Abstract
The mechanism by which micro (mi)RNAs control their target gene expression is now well understood. It is however less clear how the level of miRNAs themselves is regulated. Under specific conditions, abundant and highly complementary target RNA can trigger miRNA degradation by a mechanism involving nucleotide addition and exonucleolytic degradation. One such mechanism has been previously observed to occur naturally during viral infection. To date, the molecular details of this phenomenon are not known. We report here that both the degree of complementarity and the ratio of miRNA/target abundance are crucial for the efficient decay of the small RNA. Using a proteomic approach based on the transfection of biotinylated antimiRNA oligonucleotides, we set to identify the factors involved in target-mediated miRNA degradation. Among the retrieved proteins, we identified members of the RNA-induced silencing complex, but also RNA modifying and degradation enzymes. We further validate and characterize the importance of one of these, the Perlman Syndrome 3′-5′ exonuclease DIS3L2. We show that this protein interacts with Argonaute 2 and functionally validate its role in target-directed miRNA degradation both by artificial targets and in the context of mouse cytomegalovirus infection.
INTRODUCTION
Among the various classes of small regulatory RNAs, miRNAs represent one of the most studied in mammals. They act as guides to recruit Argonaute proteins (Ago) to target mRNAs, resulting in translation inhibition and reduced stability (1). These tiny regulators are involved in a wide variety of biological processes (2,3), and their aberrant expression can be the cause of genetic diseases and/or cancers (4,5). Consequently, their synthesis and turnover must be tightly controlled. Briefly, miRNAs are transcribed in the nucleus as a primary transcript (pri-miRNA) containing a hairpin structure, which is recognized and cleaved by the RNase III Drosha. This cleavage generates a precursor miRNA (pre-miRNA) which will be exported to the cytoplasm, where a second cleavage by the RNase III Dicer gives rise to a ≈22 nucleotides (nt) miRNA duplex (1). One strand of the duplex (guide strand) is loaded in the RISC (RNA-Induced Silencing Complex) and becomes the active miRNA, while the second strand (passenger strand) is often degraded.
miRNA biogenesis is regulated both transcriptionally and post-transcriptionally by different mechanisms controlling the level of pri-miRNA transcription, the activity or the accessibility of Drosha and/or Dicer or the stability of the pre-miRNA (1). One of the best described example of miRNA biogenesis regulation involves the LIN28 protein, which negatively impacts the synthesis of Let-7 miRNA (6–11). LIN28 reduces the cleavage activity of both Drosha and Dicer, at respectively the pri-Let-7 and the pre-Let-7 levels (6–8). LIN28 also recruits the Terminal-Uridylyl-Transferases TUT4/TUT7 (9,10), which uridylate pre-Let-7 leading to its subsequent degradation by the exonuclease DIS3L2 (11). More recently, TUT4 and TUT7 were also described to have a more widespread role in the control of pre-miRNA degradation via a mechanism involving the RNA exosome (12).
Mature miRNAs, which represent the active end products of this biogenesis were long thought to be very stable molecules with half-lives ranging from hours to days (13,14). But recently, several examples showed that they are also subjected to active regulation. In this case, modifications of the small RNA play essential roles to influence its stability or function. For example, miR-122 mono-adenylation by GLD-2 (TUT2) stabilizes this miRNA in mammals (15). At the opposite, miR-26a is no longer functional as a consequence of its uridylation by ZCCHC11 (TUT4) (16). In addition, accelerated miRNA turnover has been reported. This is especially true for biological situations that require rapid changes in gene expression (i.e. cell cycle, light-dark transitions) (17–19), or during viral infections (20–22). Moreover, the presence of a highly complementary target can induce miRNA degradation via a mechanism involving tailing (3′ addition of non-templated nucleotides) followed by trimming of the miRNA (13,23). From now on, we will refer to this phenomenon as target RNA-directed miRNA degradation (TDMD) according to a recent report from the Grosshans laboratory (24).
In several organisms, small RNA species are usually protected from degradation by addition of a 2’O-methyl group at their 3′ extremity by the methyltransferase HEN1 (23,25). In hen-1 mutant plants and flies, the lack of a 2’O-methylated 3′ terminal residue results in 3′ uridylation/adenylation and subsequent 3′ to 5′ degradation of small interfering (si)RNAs (and plant miRNAs) (23,26). As opposed to siRNAs or plant miRNAs, Drosophila and mammals miRNAs are not 3′ protected and usually present only a partial complementarity with their target RNAs. This explains why a near-perfect miRNA/target complementarity (involving an extensive pairing of the 3′ region of the miRNA) coupled to a high abundance of the target seems to trigger miRNA degradation rather than mRNA regulation (23). Therefore, TDMD could represent a potential and powerful spatiotemporal way to regulate mature miRNA accumulation. Accordingly, TDMD has been observed in vivo in the context of mouse cytomegalovirus (MCMV) infection. Indeed, MCMV expresses an abundant viral transcript (m169), which localizes to the cytoplasm and induces degradation of the cellular miR-27a and b (20,21). To date, the molecular details of the process and the cellular enzymes involved in this mechanism are unknown.
In this study, we developed a biochemical approach that allowed us to induce TDMD in cell lines and to uncover the identity of the cellular factors responsible for the target-induced degradation of mature miRNAs. We retrieved known components of the RISC as well as several putative candidates for the miRNA modification and degradation. Among those, we identified the Terminal-Uridylyl-Transferase TUT1 and the 3′-5′ exoribonuclease DIS3L2. We confirmed that these two proteins interact with Argonaute 2 and together in an RNA-dependent manner. Although we could not assign a direct role to TUT1 in the TDMD process, most likely because of the redundancy with other TUTases, we could observe that impairing DIS3L2 activity leads to a reduced degradation of miRNAs in human cells, but also in mouse cells infected with MCMV, a natural inductor of TDMD.
MATERIALS AND METHODS
Plasmids, cloning and mutagenesis
DIS3L2 was amplified from HeLa total cDNA whereas cDNA clones (Thermo Scientific) were used as template for TUT1 (Accession BC128263), mouse mTUT1 (Accession BC025499), mouse mDIS3L2 (Accession BC036177) and mouse mAGO2 (Accession BC129922). TUT2, TUT4 and TUT7 were amplified from the plasmids Flag-TEVAP-TUT2, -TUT4 and -TUT7 provided by Narry Kim. Human AGO2 was re-amplified from the pIRESneo-FLAG/HA-AGO2 vector (Addgene). The vector expressing GFP-MBP was a gift from E. Izaurralde. To generate GFP fusion proteins, TUT1 (BglII/EcoRI), DIS3L2 (BglII/BamHI), mTUT1 (BglII/EcoRI), mDIS3L2 (EcoRI/KpnI), TUT2 (EcoRI/BamHI), TUT4 (XhoI/SmaI) and TUT7 (XhoI/BamHI) were each cloned in the pEGFP-C2 expression vector (Clontech) using the restriction sites indicated in parentheses. For HA-tagged proteins, the pcDNA3.1(+) vector (Invitrogen) was first modified by mutagenesis to insert the coding sequence of the HA-tag 5′ of the MCS cassette. Then, AGO2 (EcoRI/NotI), mAGO2 (EcoRI/NotI), TUT1 (EcoRI/NotI), mTUT1 (EcoRI/NotI), DIS3L2 (BamHI/NotI) and mDIS3L2 (EcoRI/NotI) were inserted between the indicated restriction sites. The catalytic mutants of DIS3L2 (D391N, D392N) and mDIS3L2 (D389N) were generated by mutagenesis. For the luciferase reporters, m169–3′ UTR or SH fragment was cloned in the pcDNA3.1 vector using EcoRV restriction site. Mutation in the seed region was then done by mutagenesis. To concatamerize the SH region, SH fragments were amplified with the insertion of different cloning sites, then treated for multiple ligation before being re-amplified and cloned as described above. Finally, the F-Luc coding sequence was inserted in pcDNA-m169 versions using NheI/KpnI. All the sequences of the cloning and mutagenesis primers are available in Supplementary Table S2.
Cell culture and transfection
HeLa cells, Hepa 1.6 cells and HEK293 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (v/v) fetal calf serum at 37°C in a humidified 5% CO2 atmosphere. For co-immunoprecipitation, HeLa and Hepa cells were transfected with a mixture containing plasmids expressing HA-tagged and GFP-tagged protein using the Turbofect reagent (Thermo Scientific). For RNA-immunoprecipitation, HeLa cells (15 × 106 cells in 15-cm plates) were transfected with 60 μg of plasmid expressing GFP-tagged protein. The day after, cells were divided and re-seeded in two 15-cm plates. After 10 h, cells were retransfected with either control antimiRNA directed against the Caenorhabditis elegans specific miR-67 (antimiR-67) or antimiR-27 (or -16) at a final concentration of 10 nM using lipofectamine 2000 (Invitrogen). The immunoprecipitation was performed 16 h after the last transfection. For RNAi (DIS3L2 and TUT1), HeLa cells were transfected twice (at day 1 and day 2) with 100 nM siRNA using Dharmafect-3 (Dharmacon) (siRNA sequences are listed in Supplementary Table S2) or 50 nM each when co-knocked-down. On day 3, cells were re-seeded to be at 50% confluence and allowed to adhere. Then, HeLa cells were transfected with 5 nM antimiRNAs using lipofectamine 2000. Samples were collected 16 h after the antimiRNA transfection. For overexpression experiment, HEK293 cells were first transfected with 5 μg of the indicated plasmid using Lipofectamine 2000. The day after, cells are re-seeded, retransfected with the antimiRNA oligoribonucleotides and collected as described above. For luciferase assay, HeLa cells were seeded in 48-well plates and transfected with a mixture containing 25 ng of both R-Luc and F-Luc reporters using Lipofectamine 2000. To overexpress the luciferase reporters, cells were seeded in 12-well plates, and transfected with mixtures containing 100 ng of the R-Luc transfection control, 0.375, 0.750 or 1.5 μg of the F-Luc reporters and empty plasmid to have equal amounts of DNA. Samples were analyzed 24 hours post-transfection.
Streptavidin affinity chromatography
For the mass spectrometry analysis, HeLa cells were grown in 500 cm2 plates to reach 50–60% confluence and then biotinylated antisense 2’O methylated oligoribonucleotides were transfected using the calcium-phosphate method at the indicated concentrations. Up to five independent experiments were performed using 4 plates per samples. 48 hpi, cells were collected and resuspended in lysis buffer (50 mM Tris HCl [pH 7.4], 150 mM NaCl, 0.02% sodium azide, 100 μg/ml PMSF, 1% NP40), supplemented with Complete-EDTA-free Protease Inhibitor Cocktail (Roche). After lysis, the samples were cleared by 15 min centrifugation at 4000 g and 4°C. Then, 200 μl of streptavidin-beads (Dynabeads MyOne Streptavidin C1, Invitrogen) were prepared in RNase-free conditions according to the manufacturer's protocol and added to each sample. Samples were incubated for 30 min at 4°C under rotation and then, beads were washed 5 times in 0.1× SSC. Finally, 1/5 of the sample was treated with phenol-chloroform to extract total RNA, 1/5 was used for western blot and 3/5 served for mass spectrometry analysis by nanoLC-MS/MS.
For smaller scale experiments, the protocol remained the same except that HeLa cells were seeded in two 15-cm plates, lysed in 1 ml lysis buffer and incubated with 40 μl of streptavidin beads. 50% of the sample were dedicated to RNA analysis whereas the second half served for western blotting.
Immunoprecipitation
HeLa cells were harvested, washed twice with ice-cold phosphate-buffered saline (1× PBS), and resuspended in 1 ml of NET buffer (50 mM Tris HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 0.1% Triton), supplemented with Complete-EDTA-free Protease Inhibitor Cocktail (Roche). Cells are lysed by three 15 s sonication followed by 30 min incubation on ice and debris are removed by 15 min centrifugation at 16 000 g and 4°C. If required, lysates are treated 30 min with 100 μg/ml of RNase A and then re-cleared by centrifugation. An aliquot of the cleared lysates (50 μl) is kept aside as protein Input and 50 μl additional aliquot for RNA Input when needed. Polyclonal anti-GFP antibodies (a kind gift from E. Izaurralde) or AGO2 monoclonal antibodies (kindly provided by G. Meister) are added and samples are incubated for 1 h at 4°C under rotation (18 rpm). Then, 50 μl of Protein-G-Agarose beads (Roche) were added and the samples were rotated similarly for 1 h. After three washes in NET buffer and a fourth wash in NET buffer without Triton, bound proteins are eluted with 100 μl of protein sample buffer (100 mM Tris HCl [pH 6.8], 4% SDS, 20% glycerol, 0.2 M DTT, 0.5% bromophenol blue), and/or bound RNA by phenol/chloroform extraction.
Nucleocytoplasmic fractionation and subsequent immunoprecipitation
The cytoplasmic and nuclear fractions were separated following the protocol established by Lim and collaborators (27). Briefly, 10 × 106 HeLa cells were harvested, washed twice with ice-cold 1× PBS, resuspended in 500 μl of lysis buffer (50 mM Tris HCl [pH 7.4], 140 mM NaCl, 1.5 mM MgCl2, 0.1% Igepal CA-630) and incubated for 5 min on ice. After 10 min of centrifugation at 2000 g and 4°C, the supernatant (cytoplasmic fraction) was collected and put aside. The pellet was then washed twice with wash buffer (50 mM Tris HCl [pH 7.4], 140 mM NaCl, 1.5 mM MgCl2) at 2200 g for 5 min at 4°C. After resuspension in 300 μl of lysis buffer followed by sonication, the samples were spun at 16 000 g for 15 min at 4°C. The supernatant (nuclear fraction) was then collected. The concentration of each sample was measured using the DC protein assay kit (Bio-rad).
When the cytoplasmic fraction was followed by immunoprecipitation, an aliquot was kept aside as input and the volume of the fraction was increased to 1 ml. Then the samples were rotated with 25 μl of GFP-Trap-A beads (Chromotek) during 45 min at 4°C. After three washes with the lysis buffer and one additional with the wash buffer, bound proteins were eluted with 100 μl of protein sample buffer.
Luciferase assay
HeLa cells, tranfected with Luciferase reporters 24 h before, were washed in PBS, lysed with passive lysis buffer (Promega) and assayed for firefly and Renilla luciferase activities, using the dual-luciferase reporter assay system (Promega) and a luminescence module (Glomax, Promega) according to the manufacturer's instructions. The relative F-Luc reporter activity was obtained by first normalizing to the transfection control (Renilla activity), and then, to the firefly activity obtained for the reporter without m169-UTR, which was arbitrarily set to 100.
RNA extraction and northern blot
Total RNA was extracted using Tri-Reagent Solution (MRC, Inc) according to the manufacturer's instructions except the precipitation step, which was done in presence of 3 volumes absolute ethanol and 0.05 volumes 3 M NaAc [pH 5].
Northern blotting was performed on 10 to 15 μg of total RNA. RNA was resolved on a 17.5% urea-acrylamide gel of 20 cm in length, transferred onto Hybond-NX membrane (GE Healthcare). RNAs were then chemically cross-linked to the membrane during 90 min at 65°C using 1-ethyl-3-[3-dimethylaminopropyl]carbodiimide hydrochloride (EDC) (Sigma). Membranes were prehybridized for 1 h in PerfectHyb™ plus (Sigma) at 50°C. Probes consisting of oligodeoxyribonucleotides (containing Locked Nucleic Acids for the detection of miR-27 and miR-16 tailed/trimmed isoforms) (see Supplementary Table S2) were 5′-end labeled using T4 polynucleotide kinase (Fermentas) with 25 μCi of [γ-32P]dATP. The labeled probe was hybridized to the blot overnight at 50°C. The blot was then washed twice at 50°C for 20 min (5× SSC/0.1% SDS), followed by an additional wash (1× SSC/0.1% SDS) for 5 min. Northern blots were exposed to phosphorimager plates and scanned using a Bioimager FLA-5100 (Fuji).
Antibodies for western blot analysis
Antibodies were used at the following dilutions: mouse anti-GFP antibodies (1:2000) and anti-HA-Peroxidase (1:5000) were purchased from Roche. Rat anti-AGO2 monoclonal antibody (1:200), rat anti-AGO1 monoclonal antibody (1:10) and rat anti-TNRC6B antibody (1:20) were kindly provided by G. Meister. Rabbit anti-DIS3L2 (1:1000) is a gift from A. Dziembowski and rabbit anti-STARPAP (TUT1) was kindly provided by R.A. Anderson and used at 1:3300. Rabbit anti-DIS3 (1:1000), rabbit anti-TUT7 (1:500), mouse anti-tubulin (1:5000), anti-mouse, anti-rat and anti-rabbit secondary antibodies (1:10000) were purchased from Sigma-Aldrich. Rabbit anti-XRN2 (1:500), anti-EXOSC3 (1:500), anti-RRP6 (1:3000) and mouse anti-Histone H3 (1:5000) were purchased from Abcam. Rabbit anti-XRN1 (1:2000) is from Bethyl laboratories.
MCMV infection of Hepa 1.6 cells
Hepa 1.6 cells were seeded in 6-well plates (5 × 105 cells/well) at day 0, and then transfected at day 1 with 4 μg of indicated plasmids using lipofectamine 2000. On day 2, cells were counted and infected accordingly with MCMV at MOI 1 after centrifugal enhancement at 18°C and 800 g for 30 min. Media was replaced after 1 h of infection. RNA samples were then collected at the indicated time points.
Real time RT-PCR analysis
The quantitative real-time PCR analysis was performed using Roche LightCycler 480 II. Prior to reverse transcription reaction, 1 μg of total RNA was treated with DNase I (Fermentas) according to the provider's instructions. Reverse transcription reaction was performed using miScript reverse transcription II kit (Qiagen) according to the manufacturer's instructions. The resulting cDNA was PCR amplified with Maxima SYBR green kit (Fermentas) in 10 μl reaction volume. For the analysis of miRNA expression, the mature sequence of each miRNA was used to design the forward primer and the miScript universal primer (Qiagen) was used as reverse primer. Forward and reverse primers were used respectively at 0.5 μM and 0.7×. For mRNA analysis 0.25 μM of each forward and reverse primers were used. The PCR program was composed of an initial denaturation step at 95°C for 10 min followed by 44 cycles at 95°C for 15 s, 55°C for 30 s and 72°C for 30 s including melting curve analysis. Data were analyzed as described previously (28).
RESULTS
Target RNA induces miRNA decay in a non-cooperative manner
The ectopic expression of the m169 transcript using an adenoviral vector is sufficient to induce degradation of both miR-27a and miR-27b (20), thus indicating that no other MCMV-encoded factor is needed to tail and trim these miRNAs. Because it appears that both miRNAs are targeted indifferently, and since we will extensively refer to these miRNAs in the manuscript, we will refer to both miR-27a and b using the generic term miR-27.
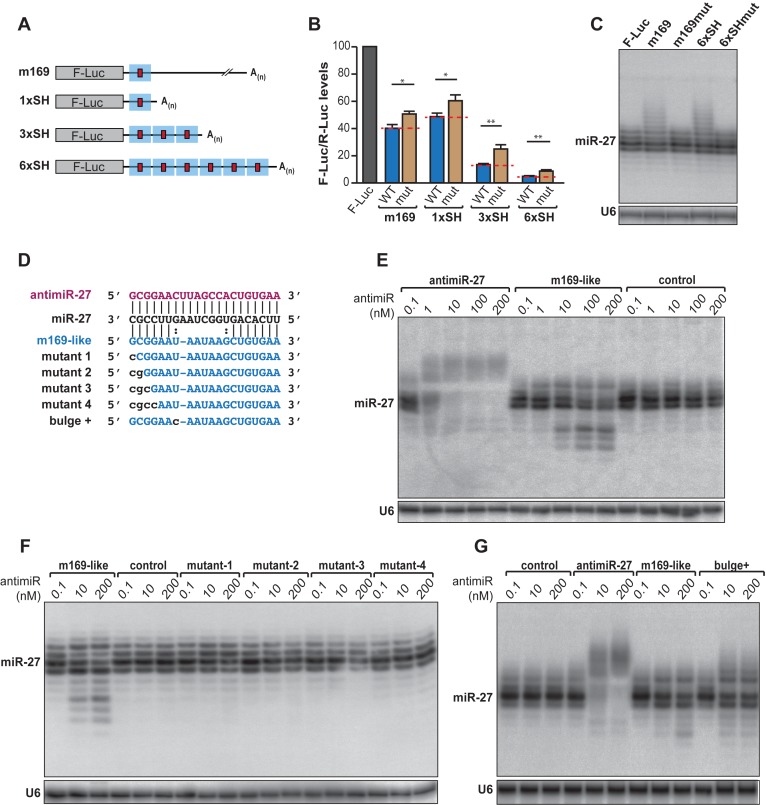
It was clearly shown that the two main features required to induce TDMD of a miRNA are an extensive pairing and an abundant target (23). To gain understanding in the sensitivity of the balance between a classical miRNA response and the triggering of TDMD, we built luciferase reporters carrying the m169 transcript full 3′ UTR, a shorter version of it (termed SH and comprising ≈200 nucleotides surrounding miR-27 binding site), or repetitions of the SH sequence to increase the number of target sites (Figure 1A). All versions of the luciferase reporter displayed translation inhibition (Figure 1B, Supplementary Figure S1A). Furthermore, the level of repression increased with the number of repetitions of the SH sequence and thus with the number of miR-27 binding sites, due to a cooperative effect of miRNAs as previously reported (29–31). This effect could be slightly but significantly reverted by inserting mutations in the miR-27 seed-match as shown previously (20) and reflects the contribution of miR-27 in the repression of the reporters. Indeed, the m169 transcript possesses several putative binding sites for other miRNAs, which could explain that the reporters bearing mutation in the miR-27 binding site are never completely derepressed. Transfecting increasing amounts of reporters did not alter the efficiency of miRNA-mediated repression (Supplementary Figure S1B), while overexpression of the reporter constructs allows the induction of miR-27 tailing as assessed by the detection of bands migrating slower than the four majors isoforms of miR-27 (Figure 1C, Supplementary Figure S1C). Nevertheless, increasing the number of miRNA binding sites did not cause a greater impact on miR-27 tailing than the single binding site contained in m169 (Figure 1C). Thus, as opposed to miRNA-mediated repression, which exhibits a cooperative effect, TDMD appears to be a non-cumulative phenomenon.
Figure 1.
Characterization of m169-miR27 features triggering TDMD. (A) Reporters containing the 3′ UTR of m169 transcript or single/multiple shorter (SH) region (blue square) containing miR-27 binding site (red square). (B) Relative expression of the reporters described in (A) and containing either an intact miR-27 binding site (WT) or mutations in the seed (mut) by luciferase assay. Each F-Luc coding reporter was transfected into HeLa cells together with a plasmid coding for the Renilla luciferase (R-Luc) as transfection control. The F-Luc activities were normalized to those of Renilla and arbitrarily set at 100 in absence of 3′ UTR (F-Luc). Mean values +/− standard deviations from three independent experiments are shown. (Student t-test *P < 0.05, **P < 0.01, n = 3 biological replicates). (C) Analysis of miR-27 expression by northern blot when the reporters are overexpressed. U6 serves as loading control. (D) Sequences and pairing between miR-27 and the antimiRNAs used in this study. AntimiR-27 is in purple and m169-like is in blue. Modifications introduced in the pairing are in lowercase. (E–G) Northern blots analyzing miR-27 accumulation profil upon antimiRNA transfection in HeLa cells. Control: antimiR-67. U6 serves as loading control.
A high complementarity between the miRNA and its target is a prerequisite to induce TDMD and it was previously shown in Drosophila that only little bulges (≈3 nt) can be tolerated (23). To understand how a m169-like binding site can trigger tailing-trimming with a 6-nt bulge, we transfected HeLa cells with increasing amounts of 2’O-methylated antisense oligoribonucleotides (antimiRNAs) presenting either a perfect complementarity with miR-27 (antimiR-27) or mimicking the interaction with m169 (m169-like, Figure 1D) since it was described that antimiRNAs are sufficient to recapitulate TDMD in cells (23). Northern blot analysis of miR-27 expression showed that a 100-fold more of the m169-like compared to antimiR-27 oligoribonucleotide was needed to induce TDMD (Figure 1E).
In addition, we confirmed the importance of a strong pairing in the 3′ of the miRNA by inserting mismatches in the antimiRNA oligoribonucleotide (Figure 1D). A single mismatch in the position exactly opposite the 3′ proximal nucleotide in the miRNA was sufficient to abrogate TDMD (Figure 1F). In addition, increasing the pairing strength 3′ of the bulge (position 15) by replacing a G-U by a G-C pair is enough to increase TDMD efficiency (Figure 1G). Thus, TDMD in mammals requires an extensive 3′ pairing between the miRNA and its target. However, the ratio between the miRNA and its target is important and this probably restrains the number of naturally occurring targets triggering miRNA tailing-trimming. Indeed, even if an m169-like interaction can trigger TDMD, the size of the central bulge limits its efficacy and only its expression at high levels such as during viral infection probably renders such interactions functional.
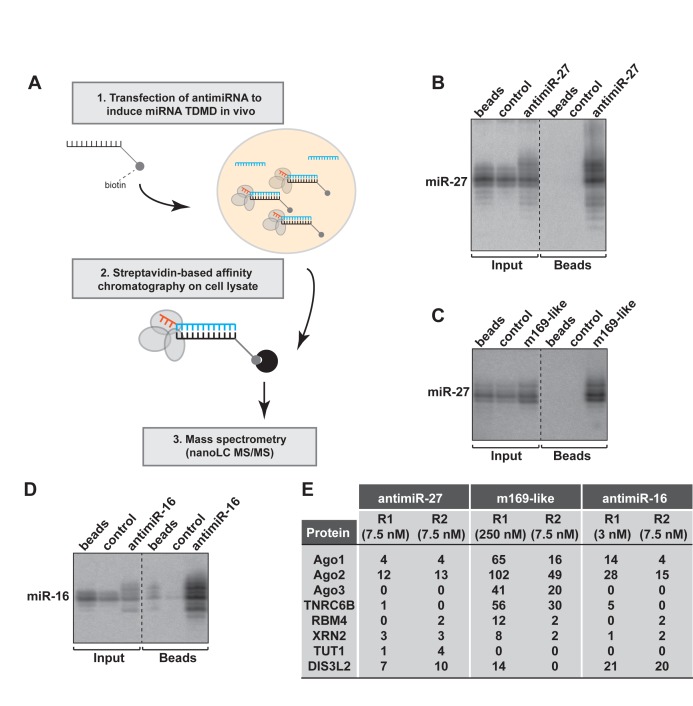
Isolation of protein complexes involved in TDMD induction
Since it is possible to purify RISC complexes from crude extract using biotinylated 2’O-methylated oligoribonucleotides (32), we used a similar approach to capture the TDMD nucleoprotein complexes by transfecting antimiRNAs in HeLa cells (Figure 2A). To this end, we used antimiR-27, m169-like and antimiR-16 2’O-methylated oligoribonucleotides carrying a triethylene glycol spacer with a biotin group on the 3′ terminal residue. We first determined the optimal concentration required to induce tailing-trimming without extensive degradation of the targeted miRNA, and found that 7.5, 250 and 3 nM were optimal for biotinylated antimiR-27, m169-like and antimiR-16 respectively. After collection of total cell lysates from transfected cells and incubation with paramagnetic-streptavidin beads, the analysis of retrieved RNAs revealed specific binding of miR-27 with both antimiR-27 and m169-like (Figure 2B,C) as well as miR-16 with its corresponding antimiRNA (Figure 2D). No signal was detected in non-transfected cells (beads) or cells transfected with a control oligoribonucleotide (control = antimiR-67). More importantly, we could also detect tailed and trimmed miRNA isoforms with this method.
Figure 2.
Identification of the cellular factors involved in TDMD. (A) Schematic representation of the biochemical approach to induce and capture the factors involved in TDMD. (B–D) HeLa cells were transfected with antimiRNA at a concentration inducing miR-27 or miR-16 TDMD. The complexes bound to antimiRNA were pulled-down using streptavidin-beads. The specific pull-down of miR-27 and miR-16 using antimiR-27 (replicate R1) (B), m169-like (C) and antimiR-16 (D) was verified by northern blot. A dashed line indicates discontinuous lanes but Input and IP fractions were systematically analyzed on a same gel. (E) Table showing the number of spectral counts detected by nanoLC-MS/MS for the indicated proteins found specifically associated with each antimiRNA. The concentration of antimiRNA used in each replicate is indicated in parentheses. Control: antimiR-67.
We then increased the scale of the experiment and analyzed the bound proteins by nanoLC-MS/MS. The data were filtered to consider only proteins specifically bound to the antimiRNAs and not to the control oligoribonucleotide or to the beads (Supplementary Figure S2A–C). Among the factors identified, we could retrieve known components of RISC complexes such as AGO1, 2 and 3, TNRC6B, and RBM4 (Figure 2E). We also identified exoribonucleases such as XRN2 or the 3′-5′ exonuclease DIS3L2, recently shown to be involved in mRNA and let-7 precursor decay (11,33) with the three capture-oligoribonucleotides. Finally, the Terminal-Uridylyl-Transferase (TUT)-1 (Star-PAP/RBM21) was pulled-down specifically with antimiR-27 in our two replicates, albeit at relatively low levels.
To increase the confidence in our mass spectrometry analysis, we performed additional replicate experiments (up to five with the antimiR-27) and decided to focus on proteins that were present in at least three of the replicates, while being absent in their respective negative control experiments (control oligoribonucleotide or beads alone) (Supplementary Table S1). This allowed us to significantly reduce the number of false positives and to narrow down the list of putative candidates. With this approach, we found that DIS3L2 was reproducibly pulled-down both with antimiR-27 and antimiR-16. This enzyme belongs to the Dis3 family together with two other homologs, DIS3 and DIS3L but with the difference that it is not associated with the RNA exosome. It is a highly processive enzyme, also capable of degrading dsRNA with little (2 nt) or no 3′ overhangs (33,34). More interestingly, DIS3L2 was also detectable with m169-like oligoribonucleotide only when it was transfected at 250 nM (Figure 2E, R1), a concentration that triggers TDMD. This re-enforces the idea that DIS3L2 is specifically recruited for miRNA trimming.
The replicate experiments also confirmed that TUT1 was only retrieved with the antimiR-27 oligonucleotide. However, it was the only of the seven human TUTases to be identified in our mass spectrometry analysis. This enzyme belongs to the family of ribonucleotidyl transferases known among others to contribute to miRNA 3′ heterogeneity (35) and it possesses both a non canonical poly(A) polymerase and a terminal uridylyl transferase activities (36–38).
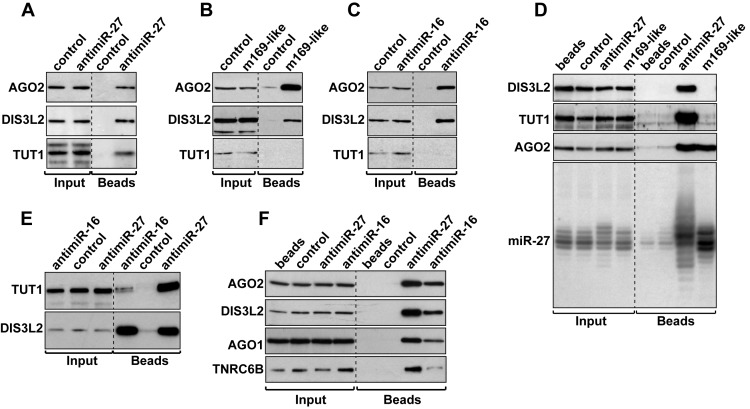
We then confirmed that DIS3L2 and TUT1 could indeed be detected in our pull-down experiments. Western blot analysis of proteins bound to antimiR-27 revealed a specific binding of both proteins, as well as of AGO2 (Figure 3A). Similarly, both DIS3L2 and AGO2 could also be detected by western blot associated to m169-like or antimiR-16 oligoribonucleotides (Figure 3B,C). The comparative analysis of proteins and RNA bound to antimiR-27 and m169-like oligoribonucleotides, when the two biotinylated-oligoribonucleotides are transfected at similar amounts, shows they both associate with similar amounts of miR-27 and are equally bound by RISC as assessed by the presence of AGO2 (Figure 3D). On the contrary, TUT1 and DIS3L2 are only associated with antimiR-27, the sole target capable to trigger TDMD at this concentration. Similarly, even though TUT1 was not found associated to antimiR-16 in the mass spectrometry data, we managed to detect it by western blot analysis when we increased the concentration of the oligoribonucleotide (Figure 3E). Under these conditions, we could also pull-down with both antimiR-27 and antimiR-16 oligonucleotides the AGO1 protein and TNRC6B, a GW182 family member essential for miRNA-mediated gene silencing (39) (Figure 3F).
Figure 3.
AntimiRNAs associate with DIS3L2, TUT1 and RISC components. (A–C) The presence of AGO2, DIS3L2 and TUT1 with antimiR-27 (A), m169-like (B) or antimiR-16 (C) was confirmed by western blot. (D) Comparative analysis of DIS3L2, TUT1, AGO2 and miR-27 pulled-down with antimiR-27 or m169-like oligonucleotides at 10 nM. (E) TUT1 and DIS3L2 detection after pull-down of antimiR-27 and antimiR-16 both transfected at 10 nM to induce TDMD. (F) AGO1 and TNRC6B are also associated with antimiR-27 and antimiR-16 both transfected at 10 nM. A dashed line indicates discontinuous lanes but Input and IP fractions were systematically analyzed on a same gel (A-F). Control: antimiR-67.
We also tested the possibility of retrieving other related enzymes with antimiR-27 or antimiR-16 oligonucleotides even if we never or rarely detected them in our MS analysis. For this purpose, we tested the 5′-3′ exoribonucleases XRN1 and XRN2 (40), the exosome components EXOSC3 (RRP40), RRP6 and DIS3 (41), and finally TUT7 (ZCCHC6), which was recently shown to participate to mature miRNA mono-uridylation (42). None of these proteins were found specifically associated with antimiR-27 and antimiR-16 after TDMD induction (Supplementary Figure S3). Only XRN2 and DIS3 were detectable but without any specificity for TDMD since they both co-purified equally well with our control oligoribonucleotide.
Altogether, these results indicate that DIS3L2 and TUT1 are good candidates for TDMD. We thus decided to proceed with their functional characterization.
TDMD is induced in the RISC complex and occurs in the cytoplasm
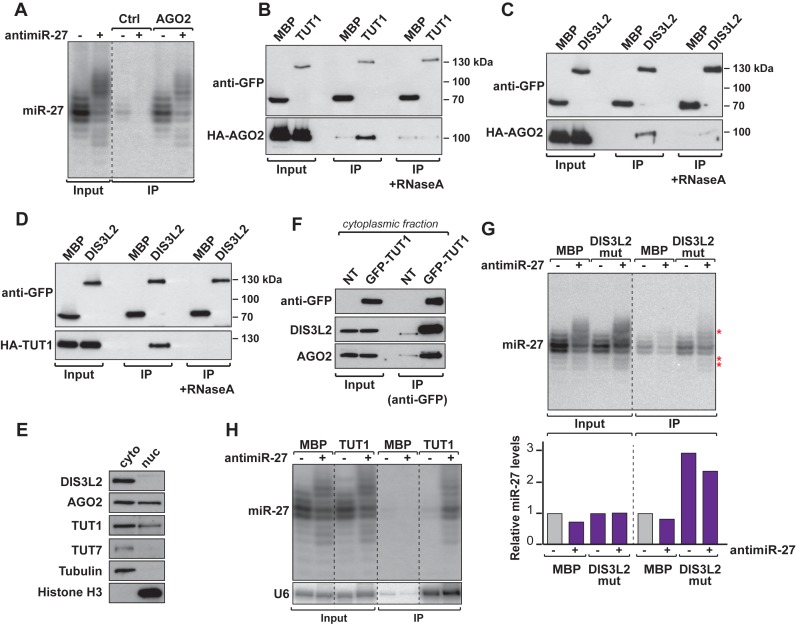
We first sought to answer the question of how the mechanism is initiated. TDMD depends on a high degree of complementarity between the miRNA and its target, raising the possibility that the miRNA tailing and trimming is induced within the RISC. Alternatively, the miRNA could also be marked for degradation after its release from the AGO protein. We therefore analyzed small RNAs by northern blot after AGO2 immunoprecipitation, which allowed us to show that the RISC effector interacted not only with the major isoforms, but also with tailed and trimmed isoforms of miR-27 when TDMD is induced (Figure 4A). The tailed and trimmed isoforms detectable by northern blot were comprised between 16 and 26 nucleotides in length (Supplementary Figure S4A). Similar results were obtained with miR-16 (Supplementary Figure S4B) or when miR-27 TDMD was induced by transient expression of a piece of the m169 transcript (Supplementary Figure S4C). Thus, although it is unclear whether the miRNA undergoing TDMD is eventually ejected from RISC, both tailing and trimming of miRNA seem to occur, at least partially, within AGO2.
Figure 4.
TDMD is induced in the RISC complex by recruitment of TUT1 and DIS3L2. (A) AGO2-associated tailed and trimmed isoforms of miR-27 were analyzed by northern blot. A space indicates discontinuous lanes from the same gel (also applicable to panels E and F). (B–D) Co-immunoprecipitation (anti-GFP) experiments show interaction between GFP-TUT1 and HA-AGO2 (B), GFP-DIS3L2 and HA-AGO2 (C) and GFP-DIS3L2 and HA-TUT1 (D). GFP-MBP serves as negative control. (E) Subcellular location of TUT1, DIS3L2 and AGO2 in cells. The relative amount of TUT1, DIS3L2 and AGO2 proteins in the nucleus (nuc) and the cytoplasm (cyto) were analyzed by western blot. TUT7 and Tubulin serve as cytoplasmic markers while Histone H3 is used as nuclear marker. (F) GFP-TUT1 interaction with endogenous DIS3L2 and AGO2 was assayed by co-immunoprecipitation (anti-GFP) performed on the cytoplasmic fraction. Non-transfected cells (NT) serve as negative control. (G) miR-27 isoforms co-precipitate together with GFP-DIS3L2 mutant. miR-27 enrichment was measured using miR-27 signal (normalized to tRNA in the input fraction) arbitrarily set at 1 for GFP-MBP in absence of antimiR-27. Asterisks highlight tailed and trimmed isoforms. (H) Northern blot showing TUT1 interacts with miR-27 isoforms only when TDMD is induced. U6, a known substrate of TUT1, serves as positive control. A dashed line indicates discontinuous lanes from the same gel (A, E, G, H).
More importantly, these observations suggest that both TUT1 and DIS3L2 might be recruited to the RISC. To test this hypothesis, we performed co-immunoprecipitation of DIS3L2, TUT1 and AGO2 after transfection of HeLa cells with plasmids expressing tagged-versions of the proteins. We observed that both TUT1 and DIS3L2 interact with AGO2 (Figure 4B,C) but also together (Figure 4D). These interactions are RNA-mediated (putatively miRNA-mediated), since they were sensitive to RNaseA treatment (Figure 4B–D). So, TUT1, DIS3L2 and AGO2 seem to be part of the same complex even if it is probably transient since target-induced miRNA decay is not a predominant mechanism.
The previous results raise the question of the formation of a complex comprising AGO2, DIS3L2 and TUT1, since TDMD is most likely to occur in the cytoplasm and earlier reports showed that TUT1 was a nuclear protein (37). However, recent work indicates that this might not be its sole localization, as it could be observed both in the nucleus and the cytoplasm (43). We thus wanted to verify the relative amount of TUT1 in the cytoplasm and in the nucleus in our experimental setup, and to confirm that the cytoplasmic TUT1 interacts with DIS3L2 and AGO2. We therefore performed a subcellular fractionation and analyzed the distribution of the different proteins (Figure 4E). Using tubulin as a cytoplasmic marker and histone H3 as a nuclear marker, we found that DIS3L2 was only present in the cytoplasm as expected (33,34), similar to TUT7 (27), AGO2 could be observed in both compartments, as previously reported (44), and we also found TUT1 both in the cytoplasm and the nucleus (Figure 4E) confirming that this TUTase is indeed a nucleo-cytoplasmic protein. We then immunoprecipitated TUT1 from a cytoplasmic extract and could observe that it interacted with endogenous DIS3L2 and AGO2, thereby confirming that these interactions naturally occur in the cytoplasm (Figure 4F).
We showed earlier that DIS3L2 and TUT1 could be pulled-down with the antimiRNA oligoribonucleotide only under conditions where TDMD was induced (Figure 3D). We therefore looked whether we could confirm the interaction by immunoprecipitating the proteins and detecting the miRNA. For DIS3L2, we used the catalytic mutant because the wild type protein association with the targeted RNA would be too transient. The immunoprecipitation of DIS3L2mut shows that the exonuclease seems to interact weakly with miR-27 but independently of TDMD (Figure 4G). DIS3L2 might therefore have additional roles in miRNA decay that are independent of TDMD. Nonetheless, the mutant protein interacted more strongly than the wild-type exonuclease with tailed/trimmed isoforms of miR-27 (Supplementary Figure S4D) confirming its role in miRNA trimming.
Finally, we analyzed small RNAs after immunoprecipitation of TUT1. We confirmed that the pull-down was efficient, since we could retrieve its known substrate U6 (38). The northern blot analysis also revealed that it exclusively interacted with miR-27 isoforms when TDMD was induced with antimiR-27 oligonucleotide (Figure 4H). In addition, we also performed northern blot analysis of miR-27 after immunoprecipitation of other TUTases (TUT4, TUT7 and TUT2), but could not detect the miRNA coming down with any of these, be it in the presence or not of the antimiRNA oligonucleotide (Supplementary Figure S4E).
DIS3L2 is implicated in miRNA trimming
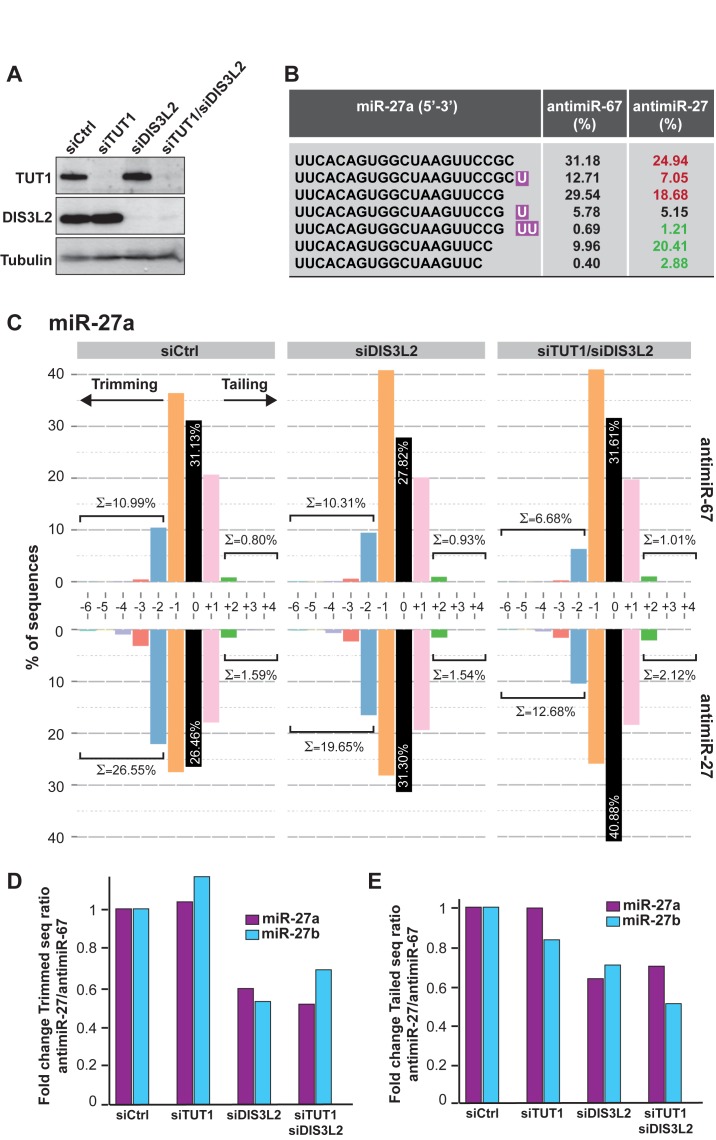
We then aimed at validating the involvement of TUT1 and DIS3L2 in TDMD by performing knockdown experiments of either each factor separately or both of them together and we monitored the level of TDMD induced by antimiRNA transfection. We observed an efficient depletion of these two factors at the protein level in HeLa cells (Figure 5A). Nonetheless, the efficiency of TDMD was not altered as assessed by northern blot analysis of miR-27 (Supplementary Figure S5A) and miR-16 (Supplementary Figure S5B) compared to the cells transfected with the control siRNA (siCtrl).
Figure 5.
Functional implication of DIS3L2 in miRNA degradation. (A) Efficiency of TUT1 and DIS3L2 knockdowns by western blot. The control siRNA (siCtrl) is targeting the Renilla luciferase. Tubulin serves as loading control. (B) Representative examples of the most abundant miR-27a isoforms detected in one of the libraries treated with the control siRNA and control antimiRNA (antimiR-67) or antimiR-27. Tailed nucleotides are highlighted in purple. The relative abundance of each isoform (%) is given before (antimiR-67) or after (antimiR-27) TDMD induction by the transfection of the corresponding antimiRNAs. The red color indicates a decrease and the green an increase in abundance of the respective sequence. (C) Distribution in percentage of miR-27a tailed and trimmed isoforms as assessed by small RNAs deep-sequencing of cells treated with the indicated siRNA and antimiRNA oligonucleotides. The ‘0’ indicates the wild-type (WT) isoform of 21 nt. The sum of all trimmed (≤-2) or tailed (≥+2) sequences is indicated. (D) The efficiency of the trimming was evaluated in each knockdown condition and antimiRNA treatment. The graph represents the ratio of miR-27a (purple) or miR-27b (blue) trimmed versus WT (‘0’ or mature sequence) reads after TDMD induction (antimiR-27) normalized to those observed without TDMD (antimiR-67) using the values shown in C. The values were arbitrarily set at 1 in cells transfected with the control siRNA (siCtrl). (E) The efficiency of the tailing in each knockdown condition was analyzed as explained in D, except that the graph represents the ratio of tailed isoforms versus WT (‘0’) reads.
This result could be explained by the tendency of TUTases to complement each other in vivo (45) and/or the high processivity of DIS3L2 (34). To evaluate the contribution of the other TUTases, we performed multiple knockdowns in HeLa cells and again tested the efficiency of the TDMD. To this end, we separated the TUTases in two groups. In addition to TUT1, the first group (TUT1+2+4+7) comprises TUT2 (also called GLD2 and PAPD4), TUT4 (ZCCHC11) and TUT7 (ZCCHC6) as they are known to act redundantly in the pre-let-7 mono-uridylation (45). The second group (TUT1+3+5+6) is composed of TUT1, TUT3 (PAPD5), TUT5 (PAPD7) and TUT6 (MTPAP). However, we did not manage to rescue miRNA stability with any of the two siRNA mixes (data not shown). So, we cannot exclude that other combinations of TUTases, or more than four of them might act redundantly in this process. Similarly, we also tried to knockdown other exoribonucleases such as XRN1 and XRN2 alone or in combination with DIS3L2, but again could not observe an effect on miR-27 degradation induced by antimiRNA oligonucleotides (data not shown).
It could also be that the northern blot analysis approach we used to measure the effect of TUT1 and DIS3L2 knockdowns on miRNA levels presents limitations when it comes to assess subtle differences. We thus decided to perform small RNA cloning and sequencing in order to verify if some changes that could not be observed by other means occur after TUT1 and/or DIS3L2 depletion. We first monitored the impact of antimiR-27 transfection on miR-27 isoforms, which allowed us to see that the most abundant tailed isoforms of miR-27a present an addition of one or two U residues at their 3′ end (Figure 5B). We could also detect a low number of miR-27 sequences presenting additional A residues, or a mix of A and U residues (data not shown). We calculated the respective abundance of the most abundant isoforms of miR-27a (in percentage relative to the total number of miR-27a in each sample) in presence or absence of TDMD induction by antimiR-27 and we observed an increase in the abundance of tailed and trimmed isoforms and a decrease of the major isoforms of miR-27a or b in presence of the antimiR-27 (respectively in green and in red in Figure 5B).
We then determined more precisely the distribution of miR-27a isoforms in libraries generated from cells treated with a control siRNA or siRNAs directed against TUT1, DIS3L2 or both and transfected with an antimiR-27 or control (antimiR-67) oligonucleotide. For each library, we calculated the number of wild-type (after miRBase) sequences of miR-27a that we set as the ‘0’ modification, as well as the number of sequences tailed or trimmed by 1, 2 or more nucleotides (respectively +1, +2… and −1, −2…). The deep-sequencing analysis confirmed that miR-27a exists mainly as three isoforms of 21 (0), 20 (−1) and 22 (+1) nucleotides. We therefore considered as trimmed and tailed sequences, all isoforms presenting with an addition or deletion of more than 2 nucleotides. As shown in Figure 5C for miR-27a, in cells treated with the control siRNA the percentage of trimmed sequences indeed increased in the presence of antimiR-27 (bottom graph) compared to antimiR-67 (top graph). We also observed a modest increase in tailing upon transfection of the specific antimiRNA. This increase in trimming seems to be less pronounced when DIS3L2, TUT1 or both DIS3L2 and TUT1 were knocked-down (Figure 5C and Supplementary Figure S5C). We did the same calculation for miR-27b and roughly observed the same effects, although less pronounced, which might be due to the fact that the antimiRNA oligonucleotide we transfected was directed against miR-27a (Supplementary Figure S6).
To quantify these differences, we then calculated the efficiency of trimming (Figure 5D) and tailing (Figure 5E) in each knockdown condition and for both miR-27a and miR-27b. To this end, we first determined the ratio of trimmed (or tailed) to wild-type (mature) sequences in antimiR-27 versus antimiR-67 in each condition, and then normalized it to the ratio found in the control cells, which was arbitrarily set to 1. TUT1 depletion did not seem to affect the efficiency of trimming of both miR-27a and b (Figure 5D and Supplementary Figure S5C). However, we observed a 40–50% reduction in trimming of miR-27a and b in cells where DIS3L2 or DIS3L2 and TUT1 were knocked-down (Figure 5D). The effect on tailing was more difficult to assess given the weaker induction of tailing measured by deep-sequencing. The extent of tailing appeared to be quite limited and did not expend much further than 2 nt at most. This might be due to the timing at which the RNA was isolated after TDMD induction, and that was a bit too late to fully capture tailed miRNAs. Alternatively, it might also be explained by the experimental procedure used for the small RNA library preparation and by a sub-optimal size selection of the PCR products prior to their sequencing. Nevertheless, it seems that TUT1 knock-down had no effect on tailing, while DIS3L2 or DIS3L2 and TUT1 knock-down had a mild negative impact (Figure 5E).
Overexpression of a catalytic mutant of DIS3L2 impairs TDMD
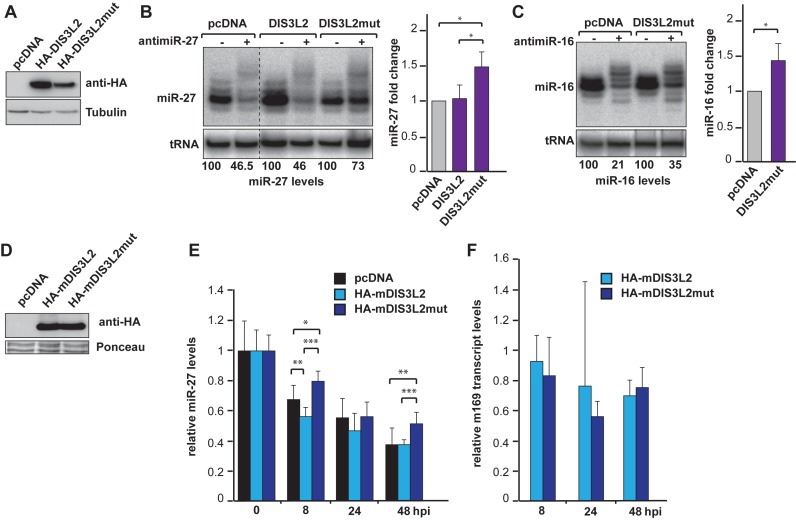
To further prove the involvement of DIS3L2 in the mechanism, we also used another strategy consisting in the expression of a catalytically inactive mutant protein. Indeed, knockdown by siRNA will never achieve a complete removal of the targeted protein, and the remaining DIS3L2 might still be capable of degrading its substrate RNA. The mutant DIS3L2 (DIS3L2mut) was therefore generated by replacing two residues from the catalytic site (D391N, D392N), a mutation known to affect DIS3L2 activity without impairing its ability to bind RNA (34,46). We then overexpressed HA-DIS3L2, HA-DIS3L3mut or an empty plasmid (pcDNA) in HEK293 cells and induced TDMD by transfection of antimiRNA oligonucleotides.
We first verified that the mutation in DIS3L2 did not affect its expression (Figure 6A). The RNA samples were then analyzed by northern blot and we measured the fold change accumulation of the targeted miRNAs. If we were expecting that the over-accumulation of HA-DIS3L2 would either stimulate TDMD or induces a dominant negative effect, its overexpression did not alter the efficiency of the TDMD of miR-27 compared to the control (Figure 6B). This might be due to the fact that the activity of the endogenous DIS3L2 was already at saturating level. On the contrary, the overexpression of DIS3L2mut led to a stabilization of miR-27 isoforms most likely due to a dominant negative effect (Figure 6B). This effect was reproducibly observed and resulted on average in a mild but significant 1.5-fold increase in miR-27 level (Figure 6B). A similar stabilization was observed for miR-16, as transfection of DIS3L2mut impaired TDMD on this miRNA as well (Figure 6C). Thus, this result confirms the involvement of DIS3L2 in TDMD.
Figure 6.
Overexpression of a catalytic mutant DIS3L2 inhibits TDMD. (A) Expression levels of HA-DIS3L2 and HA-DIS3L2mut in HEK293 cells by western blot. Tubulin serves as loading control. (B) miR-27 degradation is inhibited in HEK293 cells overexpressing the catalytically inactive version of DIS3L2 (DIS3L2mut) as shown by northern blot and after quantification. A dashed line indicates discontinuous lanes from the same gel. The bar graph shows the quantification of miR-27 fold change normalized to the empty vector (pcDNA), which was arbitrarily set to 1. Error bars indicate SDs (Student t-test: *P < 0.05, n = 4 biological replicates). (C) Same analysis as in (B), but for miR-16 degradation. (D) Expression levels of HA-mDIS3L2 and HA-mDIS3L2mut by western blot at 48 hpi. Ponceau S staining serves as loading control. (E) MiR-27 accumulation was monitored in mouse Hepa 1.6 cells during a time course of infection by the MCMV. Prior to infection, Hepa 1.6 cells were transfected with an empty plasmid (black), a plasmid coding for HA-mDIS3L2 (light blue) or HA-mDIS3L2mut (dark blue). The graph represents the relative miR-27 levels measured by qRT-PCR at 0, 8, 24 and 48 hpi. MiR-27 levels were normalized to those of miR-24 which serves as reference and arbitrarily set at 1 at 0 hpi for each condition. Error bars indicate SDs performed on three independent experiments. (Student t-test *P < 0.05, **P < 0.01, ***P < 0.001; n = 3 biological replicates). (F) The level of the viral m169 transcript was measured by qRT-PCR in the samples used in (E) at 8, 24 and 48 hpi. The transcript coding for PPIA (Peptidylprolyl isomerase A) was used as reference. For each time point, the expression of m169 transcript in HA-mDIS3L2 and HA-mDIS3L2mut expressing cells was normalized to the samples containing the empty vector (pcDNA) which was set at 1.
We have shown previously that MCMV degrades endogenous miR-27 via the viral transcript m169 (20). We therefore wanted to test whether DIS3L2 could also be involved in TDMD naturally induced by this virus. We infected cells transiently overexpressing the mouse (m)DIS3L2 or its catalytic mutant with MCMV and investigated the impact on miR-27 accumulation. Both HA-mDIS3L2 and HA-mDIS3L2mut were expressed at similar levels (Figure 6D). The levels of miR-27 (Figure 6E) and of the viral transcript m169 (Figure 6F) were monitored by qRT-PCR at different time points of infection and in three independent experiments. We first observed that the overexpression of mDIS3L2 does not globally impact miR-27 degradation compared to the control cells expressing the empty vector (Figure 6E, HA-mDIS3L2 versus pcDNA) as observed in human cells. We have only a slight enhancement of miR-27 degradation at 8 hpi, but this effect seems to disappear over time. On the contrary, we could observe a mild but significant stabilization of miR-27 in cells expressing mDIS3L2mut compared to cells transfected with mDIS3L2 or empty pcDNA at 8 and 48 hpi (Figure 6E; HA-mDIS3L2mut versus HA-mDIS3L2 and HA-mDIS3L2mut versus pcDNA).
This stabilization was confirmed by northern blot (Supplementary Figure S7), which allowed us to visualize the different isomiRs, when the qRT-PCR only measures the abundance of the whole population of a given miRNA. Indeed, we could observe a stronger accumulation of the major isoforms of miR-27 in the presence of mDIS3L2mut at 48 hpi. To verify that this difference was not solely due to a defect in the infection efficiency, or in the production of the m169 transcript, we measured its accumulation by qRT-PCR in the experiments described above. This transcript starts to be expressed 2–3 h after infection (20), we thus arbitrarily decided to normalize its relative expression to the 8 hpi time point (and not 0 hpi) as we are sure that the signal detected is due to de novo synthesis of the m169 transcript. As shown in Figure 6F, we could appreciate that m169 transcript accumulates at similar level in cells expressing mDIS3L2 and mDIS3L2mut, no significant differences were observed. Thus, the differential accumulation of miR-27 in those cells are only due to the overexpression or not, of the inactive form of mDIS3L2. Taken together, these results indicate that DIS3L2 is also involved in TDMD when this mechanism is naturally induced by MCMV.
DISCUSSION
The stability and function of a miRNA is tightly linked to its interaction with other RNA molecules. Thus, it has been demonstrated that it was possible to prevent miRNA loading into the RISC by the use of artificial decoy or sponge RNAs (47). This observation was later confirmed to naturally exist with the discovery of circular RNAs that can titer miRNAs such as miR-7 (48). Pairing with its RNA target will usually result in the stabilization of the miRNA (49), a mechanism that has been referred to as target-mediated miRNA protection. However, in other cases, the miRNA-target pairing can have the opposite effect and lead to the miRNA decay via the TDMD mechanism we studied here. In addition to the MCMV m169 transcript, the herpesvirus saimiri HSUR RNA can also target miR-27 for degradation (22). Similarly, the human cytomegalovirus encodes a bicistronic mRNA that triggers degradation of the cellular miRNA family miR-17/miR-20a (50). The molecular mechanisms at play in this particular mode of regulation have not been elucidated to date.
We confirmed here previous observations (23,24) that showed that TDMD is governed by the miRNA/target ratio as well as the degree of complementarity. Nevertheless, we also observed that TDMD operates in a non-cooperative manner in contrast to the classical miRNA-mediated target regulation. The cellular factors implicated in this particular miRNA decay mechanism were not known. Using a powerful proteomic approach, we characterized the protein complexes involved in miRNA tailing and trimming. Among the proteins we identified, we focused our analysis on two promising candidates: TUT1 and DIS3L2. As opposed to other TUTases, TUT1 had never been implicated in miRNA biogenesis. In addition, the most studied nucleotidyl transferases TUT4 and TUT7 act at the pri- or pre-miRNA level and never on the mature miRNA (51). In plants, the HESO1 protein has been implicated in uridylation of unmethylated miRNAs (52), but multiple terminal nucleotidyl transferases are required for this process to be fully active, including uncharacterized ones (53).
We did not manage to functionally implicate TUT1 in TDMD in our conditions, which might also indicate that this is a complicated synergistic process. Nonetheless, we could confirm the interaction between TUT1 and AGO2. We also show here that TUT1 interacts with tailed and trimmed isoforms of miR-27 only when TDMD is induced. In addition we also retrieved peptides for the SART3 protein in our mass spectrometry analysis (Supplementary Table S1). SART3, also known as Tip110, associates with U6 snRNP (54) and has been recently confirmed to interact directly with the C. elegans homolog of TUT1, USIP1, to participate in U6 snRNA recycling (55). Most importantly, it has been identified as a direct interactor of both AGO1 and AGO2 in HEK293 cells (56). Therefore, we have a link explaining how TUT1 could be recruited to AGO2 when TDMD is induced. This also confirms that both TUT1 and SART3 must interact with AGO proteins in the cytoplasm, as we could show for TUT1 and AGO2. Our results also indicate that TUT1 might not bind with the same efficiency to all miRNAs, since we could only retrieve it by mass spectrometry with antimiR-27 oligonucleotides, and its association with miR-16 seemed to be much weaker. Recently, it was demonstrated that TUT4 (ZCCHC11) and TUT7 (ZCCHC6) selectively mono-uridylate a subset of miRNAs harboring a specific motif (42). This observation strongly suggests that similar specificity for given miRNAs will govern the choice of TUTases recruited for the TDMD of other miRNAs. More work will be needed to definitely know which of the seven TUTases are involved in TDMD.
Our results are more conclusive when it comes to the other factor involved in TDMD. Indeed, we could functionally implicate DIS3L2 in this process both using knockdown followed by small RNA sequencing analysis and overexpression of a catalytic mutant showing a dominant negative effect. We did not manage to fully restore miRNA stability, which might either indicate that other enzymes could be implicated or that the high processivity of DIS3L2 makes it difficult to fully inactivate it be it by RNAi or overexpression of a dominant negative mutant.
It could also be that DIS3L2 would only be involved in the degradation of miR-27 presenting with additional U residues, as it was shown to preferentially target U-tailed substrates (46). The recently obtained crystal structure of DIS3L2 revealed that it could accommodate an artificial 12-mer oligonucleotides solely composed of Us (57), but our sequencing analysis revealed that the vast majority of miR-27 sequences appear to present with only few additional U residues at their 3′ extremity. In addition, a few sequences also had A residues. So, DIS3L2 might also be active on short RNAs presenting as little as two Us and/or As. This is in agreement with characterization of its in vitro activity, which indicates that the enzyme can target sequences with overhanging As (34).
Alternatively, miRNAs presenting A additions might be degraded by another enzyme. In that respect, miR-27a was shown to be adenylated by PAPD4 (TUT2), but this did not seem to affect its stability (58). Similarly, PAPD5 (TUT3) was also implicated in the adenylation of other miRNAs, such as miR-21, and in this case this resulted in its degradation by PARN (59). To verify that the enzymes that were studied in these two reports had no impact on miR-27a and b, we reanalyzed the small RNA libraries that were generated in these manuscripts, but did not find evidence of differential accumulation, or of an impact on the tailed and trimmed forms of these miRNAs (data not shown). Altogether our results therefore allowed us to identify DIS3L2 as the exoribonuclease involved in one specific mature miRNA degradation pathway.
ACCESSION NUMBERS
The sequencing data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus (60) and are accessible through GEO Series accession number GSE73210 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE73210).
Supplementary Material
Acknowledgments
We are grateful to E. Izaurralde, N. Kim, G. Meister, A. Dziembowski and R. A Anderson for the gift of plasmids and antibodies. We also thank the IGBMC Microarray and Sequencing platform, member of the France Genomique program, for the sequencing of our libraries. We thank all members of the Pfeffer laboratory for discussion, and Sophie Cooke for technical assistance.
Author contributions: G.H., S.C., M.M. and O.T. performed the experiments. J.C., L.K. and P.H. performed the MS procedure, B.C.W.M. and J.C. analyzed the MS data and B.C.W.M. compiled all MS results. B.C.W.M. performed the deep sequencing analysis. G.H., S.C. and S.P. analyzed the data. G.H. and S.P. designed the experiments and wrote the paper. S.P. supervised the project.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was funded by the European Research Council [ERC starting grant ncRNAVIR 260767] and has been published under the framework of the LABEX: ANR-10-LABX-0036_NETRNA and benefits from a funding from the state managed by the French National Research Agency as part of the Investments for the future program. Funding for open access charge: European Research Council [ERC starting grant ncRNAVIR 260767].
Conflict of interest statement. None declared.
REFERENCES
- 1.Krol J., Loedige I., Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 3.Zhang B., Wang Q., Pan X. MicroRNAs and their regulatory roles in animals and plants. J. Cell. Physiol. 2007;210:279–289. doi: 10.1002/jcp.20869. [DOI] [PubMed] [Google Scholar]
- 4.Farazi T.A., Hoell J.I., Morozov P., Tuschl T. MicroRNAs in human cancer. Adv. Exp. Med. Biol. 2013;774:1–20. doi: 10.1007/978-94-007-5590-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang T.-C., Mendell J.T. microRNAs in vertebrate physiology and human disease. Annu. Rev. Genomics Hum. Genet. 2007;8:215–239. doi: 10.1146/annurev.genom.8.080706.092351. [DOI] [PubMed] [Google Scholar]
- 6.Rybak A., Fuchs H., Smirnova L., Brandt C., Pohl E.E., Nitsch R., Wulczyn F.G. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat. Cell Biol. 2008;10:987–993. doi: 10.1038/ncb1759. [DOI] [PubMed] [Google Scholar]
- 7.Newman M.A., Thomson J.M., Hammond S.M. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viswanathan S.R., Daley G.Q., Gregory R.I. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heo I., Joo C., Kim Y.-K., Ha M., Yoon M.-J., Cho J., Yeom K.-H., Han J., Kim V.N. TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Cell. 2009;138:696–708. doi: 10.1016/j.cell.2009.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Heo I., Joo C., Cho J., Ha M., Han J., Kim V.N. Lin28 mediates the terminal uridylation of let-7 precursor MicroRNA. Mol. Cell. 2008;32:276–84. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 11.Chang H.-M., Triboulet R., Thornton J.E., Gregory R.I. A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway. Nature. 2013;497:244–248. doi: 10.1038/nature12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu X., Zheng Q., Vrettos N., Maragkakis M., Alexiou P., Gregory B.D., Mourelatos Z. A MicroRNA precursor surveillance system in quality control of MicroRNA synthesis. Mol. Cell. 2014;55:868–879. doi: 10.1016/j.molcel.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baccarini A., Chauhan H., Gardner T.J., Jayaprakash A.D., Sachidanandam R., Brown B.D. Kinetic analysis reveals the fate of a microRNA following target regulation in mammalian cells. Curr. Biol. 2011;21:369–376. doi: 10.1016/j.cub.2011.01.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gantier M.P., McCoy C.E., Rusinova I., Saulep D., Wang D., Xu D., Irving A.T., Behlke M.A., Hertzog P.J., Mackay F., et al. Analysis of microRNA turnover in mammalian cells following Dicer1 ablation. Nucleic Acids Res. 2011;39:5692–5703. doi: 10.1093/nar/gkr148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Ambrogio A., Gu W., Udagawa T., Mello C.C., Richter J.D. Specific miRNA stabilization by Gld2-catalyzed monoadenylation. Cell Rep. 2012;2:1537–1545. doi: 10.1016/j.celrep.2012.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones M.R., Quinton L.J., Blahna M.T., Neilson J.R., Fu S., Ivanov A.R., Wolf D.A., Mizgerd J.P. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat. Cell Biol. 2009;11:1157–1163. doi: 10.1038/ncb1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rüegger S., Großhans H. MicroRNA turnover: when, how, and why. Trends Biochem. Sci. 2012;37:436–446. doi: 10.1016/j.tibs.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 18.Krol J., Busskamp V., Markiewicz I., Stadler M.B., Ribi S., Richter J., Duebel J., Bicker S., Fehling H.J., Schubeler D., et al. Characterizing light-regulated retinal microRNAs reveals rapid turnover as a common property of neuronal microRNAs. Cell. 2010;141:618–631. doi: 10.1016/j.cell.2010.03.039. [DOI] [PubMed] [Google Scholar]
- 19.Rissland O.S., Hong S.-J., Bartel D.P. MicroRNA destabilization enables dynamic regulation of the miR-16 family in response to cell-cycle changes. Mol. Cell. 2011;43:993–1004. doi: 10.1016/j.molcel.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marcinowski L., Tanguy M., Krmpotic A., Rädle B., Lisnić V.J., Tuddenham L., Chane-Woon-Ming B., Ruzsics Z., Erhard F., Benkartek C., et al. Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo. PLoS Pathog. 2012;8:e1002510. doi: 10.1371/journal.ppat.1002510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Libri V., Helwak A., Miesen P., Santhakumar D., Borger J.G., Kudla G., Grey F., Tollervey D., Buck A.H. Murine cytomegalovirus encodes a miR-27 inhibitor disguised as a target. Proc. Natl. Acad. Sci. U. S. A. 2012;109:279–284. doi: 10.1073/pnas.1114204109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cazalla D., Yario T., Steitz J.A., Steitz J. Down-regulation of a host microRNA by a Herpesvirus saimiri noncoding RNA. Science. 2010;328:1563–1566. doi: 10.1126/science.1187197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ameres S.L., Horwich M.D., Hung J.H., Xu J., Ghildiyal M., Weng Z., Zamore P.D. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534–1539. doi: 10.1126/science.1187058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de la Mata M., Gaidatzis D., Vitanescu M., Stadler M.B., Wentzel C., Scheiffele P., Filipowicz W., Großhans H. Potent degradation of neuronal miRNAs induced by highly complementary targets. EMBO Rep. 2015;16:500–511. doi: 10.15252/embr.201540078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji L., Chen X. Regulation of small RNA stability: methylation and beyond. Cell Res. 2012;22:624–636. doi: 10.1038/cr.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J., Yang Z., Yu B., Liu J., Chen X. Methylation Protects miRNAs and siRNAs from a 3′-End Uridylation Activity in Arabidopsis. Curr Biol. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim J., Ha M., Chang H., Kwon S.C., Simanshu D.K., Patel D.J., Kim V.N. Uridylation by TUT4 and TUT7 Marks mRNA for Degradation. Cell. 2014;159:1365–1376. doi: 10.1016/j.cell.2014.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buck A.H., Perot J., Chisholm M.A., Kumar D.S., Tuddenham L., Cognat V., Marcinowski L., Dolken L., Pfeffer S. Post-transcriptional regulation of miR-27 in murine cytomegalovirus infection. RNA. 2010;16:307–315. doi: 10.1261/rna.1819210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Broderick J.A., Salomon W.E., Ryder S.P., Aronin N., Zamore P.D. Argonaute protein identity and pairing geometry determine cooperativity in mammalian RNA silencing. RNA. 2011;17:1858–1869. doi: 10.1261/rna.2778911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saetrom P., Heale B.S.E., Snøve O., Aagaard L., Alluin J., Rossi J.J. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 2007;35:2333–2342. doi: 10.1093/nar/gkm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grimson A., Farh K.K.-H., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell. 2007;27:91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flores-Jasso C.F., Salomon W.E., Zamore P.D. Rapid and specific purification of Argonaute-small RNA complexes from crude cell lysates. RNA. 2013;19:271–279. doi: 10.1261/rna.036921.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malecki M., Viegas S.C., Carneiro T., Golik P., Dressaire C., Ferreira M.G., Arraiano C.M. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J. 2013;32:1842–1854. doi: 10.1038/emboj.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lubas M., Damgaard C.K., Tomecki R., Cysewski D., Jensen T.H., Dziembowski A. Exonuclease hDIS3L2 specifies an exosome-independent 3′-5′ degradation pathway of human cytoplasmic mRNA. EMBO J. 2013;32:1855–1868. doi: 10.1038/emboj.2013.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wyman S.K., Knouf E.C., Parkin R.K., Fritz B.R., Lin D.W., Dennis L.M., Krouse M.A., Webster P.J., Tewari M. Post-transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Res. 2011;21:1450–1461. doi: 10.1101/gr.118059.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mellman D.L., Anderson R.A. A novel gene expression pathway regulated by nuclear phosphoinositides. Adv. Enzyme Regul. 2009;49:11–28. doi: 10.1016/j.advenzreg.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mellman D.L., Gonzales M.L., Song C., Barlow C.A., Wang P., Kendziorski C., Anderson R.A. A PtdIns4, 5P2-regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature. 2008;451:1013–1017. doi: 10.1038/nature06666. [DOI] [PubMed] [Google Scholar]
- 38.Trippe R., Guschina E., Hossbach M., Urlaub H., Lührmann R., Benecke B.-J. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA. 2006;12:1494–1504. doi: 10.1261/rna.87706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfaff J., Meister G. Argonaute and GW182 proteins: an effective alliance in gene silencing. Biochem. Soc. Trans. 2013;41:855–860. doi: 10.1042/BST20130047. [DOI] [PubMed] [Google Scholar]
- 40.Nagarajan V.K., Jones C.I., Newbury S.F., Green P.J. XRN 5′→3′ exoribonucleases: structure, mechanisms and functions. Biochim. Biophys. Acta. 2013;1829:590–603. doi: 10.1016/j.bbagrm.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chlebowski A., Lubas M., Jensen T.H., Dziembowski A. RNA decay machines: the exosome. Biochim. Biophys. Acta. 2013;1829:552–560. doi: 10.1016/j.bbagrm.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 42.Thornton J.E., Du P., Jing L., Sjekloca L., Lin S., Grossi E., Sliz P., Zon L.I., Gregory R.I. Selective microRNA uridylation by Zcchc6 (TUT7) and Zcchc11 (TUT4) Nucleic Acids Res. 2015;42:11777–11791. doi: 10.1093/nar/gku805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohan N., Ap S., Francis N., Anderson R., Laishram R.S. Phosphorylation regulates the Star-PAP-PIPKIα interaction and directs specificity toward mRNA targets. Nucleic Acids Res. 2015;43:7005–7020. doi: 10.1093/nar/gkv676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ameyar-Zazoua M., Rachez C., Souidi M., Robin P., Fritsch L., Young R., Morozova N., Fenouil R., Descostes N., Andrau J.-C., et al. Argonaute proteins couple chromatin silencing to alternative splicing. Nat. Struct. Mol. Biol. 2012;19:998–1004. doi: 10.1038/nsmb.2373. [DOI] [PubMed] [Google Scholar]
- 45.Heo I., Ha M., Lim J., Yoon M.-J., Park J.-E., Kwon S.C., Chang H., Kim V.N. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell. 2012;151:521–532. doi: 10.1016/j.cell.2012.09.022. [DOI] [PubMed] [Google Scholar]
- 46.Ustianenko D., Hrossova D., Potesil D., Chalupnikova K., Hrazdilova K., Pachernik J., Cetkovska K., Uldrijan S., Zdrahal Z., Vanacova S. Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let-7 miRNAs. RNA. 2013;19:1632–1638. doi: 10.1261/rna.040055.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ebert M.S., Sharp P.A. Emerging roles for natural microRNA sponges. Curr. Biol. 2010;20:R858–R861. doi: 10.1016/j.cub.2010.08.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 49.Chatterjee S., Fasler M., Büssing I., Grosshans H. Target-mediated protection of endogenous microRNAs in C. elegans. Dev. Cell. 2011;20:388–396. doi: 10.1016/j.devcel.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 50.Lee S., Song J., Kim S., Kim J., Hong Y., Kim Y., Kim D., Baek D., Ahn K. Selective degradation of host MicroRNAs by an intergenic HCMV noncoding RNA accelerates virus production. Cell Host Microbe. 2013;13:678–690. doi: 10.1016/j.chom.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 51.Ha M., Kim V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 52.Zhao Y., Yu Y., Zhai J., Ramachandran V., Dinh T.T., Meyers B.C., Mo B., Chen X. The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr. Biol. 2012;22:689–694. doi: 10.1016/j.cub.2012.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X., Zhang S., Dou Y., Zhang C., Chen X., Yu B., Ren G. Synergistic and independent actions of multiple terminal nucleotidyl transferases in the 3′ tailing of small RNAs in Arabidopsis. PLoS Genet. 2015;11:e1005091. doi: 10.1371/journal.pgen.1005091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Licht K., Medenbach J., Luhrmann R., Kambach C., Bindereif A. 3′-cyclic phosphorylation of U6 snRNA leads to recruitment of recycling factor p110 through LSm proteins. RNA. 2008;14:1532–1538. doi: 10.1261/rna.1129608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ruegger S., Miki T.S., Hess D., Grosshans H. The ribonucleotidyl transferase USIP-1 acts with SART3 to promote U6 snRNA recycling. Nucleic Acids Res. 2015;43:3344–3357. doi: 10.1093/nar/gkv196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hock J., Weinmann L., Ender C., Rudel S., Kremmer E., Raabe M., Urlaub H., Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Faehnle C.R., Walleshauser J., Joshua-Tor L. Mechanism of Dis3l2 substrate recognition in the Lin28-let-7 pathway. Nature. 2014;514:252–256. doi: 10.1038/nature13553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burroughs A.M., Ando Y., de Hoon M.J.L., Tomaru Y., Nishibu T., Ukekawa R., Funakoshi T., Kurokawa T., Suzuki H., Hayashizaki Y., et al. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res. 2010;20:1398–1410. doi: 10.1101/gr.106054.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boele J., Persson H., Shin J.W., Ishizu Y., Newie I.S., Søkilde R., Hawkins S.M., Coarfa C., Ikeda K., Takayama K., et al. PAPD5-mediated 3′ adenylation and subsequent degradation of miR-21 is disrupted in proliferative disease. Proc. Natl. Acad. Sci. U. S. A. 2014;111:11467–11472. doi: 10.1073/pnas.1317751111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.