Abstract
Most RNAs generated by the human genome have no protein-coding ability and are termed non-coding RNAs. Among these include circular RNAs, which include exonic circular RNAs (circRNA), mainly found in the cytoplasm, and intronic RNAs (ciRNA), predominantly detected in the nucleus. The biological functions of circular RNAs remain largely unknown, although ciRNAs have been reported to promote gene transcription, while circRNAs may function as microRNA sponges. We demonstrate that the circular RNA circ-Foxo3 was highly expressed in non-cancer cells and were associated with cell cycle progression. Silencing endogenous circ-Foxo3 promoted cell proliferation. Ectopic expression of circ-Foxo3 repressed cell cycle progression by binding to the cell cycle proteins cyclin-dependent kinase 2 (also known as cell division protein kinase 2 or CDK2) and cyclin-dependent kinase inhibitor 1 (or p21), resulting in the formation of a ternary complex. Normally, CDK2 interacts with cyclin A and cyclin E to facilitate cell cycle entry, while p21works to inhibit these interactions and arrest cell cycle progression. The formation of this circ-Foxo3-p21-CDK2 ternary complex arrested the function of CDK2 and blocked cell cycle progression.
INTRODUCTION
Non-coding RNAs represent the majority of transcripts in a cell. Circular RNAs are a large class of non-coding RNAs that are circularized by joining the 3′ end of the RNA to the 5′ end, forming a circular structure (1–7). Although circular RNAs were detected decades ago, their functions in mammalian cells are only recently emerging. Most of the circular RNAs reported so far are exon-containing circular RNAs and are detected in the cytoplasm. Some of these circular RNAs possess microRNA (miRNA) binding sites and function as sponges to arrest miRNA functions (6,7). For example, the circular RNA CiRS-7 contains many binding sites for the microRNA miR-7, and can function as a sponge of miR-7 (6,7). Another circular RNA called SRY, contains many binding sites for miR-138 and functions as a miR-138 sponge (7,8). Due to their abundance and stability, circular RNAs are believed to be more effective relative to non-circular RNAs in sponging miRNA (1,7,9). Since miRNAs are important in regulating protein expression and cellular physiology, circular RNAs may thus exert roles in modulating cellular physiology such as cell proliferation and differentiation. This has not been reported and our study was designed to explore this hypothesis. We explored the potential role of a circular RNA circular Foxo3 in regulating cell cycle progression.
Both circular Foxo3 (circ-Foxo3) and linear Foxo3 (Foxo3 mRNA) are encoded by the Foxo3 gene (10). Deregulation of Foxo3 is associated with cancer development (11), which appears to be the consequence of increased Akt activity or Phosphatase and tensin homolog (PTEN) inactivation and Foxo3 is thus classified as a tumor suppressor gene (11,12). Our previous study showed that the upregulation of Foxo3 was linked to decreased cellular senescence (13), which might be associated with cell cycle progression. Cyclins and cyclin-dependent kinases (CDKs) are two classes of regulators for cell cycle progression. As a member of the cyclin-dependent kinase family, CDK2 is a Ser/Thr protein kinase. Its activity is restricted to the G1-S phase in cell cycle progression, and is essential for the G1/S transition. In the G1 phase, CDK2 forms a complex with cyclin E. The cyclin complex phosphorylates retinoblastoma protein (Rb) and promotes gene expression leading to the progression of cells from the G1 to S phase (14). The cyclin E/CDK2 complex also phosphorylates p27 and promotes p27 degradation, thus increasing cyclin A expression, facilitating G1 to S transition. Known CDK inhibitors include p21 and p27 (15). p21 can bind CDK2 and inhibit CDK2 activity (16), therefore functioning as a regulator of cell cycle progression at the G1 and S phase (17). Our study showed that circ-Foxo3 could interact with both p21 and CDK2 forming a ternary complex, resulting in the inhibition of cell cycle progression.
MATERIALS AND METHODS
Materials
The monoclonal or polyclonal antibodies against cyclin A, cyclin B, cyclin C, cyclin D, cyclin E, CDK2, CDK4, CDK6, p16, p18 and p27 were purchased from Santa Cruz Biotechnology. The monoclonal antibodies against p21 and p57 were obtained from BD Biosciences. Horseradish peroxidase-conjugated goat anti-mouse IgG and horseradish peroxidase-conjugated goat anti-rabbit IgG were obtained from Bio-Rad. RNA and DNA extract kits, RNA RT and polymerase chain reaction (PCR) kits were obtained from Qiagen. NorthernMAX kit was from Ambion. Immunoblotting was performed using the Enhanced chemiluminescence (ECL) western blot detection kit (Amersham Biosciences). Biotin Chromogenic Detection kit was from Thermo Scientific. Protein A-Sepharose 4B Conjugate and Dynabeads MyOne Streptavidin C1 magnetic beads were obtained from Invitrogen. The cell lines used in this study were from American Type Culture Collection (ATCC).
Constructs and primers
We generated a construct expressing mouse circular RNA Foxo3 (circ-Foxo3). Briefly, the plasmids contained a Bluescript backbone, a CMV promoter driving mouse circ-Foxo3 expression or a non-related control sequence. The green fluorescent protein (GFP) expression unit was linked to the cir-Foxo3 but contained an internal ribosome entry site (IRES) allowing the GFP to be expressed separately.
To generate circ-Foxo3, a basic sequence containing circ-Foxo3 was synthesized, which contained the 3′-half-intron-exon-II fragment (splice acceptor or SA) of bacteriophage T4 td gene, a small space sequence (SSS), a exon-I splice donor (SD)-5′-half-intron segment. The SSS contained two restriction enzyme sites HindIII and SalI for insertion of circular RNA fragment (circ-Fox). In addition, the basic sequence also contained an IRES. The basic sequence was cloned into the multiple cloning sites of pEGFP-N1, which allowed generation of circular RNA and GFP by replacing SSS with the circular Foxo3, producing circ-Fox-GFP (Supplementary Figure S1a). To generate the control vector circ-RS, a random sequence was synthesized and cloned into the HindIII-SalI sites of circ-Fox-GFP.
All primer sequences used are listed in Supplementary Figure S1b. The anti-circ-Foxo3 siRNA and DNA oligo probes against endogenous or ectopic expression of circ-Foxo3, labeled with biotin or Cy5, were obtained from Integrated DNA Technologies (Supplementary Figure S1c).
Cell proliferation assay
Cells (4 × 104) were seeded onto 6-well dishes in 10% Fetal Bovine Serum (FBS)/DMEM (Dulbecco's modified Eagle's medium) medium and maintained at 37°C overnight. Cells were cultured in DMEM with different concentrations of FBS and harvested daily. Cell number was determined by a coulter counter as described (18,19).
FACS analysis
Cells were washed and resuspended in cold phosphate bufferedsaline (PBS) and incubated in ice-cold 70% ethanol for 3 h. The cells were then centrifuged at 1500 rpm for 10 min and resuspended in propidium iodide (PI) master mix (40 mg/ml PI and 100 mg/ml RNase in PBS) at a density of 5 × 105 cells/ml and incubated at 37°C for 30 min before analysis with flow cytometry as described (20,21).
Western blot analysis
Cells were lysed in lysis buffer (20 mM Hepes, pH 7.2, 10 mM KCl, 2 mM MgCl2 and PI), and the protein samples were subject to western blot as described (22,23).
Native gel analysis of the p21/CDK2 complex
About 7–15% gradient polyacrylamide gels containing no sodium dodecyl sulphate (SDS) were prepared and used with a non-SDS running buffer containing 25 mM Tris-Cl, 190 mM glycine and 1 mM DTT adjusted to pH 8.0. The running buffer, the native gels, and gel running equipment were cooled to 4°C. The gels were subject to pre-electrophoresis at 100 V for 1 h before sample loading. Cells were lysed, and one part of the protein sample was mixed with one part of the native sample loading buffer (62.5 mM Tris-Cl, pH 6.8, 40% glycerol, 0.01% Bromophenol Blue). The diluted samples were loaded onto the gels, and electrophoresis was carried out using 80 V at 4°C until the dye migrated to the bottom of the gels. The gels were then subject to western blotting.
RT-PCR and real-time PCR
This was performed as described previously (24). In brief, 2 × 106 cells were harvested, and total RNAs were extracted with the Qiagen RNeasy mini kit. Two micrograms of total RNAs were used to synthesize cDNA, a portion of which (1 μl, equal to 0.2 μg cDNA) was used in a PCR with two appropriate primers. Real-time PCR was performed with miScriptSYBR GreenPCR Kit (Qiagen) using 1 μl cDNA as templates. The primers used for real-time PCR internal controls were mouse-U6RNAf and mouse-U6RNAr.
Immunoprecipitation assay
107 cells were washed in ice-cold phosphate-buffered saline, and lysed in 500 μl co-IP buffer (20 mM Tris-CL, pH 7.5, 150 mM NaCl, 1 mM ethylenediaminetetraacetic acid, 0.5% NP-40, and 5 μg/ml aprotinin). Equal amounts of proteins were incubated with 5 μg of primary antibody and 50 μl of 50% slurry of protein A-Sepharose at 4 °C for 4 h. The pellet was washed 3× with PBS and was resuspended in 2× Laemmli buffer (0.125 M Tris–HCl, 4% SDS, 20% glycerol, 10% 2-mercaptoethanol, 0.004% bromphenol blue, pH6.8), followed by western blot analysis.
RNA binding protein immunoprecipitation assay (RIP)
In functional assays, cells were harvested when they reached 70–80% confluence. In brief, 107 cells were washed in ice-cold PBS, lysed in 500 μl co-IP buffer and incubated with 5 μg of primary antibody at 4 °C for 2 h. A total of 40 μl of 50% slurry of protein A-Sepharose was added to each sample, and the mixtures were incubated at 4 °C for 4 h. The pellets were washed 3× with PBS and resuspended in 0.5 ml Tri Reagent (Sigma-Aldrich). The eluted co-precipitated RNA in the aqueous solution was subject to qRT-PCR analysis to demonstrate the presence of the binding products using respective primers.
RNA pull-down assays
RNA pull-down assays were performed as described (25,26). In brief, 107 cells were washed in ice-cold phosphate-buffered saline, lysed in 500 μl co-IP buffer, and incubated with 3 μg biotinylated DNA oligo probes against endogenous or ectopically expressed circ-Foxo3, at room temperature for 2 h. A total of 50 μl washed Streptavidin C1 magnetic beads (Invitrogen) were added to each binding reaction and further incubated at room temperature for another hour. The beads were washed briefly with co-IP buffer for five times. The bound proteins in the pull-down materials were analyzed by western blotting.
Northern blot
RNAs were isolated with RNA extract kit. Northern blot analysis was performed with northern blot kit (Ambion) as described (27). Briefly, the preparation of total RNAs (30 μg) was denatured in formaldehyde and then electrophoresed in a 1% agarose–formaldehyde gel. The RNAs were then transferred onto a Hybond-N+nylon membrane (Amersham) and hybridized with biotin-labeled DNA probes. Biotin Chromogenic Detection kit (Thermo Scientific) was used to develop the bound RNAs.
Statistical analysis
All experiments were performed in triplicate and numerical data were subject to independent sample t-test. The levels of significance were set at *P < 0.05 and **P < 0.01.
RESULTS
Expression of circ-Foxo3 repressed cell cycle progression
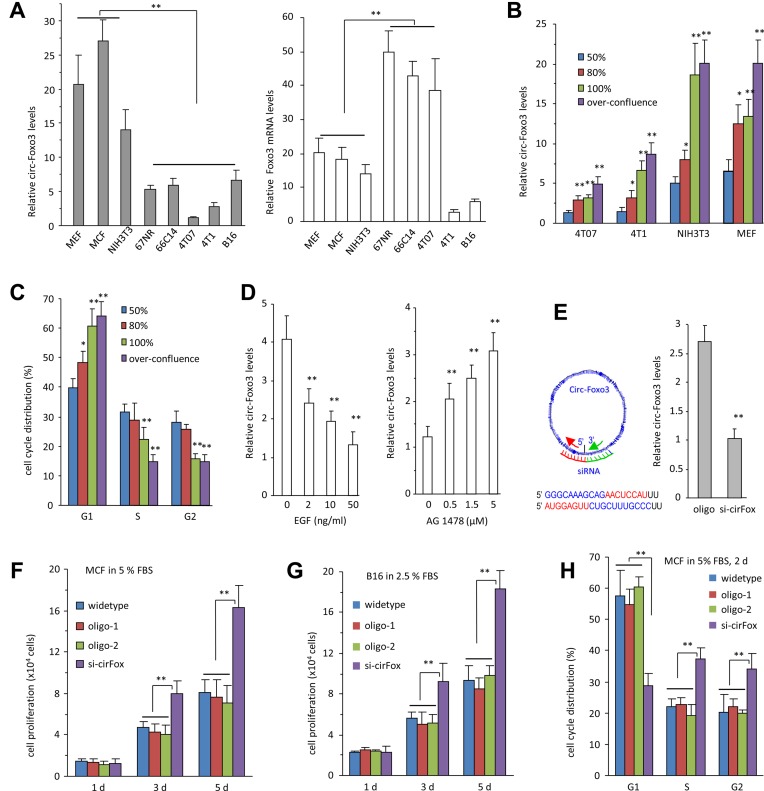
It has been proposed that circular RNAs are conserved across species (28,29). We analyzed the sequence of human circ-Foxo3 and mouse circ-Foxo3 and found that they were 91% homologous (Supplementary Figure S1d). We analyzed circ-Foxo3 and Foxo3 levels in mouse cancer cells and non-cancer cell lines by RT-PCR and real-time PCR. The levels of circ-Foxo3 and Foxo3 were inversely correlated in non-cancer and cancer cell lines, although two cancer cells line 4T1 and B16 expressed low levels of both transcripts (Figure 1A, Supplementary Figure S2a). RNAs isolated from the above cell lines were subject to northern blotting with a probe specific for circ-Foxo3, which confirmed the detection of circ-Foxo3 in the cells (Supplementary Figure S2b). These results suggested that expression of circ-Foxo3 was associated with cell cycle progression and proliferation. We cultured the cell lines MEF, NIH3T3, 4T07 and 4T1 to the confluency of 50, 80, 100% and over confluence, then measured cell cycle distribution and circ-Foxo3 levels. We found that circ-Foxo3 levels were significantly upregulated (Figure 1B) when cell cycle was arrested (Figure 1C, Supplementary Figure S2c and d). The levels of circ-Foxo3 decreased when the cells were treated with cell proliferating factor Epidermal growth factor (EGF) but increased when the cells were treated with EGF inhibitor AG1478 (Figure 1D).
Figure 1.
The effect of circ-Foxo3 on cell proliferation. (A) Left, Real-time PCR showed that circ-Foxo3 was highly expressed in the non-cancer cell lines of mouse embryo fibroblast (MEF), mouse cardiac fibroblast (MCF) and NIH3T3, as compared with the cancer cell lines 67NR, 66C14, 4T07, 4T1 and B16. Right, The levels of Foxo3 linear mRNA were not correlated with circ-Foxo3. Asterisks indicate significant differences. **P < 0.001, Error bars, SD (n = 4). (B and C) Different cell lines as indicated (B) or NIH3T3 fibroblasts (C) were seeded at the cell density of 1 × 105 cells/well on 6-well dishes in 10% FBS/DMEM medium until 50, 80, 100% or over-confluence based on the coverage of the surface of the tissue culture plates, followed by determination of circ-Foxo3 levels and cell cycle distribution. Increased cell densities expressed higher levels of circ-Foxo3 (B) and more cells were detected in the G1 phase (C). (D) NIH3T3 fibroblasts were incubated in basal medium with EGF (0, 2, 10 and 50 ng/ml, left) or AG1478 (0, 0.5, 1.5 and 5 μM, right) for 24 h. Real-time PCR showed circ-Foxo3 expression decreased after EGF treatment but increased after AG1478 treatment. **P < 0.001, Error bars, SD (n = 4). (E) Left, A siRNA was designed to specifically target circ-Foxo3. Right, Cells were transfected with siRNA targeting circ-Foxo3 or a control oligo. RNAs isolated were subject to real-time PCR to confirm downregulation of circ-Foxo3 in the siRNA-transfected cells. (F) MCF cells transfected without (wild-type) or with circ-Foxo3 siRNA or 2 oligos with random sequences were cultured in DMEM supplemented with 5% FBS for up to 5 days. The siRNA-transfected cells grew fast compared to the controls. **P < 0.001, Error bars, SD (n = 4). (G) B16 cells transfected without or with circ-Foxo3 siRNA or the control oligos were cultured in DMEM with 2.5% FBS for up to 5 days. Cell proliferation assays showed that circ-Foxo3 siRNA-transfected cells grew fast compared to the controls. **P < 0.001, Error bars, SD (n = 4). (H) Silencing circ-Foxo3 decreased the number of cells in G1 phase, but increased the number of cells in S and G2 phase. **P < 0.01. Error bars, SD (n = 4).
To validate the essential roles of circ-Foxo3 in cell cycle progression, we designed siRNA specifically targeting circ-Foxo3 to knockdown circ-Foxo3 function (Figure 1E, left). Transfection with this siRNA effectively silenced circ-Foxo3 levels (Figure 1E, right, Supplementary Figure S2e). Furthermore, mouse cardiac fibroblasts that express high levels of circ-Foxo3 were transfected with siRNA specifically targeting circ-Foxo3 or two oligos with random sequences. Silencing circ-Foxo3 was confirmed by real-time PCR in the siRNA-transfected cells relative compared with wild-type cells or the cells transfected with the oligos (Supplementary Figure S2f). Cell proliferation assay showed that the cells transfected with circ-Foxo3 siRNA had an increased proliferative capacity relative to cells transfected with the control oligos and the untransfected wild-type cells, suggesting a role of endogenous circ-Foxo3 in cell proliferation (Figure 1F). In B16 cells, we also detected an increase in cell proliferation compared with the controls (Figure 1G).
Cell cycle analysis revealed that fewer siRNA-transfected MCF cells were detected in the G1 phase, but more were detected in S and G2 phases as compared with the controls (Figure 1H, Supplementary Figure S2g), which suggested an increase in cell cycle progression.
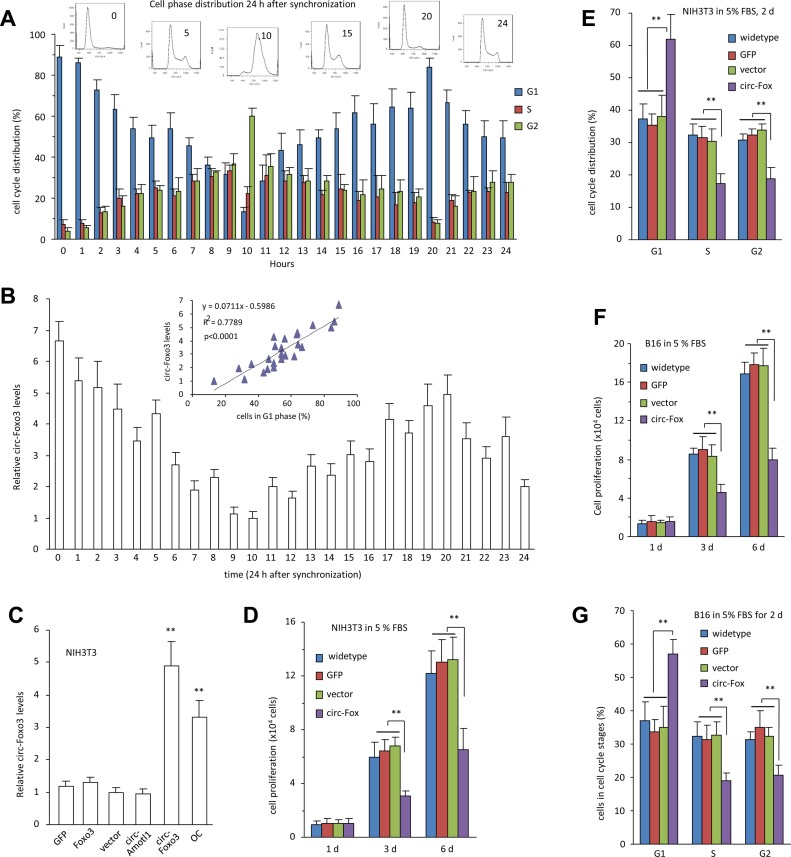
We further examined the relationship between cell cycle progression and circ-Foxo3 expression. NIH3T3 cells were cultured in serum-free medium for 48 h, then changed with medium containing 10% FBS. Cells were collected hourly for up to 24 h, followed by cell cycle analysis (Figure 2a) and circ-Foxo3 expression (Figure 2b). Plotting of both revealed a strong correlation between cell cycle progression and circ-Foxo3 expression (Figure 2B, inset).
Figure 2.
circ-Foxo3 repressed cell cycle entry. (A) NIH3T3 cells were cultured in serum-free medium for 48 h, then changed with medium containing 10% FBS for cell cycle synchronization. Cells were collected each hour for up-to 24 h and subject to flow cytometry for cell cycle analysis. Error bars, SD (n = 3). (B) Circ-Foxo3 levels were determined by real-time PCR. Inset, the correlation between circ-Foxo3 levels and percentage of cells in G1 phase was analyzed by Graph Prism 4. It showed that circ-Foxo3 levels were highly correlated with the percentages of cells in G1 phase. R squares, 0.778; P < 0.001, n = 25. (C) Levels of circ-Foxo3 were determined by real-time PCR in NIH3T3 cells transfected with GFP vector, Foxo3, mock control, circ-Amotl1 and circ-Foxo, or in over confluence (OC) cells. **P < 0.01. Error bars, SD (n = 4). (D) NIH3T3 fibroblasts transfected without or with circ-Foxo3, the control vector, or a GFP plasmid were cultured in DMEM with 5% FBS. Cell proliferation assays showed that circ-Foxo3 expressing cells grew slowly compared to the controls. **P < 0.01. Error bars, SD (n = 4). (E) The cells were also cultured in DMEM supplemented with 5% FBS for 2 days, and then processed to flow cytometry. Expression of circ-Foxo3 significantly increased the number of cells in the G1 phase, and decreased the number of cells in the S and G2 phases. **P < 0.01. Error bars, SD (n = 4). (F) B16 cells were transfected as above and cultured in DMEM with 5% FBS. Cell proliferation assays showed that circ-Foxo3 expressing cells grew slowly compared to the controls. **P < 0.01. Error bars, SD (n = 4). (G) The cells were also subject to flow cytometry. Expression of circ-Foxo3 significantly increased the number of cells in G1 phase and decreased the number of cells in S and G2 phases. **P < 0.01. Error bars, SD (n = 4).
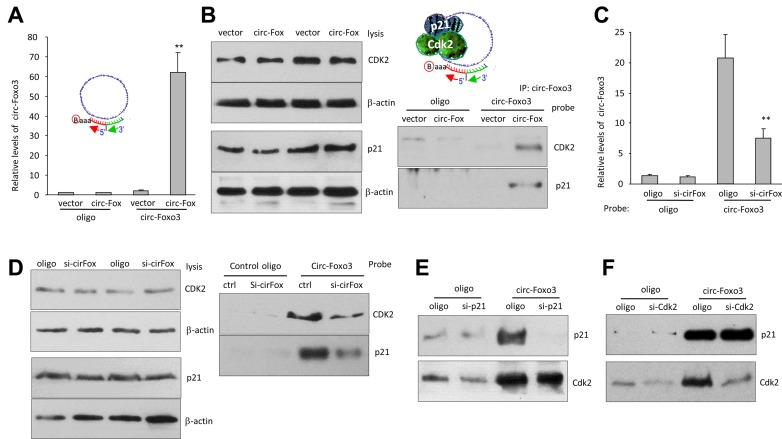
We examined the role of circ-Foxo3 in mediating cell activity by stably transfecting NIH3T3 fibroblasts with circ-Foxo3 expression construct, a control vector or other expression plasmids. Expression of circ-Foxo3 was confirmed by RT-PCR (Supplementary Figure S3a), real-time PCR (Supplementary Figure S3b) and northern blotting (Figure 2C, Supplementary Figure S3c). The RNA samples were treated with RNAse-R prior to real-time PCR. Only RNA isolated from the circ-Foxo3-transfected cells displayed resistance to RNAse-R cleavage, confirming circularization of circ-Foxo3 (Supplementary Figure S3d).
Circ-Foxo3 and vector-transfected NIH3T3 cells were cultured in DMEM 5% FBS. Cell proliferation assays showed that cells expressing circ-Foxo3 proliferated less rapidly when compared with cells transfected without or with the vector or GFP plasmid (Figure 2D, Supplementary Figure S3e). Expression of circ-Foxo3 was confirmed by real-time PCR (Supplementary Figure S3f). Cell cycle analysis indicated that there were more circ-Foxo3 cells detected in the G1 phase but fewer in S and G2 phases compared to the controls (Figure 2E, Supplementary Figure S4a). We also tested the role of circ-Foxo3 in the cancer cell line B16. Expression of circ-Foxo3 was confirmed by RT-PCR and real-time PCR (Supplementary Figure S4b). Similarly, the circ-Foxo3-transfected cells grew slower than the controls (Figure 2f, Supplementary Figure S4c) and more cells were found in the G1 phase but fewer cells in S and G2 phases (Figure 2G).
Formation of ternary complexes by circ-Foxo3, CDK2 and P21
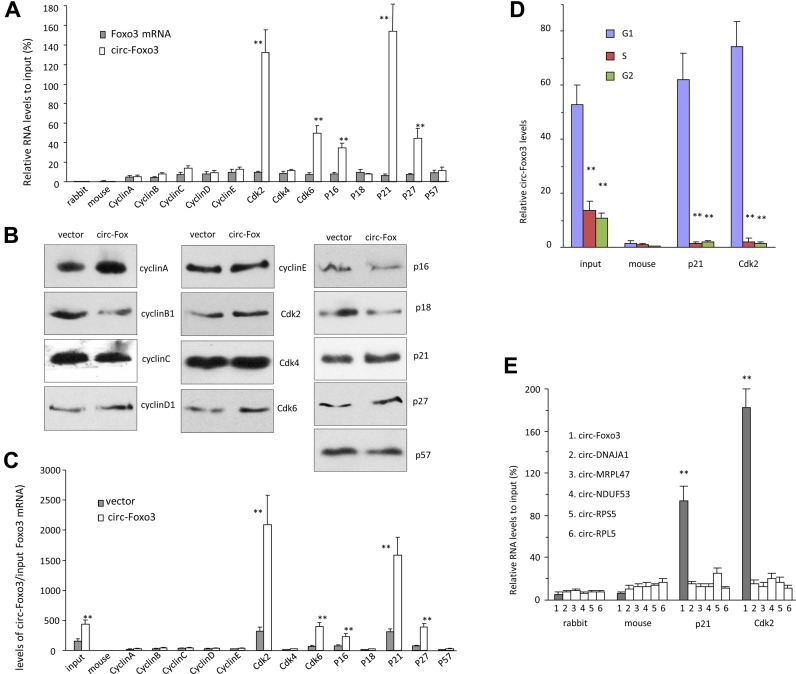
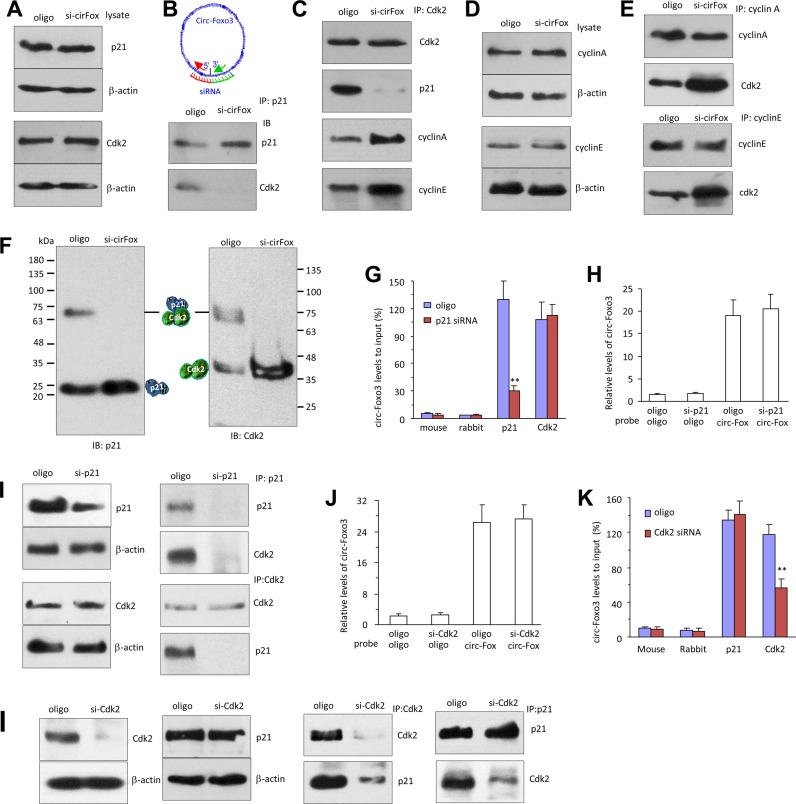
We tested potential interactions of circ-Foxo3 with cell cycle associated proteins. Cell lysis prepared from NIH3T3 cells were subject to immuno-precipitation (IP) with anti-rabbit IgG, mouse IgG, cyclin A, cyclin B, cyclin C, cyclin D, cyclin E, CDK2, CDK4, CDK6, p16, p18, p21, p27 and p57 antibodies, followed by real-time PCR with primers specific for the linear Foxo3 mRNA or circ-Foxo3. The experiment showed that circ-Foxo3 was pulled-down by immunoprecipitation experiments with antibodies against CDK2, CDK6, p16, p21and p27, which did not pull-down linear Foxo3 mRNA (Figure 3A). Cell lysates prepared from the vector- and circ-Foxo3-transfected cells were subject to IP assay with the antibodies indicated above. We validated the function of all antibodies (Figure 3B), but found that only antibodies against CDK2, CDK6, p16, p21and p27 were able to pull-down significantly higher levels of circ-Foxo3 from the circ-Foxo3 cells than those from the control cells (Figure 3c). It was noted that the CDK2 and p21 showed the greatest difference in our experiments, suggesting a critical role for CDK2 and p21 in circ-Foxo3-mediated cell cycle progression. Cells were sorted by flow cytometry to obtain G1-, S- and G2-phase cells for lysate preparation. Anti-CDK2 and anti-p21 antibodies pulling-down circ-Foxo3 mainly occurred mainly in G1 phase (Figure 3D). This was consistent with our results that ectopic expressed circ-Foxo3 arrested cells in G1 phase.
Figure 3.
circ-Foxo3 interacted with CDK2 and P21. (A) Cell lysis prepared from NIH3T3 cells were subject to immuno-precipitation (IP) with antibodies against rabbit IgG, mouse IgG, cyclin A, cyclin B, cyclin C, cyclin D1, cyclin E, CDK2, CDK4, CDK6, p16, p18, p21, p27 and p57, followed by real-time PCR with primers specific for linear Foxo3 mRNA or circ-Foxo3. Anti-CDK2, CDK6, p16, p21and p27 antibodies pulled-down circ-Foxo3, but not linear Foxo3 mRNA. It was especially obvious that precipitating CDK2 or P21 pulled-down circ-Foxo3. **P < 0.01. Error bars, SD (n = 4). (B) Cell lysates prepared from NIH3T3 cells transfected with circ-Foxo3 or mock control were subject to immunoprecipitation with anti-rabbit IgG, mouse IgG, cyclin A, cyclin B, cyclin C, cyclin D, cyclin E, CDK2, CDK4, CDK6, p16, p18, p21, p27 and p57 antibodies. Western blot showed that immunoprecipitation pulled down similar amount of proteins in both control and circ-Foxo3-transfected cells. (C) The immuno-precipitated mixtures were also subject to real-time PCR with primers specific for circ-Foxo3. Anti-CDK2, CDK6, p16, p21and p27 antibodies pulled-down more circ-Foxo3 from NIH3T3 cells transfected with circ-Foxo3 than from mock control. *P < 0.01. Error bars, SD (n = 4). (D) NIH3T3 cells were subject to flow cytometry to sort cells in G1, S and G2 phases, followed by immunoprecipitation with anti-rabbit IgG, mouse IgG, p21 and Cdk2 antibodies and real-time PCR with primers specific for circ-Foxo3. Antibodies against p21 and Cdk2 precipitated significantly more circ-Foxo3 in G1 phase than in G2 an S phases. **P < 0.01. Error bars, SD (n = 4). (E) Cell lysates prepared were subject to immunoprecipitation with anti-rabbit IgG, mouse IgG, p21 and Cdk2 antibodies, followed by real-time PCR with primers specific for circ-Foxo3, circ-DNAJA1, circ-MRPL47, circ-NDUF53, circ-RPS5 and circ-RPF5. Antibodies against p21 and Cdk2 pulled-down circ-Foxo3, but not the other circular RNAs. **P < 0.01. Error bars, SD (n = 4).
To analyze the specificity of the interaction, we examined the levels of other circular RNAs including circ-DNSJA1, circ-MRPL47, circ-NDUF53, circ-RPS5 and circ-PRL5. We found that antibodies against p21 and Cdk2 did not pull-down these circular RNAs (Figure 3E), suggesting specific interaction between circ-Foxo3 and these two proteins.
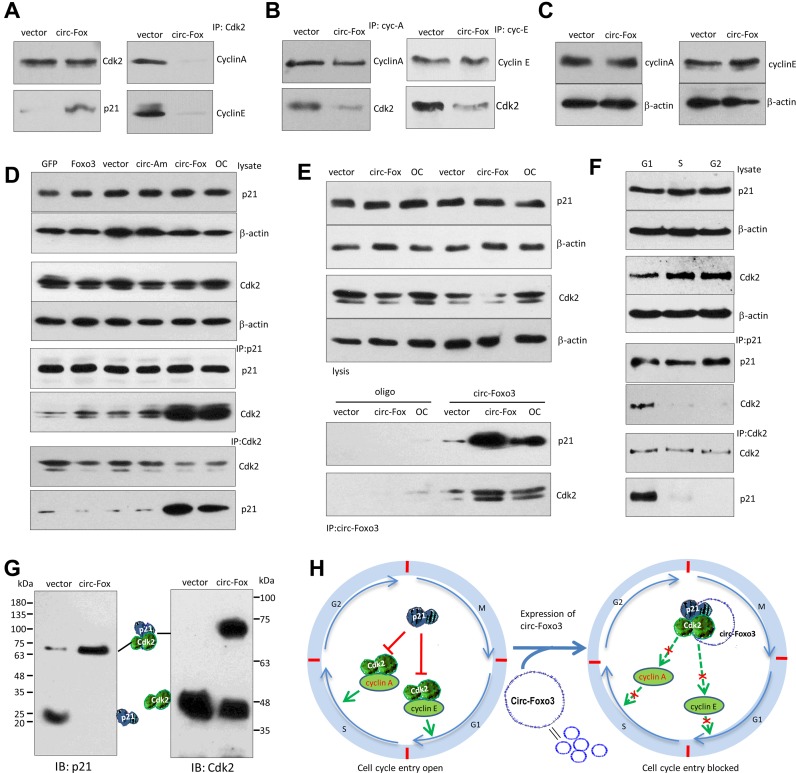
We tested the interactions between circ-Foxo3 with CDK2 and p21. Western blot analysis indicated that anti-CDK2 antibody immuno-precipitated higher levels of p21, but lower levels of cyclin A and cyclin E, in the circ-Foxo3-transfected cells (Figure 4A). However, in the control group, anti-CDK2 antibody did in fact pull-down cyclin A and cyclin E, but not p21, since CDK2 is known to interact with cyclin A and cyclin E (30–32). In the circ-Foxo3 transfected cells, anti-cyclin A and anti-cyclin E antibodies pulled-down less CDK2 compared with the control cells (Figure 4B). Nevertheless, expression of cyclin A and cyclin E was not affected by circ-Foxo3 transfection (Figure 4C).
Figure 4.
circ-Foxo3 enhanced the interaction between p21 and CDK2. (A) Lysates prepared from NIH3T3 cells transfected with circ-Foxo3 or mock control were subject to IP with anti-CDK2 antibody, followed by Western blotting. CDK2 precipitation pulled down more p21, and less cyclin A and cyclin E in the circ-Foxo3-transfected cells than the control. (B) Left, the lysates were subject to IP with anti-cyclin A antibody, followed by Western blotting. Cyclin A precipitation pulled down less CDK2 in the circ-Foxo3-transfected cells than in the control. Right, the lysates were subject to IP with anti-cyclin E antibody, followed by western blotting. Cyclin E precipitation pulled-down less CDK2 in the circ-Foxo3-transfected cells than in the control. (C) Circ-Foxo3 transfection did not change expression of cyclin A and cyclin E. (D) Cell lysates prepared from NIH3T3 cells transfected with GFP vector, Foxo3, circular RNA vector, circ-Amotl1 and circ-Foxo3, or over-confluence culture were subject to western blot probed with antibodies against CDK2, p21 and β-action. The lysates were also subject to immunoprecipitation with antibody against p21 or CDK2. Anti-p21 antibody pulled-down more Cdk2 and anti-CDK2 antibody pulled-down more p21 in the cells transfected with circ-Foxo3 or overgrown. (E) Lysates prepared from NIH3T3 cells transfected with circ-Foxo3 and a vector, or over-grown cells were mixed with biotinylated probes against circ-Foxo3 or an oligo. Western blotting showed that levels of CDK2 and p21 were not affected (upper). However, the circ-Foxo3 probe pulled down more CDK2 and p21 in the cells transfected with circ-Foxo3 or over-grown relative to the controls (lower). (F) Lysates prepared from NIH3T3 cells sorted into G1, S or G2 phase were subject to Western blotting. The levels of p21 and CDK2 were similar in cells of different phases (upper). Anti-p21 and anti-CDK2 antibodies pulled down more CDK2 and p21, respectively, in the G1 phase cells. (G) The lysates prepared from circ-Foxo3- and mock-transfected cells were subject to native gradient gel electrophoresis followed by western blotting probed with antibodies against p21 or CDK2. (H) Diagram of our hypothesis showing the effect of circ-Foxo3 on cell cycle progression. Circ-Foxo3 retards cell cycle entry via enhancing interaction between p21 and CDK2, which repressed CDK2–cyclin complex formation.
We tested the specificity of circ-Foxo3 in mediating the interaction of p21 and CDK2. NIH3T3 fibroblasts were transfected with a number of expression constructs (Foxo3, circ-Amotl1 and circFoxo3) and control vectors. The cells were also over grown to maintain high levels of circ-Foxo3. Western blot analysis showed that expression of p21 and CDK2 was not affected by transfection of the constructs nor affected by cell over growth (Figure 4D). However, in immunoprecipitation assays, anti-p21 antibody only pulled-down high levels of CDK2 in the circ-Foxo3-transfected cells or when NIH3T3 fibroblasts were overgrown (Figure 4D). Similarly, anti-CDK2 antibody only pulled-down p21 in the same conditions while circ-Foxo3 levels were high. In the cells transfected with circ-Foxo3 or over grown, we confirmed that while expression of p21 and CDK2 was not affected, the circ-Foxo3 probe pulled-down high levels of p21 and CDK2 relative to the control oligo (Figure 4E).
Since circ-Foxo3 was found to stay mainly in G1 phase (Figure 3D), we tested if the interaction of p21 and CDK2 mediated by circ-Foxo3 mainly occurred in G1 phase. We confirmed that the anti-p21 antibody was able to pull-down high levels of CDK2, and that anti-CDK2 antibody could pull-down high levels of p21 in G1 phase (Figure 4F).
These results suggested that circ-Foxo3, CDK2 and p21 formed ternary complexes. To validate this, we resolved protein lysates prepared from the vector and circ-Foxo3-transfected cells in native gradient gels, followed by western blotting probed with antibodies against p21 and CDK2. In the circ-Foxo3-transfected cells, p21 mainly migrated as a single band with a size slightly larger than the sum of CDK2 and p21 (∼70 kDa, Figure 4G, left). In the vector-transfected cells, a large portion of p21 migrated as a monomer, and a small quantity of p21 migrated with a mass of 70 kDa. Similarly, antibody against CDK2 detected CDK2 monomer and hetero-dimer with p21 (Figure 4G, right). The above results are summarized in the diagram in Figure 4H.
Disruption of the ternary complex increased cell cycle entry
We validated the effect of circ-Foxo3 in mediating the interaction of CDK2 and p21. Asynchronized NIH3T3 fibroblasts transfected with siRNA targeting circ-Foxo3 were confirmed to retain their capacity in expression of CDK2 and p21 (Figure 5A). IP assay showed that the association of p21 with CDK2 was reduced in the siRNA-transfected cells (Figure 5B). Similarly, CDK2 association with p21 decreased in the siRNA-transfected cells after IP with anti-CDK2 antibody, but the capacity of CDK2 binding with cyclin A and cyclin E was recovered (Figure 5C). Furthermore, while the expression of cyclin A and cyclin E were not affected by siRNA transfection (Figure 5D), both antibodies against cyclin A and cyclin E pulled-down more CDK2 in the cells transfected with circ-Foxo3 siRNA compared with the control oligo (Figure 5E).
Figure 5.
Silencing circ-Foxo3 enhanced cell cycle entry and decreased the interaction between p21 and CDK2. (A) Lysates prepared from subconfluence NIH3T3 cells transfected with circ-Foxo3 siRNA or a control oligo were subject to western blotting. Silencing circ-Foxo3 did not change expression of p21 and CDK2. (B) The lysates were subject to IP with anti-p21 antibody followed by western blotting. p21 precipitation pulled-down less CDK2 in the circ-Foxo3 siRNA-transfected cells relative to the control. (C) The lysates were subject to IP with anti-CDK2 antibody, followed by western blotting. CDK2 precipitation pulled-down less p21, but more cyclin A and cyclin E in the siRNA-transfected cells compared to the control. (D) Silencing circ-Foxo3 did not change expression of cyclin A and cyclin E. (E) Cyclin A and cyclin E precipitations pulled down more CDK2 in the circ-Foxo3 siRNA-transfected cells. (F) The lysates were subject to native gradient gel electrophoresis followed by western blotting probed with antibodies against p21 or CDK2. (G) Cell lysates prepared from p21 siRNA- and a control oligo-transfected NIH3T3 cells were subject to immunoprecipitation with anti-rabbit IgG, mouse IgG, p21 and Cdk2 antibodies, followed by real-time PCR. Anti-p21 antibody pulled-down less circ-Foxo3 in the p21 siRNA-transfected cells. **P < 0.01. Error bars, SD (n = 4). (H) The lysates were hybridized with circ-Foxo3 probe or a control oligo for RNA pull-down assays. Real-time PCR showed that the circ-Foxo3 probe pulled down same levels of circ-Foxo3 in the cells transfected with p21 siRNA. Error bars, SD (n = 4). (I) Silencing p21 did not affect CDK2 expression. However, anti-p21 antibody pulled-down less Cdk2 and p21 in the p21 siRNA-transfected cells. Anti-CDK2 antibody pulled-down less p21, but the same amount of Cdk2 in the p21 siRNA-transfected cells. (J) Silencing CDK2 did not affected circ-Foxo3 levels. (K) Anti-CDK2 antibody pulled-down less circ-Foxo3 in the Cdk2 siRNA-transfected cells. **P < 0.01. Error bars, SD (n = 4). (L) Silencing CDK2 did not affect p21 expression. However, anti-CDK2 antibody pulled-down less Cdk2 and p21 in the CDk2 siRNA-transfected cells. Anti-p21 antibody pulled-down less CDK2, but the same amount of p21 in the CDK2 silencing cells.
We also validated the effect of circ-Foxo3 on the formation of the ternary complexes. Protein lysates prepared from siRNA- and control oligo-transfected cells were resolved in native gradient gels, followed by Western blotting probed with antibodies against p21 and CDK2. In the circ-Foxo3 siRNA-transfected cells, p21 mainly migrated as a single band of p21 monomer (∼20 kDa, Figure 5F, left). In the control, some endogenous circ-Foxo3 appeared to facilitate the interaction of p21 and CDK2. Moreover, anti-CDK2 antibody detected a CDK2 monomer in the siRNA-transfected cells, and it also detected the hetero-dimmer with p21 (Figure 5F, right).
We further examined whether silencing p21 and CDK2 affected the interaction of circ-Foxo3 with these proteins. In cells transfected with p21 siRNA, anti-p21 antibody pulled-down decreased levels of circ-Foxo3 relative to cells transfected with the control oligo (Figure 5G), while the total levels of circ-Foxo3 appeared unaffected as shown by circ-Foxo3 pull-down assay using circ-Foxo3 probe (Figure 5H). The anti-p21 antibody pulled-down trace amounts of p21 and CDK2 (Figure 5I).
Similarly, silencing CDK2 did not affect circ-Foxo3 probe to pull-down circFoxo3 (Figure 5j), but resulted in pulling down decreased levels of circ-Foxo3 using an anti-CDK2 antibody (Figure 5K). In the CDK2 siRNA-transfected cells, anti-CDK2 antibody precipitated decreased levels of CDK2 and p21, while anti-p21 antibody only precipitated decreased levels of CDK2 (Figure 5L).
Pulling down circ-Foxo3 bound both p21 and CDK2
We tested whether the probe used for Northern blot could pull-down p21 and CDK2. Cell lysate prepared from NIH3T3 fibroblasts transfected with circ-Foxo3 or the mock control was subject to the pull-down assay. While we confirmed that the levels of circ-Foxo3 were similar in the lysates mixed with the probe or the control oligo (Supplementary Figure S4d, left), we found that the probe significantly pulled-down more circ-Foxo3 than the control oligo (Figure 6A). In the protein precipitation assay, after confirming equal amounts of p21 and CDK2 in the mixture containing the probe or the control oligo (Figure 6B, left), we found that both p21 and CDK2 were pulled-down by the probe but not by the control oligo in the circ-Foxo3-transfected cells, but this did not occur in the vector-transfected cells (Figure 6B, right).
Figure 6.
Pulling down circ-Foxo3 precipitated both p21 and CDK2. (A) Lysates prepared from NIH3T3 cells transfected with circ-Foxo3 or a control vector, were subject to RNA pull-down assays. Real-time PCR showed that the circ-Foxo3 probe pulled down high levels of circ-Foxo3 in the cells transfected with circ-Foxo3. **P < 0.01. Error bars, SD (n = 4). (B) Left: the lysates were mixed with biotinylated probes against circ-Foxo3 or a control oligo and subject to western blot with antibodies against CDK2, p21 and β-action. Right: the pull-down mixture was subject to Western blotting. Pulling down circ-Foxo3 also pulled down CDK2 and p21 in cells transfected with circ-Foxo3. (C) Lysates prepared from NIH3T3 cells transfected with circ-Foxo3 siRNA or a control oligo, were subject to RNA pull down assays. Real-time PCR showed that the probe pulled down less circ-Foxo3 in the cells transfected with circ-Foxo3 siRNA. **P < 0.01. Error bars, SD (n = 4). (D) Left, transfection with circ-Foxo3 siRNA did not affect expression of CDK2 and p21. Right, Pulling down circ-Foxo3 precipitated less CDK2 and p21 in the cells transfected with circ-Foxo3 siRNA than those transfected with the control oligo. (E) Silencing p21 had little effect on circ-Foxo3 pulling down CDK2. (F) Silencing Cdk2 had little effect on circ-Foxo3 pulling down p21.
We also tested the effect that circ-Foxo3 knock-down would have on the pulling-down p21 and CDK2. While we confirmed that the siRNA targeting circ-Foxo3 knocked-down significant levels of circ-Foxo3 (Supplementary Figure S4d, right), we found that the probe pulled-down significantly lower levels of circ-Foxo3 in the siRNA-transfected cells compared with the control (Figure 6C). Consistent with this result, lower protein levels of p21 and CDK2 were pulled-down by the probe compared with the control oligo (Figure 6D).
In the p21 siRNA-transfected cells, the circ-Foxo3 probe could only pull down trace amount of p21 but pulled down equal levels of CDK2 in the p21 siRNA-transfected cells relative to the oligo-transfected cells (Figure 6E). Reduction of p21 did not disturb the interaction of circ-Foxo3 with CDK2, suggesting the exiting of circ-Foxo3–CDK2 complex in addition to the ternary complex. In the CDK2 siRNA-transfected cells, the circ-Foxo3 probe could pull down equal levels of p21 but pulled down decreased levels of CDK2 (Figure 6F), suggesting presence of the circ-Foxo3–p21 complex.
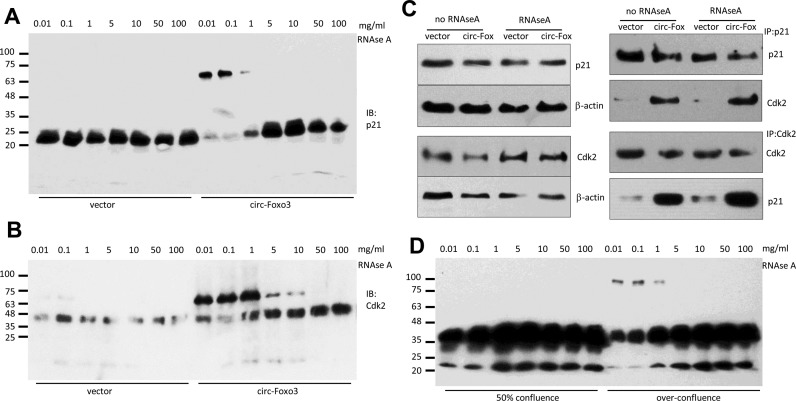
Protection of the p21 and CDK2 binding site
We further confirmed the formation of the ternary complex. Cell lysates prepared from the circ-Foxo3- and vector-transfected cells were incubated with RNAse A at different concentrations. The lysates were separated in native gradient gels, followed by western blotting probed with antibodies against p21 and CDK2. In the vector-transfected cells, p21 migrated as a single band (Figure 7A). However, in the circ-Foxo3-transfected cells, two bands, one monomer and one with size slightly larger than the sum of CDK2 and p21, were detected when the lysates were treated with lower concentrations of RNAse A. This suggests that the binding site for p21 and CDK2 on the circ-Foxo3 was protected from being degraded by RNAse A. It also confirmed the formation of a ternary complex formed by circ-Foxo3, p21 and CDK2. However, at high concentrations of RNAse A, the enzyme was too potent which caused it to destroy the complex, resulting in the presence of only the monomer. Similarly, antibody against CDK2 detected CDK2 monomer and the heterodimer with p21 when the lysates of circ-Foxo3-transfected cells were treated with low concentrations of RNAse A (Figure 7B). At high concentrations of RNAse A, the anti-CDK2 antibody again only detected monomer. Nevertheless, anti-p21 antibody not only pulled down p21, but also pulled down high levels of CDK2 in the circ-Foxo3-transfected cells with or without RNAse A treatment (Figure 7C), suggesting protection of the p21 and CDK2 binding sites in circ-Foxo3 forming the ternary complexes. As well, anti-CDK2 antibody not only pulled down CDK2, but also pulled down high levels of p21 in the circ-Foxo3-transfected cells with or without RNAse A treatment.
Figure 7.
CDK2 and p21 protected the circ-Foxo3 binding site. (A and B) The lysates prepared from circ-Foxo3- and mock-transfected cells were incubated with different concentrations of RNAse A followed by native gradient gel electrophoresis. The gels were subject to Western blotting probed with antibodies against p21 (A) or CDK2 (B). (C) Cell lysates from NIH3T3 cells transfected with circ-Foxo3 and mock control were treated with or without RNAse A (0.1 mg/ml), and subject to western blotting. While treatment with RNAse A did not affect levels of CDK2 and p21 (left), anti-p21 antibody was able to pull-down Cdk2 and anti-CDK2 antibody was able to pull down p21 in the circ-Foxo3-transfected cells with or without RNAse A treatment. (D) The lysates prepared from NIH3T3 cells grown to 50% conference and over-confluence were incubated with different concentrations of RNAse A followed by native gradient gel electrophoresis. The gels were subject to western blot probed with antibody against p21 and then against Cdk2.
We also prepared lysates from cells at 50% confluence or when the cells were over grown and incubated the lysates with RNAse A at different concentrations. After being separated in native gradient gels, protein bands were probed with a mix of p21 and CDK2 antibodies. We detected the larger bands only the over growth cells when the lysates were treated with low concentrations of RNAse A (Figure 7D).
DISCUSSION
In this study, we found that expression of the circular RNA circ-Foxo3 repressed cell proliferation and cell cycle progression. This conclusion was obtained based on a number of experiments. When cells were grown beyond confluency, levels of circ-Foxo3 increased and more cells were arrested at G1 phase, with a positive correlation between both circ-Foxo3 and cells in G1 phase. Cell treated with mitogen EGF had decreased levels of circ-Foxo3, while cells treated with EGF inhibitor produced increased levels of circ-Foxo3. Silencing endogenous circ-Foxo3 promoted cell proliferation and decreased the number of cells in G1 phase. It should be noted that the siRNA target site in circ-Foxo3 can only be located in the junction of circ-Foxo3, because siRNAs targeting other areas would also silence linear Foxo3 mRNA. To ensure that the silencing siRNA produced the desired effects, it was necessary to include more that one negative control. We included two control oligos with random sequences, as well as non-transfected cell cultures in the functional assays. Similarly, we also included two unrelated vectors and a non-transfected cell culture in the ectopic expression experiments. Taken together, we believe that this demonstrates the ectopic expression of circ-Foxo3 could inhibit cell proliferation and cell cycle progression.
To understand how circ-Foxo3 functioned in regulating cell proliferation, we explored the possibility that circ-Foxo3 might interact with cell cycle associated proteins and found that CDK2 and p21 could bind to circ-Foxo3. Formation of the circ-Foxo3–p21–CDK2 ternary complex hijacked CDK2 together with p21 avoiding the formation of cyclin E/CDK2 complex, thus blocking the transition from G1 to S phase. It also abolished the inhibitory effect of p21 on cyclin A/CDK2 complex, therefore blocking the progression of the cell cycle in S phase. As a result, cell cycle progression was arrested in the G1 phase and unable to transit to S phase.
CDK2 is an essential regulator in the G1-S phase transition. CDK2 binding to cyclin E initiates cell transition from G1 into S phase. This interaction can be inhibited by p21 binding to CDK2. Our results showed that circ-Foxo3 interacted with both p21 and CDK2. This enhanced the interaction of p21 with CDK2 allowing for a more efficient inhibition of CDK2 by p21. The formation of circ-Foxo3–p21–CDK2 ternary complex would block the interaction of CDK2 with cyclin E. Without the formation of the complex with CDK2, cyclin E could no longer phosphorylate p27 nor enhance cyclin A expression. As a result, the initiation of the transition from G1 to S phase was inhibited by overexpression of circ-Foxo3.
In normal cell cycle progression during the G1-S transition initiated by the CDK2–cyclin E complex, cyclin A takes cyclin E's position to bind CDK2. When the level of cyclin A/CDK2 complex reaches a threshold, it dissociates the assembly of the cyclin E/CDK2 complex. As the level of cyclin A/CDK2 complex continues to increase, DNA replication is initiated by the complex (33). The formation of the cyclin A/CDK2 complex is required for the cells to progress through the entire S phase (33,34). Overexpression of circ-Foxo3 would hijack CDK2 together with p21. As such, CDK2 could no longer form a complex with cyclin A. There would no longer be a sufficient level of cyclin A/CDK2 complex to dissociate the cyclin E/CDK2 complex for the G1-S transition. Therefore, the cells would then stay in the transition position between the G1 and S phase. Without sufficient level of cyclin A/CDK2 complex, DNA replication would not be triggered, and the cells could not pass through the S phase. This explains why we detected an increased number of the circ-Foxo3-transfected cells in G1 phase, and subsequently fewer of these cells in the S and G2 phases, as compared to the vector-transfected cells. Similarly, when the cells were overgrown, in which the levels of circ-Foxo3 were high, a greater proportion of cells were detected in the G1 phase. When analyzing the sorted population of G1 phase cells, we found that expression of circ-Foxo3 was significantly higher than cells in the other phases.
It appears that in the circ-Foxo3–p21–CDK2 ternary complexes, both p21 and CDK2 interacted with circ-Foxo3 on adjacent sites. When the ternary complex was treated with RNAse A, followed by a pulldown experiment with an antibody against p21, CDK2 was detected in the subsequent western blot. This suggests that the binding by p21 and CDK2 protected the binding sites of circ-Foxo3 from being cleaved by RNAse A. On the other hand, after treatment with RNAse A and immunoprecipitation with an antibody against CDK2, we detected p21 in the precipitated complex, confirming the protection of circ-Foxo3 binding sites by p21 and CDK2. However, if the cells were transfected with siRNA, specifically targeting circ-Foxo3, neither anti-p21 antibody nor anti-CDK2 antibody was able to pull-down the other binding partner. This confirms the essential role of circ-Foxo3 in the formation of the circ-Foxo3–p21–CDK2 ternary complex.
Our study focused on the interaction of circ-Foxo3 with CDK2 and p21, since both proteins appeared to possess a high binding affinity for circ-Foxo3. However, we also noted that circ-Foxo3 could interact with other cell cycle-associated proteins including CDK6, p16 and p27. Since these proteins are also important regulators of cell cycle progression, it is possible that this interaction could also regulate cell cycle progression. While the interaction of circ-Foxo3 with CDK2 and p21 exerted a strong function in the G1-S transition, the interaction of circ-Foxo3 with the other proteins may play roles in other steps of the cell cycle, or exert a more subtle regulatory function. This awaits further investigation.
It is interesting to note that the levels of circ-Foxo3 were not correlated to the levels of Foxo3 linear mRNA, although both appeared to have tumor suppressive activity. In a few cancer cell lines including 67NR, 66C14 and 4T07, the levels of Foxo3 mRNA were even significantly higher than the non-cancer cell lines. The expression and relevance of circ-Foxo3 and other endogenous circular RNAs await further investigation both in vitro and in vivo.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Discovery Grant from the Natural Sciences and Engineering Research Council of Canada [NSERC; 227937-2012 to B.B.Y.]; Heart and Stroke Foundation of Ontario Career Investigator Award [CI 7418 to to B.B.Y.]; Breast Cancer Foundation of Ontario Postdoctoral Fellowship (to W.W.D.). Funding for open access charge: CIHR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Jeck W.R., Sharpless N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014;32:453–461. doi: 10.1038/nbt.2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.AbouHaidar M.G., Venkataraman S., Golshani A., Liu B., Ahmad T. Novel coding, translation, and gene expression of a replicating covalently closed circular RNA of 220 nt. Proc. Natl. Acad. Sci. U.S.A. 2014;111:14542–14547. doi: 10.1073/pnas.1402814111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haimovich G., Medina D.A., Causse S.Z., Garber M., Millan-Zambrano G., Barkai O., Chavez S., Perez-Ortin J.E., Darzacq X., Choder M. Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell. 2013;153:1000–1011. doi: 10.1016/j.cell.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 4.Wilusz J.E., Sharp P.A. Molecular biology. A circuitous route to noncoding RNA. Science. 2013;340:440–441. doi: 10.1126/science.1238522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ivanov A., Memczak S., Wyler E., Torti F., Porath H.T., Orejuela M.R., Piechotta M., Levanon E.Y., Landthaler M., Dieterich C., et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–177. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 6.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 7.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 8.Capel B., Swain A., Nicolis S., Hacker A., Walter M., Koopman P., Goodfellow P., Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- 9.Li F., Zhang L., Li W., Deng J., Zheng J., An M., Lu J., Zhou Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6:6001–6013. doi: 10.18632/oncotarget.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson M.J., Viars C.S., Czekay S., Cavenee W.K., Arden K.C. Cloning and characterization of three human forkhead genes that comprise an FKHR-like gene subfamily. Genomics. 1998;47:187–199. doi: 10.1006/geno.1997.5122. [DOI] [PubMed] [Google Scholar]
- 11.Myatt S.S., Lam E.W. The emerging roles of forkhead box (Fox) proteins in cancer. Nat. Rev. Cancer. 2007;7:847–859. doi: 10.1038/nrc2223. [DOI] [PubMed] [Google Scholar]
- 12.Cho E.C., Kuo M.L., Liu X., Yang L., Hsieh Y.C., Wang J., Cheng Y., Yen Y. Tumor suppressor FOXO3 regulates ribonucleotide reductase subunit RRM2B and impacts on survival of cancer patients. Oncotarget. 2014;5:4834–4844. doi: 10.18632/oncotarget.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du W.W., Yang W., Fang L., Xuan J., Li H., Khorshidi A., Gupta S., Li X., Yang B.B. miR-17 extends mouse lifespan by inhibiting senescence signaling mediated by MKP7. Cell Death Dis. 2014;5:e1355. doi: 10.1038/cddis.2014.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinds P.W., Mittnacht S., Dulic V., Arnold A., Reed S.I., Weinberg R.A. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell. 1992;70:993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- 15.Levkau B., Koyama H., Raines E.W., Clurman B.E., Herren B., Orth K., Roberts J.M., Ross R. Cleavage of p21Cip1/Waf1 and p27Kip1 mediates apoptosis in endothelial cells through activation of Cdk2: role of a caspase cascade. Mol. Cell. 1998;1:553–563. doi: 10.1016/s1097-2765(00)80055-6. [DOI] [PubMed] [Google Scholar]
- 16.Harper J.W., Adami G.R., Wei N., Keyomarsi K., Elledge S.J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 17.Gartel A.L., Radhakrishnan S.K. Lost in transcription: p21 repression, mechanisms, and consequences. Cancer Res. 2005;65:3980–3985. doi: 10.1158/0008-5472.CAN-04-3995. [DOI] [PubMed] [Google Scholar]
- 18.Jeyapalan Z., Deng Z., Shatseva T., Fang L., He C., Yang B.B. Expression of CD44 3′-untranslated region regulates endogenous microRNA functions in tumorigenesis and angiogenesis. Nucleic Acids Res. 2011;39:3026–3041. doi: 10.1093/nar/gkq1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Y., Zhang Y., Cao L., Chen L., Lee V., Zheng P.S., Kiani C., Adams M.E., Ang L.C., Paiwand F., et al. Identification of the motif in versican G3 domain that plays a dominant-negative effect on astrocytoma cell proliferation through inhibiting versican secretion and binding. J. Biol. Chem. 2001;276:14178–14186. doi: 10.1074/jbc.M100618200. [DOI] [PubMed] [Google Scholar]
- 20.He J., Wu J., Xu N., Xie W., Li M., Li J., Jiang Y., Yang B.B., Zhang Y. MiR-210 disturbs mitotic progression through regulating a group of mitosis-related genes. Nucleic Acids Res. 2013;41:498–508. doi: 10.1093/nar/gks995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang B.L., Cao L., Kiani C., Lee V., Zhang Y., Adams M.E., Yang B.B. Tandem repeats are involved in G1 domain inhibition of versican expression and secretion and the G3 domain enhances glycosaminoglycan modification and product secretion via the complement-binding protein-like motif. J. Biol. Chem. 2000;275:21255–21261. doi: 10.1074/jbc.M001443200. [DOI] [PubMed] [Google Scholar]
- 22.Cao L., Yao Y., Lee V., Kiani C., Spaner D., Lin Z., Zhang Y., Adams M.E., Yang B.B. Epidermal growth factor induces cell cycle arrest and apoptosis of squamous carcinoma cells through reduction of cell adhesion. J. Cell Biochem. 2000;77:569–583. [PubMed] [Google Scholar]
- 23.Shan S.W., Fang L., Shatseva T., Rutnam Z.J., Yang X., Du W., Lu W.Y., Xuan J.W., Deng Z., Yang B.B. Mature miR-17–5p and passenger miR-17–3p induce hepatocellular carcinoma by targeting PTEN, GalNT7 and vimentin in different signal pathways. J. Cell Sci. 2013;126:1517–1530. doi: 10.1242/jcs.122895. [DOI] [PubMed] [Google Scholar]
- 24.Lopez de Silanes I., Stagno d'Alcontres M., Blasco M.A. TERRA transcripts are bound by a complex array of RNA-binding proteins. Nat. Commun. 2010;1:33–34. doi: 10.1038/ncomms1032. [DOI] [PubMed] [Google Scholar]
- 25.Wang P., Xue Y., Han Y., Lin L., Wu C., Xu S., Jiang Z., Xu J., Liu Q., Cao X. The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science. 2014;344:310–313. doi: 10.1126/science.1251456. [DOI] [PubMed] [Google Scholar]
- 26.Yang W., Du W.W., Li X., Yee A.J., Yang B.B. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2015 doi: 10.1038/onc.2015.460. doi:10.1038/onc.2015.460. [DOI] [PubMed] [Google Scholar]
- 27.Salzman J., Gawad C., Wang P.L., Lacayo N., Brown P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7:e30733. doi: 10.1371/journal.pone.0030733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang P.L., Bao Y., Yee M.C., Barrett S.P., Hogan G.J., Olsen M.N., Dinneny J.R., Brown P.O., Salzman J. Circular RNA is expressed across the eukaryotic tree of life. PLoS One. 2014;9:e90859. doi: 10.1371/journal.pone.0090859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merrick K.A., Larochelle S., Zhang C., Allen J.J., Shokat K.M., Fisher R.P. Distinct activation pathways confer cyclin-binding specificity on Cdk1 and Cdk2 in human cells. Mol. Cell. 2008;32:662–672. doi: 10.1016/j.molcel.2008.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rottmann S., Menkel A.R., Bouchard C., Mertsching J., Loidl P., Kremmer E., Eilers M., Luscher-Firzlaff J., Lilischkis R., Luscher B. Mad1 function in cell proliferation and transcriptional repression is antagonized by cyclin E/CDK2. J. Biol. Chem. 2005;280:15489–15492. doi: 10.1074/jbc.C400611200. [DOI] [PubMed] [Google Scholar]
- 31.Mitra J., Enders G.H., Azizkhan-Clifford J., Lengel K.L. Dual regulation of the anaphase promoting complex in human cells by cyclin A-Cdk2 and cyclin A-Cdk1 complexes. Cell Cycle. 2006;5:661–666. [PubMed] [Google Scholar]
- 32.Coverley D., Laman H., Laskey R.A. Distinct roles for cyclins E and A during DNA replication complex assembly and activation. Nat. Cell Biol. 2002;4:523–528. doi: 10.1038/ncb813. [DOI] [PubMed] [Google Scholar]
- 33.Yam C.H., Fung T.K., Poon R.Y. Cyclin A in cell cycle control and cancer. Cell Mol. Life Sci. 2002;59:1317–1326. doi: 10.1007/s00018-002-8510-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L., et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.