Abstract
Type IB DNA topoisomerases can eliminate torsional stresses produced during replication and transcription. These enzymes are found in all eukaryotes and a short version is present in some bacteria and viruses. Among prokaryotes, the long eukaryotic version is only observed in archaea of the phylum Thaumarchaeota. However, the activities and the roles of these topoisomerases have remained an open question. Here, we demonstrate that all available thaumarchaeal genomes contain a topoisomerase IB gene that defines a monophyletic group closely related to the eukaryotic enzymes. We show that the topIB gene is expressed in the model thaumarchaeon Nitrososphaera viennensis and we purified the recombinant enzyme from the uncultivated thaumarchaeon Candidatus Caldiarchaeum subterraneum. This enzyme is active in vitro at high temperature, making it the first thermophilic topoisomerase IB characterized so far. We have compared this archaeal type IB enzyme to its human mitochondrial and nuclear counterparts. The archaeal enzyme relaxes both negatively and positively supercoiled DNA like the eukaryotic enzymes. However, its pattern of DNA cleavage specificity is different and it is resistant to camptothecins (CPTs) and non-CPT Top1 inhibitors, LMP744 and lamellarin D. This newly described thermostable topoisomerases IB should be a promising new model for evolutionary, mechanistic and structural studies.
INTRODUCTION
DNA topoisomerases are essential enzymes found in all organisms [for reviews see (1–6)]. They modify DNA topology by introducing reversible breaks into the DNA phosphodiester backbone. Topoisomerases accomplish their task either by cleaving one strand of the DNA duplex and passing the intact complementary strand through the nick (type I topoisomerase), or by cleaving both strands and passing an intact duplex segment through the double-strand break (type II topoisomerase). Type I enzymes are classified into three families: types IA, IB and IC, each of them characterized by specific combinations of non-homologous domains (7). Type IB enzymes (TopIB) are distantly related to tyrosine recombinases (8). These enzymes can relax both positive and negative superturns in vitro. They form a tyrosyl-3′-phosphodiester linkage and the cleavage generates a transient swivel allowing DNA strands to rotate freely around each other until religation occurs [for type IB topoisomerases structures and mechanisms see (3,6,9–12)].
The type IB enzyme (Top1) was the first DNA topoisomerase discovered in eukaryotes by James Champoux in 1972 (13). Since TopIB enzymes are capable of relaxing positive superturns, it was anticipated that these enzymes, as well as type II DNA topoisomerases, are likely to play a major role in the elimination of torsional stress that accumulates as positive superturns in front of replication forks or of transcription bubbles (14–17). Indeed, genetic analyses and the use of specific DNA topoisomerase inhibitors have not only shown that type IB and IIA DNA topoisomerases probably cooperate in removing torsional stress during replication and transcription, but also in chromatin formation and chromosome condensation (2,18–23). In line with its critical role in cell physiology, human DNA topoisomerase IB is an important drug target in antitumoral treatments, since cancerous cells are very sensitive to poisons that inhibit this enzyme (6,23). The TopIB inhibitors reversibly stabilize the intermediate covalent TopIB–DNA complex that is formed during the relaxation reactions, preventing the religation (22,24,25). This is why the human DNA topoisomerase IB and a few other enzymes of this family, mainly the one of Vaccinia virus (26) and of the bacterium Deinococcus radiodurans (27), have been under investigation by several laboratories during the last four decades.
topIB genes have been found in all eukaryotic genomes sequenced so far. In vertebrates, a specific TopIB with a much shorter N-terminal sequence is also present in mitochondria (28,29). Homologs of TopIB that are smaller versions of the eukaryotic ones have been also detected in Poxviruses, Mimivirus and in several bacterial genomes (26,27,30,31). They are quite different from their eukaryotic counterparts, since they harbor a specific domain (virDNA-Topo-I_N) in their N-terminus, instead of the long Topoisom_I_N domain found in eukaryotic homologs (32).
For a long time, it was thought that the type IB enzyme was not present in the archaeal domain. However, a gene encoding a large version of a DNA topoisomerase IB, very similar to the eukaryotic enzyme, was ultimately detected in the genome of the mesophilic archaeon Cenarchaeum symbiosum, a sponge symbiont and of Nitrosopumilus maritimus, the first thaumarchaeon successfully cultivated in the laboratory (32). These species turned out to be representative of a third major archaeal phylum, the Thaumarchaeota, alongside the two other previously described archaeal phyla, the Euryarchaeota and the Crenarchaeota (33). The Thaumarchaeota are widespread in all types of environments and most of them play an important role in the nitrogen cycle (34). Therefore, since their discovery, Thaumarchaeota have been extensively studied, notably because of their ecological importance.
Phylogenetic analyses have suggested that the type IB enzyme from Thaumarchaeota was present in the last common ancestor of Archaea and Eukarya and was later on lost in Euryarchaeota and Crenarchaeota (32). However, these studies were based on the in silico analyses of complete genomes of only two species of Thaumarchaeota. These last years, genes encoding type IB enzymes have been detected in the genomes of all other characterized Thaumarchaeota whose genomes have been sequenced, as well as in the genome of the uncultivated species Ca. Caldiarchaeum subterraneum. It has been proposed to consider Ca. C. subterraneum as a member of a new phylum, Aigarchaeota (35). However, all the features first used to define the Thaumarchaeota (33,36) are present in Ca. C. subterraneum and this organism forms a monophyletic group with the Thaumarchaeota in most phylogenetic analyses (37–40), suggesting that Ca. C. subterraneum can be considered as a deep-branching thaumarchaeon (41).
The ubiquity of type IB enzymes in Thaumarchaeota suggests that the genes encoding these enzymes can be considered as a hallmark of this major archaeal phylum. Yet, the phylogenetic position of all thaumarchaeal type IB enzymes, their enzymatic activities and their expression in vivo remained to be clarified (10). In this study, we establish that all Thaumarchaeota studied so far contain a type IB enzyme that forms a monophyletic group, closely related to eukaryotic enzymes in a global type IB phylogeny. We show that the topIB gene is expressed in Nitrososphaera viennensis, a cultivated thaumarchaeon isolated from a soil in Vienna (42,43). Finally, we purified the enzyme and characterized the activities of the TopIB enzyme from Ca. C. subterraneum (Cs-TopIB) and compared it with nuclear and mitochondrial human topoisomerases IB [Hs-Top1 (Top1) and Hs-Top1mt (Top1mt)]. Our results demonstrate that the Cs-TopIB enzyme is functional, with all the known activities of eukaryotic TopIB proteins. However, in contrast to the human DNA topoisomerases IB, the archaeal enzyme is resistant to the antitumoral drug camptothecin (CPT) (23). Our results are also in good agreement with the thermophilic nature of Ca. C. subterraneum, since its type IB enzyme is active in a wide range of temperatures, from 10 to 80°C. Taken together, our results suggest that the type IB enzymes of Thaumarchaeota could become promising new models to study the biochemistry and evolution of this important class of enzymes.
MATERIALS AND METHODS
Plasmids and reagents
Negatively supercoiled plasmid pBR322 DNA was purchased from Fermentas Life Science and kinetoplast DNA (kDNA) from Topogen. The reverse-gyrase enzyme was kindly given by Prof. Marc Nadal (Université Paris Diderot). Primers were synthesized by MWG-Operon. CPT was from Developmental Therapeutics Program (DTP), DCTD, NCI, NIH and dissolved in dimethylsulfoxide (DMSO) at 10 mM final concentration as a stock solution.
Bacterial and archaeal strains and culture media
Escherichia coli strains XL10-Gold and BL21(DE3) were used for cloning and expressing topIB, respectively. These strains were grown in Lysogeny Broth (LB) medium. N. viennensis strain EN76 (42,43) was used to detect the expression of topIB and was cultivated in fresh water medium (FWM) supplemented as described in (43).
Phylogenetic analysis
Homologs of TopIB were gathered from the nr (non-redundant) amino acid sequence databank using PSI-BLAST (44) with different distantly related queries (i.e. Eucarya Homo sapiens NP_003277, Bacteria Rhodobacter sphaeroides YP_354029, Archaea C. symbiosum WP_013481455 and Megavirales Acanthamoeba polyphaga mimivirus YP_003986690 sequence queries). A representative sequence subset was extracted and aligned with MAFFT (45). Well-suited characters were selected with BMGE (46) and used to infer an ML phylogenetic tree with PhyML [evolutionary model LG + G4 + I and 1000 bootstrap replicates (47,48)].
RNA extraction
Batches of 10 ml N. viennensis cultures were grown to mid-exponential phase at 37°C without agitation in FWM medium containing 1 mM NH4Cl (43). Growth was monitored by dosing the concentration of nitrite formed over time in the culture medium. For all subsequent steps, solutions were prepared with nuclease-free water and, when possible, DEPC-treated overnight and autoclaved. A total of 80 ml of cultures were harvested by centrifugation at 8000 g, for 25 min at 4°C. The cell pellet was resuspended by pipetting in 400 μl of Acidic Buffer (50 mM CH3COONa, [pH 5.3] and 10 mM ethylenediaminetetraacetic acid (EDTA)). Forty microliters of a 20% sodium dodecyl sulfate (SDS) solution was added and the tube mixed by vortexing. One volume (440 μl) of phenol equilibrated with Acidic Buffer, preheated to 65°C, was added to the sample, which was incubated for a further 4 min at 65°C, before freezing in liquid nitrogen and storing overnight at −80°C. After centrifugation at 16 000 g for 15 min at room temperature, one volume of phenol/chloroform was added to the aqueous phase and centrifuged at 16 000 g for 5 min at room temperature. After precipitation of the nucleic acids present in the aquaeous phase, the pellet was resuspended in 15 μl of nuclease-free water. Two DNAse treatments and purification using the RNEasy MinElute Kit (Qiagen) were then performed subsequently to obtain RNA free of DNA traces. The final RNA yield was about 200 ng.
Endpoint RT-PCR
Forty nanogram of total RNA were mixed with 1 μl of 2 μM Nv-TopIB specific reverse primer R2 (5′-TCTTGCGAGTTCCTGTCCAC), 1 μM of 10 mM dNTPs and the final volume adjusted to 10 μl with nuclease-free water. RNAs were denatured by heating at 65°C for 5 min and then placed on ice for 2 min. Ten microliters of Superscript III (Promega) reverse-transcription mixture (2 μl 10× RT buffer, 4 μl 25 mM MgCl2, 2 μl 0.1M DTT, 1 μl RNAseOut and 1 μl Superscript III) were added and the reverse-transcription performed at 50°C for 50 min. The reaction was terminated by incubating at 85°C for 5 min and subsequent chilling on ice. RNAs were digested by the addition of 1 μl of RNAse H and incubating at 37°C for 20 min. Four microliters of cDNAs from the RT reaction were used for polymerase chain reaction (PCR) amplification with the GoTaq2 PCR kit (Promega), using 2 μl of the 10 μM reverse primer R2 and forward primer F2 (5′-CGGCAGGGCAATGATAAAGC) in a final 50 μl reaction. An initial denaturation step of 2 min at 94°C was followed by 33 PCR cycles (94°C, 30 s/55°C, 40 s/72°C, 1 min 30 s). To control for possible DNA contaminations in working solutions, a ‘H2O’ control was performed with nuclease-free water. To control for complete DNA digestion of the RNA preparation a ‘-RT’ control was performed, in which Superscript III was substituted with 1 μl of nuclease-free water.
Protein structure predictions
Structural predictions were performed with the Phyre2 web server (49). Comparisons between different topoisomerases models were conducted with the PyMOL Molecular Graphics System (http://www.pymol.org).
Recombinant protein expression and purification
The topIB gene from Candidatus C. subterraneum (gene ID: 557694258) was synthesized by GeneArt Gene Synthesis (Life Technologies). The codon usage for expression in E. coli was optimized using the GeneArt Gene Synthesis built-in application. The synthetic gene was cloned with an N-terminal His-Tag into the pETM-11 vector. The plasmid was transformed into E. coli and the cultures were grown subsequently in 1–4 l of LB medium containing kanamycin (50 μg/ml). Induction of the expression of Ca. C. subterraneum topoisomerase IB (Cs-TopIB) was carried out for 1 h at 37°C, after the addition of 1 mM isopropyl-D-1-thiogalactopyranoside when cell cultures reached an OD600 nm of 0.8. Cells were then harvested by centrifugation, stored overnight at −80°C, and then suspended in a 50 mM Tris–HCl [pH 8] and 1 M KCl buffer containing a protease cocktail (Sigma Aldrich). Cell lysis was completed by sonication. The lysed cells were then incubated at 75°C for 15 min and centrifuged for 60 min at 15 000 rpm at 4°C. The supernatant was filtered through a 0.2 μm filter membrane and loaded onto a Ni-NTA column (His-Select HF Nickel Affinity Gel, Sigma-Aldrich). The fractions were eluted with 500 mM imidazole in the presence of 50 mM Tris–HCl [pH 8] and 150 mM NaCl. Cs-TopIB enzyme was assessed for purity on a Sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE). Fractions containing the TopIB polypeptide were pooled and concentrated with Amicon 3 kDa cut-off concentrators (Millipore). The protein concentration was determined by UV detection method at 280 nm or a Bradford assay. The concentrated fractions were dialyzed against 50 mM Tris HCl [pH 8], 1M NaCl and 50% glycerol (v/v), aliquoted and stored at −20 or −80°C.
In vitro mutagenesis
The plasmid bearing Cs-topIBY477F gene was generated with the QuikChange Lightning Site-Directed Mutagenesis kit (Agilent Technologies) using primers TpIb_a1430t (5′-ACCAGCCTGCGTAACTtTATTGATCCGCGTGTG) and TpIb_a1430t_a (5′-CACACGCGGATCAATAaAGTTACGCAGGCTGGT). Mutations in the primer sequences compared to the Ca. C. subterraneum reference sequence are indicated by lowercase letters. After mutagenesis, the plasmids were purified with NucleoSpin Plasmid (Macherey Nagel).
Positively supercoiled pBR322 preparation
Positively supercoiled pBR322 was prepared by incubation of a negatively supercoiled pBR322 plasmid for 30 min at 85°C in the presence of Sulfolobus solfataricus reverse-gyrase in buffer containing 40 mM Tris–HCl [pH 8], 20 mM MgCl2, 1 mM adenosine triphosphate (ATP) and 1 mM DTT. The reaction was stopped by adding 10 mM EDTA and 200 mM NaCl (M. Nadal, personal communication). The positively supercoiled plasmid was purified with the NucleoSpin Gel and PCR Clean-up kits (Macherey-Nagel).
DNA relaxation, decatenation and supercoiling assays
Relaxation assays were performed in a final volume of 20 μl with negatively, positively or relaxed supercoiled pBR322 plasmid or kDNA (200 ng) and the indicated amount of enzyme, in the Cs-TopIB buffer containing 40 mM Tris–HCl [pH 8], 2 mM DTT, 1 mM spermidine and 100 mM KCl or in the human-TopIB buffer containing 10 mM Tris–HCl [pH 8.5], 0.1 mM EDTA, 15 μg/ml bovine serum albumin (BSA) and 50 mM KCl. The reactions were performed for 0–30 min at different temperatures depending on the assay and were stopped by adding 0.5% SDS on ice. A total of 2 μl of proteinase K (1 mg/ml) were then added for an additional incubation of 30 min at 55°C. The reactions were stopped with the blue charged electrophoretic buffer. All the reactions were loaded on a 1% agarose gel prepared in 0.5 × Tris-Acetate-EDTA (TAE) buffer (45 mM Tris–base, [pH 8.5] and 1 mM EDTA). The gels were run at 50 V for 4 h, then stained with ethidium bromide and visualized with a Gel Imaging system (Vilber). Quantification of the pBR322 relaxation was done by estimating the disappearance of the supercoiled DNA using the NIH ImageJ application.
DNA cleavage assays
Site cleavage specificity of TopIB was determined with linear pSK DNA (50). Briefly, a 3′-end labeled double stranded 118-bp DNA substrate was incubated with TopIB in 10 μl reaction buffer [10 mmol/l Tris–HCl (pH7.5), 50 mmol/l KCl, 5 mmol/l MgCl2, 0.1 mmol/l EDTA and 15 mg/ml BSA] at 25°C for 20 min in the presence of the drugs specified in the figures. Reactions were terminated by adding SDS (0.5% final concentration) followed by the addition of two volumes of loading dye (80% formamide, 10 mmol/l sodium hydroxide, 1 mmol/l sodium EDTA, 0.1% xylene cyanol and 0.1% bromphenol blue). Aliquots of each sample were subjected to 20% denaturing PAGE. Gels were dried. The cleavage patterns were visualized using a Phosphoimager and ImageQuant software (Molecular Dynamics). Cleavage sites were numbered as previously described (51).
In vitro assays with camptothecin
Activities of TopIB in the presence of CPT were tested with relaxation kinetics assays. The assays were realized following the same protocols as described in the relaxation assays, with addition of CPT at a concentration of 50 μM or with 5% DMSO as control.
RESULTS
Phylogenetic analysis of type IB DNA topoisomerases
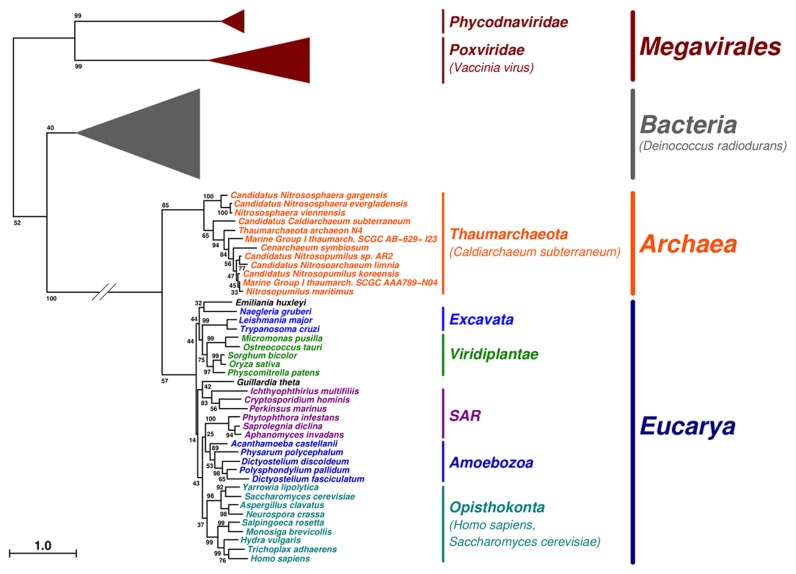
Our search for type IB DNA topoisomerases in archaeal genomes demonstrates that all completely sequenced genomes of Thaumarchaeota contain this protein-coding gene. Phylogenetic analysis using representative subsets of sequences for Thaumarchaeota, Eukarya, Bacteria and Nucleo-Cytoplasmic Large DNA Viruses (NCLDV) (Figure 1; for the complete tree, see Supplementary Figure S1) shows that all thaumarchaeal sequences form a monophyletic group, including Ca. C. subterraneum. Surprisingly, Ca. C. subterraneum branch within Thaumarchaeota and not as a sister group of other Thaumarchaeota, as observed in most phylogenies. However, the confidence support of the corresponding node is quite low (i.e. <70%), suggesting that this emerging position is not robustly supported by the data. Thaumarchaeal type IB enzymes are closely related to eukaryal sequences (long form) in the phylogenetic tree, being very distant from bacterial and NCLDV sequences (short forms). However, they are clearly separated from Eukarya. Notably, we recover the monophyly of most eukaryal divisions in our analysis, suggesting that the root of the eukaryal part of the tree indeed corresponds to the last eukaryal common ancestor.
Figure 1.
Schematic representation of the unrooted phylogenetic tree of TopIB homologous sequences. Specified taxon names are colored according to their membership, i.e. Thaumarchaeota (orange), Excavata (blue), Viridiplantae (green), SAR (dark magenta), Amoebozoa (dark blue), Opisthokonta (cyan). Remaining taxon names (black) each are the only member of their respective phylum, i.e. Emiliania huxleyi (Hacrobia) and Guillardia theta (Cryptophyta). Terminal branches corresponding to Bacteria (gray), Phycodnaviridae and Poxviridae (dark red) clades are shown as triangles of depth proportional to internal diversity (for more details, see Supplementary Figure S1). Bootstrap-based branch supports are indicated at branches. The scale bar represents the average number of substitutions per character.
The topIB gene is expressed in Nitrososphaera viennensis
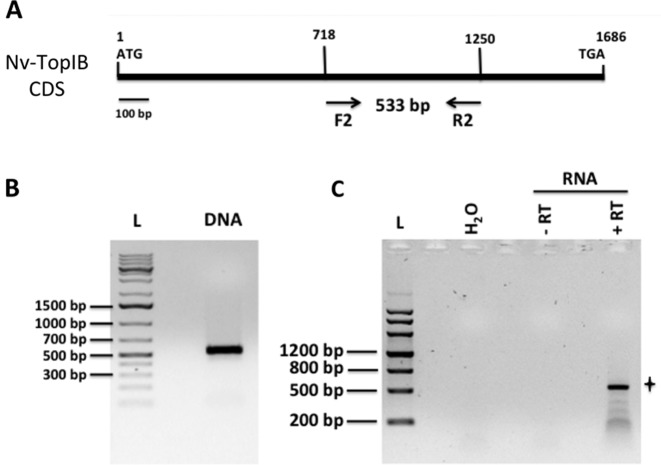
The topoisomerase IB gene is found in all thaumarchaeal genomes sequenced to date, but up to now nothing was known neither about the role and the activities of the encoded protein nor about the expression of the corresponding gene. We first determined by endpoint RT-PCR if the topIB gene is expressed in Thaumarchaeota using as a model the mesophilic thaumarchaeon N. viennensis, one of the few cultivated Thaumarchaeota to date (42,43). We chose to amplify a 533-bp fragment spanning from the middle toward the 3′ end of the 1686 bp predicted topIB gene (Figure 2A and B). Because in eukaryotes TopIB enzymes have major roles in replicating cells, we prepared RNA samples from N. viennensis actively growing (generation time of ca. 27 h) by collecting the cells in the late part of the exponential growth phase. RT-PCR results showed the expected band at around 533 bp (Figure 2C). Gel excision, purification and sequencing of the obtained PCR product confirmed that it corresponds to the topIB gene of N. viennensis. Faint lower molecular weight bands, possibly due to RNA secondary structures and/or an imperfect RT reaction can be seen below the major band (Figure 2C). These results indicate that the topIB gene is expressed in N. viennensis.
Figure 2.
Expression of topIB in Nitrososphaera viennensis. (A) Positions of the forward and reverse primers F2 and R2 on the 1686 bp coding sequence of the N. viennnensis Nv-topIB gene. (B) Amplification using primers F2 and R2 on DNA from 1 μl of a growing N. viennensis culture, yielding the expected 533-bp fragment. (C) Endpoint RT-PCR using primer R2 for the reverse transcription and primers F2 and R2 for the PCR amplification. ‘H2O’: RNA was substituted with nuclease-free water in the RT reaction. ‘−RT’: 1 μl of nuclease-free water was used instead of Superscript III reverse transcriptase in the RT reaction. Using 40 ng of total RNA, the expected band at 533 bp could be amplified by RT-PCR (panel ‘+RT’, indicated by a star). The 533-bp band was excised from the gel and the corresponding DNA purified and sequenced, matching the publicly available sequence of the N. viennensis Nv-topIB coding gene.
Identification and purification of Candidatus Caldiarchaeum subterraneum TopIB protein
Although this thaumarchaeon has not yet been isolated and thus cannot be cultivated, we chose the thermophilic species Ca. C. subterraneum for the biochemical characterization of the archaeal topoisomerase IB. As indicated by its name, the genomic DNA of this organism originated from a subsurface gold mine at a temperature of 70°C, and genome analysis showed the presence of a gene coding for a reverse gyrase. This type IB enzyme would thus be the first thermoresistant DNA topoisomerase IB characterized, which might facilitate its purification and further structural analyses. We retrieved the Ca. C. subterraneum topoisomerase IB (Cs-TopIB) protein sequence (GenBank: BAJ49459.1) from NCBI. The protein is 539 amino-acid residues long. The two characteristic topoisomerase IB domains (Topoisom_I_N and Topoisom_I) are present and are separated by a linker domain. As already observed for other thaumarchaeal TopIB proteins (32), Cs-TopIB does not possess the variable charged N-terminal region of the protein observed in the nuclear eukaryotic enzymes (29) and the linker domain is smaller (53 amino acids instead of 76 for human TopIB) (Figure 3A). Alignment of Cs-TopIB with the human nuclear ortholog shows 37% identity (57% similarity) for the core domain and 43% (52% similarity) for the C-terminus domain. Alignement of Cs-TopIB with the mitochondrial ortholog (Top1mt) shows 34% identity (54% similarity) for the core domain and 38% (52% similarity) for the C-terminus domain. (Supplementary Figure S2).
Figure 3.
Conserved domains and catalytic residues of Thaumarcheal TopIB. (A) Domain organization of human Top1 (Hs-Top1) and Cs-TopIB. In purple the non-conserved eukaryotic N-terminal domain, in green the conserved Core domain, in red the Linker domain, in blue the C-terminal domain with the catalytic tyrosine and in light green a small Cs-TopIB additional C-terminal domain. (B) Multiple amino-acid sequence alignments of topoisomerase IB in proximity of the active site. Amino acids in the active site (R488, K532, R590, H632, Y723) are marked with a + and their numbering is based on the human protein. The aligned sequences are from Candidatus Caldiarchaeum subterraneum (C.sub), Cenarchaeum symbiosum (C.sym), Nitrosopumilus maritimus (N.m), Nitrososphaera viennensis (N.v), Homo sapiens (H.s), Mus musculus (M.m) and Arabidopsis thaliana (A.t). Residues conserved in all sequences are highlighted in yellow. The tyrosine 477 of Ca. C. subterraneum mutated to a phenylalanine corresponds to Y723 of the human protein. (C) Ribbon representation of the crystal structure of the human Top1 in yellow (PDB ID: 1K4T) and the Cs-TopIB molecular model in blue. Camptothecin (CPT) and catalytic tyrosine are rainbow colored.
A multiple sequence alignment of Cs-TopIB with other thaumarchaeal and eukaryotic topoisomerases IB shows that the active site residues (the catalytic tetrad R488, K532, R590 and Y723) are conserved, suggesting the functionality of the thaumarchaeal proteins (Figure 3B). Cs-TopIB share 52, 53 and 49% identity with the TopIB ortholog of C. symbiosum, N. maritimus and N. viennensis, respectively. We compared the human TopIB structure (PDB ID: 1T8I) with the 3D predicted structure of the Cs-TopIB obtained with the Phyre2 web-server, using the molecular visualization system PyMol (Schrödinger) (Figure 3C). The two proteins show 95% of structural identity and can be superimposed, except for parts of their linker domains (Figure 3C).
The Candidatus Caldiarchaeum subterraneum TopIB protein has typical type IB topoisomerase enzymatic activities
The Cs-TopIB protein expressed with an N-terminal His-tag, was purified after expression in E. coli from a codon usage optimized Cs-topIB synthetic gene. The Cs-TopIB protein migrates on a SDS-PAGE gel at about 60 kDa, close to the theoretical mass of 63 kDa (Supplementary Figure S3A).
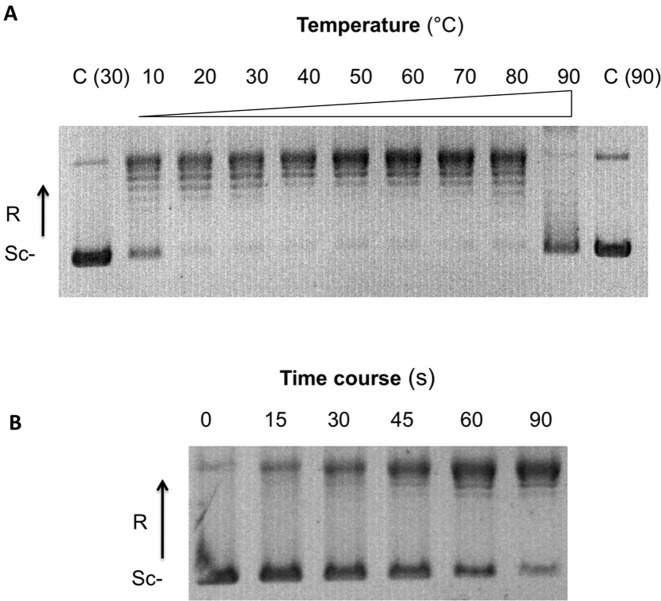
The purified Cs-TopIB turned out to be active in vitro. The enzyme relaxed negatively supercoiled plasmid pBR322 in the presence of monovalent salt (NaCl or KCl). As expected for a typical topoisomerase IB enzyme, the observed activity was ATP- and magnesium-independent. The in vitro activity was optimal from 150 to 250 mM KCl (Supplementary Figure S3B), which is higher than the KCl concentration (50 mM) used for in vitro human topoisomerases IB relaxation assays (26). The enzyme was active across an unprecedentedly wide temperatures range (from 10 to 80°C), with optimal activities between 40 and 70°C (Figure 4A). Time course Cs-TopIB relaxation assay at 70°C shows than the enzyme is processive, as its human counterparts (Top1 and Top1mt), as fully relaxed DNA started to accumulate during the first minute of the reaction, whereas most of the substrate was still fully negatively supercoiled. Topoisomers with intermediate supercoiling density never appeared in the course of the reaction (Figure 4B).
Figure 4.
Cs-TopIB supercoiled DNA relaxation assays over broad temperature range. (A) Effect of the temperature: assays were carried out for 2 min using 200 ng of pBR322 as a substrate of negative supercoiled DNA and 50 nM of Cs-TopIB protein at the indicated temperatures (°C). (B) Time courses assays were performed using 200 ng of pBR322 as a substrate of negative supercoiled DNA and 50 nM of Cs-TopIB protein at the indicated times (s). In both panel, Sc−: negatively supercoiled DNA; R: relaxed DNA; C: control reaction without enzyme.
To control for potential contamination of Cs-TopIB preparation with other DNA topoisomerases that could relax the DNA substrate, we constructed a mutant of Cs-TopIB in which the predicted catalytic tyrosine 477 (Y477) in Cs-TopIB, which corresponds to the tyrosine 723 in the human protein (Figure 3B), was changed to a phenylalanine. This substitution, which does not change the predicted overall protein structure eliminates the potential phoshodiester bond formation because of the absence of the phenolic hydroxyl group. Relaxation assays at 30°C using the Cs-TopIBY477F mutant protein and negatively supercoiled pBR322 plasmid DNA as a substrate showed no relaxation activities by comparison with the wild-type enzymes (Supplementary Figure S3C), demonstrating that the relaxation observed with the wild-type Cs-TopIB preparation was not due to a contamination. In addition, we tested Cs-TopIB decatenation and supercoiling activities. As expected for a typical topoisomerase IB enzyme, the results showed that Cs-TopIB has neither of these activities (Supplementary Figure S3D).
Candidatus Caldiarchaeum subterraneum TopIB is thermoresistant
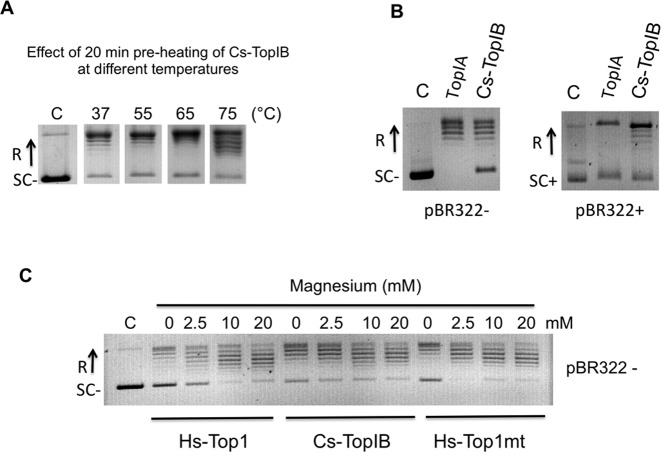
The purified Cs-TopIB was pre-heated for 20 min at different temperatures (from 37 to 75°C) before testing its relaxation activity at 65°C. Figure 5A shows that the enzyme was still active after preincubation for 20 min at 75°C. This thermoresistance was then used to facilitate the purification of the protein by heating of the E. coli cell lysate at 75°C for 15 min before the Ni-NTA column step. These results, together with the observation that the enzyme is still fully active at 80°C are in agreement with the thermophilic nature of Ca. C. subterraneum and make Cs-TopIB the first thermoresistant topoisomerase IB characterized to date.
Figure 5.
Cs-TopIB thermoresistance, relaxation of both positively and negatively supercoiled DNA and activity in the absence of magnesium. (A) Cs-TopIB was pre-heated at the indicated temperatures for 20 min, then assayed for pBR322 relaxation for 20 min at 65°C using 200 ng of pBR322 and 130 nM of Cs-TopIB protein. (B) Assays on negatively and positively supercoiled DNA: negatively (SC−) and positively (SC+) supercoiled pBR322 were assayed for relaxation with Escherichia coli TopIA and Cs-TopIB enzymes at 30°C for 30 min in their respective reaction buffers (see panel C and Supplementary Figure S4 for time-course experiments). (C) Assays were carried out for 30 min at 30°C with 0.8 nM of Hs-Top1, 2 nM of Cs-TopIB and 6.8 nM of Hs-Top1mt and 200 ng of pBR322 as a negatively supercoiled substrate. Mg2+ was added at the indicated final concentrations. ‘C’: reaction without enzyme. In all panels, SC−: negatively supercoiled DNA; SC+: positively supercoiled DNA; R: relaxed DNA.
In contrast to the type IA topoisomerases, TopIB enzymes are known to relax both negatively and positively supercoiled DNA. This is indeed the case for Cs-TopIB. As shown in Figure 5B, Cs-TopIB relaxed positively supercoiled pBR322 whereas the E. coli Topo IA was inactive on this substrate. We observed that at 30°C, Cs-TopIB relaxed negatively and positively supercoiled pBR322 plasmid DNA at the same rate and was processive on both substrates (Supplementary Figure S4). Adding the divalent cation Mg2+ stimulated slightly the relaxation activity of Cs-TopIB with negative DNA substrate, consistent with the Hs-Top1 or Hs-Top1mt enzymes (Figure 5C) (28,52). Comparison of the Cs-TopIB DNA relaxation activity under different DNA/protein ratio confirmed that Cs-TopIB is processive under our reaction conditions similarly to the human DNA relaxation reactions (Supplementary Figure S3C, Cs-TopIBwt).
Candidatus Caldiarchaeum subterraneum TopIB is resistant to camptothecin and exhibits a different cleavage complex selectivity
The antitumoral drug CPT reversibly traps the eukaryotic topoisomerase IB–DNA complex, inhibiting or slowing down the religation of the cleaved DNA strand (22,53). As a consequence, this drug both inhibits the relaxation activity of the enzyme and stimulates DNA cleavage. Eukaryotic nuclear type IB topoisomerase (Hs-Top1) is very sensitive to molecules from the CPT family, while mitochondrial type IB topoisomerase (Hs-Top1mt) is also sensitive, but to a lesser extent (28).
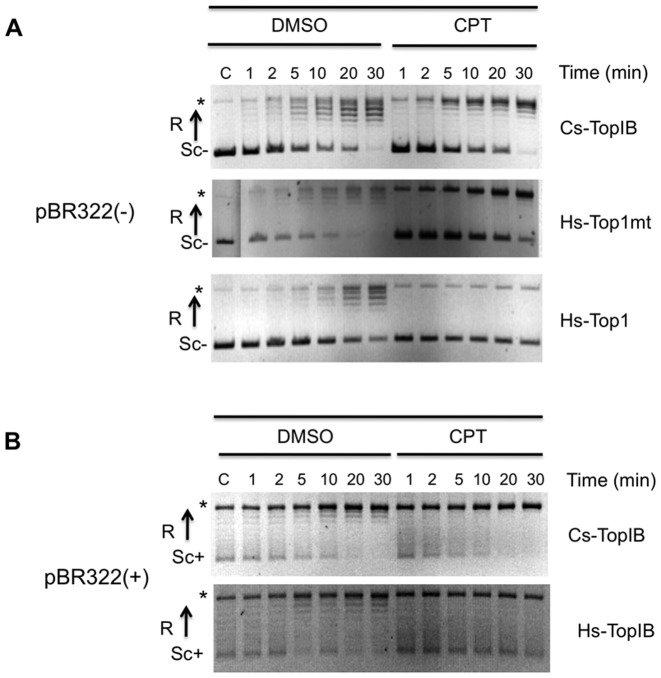
We tested the effect of CPT on the kinetics of relaxation by Cs-TopIB, Hs-Top1 and Hs-Top1mt at a concentration of 50 μM and at 30°C, usually used for this type of analyses (50). As shown in Figure 6A, CPT had no effect on the relaxation activity of Cs-TopIB on negatively supercoiled DNA substrate under conditions where it inhibited the relaxation activities of the Hs-Top1 and to a lesser extent of the Hs-Top1mt. Similarly, the presence of CPT does not affect the activity of TopIB on the positively supercoiled DNA (Figure 6B).
Figure 6.
CPT resistance of Cs-TopIB on DNA relaxation. (A) Time courses assays were carried out from 1 to 30 min at 30°C in the presence of 5% DMSO or 50 mM of CPT and 200 ng of a negatively supercoiled pBR322. (B) Time courses assays were carried out from 1 to 30 min at 30°C in the presence of 5% DMSO or 50 mM of CPT and 200 ng of a positively supercoiled pBR322. Assays in (A) or (B) were done with addition of 10, 6 and 5 nM of Cs-TopIB, Hs-Top1mt and Hs-Top1 respectively. SC−: negatively supercoiled DNA; SC+: positively supercoiled DNA; R: relaxed DNA; C:control reaction without enzyme. Asterisk represents the open circular form in the control and a mix of the open form and the entirely relaxed form of the plasmids for the assays with enzymes.
To further evaluate the lack of response of Cs-TopIB to CPT, we analyzed the patterns of DNA cleavage of Cs-TopIB in comparison with Hs-Top1 and Hs-Top1mt on linear DNA in the absence and presence of CPT (50). While the two human enzymes produced the same patterns of DNA cleavage and responded to CPT [albeit the Hs-Top1mt less than Hs-Top1 (28)] (prominent cleavage sites labeled 18, 44, 66 and 93 in Figure 7A), the archaeal enzyme only produced three weak cleavage sites (labeled 72, 80 and 93). The first two were independent of CPT and did not match the sites induced by the vertebrate enzymes (Figure 7A). Cs-TopIB cleavage was also not stimulated by two non-CPT topoisomerase IB inhibitors, the indenoisoquinoline LMP744 and lamellarin D (22,54,55) (Figure 7B). Testing the Cs-TopIB-Y477F catalytic mutant (see Supplementary Figure S3C) confirmed that the unique cleavage sites induced by Cs-TopIB were specific (Figure 7B). The DNA sequence of the three cleavage sites detected in the pSK-DNA were different, thereby showing no specific consensus cleavage sequence, and the one that had weak sensitivity to CPTs (site 93) (Figure 7B) showed a guanine +1, which is usually the case for the eukaryotic TopIB enzymes (56).
Figure 7.
Sequence selectivity and cleavage complexes induced by Cs-TopIB, Hs-Top1mt and Hs-Top1 in the absence and presence of Top1 inhibitors. (A) 3′-end 32P-end labeled pSK-DNA was incubated with Cs-TopIB, Hs-Top1 or Hs-Top1mt at 25°C for 20 min in the presence of the indicated concentrations of CPTs (CPT in μM). Cleavage sites are indicated to the left with corresponding sequences to the right (cleavage indicated by caret). (B) Additional experiments with the catalytic dead (see Supplementary Figures S2–S3) Cs-TopIB-Y477F enzyme compared to the wild-type enzyme and with additional Top1 inhibitors: the indenoisoquinoline LMP744 and the marine alkaloid lamellarin D (LamD).
DISCUSSION
Up to now, more than 50 genes encoding Type IB enzymes have been identified in cultured and uncultured Thaumarchaeota. Our phylogenetic analysis show that all type IB enzymes from Thaumarchaeota form a monophyletic group that branch as sister group of the eukaryotic DNA topoisomerases IB. We found that the gene encoding the type IB DNA topoisomerase of the thaumarchaeon N. viennensis is expressed and that the enzyme isolated from Ca. C. subterraneum exhibits canonical characteristics of type IB topoisomerases, apart from its temperature stability, sequence selectivity and resistance to CPTs and non-CPT Top1 inhibitors. The confirmation of the topoisomerase IB typical biochemical activity pattern for the purified thaumarchaeal TopIB enzyme and the detection of transcriptional activity of the corresponding gene in an available laboratory strain strongly suggest that type IB DNA topoisomerases are active in Thaumarchaeota and confirm that topIB genes could be considered as hallmark genes for Thaumarchaeota.
The phylogenetic tree of Figure 1 suggests that a type IB enzyme was present in the common ancestor of Archaea and Eukarya and was later on lost in Euryarchaea and Crenarchaeota, as previously proposed (32). However, several authors have suggested these last years that Eukarya and Archaea do not share a last common ancestor but that Eukarya are sister group of an archaeal ‘TACK’ superphylum grouping Thaumarchaeota, Aigarchaeota (Ca. C. subterraneum), Crenarchaeota and Korarchaeota (57). If this were the case, the long form of type IB enzyme should have been introduced in Archaea during the diversification of this domain. Recently, it has been proposed that Eukarya are a sister group of a newly proposed archaeal phylum, Lokiarchaeota, another putative member of the ‘TACK’ superphylum (39). We failed to detect a type IB topoisomerase in published sequences of Lokiarchaea. However, since genomes of Lokiarchaea are presently incomplete, it is not yet possible to draw a definitive conclusion on this matter. In fact, the exact topology of the tree of life and the evolutionary relationships between Archaea and Eukarya is still a controversial issue (58). In any scenario, the origin of type IB enzymes remains mysterious. The short form is present in only around half of bacterial phyla and, in these phyla, is present in between 2 and 25% of sequenced genomes (our unpublished observation), raising doubt on the presence of these enzymes in the ancestor of bacteria. Type IB enzymes might have been transferred in modern organisms from now extinct cellular or viral lineages, the viral scenario being supported by homology between type IB enzymes and viral tyrosine recombinases (59).
The enzyme isolated from the putative thermophilic uncultivated strain Ca. C. subterraneum exhibits the canonical activities of type IB enzymes, relaxing both positive and negative supercoiled DNA without divalent metal (Figure 5). This result is not surprising considering its high sequence and structural similarities with the eukaryotic enzymes (Figure 3). We found that the gene encoding the type IB DNA topoisomerase of the thaumarchaeon N. viennensis is expressed when N. viennensis is growing in exponential phase, suggesting a role for the protein in rapidly growing cells (Figure 2). As in Eukaryotes, the thaumarcheal TopIB is a plausible candidate to eliminate positive supercoils produced ahead of DNA replication forks, as well as the positive and negative supercoiled DNA produced in the course of transcription (6,10,60,61). However, these tasks could also be realized by the canonical archaeal DNA topoisomerase VI (type IIB), which is also present in the genomes of all Thaumarchaea analyzed so far. Interestingly, some Thaumarchaea lack a gene encoding a type IA enzyme (32). This is striking because all other organisms from the three domains of life without exception encode either one or several type IA enzymes (4). This indicates that the type IB enzyme should be able to replace some critical function of type IA enzymes in these Thaumarchaeota. Further characterization of the role of the type IB enzyme in the physiology of the Thaumarchaeota will require developing genetic tools for these fascinating microorganisms.
Type IB DNA topoisomerases are very important targets for antitumoral drugs in human (22). In particular, drugs of the CPT family, like topotecan and irinotecan, are commonly used in chemotherapy. We report here that CPT only interacts weakly with the thaumarchaeal TopIB by stabilizing one of the cleavage complexes and inducing DNA cleavage at the preferred guanosine +1 sequence (56) (Figure 7). However, in contrast to the situation observed with the human type IB enzymes, CPT had no effect on the relaxation activities of the archaeal enzyme, despite the conservation in the Cs-TopIB of the catalytic tetrad (in human : R488, K532, R590 and H632), and of the amino acids implicated with the CPT resistance (in human : F361, R364, D533, N722) (62). This is likely explained by the fact that CPTs and the other non-CPT Top1 inhibitors tested (the indenoisoquinoline LMP744 and the natural product lamellarin D) act as interfacial inhibitors and require specific DNA sequences for trapping the Top1 cleavage complexes (22). Drug resistance can also be related to protein sequence differences (see Figure 3 and Supplementary Figure S2) as the drugs require specific interactions with amino acid residue for trapping topoisomerase IB enzymes (22,63). The difference in DNA sequence selectivity between the Cs-TopIB and Hs-Top1 are likely to explain the resistance of Cs-TopIB to CPTs and other anticancer Top1 inhibitors.
Finally, the thermoresistance of Cs-TopIB makes the enzyme stable and easy to purify in E. coli (Supplementary Figure S3A). Cs-TopIB can work over a wide range of temperatures (Figure 4A), whereas the vertebrate Top1 are inactivated at 65°C (50), a property that could be useful for biochemical characterization and biotechnological applications. Importantly, the archaeal type IB topoisomerases are much more similar to the eukaryotic enzymes than the Vaccinia virus type IB topoisomerase, which has been widely used as model system to understand human enzyme (Figure 1). These characteristics suggest that the Cs-TopIB or others thaumarchaeal TopIB might be promising new models for evolutionary, mechanistic and structural studies of the type IB DNA topoisomerases.
Supplementary Material
Acknowledgments
The authors wish to thank Christa Schleper (University of Vienna) and her laboratory for the kind gift of N. viennensis and their advices on the growth of this thaumarchaeon, the Francis-André Wollman laboratory (UMR 7141, CNRS-Université Paris 6), for providing help for the RNA analysis and Marc Nadal (Institut Jacques Monod) for his advices and the gift of purified reverse gyrase. We also wish to thank Dr Keli Agama, DTB, NCI-NIH for running the DNA cleavage complex assays.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
LabeX ‘Initiative d'Excellence’ program [Grant ‘DYNAMO,’ ANR-11- LABX-0011-01]; NCI Intramural Program, Center for Cancer Research [BC-006161]. Funding for open access charge: Institut Pasteur.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wang J.C. DNA topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 2.Champoux J.J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 3.Corbett K.D., Berger J.M. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 4.Forterre P., Gribaldo S., Gadelle D., Serre M.C. Origin and evolution of DNA topoisomerases. Biochimie. 2007;89:427–446. doi: 10.1016/j.biochi.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Forterre P. In: DNA Topoisomerases and Cancer. Pommier Y, editor. Humana Press; 2012. pp. 1–52. [Google Scholar]
- 6.Pommier Y., Leo E., Zhang H., Marchand C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 2010;17:421–433. doi: 10.1016/j.chembiol.2010.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forterre P. DNA topoisomerase V: a new fold of mysterious origin. Trends Biotechnol. 2006;24:245–247. doi: 10.1016/j.tibtech.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 8.Cheng C., Kussie P., Pavletich N., Shuman S. Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases. Cell. 1998;92:841–850. doi: 10.1016/s0092-8674(00)81411-7. [DOI] [PubMed] [Google Scholar]
- 9.Schoeffler A.J., Berger J.M. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 10.Chen S.H., Chan N.L., Hsieh T.S. New mechanistic and functional insights into DNA topoisomerases. Annu. Rev. Biochem. 2013;82:139–170. doi: 10.1146/annurev-biochem-061809-100002. [DOI] [PubMed] [Google Scholar]
- 11.Stewart L., Redinbo M.R., Qiu X., Hol W.G., Champoux J.J. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 12.Redinbo M.R., Champoux J.J., Hol W.G. Novel insights into catalytic mechanism from a crystal structure of human topoisomerase I in complex with DNA. Biochemistry. 2000;39:6832–6840. doi: 10.1021/bi992690t. [DOI] [PubMed] [Google Scholar]
- 13.Champoux J.J., Dulbecco R. An activity from mammalian cells that untwists superhelical DNA–a possible swivel for DNA replication (polyoma-ethidium bromide-mouse-embryo cells-dye binding assay) Proc. Natl. Acad. Sci. U.S.A. 1972;69:143–146. doi: 10.1073/pnas.69.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu L.F., Wang J.C. Supercoiling of the DNA template during transcription. Proc. Natl. Acad. Sci. U.S.A. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giaever G.N., Wang J.C. Supercoiling of intracellular DNA can occur in eukaryotic cells. Cell. 1988;55:849–856. doi: 10.1016/0092-8674(88)90140-7. [DOI] [PubMed] [Google Scholar]
- 16.Wu H.Y., Shyy S.H., Wang J.C., Liu L.F. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53:433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- 17.Wang J.C. Moving one DNA double helix through another by a type II DNA topoisomerase: the story of a simple molecular machine. Q. Rev. Biophys. 1998;31:107–144. doi: 10.1017/s0033583598003424. [DOI] [PubMed] [Google Scholar]
- 18.Belmont A.S. Mitotic chromosome structure and condensation. Curr. Opin. Cell Biol. 2006;18:632–638. doi: 10.1016/j.ceb.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 19.Salceda J., Fernandez X., Roca J. Topoisomerase II, not topoisomerase I, is the proficient relaxase of nucleosomal DNA. EMBO J. 2006;25:2575–2583. doi: 10.1038/sj.emboj.7601142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nitiss J.L. DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer. 2009;9:327–337. doi: 10.1038/nrc2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teves S.S., Henikoff S. Transcription-generated torsional stress destabilizes nucleosomes. Nat. Struct. Mol. Biol. 2014;21:88–94. doi: 10.1038/nsmb.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pommier Y. DNA topoisomerase I inhibitors: chemistry, biology, and interfacial inhibition. Chem. Rev. 2009;109:2894–2902. doi: 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pommier Y. Drugging topoisomerases: lessons and challenges. ACS Chem. Biol. 2013;8:82–95. doi: 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsiang Y.H., Hertzberg R., Hecht S., Liu L.F. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J. Biol. Chem. 1985;260:14873–14878. [PubMed] [Google Scholar]
- 25.Hsiang Y.H., Liu L.F. Identification of mammalian DNA topoisomerase I as an intracellular target of the anticancer drug camptothecin. Cancer Res. 1988;48:1722–1726. [PubMed] [Google Scholar]
- 26.Shuman S. Vaccinia virus DNA topoisomerase: a model eukaryotic type IB enzyme. Biochim. Biophys. Acta. 1998;1400:321–337. doi: 10.1016/s0167-4781(98)00144-4. [DOI] [PubMed] [Google Scholar]
- 27.Krogh B.O., Shuman S. A poxvirus-like type IB topoisomerase family in bacteria. Proc. Natl. Acad. Sci. U.S.A. 2002;99:1853–1858. doi: 10.1073/pnas.032613199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang H., Barcelo J.M., Lee B., Kohlhagen G., Zimonjic D.B., Popescu N.C., Pommier Y. Human mitochondrial topoisomerase I. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10608–10613. doi: 10.1073/pnas.191321998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H., Meng L.H., Zimonjic D.B., Popescu N.C., Pommier Y. Thirteen-exon-motif signature for vertebrate nuclear and mitochondrial type IB topoisomerases. Nucleic Acids Res. 2004;32:2087–2092. doi: 10.1093/nar/gkh525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bauer W.R., Ressner E.C., Kates J., Patzke J.V. A DNA nicking-closing enzyme encapsidated in vaccinia virus: partial purification and properties. Proc. Natl. Acad. Sci. U.S.A. 1977;74:1841–1845. doi: 10.1073/pnas.74.5.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benarroch D., Claverie J.M., Raoult D., Shuman S. Characterization of mimivirus DNA topoisomerase IB suggests horizontal gene transfer between eukaryal viruses and bacteria. J. Virol. 2006;80:314–321. doi: 10.1128/JVI.80.1.314-321.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brochier-Armanet C., Gribaldo S., Forterre P. A DNA topoisomerase IB in Thaumarchaeota testifies for the presence of this enzyme in the last common ancestor of Archaea and Eucarya. Biol. Direct. 2008;3:54–62. doi: 10.1186/1745-6150-3-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brochier-Armanet C., Boussau B., Gribaldo S., Forterre P. Mesophilic Crenarchaeota: proposal for a third archaeal phylum, the Thaumarchaeota. Nat. Rev. Microbiol. 2008;6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- 34.Offre P., Spang A., Schleper C. Archaea in biogeochemical cycles. Annu. Rev. Microbiol. 2013;67:437–457. doi: 10.1146/annurev-micro-092412-155614. [DOI] [PubMed] [Google Scholar]
- 35.Nunoura T., Takaki Y., Kakuta J., Nishi S., Sugahara J., Kazama H., Chee G.J., Hattori M., Kanai A., Atomi H., et al. Insights into the evolution of Archaea and eukaryotic protein modifier systems revealed by the genome of a novel archaeal group. Nucleic Acids Res. 2011;39:3204–3223. doi: 10.1093/nar/gkq1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spang A., Hatzenpichler R., Brochier-Armanet C., Rattei T., Tischler P., Spieck E., Streit W., Stahl D.A., Wagner M., Schleper C. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol. 2010;18:331–340. doi: 10.1016/j.tim.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 37.Brochier-Armanet C., Forterre P., Gribaldo S. Phylogeny and evolution of the Archaea: one hundred genomes later. Curr. Opin. Microbiol. 2011;14:274–281. doi: 10.1016/j.mib.2011.04.015. [DOI] [PubMed] [Google Scholar]
- 38.Raymann K., Forterre P., Brochier-Armanet C., Gribaldo S. Global phylogenomic analysis disentangles the complex evolutionary history of DNA replication in archaea. Genome Biol. Evol. 2014;6:192–212. doi: 10.1093/gbe/evu004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spang A., Saw J.H., Jorgensen S.L., Zaremba-Niedzwiedzka K., Martijn J., Lind A.E., van Eijk R., Schleper C., Guy L., Ettema T.J. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. 2015;521:173–179. doi: 10.1038/nature14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raymann K., Brochier-Armanet C., Gribaldo S. The two-domain tree of life is linked to a new root for the Archaea. Proc. Natl. Acad. Sci. U.S.A. 2015;112:6670–6675. doi: 10.1073/pnas.1420858112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Forterre P. A new fusion hypothesis for the origin of Eukarya: better than previous ones, but probably also wrong. Res. Microbiol. 2011;162:77–91. doi: 10.1016/j.resmic.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 42.Stieglmeier M., Klingl A., Alves R.J., Rittmann S.K., Melcher M., Leisch N., Schleper C. Nitrososphaera viennensis gen. nov., sp. nov., an aerobic and mesophilic, ammonia-oxidizing archaeon from soil and a member of the archaeal phylum Thaumarchaeota. Int. J. Syst. Evol. Microbiol. 2014;64:2738–2752. doi: 10.1099/ijs.0.063172-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tourna M., Stieglmeier M., Spang A., Konneke M., Schintlmeister A., Urich T., Engel M., Schloter M., Wagner M., Richter A., et al. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc. Natl. Acad. Sci. U.S.A. 2011;108:8420–8425. doi: 10.1073/pnas.1013488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Criscuolo A., Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 2010;10:210–231. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Le S.Q., Gascuel O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 48.Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 49.Kelley L.A., Sternberg M.J. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 50.Dexheimer T.S., Pommier Y. DNA cleavage assay for the identification of topoisomerase I inhibitors. Nat. Protoc. 2008;3:1736–1750. doi: 10.1038/nprot.2008.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Antony S., Agama K.K., Miao Z.H., Takagi K., Wright M.H., Robles A.I., Varticovski L., Nagarajan M., Morrell A., Cushman M., et al. Novel indenoisoquinolines NSC 725776 and NSC 724998 produce persistent topoisomerase I cleavage complexes and overcome multidrug resistance. Cancer Res. 2007;67:10397–10405. doi: 10.1158/0008-5472.CAN-07-0938. [DOI] [PubMed] [Google Scholar]
- 52.Bauer W.R., Ressner E.C., Kates J., Patzke J.V. A DNA nicking-closing enzyme encapsidated in vaccinia virus: partial purification and properties. Proc. Natl. Acad. Sci. U.S.A. 1977;74:1841–1845. doi: 10.1073/pnas.74.5.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hsiang Y.H., Hertzberg R., Hecht S., Liu L.F. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J. Biol. Chem. 1985;260:14873–14878. [PubMed] [Google Scholar]
- 54.Antony S., Jayaraman M., Laco G., Kohlhagen G., Kohn K.W., Cushman M., Pommier Y. Differential induction of topoisomerase I-DNA cleavage complexes by the indenoisoquinoline MJ-III-65 (NSC 706744) and camptothecin: base sequence analysis and activity against camptothecin-resistant topoisomerases I. Cancer Res. 2003;63:7428–7435. [PubMed] [Google Scholar]
- 55.Khiati S., Seol Y., Agama K., Rosa I.D., Agrawal S., Fesen K., Zhang H., Neuman K.C., Pommier Y. Poisoning of mitochondrial topoisomerase I by lamellarin D. Mol. Pharmacol. 2014;86:193–199. doi: 10.1124/mol.114.092833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jaxel C., Capranico G., Kerrigan D., Kohn K.W., Pommier Y. Effect of local DNA sequence on topoisomerase I cleavage in the presence or absence of camptothecin. J. Biol. Chem. 1991;266:20418–20423. [PubMed] [Google Scholar]
- 57.Guy L., Ettema T.J. The archaeal ‘TACK’ superphylum and the origin of eukaryotes. Trends Microbiol. 2011;19:580–587. doi: 10.1016/j.tim.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 58.Forterre P. The universal tree of life: an update. Front. Microbiol. 2015;6:717. doi: 10.3389/fmicb.2015.00717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Forterre P., Gadelle D. Phylogenomics of DNA topoisomerases: their origin and putative roles in the emergence of modern organisms. Nucleic Acids Res. 2009;37:679–692. doi: 10.1093/nar/gkp032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J.C. DNA topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 61.Champoux J.J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 62.Staker B.L., Hjerrild K., Feese M.D., Behnke C.A., Burgin A.B., Jr, Stewart L. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc. Natl. Acad. Sci. U.S.A. 2002;99:15387–15392. doi: 10.1073/pnas.242259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pommier Y., Kiselev E., Marchand C. Interfacial inhibitors. Bioorg. Med. Chem. Lett. 2015;25:3961–3965. doi: 10.1016/j.bmcl.2015.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.