Abstract
Gene fusions and their products (RNA and protein) were once thought to be unique features to cancer. However, chimeric RNAs can also be found in normal cells. Here, we performed, curated and analyzed nearly 300 RNA-Seq libraries covering 30 different non-neoplastic human tissues and cells as well as 15 mouse tissues. A large number of fusion transcripts were found. Most fusions were detected only once, while 291 were seen in more than one sample. We focused on the recurrent fusions and performed RNA and protein level validations on a subset. We characterized these fusions based on various features of the fusions, and their parental genes. They tend to be expressed at higher levels relative to their parental genes than the non-recurrent ones. Over half of the recurrent fusions involve neighboring genes transcribing in the same direction. A few sequence motifs were found enriched close to the fusion junction sites. We performed functional analyses on a few widely expressed fusions, and found that silencing them resulted in dramatic reduction in normal cell growth and/or motility. Most chimeras use canonical splicing sites, thus are likely products of ‘intergenic splicing’. We also explored the implications of these non-pathological fusions in cancer and in evolution.
INTRODUCTION
Genes and their encoded products (RNAs and proteins), are not expected to intermingle except in pathological situations, i.e. cancer. This conventional wisdom is further supported by a group of fusion genes that have been successfully used as cancer diagnostic markers and therapeutic targets (1,2). On the other hand, RNA-Sequencing analyses from normal margins of cancer patients revealed that fusion RNAs can also exist in histologically non-neoplasia tissues (3–6). However, because of the nature of the samples, it is not clear whether the detected fusion RNAs truly exist in non-cancer patients. A few isolated studies have reported the existence of fusion RNAs in non-pathological situations (3,7–9). Recently, a database is established to incorporate 29 000 chimeric RNAs data-mined from Genbank and other RNA collections. However, the validation for the vast number of fusions is limited to RNA-Seq reads for only a few cell lines (10). Due to the lack of validation and functional relevance, the chimeras in non-cancer situations have been viewed largely as ‘junks’, or transcription noise. In fact, some other studies have also attributed many chimeras to template switching by reverse transcriptase during cDNA preparation in vitro (11,12), raising questions about whether these chimeras are truly real.
We performed, curated and analyzed around 300 RNA sequencing libraries covering 27 different non-neoplastic human tissues, 15 mouse tissues, human embryonic stem cells, mesenchymal stem cells induced for muscle differentiation and MCF10A breast epithelial cells. Over 10 000 fusion events involving 9778 fusions were found in these non-cancer samples. The majority of the fusions are seen in only one tissue/cell sample. To minimize false discoveries due to library construction and sequencing errors, and to uncover recurrent fusion RNAs, we focused on the group of fusions that are present in more than one tissue/cell line. A total of 291 recurrent fusions involving 238 gene pairs were found. We used several approaches to validate sub-populations of the fusions at RNA and protein levels. Fusions are then characterized according to their parental genes’ chromosomal location, junction position relative to exons, expression level, 3′ UTR size and the fusions’ protein-coding potential. A few fusions that are widely expressed seem to serve basic cell maintenance roles. Most of the recurrent fusions use canonical splicing sites, are thus likely to be products of cis-splicing between neighboring genes or RNA trans-splicing. When focused on evolutionarily conserved recurrent fusions, we found only a small overlap between the fusion RNA profiles of human and mouse, suggesting that forming chimeric fusion RNAs may be a way to expand functional genome. We also found some overlaps between the normal fusion pool and documented fusions in cancer, raising questions about their cancer-specificity.
MATERIALS AND METHODS
Cell lines and culture conditions
Mesenchymal stem cells were obtained from the Tulane University Center for Gene Therapy. They were maintained in MEM alpha medium with 20% fetal bovine serum (FBS). RWPE1 cells were maintained in RPMI 1640 medium containing 10% of FBS. Primary endometrial stromal fibroblasts and foreskin fibroblasts were maintained in DMEM with 10% FBS. Immortalized astrocytes were maintained in DMEM/F12 with 10% FBS, and supplemented with glucose. All the above media were supplemented with 1% of pen-strep and 1% of L-Glutamine. MCF10A cells were grown in DMEM/F12 supplemented with 5% horse serum, EGF, hydrocortisone, cholera toxin and insulin, as described before (13). For wound healing assay, cells transfected with si-negative control, or siRNAs against tested fusions were cultured for 3 days to obtain 80–90% monolayer confluency. A wound was created by scraping the cells using a 10 μl plastic pipette tip, and the medium was replaced with fresh medium. Images were captured immediately after the scratch and 6 h later. Cell migration was qualitatively assessed by the size of the wounds at the end of the experiment.
Clinical samples
Different tissues from non-cancer donors were collected under approved IRB protocol through the Biorepository and Tissue Research Facility at the University of Virginia, USA.
RNA extraction and sequencing
Different primary tissues were first homogenized with mortar-pestle grinding in the presence of liquid N2. RNAs were extracted with TRIzol reagent (Life Technologies) following the manufacturer's instruction. mRNA from total RNA was used as template for the subsequent validation processes. To assure the high quality RNA for next generation sequencing, RNAs were further cleaned using the RNeasy kit (Qiagen). mRNA in total RNA was converted into a library of template molecules suitable for subsequent cluster generation using the reagents provided in the Illumina TruSeq TM RNA Sample Preparation Kit. Millions of unique clusters on flow cells were loaded into the Hiseq 2000 platform and processed for RNA sequencing. RNA sequencing was performed by Axeq of Macrogen.
RT-PCR and Sanger sequencing
The presence of fusions candidates generated by the SOAPfuse algorithm from analyzed RNA-Seq data were confirmed by RT-PCR. All of the RNA samples used in this study were treated with DNase I (NEB, M0303), followed by standard Reverse Transcription using AMV RT (NEB, M0277). Real-time PCR experiment was performed using the ABI StepOne Plus system (Life Technologies) with Absolue Blue QPCR mix (Thermal Fisher, AB-4322). Following RT-PCR and gel electrophoresis, all purified bands were submitted for sequencing.
RNAi
siRNAs were synthesized by Life Technology. Their targeting sequences are: si-negative, CGTACGCGGAATACTTCGA; siCTNNBIP1, GGAAGAGTCCGGAGGAGAT; siCTNNBIP1-CLSTN1, TGCTTGTTAACCTGGTCGA; and siCTBS-GNG5, ATAACTATAAAGTTTCCCA.
Bioinformatics analysis
Chimeric RNA identification: Deep sequencing data were mapped to Human genome version hg19 and analyzed using software SOAPfuse (14). The output generated from multiple SOAPfuse running is represented by putative fusions lists derived from the union of two parental genes. These fusions were analyzed either in the context of one tissue or, per assemble, for the whole human organism. The fusion landscapes, by tissue, were presented using Circos generated plots (15).
Chimeric peptide identification: The files containing putative nucleotide fusion sequences generated through each RNA-Seq data analysis were unified and the three-frame translation of the sequences was created. The amino acid sequences underwent further processing by performing an in silico tryptic digestion. Out of all pieces generated through digestion only the pieces that position over the bridge between the two parental genes, and with a length longer than 20 amino acids on each side were retained. These were our input for the MS identification.
Motif finding: The MEME motif discovery tool, GLAM2 was run on four sets of data in total, categorized by the following distinctions: 5′/3′ genes, and upstream/downstream of fusion junction. The motifs presented are the highest scoring for each data set.
Mass spectrometry identification/validation for chimeric peptides
For MCF10A LCMS validation, the protein extract was denatured with urea (8 M), reduced with DTT (4.5 mM, 1 h, 60°C), diluted 1:10 with NH4HCO3 (50 mM) and digested with trypsin at a substrate:enzyme ratio of 50:1 w/w (24 h, 37°C). The digest was quenched with glacial acetic acid and subjected to C-18/SCX clean-up for salt removal. Ultimately, the protein digest was re-suspended in CH3CN/H2O/TFA (5:95:0.01) at a concentration of 2 μg/μl, and subjected to liquid chromatography (LC)-mass spectrometry (MS) analysis using an Agilent 1100 micro-LC system (Palo Alto, CA, USA) interfaced to a Thermo LTQ mass spectrometer (San Jose, CA, USA). LC separations were performed on nano-LC columns (100 μm i.d. × 12 cm, 5 μm Zorbax SB-C18 particles) fabricated in-house using a 3 h long LC gradient (0–100% B) at 180 nL/min. Solvent A was H2O:CH3CN:TFA (95:5:0.01 v/v) and solvent B was H2O:CH3CN:TFA 20:80:0.01 v/v (16). The LTQ-MS was operated using a data-dependent acquisition method, each MS scan being followed by zoom/MS2 scans on the five most intense peaks. The Discoverer 1.4 software package (Thermo) was used for the interpretation of tandem mass spectra using a UniProt database downloaded on Jan 15, 2015 (∼21 000 entries) appended with MCF10A fusion in silico translated sequences. Only fully tryptic fragments were allowed in the search (maximum two missed tryptic cleavages), using mass tolerances of 2 Da and 1 Da, at the MS and MS2 levels, respectively. False Discovery Rates (FDRs) (<3%) were evaluated by using a forward-reverse human protein database.
Visualization
For the heat map generation and clustering, MultiExperimentViewer (MeV) software, a component of the TM4 suite, was used. MeV is an open source Java application and is hosted at SourceForge (17). The method used was hierarchical clustering with complete linkage as the agglomeration rule. Pearson correlation coefficient was used to measure the similarities between gene profiles and Spearman rank to measure the similarity between samples.
Statistical analysis
The level of statistical significance was set at P < 0.05. Independent paired t-test and Pearson's correlation coefficient were used for different data sets to test for group differences. Fisher's Exact test was used for calculating statistical significance between the differences of in-frame versus frame-shift chimeric peptides.
Data access
The accession numbers of the 27 normal tissues, ESC, MSC, MCF10A and mouse tissue data sets from different donors are listed in Supplementary Table S1.
RESULTS
Chimeric transcriptome
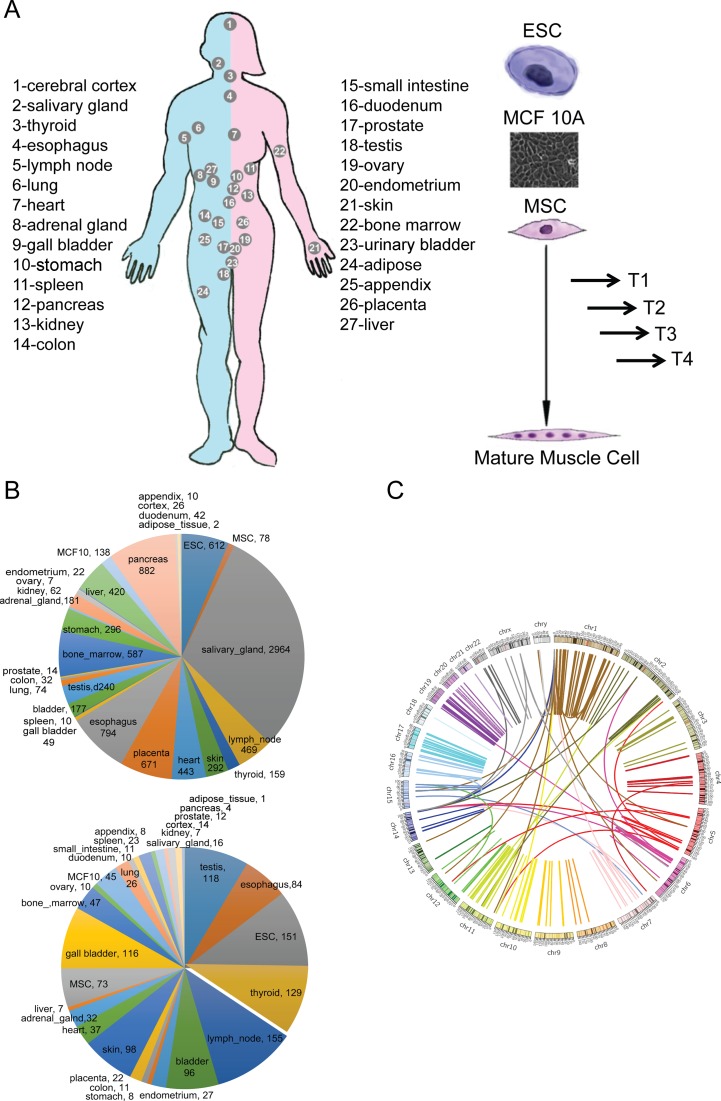
We attempted to capture chimeric fusion RNAs in various non-cancer samples. We also wanted to uncover ‘normal’ chimeric RNAs that were reported before in cancer samples. Suspecting that some of the chimeras may be developmentally transient events, we included data sets from embryonic stem cells, and from time points along the mesenchymal stem cell differentiation process. Finally, for the ease of MS validation, we included a non-malignant breast epithelial cell line, MCF10A (Supplementary Table S1). For human, we used the SOAPfuse software (14) to analyze 210 paired-end RNA sequencing libraries from 171 non-neoplastic tissue samples covering 27 different tissues (18), embryonic stem cells (19), our own libraries of four muscle differentiation time points starting from human mesenchymal stem cells and MCF10A sequencing from different labs (20–22) (Figure 1A). Contradictory to the conventional wisdom that chimeric RNAs are rare events in non-cancer samples, a total of 11 531 candidate chimeric RNA fusion events, and 9778 candidate fusion transcripts were uncovered (Table 1) (Supplementary Table S2). This involves 4408 gene pairs, 1536 genes as the 5′ portion of the fusions, 1578 as the 3′ portion.
Figure 1.
Identification of chimeric fusion RNAs in various tissues and cell types. (A) The sample sources include 27 different adult tissues on a human body map, ES cells, four time points collected along MSC muscle differentiation process, and MCF10A cells. (B) Distribution of fusions among different tissues/cells. Number of fusions in each sample type is also annotated. Upper: all the fusions; Lower: recurrent fusions. (C) Recurrent fusions were shown by a Circos plot. The fused genes are illustrated here as a line that connects two parental genes.
Table 1. Summary of fusion events, fusions and parental gene pairs in all and recurrent (more than one sample) groups.
| Total | Recurrent | |
|---|---|---|
| fusion events | 11531 | 1399 |
| fusions | 9778 | 291 |
| parental gene pairs | 4408 | 238 |
A fusion event refers to the detection of a particular fusion RNA in certain sample. A fusion is a unique fusion RNA. Note that same parental gene pairs can have multiple fusions due to different junction positions.
Fusion RNA profiles in each tissue or cell type were very different, with some tissues having only a few events (adipose tissue), and some having hundreds (pancreas and salivary gland) (Figure 1B). The majority of the fusions were seen once, while about 10% of fusions were seen in more than one sample (Supplementary Figure S1). To minimize false discoveries due to library construction and sequencing errors, and to uncover recurrent fusion RNAs, we decided to focus on the fusions that are detected in more than one sample. In total, there are 291 recurrent fusions involving 238 gene pairs. A total of 433 genes participate in forming these fusion transcripts (Figure 1C) (Supplementary Table S3). Out of the 238 gene pairs in the recurrent group, 51 were found in more than 5 tissues (Supplementary Figure S1 and Table S4). For instance, CTBS-GNG5 was found in 70 samples from 15 different tissues and cell lines by RNA-Sequencing. TIMM23B-LINC00843 was observed in 26 different tissues and cell lines.
Validation
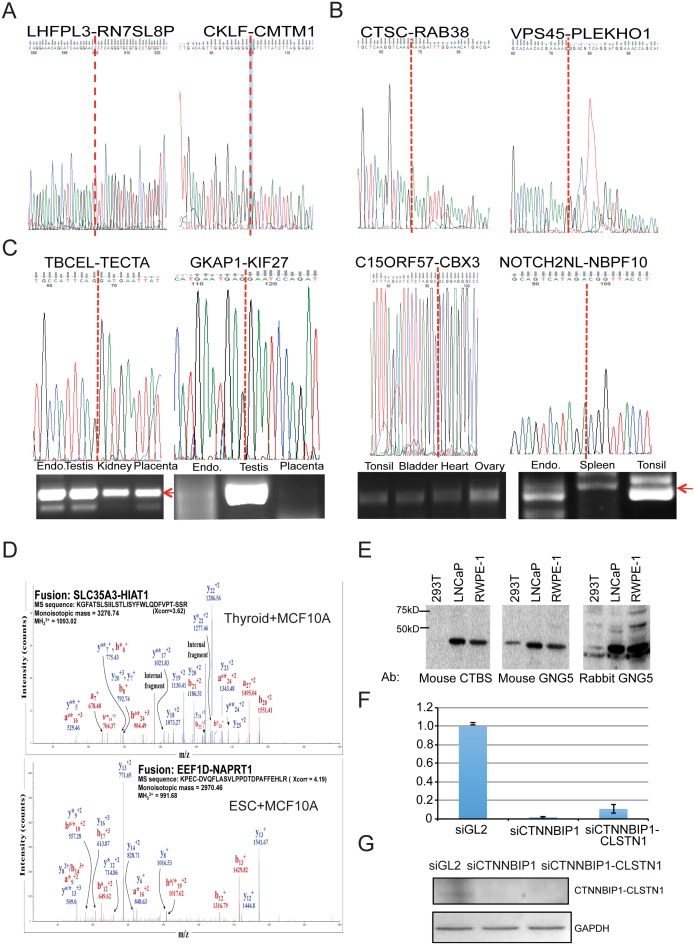
Candidate fusions unique to one tissue or cell type had lower validation rates, presumably due to the difference in sample source, heterogeneity of tissues and variable factors involved in cell culture. Nonetheless, many tissue specific fusions were validated in various types of tissues by RT-PCR and Sanger sequencing (two examples in Figure 2A). We also used the same RNA samples that were processed for sequencing MSC muscle differentiation time points, randomly selected 40 candidate fusion transcripts and successfully validated 30 fusions by RT-PCR and traditional Sanger sequencing (two examples in Figure 2B). We reasoned that the fusion candidates found in multiple tissue/cell types should have a higher chance of being detected when a different samples were used for validation. Indeed, 30 out of 35 randomly selected such fusions were confirmed by RT-PCR and Sanger sequencing (four examples in Figure 2C).
Figure 2.
Validation of the fusions. (A) Sanger sequencing of RT-PCR products for two tissue-specific fusions. LHFPL3-RN7SL8P in lung (M/E) and CKLF-CMTM in testis (E/E). Red dotted lines indicate the fusion junction. (B) Sanger sequencing of RT-PCR product for two fusions identified during MSC muscle differentiation process. (C) RT-PCR and Sanger sequencing for four recurrent fusions. TBCEL-TECTA were detected in endometrium (endo.), testis, kidney and placenta (E/E, INTRACHR-SS-0GAP), GKAP1-KIF27 in endometrium, testis and placenta (E/E, INTRA-OTHERS), C15ORF57-CBX3 in tonsil, bladder, heart, ovary (E/M, INTERCHR) and NOTCH2NL-NBPF10 in endometrium, and tonsil (E/M, INTRACHR-SS-0GAP fusion). Red arrows point to the correct PCR product. (D) Two examples of chimeric peptides supported by LCMS in MCF10A cells. The superscripts ‘o’ and ‘*’ represent H2O and NH3 losses, respectively. (E) Western blot analyses using a CTBS antibody, and two GNG5 antibodies detecting CTBS-GNG5 protein. (F) qRT-PCR of CTNNBIP1-CLSTN1 normalized against GAPDH. Human foreskin fibroblast cells were transfected with si-negative control, siCTNNBIP1 and siCTNNBIP1-CLSTN1. (G) Western blot analyses of protein extracts from the same three samples as above. Upper: CLSTN1 antibody. Lower: GAPDH antibody.
To provide more support for the existence of the chimeric RNA and potential protein products, we performed in silico translation around the fusion junction sequence in three reading frames. We then filtered off the putative peptides, if either side of the junction reached a stop codon, resulting peptides with less than 20 amino acids. Since most MS experiments were conducted with samples digested with trypsin, we performed an in silico tryptic digestion of the putative fusion peptides to the nearest Lysine (K) or Arginine (R) from the junction points (Supplementary Figure S2). We then used LCMS to validate the predicted recurrent chimeric fusion peptides in MCF10A cells. Out of the list of 291 recurrent fusion RNAs, 40 were also found in MCF10A. LC-MS analysis enabled the identification of matches to 8 chimeric peptide sequences that span across fusion junctions (FDR ≤ 3%), (Supplementary Table S5). Two of the best hits are shown in Figure 2D.
We also used traditional Western blot analyses to confirm two fusions that were predicted to generate in-frame fusion proteins. For CTBS-GNG5, we were able to obtain one antibody against the N-terminal of CTBS, and two antibodies against the C-terminal of GNG5. In all three Western blots, we found the same band that was the correct predicted size for the fusion protein (Figure 2E). For CTNNBIP1-CLSTN1, only the antibody against C-terminal CLSTN1 is available. We thus designed one siRNA targeting the CTNNBIP1 5′ part which silenced both the wild-type CTNNBIP1 and the CTNNBIP1-CLSTN1 fusion transcripts, and one siRNA targeting the fusion (Figure 2F). Consistent with the reduction of the fusion transcript, the signal at the predicted protein size in Western blot analysis was also reduced when cells were transfected with these two siRNAs (Figure 2G).
Characterizing the recurrent fusion RNAs
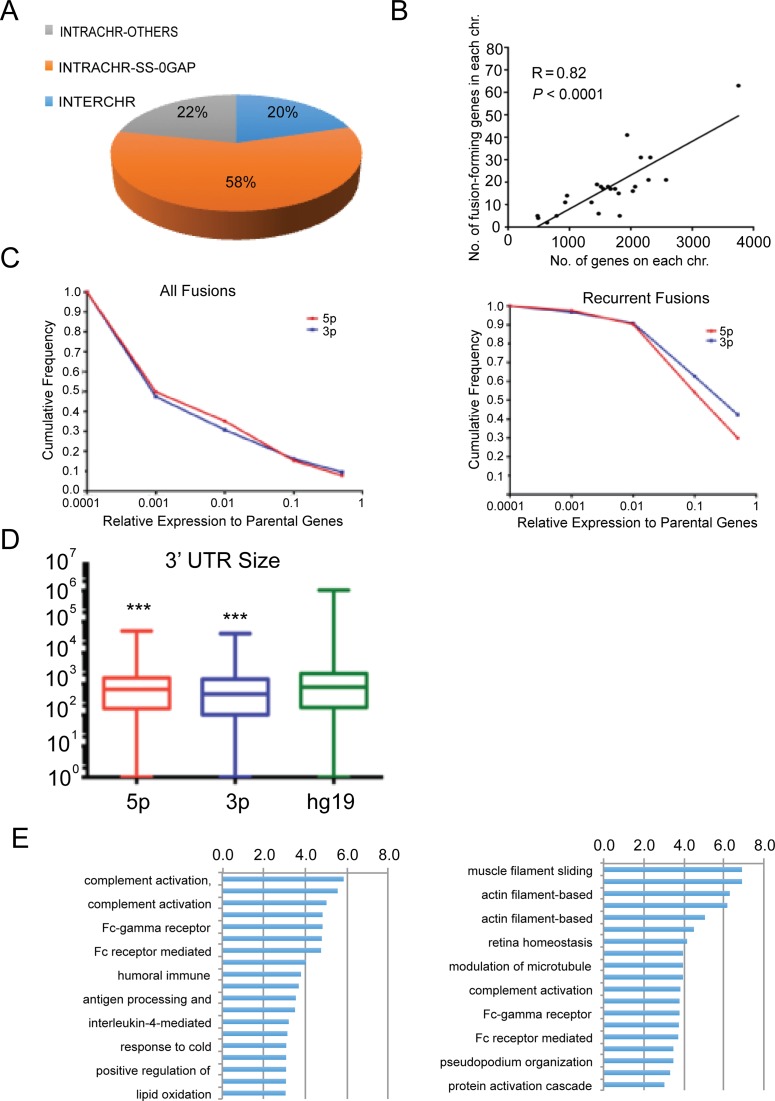
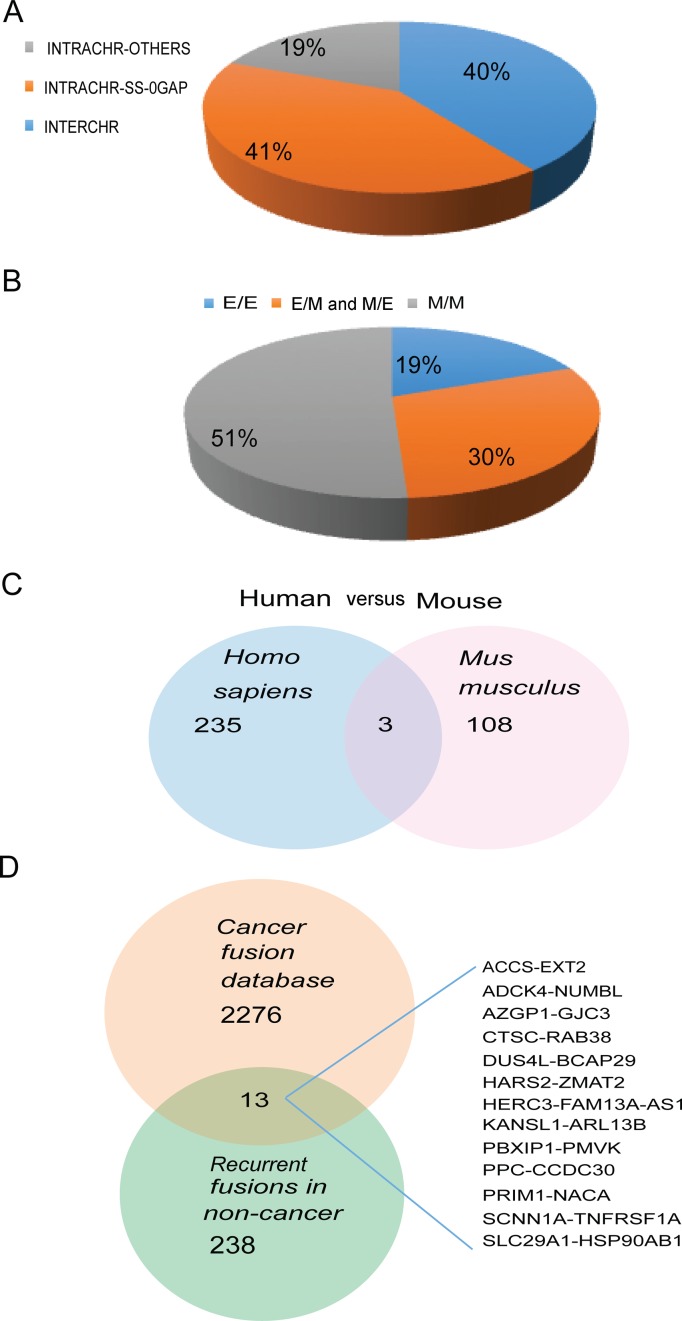
We then characterized the fusions according to the chromosomal locations of their parental genes: parental genes located on different chromosomes (INTERCHR), neighboring genes transcribing the same strand (INTRACHR-SS-0GAP) and other fusions with parental genes on the same chromosome (INTRACHR-OTHER). INTRACHR-SS-0GAP is the biggest group (58%) in these recurrent fusions (Figure 3A).
Figure 3.
Characteristics of the recurrent fusions. (A) Distribution of fusions according to the chromosomal location of the parental genes. INTERCHR: fusions involving parental genes located on different chromosomes; INTRACHR-SS-0GAP: fusions involving neighboring genes transcribing the same strand; and INTRACHR-OTHER: other fusions with parental genes on the same chromosome. (B) The density of genes participating in fusion formation correlates to the overall gene density on individual chromosomes. (C) Cumulative frequency of the relative expression of the fusion transcripts to their parental genes. Left: all the candidate fusions. Right: recurrent fusions. The parental gene expression was based on FPKM. The fusion RNA expression was converted into FPKM from the sum of junction and split reads number. (D) Box plot depicting the comparison between the 3′ UTR size for the parental 5′ and 3′ genes involved in fusions, and all genes known to date in hg19. (E) Gene ontology terms enriched in 5′ parental genes and 3′ parental genes involved in recurrent fusions. Plotted are statistical significance (-Log10(P value)) of each term.
The overall distribution of fusion-forming genes to individual chromosome correlated with the total gene numbers on each chromosome (Pearson's correlation r(22) = 0.82, P < 0.001 (Figure 3B). The correlation became even stronger (Pearson's correlation r(22) = 0.93, P < 0.001 0.93), when we examined the density of fusion parental genes against the density of genes on each chromosome (Supplementary Figure S3). This observation suggests that the fusion-forming genes are distributed throughout the genome similarly to other genes.
We examined the expression levels of the fusion RNAs against those of parental genes. When we looked at all the 9778 candidate fusions, we found that they tend to be expressed at lower levels relative to their parental genes (Figure 3C). Less than 20% of fusions were expressed above 10% of the level of their parental genes. However, when we focused on the recurrent fusions, they are expressed at a significantly higher levels. Around 40% (relative to 5′ parental) or 30% (relative to 3′ parental) were expressed above 50% of the level of their parental genes.
Since most fusions will result in replacement of 3′ UTR of the upstream parental gene with the downstream parental gene, we wondered whether the fusion formation could be a mechanism to escape, or acquire additional regulation by microRNAs (mRNAs). We used the length of 3′ UTR as a proxy. Overall, no statistical difference was observed between 5′ genes and 3′ genes (P = 0.23). However, both groups had significantly shorter 3′ UTR than an average gene in the genome (5′ versus hg19, P = 2.3E-23; 3′ versus hg19, p = 1.4E-39) (Figure 3D).
We searched gene ontology terms for the parental genes using Gorilla (23). In contrast to the tissue-specific fusions, where we found over 300 enriched GO terms (P < 0.001), around 20 GO terms were found for both 5′ and 3′ parental genes of the recurrent fusions (Figure 3E) (Supplementary Table S6). A few terms related to ‘response to cold’ and ‘cell movement’ are found unique to the recurrent fusions.
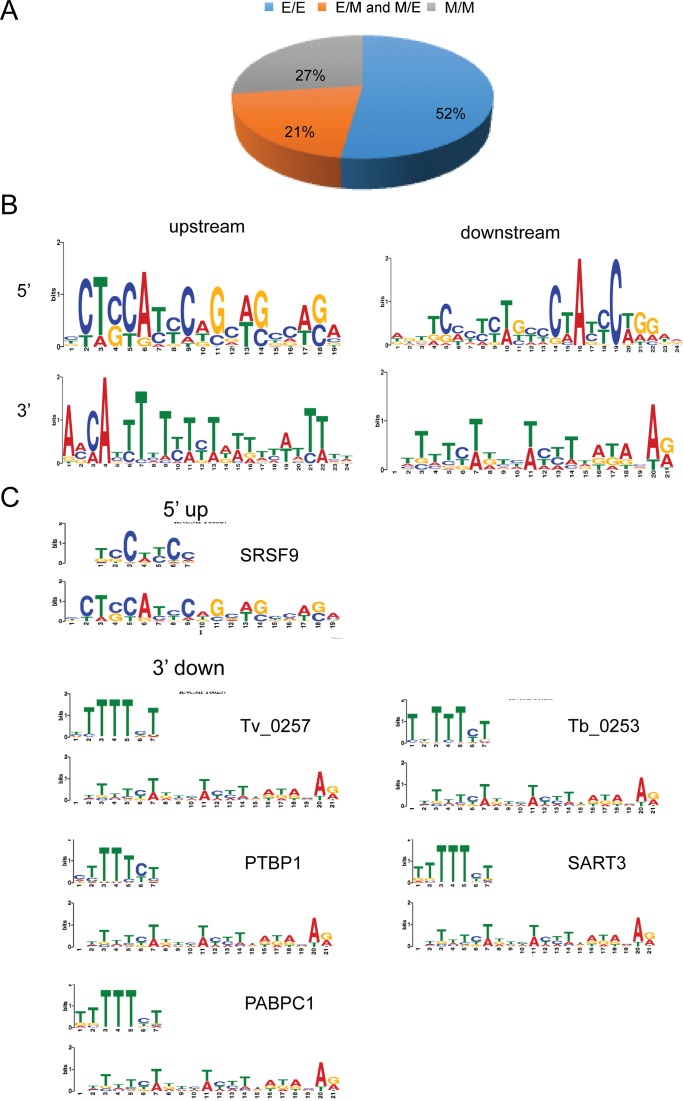
To probe into the generating mechanism of the fusion RNAs, we subdivided the fusions according to the junction position relative to the exon of the parental genes (Supplementary Figure S4): both sides being known exon boundaries (E/E); one side being exon boundary, the other not (E/M or M/E); both sides falling into the middle of exons (M/M). Consistent with an intergenic splicing mechanism, the biggest portion in these recurrent fusions are E/E fusions (Figure 4A). For the M/M fusions not using canonical splicing sites, it is possible that they are artifacts due to template switching during reverse transcription and sequencing steps mediated by short homologous sequences (SHS) (24). We searched for such homologous sequences shared by the fragments of parental genes around the junction sites in the 61 M/M fusions. Twenty percent of the fusions have no such SHS at the junction, thus potentially are true fusions utilizing non-canonical splicing sites. Forty two percent of the M/M fusions have SHS over 5 base pairs long. Thirty eight percent have 1–5 bp homologous sequences between the two fragments (Supplementary Figure S5). Considering the potential false positives associated with M/M fusions, we reexamined some of characterizations of the fusions. The 3′ UTR size difference between the parental genes and the whole genome as well as the enriched GO terms, still persist, even when we eliminated the M/M fusions.
Figure 4.
Further characterization of the recurrent fusions. (A) Distribution of fusions according to the junction position relative to the parental exons. E/E: both 5′ and 3′ using known exon boundaries; E/M or M/E: one side using known exon boundary, the other not; M/M: both sides fall into the middle of known exons. (B) Motif scanning using sequences 200 bp immediately upstream or downstream to the fusion junction site of both 5′ genes and 3′ genes. Shown are the motifs found having the highest GLAM2 scores. (C) Known RNA-binding motifs matching the motifs found through MEME. Using P = 0.001 as cutoff, one known RNA-binding motif was found similar to the 5′ gene downstream motif. Five RNA-binding motifs were found similar to the 3′ gene downstream motif.
We obtained 200 bp sequences upstream and downstream of fusion junction sites of both 5′ and 3′ parental genes. We then use the MEME motif discovery tool, Gapped Local Alignment, to look for sequence motifs enriched in these fragments. We didn't include the M/M fusions for this analysis to avoid potential influence of homologous recombination. The motifs presented in Figure 4B are the ones with the highest scoring for each data set. We then used the Tomtom motif comparison tool to search for RNA-binding protein motifs reported in RBP compendium (25). Using P = 0.001 as cut off, we found one RNA-binding motif similar to the 5′ upstream motif (SRSF9), and five similar to the 3′ downstream motif (Tv_0257, Tb_0253, PTBP1, SART3 and PABPC1) (Figure 4C). Among these RNA binding proteins, SRSF9 is known to play a role in exon inclusion (26). Polypyrimidine tract-binding protein (PTBP1), also known as heterogeneous nuclear ribonucleoprotein type I (hnRNP I), has been implicated in pre-mRNA splicing (27). SART3, also known as TIP110, has been found to interact with U6 and U6/U4 snRNPs (28). PABPC1 is a poly(A)-binding protein (PABP). It is found complexed to the 3-prime poly(A) tail of eukaryotic mRNA, and is required for poly(A) shortening and translation initiation (29). How these factors may contribute to potential intergenic splicing is not clear. We also searched for known sequence motifs in the JASPAR Vertebrates and UniPROBE Mouse databases. Five known motifs similar to the 5′ downstream motif (PPARG::RXRA, ESR2, NR3C1, LEF1 and ASCL2), and one known motif similar to the 3′ upstream motif (MTF1) were found (Supplementary Figure S6).
Functional relevance
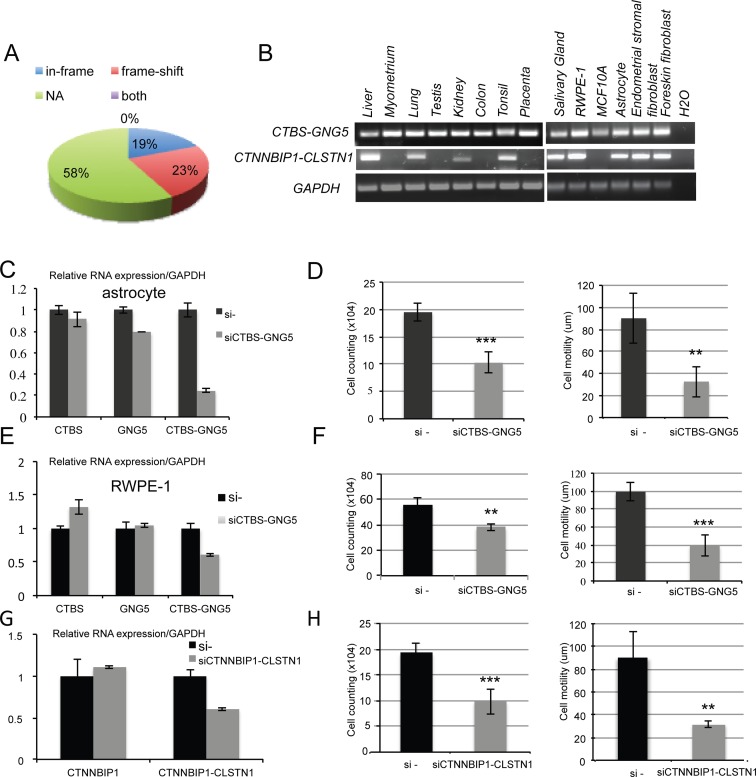
We categorized the fusions according to the reading frames: the known protein coding sequence of the 3′ gene uses a different reading frame than the 5′ gene (frame-shift); the known reading frame of the 3′ gene is the same as the 5′ gene (in-frame); no effect on the reading frame of the parental genes (NA). NA could occur when the fusion junction falls into the 3′ UTR of the 3′ parental gene, or no known protein coding sequence for the 3′ gene, or the 5′ part of the fusion will not affect the CDS of the 3′ gene; a very small population of fusions fell into the ‘both’ category, which could be in-frame or frame-shift depending on the alternative splicing isoforms of the parental genes. About 70% of all the fusions fell into the NA category (Figure 5A).
Figure 5.
Functional relevance of the fusions. (A) Distribution of the fusions according to their protein-coding potential: the known protein coding sequence of the 3′ gene uses a different reading frame than the 5′ gene (frame-shift); the known reading frame of the 3′ gene is the same as the 5′ gene (in-frame); no effect on the reading frame of the parental genes (NA). (B) CTBS-GNG5 and CTNNBIP1-CLSTN1 are widely expressed among tissues and cell lines. (C) An siRNA targeting CTBS-GNG5 resulted in significant reduction of the fusion transcript in immortalized astrocytes. (D) Knocking down CTBS-GNG5 using the fusion-targeting siRNA resulted in significance growth suppression and reduction in cell motility. (E) The siRNA also specifically silenced CTBS-GNG5 fusion transcript in RWPE-1 cells. (F) Knocking down CTBS-GNG5 in RWPE cells also resulted in reduced cell growth and motility. (G) An siRNA targeting CTNNBIP1-CLSTN1 resulted in significant reduction of the fusion transcript, but not wild-type CTNNBIP1 in immortalized astrocytes. Wild-type CLSTN1 was undetectable in these cells. (H) Knocking down CTNNBIP1-CLSTN1 in these astrocyte cells resulted in reduced cell growth and motility. **P < 0.005. ***P < 0.001.
The fact that some of the fusions were found in multiple tissues and cell types, supports the possibility that they may play some basic cellular maintenance roles. We chose two such candidates that belong to the ‘in-frame’ category for further functional study. Using RT-PCR, we could detect CTBS-GNG5 in nearly all of the tissues and cell lines we tested (Figure 5B). It was also found by RNA-Sequencing of cancer cells and seems to be conserved in mouse (30–32). CTBS encodes chitobiase; GNG5 encodes the di-N- acetyl-binding and guanine-nucleotide-binding proteins. The fusion creates an in-frame chimeric protein, which contains the chitinase catalytic domain from CTBS and most of the C-terminal GNG5, including its Gβ-binding interface. We designed an siRNA that specifically silenced the fusion transcript (Figure 5C and E). In two non-neoplastic cell lines we tested, we observed a significant reduction in cell growth and motility (Figure 5D and F).
Similarly, CTNNBIP1-CLSTN1 was detected in 9 out of 14 samples we tested. CTNNBIP1 encodes Beta-Catenin-Interacting Protein I. CLSTN1, also called Cadherin-Related Family Member 12, encodes Calsyntenin 1. The fusion contains the first 62 amino acids of the ICAT domain from CTNNBIP1, and the majority of CLSTN1 proteins. Transfection of an siRNA targeting specifically the fusion in immortalized astrocyte cells (wild-type CLSTN1 was undetectable) (Figure 5G) also resulted in significant reduction in cell growth and cell motility (Figure 5H).
Conservation and cancer implications
We then obtained 81 RNA-Sequencing libraries covering 15 different mouse tissue types. Using the same pipeline, we found 210 recurrent fusions, involving 111 pairs of mouse genes. The distribution of the fusions is similar to that in humans, with 41% of fusions being INTRACHR-SS-0GAP (Figure 6A). The percent of E/E fusions was less than that in human, only 19% (Figure 6B). Of interest, only three recurrent fusions were found in both human and mouse samples by RNA-Sequencing (Figure 6C) (Supplementary Table S7).
Figure 6.
Conservation and cancer implications. (A) Fusion RNAs of 81 samples from 15 mouse tissues. Similar to that in human, INTRACHR-SS-0GAP also constituted the biggest portion. (B) Distribution of fusions according to the junction position relative to the parental exon. (C) Venn diagram showing the similarities/differences of fusions in Homo sapiens versus Mus musculus. (D) Venn diagram showing the common fusions in the normal tissues/cells versus the Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. Thirteen common fusions are listed.
Given that many cancers arise by the dysregulated recapitulation of processes in normal development, we hypothesized that comparable chimeric fusions may exist in normal cells. At the time this manuscript was prepared, there were 2276 fusions documented in the Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer in the Cancer Genome Anatomy Project (33). Thirteen fusions were common to this list and the list of fusions we found in normal tissues/cells, including ADCK4-NUMBL, CTSC-RAB38, PBXIP1-PMVK and SCNN1A-TNFRSF1A reported in glioblastoma (34), HARS2-ZMAT2, HERC3-FAM13A-AS1, KANSL1-ARL17B, PPCS-CCDC30 and CTSC-RAB38 in leukemia (35,36), ADCK4-NUMBL, AZGP1-GJC3 and DUS4L-BCAP29 in prostate cancer (31,37) (Figure 6D) (Supplementary Table S8). The fact that these fusions are present in non-neoplastic samples raised concerns for their potential as biomarkers. Of note, all of these 13 fusions were recently discovered through deep-sequencing approaches, and none are signature fusions that have been experimentally validated as cancer-driving events (3). However, that is not to say that ‘cancer-signature’ fusion do not exist in normal cells. Such fusions may be tissue/cell lineage specific, and may only be transiently expressed during development, as demonstrated before (8).
We also performed gene ontology term analysis for the parental genes using Gorilla (23). For this analysis, we grouped parental genes that are only involved in ‘non-cancer’ fusion, genes that are only involved in ‘cancer’ fusions listed on the Mitelman list, and genes involved in both (Supplementary Table S9). Interestingly, the top GO terms for ‘non-cancer’ fusion genes are related to protein targeting, top terms for ‘cancer’ fusions are positive regulations related to cell proliferation and top terms for ‘both’ are more related to basic cell process.
DISCUSSION
Profiling fusion RNAs in normal tissues and cells have important implications in basic biology, evolution, cancer diagnosis and treatment. Human and other primates are known to have about twice as much alternative splicing as mice (38). We also found many more chimeric RNAs in humans than in mice, and the two species share only a few common fusions. Given the over 95% similarity between mouse and human genome, forming chimeric transcripts may be another way to expand functional genome, in addition to alternative splicing.
Gene fusions generated by recurrent chromosomal rearrangement is considered one of the hallmarks of neoplasia. Like fingerprints, the fusions and their products are often the signatures of distinctive types of tumors and tend to be also driver mutations for those tumors (1–2,39–41). Ever since the discovery of Philadelphia chromosome and BCR-ABL fusion, gene fusions have been heavily sought after. With many efforts devoted to data mining the Cancer Genome Atlas and other large cancer data sets, the fusion RNAs we identified in the normal samples should be considered ‘background’ and subtracted for cancer-specific fusion discovery. On the flip side, it is likely that at least some previously believed cancer-specific fusions do happen during development or specific physiological conditions, but may be products of intergenic splicing instead of chromosomal rearrangement. Indeed, certain cancer-signature fusions have been identified transiently during stem cell differentiation (8).
The pool of fusions identified through current RNA-Seq is far from saturation. In addition, the majority of fusions tend to exist in unique tissues and cell types, and are expressed at relatively low levels. SOAPfuse software was selected in this study because of its high validation rate (42). This choice is further supported by a recent study, when 15 popular fusion-mining tools were compared (43). However, none of the current software tools is inclusive. In fact, we found small overlaps between six most commonly used tools (manuscript under review). All of these factors will lead to an underestimate of the total number of fusions existing in various types of tissues and cells. Many more fusions may be uncovered by deeper sequencing reads, more sensitive fusion-mining software, enrichment of target cells and unbiased collection of time points throughout developmental processes. On the other hand, we cannot completely filter out the false positives generated during the process of cDNA library construction, sequencing and data mining. In this study, we purposely selected recurrent fusions to minimize false discoveries. However, we should not ignore all the ‘rare’ fusions. In fact, a large number of individualized fusions are consistent with a recent GTEx study, where many alternative splicing variants were observed inter-individually (44).
Instead of being ‘junk’ or ‘transcriptional noise’, at least a subset of the chimeric RNAs are functional. Some will translate into chimeric proteins, evidenced by MS and Western blot analyses. The further use of sensitive, high mass-accuracy instrumentation could lead to discovery of more chimeric proteins and more unambiguous assignment of matched sequences. Besides the in-frame fusions, a large population of fusions may function as non-coding RNAs. Supporting this idea, a recent study identified over 58 000 transcripts as long non-coding RNAs (45). Furthermore, a large portion of fusions will have no new function, but only affect parental gene expression. High throughput approaches are needed to study the functions of this large number of fusions.
A subpopulation of M/M fusions could be artifacts produced by short homologous sequences, but they could also be the products of non-allelic homologous recombination happening at the DNA level, as maybe in the case of globin genes (46). The INTRACHR-SS-0GAP fusions are most likely the products of read through/cis-SAGe (cis-splicing between adjacent genes) (31,42,47). We suspect that the rest of interchromosomal and other intrachromosomal fusions are the products of RNA trans-splicing (48–50). They are unlikely the products of chromosomal rearrangement, since the samples are all non-neoplastic tissues or cells. However, each fusion has to be examined individually to formally rule out the possibility of chromosomal rearrangement.
In conclusion, fusion RNAs in non-cancer tissues and cells are widely spread. They are not unique features of cancer. At least some of the fusions are functional. Evolutionally, human and mouse share a few common fusions, suggesting that forming fusion RNAs may be means to expand the repertoire of functional genome. Some fusions previously reported in cancer are not cancer-specific. More efforts are needed to study the functions and mechanisms of these physiological fusion transcripts.
ACCESSION NUMBER
Raw and processed RNA sequencing data for MSC muscle differentiation are available at GEO (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE64032.
Supplementary Material
Acknowledgments
We thank Dr Roger Abounader for providing immortalized astrocyte cell line, Dr Kevin Janes for MCF10A cells and Sung-min Park for his help on in silico translation of chimeric sequences.
Footnotes
Present address: Mihaela Babiceanu, Sanofi Pasteur, Orlando, FL 32826, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NCI [CA190713]; American Cancer Society [Research Scholar Grant 126405-RSG-14–065–01-RMC to H.L.]; St. Baldrick's V Scholar. Funding for open access charge: NCI [CA190713].
Conflict of interest statement. None declared.
REFERENCES
- 1.Rowley J.D. The role of chromosome translocations in leukemogenesis. Semin. Hematol. 1999;36:59–72. [PubMed] [Google Scholar]
- 2.Heim S., Mitelman F. Molecular screening for new fusion genes in cancer. Nat. Genet. 2008;40:685–686. doi: 10.1038/ng0608-685. [DOI] [PubMed] [Google Scholar]
- 3.Chase A., Ernst T., Fiebig A., Collins A., Grand F., Erben P., Reiter A., Schreiber S., Cross N.C. TFG, a target of chromosome translocations in lymphoma and soft tissue tumors, fuses to GPR128 in healthy individuals. Haematologica. 2010;95:20–26. doi: 10.3324/haematol.2009.011536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ren G., Zhang Y., Mao X., Liu X., Mercer E., Marzec J., Ding D., Jiao Y., Qiu Q., Sun Y., et al. Transcription-mediated chimeric RNAs in prostate cancer: time to revisit old hypothesis. OMICS. 2014;18:615–624. doi: 10.1089/omi.2014.0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoshihara K., Wang Q., Torres-Garcia W., Zheng S., Vegesna R., Kim H., Verhaak R.G. The landscape and therapeutic relevance of cancer-associated transcript fusions. Oncogene. 2015;34:4845–4854. doi: 10.1038/onc.2014.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stransky N., Cerami E., Schalm S., Kim J.L., Lengauer C. The landscape of kinase fusions in cancer. Nat. Commun. 2014;5:4846–4855. doi: 10.1038/ncomms5846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu C.S., Yu C.Y., Chuang C.Y., Hsiao M., Kao C.F., Kuo H.C., Chuang T.J. Integrative transcriptome sequencing identifies trans-splicing events with important roles in human embryonic stem cell pluripotency. Genome Res. 2014;24:25–36. doi: 10.1101/gr.159483.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan H., Qin F., Movassagh M., Park H., Golden W., Xie Z., Zhang P., Sklar J., Li H. A chimeric RNA characteristic of rhabdomyosarcoma in normal myogenesis process. Cancer Discov. 2013;3:1394–1403. doi: 10.1158/2159-8290.CD-13-0186. [DOI] [PubMed] [Google Scholar]
- 9.Ma L., Yang S., Zhao W., Tang Z., Zhang T., Li K. Identification and analysis of pig chimeric mRNAs using RNA sequencing data. BMC Genomics. 2012;13:429–439. doi: 10.1186/1471-2164-13-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frenkel-Morgenstern M., Gorohovski A., Vucenovic D., Maestre L., Valencia A. ChiTaRS 2.1–an improved database of the chimeric transcripts and RNA-Seq data with novel sense-antisense chimeric RNA transcripts. Nucleic Acids Res. 2015;43:D68–D75. doi: 10.1093/nar/gku1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Houseley J., Tollervey D. Apparent non-canonical trans-splicing is generated by reverse transcriptase in vitro. PLoS One. 2010;5:e12271. doi: 10.1371/journal.pone.0012271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McManus C.J., Duff M.O., Eipper-Mains J., Graveley B.R. Global analysis of trans-splicing in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2010;107:12975–12979. doi: 10.1073/pnas.1007586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Isakoff S.J., Engelman J.A., Irie H.Y., Luo J., Brachmann S.M., Pearline R.V., Cantley L.C., Brugge J.S. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005;65:10992–11000. doi: 10.1158/0008-5472.CAN-05-2612. [DOI] [PubMed] [Google Scholar]
- 14.Jia W., Qiu K., He M., Song P., Zhou Q., Zhou F., Yu Y., Zhu D., Nickerson M.L., Wan S., et al. SOAPfuse: an algorithm for identifying fusion transcripts from paired-end RNA-Seq data. Genome Biol. 2013;14:R12. doi: 10.1186/gb-2013-14-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tenga M.J., Lazar I.M. Proteomic snapshot of breast cancer cell cycle: G1/S transition point. Proteomics. 2013;13:48–60. doi: 10.1002/pmic.201200188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saeed A.I., Sharov V., White J., Li J., Liang W., Bhagabati N., Braisted J., Klapa M., Currier T., Thiagarajan M., et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 18.Fagerberg L., Hallstrom B.M., Oksvold P., Kampf C., Djureinovic D., Odeberg J., Habuka M., Tahmasebpoor S., Danielsson A., Edlund K., et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell Proteomics. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sigova A.A., Mullen A.C., Molinie B., Gupta S., Orlando D.A., Guenther M.G., Almada A.E., Lin C., Sharp P.A., Giallourakis C.C., et al. Divergent transcription of long noncoding RNA/mRNA gene pairs in embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 2013;110:2876–2881. doi: 10.1073/pnas.1221904110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen A., Beetham H., Black M.A., Priya R., Telford B.J., Guest J., Wiggins G.A., Godwin T.D., Yap A.S., Guilford P.J. E-cadherin loss alters cytoskeletal organization and adhesion in non-malignant breast cells but is insufficient to induce an epithelial-mesenchymal transition. BMC Cancer. 2014;14:552–565. doi: 10.1186/1471-2407-14-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asmann Y.W., Hossain A., Necela B.M., Middha S., Kalari K.R., Sun Z., Chai H.S., Williamson D.W., Radisky D., Schroth G.P., et al. A novel bioinformatics pipeline for identification and characterization of fusion transcripts in breast cancer and normal cell lines. Nucleic Acids Res. 2011;39:e100. doi: 10.1093/nar/gkr362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang B.H., Jensen K.J., Hatch J.A., Janes K.A. Simultaneous profiling of 194 distinct receptor transcripts in human cells. Sci. Signal. 2013;6:rs13. doi: 10.1126/scisignal.2003624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eden E., Navon R., Steinfeld I., Lipson D., Yakhini Z. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics. 2009;10:1–7. doi: 10.1186/1471-2105-10-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X., Zhao L., Jiang H., Wang W. Short homologous sequences are strongly associated with the generation of chimeric RNAs in eukaryotes. J. Mol. Evol. 2009;68:56–65. doi: 10.1007/s00239-008-9187-0. [DOI] [PubMed] [Google Scholar]
- 25.Ray D., Kazan H., Cook K.B., Weirauch M.T., Najafabadi H.S., Li X., Gueroussov S., Albu M., Zheng H., Yang A., et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499:172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Young P.J., DiDonato C.J., Hu D., Kothary R., Androphy E.J., Lorson C.L. SRp30c-dependent stimulation of survival motor neuron (SMN) exon 7 inclusion is facilitated by a direct interaction with hTra2 beta 1. Hum. Mol. Genet. 2002;11:577–587. doi: 10.1093/hmg/11.5.577. [DOI] [PubMed] [Google Scholar]
- 27.Ghetti A., Pinol-Roma S., Michael W.M., Morandi C., Dreyfuss G. hnRNP I, the polypyrimidine tract-binding protein: distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 1992;20:3671–3678. doi: 10.1093/nar/20.14.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bell M., Schreiner S., Damianov A., Reddy R., Bindereif A. p110, a novel human U6 snRNP protein and U4/U6 snRNP recycling factor. EMBO J. 2002;21:2724–2735. doi: 10.1093/emboj/21.11.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kahvejian A., Svitkin Y.V., Sukarieh R., M'Boutchou M.N., Sonenberg N. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 2005;19:104–113. doi: 10.1101/gad.1262905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moon A.M., Stauffer A.M., Schwindinger W.F., Sheridan K., Firment A., Robishaw J.D. Disruption of G-protein gamma5 subtype causes embryonic lethality in mice. PLoS One. 2014;9:e90970. doi: 10.1371/journal.pone.0090970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nacu S., Yuan W., Kan Z., Bhatt D., Rivers C.S., Stinson J., Peters B.A., Modrusan Z., Jung K., Seshagiri S., et al. Deep RNA sequencing analysis of readthrough gene fusions in human prostate adenocarcinoma and reference samples. BMC Med. Genomics. 4:11–32. doi: 10.1186/1755-8794-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plebani R., Oliver G.R., Trerotola M., Guerra E., Cantanelli P., Apicella L., Emerson A., Albiero A., Harkin P.D., Kennedy R.D., et al. Long-range transcriptome sequencing reveals cancer cell growth regulatory chimeric mRNA. Neoplasia. 2013;14:1087–1096. doi: 10.1593/neo.121342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitelman F., J.B.A.M.F.E. Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. 2015.
- 34.Frattini V., Trifonov V., Chan J.M., Castano A., Lia M., Abate F., Keir S.T., Ji A.X., Zoppoli P., Niola F., et al. The integrated landscape of driver genomic alterations in glioblastoma. Nat. Genet. 2013;45:1141–1149. doi: 10.1038/ng.2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wen H., Li Y., Malek S.N., Kim Y.C., Xu J., Chen P., Xiao F., Huang X., Zhou X., Xuan Z., et al. New fusion transcripts identified in normal karyotype acute myeloid leukemia. PLoS One. 2012;7:e51203. doi: 10.1371/journal.pone.0051203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Atak Z.K., Gianfelici V., Hulselmans G., De Keersmaecker K., Devasia A.G., Geerdens E., Mentens N., Chiaretti S., Durinck K., Uyttebroeck A., et al. Comprehensive analysis of transcriptome variation uncovers known and novel driver events in T-cell acute lymphoblastic leukemia. PLoS Genet. 2013;9:e1003997. doi: 10.1371/journal.pgen.1003997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mani R.S., Tomlins S.A., Callahan K., Ghosh A., Nyati M.K., Varambally S., Palanisamy N., Chinnaiyan A.M. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barbosa-Morais N.L., Irimia M., Pan Q., Xiong H.Y., Gueroussov S., Lee L.J., Slobodeniuc V., Kutter C., Watt S., Colak R., et al. The evolutionary landscape of alternative splicing in vertebrate species. Science. 2013;338:1587–1593. doi: 10.1126/science.1230612. [DOI] [PubMed] [Google Scholar]
- 39.Rabbitts T.H. Chromosomal translocations in human cancer. Nature. 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 40.Rowley J.D. Letter: a new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 41.Tomlins S.A., Rhodes D.R., Perner S., Dhanasekaran S.M., Mehra R., Sun X.W., Varambally S., Cao X., Tchinda J., Kuefer R., et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 42.Qin F., Song Z., Babiceanu M., Song Y., Facemire L., Singh R., Adli M., Li H. Discovery of CTCF-sensitive cis-Spliced fusion RNAs between adjacent genes in human prostate cells. PLoS Genet. 2015;11:e1005001. doi: 10.1371/journal.pgen.1005001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu S., Tsai W.H., Ding Y., Chen R., Fang Z., Huo Z., Kim S., Ma T., Chang T.Y., Priedigkeit N.M., et al. Comprehensive evaluation of fusion transcript detection algorithms and a meta-caller to combine top performing methods in paired-end RNA-Seq data. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1234. doi:10.1093/nar/gkv1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mele M., Ferreira P.G., Reverter F., DeLuca D.S., Monlong J., Sammeth M., Young T.R., Goldmann J.M., Pervouchine D.D., Sullivan T.J., et al. Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348:660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y., Barrette T.R., Prensner J.R., Evans J.R., Zhao S., et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaudry M.J., Storz J.F., Butts G.T., Campbell K.L., Hoffmann F.G. Repeated evolution of chimeric fusion genes in the beta-globin gene family of laurasiatherian mammals. Genome Biol. Evol. 2014;6:1219–1234. doi: 10.1093/gbe/evu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y., Gong M., Yuan H., Park H.G., Frierson H.F., Li H. Chimeric transcript generated by cis-Splicing of adjacent genes regulates prostate cancer cell proliferation. Cancer Discov. 2012;2:598–607. doi: 10.1158/2159-8290.CD-12-0042. [DOI] [PubMed] [Google Scholar]
- 48.Gingeras T.R. Implications of chimaeric non-co-linear transcripts. Nature. 2009;461:206–211. doi: 10.1038/nature08452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H., Wang J., Ma X., Sklar J. Gene fusions and RNA trans-splicing in normal and neoplastic human cells. Cell Cycle. 2009;8:218–222. doi: 10.4161/cc.8.2.7358. [DOI] [PubMed] [Google Scholar]
- 50.Zaphiropoulos P.G. Trans-splicing in higher eukaryotes: implications for cancer development. Front. Genet. 2012;2:92. doi: 10.3389/fgene.2011.00092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.