Abstract
The RNase P-mediated endonucleolytic cleavage plays a crucial role in the 3′ end processing and cellular accumulation of MALAT1, a nuclear-retained long noncoding RNA that promotes malignancy. The regulation of this cleavage event is largely undetermined. Here we characterize a broadly expressed natural antisense transcript at the MALAT1 locus, designated as TALAM1, that positively regulates MALAT1 levels by promoting the 3′ end cleavage and maturation of MALAT1 RNA. TALAM1 RNA preferentially localizes at the site of transcription, and also interacts with MALAT1 RNA. Depletion of TALAM1 leads to defects in the 3′ end cleavage reaction and compromises cellular accumulation of MALAT1. Conversely, overexpression of TALAM1 facilitates the cleavage reaction in trans. Interestingly, TALAM1 is also positively regulated by MALAT1 at the level of both transcription and RNA stability. Together, our data demonstrate a novel feed-forward positive regulatory loop that is established to maintain the high cellular levels of MALAT1, and also unravel the existence of sense-antisense mediated regulatory mechanism for cellular lncRNAs that display RNase P-mediated 3′ end processing.
INTRODUCTION
Long noncoding RNAs (lncRNAs) function in every step of gene regulation by modulating the activity of several protein-coding genes (1,2). Most lncRNAs are transcribed by RNA polymerase II (RNA Pol II), and they share similar features with mRNAs, including having a 5′ cap structure and 3′ poly(A) tails (1). The 3′ end processing of RNA is critical for the termination of RNA Pol II and also determines the stability, subcellular localization and eventually the functionality of the mature RNA (3,4). Recent studies have reported the existence of non-canonical 3′ end processing mechanisms at several lncRNA loci that generate non-polyadenylated RNAs (5–7). Among them, metastasis-associated lung adenocarcinoma transcript 1 (MALAT1; also known as NEAT2) and multiple endocrine neoplasia-β (MENβ; also known as NEAT1) RNAs are unique as they are highly abundant linear transcripts. For example, MALAT1 exists in ≈2500–3000 copies/cell (8), with a half-life up to 16.5 h (9). In contrast, other non-polyadenylated lncRNAs like enhancer RNAs and circular RNAs are present in low abundance and exhibit less stability (5).
The strikingly high cellular accumulation of MALAT1 and MENβ has been attributed to an unusual processing and maturation event at their ultra-conserved 3′ ends (10–13): the primary transcripts of both MALAT1 and MENβ harbor a tRNA-like structure at their 3′ end, which are recognized and cleaved by tRNA processing enzymes RNase P and RNase Z. This process generates a cytoplasmic tRNA-like small RNA, known as mascRNA for MALAT1 and menRNA for MENβ, along with mature nuclear retained MALAT1 and MENβ lncRNAs. The mature MALAT1 and MENβ lncRNAs lack canonical poly(A) tail, but form bipartite triple helical structures at their 3′ ends, which confer resistance against exonuclease-mediated RNA degradation (12,13). This triple helix is formed through engaging two upstream U-rich motifs and a 3′ terminal genome-encoded A-rich tract (13). Based on the sequence, structure and function, the two highly conserved U-rich motifs are proposed to be the cellular homolog of the Expression and Nuclear retention Element (ENE) discovered in viral lncRNAs. The viral ENE is known to protect viral lncRNAs from rapid nuclear deadenylation-dependent decay pathway, through clamping the poly(A) tail of viral lncRNA into a triple helical structure similar to that of MALAT1 and MENβ (10–14). Slightly different from the ENE-containing viral lncRNAs, the stable formation of the triple helix in MALAT1 and MENβ requires the mascRNA/menRNA cleavage reaction to generate a blunt end in the triple helix (12,13,15). The fully processed mature MALAT1 is localized in nuclear speckles, where it plays crucial roles in regulating the organization and activities of several pre-mRNA splicing factors and transcription factors (8,16–19). In accordance with its pro-proliferative molecular functions, upregulation of MALAT1 has been identified as a hallmark of tumor progression and metastasis in multiple cancers (9).
Although the post-transcriptional processing of MALAT1 3′ end has been recognized to be crucial for the enhanced stability of MALAT1, how this processing event is regulated is not well understood. In the present study, we identified a natural antisense transcript (NAT) at the MALAT1 locus, which we named as TALAM1. TALAM1 contributes to the stability of MALAT1 by promoting the 3′ end cleavage and maturation of MALAT1. Meanwhile, MALAT1 RNA positively regulates the transcription and stability of TALAM1, and thereby establishes a feed-forward positive regulatory loop at the MALAT1 locus to achieve high cellular levels of MALAT1.
MATERIALS AND METHODS
Strand-specific RT-qPCR and Taqman small RNA assay
For strand-specific RT-qPCR, reverse transcription was performed using gene specific reverse transcription primers with a linker sequence at 5′ end, and then qPCR was performed using gene specific forward primer and the linker as reverse primer. Sequences of primers are listed in supplementary methods. qPCR analysis of human mascRNA was performed using Taqman small RNA assay (Life technologies, Assay ID: CSCSU4N), with U6 snRNA as internal loading control (Assay ID: 001973).
RNA/DNA FISH
A set of custom Stellaris® FISH probes comprising 17 single-labeled oligonucleotides designed for MALAT1 were purchased from Biosearch Technologies. TALAM1 probe was generated via nick-translation cDNA kit (Abbott Molecular, USA), using a pGEM-Teasy construct containing the 5′ 1 kb unique region of TALAM1 as template. For RNA/DNA FISH, DNA FISH probe was generated via nick translation from BAC clone RP11–1104L6. For dual RNA FISH, the fixation and hybridization of HeLa cells were performed as described previously (20). For RNA/DNA FISH, the RNA FISH probe against TALAM1 was labeled with digoxigenin-11-dUTP using DIG nick translation mix (Roche), and fixation and hybridization of HeLa cells were performed as described previously (21).
Antisense oligonucleotide, 2′ MOE
Phosphorothioate internucleosidic linkage-modified DNA antisense oligonucleotides were used to deplete human MALAT1 and TALAM1. Uniform 2′-O-methoxy ethyl substituted oligonucleotides with a full phosphorothioate backbone and AGTmC were used to inhibit the mascRNA cleavage. These reagents were designed and synthesized by Ionis Pharmaceuticals, and their sequences are listed in supplementary methods.
RNA cutoff assay
To specifically detect the levels of uncleaved and cleaved MALAT1, RNA samples were subject to PolyG tailing, before being reverse transcribed with Oligo dC primers tagged with an adaptor sequence. Poly G tailing was performed using yeast poly(A) polymerase (Affymetrix) and rGTP. After reverse transcription, qPCR was performed using the adaptor as common reverse primer, and gene-specific forward primers located upstream and downstream of mascRNA site to detect cleaved and uncleaved MALAT1, respectively. For internal control of RNA loading and reaction efficiency, a forward primer located at the 3′ end of GAPDH mRNA was used. Sequences of primers are listed in supplementary methods.
RESULTS
Identification of TALAM1, an NAT at the MALAT1 locus
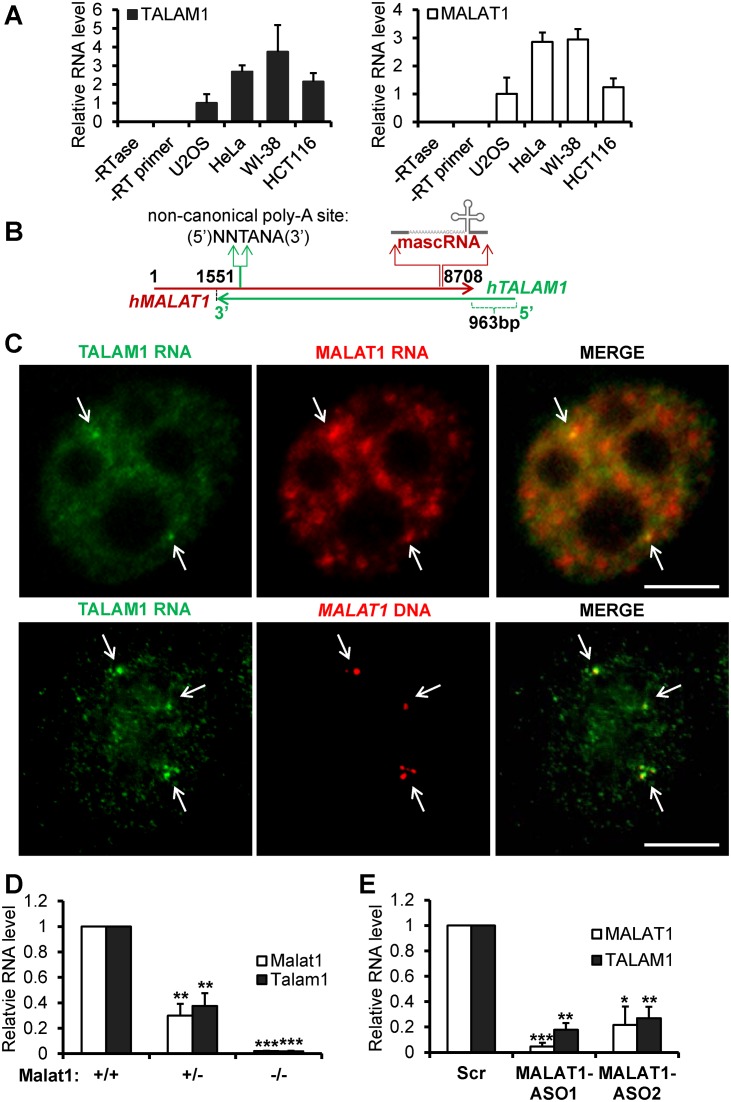
An NAT of ≈6 kb long at the mouse Malat1 locus was initially reported in mouse embryonic stem cells (22). To verify the existence of this NAT in other species and cell types, we examined the presence and relative abundance of this NAT in various human cell types, including human diploid fibroblasts (WI-38) and multiple cancer cell lines. The conventional RT-qPCR strategy, utilizing gene specific RT primer and PCR primer pairs (without linkers; Supplementary Figure S1Aa), does not always provide strand specificity, and detects both sense and antisense transcripts (Supplementary Figure S1Ab and see legends for details; 23,24). To specifically detect and quantify the NAT and MALAT1 sense transcript, we applied a strand-specific reverse transcription and real time PCR strategy (ssRT-qPCR) that was previously used to strand-specifically detect viral transcripts(Supplementary Figure S1B and see also Materials and Methods; 24). Specific detection of NAT using ssRT-qPCR in various cell types indicated that NAT from MALAT1 locus is widely expressed in several human cell lines (Figure 1A). We named this NAT as TALAM1, since it is transcribed from the complementary strand of MALAT1 gene. RNA expression analyses in different cell types revealed a positive correlation in the levels of TALAM1 and MALAT1, indicating potential co-regulation of these transcripts (Figure 1A). Further, copy number analysis in HeLa cells revealed TALAM1 is ≈290-fold less abundant than the highly abundant MALAT1 RNA (Supplementary Figure S2).
Figure 1.
TALAM1 is a NAT at the MALAT1 locus. (A) Expression analysis of TALAM1 (left panel) and MALAT1 (right panel) in various human cell types via ssRT-qPCR. The levels of TALAM1 and MALAT1 in different cell types were individually normalized against the levels in U2OS. (B) Schematic diagrams of TALAM1 and MALAT1 gene positions in human genome. (C) Upper panel: dual RNA-FISH in HeLa cells using probes against the 5′ unique region of TALAM1 (green) and single molecule FISH probes against MALAT1 (red); Bottom panel: TALAM1 RNA-FISH (green), with DNA-FISH against MALAT1 (red) genomic locus (Scale bar, 5 μm). In HeLa cells DNA FISH targeting MALAT1 genomic loci generally showed 2–4 foci per cell, which is due to the presence of multiple copies of chromosome 11, harboring MALAT1 locus in these cells. (D) ssRT-qPCR analyses to quantify mouse Malat1 and Talam1 levels in Malat1 Wild type (+/+), heterozygous (+/−) and homozygous (-/-) knockout primary mouse embryonic fibroblasts. (E) ssRT-qPCR analyses of human MALAT1 and TALAM1 levels in HeLa cells treated with Control-Scr, MALAT1-ASO1, and MALAT1-ASO2. Bar data are mean values with SD, N = 3 (biological replicates, *P < 0.05, **P < 0.01 and ***P < 0.001, two-tailed paired Student t-test).
We next performed rapid amplification of cDNA ends (RACE) and directional sequencing to determine the 5′ and 3′ ends of TALAM1. RACE analyses identified human TALAM1 as a ≈8.1 kb long transcript (Figure 1B, Supplementary Figure S3). This includes a 7.1 kb region that completely overlaps with MALAT1 sense transcript, and a 5′ end ≈1 kb unique region that extends beyond the 3′ end of MALAT1 gene (Figure 1B). Northern blot analysis using a probe that mostly spans the unique 5′ end of TALAM1 detected specific TALAM1 signal at the expected 8 kb position (Supplementary Figure S4). Analysis of the 5′ end sequence of TALAM1 in UCSC (University of Santa-Cruz) genome browser revealed signatures of an active promoter, including chromatin immunoprecipitation (ChIP) signals of RNA Pol II, numerous transcription factors, as well as histone modifications associated with active transcription (H3K4me3, H3K4me1, H3K27Ac; Supplementary Figure S5A). RNA fractionation using Oligo-dT columns, together with the 3′ RACE and northern blot data, confirmed TALAM1 as a polyadenylated transcript (Supplementary Figure S5B). These results together indicate TALAM1 as a novel NAT from the MALAT1 locus, which is subject to active RNA Pol II transcriptional regulation.
To determine the cellular localization of TALAM1, RNA-fluorescent in situ hybridization (RNA-FISH) was performed in HeLa cells (Figure 1C, upper panel). In contrast to MALAT1, which is primarily enriched in nuclear speckles, TALAM1 displayed weak homogenous nuclear distribution, with preferential enrichment at 2–4 nuclear foci where it co-localizes with MALAT1 RNA (see arrows in Figure 1C, upper panel). These foci were further demonstrated to be the transcription sites of the MALAT1 gene loci, as they completely co-localized with the DNA FISH signals targeting the MALAT1 genomic DNA loci (see arrows in Figure 1C, bottom panel).
To address the potential co-regulation between MALAT1 and TALAM1, we first examined the level of TALAM1 in wild type (WT) and Malat1 knockout (KO) mouse embryonic fibroblast (MEF) cells. This Malat1 KO mouse was generated by inserting a LacZ-polyadenylation signal cassette at 69bp downstream of the Malat1 promoter, thereby disrupting the transcription from the Malat1 promoter, without changing any sequences of the Malat1 gene locus, including the region corresponding to Talam1 gene (25). TALAM1 levels were consistently low in both heterozygous and homozygous Malat1 null MEFs (Figure 1D), suggesting that the TALAM1 levels are dependent on either the transcription of MALAT1 or the presence of MALAT1 transcripts. To distinguish between these two scenarios, we utilized MALAT1-specific antisense oligos (ASO) to deplete MALAT1 transcripts, without affecting the transcription of MALAT1 (8,26). In MALAT1-depleted human cells, TALAM1 level was found to decrease dramatically (Figure 1E), indicating that the maintenance of TALAM1 levels in cells requires the presence of MALAT1 transcripts.
MALAT1 RNA positively regulates the transcription and stability of TALAM1 RNA
The cellular pool of RNAs could be regulated at the level of both transcription as well as post-transcriptional processing and stability. We first examined whether MALAT1 positively regulates TALAM1 through promoting its transcription. RNA Pol II occupancy at the TALAM1 promoter showed ≈50% reduction upon MALAT1 depletion, indicating that active transcription of TALAM1 requires the presence of MALAT1 RNA (Figure 2A,B). To assess whether the stability of TALAM1 RNA is also dependent on MALAT1, we determined TALAM1 RNA stability in control versus MALAT1-depleted cells, via Actinomycin D chase experiment. Actinomycin D is an RNA Pol II inhibitor that blocks nascent RNA synthesis. In control cells, TALAM1 was a relatively stable RNA, with a half-life of ≈8 h. However, in cells depleted of MALAT1, the half-life of TALAM1 reduced dramatically to ≈2–3 h (Figure 2C and Supplementary Figure S6). To identify whether this regulation of TALAM1 by MALAT1 acts in cis or in trans, we utilized a MALAT1 construct with point mutations conferring resistance to ASO knockdown, and observed that the exogenously expressed, ASO-resistant MALAT1 transcripts could rescue in trans the levels of endogenous TALAM1 in cells that were depleted of endogenous MALAT1 (Supplementary Figure S7A). To further analyze the requirement of sequence complementarity between MALAT1 and TALAM1 in conferring the stability, we overexpressed human MALAT1 in Malat1 KO transformed MEFs and assessed whether exogenously expressed human MALAT1 could rescue the levels of endogenous mouse Talam1. Our results revealed that the exogenously expressed human MALAT1 transcripts could not rescue the endogenous mouse Talam1 (Supplementary Figure S7B), suggesting that the rescue may require higher level of sequence complementarity between these two transcripts at certain crucial sequence motifs. Collectively, our data demonstrate that MALAT1 RNA positively regulates the transcription as well as the stability of TALAM1 RNA.
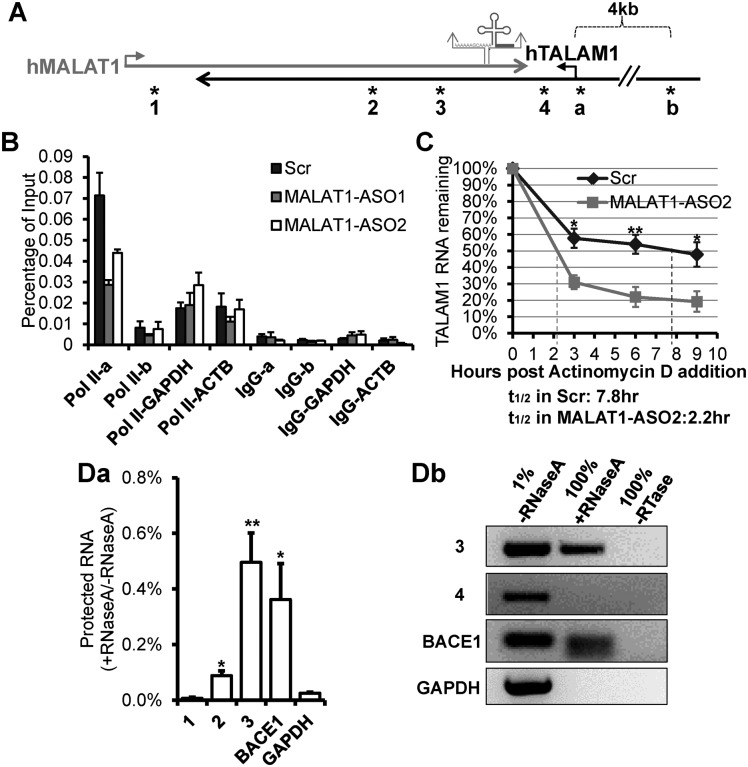
Figure 2.
MALAT1 positively regulates the transcription and stability of TALAM1, and forms RNA duplex with TALAM1 in vivo. (A) Schematic diagrams depicting MALAT1 genomic locus. Relative positions of the primers used in ChIP experiment in (B), are indicated by asterisks labeled as ‘a’ and ‘b’: primer set ‘a’ is at TALAM1 promoter, ‘b’ is at upstream 4 kb of TALAM1 TSS. Relative positions of the primers analyzed in RNase protection assay in (D), are indicated by asterisks labeled as 1 to 4. (B) ChIP-qPCR of RNA Pol II at TALAM1 promoter region, in HeLa cells treated with Control-Scr, MALAT1-ASO1 and ASO2. Primers from 4 kb upstream of TALAM1 TSS, GAPDH and ACTB promoters were used as negative controls. (C) TALAM1 stability in HeLa cells treated with Scr or MALAT1-ASO2. Dotted lines represent the half-life of TALAM1. (D) In vivo RNase protection assay. HeLa cells were lysed in absence or presence of RNaseA, then RNA was extracted and analyzed via (Da) RT-qPCR and (Db) RT-PCR, using primers illustrated in (A). Data in (B) and (C) are mean values with SEM, NB = 3 (3 technical replicates of representative biological replicates), NC = 4; Data in (Da) is mean value with SD, N = 3 (biological replicates, *P < 0.05, **P < 0.01, two-tailed paired Student t-test).
The positive regulation of NATs by their sense transcripts could be achieved via the formation of RNA duplex, which alters the secondary or tertiary structure of RNA, thereby influencing RNA stability (27–30). To test the existence of RNA duplex formed between MALAT1 and TALAM1, an in vivo RNase protection assay was performed (31). While non-overlapping regions of MALAT1 and TALAM1 were almost completely degraded by RNase A (Figure 2A,Da,Db, primer sets 1 and 4), multiple sites in the overlapping regions were protected from degradation (Figure 2A,Da,Db, primer sets 2 and 3). The RNase protection ratio of 0.1–0.5% is consistent with the previous copy number analysis that TALAM1 is ≈290-fold less abundant than MALAT1 (Supplementary Figure S2). This means that for every 1 molecule of TALAM1, there will be 290 molecules of MALAT1, and therefore, 1 RNA duplex can be formed between TALAM1 and MALAT1, which can protect the RNA from RNaseA-mediated degradation. Thus, the protection ratio will be 1/(290+1) ≈ 0.34%. As a positive control, a similar protection was observed in case of BACE1 transcript, which is known to form RNA duplexes with its NAT in vivo (27; Figure 2D). Our results indicate that significant proportions of TALAM1 and MALAT1 indeed form RNA duplex in vivo, possibly occurring at the sites of their transcription, which in turn could positively influence the stability of both the transcripts.
TALAM1 is required for the efficient 3′ end processing and cellular accumulation of MALAT1 lncRNA
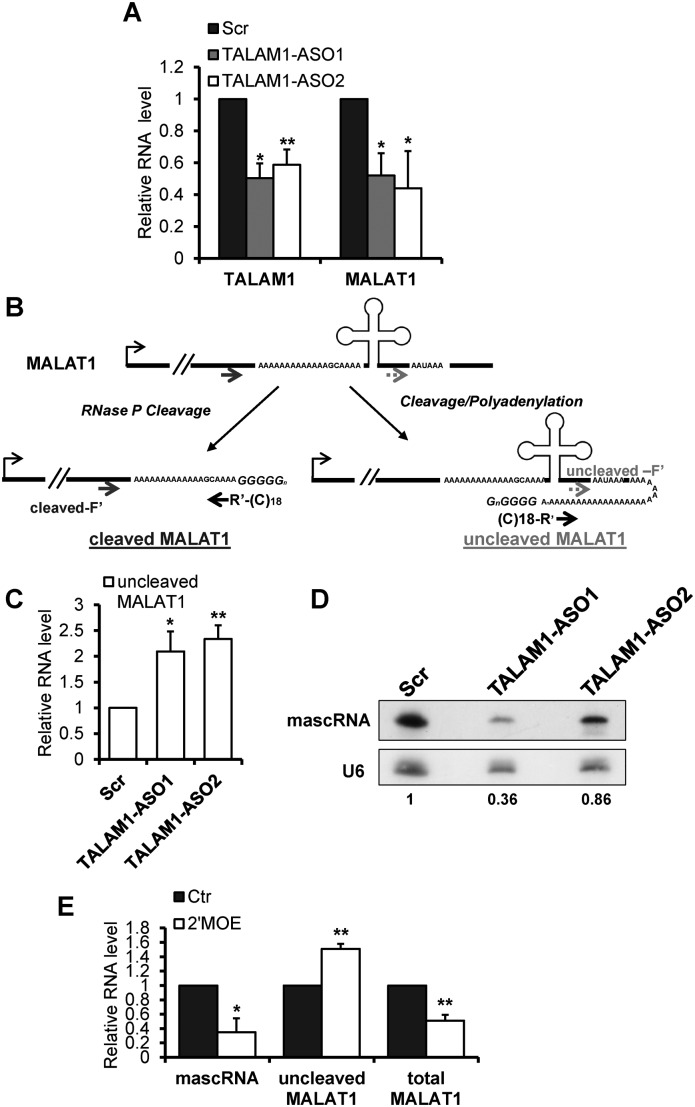
Next, to determine the cellular function of TALAM1, we assessed whether TALAM1 could also regulate MALAT1. First, to deplete TALAM1, we screened a set of strand-specific ASOs targeting the 5′ unique region of TALAM1. We consistently achieved ≈50% of TALAM1 depletion using two independent ASOs (Figure 3A). Remarkably, TALAM1-depleted cells showed a concomitant reduction in MALAT1 levels (Figure 3A) as determined by ssRT-PCR, suggesting TALAM1 RNA also positively influences the cellular levels of MALAT1.
Figure 3.
MALAT1 cellular accumulation and 3′ end processing are compromised in TALAM1-depleted cells. (A) ssRT-qPCR analyses of TALAM1 and MALAT1 levels in HeLa cells treated with Control-Scr, and two TALAM1-ASOs. (B) Schematic diagrams illustrating the RNA cutoff assay. Please also refer to Materials and Methods for details. R’ represents the adaptor sequence. Gene-specific forward primers upstream and downstream of the mascRNA cleavage site (cleaved-F’ and uncleaved-F’) are used to detect cleaved MALAT1 and uncleaved MALAT1, respectively. (C) Determination of uncleaved MALAT1 levels in Control-Scr and TALAM1-ASOs treated HeLa cells, using cutoff assay. (D) Northern blot analysis to detect mascRNA after TALAM1 knockdown. U6 snRNA was used as internal loading control. The reduction of mascRNA after TALAM1-ASO1 and ASO2 treatment was estimated by quantifying and normalizing the signal intensity of mascRNA to U6, via ImageJ software (http://rsb.info.nih.gov/ij/index.html). (E) qPCR analyses of HeLa samples treated with Control-2′MOE (Ctr) and MALAT1–2′MOE targeting human mascRNA site (2′MOE). Bar data are mean values with SD, NA = 4, NC = 4, NE = 3 (biological replicates, *P < 0.05, **P < 0.01, two-tailed paired Student t-test).
Based on recent studies, the cellular accumulation of MALAT1 depends on its efficient 3′ end cleavage and the formation of the 3′ end triple helix structure (12,13). Our observation that TALAM1 depletion results in a decrease in the total cellular pools of MALAT1, prompted us to explore whether TALAM1, with its ability to form duplex with MALAT1, could regulate the 3′ end processing of MALAT1. The cleaved mature form of MALAT1 was reported to be the dominant form, while the uncleaved precursor exists at a very low level (10). To distinguish these two forms of MALAT1 RNA, and to quantitatively detect the potential changes in their levels, we modified a RNA cutoff assay, a technique that was previously used to determine pre-microRNA processing in cells (32,33; Figure 3B and see also Materials and Methods). In cells where TALAM1 has been reduced to ≈50% (Figure 3A, TALAM1-ASO-treated sample), we observed an increase in the level of uncleaved MALAT1 (Figure 3C), together with a concomitant decrease in the level of mascRNA (Figure 3D), suggesting a compromised efficiency of MALAT1 3′ end processing event (see also Supplementary Figure S8). 2′ MOE-modified DNA ASOs with sequence complementary to mascRNA site and 2′-O-methoxy-ethyl ribose phosphorothioate backbone, which imparts high affinity for targeted RNA but does not activate RNase H-mediated target RNA cleavage, has been previously applied to inhibit MALAT1 3′ end cleavage in mouse cells (10). We utilized a similar approach, and treated human cells with 2′ MOE ASO complementary to the human mascRNA cleavage site. The human 2′MOE ASO successfully inhibited MALAT1 cleavage, as reflected by the decrease in mascRNA level, and a concomitant increase in the level of uncleaved precursor MALAT1 (Figure 3E), confirming the specificity of our RNA cutoff assay. In addition, similar to TALAM1-ASO-treated samples (Figure 3A), the total cellular level of MALAT1 also showed a reduction in the 2′MOE-treated sample (Figure 3E). These data support the current model that efficient MALAT1 3′ end processing is required for the high cellular accumulation of MALAT1 (12,13). And our results showing similar effects exerted by TALAM1-ASO and MALAT1–2′MOE in altering the ratio of uncleaved to cleaved MALAT1 RNA as well as total MALAT1 levels strongly indicate that TALAM1 promotes the cellular accumulation of MALAT1 through facilitating its 3′ end processing and maturation event.
TALAM1 promotes the 3′ end processing of MALAT1
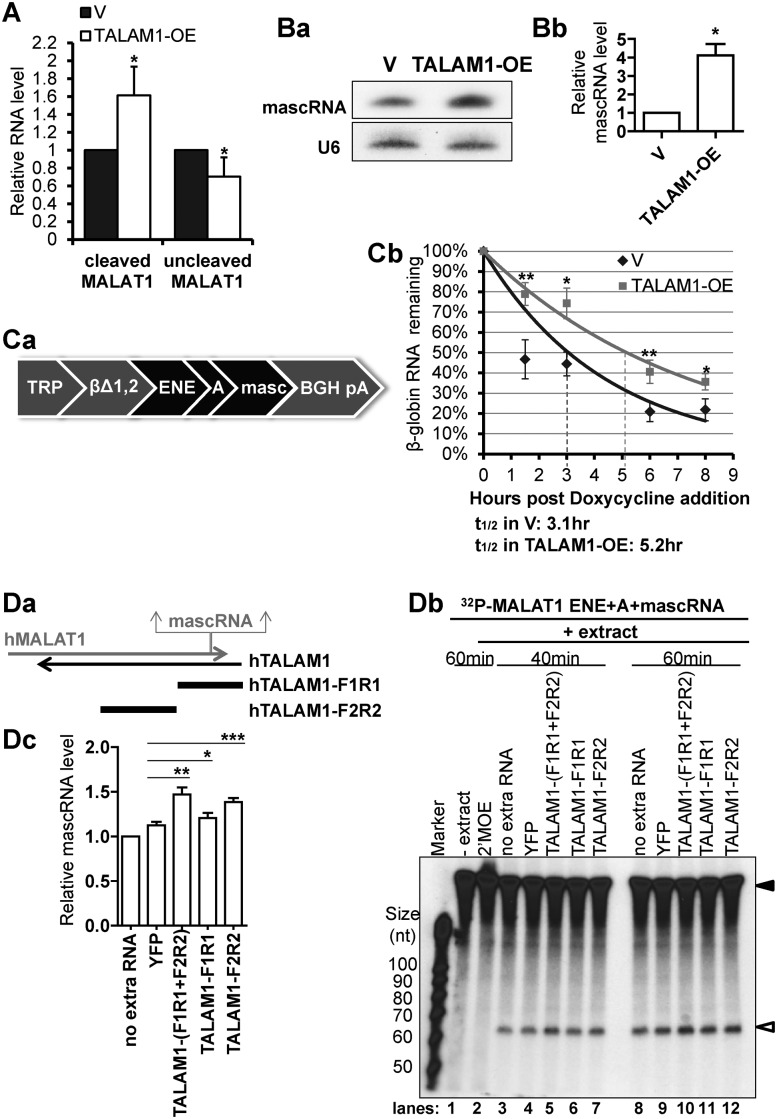
In order to gain insights into the involvement of TALAM1 in the 3′ end processing of MALAT1, we determined the effect of TALAM1 overexpression on endogenous MALAT1. Upon TALAM1 overexpression, even though the total cellular level of MALAT1 remained unaffected (Supplementary Figure S9), there was a small but consistent increase in the level of cleaved mature MALAT1, with a concomitant reduction in the level of uncleaved precursor MALAT1 (Figure 4A). In addition to assessing the levels of cleaved versus uncleaved MALAT1, mascRNA level could serve as another indicator of the efficiency of the cleavage reaction. Since TALAM1 does not influence the total level of MALAT1 (Supplementary Figure S9), we could exclude the possibility that TALAM1 enhances the expression of MALAT1, thus producing more mascRNA. In cells that overexpress TALAM1, a significant increase in mascRNA level was revealed by northern blot analyses (Figure 4Ba). MascRNA-specific Taqman small RNA RT-qPCR assay also showed consistent increase in the level of mascRNA in TALAM1 overexpressed cells (Figure 4Bb). The relative increase in the levels of mascRNA and cleaved MALAT1, together with the decrease in uncleaved MALAT1 upon TALAM1 overexpression, suggest that transiently overexpressed TALAM1 could facilitate in trans the 3′ end processing of MALAT1. It is important to note that we observed the effects on the cleavage of endogenous MALAT1 only when we overexpressed TALAM1 to ≈300-fold. However, under physiological conditions, TALAM1 is a low copy transcript, and primarily enriched at the site of transcription. Based on our results, TALAM1 may primarily function in cis, and the trans effect that we observed requires large amount of TALAM1 RNA.
Figure 4.
TALAM1 promotes the 3′ end processing of MALAT1 RNA. (A) Cutoff assay of cleaved and uncleaved MALAT1 in HeLa cells transfected with empty vector (V) or with TALAM1-overexpression construct (TALAM1-OE). (B) Analyses of mascRNA levels in V or TALAM1-OE samples by (Ba) northern blot, and (Bb) Taqman small RNA assay. (C) In vivo RNA decay assay. (Ca) Schematic representation of the βΔ1,2 construct. (Cb) Stability of the intronless β-globin transcript in U2OS Tet-off cells transfected with empty vector (V) or with TALAM1-overexpression construct (TALAM1-OE). Trendlines are single-exponential regression of the percentage of RNA remaining versus time. Dotted lines represent the half-life of reporter RNA. (D) In vitro processing assay of MALAT1 3′ end sequence. Internally 32P-labeled MALAT1 ENE+A+mascRNA RNA substrate (filled arrow in Db) was incubated with HeLa total cell extract in absence or presence of in vitro transcribed YFP, TALAM1-F1R1, F2R2 RNA or 2′MOE targeting the mascRNA site, and its processing to mascRNA (open arrow in Db) is monitored at 40 min and 60 min time points. (Da) Schematic diagrams shown for the relative positions of the TALAM1-F1R1 and TALAM1-F2R2 fragments generated by in vitro transcription. (Db) A representative autoadiogram is shown. (Dc) The quantification of the mascRNA product levels measured from independent experiments, with ImageJ software. At 40 min time point, levels of mascRNA produced under different conditions were normalized to the condition with no extra RNA, which was set as one. Bar data in (A) and (Bb) are mean values with SD, NA = 4, NBb = 3; Data in (Cb) and (Dc) are mean values with SEM, NCb = 4, NDc = 6 (biological replicates, *P < 0.05, **P < 0.01, ***P < 0.001, two-tailed paired Student t-test).
The stabilization effect of the MALAT1 3′ end sequence has been recently characterized using an in vivo reporter decay system, where the MALAT1 3′ end ENE+A+mascRNA sequence cassette stabilizes an intronless β-globin transcript (βΔ1,2) against the rapid RNA decay in vivo (12,13). We therefore exploited this reporter system to evaluate the physiological significance of TALAM1 in modulating the stabilization activity of the MALAT1 ENE+A+mascRNA cassette. The βΔ1,2 gene containing the MALAT1 ENE+A+mascRNA in its 3′ untranslated region was placed under the control of doxycycline responsive promoter (Figure 4Ca). This reporter construct was transfected along with empty vector or TALAM1 overexpression construct into a U2OS Tet-off cell line. Cells that overexpressed TALAM1 consistently showed increased stability of the βΔ1,2 transcript (Figure 4Cb). This result indicates that TALAM1 could facilitate the stability of even a reporter RNA containing the MALAT1 ENE+A+mascRNA cassette.
To address if TALAM1 directly promotes the 3′ end processing event, we set up an in vitro processing assay, using in vitro transcribed 32P-labeled MALAT1 3′ end fragment, containing the MALAT1 ENE+A+mascRNA sequence, as substrate. This substrate was incubated with HeLa cell extract, with or without the addition of in vitro transcribed TALAM1 RNA. Since it is technically challenging to in vitro transcribe ≈8000 nt long TALAM1 RNA, we generated fragments of TALAM1 to cover the 5′ half of TALAM1 full length RNA, including TALAM1-F1R1, a 2600 nt fragment from the 5′ end region of TALAM1 that harbors the complementary sequence to the 3′ end of MALAT1, and TALAM1-F2R2, a successive 3000 nt fragment from the middle region of TALAM1 overlapping with MALAT1 (Figure 4Da). When incubated with HeLa cell extract, the MALAT1 ENE+A+mascRNA fragment was processed to produce the 61 nt mascRNA (Figure 4Db, lanes 3, 8). MascRNA production was efficiently blocked by MALAT1–2′MOE ASO, confirming the specificity of this reaction (Figure 4Db, lane 2). With the addition of in vitro transcribed TALAM1-F1R1 and F2R2 together, a moderate and consistent increase in mascRNA level was observed (Figure 4Db, lane 5, 10), indicating that TALAM1 could directly promote the 3′ end processing of MALAT1 in vitro. To further map the TALAM1 sequence element, TALAM1-F1R1 or TALAM1-F2R2 was added individually to the cell extract. In comparison to F1R1 or F2R2 individually incubated extracts, TALAM1 F1R1+F2R2-incubated extract displayed highest mascRNA cleavage activity (Figure 4Db, compare lanes 5 or 10 with 6–7 or 11–12, respectively). Unexpectedly, more activity was found to reside within TALAM1-F2R2 than TALAM1-F1R1 (Figure 4Db, compare lanes 6 with 7, and 11 with 12). These modest effects were confirmed by multiple independent experiments (Supplementary Figure S10), and the changes in mascRNA levels were summarized and quantified by Image J (Figure 4Dc). The result that TALAM1-F2R2 is more potent than TALAM1-F1R1 in promoting the mascRNA cleavage in vitro, implies TALAM1 may not be functioning in a linear base-to-base manner, but instead, more complex secondary or tertiary structures may be formed between TALAM1-F2R2 and MALAT1 3′ end sequence. These interactions between TALAM1 and MALAT1 may facilitate the folding, and/or stabilize the tRNA structure to be efficiently recognized and cleaved by RNase P. It is also possible that other factors may be recruited to MALAT1 by TALAM1-F2R2 to facilitate MALAT1 cleavage reaction.
DISCUSSION
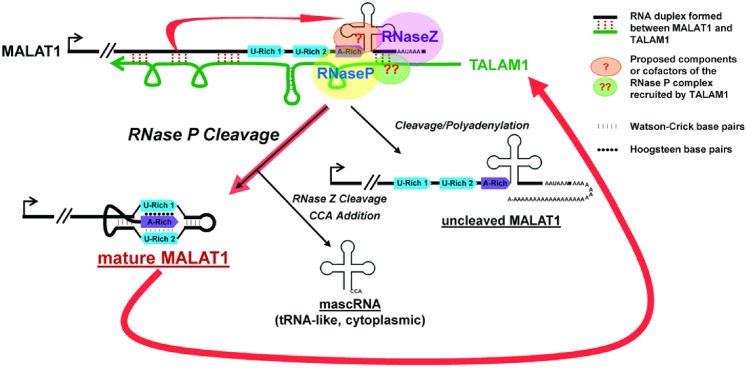
MALAT1 localizes to nuclear speckles and has been implicated in pre-mRNA splicing and transcriptional activation of genes involved in cell-growth and proliferation (8,16–19). Upregulation of MALAT1 is observed to be widely associated with hyper-proliferation and metastasis in cancer, and the oncogenic activity of MALAT1 has been documented in several types of cancers (9). In the present study, we have identified TALAM1, a NAT from the MALAT1 locus, as an important positive regulator of MALAT1 through promoting its 3′ end processing event, a step that is crucial for stabilizing MALAT1 RNA. TALAM1 was found to positively influence the cellular accumulation of MALAT1 and could promote the stabilization effect of MALAT1 3′ end sequence towards labile RNA. Further, TALAM1 itself is positively regulated by MALAT1, thereby forming a feed-forward regulatory loop to maintain the high cellular levels of MALAT1 (Figure 5).
Figure 5.
Model for the feed-forward positive regulatory loop at the MALAT1 locus to maintain the high cellular levels of MALAT1.
The cellular accumulation of MALAT1 RNA has been found to rely on the presence of the ENE+A sequence, which forms a triple helix after the 3′ end processing events (10–13). MALAT1 constructs lacking the ENE sequence, when transiently expressed in cells, showed reduced stability, indicating the requirement of ENE in stabilizing MALAT1 RNA (13). Further, crystallography studies confirmed the formation of a bipartite triple helical structure within the ENE elements and the A-rich tract. Such a structure is demonstrated to confer resistance to exonucleases (13,15). The cleavage of mascRNA from the primary MALAT1 transcript generates a blunt end in the triple helix, removing the steric hindrance, and is required for the stable formation of the triple helix. This was demonstrated by crystallography analyses as well as by mutational studies, where removal of mascRNA as the cleavage signal or creation of terminal nucleotides overhangs greatly diminished the stability of reporter constructs containing MALAT1 3′ end sequence (12,13,15). Therefore, in addition to the triple helical structure, which is encoded in the MALAT1 sequence, the stability of MALAT1 transcript is also controlled by the efficient 3′ end post-transcriptional cleavage. Our loss- and gain-of-function analyses revealed that TALAM1 positively regulates the 3′ end cleavage of MALAT1. Importantly, TALAM1 depletion phenocopies the MALAT1–2′MOE ASO treatment. Both of these treatments impair MALAT1 3′ end cleavage, thereby dramatically reducing MALAT1 cellular levels. Our data altogether support TALAM1 as an important positive regulator of MALAT1 3′ end processing, a step that is required for the stabilization of MALAT1 transcripts.
At present it is not clear how TALAM1 modulates the 3′ end cleavage of MALAT1. TALAM1 RNA concentrates predominantly at the MALAT1 gene locus and could form RNA duplex structures with MALAT1 RNA. At the same time, exogenous expression studies indicate that upon overexpression, TALAM1 could also exert its function in trans. It is possible that TALAM1, by interacting with MALAT1, modulates the secondary and tertiary structure of MALAT1 RNA, and this could influence the recognition of tRNA-structure in uncleaved MALAT1 by RNase P. Eukaryotic RNase P is a ribonucleoprotein complex, consisting of catalytic H1 RNA and multiple protein subunits. The protein subunits within the RNase P complex are involved in substrate recognition and binding, regulation of enzymatic activity as well as structural organization of the RNP complex. The differential distribution of H1 RNA and the protein subunits in distinct sub-nuclear compartments as well as the rapid mobility of some subunits within the nucleus, suggest that the assembly of the RNase P could be dynamic and subject to regulation (34–36). Earlier studies have also documented the roles of specific co-factors in regulating RNase P activity. For example, the human La antigen directly interacts with precursor tRNAs and blocks the site of cleavage, thereby preventing precursor tRNA from being processed by RNase P (34). Thus, it is also possible that TALAM1 may facilitate the recruitment of specific components of the RNase P complex or its cofactors to MALAT1 RNA, or act as a decoy to sponge inhibitory protein factors away from MALAT1 RNA (Figure 5). Future studies will probe into the mechanistic insights of TALAM1, and potentially other NATs, in RNase P-mediated cleavage of non-canonical substrates, including MALAT1 and additional cellular RNAs with ENE-like structures.
MALAT1's important role in normal cell physiology and its pathophysiological dysregulation in cancer highlight the importance of maintaining a tight and robust regulatory control of MALAT1 expression (9). A few earlier studies have described the involvement of transcriptional and post-transcriptional processes that regulate MALAT1 expression (9). Based on our results, we speculate that the levels of TALAM1 could contribute to differential stability of MALAT1. In support of this, we found TALAM1 and MALAT1 are co-expressed with a positive correlation in multiple human and mouse cell lines. However, although we have demonstrated TALAM1 as a regulator of MALAT1 3′ end processing as well as cellular accumulation, how much is this TALAM1-mediated post-transcriptional regulation contributing to the dysregulation of MALAT1 during tumorigenesis, remains to be explored. In addition, even though we find a feedback loop through which MALAT1 positively regulates TALAM1 levels, the regulation of TALAM1 expression by other cellular factors and pathways, for example, by the transcription factors, which interact with TALAM1 promoter (Supplementary Figure S5A), awaits further investigation. It would be interesting to determine whether specific oncogenic pathways regulate TALAM1 levels in order to increase MALAT1 levels. In this context, it would be fruitful to profile human cancers with upregulated MALAT1 levels, and determine whether the elevated MALAT1 levels are attributed to its enhanced stability, altered efficiency in 3′ end processing and dysregulated expression of TALAM1.
Antisense transcription has been recently recognized as a widespread feature of mammalian genome (37). Arguing against the simplistic assumption of a negative regulatory role of antisense transcription through transcriptional collision, RNA-dependent heterochromatinization or formation of sense/antisense (S/AS) hybrids, which then are processed by RNAi machinery, a growing number of recent studies describe concordant regulation by S/AS pairs (37,38). Multiple NATs have been found to be actively involved in promoting the expression of their sense transcripts, via various mechanisms, including DNA demethylation, chromatin remodeling, as well as post-transcriptional regulation of RNA stability, splicing and RNA editing (39). More specifically, stabilization of the sense RNA by NAT has been reported where the formation of RNA duplex is proposed to be responsible for stabilizing the sense RNA (27–29). Importantly, some of these NATs are exploited by pathophysiological pathways to control the levels of their sense transcripts, as observed in the case of PTENpg1 asRNA in cancer (29), as well as BACE1-AS RNA in Alzheimer's disease (27). Our study on TALAM1 provides unique insights into the utilization of NATs to modulate post-transcriptional 3′ end processing and stability of sense lncRNAs, especially for those regulatory RNAs whose vital cellular functions demand stringent expression control.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Acknowledgments
We thank Drs S. Altman, J. Brown, J. Steitz, D. Wesolowski, for reagents. We thank Drs. F. Rigo, R. MacLeod, members of S. Martinis, R. Huang and Prasanth laboratory for discussions and suggestions.
Footnotes
Present addresses:
Jingyi Fei, Department of Biochemistry and Molecular Biology, University of Chicago, IL, USA.
Taekjip Ha, Department of Biophysics and Biophysical Chemistry, Department of Biophysics and Department of Biomedical Engineering, John Hopkins University, Baltimore, MD, USA.
FUNDING
National Institute of Health [GM088252 to K.V.P., GM099669 to S.G.P.]; American Cancer Society [RSG-11-174-01-RMC to K.V.P.]; National Science Foundation career award [NSF1243372 to S.G.P.]. T.H. is a Howards Hughes Medical Institute investigator. S.M.F. is an employee of Ionis Pharmaceuticals, receives salary from the company. Funding for open access charge: American Cancer Society [RSG-11-174-01-RMC].
Conflict of interest statement. None declared.
REFERENCES
- 1.Wang K.C., Chang H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rinn J.L., Chang H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lutz C.S., Moreira A. Alternative mRNA polyadenylation in eukaryotes: an effective regulator of gene expression. Wiley Interdiscip. Rev. RNA. 2011;2:23–31. doi: 10.1002/wrna.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tian B., Manley J.L. Alternative cleavage and polyadenylation: the long and short of it. Trends Biochem. Sci. 2013;38:312–320. doi: 10.1016/j.tibs.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang Y., Yang L., Chen L.L. Life without A tail: new formats of long noncoding RNAs. Int. J. Biochem. Cell Biol. 2014;54:338–349. doi: 10.1016/j.biocel.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Wilusz J.E., Spector D.L. An unexpected ending: noncanonical 3′ end processing mechanisms. RNA. 2010;16:259–266. doi: 10.1261/rna.1907510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang L., Duff M.O., Graveley B.R., Carmichael G.G., Chen L.L. Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 2011;12:R16. doi: 10.1186/gb-2011-12-2-r16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tripathi V., Ellis J.D., Shen Z., Song D.Y., Pan Q., Watt A.T., Freier S.M., Bennett C.F., Sharma A., Bubulya P.A., et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gutschner T., Hammerle M., Diederichs S. MALAT1 – a paradigm for long noncoding RNA function in cancer. J. Mol. Med. (Berl.) 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 10.Wilusz J.E., Freier S.M., Spector D.L. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilusz J.E., JnBaptiste C.K., Lu L.Y., Kuhn C.D., Joshua-Tor L., Sharp P.A. A triple helix stabilizes the 3′ ends of long noncoding RNAs that lack poly(A) tails. Genes Dev. 2012;26:2392–2407. doi: 10.1101/gad.204438.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown J.A., Valenstein M.L., Yario T.A., Tycowski K.T., Steitz J.A. Formation of triple-helical structures by the 3′-end sequences of MALAT1 and MENbeta noncoding RNAs. Proc. Natl. Acad. Sci. U.S.A. 2012;109:19202–19207. doi: 10.1073/pnas.1217338109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown J.A., Bulkley D., Wang J., Valenstein M.L., Yario T.A., Steitz T.A., Steitz J.A. Structural insights into the stabilization of MALAT1 noncoding RNA by a bipartite triple helix. Nat. Struct. Mol. Biol. 2014;21:633–640. doi: 10.1038/nsmb.2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitton-Fry R.M., DeGregorio S.J., Wang J., Steitz T.A., Steitz J.A. Poly(A) tail recognition by a viral RNA element through assembly of a triple helix. Science. 2010;330:1244–1247. doi: 10.1126/science.1195858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conrad N.K. The emerging role of triple helices in RNA biology. Wiley Interdiscip. Rev. RNA. 2014;5:15–29. doi: 10.1002/wrna.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zong X., Tripathi V., Prasanth K.V. RNA splicing control: yet another gene regulatory role for long nuclear noncoding RNAs. RNA Biol. 2011;8:968–977. doi: 10.4161/rna.8.6.17606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tripathi V., Shen Z., Chakraborty A., Giri S., Freier S.M., Wu X., Zhang Y., Gorospe M., Prasanth S.G., Lal A., et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013;9:e1003368. doi: 10.1371/journal.pgen.1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang L., Lin C., Liu W., Zhang J., Ohgi K.A., Grinstein J.D., Dorrestein P.C., Rosenfeld M.G. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li L., Feng T., Lian Y., Zhang G., Garen A., Song X. Role of human noncoding RNAs in the control of tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12956–12961. doi: 10.1073/pnas.0906005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tripathi V., Fei J., Ha T., Prasanth K.V. RNA fluorescence in situ hybridization in cultured mammalian cells. Methods Mol. Biol. 2015;1206:123–136. doi: 10.1007/978-1-4939-1369-5_11. [DOI] [PubMed] [Google Scholar]
- 21.Rose Tam L.S.S., Carol V. Johnson, McNeil John A., Lawrence Jeanne B. FISH: A Practical Approach. Oxford University Press; 2002. [Google Scholar]
- 22.Zhao J., Ohsumi T.K., Kung J.T., Ogawa Y., Grau D.J., Sarma K., Song J.J., Kingston R.E., Borowsky M., Lee J.T. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tuiskunen A., Leparc-Goffart I., Boubis L., Monteil V., Klingstrom J., Tolou H.J., Lundkvist A., Plumet S. Self-priming of reverse transcriptase impairs strand-specific detection of dengue virus RNA. J. Gen. Virol. 2010;91:1019–1027. doi: 10.1099/vir.0.016667-0. [DOI] [PubMed] [Google Scholar]
- 24.Vashist S., Urena L., Goodfellow I. Development of a strand specific real-time RT-qPCR assay for the detection and quantitation of murine norovirus RNA. J. Virol. Methods. 2012;184:69–76. doi: 10.1016/j.jviromet.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakagawa S., Ip J.Y., Shioi G., Tripathi V., Zong X., Hirose T., Prasanth K.V. Malat1 is not an essential component of nuclear speckles in mice. RNA. 2012;18:1487–1499. doi: 10.1261/rna.033217.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zong X., Huang L., Tripathi V., Peralta R., Freier S.M., Guo S., Prasanth K.V. Knockdown of nuclear-retained long noncoding RNAs using modified DNA antisense oligonucleotides. Methods Mol. Biol. 2015;1262:321–331. doi: 10.1007/978-1-4939-2253-6_20. [DOI] [PubMed] [Google Scholar]
- 27.Faghihi M.A., Modarresi F., Khalil A.M., Wood D.E., Sahagan B.G., Morgan T.E., Finch C.E., St Laurent G. 3rd, Kenny P.J., Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan S.X., Tao Q.F., Wang J., Yang F., Liu L., Wang L.L., Zhang J., Yang Y., Liu H., Wang F., et al. Antisense long non-coding RNA PCNA-AS1 promotes tumor growth by regulating proliferating cell nuclear antigen in hepatocellular carcinoma. Cancer Lett. 2014;349:87–94. doi: 10.1016/j.canlet.2014.03.029. [DOI] [PubMed] [Google Scholar]
- 29.Johnsson P., Ackley A., Vidarsdottir L., Lui W.O., Corcoran M., Grander D., Morris K.V. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat. Struct. Mol. Biol. 2013;20:440–446. doi: 10.1038/nsmb.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mahmoudi S., Henriksson S., Corcoran M., Mendez-Vidal C., Wiman K.G., Farnebo M. Wrap53, a natural p53 antisense transcript required for p53 induction upon DNA damage. Mol. Cell. 2009;33:462–471. doi: 10.1016/j.molcel.2009.01.028. [DOI] [PubMed] [Google Scholar]
- 31.Ogawa Y., Sun B.K., Lee J.T. Intersection of the RNA interference and X-inactivation pathways. Science. 2008;320:1336–1341. doi: 10.1126/science.1157676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Legnini I., Morlando M., Mangiavacchi A., Fatica A., Bozzoni I. A feedforward regulatory loop between HuR and the long noncoding RNA linc-MD1 controls early phases of myogenesis. Mol. Cell. 2014;53:506–514. doi: 10.1016/j.molcel.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin G., Keller W. Tailing and 3′-end labeling of RNA with yeast poly(A) polymerase and various nucleotides. RNA. 1998;4:226–230. [PMC free article] [PubMed] [Google Scholar]
- 34.Jarrous N. Human ribonuclease P: subunits, function, and intranuclear localization. RNA. 2002;8:1–7. doi: 10.1017/s1355838202011184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jarrous N., Reiner R. Human RNase P: a tRNA-processing enzyme and transcription factor. Nucleic Acids Res. 2007;35:3519–3524. doi: 10.1093/nar/gkm071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jarrous N., Gopalan V. Archaeal/eukaryal RNase P: subunits, functions and RNA diversification. Nucleic Acids Res. 2010;38:7885–7894. doi: 10.1093/nar/gkq701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Katayama S., Tomaru Y., Kasukawa T., Waki K., Nakanishi M., Nakamura M., Nishida H., Yap C.C., Suzuki M., Kawai J., et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 38.Villegas V.E., Zaphiropoulos P.G. Neighboring gene regulation by antisense long non-coding RNAs. Int. J. Mol. Sci. 2015;16:3251–3266. doi: 10.3390/ijms16023251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faghihi M.A., Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.