Abstract
Fever is commonly used to diagnose disease and is consistently associated with increased mortality in critically ill patients. However, the molecular controls of elevated body temperature are poorly understood. We discovered that the expression of RNA-binding motif protein 3 (RBM3), known to respond to cold stress and to modulate microRNA (miRNA) expression, was reduced in 30 patients with fever, and in THP-1-derived macrophages maintained at a fever-like temperature (40°C). Notably, RBM3 expression is reduced during fever whether or not infection is demonstrable. Reduced RBM3 expression resulted in increased expression of RBM3-targeted temperature-sensitive miRNAs, we termed thermomiRs. ThermomiRs such as miR-142–5p and miR-143 in turn target endogenous pyrogens including IL-6, IL6ST, TLR2, PGE2 and TNF to complete a negative feedback mechanism, which may be crucial to prevent pathological hyperthermia. Using normal PBMCs that were exogenously exposed to fever-like temperature (40°C), we further demonstrate the trend by which decreased levels of RBM3 were associated with increased levels of miR-142–5p and miR-143 and vice versa over a 24 h time course. Collectively, our results indicate the existence of a negative feedback loop that regulates fever via reduced RBM3 levels and increased expression of miR-142–5p and miR-143.
INTRODUCTION
Since antiquity, fever has been used as an indicator for diseases. Fever is defined as a regulated increase in body temperature above normal fluctuations, and is associated with various immune stressors from infectious and non-infectious sources. The increase in body temperature is initiated and regulated by numerous cytokines that act either as pyrogens or antipyretics (1). These constitute a complex circuitry that resets the temperature balance point of the body through a humoral or neural response (2).
There are long-standing arguments for and against treating elevated body temperature depending mainly on the presence of specific acute neurological injuries. Fever is associated with a worse outcome for patients with stroke and neurologic injury (3) and antipyretic treatment is thus recommended in these cases. However letting a fever run its course can be beneficial in sepsis where an elevated temperature in the first 24 h is associated with decreased mortality in severe infections (4).
MicroRNAs (miRNAs) are short RNAs (≈22 nucleotides) that reduce gene expression, usually by binding to the 3′ untranslated region (UTR) of target mRNAs. miRNAs guide an RNA-induced silencing complex (RISC) to specific mRNA target sites called miRNA responsive elements (mREs) to trigger translation inhibition and/or mRNA degradation (5). The first eight nucleotides of a miRNA, now called the seed region, may be complementary to motifs that determine their ability to regulate gene expression (6). Over 1000 miRNAs have been identified in humans, hundreds of which are associated with major biological processes including cell proliferation and differentiation, development and diseases (7,8). Consequently, miRNAs are arguably one of the most important classes of functional RNAs.
Specific genes and miRNAs associated with the febrile response may impact patient outcomes after infection (9,10). The effects of isolated temperature elevation however have not been examined at a molecular level. Our in silico analysis showed decreased levels of mRNA encoding a cold-shock protein, RBM3 in febrile patients that is dependent on the presence of fever but not infection. We also identified differentially expressed mRNAs and miRNAs in THP-1-derived macrophages at normal (37°C) and fever-like temperatures (40°C). As expected, mRNAs encoding RBM3 were the most significantly downregulated at 40°C. Small RNA sequencing and confirmation by quantitative polymerase chain reaction (PCR) assays revealed upregulation of temperature-sensitive miRNAs, we termed thermomiRs, including miR-10a, miR-10b, miR-151–5p, miR-151–3p, miR-125a, miR-98, miR-142–5p and miR-143 in THP-1-derived macrophages at 40°C compared to 37°C. Two thermomiRs, miR-142–5p and miR-143 were significantly increased following RBM3 knockdown in THP-1-derived macrophages; confirming the role of RBM3 in the regulation of these miRNAs at fever-like temperatures. Quantitation of target mRNA levels following knockdown and overexpression of miR-142–5p and miR-143 confirmed their roles in the regulation of pyrogen expression. In peripheral blood mononuclear cells (PBMCs) exposed to 40°C over a time course of 24 h (n = 5), we observed a trend whereby RBM3 levels increased when miR-142–5p and miR-143 decreased and vice versa. Our data indicate the existence of temperature-sensitive miRNAs, miR-142–5p and miR-143, which are regulated by RBM3 in a negative feedback mechanism established in response to fever.
MATERIALS AND METHODS
Cell Lines and primary samples
Human THP-1 cells were maintained in RPMI media supplemented with 10% (v/v) FCS, 0.05 mM 2-mercaptoethanol, 0.1 mg/ml penicillin/streptomycin and 2 mM L-glutamine. Differentiation of THP-1 into macrophages was performed as previously described (11). Briefly, cells were plated at a density of 1.5 × 107 cells per 75 cm2 culture flask containing supplemented RPMI with 100 ng/ml phorbol-12-myristate 13-acetate (PMA) and 50 μM 2-mercaptoethanol for 48 h. Undifferentiated cells that did not exhibit plastic-adherence were removed. Differentiated THP-1 cells, which adhered to tissue culture flasks, were then maintained at normal (37°C) or fever-like (40°C) temperatures for up to 24 h. Cells maintained at 40°C were harvested at 2, 8 and 24 h for subsequent small RNA and transcriptomic analyses. Human K562 cells were maintained in RPMI 1640 supplemented with 10% (v/v) FCS and antibiotics.
With ethics approval from the Human Research Ethics Committee of the Royal Prince Alfred Hospital (HREC/08/RPAH/222) and informed consent, whole blood samples were obtained from five healthy individuals (N1, N2, N3, N4 and N5). PBMCs were isolated using PolymorphPrep (Axis-Shield) according to the manufacturer's instructions, and maintained in supplemented RPMI media (used for THP-1) at fever-like (40°C) temperatures for 0, 2, 8, 16 and 24 h followed by RNA extraction and RT-qPCR analyses.
Flow cytometry analysis
Flow cytometry analysis to confirm the differentiation of THP-1 into macrophages was performed with anti-CD11b and -CD44 staining. Cell viability following exposure to 40°C was determined by measuring the percentages of necrotic and apoptotic cells stained with propidium iodide (PI) and Annexin V. All analysis was performed in triplicate on an LSR Fortessa (BD Biosciences).
Trypan blue staining and caspase activity assay
Differentiated THP-1 cells were stained with 0.4% (w/v) trypan blue and counted under the microscope to assess viability (250 - 300 cells for each culture condition). Caspase activity assay was performed using the Caspase-Glo®3/7 Kit (Promega) according to the manufacturer's instructions. Positive control THP-1 cells were irradiated in a UV Stratalinker 2400 (Stratagene) with a 400 000 μJ UVC dosage and incubated in fresh media for 16 h.
RNA extraction, RT-qPCR and miRNA detection assays
RNA extraction was performed using Trizol (Invitrogen) according to the manufacturer's instructions. RT-qPCR was performed on cDNA generated from 1 μg DNaseI-treated total RNA using SuperScript® III First-Strand Synthesis System (Invitrogen), according to the manufacturers’ instructions. RT-qPCR reactions were prepared in 20 μl volumes containing 1× IQ SyberGreen supermix and 0.3 μM of the respective forward and reverse primers. Samples were amplified and analysed using the CFX96(TM) Real Time PCR Machine (Bio-Rad).
The expression of known miRNAs was measured using TaqMan® MiRNA assays (Applied Biosystems) according to the manufacturer's instructions. Normalisation for cDNA input was performed using a stably expressed reference snoRNA, RNU24.
Sequencing and Bioinformatics analysis
Small RNA libraries were prepared from 5 μg of RNA using the Small RNA Sample Preparation Alternative v1.5 Protocol (Illumina) according to the manufacturers’ instructions and sequenced at Geneworks (Adelaide) using an Illumina GAIIx platform. Reads were aligned with Bowtie2 (12) on hg19 and miRNA expression was quantified using SeqMonk (http://www.bioinformatics.babraham.ac.uk/projects/seqmonk/) with a loess normalisation. Novel miRNAs were investigated using a sliding window of 20 bp across the aligned reads on the genome. Contigs of length between 17 and 25 bp with a coverage of at least 10 reads were then evaluated by a support vector machine for characteristics of a miRNA hairpin (13).
Western blot
Total protein lysates were loaded onto precast SDS-PAGE gels (Invitrogen) and subjected to electrophoresis before being transferred onto PVDF membranes. Membranes were blocked with 5% (w/v) BSA in TBS for 2 h at room temperature and incubated overnight with a mouse monoclonal anti-RBM3 antibody (1:5000; Atlas Antibodies) or a rabbit polyclonal anti-actin antibody (1:5000; Sigma). Following washes, membranes were incubated with HRP-conjugated secondary anti-mouse or anti-rabbit antibody (1:5000; Chemicon), and exposed using enhanced chemiluminescence reagents (Pierce) on a Kodak Imager (Kodak). Bands were quantified using ImageJ.
Knockdown of RBM3
Knockdown of RBM3 in differentiated THP-1 cells was performed by transfecting small interfering RNAs (siRNAs) into cells using nucleofection (11). A total of three siRNAs against RBM3 (s118 #58, #59 and #60; Applied Biosystems) and a negative control siRNA (Applied Biosystems) were used.
nCounter miRNA quantification
Total RNA (100 ng each) was analysed using the nCounter Human miRNA Expression Assay Kit Version 2.0 (NanoString Technologies) according to the manufacturers’ instructions. Data were analysed using the nSolver software, with normalisation performed using the top 100 highly expressed miRNAs.
Nucleofection of cells with anti-miRNA and mimic
THP-1 cells were differentiated into macrophages at 40°C, harvested and nucleofected with the custom designed 2′-O-methylated miRIDIAN miRNA Hairpin Inhibitor (Dharmacon) directed against hsa-miR-142–5p, hsa-miR-143–3p or non-human miRNA control. These inhibitors do not necessarily direct degradation of miRNAs but would induce loss-of-function as previously published (14). Cells were co-transfected with pmaxGFP vector and GFP-expressing cells were purified 24 h post nucleofection using fluorescence-activated cell sorting on the InfluxTM Cell Sorter (BD Biosciences). Nucleofection of miR-142 and miR-143 mimics (Dharmacon) and the miRIDIAN microRNA Mimic Negative Control #1 (Dharmacon) was performed using the same protocol. 300nM of miRNA inhibitor or mimic was used in each nucleofection reaction.
Analysis of gene expression in febrile patients and controls
Data from GSE40396 were normalised using the cyclic lowess algorithm (15) from the Affymetrix package in R (3 cycles). Pseudogenes and predicted genes were removed based on Gencode annotation (http://www.gencodegenes.org) before analysis of differential gene expression using GENE-E (Broad Institute). Individual patients were labelled for infection (presence/type or absence) and fever (presence or absence). A heat map for the top 100 genes (50 increased and 50 decreased; signal to noise) was determined using Marker Selection for patient samples with fever versus no fever.
Gene expression array
Differential expression of mRNAs was assessed using the GeneChip® Prime View™ Human Gene Expression Array (Affymetrix) at the Ramaciotti Centre for Genomics (Sydney) according to the manufacturer's instructions. The arrays were normalised using the justRMA suite of algorithms from Bioconductor (http://www.bioconductor.org) and analysed as previously described (16). Raw data were deposited in the Gene Expression Omnibus database (accession number GSE69343). Gene function enrichment was performed using the DAVID functional enrichment webtool (17). PrimeView gene set was used as background.
Statistics
The significance of fold-change in mRNA and miRNA expression between treatment and control cases were determined using two-tailed Student's t-test. Values are shown as mean ± SEM. Analyses were performed using GraphPad Prism v.6 (La Jolla, CA, USA). Results were considered significant when P-value < 0.05.
RESULTS
Reduced RBM3 expression is dependent on the presence of fever but not infection
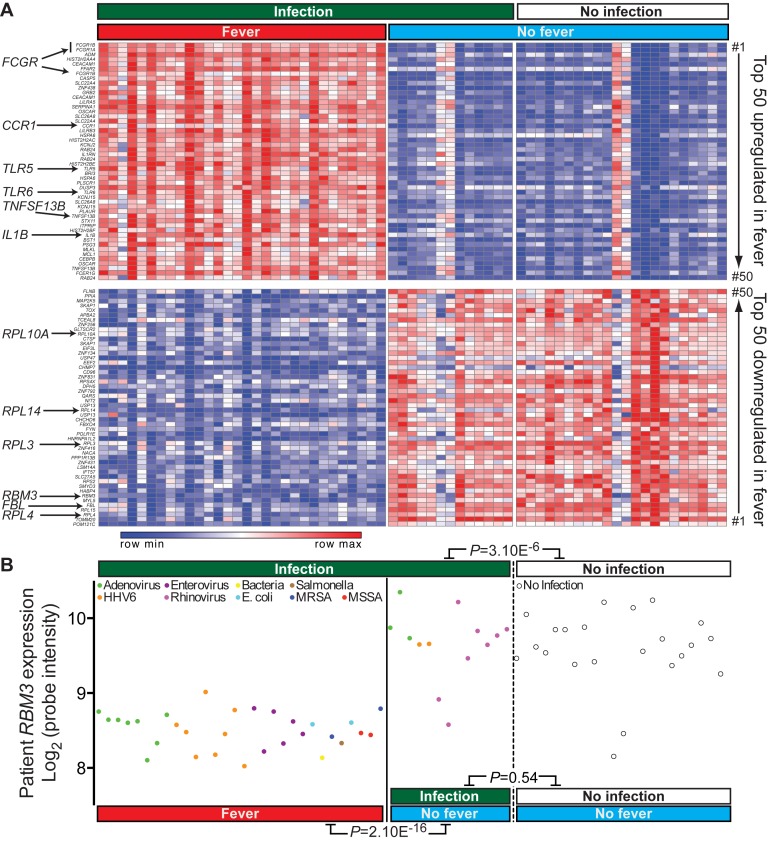
To determine fever-related gene expression changes, we re-analysed whole genome microarray data performed on whole blood from 30 febrile and 35 non-febrile children, classified on the basis of infection by diverse viruses and bacteria (18). As expected, transcripts encoding immune and inflammatory response genes including FCGR1 family members (19), CCR1 (20), TLR5 (21), TLR6 (22), TNFSF13B (23) and IL1B (24) were highly enriched amongst the top 50 upregulated transcripts (Figure 1A). Many mRNA processing and ribosomal protein synthesis related genes were downregulated, including FBL, RPL3, RPL4, RPL10A and RPL14 (Figure 1A), consistent with the widespread inhibition of RNA processing and ribosomal protein synthesis following heat-shock (25–27). Amongst the most downregulated genes was RBM3 (signal-to-noise; 2.2-fold, Figure 1A), which encodes a cold-shock protein known to be induced under conditions of mild hypothermia and also to alter miRNA levels by binding to the precursor transcript (28,29). We determined whether RBM3 expression was more relevant to fever or infections in patients (Figure 1B). RBM3 levels were significantly lower in the presence of fever regardless of the type of infection (P = 2.1E-16). We found no significant difference in RBM3 levels between patients with infection but no fever and patients with no infection (P = 0.54, t-test). Thus, the presence of fever, not infection, determined lower RBM3 levels.
Figure 1.
Differential gene expression in febrile and non-febrile patients. (A) Heatmap showing top 50 most upregulated (red) and downregulated (blue) genes in 30 febrile patients compared to 35 non-febrile controls. (B) Re-analysis of RBM3 expression from Hu et al., 2013 (18) in 30 febrile and 35 non-febrile patients.
Fever-like temperature alters the expression of diverse genes including RBM3
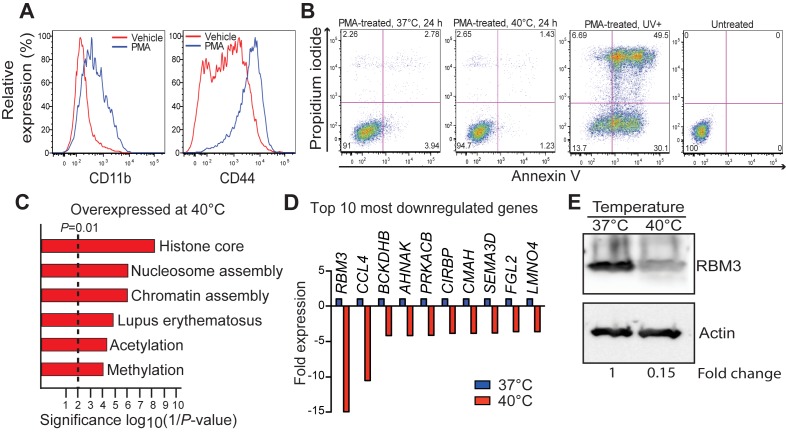
In order to further understand the role of RBM3 during fever, we performed in vitro studies using THP-1-derived macrophages; a model used extensively for studying the immune response (30). Following 48 h exposure to PMA, THP-1 cells exhibited plastic-adherence and expressed higher levels of CD11b and CD44 compared to untreated cells, confirming macrophage differentiation (Figure 2A). THP-1-derived macrophages were exposed to fever-like temperature (40°C) in vitro. In order to confirm that these cells were viable at the elevated temperature, and therefore appropriate for transcriptome and miRNAome analyses, Annexin V/propidium iodide (PI) staining (Figure 2B), trypan blue staining (Supplementary Figure S1A) and caspase activity assays (Supplementary Figure S1B) were performed. All three approaches demonstrated that exposure of THP-1-derived macrophages to 40°C for up to 24 h did not result in significant cell death compared to cells maintained at 37°C (Figure 2B, Supplementary Figure S1A,B). Viability of >90% was maintained at 40°C (Figure 2B and Supplementary Figure S1A).
Figure 2.
Expression of the cold-shock protein RBM3 is altered between 37°C and 40°C in THP-1-derived macrophages. (A) Confirmation of THP-1 differentiation into macrophages as shown by increased expression of CD11b and CD44 using flow cytometry. (B) Viability of THP-1 cells incubated at 40°C compared to controls (37°C) as measured by flow cytometry following Annexin V and propidium iodide (PI) staining. Ultraviolet-exposed cells (UV+) were used as positive control. (C) Gene function enrichment of genes overexpressed at 40°C. (D) Top ten most downregulated genes at 40°C. (E) Western blot for RBM3 expression following incubation at 37°C and 40°C with actin used as the loading control. PMA, phorbol-12-myristate 13-acetate.
Using whole genome expression array, we found 309 protein-coding genes that changed significantly (P < 0.01; fold-change > 2) between 37°C and 40°C in THP-1-derived macrophages (Supplementary Table S1). Of these, 132 genes had increased expression at 40°C and 177 had reduced expression. Upregulated genes were enriched in histone, nucleosome and acetylation functions (Figure 2C). This enrichment was anticipated by recent reports that an increase in expression of acetylation genes such as HDAC and histones are vital to the immune response (31). Downregulated genes were not significantly enriched in any category (P < 1.0E-3). RBM3 showed the highest reduction at 40°C (≈15-fold decrease), consistent with our observation in febrile patients (Figure 2D). While RBM3 levels have never been investigated in hyperthermia, our model of fever demonstrated that RBM3 mRNA levels were the most destabilised with a concomitant ≈6-fold decrease in RBM3 protein levels (Figure 2E). Because our focus is the temperature component of fever and RBM3 is a temperature-sensitive protein known to regulate miRNA levels (28,29), we further investigated the link between RBM3 and miRNA regulation in our model.
Fever-like temperatures alter the expression of miRNAs in THP-1-derived macrophages
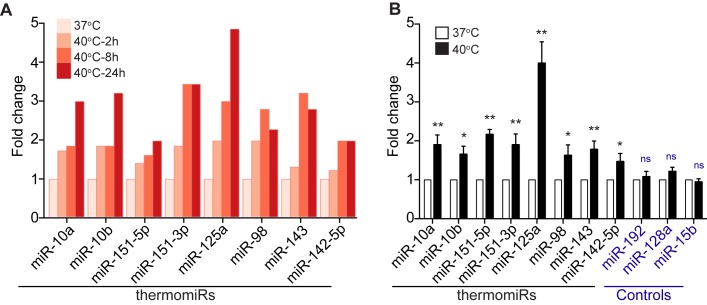
We performed small RNA sequencing on THP-1-derived macrophages at 37°C and on cells incubated at 40°C for 2, 8 and 24 h. No miRNA showed significantly lower expression after incubation at 40°C. Eight miRNAs exhibited a significantly higher expression after 24 h of incubation at 40°C (P < 0.05; fold-change > 2; Figure 3A), which we called thermomiRs. Importantly thermomiRs showed a consistent increase in expression starting at 2 h. This increase in expression after 24 h was validated using RT-qPCR on all eight miRNAs and three control miRNAs that did not show any change in the sequencing data (Figure 3B). Many of these miRNAs are known to have roles in the immune response. For example, miR-10a is a key mediator of regulatory T cell differentiation (32), miR-125a controls the inflammatory response in macrophages (33) and in autoimmune disease (34), miR-98 regulates Fas-ligand apoptosis that plays a crucial role in multiple immune functions, miR-142–5p mediates T cell activation (35) and miR-143 regulates Toll-like receptors (36).
Figure 3.
ThermomiR expression increases at 40°C in THP-1-derived macrophages. (A) Expression of temperature-sensitive miRNAs (thermomiRs) measured by small RNA sequencing at 37°C and after 2, 8 and 24 h of incubation at 40°C. (B) Expression of thermomiRs and controls measured by RT-qPCR after incubation for 24 h. Data are from three independent experiments each in triplicate and show mean ± SEM. Two-tailed Student's t-test was used to determine significance, denoted by * (P < 0.05) and ** (P < 0.001). ns, not significant.
RBM3 regulates expression of miR-142–5p and 143
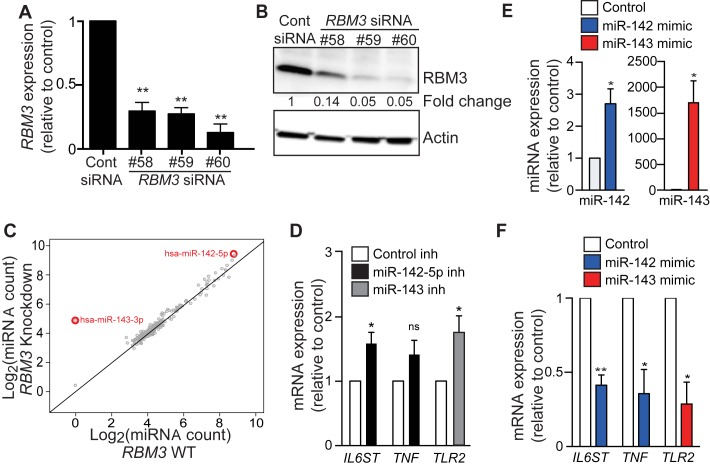
As RBM3 controls miRNA expression through an RNA-binding motif, we examined the effects of reducing RBM3 levels on miRNAs in THP-1-derived macrophages using three different siRNAs (37). RT-qPCR and Western blot analyses confirmed reduced RBM3 expression in cells transfected with each of these three siRNAs by ≈3–20-fold (Figure 4A,B). Using the NanoString nCounter analysis, we found 40 miRNAs for which the expression increased consistently for all three RBM3 knockdowns (Supplementary Table S2). Only one miRNA, miR-221 showed a decrease after RBM3 knockdown. Of the 40 miRNAs affected by RBM3 knockdown, miR-142–5p and miR-143 were outliers in terms of change in expression (Figure 4C). miR-143 was only detectable after RBM3 knockdown. miR-142–5p was most highly expressed amongst miRNAs with increased expression after RBM3 knockdown. Notably, these miRNAs were also amongst those that increased in expression following elevated temperature treatments (Figure 3A,B). These miRNAs may be two key modulators of pyrogens during a feedback loop that involves elevated temperature and reduced RBM3 levels.
Figure 4.
RBM3 siRNA knockdown increases the expression of thermomiRs. (A) Expression of RBM3 mRNA by RT-qPCR, and (B) RBM3 protein levels by Western blot in THP-1-derived macrophages nucleofected with negative control siRNA and each of the three siRNAs against RBM3: #58, #59 and #60. (C) Average counts of individual miRNAs from the three RBM3 knockdown experiments as measured using NanoString nCounter. Two miRNAs miR-142 and miR-143 showed the greatest change in expression. (D) Expression of pyrogens in THP-1-derived macrophages: IL6ST, TNF and TLR-2 following inhibition of miR-142–5p and miR-143 (relative to control inhibitor). (E) Expression of miR-142 and miR-143 in THP-1-derived macrophage at 24 h after nucleofection with respective miRNA mimics (relative to control). (F) Expression of IL6ST, TNF and TLR- in THP-1-derived macrophage at 24 h following nucleofection of miR-142 and miR-143 mimic (relative to control). Data are from three independent experiments each in triplicate and show mean ± SEM. Two-tailed Student's t-test was used to determine significance, denoted by * (P < 0.05) and ** (P < 0.001). inh, inhibitor; ns, not significant.
ThermomiRs miR-142–5p and miR-143 target important immune genes and genes in the fever-response pathway
miR-143 is involved in the regulation of multiple pyrogens including TLR2 (36), PGE2 (38) and IL-6 (39), as is miR-142 (40). We inhibited miR-142–5p and miR-143 in THP-1 cells using antagomir oligonucleotides and measured the expression of six pyrogens: IL6ST, IFNA1, TNF, TLR2, IL6 and PGE2 by RT-qPCR (Figure 4D). Of these, three targets IL6, IFNA1 and PGE2, were expressed at levels below the detection limit of RT-qPCR pre- and post-knockdown of either miRNA. We confirmed that miR-142–5p and miR-143 regulated the expression of IL6ST (P = 0.0279) and TLR2 (P = 0.0476) respectively (Figure 4D). Although inhibition of miR-142–5p resulted in the increased expression of TNF by 1.4-fold, this did not reach statistical significance (P = 0.1304). Increased expression of miR-142 and miR-143 via nucleofection with miRNA mimics (Figure 4E) resulted in significantly reduced expression of IL6ST, TNF and TLR2 (Figure 4F), confirming that these pyrogens are regulated by miR-142 or miR-143. Our data in addition to the previously described validated targets of miR-142 and miR-143 show that these miRNAs regulate major pyrogens (41).
RBM3 levels decreased with increased miR-142–5p and miR-143 levels and vice versa in primary human PBMCs
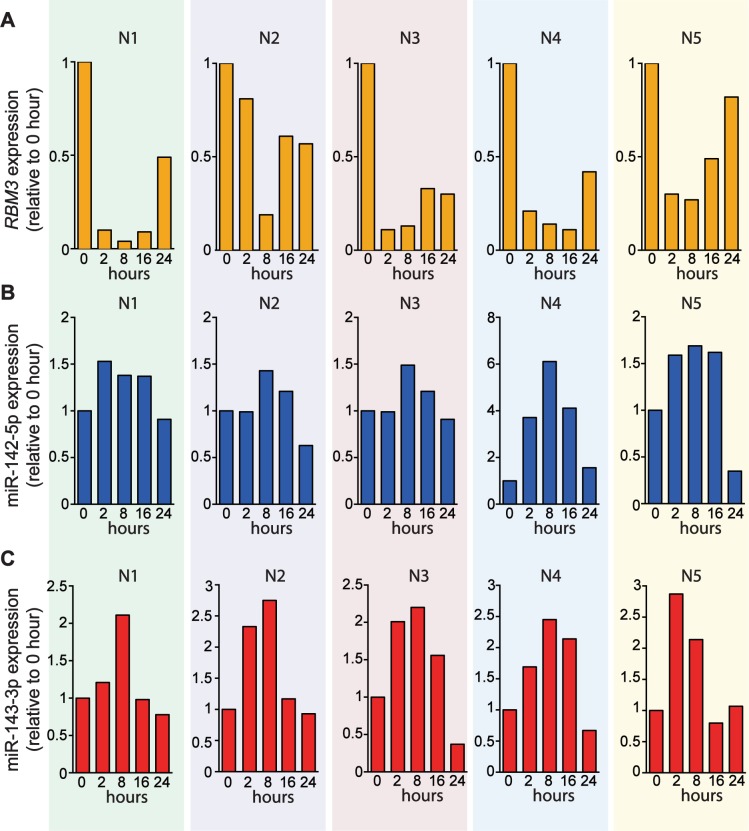
In order to confirm the association between RBM3, miR-142–5p and miR-143 in primary cells, we measured their expression in PBMCs from healthy individuals (n = 5) subjected to 40°C over a 24 h time course. Exogenous exposure of PBMCs to 40°C resulted in decreasing RBM3 expression starting from 2 up to 8 h before the levels increased at 16–24 h (Figure 5A). In contrast, the levels of miR-142–5p and miR-143 increased and predominantly peaked at 8 h before dropping again at 16–24 h (Figure 5B and C). This trend of increasing RBM3 expression in conjunction with decreasing miR-142–5p and 143 levels, and vice versa, supports a feedback mechanism involving RBM3 and these miRNAs in response to increased temperature.
Figure 5.
Temporal changes of RBM3, miR-142–5p and miR-143 expression in PBMCs exposed to 40°C over a 24 h time course. (A) Expression of RBM3 mRNA in PBMCs by RT-qPCR (n = 5). (B) Expression of miR-142–5p and (C) miR-143 in PBMCs by Taqman miRNA assays (n = 5). Fold change was measured relative to 0 h. N1–N5 denote five healthy individuals recruited for this experiment.
DISCUSSION
During infection, lizards will move to a warmer environment. Preventing them from doing so can result in a mortality rate of up to 70% (42). This simple experiment demonstrates that fever is a crucial part of the response to infection. Indeed, fever is an adaptive mechanism that evolved over 200 million years ago. All vertebrates elevate their body temperature in response to infection and fever confers a survival advantage to all vertebrates studied. Curing paralysis caused by syphilis by inducing fever from malaria infection was the subject of the 1927 Nobel prize in medicine. Despite a growing body of evidence pointing towards a beneficial effect of fever on patient outcome, antipyretic drugs are still commonly used to treat fever at home and in the intensive care unit (ICU) and no study has investigated the molecular consequences of fever at a cellular level.
Our findings define a novel role for RBM3 as a gauge and potential mediator of pyrexia with crucial downstream functions effected via miRNAs. miRNAs have previously been implicated in diverse homeostatic and metabolic pathways (43) and more recently have been used to predict outcome in critically ill patients and sepsis (44). In our model, temperature–sensitive miR-142–5p and miR-143 were shown to be key regulators of pyrexia. When RBM3 levels decrease, the expression of these ‘thermomiRs’ increases dramatically. In turn these ‘thermomiRs’ target the pyrogens that cause the increase in body temperature. We present here a negative feedback loop to regulate body temperature that involves reduced expression of RBM3 and increased expression of ‘thermomiRs’ to fine-tune levels of pyrogens (Figure 6). Our microarray data demonstrate lack of changes in pyrogen expression in THP-1-differentiated macrophages following 40°C exposure (Supplementary Table S1). In contrast, knockdown of miR-142–5p or 143 in the absence of elevated temperature exposure resulted in a significant increase in the expression of two pyrogens (Figure 4D). These results support our model that miR142 and miR-143 counteract the natural increase of pyrogens following increased temperature.
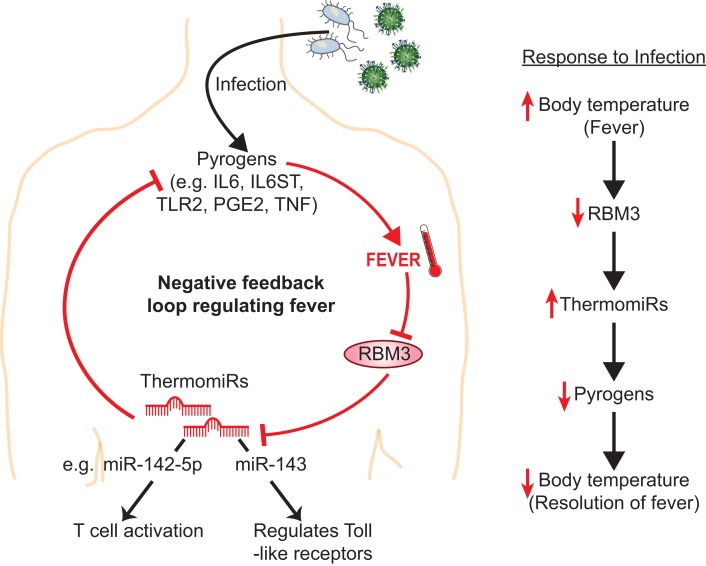
Figure 6.
Proposed negative feedback mechanism regulating body temperature when fever occurs. Triggering an immune response (e.g. via infection), increases the expression of endogenous pyrogens, leading to fever. Fever will result in decreased expression of RBM3, which in turn leads to increased expression of thermomiRs normally targeted by RBM3, thereby fine-tuning expression of endogenous pyrogens. The thermomiRs themselves play integral roles in coordinating the response to fever and infection. This negative feedback loop attenuates excessive increases in body temperature.
Consistent with this model, we demonstrate in primary PBMCs exposed to fever-like temperature that RBM3 levels anti-correlate with miR-142–5p and miR-143 levels, potentially in order to maintain homeostasis during unfavourable temperature conditions. This mechanism may be crucial to prevent an excessive increase in body temperature and related molecular response leading to pathological hyperthermia.
Our data suggest that most temperature-sensitive miRNAs are not regulated by RBM3 (Figure 4C). These miRNAs may respond directly to temperature changes or may be regulated by other heat responsive protein(s). These proteins may include the cold inducible RNA binding protein, CIRBP, that was also downregulated following exposure of THP-1-differentiated macrophages to 40°C (Supplementary Table S1). Future work is required to determine whether downregulation of this protein regulates the expression of one or more thermomiRs. Notably, CIRBP and RBM3 are regulated by body temperature changes associated with the circadian rhythm (45). It would be interesting to determine if miRNA expression that changes during this process (46) is also regulated by CIRBP and RBM3.
Given the importance of the targets of thermomiRs that respond to changes in temperature, this study provides novel insight into the mechanisms that mediate fever. From the use of over the counter antipyretics to targeted temperature management used in the ICU for cardiac arrest or elevated intracerebral pressure (47), medical control of body temperature is common and can be critical to a patient's outcome. Here we demonstrate that a small change in temperature between 37°C and 40°C has a major effect on miRNAs and on their target gene expression. Given that these miRNAs and their targets are crucial to the control of pyrexia and to the immune response in general, it is essential to understand how these molecular changes mediate different types of insult. Our results are directly relevant to the clinical context given recent evidence that increased levels of IL-6, IL6ST (48), TLR2 (49), PGE2 (50) and TNF (51), each of which is regulated by miR-142–5p or miR-143, are associated with a poor outcome in patients with severe sepsis.
Supplementary Material
Acknowledgments
The authors thank Steven Allen, Suat Dervish and Frank Kao for fluorescence-activated cell sorting and Fiona Guan, Kinsha Baidya, Rajini Nagarajah, Patrick O'Young and Amy Marshall for technical advice and assistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Health and Medical Research Council [1061906 to J.E.J.R., W.R. and 1080530 to J.E.J.R., W.R., J.J-L.W.]; Tour de Cure [J.E.J.R., C.G.B.]; Cure the Future [J.E.J.R.]; an anonymous foundation [J.E.J.R.]. J.J-L.W. and W.R. hold fellowships from the Cancer Institute of NSW. J.H. is a National Breast Cancer Foundation Fellow. Funding for open access charge: Bioinformatics fund, Centenary Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Leon L.R. Invited review: cytokine regulation of fever: studies using gene knockout mice. J. Appl. Physiol. 2002;92:2648–2655. doi: 10.1152/japplphysiol.01005.2001. [DOI] [PubMed] [Google Scholar]
- 2.Ogoina D. Fever, fever patterns and diseases called ‘fever’–a review. J. Infect. Public Health. 2011;4:108–124. doi: 10.1016/j.jiph.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 3.Greer D.M., Funk S.E., Reaven N.L., Ouzounelli M., Uman G.C. Impact of fever on outcome in patients with stroke and neurologic injury: a comprehensive meta-analysis. Stroke. 2008;39:3029–3035. doi: 10.1161/STROKEAHA.108.521583. [DOI] [PubMed] [Google Scholar]
- 4.Young P., Saxena M., Beasley R., Bellomo R., Bailey M., Pilcher D., Finfer S., Harrison D., Myburgh J., Rowan K. Early peak temperature and mortality in critically ill patients with or without infection. Intensive Care Med. 2012;38:437–444. doi: 10.1007/s00134-012-2478-3. [DOI] [PubMed] [Google Scholar]
- 5.Wilczynska A., Bushell M. The complexity of miRNA-mediated repression. Cell Death Differ. 2015;22:22–33. doi: 10.1038/cdd.2014.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai E.C. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 7.Vidigal J.A., Ventura A. The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol. 2015;25:137–147. doi: 10.1016/j.tcb.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y., Kowdley K.V. MicroRNAs in common human diseases. Genomics Proteomics Bioinformatics. 2012;10:246–253. doi: 10.1016/j.gpb.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma Y., Vilanova D., Atalar K., Delfour O., Edgeworth J., Ostermann M., Hernandez-Fuentes M., Razafimahatratra S., Michot B., Persing D.H., et al. Genome-wide sequencing of cellular microRNAs identifies a combinatorial expression signature diagnostic of sepsis. PLoS One. 2013;8:e75918. doi: 10.1371/journal.pone.0075918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maslove D.M., Wong H.R. Gene expression profiling in sepsis: timing, tissue, and translational considerations. Trends Mol. Med. 2014;20:204–213. doi: 10.1016/j.molmed.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schnoor M., Buers I., Sietmann A., Brodde M.F., Hofnagel O., Robenek H., Lorkowski S. Efficient non-viral transfection of THP-1 cells. J. Immunol. Methods. 2009;344:109–115. doi: 10.1016/j.jim.2009.03.014. [DOI] [PubMed] [Google Scholar]
- 12.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ritchie W., Gao D., Rasko J.E. Defining and providing robust controls for microRNA prediction. Bioinformatics. 2012;28:1058–1061. doi: 10.1093/bioinformatics/bts114. [DOI] [PubMed] [Google Scholar]
- 14.Vermeulen A., Robertson B., Dalby A.B., Marshall W.S., Karpilow J., Leake D., Khvorova A., Baskerville S. Double-stranded regions are essential design components of potent inhibitors of RISC function. RNA. 2007;13:723–730. doi: 10.1261/rna.448107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dudoit S., Yang Y.H., Callow M.J., Speed T.P. Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Stat. Sinica. 2002;12:111–139. [Google Scholar]
- 16.Wong J., Ritchie W., Gao D., Lau K., Gonzalez M., Choudhary A., Taft R., Rasko J., Holst J. Identification of nuclear-enriched miRNAs during mouse granulopoiesis. J. Hematol. Oncol. 2014;7:42. doi: 10.1186/1756-8722-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 18.Hu X., Yu J., Crosby S.D., Storch G.A. Gene expression profiles in febrile children with defined viral and bacterial infection. Proc. Natl. Acad. Sci. U. S. A. 2013;110:12792–12797. doi: 10.1073/pnas.1302968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mancardi D.A., Albanesi M., Jönsson F., Iannascoli B., Van Rooijen N., Kang X., England P., Daëron M., Bruhns P. The high-affinity human IgG receptor FcγRI (CD64) promotes IgG-mediated inflammation, anaphylaxis, and antitumor immunotherapy. Blood. 2013;121:1563–1573. doi: 10.1182/blood-2012-07-442541. [DOI] [PubMed] [Google Scholar]
- 20.Neote K., DiGregorio D., Mak J.Y., Horuk R., Schall T.J. Molecular cloning, functional expression, and signaling characteristics of a C-C chemokine receptor. Cell. 1993;72:415–425. doi: 10.1016/0092-8674(93)90118-a. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi F., Smith K.D., Ozinsky A., Hawn T.R., Yi E.C., Goodlett D.R., Eng J.K., Akira S., Underhill D.M., Aderem A. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410:1099–1103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 22.Takeuchi O., Kawai T., Sanjo H., Copeland N.G., Gilbert D.J., Jenkins N.A., Takeda K., Akira S. TLR6: A novel member of an expanding Toll-like receptor family. Gene. 1999;231:59–65. doi: 10.1016/s0378-1119(99)00098-0. [DOI] [PubMed] [Google Scholar]
- 23.Moore P.A., Belvedere O., Orr A., Pieri K., LaFleur D.W., Feng P., Soppet D., Charters M., Gentz R., Parmelee D., et al. BLyS: Member of the tumor necrosis factor family and B lymphocyte stimulator. Science. 1999;285:260–263. doi: 10.1126/science.285.5425.260. [DOI] [PubMed] [Google Scholar]
- 24.Dinarello C.A. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood. 2011;117:3720–3732. doi: 10.1182/blood-2010-07-273417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yost H.J., Lindquist S. RNA splicing is interrupted by heat shock and is rescued by heat shock protein synthesis. Cell. 1986;45:185–193. doi: 10.1016/0092-8674(86)90382-x. [DOI] [PubMed] [Google Scholar]
- 26.Bell J., Neilson L., Pellegrini M. Effect of heat shock on ribosome synthesis in Drosophila melanogaster. Mol. Cell Biol. 1988;8:91–95. doi: 10.1128/mcb.8.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shalgi R., Hurt J.A., Lindquist S., Burge C.B. Widespread inhibition of posttranscriptional splicing shapes the cellular transcriptome following heat shock. Cell Rep. 2014;7:1362–1370. doi: 10.1016/j.celrep.2014.04.044. [DOI] [PubMed] [Google Scholar]
- 28.Dresios J., Aschrafi A., Owens G.C., Vanderklish P.W., Edelman G.M., Mauro V.P. Cold stress-induced protein Rbm3 binds 60S ribosomal subunits, alters microRNA levels, and enhances global protein synthesis. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1865–1870. doi: 10.1073/pnas.0409764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pilotte J., Dupont-Versteegden E.E., Vanderklish P.W. Widespread regulation of miRNA biogenesis at the Dicer step by the cold-inducible RNA-binding protein, RBM3. PLoS One. 2011;6:e28446. doi: 10.1371/journal.pone.0028446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chanput W., Mes J.J., Wichers H.J. THP-1 cell line: An in vitro cell model for immune modulation approach. Int. Immunopharmacol. 2014;23:37–45. doi: 10.1016/j.intimp.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 31.Roger T., Lugrin J., Le Roy D., Goy G., Mombelli M., Koessler T., Ding X.C., Chanson A.L., Reymond M.K., Miconnet I., et al. Histone deacetylase inhibitors impair innate immune responses to Toll-like receptor agonists and to infection. Blood. 2011;117:1205–1217. doi: 10.1182/blood-2010-05-284711. [DOI] [PubMed] [Google Scholar]
- 32.Kelada S., Sethupathy P., Okoye I.S., Kistasis E., Czieso S., White S.D., Chou D., Martens C., Ricklefs S.M., Virtaneva K., et al. miR-182 and miR-10a are key regulators of Treg specialisation and stability during Schistosome and Leishmania-associated inflammation. PLoS Pathog. 2013;9:e1003451. doi: 10.1371/journal.ppat.1003451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen T., Huang Z., Wang L., Wang Y., Wu F., Meng S., Wang C. MicroRNA-125a-5p partly regulates the inflammatory response, lipid uptake, and ORP9 expression in oxLDL-stimulated monocyte/macrophages. Cardiovasc. Res. 2009;83:131–139. doi: 10.1093/cvr/cvp121. [DOI] [PubMed] [Google Scholar]
- 34.Ceribelli A., Yao B., Dominguez-Gutierrez P.R., Nahid M.A., Satoh M., Chan E.K. MicroRNAs in systemic rheumatic diseases. Arthritis Res. Ther. 2011;13:229. doi: 10.1186/ar3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ding S., Liang Y., Zhao M., Liang G., Long H., Zhao S., Wang Y., Yin H., Zhang P., Zhang Q., et al. Decreased microRNA-142–3p/5p expression causes CD4+ T cell activation and B cell hyperstimulation in systemic lupus erythematosus. Arthritis Rheum. 2012;64:2953–2963. doi: 10.1002/art.34505. [DOI] [PubMed] [Google Scholar]
- 36.Guo H., Chen Y., Hu X., Qian G., Ge S., Zhang J. The regulation of Toll-like receptor 2 by miR-143 suppresses the invasion and migration of a subset of human colorectal carcinoma cells. Mol. Cancer. 2013;12:77. doi: 10.1186/1476-4598-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ehlen A., Brennan D., Nodin B., O'Connor D., Eberhard J., Alvarado-Kristensson M., Jeffrey I., Manjer J., Brandstedt J., Uhlen M., et al. Expression of the RNA-binding protein RBM3 is associated with a favourable prognosis and cisplatin sensitivity in epithelial ovarian cancer. J. Transl. Med. 2010;8:78. doi: 10.1186/1479-5876-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pham H., Rodriguez C.E., Donald G.W., Hertzer K.M., Jung X.S., Chang H.H., Moro A., Reber H.A., Hines O.J., Eibl G. miR-143 decreases COX-2 mRNA stability and expression in pancreatic cancer cells. Biochem. Biophys. Res. Commun. 2013;439:6–11. doi: 10.1016/j.bbrc.2013.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Y., Varambally S., Maher C.A., Cao Q., Chockley P., Toubai T., Malter C., Nieves E., Tawara I., Wang Y., et al. Targeting of microRNA-142–3p in dendritic cells regulates endotoxin-induced mortality. Blood. 2011;117:6172–6183. doi: 10.1182/blood-2010-12-325647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma S., Liu J., Wei J., Yuan H., Zhang T., Bishopric N.H. Repression of miR-142 by p300 and MAPK is required for survival signalling via gp130 during adaptive hypertrophy. EMBO Mol. Med. 2012;4:617–632. doi: 10.1002/emmm.201200234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dinarello C.A. Infection, fever, and exogenous and endogenous pyrogens: some concepts have changed. J. Endotoxin Res. 2004;10:201–222. doi: 10.1179/096805104225006129. [DOI] [PubMed] [Google Scholar]
- 42.Bernheim H.A., Kluger M.J. Fever and antipyresis in the lizard Dipsosaurus dorsalis. Am. J. Physiol. 1976;231:198–203. doi: 10.1152/ajplegacy.1976.231.1.198. [DOI] [PubMed] [Google Scholar]
- 43.Rottiers V., Naar A.M. MicroRNAs in metabolism and metabolic disorders. Nat. Rev. Mol. Cell Biol. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roderburg C., Luedde M., Vargas Cardenas D., Vucur M., Scholten D., Frey N., Koch A., Trautwein C., Tacke F., Luedde T. Circulating microRNA-150 serum levels predict survival in patients with critical illness and sepsis. PLoS One. 2013;8:e54612. doi: 10.1371/journal.pone.0054612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y., Hu W., Murakawa Y., Yin J., Wang G., Landthaler M., Yan J. Cold-induced RNA-binding proteins regulate circadian gene expression by controlling alternative polyadenylation. Sci. Rep. 2013;3:2054. doi: 10.1038/srep02054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Na Y.-J., Sung J.H., Lee S.C., Lee Y.-J., Choi Y.J., Park W.-Y., Shin H.S., Kim J.H. Comprehensive analysis of microRNA-mRNA co-expression in circadian rhythm. Exp. Mol. Med. 2009;41:638–647. doi: 10.3858/emm.2009.41.9.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Corry J.J. Use of hypothermia in the intensive care unit. World J. Crit. Care Med. 2012;1:106–122. doi: 10.5492/wjccm.v1.i4.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gouel-Cheron A., Allaouchiche B., Guignant C., Davin F., Floccard B., Monneret G., AzuRea G. Early interleukin-6 and slope of monocyte human leukocyte antigen-DR: a powerful association to predict the development of sepsis after major trauma. PLoS One. 2012;7:e33095. doi: 10.1371/journal.pone.0033095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rybka J., Butrym A., Wrobel T., Jazwiec B., Stefanko E., Dobrzynska O., Poreba R., Kuliczkowski K. The expression of Toll-like receptors and development of severe sepsis in patients with acute myeloid leukemias after induction chemotherapy. Med. Oncol. 2014;31:319. doi: 10.1007/s12032-014-0319-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brogliato A.R., Antunes C.A., Carvalho R.S., Monteiro A.P., Tinoco R.F., Bozza M.T., Canetti C., Peters-Golden M., Kunkel S.L., Vianna-Jorge R., et al. Ketoprofen impairs immunosuppression induced by severe sepsis and reveals an important role for prostaglandin E2. Shock. 2012;38:620–629. doi: 10.1097/SHK.0b013e318272ff8a. [DOI] [PubMed] [Google Scholar]
- 51.Lv S., Han M., Yi R., Kwon S., Dai C., Wang R. Anti-TNF-alpha therapy for patients with sepsis: a systematic meta-analysis. Int. J. Clin. Pract. 2014;68:520–528. doi: 10.1111/ijcp.12382. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.