termed YB886) bearing pBT61, containing recA under the control of its own promoter, were used to overexpress RecA (37).
A single C to T transition mutation at codon 482 in the house-keeping rpoB gene, which encodes the essential β subunit of RNA polymerase, confers resistance to rifampicin (RifR) (38). The His codon 482, of 2997 bp rpoB region, of the indicated Bacilli species or subspecies was substituted by a Tyr codon, leading to the rpoB482 mutant variant. Part of the rpoB482 gene (codon 1 to 999) of Bsu 168 (99,96% identity, 1 mismatch), Bsu W23 (97.53%, 74 mismatches), Bat 1942 (91.65%, 250 mismatches), Bam DSM7 (89.88%, 303 mismatches), Bli DSM13 (85.48%, 435 mismatches) and Bth MC28 (79.17%, 624 mismatches) were in vitro synthesized and cloned into E. coli 2710 bp pUC57 vector (GeneWiz, London, UK). The sequence of the rpoB482 Bacilli variants was confirmed by nucleotide sequencing. The null rok mutation (Δrok) was mobilized onto the B. subtilis BG214 (trpCE metA5 amyE1 rsbV37 xre1 xkdA1 attSPß attICEBs1) by SPP1-mediated generalize transduction, leading to the BG1359 strain.
Transformation assays
Competent cultures were grown as described previously (39). Competent B. subtilis BG214 (rok+) and its isogenic derivative BG1359 (Δrok) were transformed with undigested or SpeI-linearized pUC57-borne rpoB482 region (SpeI site at position 11 of the rpoB482 DNA) from different Bacilli origin with selection for RifR (8 μg/ml). The yield of RifR transformants was corrected for DNA uptake (assayed through the determination of uptake of radioactively labelled DNA into cells grown to competence through DNase I degradation of the labelled DNA), the rate of spontaneous mutations towards RifR and the values obtained were normalized relative to the presence of one single mismatch (rpoB482 from BG214 or BG1359 origin), which is taken as 100 (40,41).
Mapping of integration end-points
BG1359 competent cells were transformed with undigested rpoB482 DNA from the indicated Bacilli species or subspecies with selection for RifR. To map integration end-points, the rpoB482 DNA from the RifR transformants was amplified by polymerase chain reaction (PCR) using primers hybridizing with the region upstream and downstream of the 2997 bp rpoB482 DNA. The resulting PCR products were sequenced, and their nucleotide sequence compared with the one of the recipient BG1359 strain. The presence or the absence of the expected mismatch was used to determine the overall integration length. The maximal integration length that can be detected with rpoB482 Bsu W23 DNA is 2628 bp, because the first mismatch is located at position 350 and the last at position 2978. The maximal integration length with rpoB482 Bat 1942 is 2831 bp, Bam DSM7 is 2790 bp, Bli DSM13 is 2941 bp and Bth 2974 bp.
Enzymes, reagents, DNA and protein purification
All chemicals used in this study were of analytical grade. IPTG was from Calbiochem (Darmstadt, Germany), and polyethyleneimine, DTT, ATP, dATP and ATPγS were from Sigma (Seelze, Germany). DNA restriction enzymes, DNA ligase, etc., were supplied by Roche (Mannheim, Germany). DEAE, Q- and SP-sepharose were from GE healthcare (Madrid, Spain), hydroxyapatite from BioRad (Hercules, USA) and phosphocellulose was from Whatman (Little Chalfont, USA).
The SsbA (18.7 kDa), DprA (32.7 kDa), RecO (29,3 kDa) and RecA (38.0 kDa) proteins were purified as described, to greater than 98% homogeneity (12,28,37). The N-termini of the purified proteins were sequenced by automatic Edman degradation. The molar extinction coefficients for SsbA, DprA, RecO and RecA were calculated as 11 400, 46 500, 19 600 and 15 200 M−1 cm−1, respectively, at 280 nm, as previously described (37). The protein concentrations were determined using the aforementioned molar extinction coefficients. Quantities of RecA and DprA are expressed as moles of protein as monomers, RecO as dimers, and SsbA as tetramers. In the text the protein concentrations are expressed in their stoichiometric ratios with ssDNA, which was expressed as moles of nt, whereas in the figure legends the molar concentrations of proteins and ssDNA are presented.
Plasmid pGEM3Zf (+) was from Promega (Promega Biotech Ibérica, Madrid). A 1175 bp heterologous DNA segment was joined to pGEM3Zf (+) DNA to render the 4374 bp pGEM-1.2 dsDNA. pGEM3Zf (+) and the pGEM-1.2 dsDNA and the pGEM3Zf (+ strand) ssDNA were purified as described (28).
RecA-mediated DNA strand exchange
The 3199 bp KpnI-cleaved pGEM3Zf (+) or 4374 bp PstI-, EcoRI- or NcoI-cleaved pGEM-1.2 dsDNA (20 μM in nts) and homologous circular 3199 nt ssDNA (10 μM) were incubated with the indicated concentrations of protein or protein combinations in Buffer A (50 mM Tris-HCl (pH 7.5), 1 mM DTT, 50 mM NaCl, 10 mM MgOAc [magnesium acetate], 50 μg/ml BSA, 5% glycerol) containing 5 mM (d)ATP or ATPγS for 60 min at 37°C in a final volume of 20 μl. A (d)ATP regeneration system (8 units/ml creatine phosphokinase and 8 mM phosphocreatine) was included when (d)ATP was used. The samples were deproteinized as described (36,42), and separated by 0.8% agarose gel electrophoresis (AGE) with ethidium bromide. The signal was quantified using a Gel Doc (BioRad) system as described (29).
Scanning force microscopy (SFM) of recombination intermediates
Recombination intermediates were phenol-chloroform treated and precipitated by ethanol. Samples were diluted to 50 ng/μl in TE (10 mM Tris-HCl, pH 7.5, 1 mM EDTA). Before deposition, DNA intermediates (50 ng) were incubated with 200 nM SSBEco (Promega, Wisconsin, USA) in 10 mM Tris-HCl, pH 7.5, 50 mM NaCl, 10 mM MgCl2, and 1 mM DTT for 10 min at 37°C. The DNA (5 ng) was deposited on freshly cleaved mica in the presence of HEPES, pH 8.0, and 50 μM spermidine. After 1 min the mica was rinsed with milli Q water and dried with filtered air. Samples were imaged in air by tapping mode SFM using a Nanoscope III or IV (Digital Instruments; Santa Barbara, USA). Silicon tips (ACT-W) with resonance frequency 300 kHz were from AppNano (Mountain View, USA). Images were collected at 2 μm × 2 μm, and processed only by flattening to remove background slope.
RESULTS
Experimental design
A key feature of HR in E. coli cells is RecAEco-mediated jm formation and strand exchange between 3′-tailed duplex DNA and intact homologous daughter or sister dsDNA by a polar reaction that displace the 5′-end of the plus strand from the duplex molecule (17–20). However, during natural B. subtilis chromosomal transformation the incoming ssDNA shows no polarity (23). To address whether RecA promotes polar formation of heteroduplex joints, and if RecA-mediated recombination requires homology at each DNA end for efficient DNA strand exchange (25,26), we examined the effects of sequence divergence during natural B. subtilis chromosomal transformation.
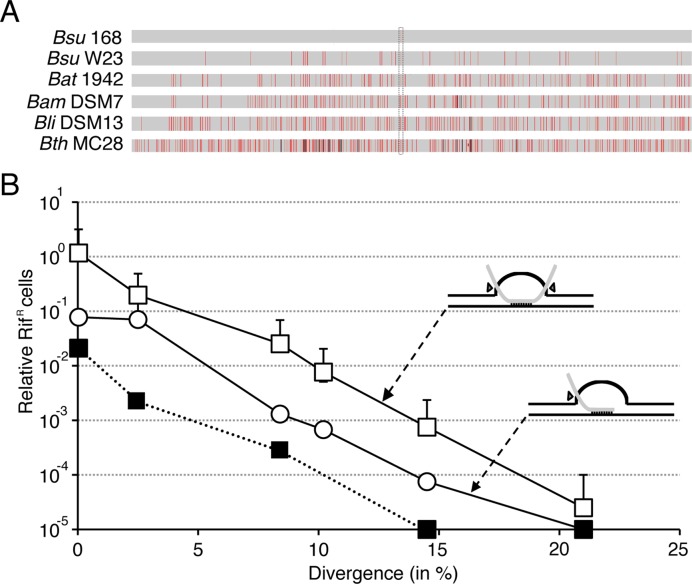
Natural transformation with less than 3% heterologous DNA occurred at very low frequency for lys3, trpC2 or metB10 auxotrophic markers, but transformation to RifR was 100- to 1000-fold more efficient that transformation to prototrophy (24). To avoid any potential expression problem and any polar effect of downstream genes within the operon the first 999 codons of the 1194 codon long rpoB482 gene were used. The RifR mutation is located at position 1443 of the 2997 bp rpoB482 DNA (Figure 1A).
Figure 1.
Average chromosomal transformation frequencies as a function of sequence divergence (in %) from recipient. (A) The rpoB482 DNA was derived from B. subtilis 168 (Bsu 168, 99.96% identity), B. subtilis W23 (Bsu W23, 97.53%), B. atrophaeus 1942 (Bat 1942, 91.65%), B. amyloliquefaciens DSM7 (Bam DSM7, 89.88%), B. licheniformis DSM13 (Bli DSM13, 85.48%) and B. thuringiensis MC28 (Bth MC28, 79.17%). The rpoB482 mutation, which confers resistance to rifampicin (RifR) is framed by dotted lines. Mismatches between RifR donor and the corresponding rpoB in the RifS recipient strain are indicated by vertical red bars and an insertion/deletions by vertical black bars. If two mismatches are located below the MEPS distance they count as a single red bar, the thickness of the bar represent the number of mismatches in the neighbourhood. (B) The undigested rpoB482 DNA (empty or filled squares) or linear SpeI-cleaved rpoB482 DNA (empty circle) was used to transform BG1359 (Δrok) (empty circles and squares) and its isogenic parental strain BG214 (filled squares) competent cells with selection for RifR. All values are means from 3 to 5 independent experiments. In the upper and lower inserts the black lines represent the recipient chromosomal DNA (a supercoiled molecule), and the grey line the homologous donor linear ssDNA. If the ssDNA was randomly linearized by the uptake machinery the transformation heteroduplex would form a double-ended D-loop structure (upper insert), but a one-ended D-loop would be observed if the DNA was in vitro linearized within the rpoB482 DNA (lower insert). The pointing triangles indicate the simultaneous incision (upper insert) or asynchronous incision (bottom insert) by a putative D-loop resolvase.
To gain insight in the natural transformation process and to warrant that only one recipient chromosome is present in the recipient the B. subtilis BG214 strain was chosen. This strain lacks restriction-modification systems, resident prophages, and conjugative elements that could work as a barrier to DNA strand exchange (3). The frequency of RifR transformants of BG214 competent cells decreased in a log-linear mode up to ≈8% divergence (200–400-fold), but the log-linear relationship broke down at higher sequence divergence (25), suggesting that the genetic exchange barrier can become ‘saturated’ and the HR machinery was insensitive to a sequence divergence higher than 8%. An alternative explanation is that this biphasic curve might simply be due to limited resolution of the system and the recombination machinery is sensitive to heterology at high sequence divergence. To test this assumption the proportion of competent cells was increased by up-regulating one of the control mechanisms of competence development, and the frequency of RifR transformants was measured.
Competence development is a bi-stable process, and only a limited number of cells accumulate the ComK competence master regulator, leading ultimately to development of competence in up to 10% of total isogenic cells (9). One way to increase the proportion of cells that become competent is to increase the level of the ComK regulator, by removing Rok, which represses comK transcription (43). The absence of Rok, which does not affect either the DNA uptake or HR, markedly increases the subpopulation of cells that became competent to take up exogenous DNA (up to 50% of total cells) (43). As expected, the chromosomal transformation frequency was 20- to 50-fold higher in the BG1359 Δrok strain (empty symbols) when compared to the parental BG214 (rok+) strain (filled squares) (Figure 1B) (43).
Competent cells take up linear ssDNA from any origin with similar efficiency (8). The DNA uptake machinery introduces a random DSB in the duplex circular rpoB482 DNA substrate. Then, one of the strands is degraded and the complementary one is internalized (3,4,9). SsbA, SsbB or SsbA and SsbB coated the internalized ssDNA. Then DprA or RecO, in concert with SsbA, nucleates and activates RecA to polymerize onto the incoming linear ssDNA (see Supplementary Figure S1B) (32,33). RecA forms a pre-synaptic filament that invades a homologous region on the bacterial genome, forming a two-ended displacement loop (D-loop) intermediate (Figure 1B, upper drawing). Simultaneous incision of donor and recipient DNA by a putative D-loop resolvase would be followed by gap filling by DNA polymerase I and joining of the donor marker by DNA ligase to generate the recombinant product (Supplementary Figure S1B, step d) (3).
The yield of RifR transformants (transformation frequency ≈1 × 10−3 or 1.1 × 105 RifR/colonies forming unit [CFU]) of BG1359 competent cells with undigested circular Bsu 168 DNA (1 single mismatch) (Figure 1B, empty squares) was 10- to 20-fold higher compared to SpeI-linearized, rpoB482 DNA (Figure 1B, empty circles and Supplementary Figure S1B, step e). The SpeI restriction site maps at position 11 of the 2997 bp rpoB482 DNA. Similar results were observed when other restriction enzymes were used to linearize the circular molecule within the rpoB482 coding sequence (data not shown).
The lower yield of transformants with SpeI-linearized rpoB482 DNA (Figure 1B, empty circles) compared to undigested DNA (Figure 1B, empty squares) it is likely due to a defect at the post-synaptic level. In the latter case ≈50% of the internalized ssDNA molecules should have heterologous region at both ends leading to a two-ended D-loop structure (Figure 1B, upper insert and Supplementary Figure S1B, step d), whereas in the SpeI-cleaved condition the uptake cleavage should reduce the heterologous sequence to only one end, and a one-ended D-loop would be formed (Figure 1B, lower insert and Supplementary Figure S1B, step e). It was postulated that a simultaneous incision of the internalized strand and the displaced strand of the same polarity (denoted by triangles, Figure 1B, upper insert and Supplementary Figure S1B), by putative resolvase, might facilitate removal of the recipient displaced strand followed by ligation of the internalize strand (Supplementary Figure S1B, step d). In contrast, in the SpeI-cleaved condition, where the heterologous sequence was present to only one DNA end, a tailed intermediated would be formed (Supplementary Figure S1B, step e). The resection of this poorly understood event was not further analysed.
Sequence divergence decrease the frequency of chromosomal transformation
To investigate the effect of nucleotide sequence divergence on B. subtilis chromosomal transformation a 2997 bp rpoB482 DNA, derived from different Bacilli species, was used to transform BG1359 (Δrok) or BG214 (rok+) competent cells (see Material and methods). The sequence divergence, mainly to synonymous substitution, derived from B. subtilis 168 (Bsu 168, 1 mismatch, the rpoB482 mutation); B. subtilis subsp. spizizenii str. W23 (Bsu W23, 74 mismatches); B. atrophaeus 1942 (Bat 1942, 250 mismatches); B. amyloliquefaciens DSM7 (Bam DSM7, 303 mismatches); B. licheniformis DSM13 (Bli DSM13, 435 mismatches) and B. thuringiensis MC28 (Bth MC28, 624 mismatches/insertion/deletion) (Figure 1A). Except Bth MC28, which shares 90% sequence identity at the amino acid level, the remaining RpoB proteins share 98–99% sequence identity.
To determine the extent to which randomly distributed sequence divergence (Figure 1A) decreased heterogamic transformation, rpoB482 DNA was used to transform Δrok or rok+ competent cells. As revealed in Figure 1B, the transformation frequency of Δrok (empty symbols) or rok+ (filled symbol) competent cells for the RifR phenotype decreased exponentially with a log linear increase in sequence divergence. The transformation efficiency of rok+ (Figure 1B, filled symbol) was low because the proportion of total competent cells was lower. Thus, the Δrok strain significantly increases the limit of detection of the transformation frequency, and was used in our experiments.
The transformation efficiency of undigested rpoB482 DNA (RifR) with a sequence divergence of 2.4% (Bsu W23) and of 8.3% (Bat 1942) decreased ≈5-fold and ≈50-fold, respectively (Figure 1B, empty squares). Sequence divergence of 10.1% (Bam DSM7), 14.5% (Bli DSM13) and 20.8% (Bth MC28) decreased transformation efficiency from ≈150-, ≈1500- and ≈45 000-fold, respectively (Figure 1B, empty squares). A similar decrease in transformation frequency with SpeI-linearized rpoB482 DNA was observed (Figure 1B, empty circles), but here transformation efficiency was lower. It is likely that: i) the genetic exchange barrier was sensitive to sequence divergence higher than 8%, and it was not saturated using the Δrok strain (see above); and ii) the transformation frequency with linear or undigested DNA showed a similar exponential decrease with a log linear increase in the sequence divergence.
The strong reduction in the transformation frequency with Bth MC28 DNA might be due to the sum of two, non-mutually exclusive, effects: first sequence divergence reduced the integration tract. Alternatively, the low transformation efficiency of the Bth MC28 donor DNA could be attributed to compromised expression of the essential rpoB gene by 6% increase in the dC + dG content of the B. thuringiensis source, poor assembly of the β subunits of the RNA polymerase due to 10% protein sequence divergence, or to both leading to decrease in the fitness of the recipient cell. We consider these hypotheses unlikely because a plasmid-borne B. thuringiensis rpoB482 gene confers RifR to the BG1359 strain without apparent fitness cost when compared to cells bearing the empty vector (data not shown).
Sequence divergence decrease the length of integration of donor DNA
To gain insight into this transformation barrier the length of integrated DNA was measured by determining the presence and absence of the mismatch at each side of the donor DNA, and indirectly the endpoint of integration can be deduced. The sequence divergence is nearly randomly distributed on the Bsu W23 or Bat 1942 rpoB482 DNA when compared with Bsu 168 rpoB482 DNA (Figure 1A). The length of integration decreased by increasing the sequence divergence (Table 1). The mean integration length of Bsu W23 and Bat 1942 rpoB482 DNA was ≈2300 bp (range from 2147 bp to 2436 bp) and ≈933 bp (range from 577 bp to 1060 bp), respectively, and under this condition 62–70 and 54–96 mismatches were still integrated with a sequence divergence ≈2.9% and ≈9.4%, respectively (Table 1). The integration length of Bam DSM7 and Bli DSM13 rpoB482 DNA was ≈349 bp (range 299–385 bp) and ≈230 bp (range 170–270 bp), and 30–42 and 24–38 mismatches were still integrated with a sequence divergence ≈10.5% and ≈14.5%, respectively. The sequence divergence of the incoming Bsu W23 DNA [2.4%], Bat 1942 DNA [8.3%]; Bam DSM7 [10.1%] and Bli DSM13 [14.5%] was similar to the observed heterology of the transformants (Table 1). It is likely that there was no mismatch correction. This is consistent with the observation that in the absence of methyl-directed mismatch repair and DNA replication, mismatch correction cannot discriminate the strand to be corrected (44).
Table 1. Length of integration of donor DNA.
| Donor DNAa | Divergence | Left end (in bp) | Right end (in bp) | Integration | Observed divergence | ||
|---|---|---|---|---|---|---|---|
| (in %) | recipient | donor | donor | recipient | (in bp) | (in %) | |
| Bsu W23 | 2.47b | 1 | 350 | 2501 | 2567 | ≈2359 | 2.5 |
| 1 | 350 | 2294 | 2567 | ≈2256 | 2.9 | ||
| 395 | 605 | 2642 | 2651 | ≈2147 | 3.0 | ||
| 395 | 605 | 2690 | 2918 | ≈2304 | 3.0 | ||
| 395 | 605 | 2924 | 2948 | ≈2436 | 2.9 | ||
| Bat 1942 | 8.35 | 479 | 608 | 1508 | 1590 | ≈1006 | 8.9 |
| 479 | 608 | 1590 | 1598 | ≈1060 | 8.9 | ||
| 932 | 941 | 2133 | 2165 | ≈1213 | 10.0 | ||
| 920 | 932 | 1733 | 1736 | ≈ 809 | 8.8 | ||
| 878 | 896 | 1463 | 1464 | ≈ 577 | 9.7 | ||
| Bam DSM7 | 10.12 | 1172 | 1184 | 1535 | 1590 | ≈ 385 | 10.6 |
| 1226 | 1238 | 1535 | 1590 | ≈ 385 | 10.0 | ||
| 1175 | 1214 | 1508 | 1590 | ≈ 355 | 8.2 | ||
| 1233 | 1245 | 1535 | 1590 | ≈ 324 | 10.0 | ||
| 1250 | 1277 | 1535 | 1590 | ≈ 299 | 10.4 | ||
| Bli DSM13 | 14.52 | 1184 | 1190 | 1577 | 1589 | ≈ 270 | 15.6 |
| 1328 | 1334 | 1451 | 1463 | ≈ 252 | 13.1 | ||
| 1211 | 1224 | 1451 | 1463 | ≈ 240 | 15.0 | ||
| 1328 | 1334 | 1556 | 1565 | ≈ 230 | 13.0 | ||
| 1328 | 1334 | 1493 | 1508 | ≈ 170 | 14.1 | ||
| Bth MC28 | 20.83 | 1439 | 1443 | 1443 | 1445 | ≈ 3 | 33.3 |
| 1439 | 1443 | 1443 | 1445 | ≈ 3 | 33.3 | ||
| 1439 | 1443 | 1443 | 1445 | ≈ 3 | 33.3 | ||
| 1442 | 1443 | 1443 | 1448 | ≈ 3 | 33.3 | ||
| 1439 | 1443 | 1451 | 1452 | ≈ 9 | 22.2 | ||
aThe length of donor DNA is 2997 bp and the RifR mutation is located at position 1443.
bThe first mismatch is at position 350 of the donor DNA (see Materials and methods).
When the transformants with the Bth MC28 rpoB482 DNA (transformation frequency ≈3 × 10−8) were analysed integration length reduced to ≈4 bp (range 9–3 bp), under this condition 1 or 2 mismatches were still integrated (Table 1). The frequency of RifR is ≈6-fold above the spontaneous mutation rate (≈5 × 10−9) and ≈30-fold higher than the integration rate of 14 bp homologous segment by illegitimate recombination (≈1 × 10−9). It is likely, therefore, that small patches of identity might help to stabilize the invading ssDNA to render a transformation frequency even 10-fold below MEPS. Previously it was reported that the MEPS requires >40 bp both in vivo (45,46) and in vitro (14,47–50).
From these data together we could assume that the sequence divergence barrier decreases the chromosome transformation frequency and the length of integration during genetic exchange.
Bidirectional recombination might facilitate genetic exchange
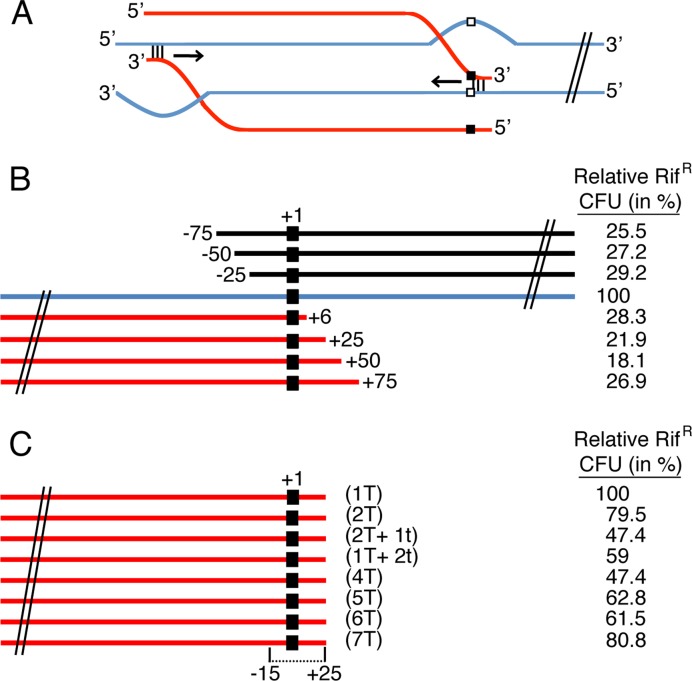
Previously it was suggested that the initiation of RecA-mediated DNA strand transfer is more sensitive to DNA mismatches than the termination step (3). By placing the rpoB482 mutation at a critical distance from one of the transforming DNA ends, the polarity of RecA-mediated DNA strand exchange, and the need for homology at one or at both DNA ends can be addressed. First, we took advantage of the length of a dsDNA molecule to find sequence identity (MEPS), and the high invasiveness of the 3′-end (51) or the 5′-end in other experiments (52) was >40 bp (see above). The rpoB482 mutation [RifR, filled square, +1 position] was placed at the proximal end (defined as Watson strand, Figure 2A) or at the 3′-end distal from the rpoB482 mutation (Crick strand, Figure 2A). For the sake of simplicity recombination initiated at the 5′-end of the internalized ssDNA was not depicted. To do so, the Bsu 168 rpoB482 DNA was PCR amplified to place the rpoB482 point mutation 6 (+6), 25 (+25), 50 (+50) or 75 bp (+75 nucleotides) from the 3′-end on the Watson strand (see Figure 2B, denoted in red). Similarly, the Bsu 168 rpoB482 DNA was PCR amplified to locate the 1 mutation 25 (−25), 50 (−50) or 75 bp (−75 nucleotides) from the 5′-end on the Watson strand (denoted in black).
Figure 2.
Average chromosomal transformation frequencies as a function of the length from 3′- or 5′-end. (A) Cartoon describing a RecA-mediated homology search and DNA pairing of the 3′-ended rpoB482 internalized linear ssDNA (red lanes) distal from the mutation or proximal to the mutation (denoted by a filled square) and the recipient duplex (blue lanes, and rpoB482 denoted as empty square). The horizontal arrow indicates the direction of strand assimilation. Following extension of the heteroduplex initiated at the 3′-end proximal from the mutation (+1 denoted as filled square) would require a 5′ → 3′ polarity, but the distal 3′-end would promote extension in the 3′ → 5′ direction that is opposite to the direction of RecA filament assembly and strand exchange. The opposite would hold with the invasive 5′-end (not depicted). (B) Relative chromosomal transformation frequencies as a function of the distance of the selectable mutation (+1 position denoted as filled square) from the 5′-end (black lane) denoted as -75 to -25 (position in nucleotides), and from the 3′-end (red lane) denoted as +6 to +75. The full-length rpoB482 linearised DNA (denoted in blue) was considered as 100% (8 × 103 CFU/ml). (C) Relative chromosomal transformation frequencies as a function of sequence divergence at the selectable mutation (+1 position) region. The number of transition (T) or transversion (t) mutations at the 3′-end is indicated. All values are means from 3 to 5 independent experiments (the results given are within a 10% standard error).
If HR initiates at the proximal 3′-end of the internalized ssDNA, chromosomal transformation would be inversely proportional to the distance from the transforming mutation (at position +1). However, if transformation only requires complementarity at the initiation site and proceeds bi-directionally, the transformation frequency should be insensitive to the distance of the RifR mutation from the ssDNA end (Figure 2A). The 2997 bp SpeI-cleaved with the +1 mutation, at the centre of the fragment (position 1443), only transformed with 2- to 3-fold higher efficiency than any of the circa 1.5-kb DNAs with the transition +1 mutation at the 3′- or the 5′-end (Figure 2B). Independent of the distance of the mutation from the 3′- or the 5′-end, the frequency of homogamic transformation was similar. To rationalise these results it was assumed that the recombination events that initiate at any end with respect to the internalized ssDNA, or at the central region, might be sufficient for bidirectional chromosomal transformation. Alternatively, the polarity observed in vivo might reflect a balance involving different reaction steps.
Nested DNA mismatches do not significantly decrease the frequency of transformation
Previously it has been shown that a single mismatch near the 5′-end of the incoming strand has a minor effect on RecAEco-mediated recombination, whereas a mismatch near the 3′-end hinders strand exchange 3- to 5-fold in vitro (53). Furthermore, it was suggested that genetic recombination in B. subtilis competent cells requires short regions of conserved sequences at both ends of the donor-recipient DNA (26). Since the DNA uptake machinery either internalizes the Watson or the Crick strand, and the sequence divergence at the distal end should not affect the transformation frequency the mutations surrounding rpoB482, up to 7 neutral mutations, were asymmetrically located, within a 16 bp interval (−15 to +1) and followed by 25 bp homologous DNA at the 3′-end (Watson strand) (Figure 2C). To test whether recombination might initiate at either end or at the central region, the rpoB482 mutation (+1) was placed within the ≈40 bp MEPS. The sequence divergence within the MEPS varied from ≈2.5% (1 mismatch) to ≈17.5% (7 mismatches) (see Figure 2C). Furthermore, a DNA segment with a 7.5% sequence divergence with 1 or 2 transition (T, the rpoB482 mutation) and 1 or 2 transversion (t) mutations was constructed.
Independent of the sequence divergence at the rpoB482 +1 region the frequency was marginally, if at all, affected (Figure 2C). The fate of the circa 1.5-kb donor DNA with 17.5% sequence divergence within the −15 to +1 minimal homology at the 3′-end (mismatches at positions −15 to +1) revealed that the 7 mismatches were integrated in the recipient strain. It is likely, therefore, that there is no significant bias in the DNA strand internalized and recombination might initiate at the 5′-end distal from the mutation and proceeds in the 5′ → 3′ direction with respect to the ssDNA as previously postulated (20). Alternatively, recombination could start at an internal region progressed in the 3′ → 5′ direction with respect to the internalized ssDNA (54). Our assay does not offer sufficient sensitivity to discriminate between these two options.
RecA-mediated bidirectional DNA strand exchange in vitro
The data of the previous section suggest that one DNA end might be sufficient for chromosomal transformation and that recombination between the internalized linear ssDNA and recipient duplex (three-stranded recombination) might be initiated at the 3′- or 5′-end. In contrast, using a three-strand recombination assay it was shown that RecAEco·ATP polymerises with a 5′ → 3′ polarity, and only catalyses unidirectional DNA strand exchange (17–20,55). Although, RecAEco polymerises with a 5′ → 3′ polarity ≈2-fold faster than in the opposite direction and catalyses bidirectional DNA recombination in the presence of ATPγS (47,50,56,57). The directionality of the recombination reaction was addressed using the three-stranded recombination reaction, purified B. subtilis proteins and different nucleotide cofactors.
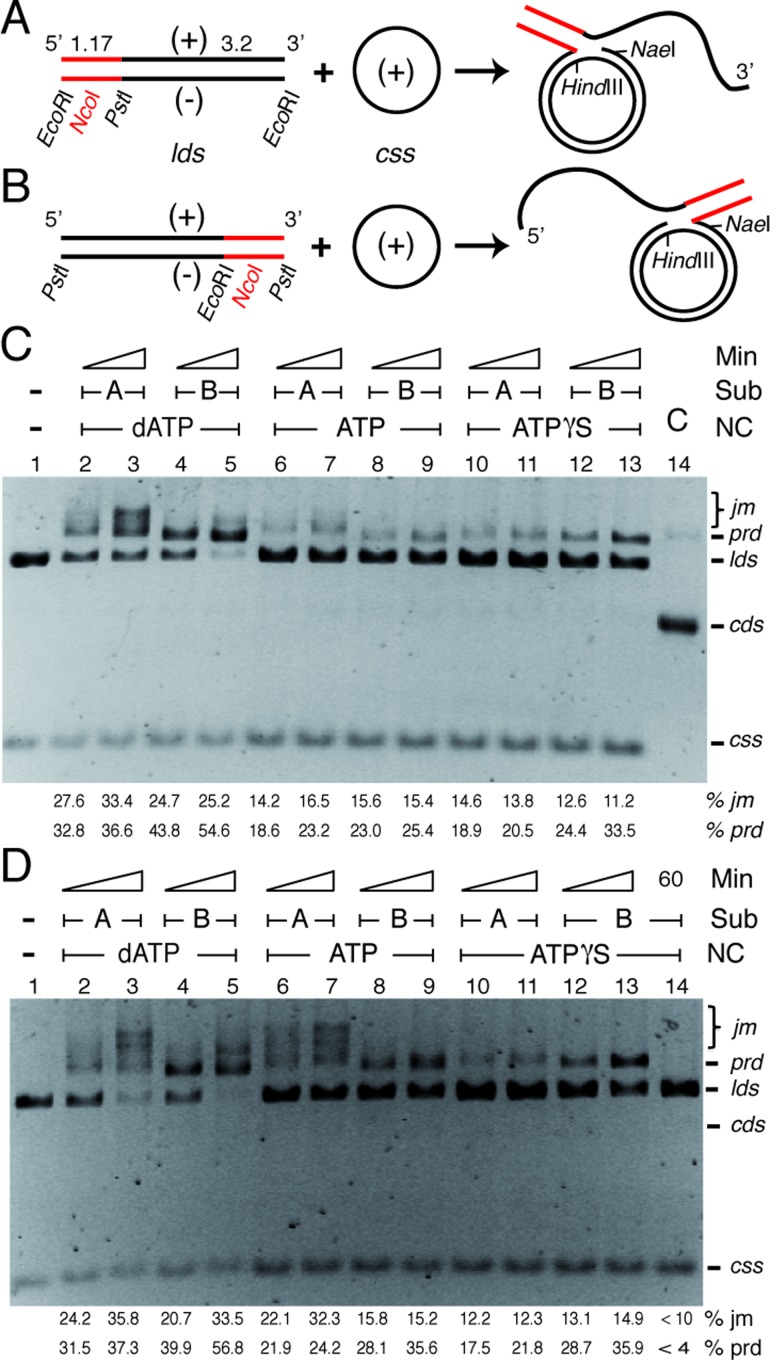
To gain insight in the polarity of RecA-mediated DNA strand exchange we used both sets of mediators (SsbA and DprA or RecO), RecA bound to a hydrolysable (dATP or ATP) or a non-hydrolysable (ATPγS) nucleotide, and a 4374 bp lds substrate, with a 1175 bp heterologous region at the 3′- (substrate A, Sub A) or 5′-end (substrate B, Sub B) (Figure 3A,B) and its complementary 3199 nt css (+ strand). The lds and css were pre-incubated with SsbA and RecO (Figure 3C) or with SsbA and DprA (Figure 3D). Then, limiting RecA (1 RecA monomer/12 nt, to avoid multiple initiation events) was added in the presence of different nucleotide cofactors and the strand exchange reaction monitored. In the presence of saturating RecA concentrations, however, the differences between the substrates were less obvious (data not shown).
Figure 3.
SsbA and RecO or SsbA and DprA contribute to RecA-mediated bidirectional recombination in the presence of different nucleotide cofactors. (A and B) Scheme of the three-strand exchange reaction between circular 3199 nt ssDNA (css, + strand in black) and the 4374 bp lds substrate with homology restricted to the 5′-end (in black, A) or the 3′-end (in black, B) of the (-) strand. The expected prd final products of RecA-mediated DNA strand exchange are illustrated. The relevant restriction sites are indicated. The relative lengths of homology and heterology (denoted in red) are indicated. (C and D) Circular ssDNA (10 μM) and 4374 bp PstI- or EcoRI-linearised dsDNA substrate (20 μM in nt) were pre-incubated with SsbA (0.3 μM) for 5 min at 37°C in buffer A containing 5 mM dATP, ATP or ATPγS, then the preformed SsbA·ssDNA complex was incubated with RecO (C, 0.2 μM) or DprA (D, 0.3 μM) for 5 min at 37°C. Finally RecA (1.2 μM) was added, and the reaction was incubated for 30 or 60 min at 37ºC and products separated by 0.8% AGE. (C) In lane 14, the supercoiled DNA substrate (cds) was treated with DNase I to generate nicked DNA (nc) that co-migrate with the prd. (D) In lane 14, SsbA was omitted and the reaction incubated by 60 min. The positions of the bands corresponding to css, lds, prd and jm are indicated. Amounts of recombination intermediates and products are indicated, and expressed as percentage of total substrate added. The results are the average value obtained from more than three independent experiments (the results given are within a 5% standard error).
In the presence of homology at the 3′-end (Sub B), RecA·dATP catalysed DNA pairing with the homologous 3′-end, the non-complementary linear (+ strand) strand is displaced from duplex DNA in a 5′→3′ direction forming a 5′-tailed jm; and then extensive strand exchange recombinant products (prd, as denoted later these are nc products with a duplex tail, Figure 3B) was observed (Figure 3C,D). Here, ≈90% of the substrate was converted to jm intermediates and recombinant prd after 60 min (Figure 3C,D, lane 5). In the presence of ATP or ATPγS, RecA efficiently catalysed DNA pairing of the 3′-complementary end (Sub B) with css (+ strand), to form 5′-tailed jm and prd products. RecA·ATP or RecA·ATPγS promoted recombination of 40–50% of the DNA substrates in the presence of the RecO and SsbA (Figure 3C, lanes 8–9 and 12–13) or DprA and SsbA (Figure 3D, lanes 8–9 and 12–13) mediators. It is likely that RecA moves along the lds (-) substrate in a 5′ → 3′ direction, heteroduplex formation would occur by a processive assimilation of the (-) strand of the lds into the css.
In the presence of homology at the 5′-end (Sub A), RecA-mediated DNA pairing (jm) and DNA strand exchange was observed in the presence of any of the nucleotide cofactors (dATP, ATP or ATPγS) and any of the two-component mediators (Figure 3C,D, lanes 2–3, 6–7 and 10–11). The initial pairing reaction did not dissociate upon deproteinization, suggesting that three-stranded synaptic complexes occurred at the 5′-complementary end where topologically interwound intermediates would accumulate. The proportion of jm intermediates converted to prd products under all conditions with Sub A varied less than 2-fold when compared with those of Sub B under equivalent conditions (see Figure 3C,D, lanes 4–5, 8–9 and 12–13 versus 2–3, 6–7 and 10–11). Conversely, RecAEco only catalyses unidirectional strand exchange in the presence of ATP, while dissociating those heteroduplex joints formed at the 5′-complementary end in the presence of SSBEco (17–19,55,58–61). Bidirectional RecAEco-mediated DNA recombination was observed only in the presence of ATPγS (56).
RecA-mediated HR in the presence of heterologous ends
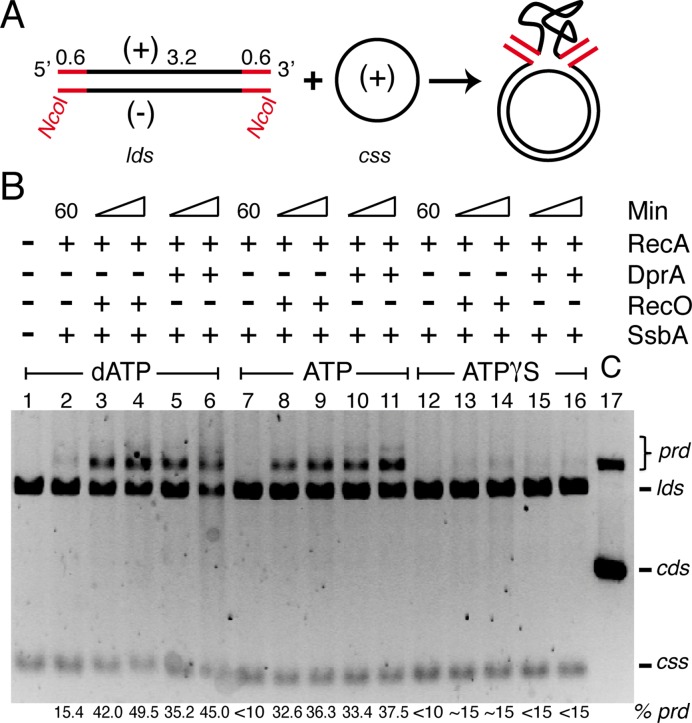
The previous section demonstrated that RecA-mediated recombination between the css and the lds substrate with complementarity at the 5′- or 3′-ends exhibited no strict polarity in the presence of any of the nucleotides tested, while the 3′ complementary end showed marginally higher efficiency independent of the accessory factors used (Figure 3C,D). Alternatively, the initial pairing between the css and the lds substrates occurs randomly and proceeded outwards from the homologous region (see 54). To test whether DNA strand exchange was underestimated when the linear substrate with a 5′-complementary end, an lds substrate containing heterologous sequences at both ends (homology at the central region) was used (Figure 4A).
Figure 4.
RecA-mediated HR in the presence of heterologous ends. (A) Scheme of the lds substrate with heterology at the 3'- and 5'-end (in red) with the css (+) substrate and the expected product (prd). (B) Circular 3199 nt ssDNA (10 μM) and 4374 bp NcoI-linearized dsDNA substrate (20 μM in nt) were pre-incubated with SsbA (0.3 μM) for 5 min at 37°C in buffer A containing 5 mM dATP, ATP or ATPγS, then the preformed SsbA·ssDNA complex was incubated or not with RecO (0.2 μM) or DprA (0.3 μM) for 5 min at 37°C. Finally RecA (1.2 μM) was added, and the reaction was incubated for 30 or 60 min at 37ºC and products separated by 0.8% AGE. In lane 17, the cds DNA substrate was treated with DNase I to generate nc DNA. The positions of the bands corresponding to css, lds, prd and jm are indicated. The symbols + and – denote the presence or absence of the indicated protein(s). Amounts of recombination intermediates and products are indicated, and expressed as percentage of total substrate added. The results are the average of more than three independent experiments (the results given are within a 5% standard error).
In the presence of ≈600 bp heterology at the 3′- and 5′-ends, RecA·dATP or RecA·ATP promoted DNA pairing, jm formation and final prd recombinant products in the presence of SsbA and RecO or SsbA and DprA (Figure 4B, lanes 3–6 and 8–11). RecA-mediated jm formation and subsequent DNA strand exchange occurred with a similar efficiency in the presence of dATP or ATP. When RecO, DprA (Figure 4B, lanes 7 and 12) or SsbA (32,33) was omitted, RecA·ATP (1 RecA/12 nt) failed to catalyse DNA recombination. In the presence of ATPγS, which contains 6–8% of contaminating ATP (62), recombinant products accumulate with very low efficiency in the presence of any of the two component mediators (Figure 4B, lanes 14 and 16), suggesting that the paired complexes stabilized by RecA·ATPγS (plus contaminating RecA·ATP) are not sufficient to accumulate significant amount of recombinant products.
From these data we proposed that RecA-mediated jm formation occurs randomly along the length of the lds DNA, and bidirectional branch migration of recombinants initiated at a central region might contribute to the accumulation of final recombination products, a full-length heteroduplex with two duplex tailed molecules (Supplementary Figure S1B, step d). Alternatively, the lack of end preference observed in the presence of dATP or ATP demonstrates that jm can be formed at both ends, but in the absence of homologous ends multiples rounds of RecA·ADP (RecA·dADP) dissociation and RecA·ATP (RecA·dATP) reassociation might correct discontinuities on the nucleoprotein filament that otherwise impede DNA strand exchange, as seen with RecAEco·ATP (63).
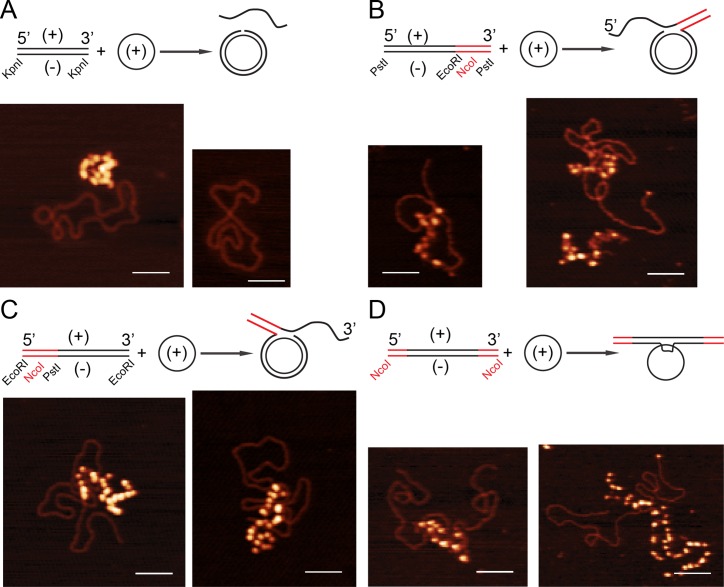
Visualization of the recombination intermediates by SFM
The mobility of the recombination products between css and lds with complementary 3′- or 5′-end (- strand), were indistinguishable from nicked circular (nc) products (Figure 3C,D, lanes 2–13 versus 14), therefore it was assumed that recombination went to completion and that nc DNA with a duplex tail (prd) was formed with substrates showing heterologous segments at the 3′ or 5′-ends (Figure 3A,B). The recombination reactions, performed in the presence of RecO, SsbA and RecA·ATP, were deproteinized and subjected to restriction analyses of the recombinant product with specific restriction enzymes (HindIII or NaeI) that cleavage at either side of the nick (see Figure 3A,B) suggested that nicked circular DNA with a duplex tail might be formed (data not shown). To confirm these results the above deproteinized reaction products were visualized by SFM in the presence of SSBEco to highlight the ssDNA (Figure 5). When a fully homologous substrate was used jm intermediates and genuine nc and lss products were observed (Figure 5A). The presence of nc with a duplex tail (prd) was detected using the lds substrate (- strand) with heterology at the 3′-end or at the 5′-end (Figure 5B,C).
Figure 5.
Visualization of the recombination intermediates by SFM. (A–D), After 60 min of incubation, in the presence of SsbA, RecO and RecA·ATP, the recombination reaction was deproteinased and the DNA precipitated. SSBEco (200 nM) was added to the resuspended DNA in TE buffer and visualized by tapping mode SFM. The heterologous regions (denoted in red) are indicated. Images shown were cropped original files collected as 2 μm × 2 μm, 512 × 512 pixels. Regions of dsDNA appear in the SFM images as thin lines, SSBEco-coated ssDNA appears as thick brighter beads-on-string. Colour indicates height (from dark to bright, 0 to 5 nm). Scale bar = 100 nm.
From the data presented in Figure 4B it was assumed that nc with double-tailed structures might be formed when the heterology was present at both DNA ends. The presence of nc products with two duplex tails were observed by SFM (Figure 5D). It is likely that RecA catalyses the formation of stable homologous joints and final prd products in the presence of SsbA and DprA and SsbA and RecO mediators during chromosomal transformation.
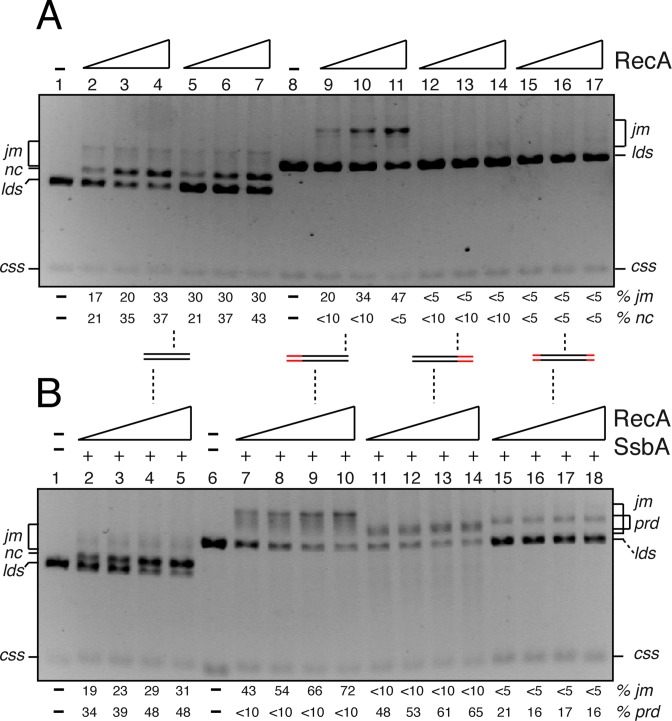
RecA·dATP facilitates strand DNA exchange with 5′-complementary ends and SsbA overcomes this bias
RecA·dATP catalyses DNA recombination even in the absence of accessory factors (29). To gain insight into the role of the mediators in the polarity of RecA, the reaction was performed in the presence of dATP and in the absence of accessory factors. Under these conditions, a RecA·dATP NPF (1 RecA monomer/12–6 nt) led to the formation of jm between the free end of lds and the css, followed by extensive DNA strand exchange to generate the nc and the linear ssDNA (lss) products (Figure 6, lanes 2–7) (12,13,29). Unexpectedly, RecA·dATP mediated the accumulation of recombination intermediates only in the presence of a linear 4374 bp duplex with the 5′-complementary end (Sub A, Figure 3A) and the css (+ strand), but final prd did not accumulate (Figure 6A, lanes 9–11). When linear substrate with a short 3′-complementary end (Sub B, Figure 3B) or with complementarity at the central region (Figure 4A) was used, RecA·dATP catalysed DNA strand exchange with low efficiency (Figure 6A, lanes 12–14 and 15–17). We can envisage that recombination occurs with the three substrates with about similar efficiency, but RecA·dATP dissociates from the heteroduplex DNA and binds to the displaced 5′-tailed duplex that might reverse the reaction to generate the initial substrates. Alternatively the jm intermediates formed between css and the lds substrate with a 3′-complementary end or with heterology at both DNA ends were unstable to deproteinisation and seen as substrates. Addition of SsbA to the reaction should compete with RecA·dATP for the 5′-end inhibiting the re-invasion of the displaced strand, and recombinant products would be accumulated, as seen with RecAEco using a D-loop assay (52).
Figure 6.
RecA·dATP-mediated recombination in the absence or presence of SsbA. (A) Circular 3199 nt ssDNA (10 μM) and the 3199 bp KnpI (In lanes 1–4) or EcoRI-linearised duplex (lanes 5–7) or the 4374 bp EcoRI- (Sub A, lanes 8–11), PstI- (Sub B, 12–14), or NcoI–linearized duplex (lanes 15–17) substrate (20 μM in nt) was incubated with RecA (0.8, 1.0 and 1.2 μM) in buffer A containing 5 mM dATP for 60 min at 37°C, and separated by 0.8% AGE. (B) Circular ssDNA and duplex substrate of part (A) were pre-incubated with SsbA (0.3 μM) in buffer A containing 5 mM dATP for 5 min at 37°C. RecA (0.8, 1.0, 1.2 and 1.4 μM) was added, the reaction was incubated for 60 min at 37ºC and then separated by 0.8% AGE. In lanes 2-5 the 3199 bp KnpI–linearized duplex DNA was used. The symbols + and – denote the presence or absence of the indicated protein(s). The positions of the bands corresponding to css, lds, cds, prd and jm are indicated. A cartoon of the lds substrate with homology (in black) and heterology (in red) with the css substrate are indicated, with the broken line at the centre of each condition. The results are the average of more than three independent experiments (the results given are within a 5% standard error).
When increasing RecA·dATP concentrations (1 RecA monomer/6–12 nt) were incubated with linear 4374 bp duplex with heterology at the 3′- (Sub B), the 5′- end (Sub A) (- strand) or at both DNA ends and SsbA-coated css DNA, strand exchange was observed in all conditions (Figure 6B). The presence of SsbA (1 SsbA tetramer/33 nt) facilitated RecA·dATP-mediated recombination with a 3′-complementary end or with heterology at both DNA ends leading to the accumulation prd products (Figure 6B, lanes 11–14 or 15–18). Since the intervening sequences are longer than ≈3100 bp we favour the hypothesis that SsbA selectively inhibited the reactivity of the displaced 5′-end with subsequent suppression of the reversion of the recombination reaction. The presence of SsbA, also favoured RecA·dATP recombination reaction by decreasing the mobility of the jm intermediates with the heterologous substrate at the 3′-end (Figure 6B, lanes 7–10). We find that: (i) RecA showed no intrinsic preference for 3′- or 5′-complementary ends, but the presence of SsbA (Figure 6B) increased the reactivity of the 3′-complementary ends; and (ii) RecO or DprA in concert with SsbA eliminate the 5′-end bias (Figure 3C,D). It is likely that the re-invasion of the displaced 5′-end (poorly coated by RecA·dATP) into nc products reversed the reaction to form the substrates, as seen with RecAEco·ATPγS in the presence of SSBEco (51,59).
DISCUSSION
HGT by natural transformation with DNA from closely related species, from the same taxonomic group, is a powerful mechanism for promoting genetic diversity, and acquisition of antibiotics resistance and virulence genes of both episomal and chromosomal origin. Although, HGT is becoming a major threat to modern medicine, it can represent a biological barrier for gene acquisition from donors of dissimilar sequences. We reported here that the frequency of chromosomal transformation was log linearly reduced up to ≈45 000-fold with sequence divergence increasing from 0.03% to 20.8%.
The sequence divergence decreased the length of the integrated DNA, from about full-length (≈2.5% divergence) to a few nucleotides (≈21% divergence), maintaining the relative proportion of sequence divergence (Table 1). When the rpoB482 (position +1) mutation, within the MEPS, was placed at the 3′- or 5′-end of the same strand or surrounded by the presence of sequence divergence at the 3′-end of the complementary strands the transformation frequency was marginally affected. It is likely that the Watson or Crick strand enters with about similar efficiency and only one conserved invasive end is necessary for chromosomal transformation. This is consistent with the observation that the frequency of chromosomal transformation with the mutation at the 3′- or 5′-end varied less than 5-fold.
Natural chromosomal transformation involves a three-strand exchange reaction between the linear ssDNA, internalized in a non-polar fashion and coated by SsbA or SsbA and SsbB, with a recipient homologous circular duplex. In vitro, using the three-strand recombination reaction, that mimics natural chromosomal transformation, purified RecA, in the presence of one of the two-component mediator (SsbA and DprA or RecO), showed no intrinsic preference for the 3′- or the 5-end. RecA·dATP, RecA·ATP or RecA·ATPγS is proficient to catalyse DNA strand exchange of the linear duplex with complementarity at the 3′- or 5-end (- strand) with the css (+ strand), the former processed with marginally (< 2-fold) higher efficiency in vitro (Figure 3C,D). These recombination products, which are genuine intertwined strands (plectonemic) resistant to deproteinization were confirmed by SFM analysis. When the homologous sequence was restricted to a central region, the ssDNA invades a duplex in a region of homology displacing the strand of the same sequence leading to D-loop formation, following by multi-step strand exchange reaction to render recombinant products (Figure 4). Here, however, RecA·ATPγS-mediated DNA strand exchange was strongly reduced and a homologous DNA end was needed to catalyse HR (Figure 4B, lanes 13–16). It is likely that multiple rounds of RecA dissociation and re-association might correct discontinuities on the nucleoprotein filament that otherwise impede DNA strand exchange. Conversely, during recombinational repair the end-processing apparatus generated 3′-tailed duplex DNA and SsbA and RecO (or RecOR in vivo) load and activate RecA to catalyse DNA strand exchange (Supplementary Figure S1A, step a) (33). It is likely that RecA has evolved to accommodate and process different types of DNA substrates (genetic recombination by chromosomal transformation and recombinational DNA repair). Conversely, RecAEco is optimized to deal with a 3′-tailed duplex, and mediates unidirectional DNA strand exchange in the presence of SSBEco and ATP hydrolysis. RecAEco, however, shows bidirectional DNA strand exchange in the presence of ATPγS (47,50,56,60,61,64).
RecA·ATP from natural competent bacteria (e.g. RecA, RecASpn or RecADra) cannot catalyse DNA strand exchange in the absence of accessory factors (27–31). RecA·ATP requires one of the two-component mediators (SsbA and DprA or RecO) to polymerize onto the internalized linear ssDNA, coated by SsbA (or SsbA and SsbB) in vivo, and to activate RecA·ATP to catalyse HR between circular ssDNA with linear duplex DNA in vitro (32,33). However, in the absence of accessory factors, RecA·dATP is proficient to catalyse DNA strand exchange between css and the lds DNA (Figure 6A) (13,28,29). RecA·dATP is proficient to catalyse jm formation with the 5′-complementary end, but the accumulation of recombination intermediates was not observed with a linear substrate with the 3′-complementary end or with a complementary central region (Figure 6B, lanes 12–14 and 15–17). It is likely that the jm formed with these substrates would displace a 5′-end tail or a central region that might promote re-invasion with subsequent reversion of the strand exchange reaction. To test this hypothesis the strand exchange reaction was performed in the presence of SsbA that might coat the displaced 5′-end tail. In the presence of SsbA, RecA·dATP is proficient to catalyse DNA strand exchange with the three substrates tested (Figure 6B, lanes 7–10, 11–14 and 15–18).
Like eukaryotic Rad51 (65,66), RecA promotes DNA strand exchange with no intrinsic polarity. What determines the RecA polarity of strand exchange? During DSB repair the long-range end processing machineries (the AddAB or RecJ-RecQ(S)-SsbA complex) generate 3′-tailed duplex and determine the 5′ → 3′ direction of filament extension and indirectly the polarity of the substrate for RecA-mediated recombinational repair (Supplementary Figure S1A) (14–16). Conversely, the DNA uptake machinery of competent cells internalized linear ssDNA in a non-polar fashion. SsbA or SsbA and SsbB would coat the lss substrates and then DrpA or RecO recruits and activates RecA·ATP to catalyse DNA strand exchange (Supplementary Figure S1B) (32,33). Indeed, RecA·ATP can nucleate on the SsbA·ssDNA·DprA or SsbA·ssDNA·RecO complexes and catalyse DNA strand with 3′- and 5′-complementary ends in vitro. Like the eukaryotic recombinases (67,68), RecA from natural competent bacteria might catalyse HR in both directions.
Supplementary Material
Acknowledgments
We thank S. Ayora for comments on the manuscript. E.S. thanks the Ministerio de Economia y Competividad, Dirección General de Investigación (MINECO-DGI) for the BES-2013-063433 fellowship. This work was partially supported by the MINECO, Dirección General de Investigación (DGI) Grants BFU2012-39879-C02-01 and BFU2015-67065-P to J.C.A. The work of H.S. and C.W. is supported by a Marie Curie Reintegration Grant (FP7-276898), and NanoNextNL, a micro and nanotechnology consortium of the Government of the Netherlands and 130 partners.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
For open access charge to MINECO-DGI BFU2012–39879-C02-01. Funding for open access charge: Ministerio de Economia y Competividad, Dirección General de Investigación [BFU2015-67065-P to J.C.A.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Coop G., Przeworski M. An evolutionary view of human recombination. Nat. Rev. Genet. 2007;8:23–34. doi: 10.1038/nrg1947. [DOI] [PubMed] [Google Scholar]
- 2.Takeuchi N., Kaneko K., Koonin E.V. Horizontal gene transfer can rescue prokaryotes from Muller's ratchet: benefit of DNA from dead cells and population subdivision. G3 (Bethesda) 2014;4:325–339. doi: 10.1534/g3.113.009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kidane D., Ayora S., Sweasy J.B., Graumann P.L., Alonso J.C. The cell pole: the site of cross talk between the DNA uptake and genetic recombination machinery. Crit. Rev. Biochem. Mol. Biol. 2012;47:531–555. doi: 10.3109/10409238.2012.729562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnston C., Campo N., Berge M.J., Polard P., Claverys J.P. Streptococcus pneumoniae, le transformiste. Trends Microbiol. 2014;22:113–119. doi: 10.1016/j.tim.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Muller H.J. The relation of recombination to mutational advance. Mutat. Res. 1964;106:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 6.Croucher N.J., Harris S.R., Fraser C., Quail M.A., Burton J., van der Linden M., McGee L., von Gottberg A., Song J.H., Ko K.S., et al. Rapid pneumococcal evolution in response to clinical interventions. Science. 2011;331:430–434. doi: 10.1126/science.1198545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanchez-Buso L., Comas I., Jorques G., Gonzalez-Candelas F. Recombination drives genome evolution in outbreak-related Legionella pneumophila isolates. Nat. Genet. 2014;46:1205–1211. doi: 10.1038/ng.3114. [DOI] [PubMed] [Google Scholar]
- 8.Chen I., Dubnau D. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2004;2:241–249. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- 9.Chen I., Christie P.J., Dubnau D. The ins and outs of DNA transfer in bacteria. Science. 2005;310:1456–1460. doi: 10.1126/science.1114021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kramer N., Hahn J., Dubnau D. Multiple interactions among the competence proteins of Bacillus subtilis. Mol. Microbiol. 2007;65:454–464. doi: 10.1111/j.1365-2958.2007.05799.x. [DOI] [PubMed] [Google Scholar]
- 11.Kidane D., Carrasco B., Manfredi C., Rothmaier K., Ayora S., Tadesse S., Alonso J.C., Graumann P.L. Evidence for different pathways during horizontal gene transfer in competent Bacillus subtilis cells. PLoS Genet. 2009;5:e1000630. doi: 10.1371/journal.pgen.1000630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yadav T., Carrasco B., Hejna J., Suzuki Y., Takeyasu K., Alonso J.C. Bacillus subtilis DprA recruits RecA onto single-stranded DNA and mediates annealing of complementary strands coated by SsbB and SsbA. J. Biol. Chem. 2013;288:22437–22450. doi: 10.1074/jbc.M113.478347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yadav T., Carrasco B., Myers A.R., George N.P., Keck J.L., Alonso J.C. Genetic recombination in Bacillus subtilis: a division of labor between two single-strand DNA-binding proteins. Nucleic Acids Res. 2012;40:5546–5559. doi: 10.1093/nar/gks173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Radding C.M. Helical interactions in homologous pairing and strand exchange driven by RecA protein. J. Biol. Chem. 1991;266:5355–5358. [PubMed] [Google Scholar]
- 15.Kowalczykowski S.C., Eggleston A.K. Homologous pairing and DNA strand-exchange proteins. Annu. Rev. Biochem. 1994;63:991–1043. doi: 10.1146/annurev.bi.63.070194.005015. [DOI] [PubMed] [Google Scholar]
- 16.Cox M.M. Motoring along with the bacterial RecA protein. Nat. Rev. Mol. Cell. Biol. 2007;8:127–138. doi: 10.1038/nrm2099. [DOI] [PubMed] [Google Scholar]
- 17.Cox M.M., Lehman I.R. Directionality and polarity in RecA protein-promoted branch migration. Proc. Natl. Acad. Sci. U.S.A. 1981;78:6018–6022. doi: 10.1073/pnas.78.10.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.West S.C., Cassuto E., Howard-Flanders P. Heteroduplex formation by recA protein: polarity of strand exchanges. Proc. Natl. Acad. Sci. U.S.A. 1981;78:6149–6153. doi: 10.1073/pnas.78.10.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kahn R., Cunningham R.P., DasGupta C., Radding C.M. Polarity of heteroduplex formation promoted by Escherichia coli recA protein. Proc. Natl. Acad. Sci. U.S.A. 1981;78:4786–4790. doi: 10.1073/pnas.78.8.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howard-Flanders P., West S.C., Stasiak A. Role of RecA protein spiral filaments in genetic recombination. Nature. 1984;309:215–219. doi: 10.1038/309215a0. [DOI] [PubMed] [Google Scholar]
- 21.Alonso J.C., Lüder G., Tailor R.H. Characterization of Bacillus subtilis recombinational pathways. J. Bacteriol. 1991;173:3977–3980. doi: 10.1128/jb.173.13.3977-3980.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pasta F., Sicard M.A. Polarity of recombination in transformation of Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U.S.A. 1999;96:2943–2948. doi: 10.1073/pnas.96.6.2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vagner V., Claverys J.P., Ehrlich S.D., Mejean V. Direction of DNA entry in competent cells of Bacillus subtilis. Mol. Microbiol. 1990;4:1785–1788. doi: 10.1111/j.1365-2958.1990.tb00557.x. [DOI] [PubMed] [Google Scholar]
- 24.Wilson G.A., Young F.E. Intergenotic transformation of the Bacillus subtilis genospecies. J. Bacteriol. 1972;111:705–716. doi: 10.1128/jb.111.3.705-716.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zawadzki P., Roberts M.S., Cohan F.M. The log-linear relationship between sexual isolation and sequence divergence in Bacillus transformation is robust. Genetics. 1995;140:917–932. doi: 10.1093/genetics/140.3.917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Majewski J., Cohan F.M. DNA sequence similarity requirements for interspecific recombination in Bacillus. Genetics. 1999;153:1525–1533. doi: 10.1093/genetics/153.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lovett C.M., Jr, Roberts J.W. Purification of a RecA protein analogue from Bacillus subtilis. J. Biol. Chem. 1985;260:3305–3313. [PubMed] [Google Scholar]
- 28.Carrasco B., Manfredi C., Ayora S., Alonso J.C. Bacillus subtilis SsbA and dATP regulate RecA nucleation onto single-stranded DNA. DNA Repair (Amst.) 2008;7:990–996. doi: 10.1016/j.dnarep.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 29.Manfredi C., Carrasco B., Ayora S., Alonso J.C. Bacillus subtilis RecO nucleates RecA onto SsbA-coated single-stranded DNA. J. Biol. Chem. 2008;283:24837–24847. doi: 10.1074/jbc.M802002200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steffen S.E., Katz F.S., Bryant F.R. Complete inhibition of Streptococcus pneumoniae RecA protein-catalyzed ATP hydrolysis by single-stranded DNA-binding protein (SSB protein): implications for the mechanism of SSB protein-stimulated DNA strand exchange. J. Biol. Chem. 2002;277:14493–14500. doi: 10.1074/jbc.M112444200. [DOI] [PubMed] [Google Scholar]
- 31.Ngo K.V., Molzberger E.T., Chitteni-Pattu S., Cox M.M. Regulation of Deinococcus radiodurans RecA protein function via modulation of active and inactive nucleoprotein filament states. J. Biol. Chem. 2013;288:21351–21366. doi: 10.1074/jbc.M113.459230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yadav T., Carrasco B., Serrano E., Alonso J.C. Roles of Bacillus subtilis DprA and SsbA in RecA-mediated genetic recombination. J. Biol. Chem. 2014;289:27640–27652. doi: 10.1074/jbc.M114.577924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carrasco B., Yadav T., Serrano E., Alonso J.C. Bacillus subtilis RecO and SsbA are crucial for RecA-mediated recombinational DNA repair. Nucleic Acids Res. 2015;43:9249–9261. doi: 10.1093/nar/gkv545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alonso J.C., Cárdenas P.P., Sanchez H., Hejna J., Suzuki Y., Takeyasu K. Early steps of double-strand break repair in Bacillus subtilis. DNA Repair (Amst.) 2013;12:162–176. doi: 10.1016/j.dnarep.2012.12.005. [DOI] [PubMed] [Google Scholar]
- 35.Ayora S., Carrasco B., Cárdenas P.P., Cesar C.E., Cañas C., Yadav T., Marchisone C., Alonso J.C. Double-strand break repair in bacteria: a view from Bacillus subtilis. FEMS Microbiol. Rev. 2011;35:1055–1081. doi: 10.1111/j.1574-6976.2011.00272.x. [DOI] [PubMed] [Google Scholar]
- 36.Ayora S., Missich R., Mesa P., Lurz R., Yang S., Egelman E.H., Alonso J.C. Homologous-pairing activity of the Bacillus subtilis bacteriophage SPP1 replication protein G35P. J. Biol. Chem. 2002;277:35969–35979. doi: 10.1074/jbc.M204467200. [DOI] [PubMed] [Google Scholar]
- 37.Carrasco B., Ayora S., Lurz R., Alonso J.C. Bacillus subtilis RecU Holliday-junction resolvase modulates RecA activities. Nucleic Acids Res. 2005;33:3942–3952. doi: 10.1093/nar/gki713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicholson W.L., Maughan H. The spectrum of spontaneous rifampin resistance mutations in the rpoB gene of Bacillus subtilis 168 spores differs from that of vegetative cells and resembles that of Mycobacterium tuberculosis. J. Bacteriol. 2002;184:4936–4940. doi: 10.1128/JB.184.17.4936-4940.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dubnau D., Davidoff-Abelson R., Scher B., Cirigliano C. Fate of transforming deoxyribonucleic acid after uptake by competent Bacillus subtilis: phenotypic characterization of radiation-sensitive recombination-deficient mutants. J. Bacteriol. 1973;114:273–286. doi: 10.1128/jb.114.1.273-286.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alonso J.C., Tailor R.H., Lüder G. Characterization of recombination-deficient mutants of Bacillus subtilis. J. Bacteriol. 1988;170:3001–3007. doi: 10.1128/jb.170.7.3001-3007.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ceglowski P., Lüder G., Alonso J.C. Genetic analysis of recE activities in Bacillus subtilis. Mol. Gen. Genet. 1990;222:441–445. doi: 10.1007/BF00633853. [DOI] [PubMed] [Google Scholar]
- 42.Ayora S., Piruat J.I., Luna R., Reiss B., Russo V.E., Aguilera A., Alonso J.C. Characterization of two highly similar Rad51 homologs of Physcomitrella patens. J. Mol. Biol. 2002;316:35–49. doi: 10.1006/jmbi.2001.5336. [DOI] [PubMed] [Google Scholar]
- 43.Maamar H., Dubnau D. Bistability in the Bacillus subtilis K-state (competence) system requires a positive feedback loop. Mol. Microbiol. 2005;56:615–624. doi: 10.1111/j.1365-2958.2005.04592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kunkel T.A., Erie D.A. DNA mismatch repair. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 45.Alonso J.C., Lüder G., Trautner T.A. Requirements for the formation of plasmid-transducing particles of Bacillus subtilis bacteriophage SPP1. EMBO J. 1986;5:3723–3728. doi: 10.1002/j.1460-2075.1986.tb04706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen P., Huang H.V. Homologous recombination in Escherichia coli: dependence on substrate length and homology. Genetics. 1986;112:441–457. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joo C., McKinney S.A., Nakamura M., Rasnik I., Myong S., Ha T. Real-time observation of RecA filament dynamics with single monomer resolution. Cell. 2006;126:515–527. doi: 10.1016/j.cell.2006.06.042. [DOI] [PubMed] [Google Scholar]
- 48.van der Heijden T., Modesti M., Hage S., Kanaar R., Wyman C., Dekker C. Homologous recombination in real time: DNA strand exchange by RecA. Mol Cell. 2008;30:530–538. doi: 10.1016/j.molcel.2008.03.010. [DOI] [PubMed] [Google Scholar]
- 49.Forget A.L., Kowalczykowski S.C. Single-molecule imaging of DNA pairing by RecA reveals a three-dimensional homology search. Nature. 2012;482:423–427. doi: 10.1038/nature10782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bell J.C., Plank J.L., Dombrowski C.C., Kowalczykowski S.C. Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA. Nature. 2012;491:274–278. doi: 10.1038/nature11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Konforti B.B., Davis R.W. The preference for a 3′ homologous end is intrinsic to RecA-promoted strand exchange. J. Biol. Chem. 1990;265:6916–6920. [PubMed] [Google Scholar]
- 52.McIlwraith M.J., West S.C. The efficiency of strand invasion by Escherichia coli RecA is dependent upon the length and polarity of ssDNA tails. J. Mol. Biol. 2001;305:23–31. doi: 10.1006/jmbi.2000.4268. [DOI] [PubMed] [Google Scholar]
- 53.Sagi D., Tlusty T., Stavans J. High fidelity of RecA-catalyzed recombination: a watchdog of genetic diversity. Nucleic Acids Res. 2006;34:5021–5031. doi: 10.1093/nar/gkl586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morel P., Stasiak A., Ehrlich S.D., Cassuto E. Effect of length and location of heterologous sequences on RecA-mediated strand exchange. J. Biol. Chem. 1994;269:19830–19835. [PubMed] [Google Scholar]
- 55.Cox J.M., Tsodikov O.V., Cox M.M. Organized unidirectional waves of ATP hydrolysis within a RecA filament. PLoS Biol. 2005;3:e52. doi: 10.1371/journal.pbio.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Konforti B.B., Davis R.W. ATP hydrolysis and the displaced strand are two factors that determine the polarity of RecA-promoted DNA strand exchange. J. Mol. Biol. 1992;227:38–53. doi: 10.1016/0022-2836(92)90680-i. [DOI] [PubMed] [Google Scholar]
- 57.Mani A., Braslavsky I., Arbel-Goren R., Stavans J. Caught in the act: the lifetime of synaptic intermediates during the search for homology on DNA. Nucleic Acids Res. 2010;38:2036–2043. doi: 10.1093/nar/gkp1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Konforti B.B., Davis R.W. 3′ homologous free ends are required for stable joint molecule formation by the RecA and single-stranded binding proteins of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1987;84:690–694. doi: 10.1073/pnas.84.3.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dutreix M., Rao B.J., Radding C.M. The effects on strand exchange of 5′ versus 3′ ends of single-stranded DNA in RecA nucleoprotein filaments. J. Mol. Biol. 1991;219:645–654. doi: 10.1016/0022-2836(91)90661-o. [DOI] [PubMed] [Google Scholar]
- 60.Burnett B., Rao B.J., Jwang B., Reddy G., Radding C.M. Resolution of the three-stranded recombination intermediate made by RecA protein. An essential role of ATP hydrolysis. J. Mol. Biol. 1994;238:540–554. doi: 10.1006/jmbi.1994.1313. [DOI] [PubMed] [Google Scholar]
- 61.Jain S.K., Cox M.M., Inman R.B. On the role of ATP hydrolysis in RecA protein-mediated DNA strand exchange. III. Unidirectional branch migration and extensive hybrid DNA formation. J. Biol. Chem. 1994;269:20653–20661. [PubMed] [Google Scholar]
- 62.Cárdenas P.P., Carzaniga T., Zangrossi S., Briani F., Garcia-Tirado E., Dehò G., Alonso J.C. Polynucleotide phosphorylase exonuclease and polymerase activities on single-stranded DNA ends are modulated by RecN, SsbA and RecA proteins. Nucleic Acids Res. 2011;39:9250–9261. doi: 10.1093/nar/gkr635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Menetski J.P., Bear D.G., Kowalczykowski S.C. Stable DNA heteroduplex formation catalyzed by the Escherichia coli RecA protein in the absence of ATP hydrolysis. Proc. Natl. Acad. Sci. U.S.A. 1990;87:21–25. doi: 10.1073/pnas.87.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Galletto R., Amitani I., Baskin R.J., Kowalczykowski S.C. Direct observation of individual RecA filaments assembling on single DNA molecules. Nature. 2006;443:875–878. doi: 10.1038/nature05197. [DOI] [PubMed] [Google Scholar]
- 65.Mazin A.V., Zaitseva E., Sung P., Kowalczykowski S.C. Tailed duplex DNA is the preferred substrate for Rad51 protein-mediated homologous pairing. EMBO J. 2000;19:1148–1156. doi: 10.1093/emboj/19.5.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Namsaraev E.A., Berg P. Rad51 uses one mechanism to drive DNA strand exchange in both directions. J. Biol. Chem. 2000;275:3970–3976. doi: 10.1074/jbc.275.6.3970. [DOI] [PubMed] [Google Scholar]
- 67.San Filippo J., Sung P., Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 68.Holthausen J.T., Wyman C., Kanaar R. Regulation of DNA strand exchange in homologous recombination. DNA Repair (Amst.) 2010;9:1264–1272. doi: 10.1016/j.dnarep.2010.09.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.