Abstract
Emerging evidence has demonstrated that regulating the length of the poly(A) tail on an mRNA is an efficient means of controlling gene expression at the post-transcriptional level. In early development, transcription is silenced and gene expression is primarily regulated by cytoplasmic polyadenylation. In somatic cells, considerable progress has been made toward understanding the mechanisms of negative regulation by deadenylation. However, positive regulation through elongation of the poly(A) tail has not been widely studied due to the difficulty in distinguishing whether any observed increase in length is due to the synthesis of new mRNA, reduced deadenylation or cytoplasmic polyadenylation. Here, we overcame this barrier by developing a method for transcriptional pulse-chase analysis under conditions where deadenylases are suppressed. This strategy was used to show that a member of the Star family of RNA binding proteins, QKI, promotes polyadenylation when tethered to a reporter mRNA. Although multiple RNA binding proteins have been implicated in cytoplasmic polyadenylation during early development, previously only CPEB was known to function in this capacity in somatic cells. Importantly, we show that only the cytoplasmic isoform QKI-7 promotes poly(A) tail extension, and that it does so by recruiting the non-canonical poly(A) polymerase PAPD4 through its unique carboxyl-terminal region. We further show that QKI-7 specifically promotes polyadenylation and translation of three natural target mRNAs (hnRNPA1, p27kip1 and β-catenin) in a manner that is dependent on the QKI response element. An anti-mitogenic signal that induces cell cycle arrest at G1 phase elicits polyadenylation and translation of p27kip1 mRNA via QKI and PAPD4. Taken together, our findings provide significant new insight into a general mechanism for positive regulation of gene expression by post-transcriptional polyadenylation in somatic cells.
INTRODUCTION
The 3′ poly(A) tails of messenger RNAs play a crucial role in the post-transcriptional control of gene expression through two major mechanisms. First, the poly(A) tail is specifically bound by the cytoplasmic poly(A) binding protein, PABPC1, which physically associates with the translation initiation factor eIF4F; because eIF4F is specifically bound to the 5′ cap structure, the net result is mRNA circularization (1). By bringing the terminating ribosome into close proximity with the initiation site, this arrangement is believed to increase the efficiency of ribosome recruitment for the next round of translation (2). Consistent with this model, a previous study demonstrated that the poly(A) tail synergistically stimulates the translation of capped mRNA in an in vitro translation system (3). Second, the poly(A) tail contributes to mRNA stability (4). In general, mRNA decay is initiated by shortening of the poly(A) tail through deadenylation (5). Because deadenylation is the rate-limiting step, regulating the length of the poly(A) tail is an efficient means to control the stability of any given mRNA (6). Thus, by influencing both the translation and stability of an mRNA, the poly(A) tail plays a pivotal role in controlling the output of any given gene.
Considerable progress has been made toward understanding the mechanism of negative regulation by deadenylation, a widespread strategy for controlling gene expression. The generalized mechanism includes a cis-acting element in the mRNA 3′ UTR that is recognized by a trans-acting RNA-binding protein, which in turn recruits a deadenylating enzyme. The most extensively studied example is the tandem CCCH zinc-finger RNA-binding protein TTP (tristetraproline), which directly binds adenine/uridine-rich elements (AREs) (7) and recruits the scaffold protein Not1 in a complex with the Caf1-Ccr4 deadenylases to accelerate deadenylation and decay of target mRNAs (8). Similarly, Roquin has been reported to recruit the Caf1-Ccr4-Not deadenylase complex, but does so via binding to a conserved class of stem-loop recognition motifs (9). Although the cytoplasmic polyadenylation element (CPE)-binding protein (CPEB) was first identified through its role in post-transcriptional polyadenylation, members of this family of RNA binding proteins have also been shown to accelerate deadenylation of some mRNAs by recruiting Caf1-Ccr4 with the help of the anti-proliferative protein Tob (10,11). Finally, recent findings have demonstrated that Caf1-Ccr4 mediates miRNA-guided degradation of target mRNAs through a mechanism involving deadenylation (12–14). Thus, negative regulation by deadenylation is a widespread and effective means of regulating gene expression.
Positive regulation through elongation of the poly(A) tail, termed cytoplasmic polyadenylation, was first discovered through studies of oocyte maturation in Xenopus and has been extensively studied in the context of germ line and early embryonic development in C. elegans (15). More recently, cytoplasmic polyadenylation has also been reported in somatic cells (16,17). In early development, CPEB facilitates the cytoplasmic polyadenylation of maternal mRNA by the non-canonical poly(A) polymerase PAPD4/Gld2 to promote translation (18). The CPEB-mediated mechanism has been well characterized; however, CPEB-independent mechanisms in which cytoplasmic polyadenylation in early development is facilitated by other RNA binding proteins have also been reported. The specificity factors implicated in these mechanisms include Musashi (19), ElrA, a member of the ELAV family of RNA binding proteins (20), hnRNPE2/αCP2/PCBP2 (21) and GLD-3/Bicaudal C (22). Similar to the mechanism through which it operates in early developmental processes, CPEB has been shown to mediate cytoplasmic polyadenylation in somatic cells via PAPD4/Gld2 or PAPD5/Gld5 to regulate processes including the cell cycle, senescence and synaptic plasticity (16). Importantly, cytoplasmic polyadenylation facilitated by RNA binding proteins other than CPEB has not yet been definitively demonstrated in somatic cells. The main reason for this is that, in contrast to early development where mRNAs transcribed during oocyte maturation have short poly(A) tails and are translationally silent until activated by cytoplasmic polyadenylation, somatic cells are actively carrying out RNA synthesis, translation and turnover. Thus, it has been difficult to distinguish whether any observed poly(A) tail elongation reflects the synthesis of new mRNA, reduced deadenylation or cytoplasmic polyadenylation.
In the study reported here, we demonstrated that the QKI RNA binding protein plays a role in cytoplasmic polyadenylation in somatic cells. This advance was made possible through development of a method to carry out transcriptional pulse-chase analysis under conditions where deadenylases are suppressed, thereby enabling specific detection of cytoplasmic polyadenylation. The QKI protein encoded by the qkI gene is a member of the STAR (signal transduction and activation of RNA) family of RNA binding proteins (23,24). Three QKI isoforms (QKI-5, QKI-6 and QKI-7), which differ only at their extreme C-terminus, are produced from a single gene by alternative splicing (25). All three isoforms share a highly conserved STAR domain of ∼200 amino acids located at their amino termini; this region contains an hnRNP K homology (KH) domain. QKI-5 has a nuclear localization signal (NLS) within its unique C terminal region and is restricted to the nucleus (26). In contrast, QKI-6 is distributed between the nucleus and cytoplasm and QKI-7 is predominatly localized to the cytoplasm. Recent findings have demonstrated that a locus including the qkI gene is associated with schizophrenia, and the expression of QKI-7 was shown to be severely downregulated in schizophrenic patients (27). We show here that QKI-7 induces cytoplasmic polyadenylation when tethered to its target mRNA. The unique C-terminus of QKI-7 is essential for this activity, and functions by specifically recruiting the cytoplasmic poly(A) polymerase PAPD4 to the target mRNA so as to induce polyadenylation and translation. Furthermore, we used siRNA-mediated downregulation of QKI and PAPD4 to confirm that they regulate the poly(A) tail length of hnRNPA1, p27kip1, β-catenin mRNAs, previously established targets of QKI. Lovastatin, an anti-mitogenic agent that induces cell cycle arrest at the G1 phase, elicits polyadenylation and translation of p27kip1 mRNA via QKI and PAPD4. In aggregate, these results indicate that QKI-7 acts as a novel cytoplasmic polyadenylation specificity factor that functions in somatic cells.
MATERIALS AND METHODS
Plasmids
To construct pCMV-5×Flag-QKI-7 and pCMV-5×Myc-QKI-7, the open reading frame (ORF) of QKI-7 was PCR-amplified using the primer pair TM-8/TM-7 and NIH3T3 cDNA library as a template. The resulting fragments were digested with EcoRI and inserted into the EcoRI and EcoRV sites of pCMV-5×Flag (28) and pCMV-5×Myc (10), respectively. pCMV-5×Flag-QKI-7(1–205), pCMV-5×Flag-QKI-7(1–235), pCMV-5×Flag-QKI-7(1–265), pCMV-5×Flag-QKI-7(1–295) and pCMV-5×Flag-QKI-7(1–311) were generated by inverse PCR using pCMV-5×Flag-QKI-7 and the primer pairs NH0145/TM-28, NH0145/TM-32, NH0145/TM-33, NH0145/TM-34 and NH0145/HM-3, respectively. To construct pCMV-5×Flag-QKI-6 and pCMV-5×Myc-QKI-6, the ORF of QKI-6 was PCR-amplified using the primer pair TM-8/HM-2 and NIH3T3 cDNA library as a template. The resulting fragments were digested with EcoRI and inserted into the EcoRI and EcoRV sites of pCMV-5×Flag and pCMV-5×Myc, respectively. To construct pCMV-5×Flag-QKI-5 and pCMV-5×Myc-QKI-5, the ORF of QKI-5 was PCR-amplified using the primer pair TM-8/HM-3 and NIH3T3 cDNA library as a template. The resulting fragments were digested with EcoRI and inserted into the EcoRI and EcoRV sites of pCMV-5×Flag and pCMV-5×Myc, respectively. To construct pCMV-5×Flag-GST, the GST fragment was ligated into the HindIII site of pCMV-5×Flag. To construct pCMV-5×Myc-GST, the GST fragment was ligated into the HindIII and XhoI sites of pCMV-5×Myc. pMS2 was generated by inverse PCR using pMS2-HA (10) and the primer pair NH0538/NH0539. To construct pCMV-5×Flag-GST-QKI-7, the GST fragment was ligated into the HindIII site of pCMV-5×Flag-QKI-7. To construct pCMV-5×Flag-GST-QKI-7(296–315), inverse PCR was performed using pCMV-5×Flag-GST-QKI-7 and the primer pair ry-110/TM-28. The resulting fragment was digested with EcoRI and circularized by self-ligation. To construct pMS2–5×Myc-GST, pMS2–5×Myc-QKI-5, pMS2–5×Myc-QKI-6 and pMS2–5×Myc-QKI-7, the cDNAs of 5×Myc-GST, 5×Myc-QKI-5, 5×Myc-QKI-6 and 5×Myc-QKI-7 were amplified using the primer pair NH540/CMV6 and pCMV-5×Myc-GST, pCMV-5×Myc-QKI-7, pCMV-5×Myc-QKI-6 and pCMV-5×Myc-QKI-5, respectively, as templates. The resulting fragments were digested with NotI, and inserted into the EcoRV and NotI sites of pMS2. pMS2–5×Myc-QKI-7(1–295) and pMS2–5×Myc-QKI-7(1–311) were generated by inverse PCR using pMS2–5×Myc-QKI-7 and the primer pairs T-6/TM34 and T-6/HM-3, respectively. To construct pHA-CMV5-Pan2 D1083A, pFlag-CMV5-Pan2 D1083A (29) was digested with EcoRI and SalI, and the Pan2 D1083A cDNA fragment was inserted into the EcoRI and SalI sites of pHA-CMV5. To construct pCMV-5×Flag-PAPD5 and pCMV-5×Flag-PAPD7, pCMV-5×Myc-PAPD5 and pCMV-5×Myc-PAPD7 l (30) were digested with EcoRI and SalI, and were then inserted into the EcoRI and SalI sites of pCMV-5×Myc, respectively. To construct pCMV-5×Flag-PAPD4 and pCMV-5×Myc-PAPD4, the ORF of PAPD4 was PCR-amplified using the primer pair hGLD2-F/hGLD2-R and U2OS cDNA library as a template. The resulting fragments were digested with EcoRI and SalI and inserted into pCMV-5×Flag and pCMV-5×Myc, respectively. To construct pCMV-5×Flag-PAPD4(1–141), the PAPD4(1–141) fragment was PCR-amplified using the primer pair CMV7/ry-107 and pCMV-5×Flag-PAPD4 as a template. The resulting fragments were digested with EcoRI and SalI and inserted into pCMV-5×Flag. To construct pCMV-5×Flag-PAPD4(142–484), the PAPD4(142–484) fragment was PCR-amplified using the primer pair ry-108/CMV6 and pCMV-5×Flag-PAPD4 as a template. The resulting fragments were digested with EcoRI and SalI and inserted into pCMV-5×Flag. To construct pCMV-5×Flag-PAPD4(1–207), pCMV-5×Flag-PAPD4 was digested with KpnI and circularized by self-ligation. To construct pFlag-CMV5/TO-BGG-hnRNPA1 3′ UTR (L), hnRNPA1 3′ UTR was PCR-amplified using NIH3T3 genomic DNA as a template and the primer pair ry-52/ry-53. The amplified DNA was inserted into the XbaI and PstI sites of pFlag-CMV5/TO-BGG. pFlag-CMV5/TO-BGG-hnRNPA1 3′ UTR (S) was generated by inverse PCR using pFlag-CMV5/TO-BGG-hnRNPA1 3′ UTR (L) and the primer pair ry-57/ry-58. pFlag-CMV5/TO-BGG-hnRNPA1 3′ UTR (S) ΔQRE was generated by inverse PCR using pFlag-CMV5/TO-BGG-hnRNPA1 3′ UTR (S) and the primer pair T-2/T-3. To construct p5×Flag-CMV5/TO-BGG-hnRNPA1 3′ UTR and p5×Flag-CMV5/TO-BGG-hnRNPA1 3′ UTR ΔQRE, p5×Flag-CMV5/TO-BGG (5) was digested by EcoRI, and 4x Flag cDNA fragments were inserted into EcoRI site of pFlag-CMV5/TO-BGG-hnRNPA1 3′ UTR (S) and pFlag-CMV5/TO-BGG-hnRNPA1 3′ UTR (S) ΔQRE, respectively. The construction of all other plasmids used in this study was described previously (10,29,31). The synthetic oligonucleotides used in this study are listed in Supplementary Table S1.
Cell culture and DNA/RNA transfection
HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Nissui) supplemented with 5% fetal bovine serum (FBS). DNA/RNA transfection was performed using Lipofectamine 2000 (Invitrogen), Lipofectamine RNAiMAX (Invitrogen) or Polyethylenimine Max (Polyscience, Inc.) as described previously (11,31). siRNAs used in this study were the following: Luciferase siRNA (5′ r (CGU ACG CGG AAU ACU UCG A)d(TT) 3′), QKI siRNA (5′ r(GACAUGUACAAUGACACAU)d(TT) 3′), PAPD4 siRNA (5′ r(CAAUAUUGUUGGAAUAAGA)d(TT) 3′), Caf1 siRNA (5′ r(CAUCUGGUAUCCAGUUUAA)d(TT) 3′), Pop2 siRNA (5′ r(GUUGCUGAUCAGUUGGAUU)d(TT) 3′).
Antibodies
The antibodies used in this study were anti-Flag (M2, Sigma), anti-Myc (9E10, Roche), anti-HA (3F10, Roche), anti-PAPD41 (N-15, SANTA CRUZ) and anti-hnRNPA1 (R196, Cell Signaling Technology). Anti-QKI and anti-GAPDH antibodies were raised against His-tagged QKI-7 and His-tagged GAPDH, respectively. The anti-PAPD42 antibody was a kind gift from Dr Shin-ichi Kashiwabara (32). The anti-Caf1 antibody was a kind gift from Dr Ann-Bin Shyu (33).
Immunoprecipitation
Transfected cells were lysed in buffer A consisting of 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, 0.1% Nonidet P-40, 1 mM dithiothreitol, 1 mM EDTA-Na (pH 8.0), 100 μM PMSF, 2 μg/ml of aprotinin, 2 μg/ml of Leupeptin and 10 μg/ml RNase A (Sigma). After centrifugation at 15 000 g for 20 min, the lysate was incubated for 1 h at 4°C with anti-Flag-agarose (Sigma), and the resin was washed three times with buffer A. Bound protein was eluted by SDS-PAGE sample buffer and analyzed by western blotting. For immunoprecipitation assay using anti-PAPD41 antibody, the lysate was incubated with either anti-PAPD4 antibody or normal goat IgG (SANTA CRUZ) and Protein G Sepharose 4 Fast Flow (GE Healthcare), and performed immunoprecipitation. PAPD4 protein was detected with anti-PAPD42 antibody.
RNA analysis
Total RNA isolation, northern blot analysis and RNase H treatment of mRNA to generate deadenylated (A0) mRNA were performed as described previously (29). HEK293T cells were co-transfected with pT7-TR, which expresses the T7-tagged tetracycline receptor (31), a reporter plasmid, a reference plasmid and the specified plasmids with or without siRNAs for transcriptional pulse-chase analysis. Twenty four hours after transfection, cells were treated with 40 ng/ml tetracycline for 2 h to induce transcription, and were washed with phosphate-buffered saline three times to remove tetracycline. Cells were harvested at the specified times after transcription shut off. Total RNA was analyzed by northern blot analysis (29).
RL-PAT assay
Total RNA was isolated at the 72 h after siRNA transfection. Two micrograms of total RNA was ligated to 50 pmol of anchor primer KO146 by using T4 RNA ligase1 (BioLabs). The ligated RNA was used for reverse transcriptase reaction, utilizing primer NH20 and SuperScript III Reverse Transcriptase (invitrogen) according to manufacturer's instructions. cDNA fragments were amplificated by semi-nested PCR using the gene-specific sense primers and anchor antisense primer KO197. The cDNA fragments were separated on 2% NuSieve 3:1 agarose (Lonza) gel and visualized by ethidium bromide staining. hnRNPA1 mRNA was amplified using primer pair ry-96/KO197 in first PCR and primer pair ry-97/KO197 in second PCR. p27kip1 mRNA was amplified using primer pair ry-100/KO197 in first PCR and primer pair ry-101/KO197. CTNNB1 mRNA was amplified using primer pair ry-102/KO197 in first PCR and primer pair ry-103/KO197 in second PCR. GAPDH mRNA was amplified using primer pair ry-104/KO197 in first PCR and primer pair ry-105/KO197.
Southern blot analysis
cDNA fragments were separated on 2% NuSieve 3:1 agarose gel. The gel was treated with denaturation buffer consisting of 1.5 M NaCl and 0.5 M NaOH for 30 min. After neutralizing by treatment with neutralizing buffer (pH 7.5) consisting of 0.5 M Tris-HCl and 3 M NaCl for 30 min, cDNA fragments were transferred to Biodyne B (Pall Life Sciences) using blotting buffer consisting of 3 M NaCl and 8 mM NaOH. cDNA fragments were detected using [33P]-labeled oliginucleotide KO197.
Real-time PCR
Total RNA was isolated at 72 h after siRNA transfection, and analyzed using the StepOne Real-Time PCR (RT-PCR) system with PowerSYBR Green PCR Master Mix (Applied Biosystems) as described previously (10,11). hnRNPA1, p27kip1 and CTNNB1 mRNAs were amplified using primer pairs ry-83/ry-84, ry-117/ry-118 and ry-115/ry-116, respectively. GAPDH mRNA was amplified as described previously (10).
RESULTS
QKI-7 specifically induces polyadenylation when tethered to a reporter mRNA
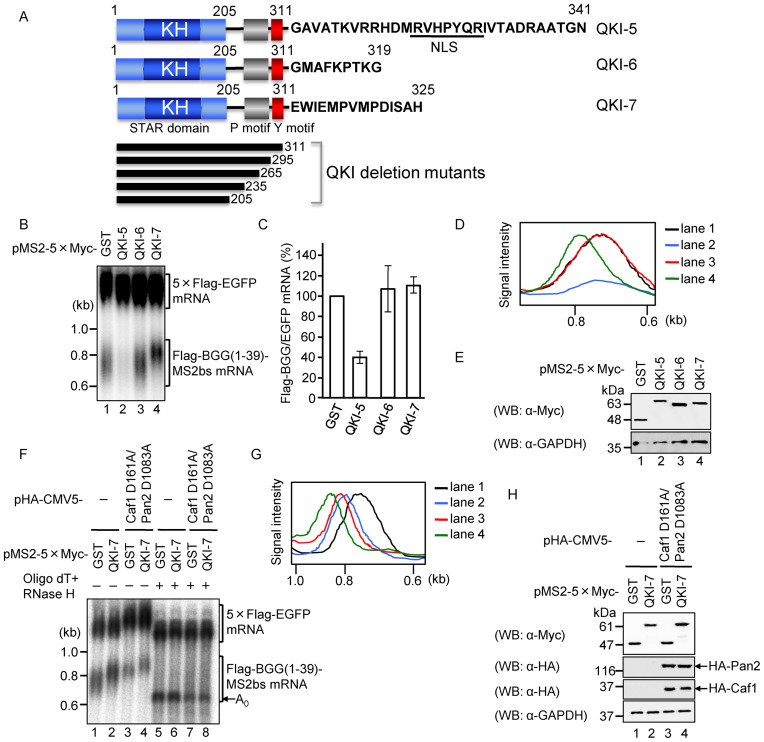
Previous work demonstrated that the mammalian QKI gene produces at least three major protein isoforms, QKI-5, QKI-6 and QKI-7, which arise through alternative splicing (25). All three isoforms contain an amino-terminal hnRNP KH domain in common but differ at their C-termini (Figure 1A). To examine the effect of the QKI isoforms on mRNA metabolism, we first used a tethering strategy in which the proteins of interest were fused to the coat protein of bacteriophage MS2 and the MS2 binding site was incorporated into a reporter mRNA. HEK293T cells were co-transfected with three plasmids: the first carried a β-globin reporter mini gene (BGG(1–39)) that produces mRNA with appended MS2-binding sites (Flag-BGG(1–39)-MS2bs) (10), the second is a reference expressing 5×Flag-EGFP as a transfection/loading control, and the third expresses either 5×Myc-tagged QKI-5, QKI-6 or QKI-7 fused to the MS2 coat protein coding sequence. Interestingly, the three isoforms showed differential effects on the amount and the length of the reporter mRNA. Tethering of QKI-5 decreased reporter mRNA levels, whereas QKI-7 increased the length of the reporter mRNA without affecting its abundance as compared to the GST control. QKI-6 had no significant effect on either mRNA length or accumulation (Figure 1B, C and D). The expression of QKI proteins was confirmed by western blotting (Figure 1E).
Figure 1.
QKI-7 specifically induces poly(A) tail lengthening when tethered to a reporter mRNA. (A) Schematic representation of the domain structure of the three QKI isoforms. All isoforms have a highly conserved STAR domain, which harbors an hnRNP K homology (KH) domain. Proline and tyrosine rich regions are represented as P and Y motifs, respectively, and the nuclear localization signal within the unique C-terminal domain of QKI-5 is marked nuclear localization signal (NLS) (26). The QKI C-terminal deletion mutants used in this study are depicted beneath the domain structures. (B) HEK293T cells were co-transfected with the pFlag-CMV5/TO-BGG (1–39)-MS2bs reporter plasmid, the pCMV-5×Flag-EGFP reference plasmid, and either pMS2–5×Myc-GST, pMS2–5×Myc-QKI-5, pMS2–5×Myc-QKI-6 or pMS2–5×Myc-QKI-7. RNA was isolated from each sample and BGG(1–39)-MS2bs mRNA was detected by northern blot analysis. (C) The amount of Flag-BGG(1–39)-MS2bs mRNA in each sample was measured and normalized to the EGFP mRNA. The mRNA level in pMS2–5×Myc-GST-transfected cells was defined as 100%. The results were derived from three independent experiments carried out as in (B) and are shown as the mean ± SD. (D) The length distribution of BGG(1–39)-MS2bs mRNA was visualized using Profile/MW Mode in the Image Gauge ver. 4.23 (FUJIFILM) software. (E) Protein expression was analyzed by Western blotting with the indicated antibodies. (F) HEK293T cells were co-transfected with the pFlag-CMV5/TO-BGG(1–39)-MS2bs reporter plasmid, the pCMV-5×Flag-EGFP reference plasmid, and a combination of either pMS2–5×Myc-GST (lanes 1, 3, 5 and 7) or pMS2–5×Myc-QKI-7 (lanes 2, 4, 6 and 8), and either pHA-CMV5 (lanes 1, 2, 5 and 6) or pHA-CMV5-Caf1 D161A plus pHA-CMV5-Pan2 D1083A (lanes 3, 4, 7 and 8). The lengths of the poly(A) tails on the BGG(1–39)-MS2bs mRNA were deduced from samples 1–4. To remove the poly(A) tails, each sample was digested with RNase H in the presence of oligo(dT) (lanes 5–8). The position of deadenylated (A0) BGG(1–39)-MS2bs mRNA is indicated by an arrow. (G) The length distribution of BGG(1–39)-MS2bs mRNAs in lanes 1–4 of Figure 1F was visualized as in Figure 1D. (H) Protein expression was analyzed by Western blotting with the indicated antibodies.
We hypothesized that the increase in reporter mRNA length induced by QKI-7 may have been due to extension of the poly(A) tail. To test this possibility, each RNA sample was treated with oligo(dT)/RNase H to remove the poly(A) tail, and the lengths of the reporter mRNAs were subsequently analyzed. This treatment collapsed each reporter mRNA to a single band that migrated at the A0 (unpolyadenylated) position. The results demonstrate that the observed size differences were indeed due to differences in the lengths of the poly(A) tails (Figure 1F and G, compare lanes 5 and 6 to lanes 1 and 2, respectively). The expression of QKI-7 proteins was confirmed by Western blotting (Figure 1H). Thus, we conclude that QKI-7 promotes lengthening of the poly(A) tail when tethered to the reporter mRNA by a specific RNA-protein interaction.
Poly(A) tail lengthening induced by QKI-7 is neither due to reduced deadenylation nor due to synthesis of new mRNAs
Two possible molecular events that could account for the observed increase in the length of the poly(A) tail are enhanced polyadenylation or inhibition of deadenylation. To distinguish these possibilities, we first tested whether the QKI-7-induced increase in poly(A) tail length could be attributed to the inhibition of mRNA deadenylation by examining the effect of expressing catalytically inactive Pan2 and Caf1 mutants (Pan2 D1083A and Caf1 D161A), which were previously shown to inhibit the deadenylation of mRNA in a dominant negative manner (29). Although the overexpression of these mutants completely repressed deadenylation of the reporter mRNA in HEK293T cells (Figure 1F, compare lane 1 and 3 and Figure 1G), under these conditions, QKI-7 still increased the length of the mRNA poly(A) tail (Figure 1F, compare lanes 3 and 4 and Figure 1G). This result indicates that the QKI-7-induced increase in mRNA poly(A) tail length was not due to the inhibition of mRNA deadenylation. Protein expression was confirmed by Western blotting (Figure 1H). By process of elimination, these results strongly suggest that QKI-7 increases mRNA poly(A) tail length via mRNA polyadenylation.
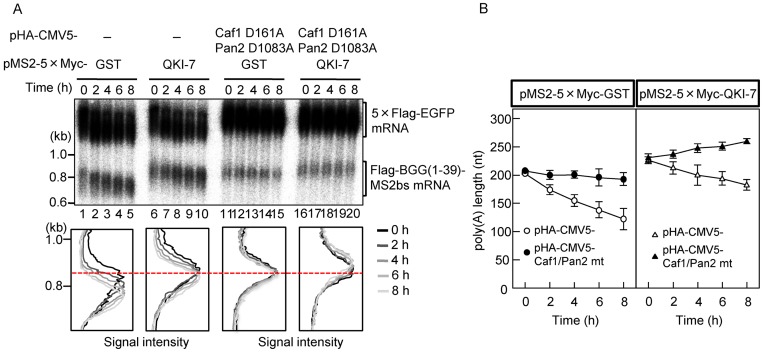
To rigorously demonstrate that the observed poly(A) tail elongation is due to cytoplasmic polyadenylation, it is critical to rule out the possibility that QKI-7 promotes synthesis of new mRNA. To this end, we monitored the deadenylation kinetics of the QKI-7-tethered mRNA using a tetracycline-based pulse-chase assay as described previously (29). Flag-BGG(1–39)-MS2bs mRNA was induced by treatment with a short burst of tetracycline, and changes due to deadenylation after transcriptional shut-off were followed over time. The results indicated that tethering of QKI-7 to the reporter mRNA slowed the rate of poly(A) tail shortening as compared to the GST control (Figure 2A, compare lanes 1–5 with 6–10). Moreover, when the deadenylase mutants (Pan2 D1083A and Caf1 D161A) were overexpressed, deadenylation of the GST-tethered control mRNA was almost completely blocked (Figure 2A, compare lanes 1–5 with 11–15, and Figure 2B Left). In contrast, the length of the poly(A) tail on QKI-7-tethered mRNA increased gradually (Figure 2A, lanes 16–20 and Figure 2B Right). These results substantiate our conclusion that the QKI-7-induced increase in mRNA poly(A) tail length was not due to the inhibition of mRNA deadenylation and instead reflects polyadenylation of pre-existing mRNA.
Figure 2.
The increase in poly(A) tail length induced by QKI-7 is neither due to reduced deadenylation nor due to synthesis of new mRNAs. (A) Pulse-chase analysis was carried out as described previously (29). HEK293T cells were co-transfected with the pFlag-CMV5/TO-BGG(1–39)-MS2bs reporter plasmid, the pCMV-5×Flag-EGFP reference plasmid, the pT7-TR plasmid expressing the T7-tagged tetracycline receptor, and a combination of either pMS2–5×Myc-GST (lanes 1–5 and 11–15) or pMS2–5×Myc-QKI-7 (lanes 6–10 and 16–20) and either pHA-CMV5 (lanes 1–10) or pHA-CMV5-Caf1 D161A plus pHA-CMV5-Pan2 D1083A (lanes 11–20). One day later, BGG(1–39)-MS2bs mRNA was induced by exposing the cells to tetracycline for 2 h. Cells were harvested at the specified time after BGG(1–39)-MS2bs mRNA synthesis was shut off. (Upper panel) BGG(1–39)-MS2bs mRNA was detected by Northern blot analysis. (Lower panel) BGG(1–39)-MS2bs mRNA length-distribution was visualized as in Figure 1D. (B) The average length of the poly(A) tails at each time point was measured from Figure 2A (Lower) using the peak search analysis in Image Gauge ver. 4.23 (FUJIFILM) software. Error bars represent the standard deviation in three independent experiments.
QKI-7 promotes mRNA translation when tethered to the mRNA
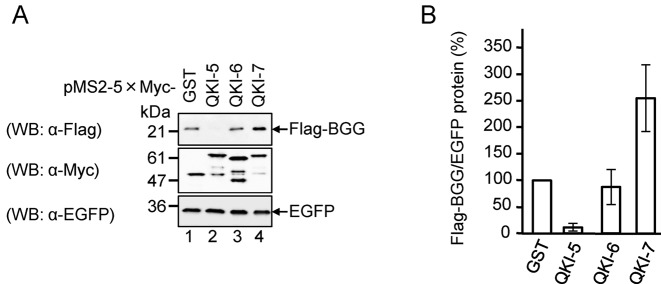
Because the poly(A) tail plays a crucial role in mRNA translation, we anticipated that the increase in poly(A) tail length of mRNA by QKI-7 would increase its protein output. To test this prediction, we examined the amount of protein produced from QKI-tethered mRNA. HEK293T cells were co-transfected with the reporter plasmid expressing the Flag-BGG-MS2bs mRNA/Flag-BGG protein, reference plasmid expressing EGFP as a transfection/loading control, and a plasmid expressing 5×Myc-tagged QKI-5, QKI-6 or QKI-7 fused to the MS2 protein. The results indicate that BGG protein levels were 2.5-fold higher with QKI-7 than with the GST control (Figure 3A and B). Under these conditions, the BGG mRNA level was not significantly affected by the tethering of QKI-7 (Supplementary Figure S1A and S1B). The translation efficiency of the BGG mRNA, estimated by normalizing the BGG protein to the mRNA level, was 2.5-fold higher with QKI-7 than with the GST control (Supplementary Figure S1C). Taken together, these results indicate that tethering of QKI-7 to the reporter mRNA induces polyadenylation to increase its translation efficiency.
Figure 3.
QKI-7 promotes translation when tethered to a reporter mRNA. (A) HEK293T cells were co-transfected with the pFlag-CMV5/TO-BGG-MS2bs reporter plasmid, the pEGFP-C1 reference plasmid and either pMS2–5×Myc-GST, pMS2–5×Myc-QKI-5, pMS2–5×Myc-QKI-6 or pMS2–5×Myc-QKI-7. The total cell lysate was analyzed by Western blotting with the indicated antibodies. (B) The amount of Flag-BGG protein in (A) was measured and normalized to the EGFP protein. The protein level in pMS2–5×Myc-GST transfected cells was defined as 100%. The results were derived from three independent experiments and shown as the means ± SD.
A C-terminal region unique to QKI-7 is necessary for the polyadenylation of its target mRNA
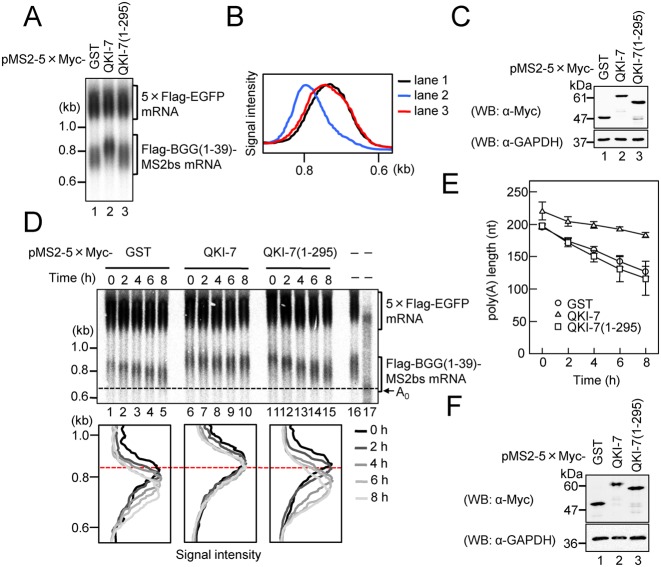
Based on our finding that the three QKI isoforms, which differ only at their C-termini, showed distinct effects on mRNA metabolism (Figure 1B), we speculated that the C-terminal region of QKI-7 is important for the induction of polyadenylation. To test this possibility, we examined the effect of deleting the C-terminus of QKI-7 on its ability to induce polyadenylation of the reporter mRNA. HEK293T cells were again co-transfected with three plasmids: a reporter expressing BGG (1–39)-MS2bs mRNA, a reference expressing 5×Flag-EGFP as a transfection/loading control, and a plasmid expressing either 5×Myc-tagged QKI-7 or QKI-7 (1–295) fused to MS2. As predicted, QKI-7-induced polyadenylation of the reporter mRNA was dramatically decreased by the C-terminal deletion (Figure 4A and B). Expression of the wild-type and QKI-7 deletion mutant proteins was confirmed by Western blotting (Figure 4C).
Figure 4.
The C-terminal region of QKI-7 is necessary for induction of polyadenylation. (A) HEK293T cells were co-transfected with the pFlag-CMV5/TO-BGG(1–39)-MS2bs reporter plasmid, the pCMV-5×Flag-EGFP reference plasmid and either pMS2–5×Myc-GST, pMS2–5×Myc-QKI-7, pMS2–5×Myc-QKI7(1–295). BGG(1–39)-MS2bs mRNA was detected by Northern blot analysis. (B) BGG(1–39)-MS2bs mRNA length distribution was visualized as in Figure 1D. (C) Protein expression was analyzed by Western blotting with the indicated antibodies. (D) HEK293T cells were co-transfected with the pFlag-CMV5/TO-BGG (1–39)-MS2bs reporter plasmid, the pCMV-5×Flag-EGFP reference plasmid, the pT7-TR plasmid and either pMS2–5×Myc-GST (lanes 1–5), pMS2–5×Myc-QKI7 (lanes 6–10) or pMS2–5×Myc-QKI7 (1–295) (lanes 11–15). One day later, synthesis of the BGG(1–39)-MS2bs mRNA was induced by treatment with tetracycline for 2 h. Cells were harvested at the specified times after BGG(1–39)-MS2bs mRNA production was shut off. (Upper panel) BGG(1–39)-MS2bs mRNA was detected by Northern blot analysis. To mark deadenylated (A0) BGG(1–39)-MS2bs mRNA, steady-state BGG(1–39)-MS2bs mRNA (lane 16) was digested with RNase H in the presence of oligo(dT) (lane 17). (Lower panel) BGG(1–39)-MS2bs mRNA length-distribution was visualized as in Figure 1D. (E) The average length of poly(A) tails at each time point was measured as in Figure 2B. Error bars represent the standard deviation of three independent experiments. (F) Protein expression was analyzed by Western blotting with the indicated antibodies.
To further verify our conclusion, we monitored the deadenylation kinetics of QKI-7-tethered mRNA using pulse-chase analysis. QKI-7 decreased the rate of deadenylation of Flag-BGG (1–39)-MS2bs mRNA (Figure 4D, compare lanes 1–5 and lanes 6–10 and Figure 4E). In this situation, the C-terminal deletion mutant QKI-7 (1–295) had no significant effect on the length of the poly(A) tail (Figure 4D, lanes 11–15). The expression of QKI-7 was confirmed by Western blotting (Figure 4F). These results bolster our conclusion that QKI-7 induces mRNA polyadenylation and provide further support for the importance of the C-terminal region of QKI-7 in the induction of polyadenylation.
QKI-7 specifically binds PAPD4 through its unique C-terminal region to induce polyadenylation of a target mRNA
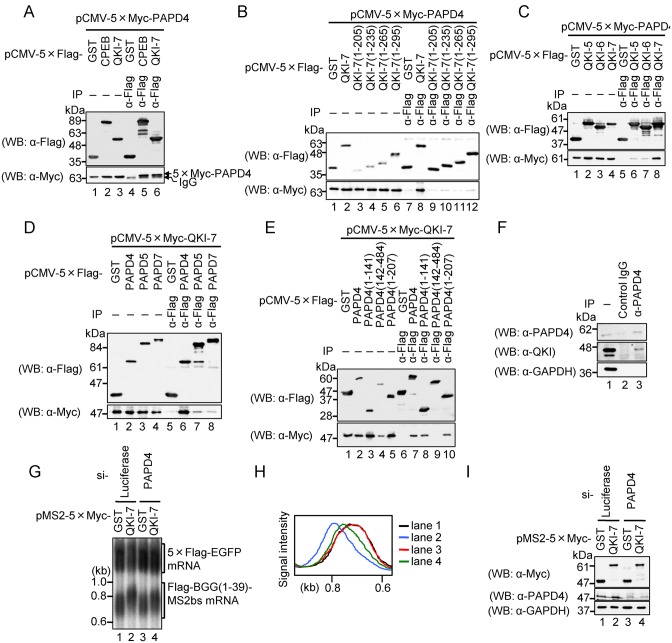
Cytoplasmic polyadenylation is carried out by a class of non-canonical poly(A) polymerases that are distinct from the nuclear enzyme that adds poly(A) tails in conjunction with transcription termination (34). Based on the above results, a reasonable hypothesis is that QKI-7 induces mRNA polyadenylation through an interaction with one of these enzymes. The most plausible candidate is PAPD4, a cytoplasmic poly(A) polymerase previously shown to function with CPEB in mammalian cells (35). To test for an interaction between QKI-7 and PAPD4, we used an immunoprecipitation assay. First, HEK293T cells were co-transfected with a plasmid expressing 5×Myc-PAPD4 and a plasmid expressing either 5×Flag-tagged GST, CPEB or QKI-7. Cell extracts were prepared and subjected to immunoprecipitation with an anti-Flag antibody. The results indicated that 5×Myc-PAPD4 co-precipitated with QKI-7 as well as CPEB (positive control), but not with GST (Figure 5A).
Figure 5.
QKI-7 specifically interacts with PAPD4 through its unique C-terminal region to induce mRNA polyadenylation. (A) HEK293T cells were co-transfected with pCMV-5×Myc-PAPD4 and either pCMV-5×Flag-GST (lanes 1 and 4), pCMV-5×Flag-CPEB (lanes 2 and 5) or pCMV-5×Flag-QKI-7 (lanes 3 and 6). Cell extracts were subjected to immunoprecipitation (IP) using the anti-Flag antibody. The immunoprecipitates (lanes 4–6) and inputs (lanes 1–3) were analyzed by Western blotting with the indicated antibodies. (B) HEK293T cells were co-transfected with pCMV-5×Myc-PAPD4 and either pCMV-5×Flag-GST (lanes 1 and 7), pCMV-5×Flag-QKI-7 (lanes 2 and 8), pCMV-5×Flag-QKI-7(1–205) (lanes 3 and 9), pCMV-5×Flag-QKI-7(1–235) (lanes 4 and 10), pCMV-5×Flag-QKI-7(1–265) (lanes 5 and 11) or pCMV-5×Flag-QKI-7(1–295) (lanes 6 and 12). Cell extracts were subjected to IP using the anti-Flag antibody. The immunoprecipitates (lanes 7–12) and inputs (lanes 1–6) were analyzed by Western blotting with the indicated antibodies. (C) HEK293T cells were co-transfected with pCMV-5×Myc-PAPD4 and either pCMV-5×Flag-GST (lanes 1 and 5), pCMV-5×Flag-QKI-5 (lanes 2 and 6), pCMV-5×Flag-QKI-6 (lanes 3 and 7) or pCMV-5×Flag-QKI-7 (lanes 4 and 8). Cell extracts were subjected to IP using the anti-Flag antibody. Immunoprecipitates (lanes 5–8) and inputs (lanes 1–4) were analyzed by Western blotting with the indicated antibodies. (D) HEK293T cells were co-transfected with pCMV-5×Myc-QKI-7 and either pCMV-5×Flag-GST (lanes 1 and 5), pCMV-5×Flag-PAPD4 (lanes 2 and 6), pCMV-5×Flag-PAPD5 (lanes 3 and 7) or pCMV-5×Flag-PAPD7 (lanes 4 and 8). Cell extracts were subjected to IP using anti-Flag antibody. Immunoprecipitates (lanes 5–8) and inputs (lanes 1–4) were analyzed by Western blotting with the indicated antibodies. (E) HEK293T cells were co-transfected with pCMV-5×Myc-QKI-7 and either pCMV-5×Flag-GST (lanes 1 and 6), pCMV-5×Flag-PAPD4 (lanes 2 and 7), pCMV-5×Flag-PAPD4(1–141) (lanes 3 and 8), pCMV-5×Flag-PAPD4(142–484) (lanes 4 and 9) or pCMV-5×Flag-PAPD4(1–207) (lanes 5 and 10). Cell extracts were subjected to IP using anti-Flag antibody. Immunoprecipitates (lanes 6–10) and inputs (lanes 1–5) were analyzed by western blotting with the indicated antibodies. (F) HEK293T cell extracts were subjected to IP using anti-PAPD4 or normal goat IgG as a non-specific control. The immunoprecipitaes (lanes 2 and 3) and input (lane 1) were analyzed by Western blotting with the indicated antibodies. (G) HEK293T cells were co-transfected with the pFlag-CMV5/TO-BGG (1–39)-MS2bs reporter plasmid, the pCMV-5×Flag-EGFP reference plasmid, either pMS2–5×Myc-GST (lanes 1 and 3) or pMS2–5×Myc-QKI7 (lanes 2 and 4), and either Luciferase siRNA (lanes 1 and 2) or PAPD4 siRNA (lane 3 and 4). BGG(1–39)-MS2bs mRNA was detected by Northern blot analysis. (H) BGG(1–39)-MS2bs mRNA length-distribution was visualized as in Figure 1D. (I) Protein expression was analyzed by Western blotting with the indicated antibodies.
Based on our finding that the C-terminal domain of QKI-7 is important for its role in lengthening of poly(A) tails, we tested whether it is this region that interacts with PAPD4 using the deletion mutants described above (Figure 1A). The results indicated that 5×Flag-tagged QKI-7, but not its deletion mutants (1–205, 1–235, 1–265 and 1–295) co-precipitated with 5×Myc-PAPD4 (Figure 5B). Thus, we conclude that the unique C-terminal region of QKI-7 is required for its interaction with PAPD4. The observed interactions do not appear to be mediated by RNA, since all of the immunoprecipitation experiments in this study were performed in the presence of RNase A.
To extend these results, we tested whether the binding was specific to QKI-7 among the QKI isoforms. To this end, 5×Myc-PAPD4 and either 5×Flag-tagged QKI-5, QKI-6 or QKI-7 were expressed in HEK293T cells and an immunoprecipitation assay was performed as above. The most significant interaction was observed between 5×Myc-PAPD4 and 5×Flag-QKI-7 as compared to the other two QKIs (Figure 5C). As QKI-5, QKI-6 and QKI-7 are localized in the nucleus, nucleus/cytoplasm and cytoplasm, respectively, we next examined if the binding of QKI-7 with PAPD4 is simply due to its cytoplasmic localization. 5×Myc-PAPD4 and 5×Flag-QKI-5, from which the NLS within its unique C terminal region (Figure 1A) was deleted (QKI-5 ΔNLS), were expressed in HEK293 cells and immunoprecipitation assays were performed as above. Deletion of the NLS did not increase the interaction between QKI-5 and PAPD4 (Supplementary Figure S2). These results strengthen our conclusion that the binding of QKI-7 with PAPD4 is not merely due to its cytoplasmic localization. Rather, the amino acid sequence of the C-terminal region that is unique to QKI-7 is necessary for the interaction with PAPD4.
To further explore the interaction between QKI-7 and other poly(A) polymerases, we used an immunoprecipitation assay. 5×Myc-QKI-7 was expressed in HEK293T cells together with either 5×Flag-tagged PAPD4, PAPD5 or PAPD7 and immunoprecipitation was performed as described above. The most significant interaction was observed between 5×Myc-QKI-7 with 5×Flag-PAPD4 as compared to the other PAPDs (Figure 5D), which indicates that QKI-7 preferentially binds to PAPD4 over the other poly(A) polymerases. To map the QKI-7-binding region in PAPD4, we examined the interaction between QKI-7 and PAPD4 deletion mutants. The results indicated that 5×Myc-QKI-7 co-precipitated with PAPD4(1–141) and PAPD4(1–207) but not with PAPD4(142–484) (Figure 5E). Thus, we conclude that the N terminal unstructured region of PAPD4 is responsible for its interaction with the unique C terminal region of QKI-7. Finally, we examined the interaction between endogenous QKI-7 and PAPD4. The lysate of HEK293T cells was subjected to immunoprecipitation with anti-PAPD4 antibody. As shown in Figure 5F, endogenous PAPD4 co-precipitated with endogenous QKI.
The above results strongly suggest that PAPD4 mediates QKI-7-induced polyadenylation. To provide further confirmation, we utilized an RNAi-mediated knockdown strategy. HEK293T cells were co-transfected with the following: a reporter plasmid expressing Flag-BGG (1–39)-MS2bs mRNA, reference plasmid expressing 5×Flag-EGFP as a transfection/loading control, a plasmid expressing either MS2-fused 5×Myc-tagged QKI-7 or GST, and either PAPD4 siRNA or its control siRNA. The results indicated that PAPD4 siRNA down-regulates PAPD4 protein levels to 44% (Figure 5I), which in turn led to the partial repression of QKI-7-induced polyadenylation without affecting the GST control (Figure 5G and H). This result provides compelling evidence that PAPD4 mediates the QKI-7-induced polyadenylation of mRNA.
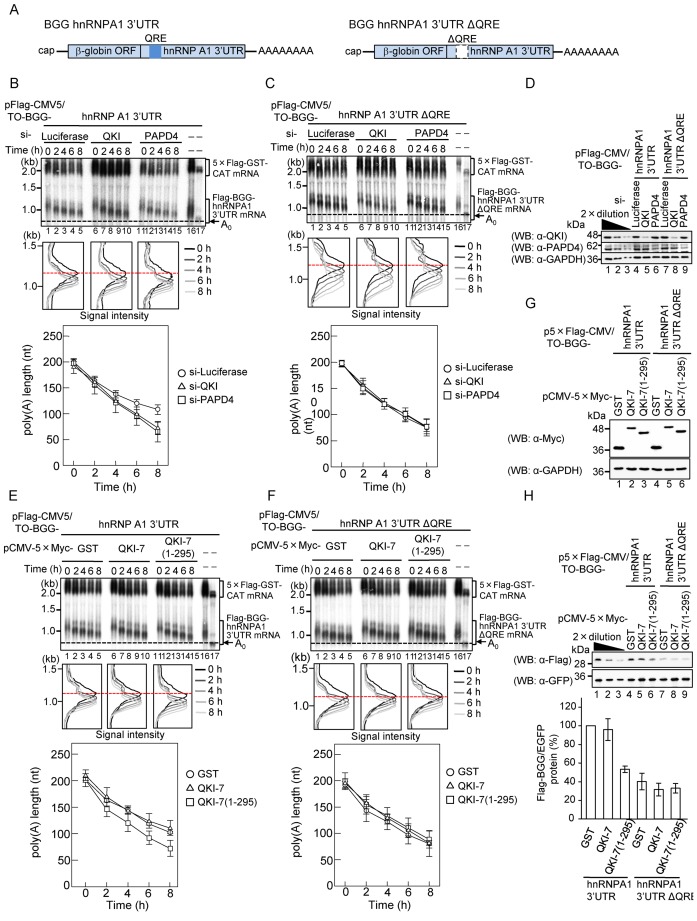
QKI and PAPD4 regulate the poly(A) tail length of hnRNPA1 mRNA in a QRE-dependent manner
A previous study demonstrated that QKI binds to the QKI response element (QRE) in the 3′ UTR to regulate its targets including hnRNPA1 mRNA (36). To determine whether QKI and PAPD4 regulate the poly(A) tail length of the QKI target mRNA through the QRE, BGG mRNA was appended with either the wild-type hnRNPA1 3′ UTR or the hnRNPA1 3′ UTR lacking the QRE (ΔQRE; Figure 6A), and the rate of its poly(A) tail shortening was analyzed using pulse-chase analysis. HEK293T cells were co-transfected with a reporter plasmid expressing either Flag-BGG hnRNPA1 3′ UTR or Flag-BGG hnRNPA1 3′ UTR ΔQRE mRNA, a reference plasmid expressing 5×Flag-GST-CAT as a transfection/loading control, and siRNA against either QKI, PAPD4 or luciferase (control). The kinetics of deadenylation were monitored as described above. The rate of poly(A) tail shortening for the BGG hnRNPA1 3′ UTR mRNA was increased by the siRNA-mediated knockdown of QKI (Figure 6B, compare lanes 1–5 and 6–10) and PAPD4 (Figure 6B, compare lanes 1–5 and 11–15). On the other hand, the knockdown of QKI and PAPD4 had no significant effect on the deadenylation rate of BGG hnRNPA1 3′ UTR ΔQRE mRNA, which lacks QRE (Figure 6C). The expression of QKI and PAPD4 were confirmed by Western blotting (Figure 6D). Thus, we conclude that QKI and PAPD4 regulate the poly(A) tail length of the mRNA in a QRE-dependent manner.
Figure 6.
QKI and PAPD4 regulate the poly (A) tail length of hnRNPA1 mRNA in a QRE-dependent manner. (A) Schematic representation of BGG hnRNPA1 reporter constructs. The β-globin ORF (BGG) was appended with either the hnRNPA1 3′ UTR, which contained the QRE, or the hnRNPA1 3′ UTR in which the QRE had been deleted. (B and C) HEK293T cells were transfected with siRNA against luciferase (lanes 1–5), QKI (lanes 6–10) or PAPD4 (lanes 11–15). Cells were co-transfected with either pFlag-CMV5/TO-BGG-hnRNPA1 3′ UTR (Figure 6B), or pFlag-CMV5/TO-BGG-hnRNPA1 3′ UTR ΔQRE (Figure 6C) pCMV-5×Flag-GST-CAT reference plasmid and pT7-TR, 24 h after siRNA transfection. Pulse-chase analysis was performed as in Figure 4D. (Upper panel) BGG-hnRNPA1 3′ UTR mRNA was detected by Northern blot analysis. (Middle panel) BGG-hnRNPA1 3′ UTR mRNA length-distribution was visualized as in Figure 1D. (Lower panel) The average length of poly(A) tails at each time point was measured from the middle panel as in Figure 2B. Error bars represent the standard deviation of three independent experiments. (D) Protein expression was analyzed by Western blotting with the indicated antibodies. (E and F) HEK29T cells were co-transfected with either the pFlag-CMV5/TO-BGG-hnRNPA1 3′ UTR (Figure 6E) or the pFlag-CMV5/TO-BGG-hnRNPA1 3′ UTR ΔQRE (Figure 6F), the pCMV-5×Flag-GST-CAT reference plasmid, pT7-TR and either pCMV-5×Myc-GST (lanes 1–5), pCMV-5×Myc-QKI-7 (lanes 6–10) or pCMV-5×Myc-QKI-7(1–295) (lanes 11–15). Pulse-chase analysis was performed as in Figure 4D. (Upper panel) BGG-hnRNPA1 3′ UTR mRNA was detected by Northern blot analysis. (Middle panel) BGG-hnRNPA1 3′ UTR mRNA length-distribution was visualized as in Figure 1D. (Lower Panel) The average length of poly(A) tails at each time point was measured from the middle panel as in Figure 2B. Error bars represent the standard deviation of three independent experiments. (G) Protein expression was analyzed by Western blotting with the indicated antibodies. (H) HEK293T cells were co-transfected with either the p5×Flag-CMV5/TO-BGG-hnRNPA1 3′ UTR or the p5×Flag- CMV5/TO-BGG-hnRNPA1 3′ UTR ΔQRE, the pEGFP-C1 reference plasmid and either pCMV-5×Myc-GST, pCMV-5×Myc-QKI-7 or pCMV-5×Myc-QKI-7(1–295). Protein expression was analyzed by Western blotting with the indicated antibodies (upper panel). The amount of Flag-BGG protein was measured and normalized to the EGFP protein. The protein level in pCMV-5×Myc-GST transfected cells was defined as 100% (lower panel). The results were derived from three independent experiments and shown as the means ± SD.
To confirm that the observed effect was mediated through an interaction between QKI and PAPD4, a dominant negative strategy was utilized. We investigated the effect of overexpressing the QKI-7 deletion mutant, QKI-7 (1–295), which cannot interact with PAPD4, on the deadenylation rate of BGG hnRNPA1 3′ UTR mRNA. HEK293T cells were co-transfected with a reporter plasmid expressing either Flag-BGG hnRNPA1 3′ UTR or Flag-BGG hnRNPA1 3′ UTR ΔQRE mRNA, a reference plasmid expressing 5×Flag-GST-CAT as a transfection/loading control, and a plasmid expressing either 5×Myc-tagged GST, QKI-7 or QKI-7 (1–295), and the deadenylation kinetics were monitored. As shown in Figure 6E, the deadenylation rate of the BGG hnRNPA1 3′ UTR mRNA was increased by QKI-7 (1–295) in a dominant negative manner as compared to the GST control, whereas wild-type QKI-7 had no significant effect. On the other hand, the deadenylation rate of BGG hnRNPA1 3′ UTR ΔQRE mRNA was not significantly affected by either QKI-7 (1–295) or QKI-7 (Figure 6F). The expression of QKI-7 proteins was confirmed by Western blotting (Figure 6G).
To provide further evidence that QKI and PAPD4 regulate the poly(A) tail length of the mRNA in a QRE-dependent manner, we constructed a reporter plasmid expressing β-globin mRNA appended with either the QRE of hnRNPA1 (BGG-2×QRE) or its complementary sequence (BGG-2×QRE complement). The rate of poly(A) tail shortening for the corresponding mRNAs was analyzed using pulse-chase analysis as described above. HEK293T cells were co-transfected with a reporter plasmid expressing either Flag-BGG-2×QRE or Flag-BGG-2×QRE complementary mRNA, a reference plasmid expressing 5×Flag-EGFP as a transfection/loading control, and siRNA directed against either QKI, PAPD4 or luciferase (control). The deadenylation kinetics were monitored as above. As expected, the poly(A) tail shortening rate of BGG-2×QRE mRNA was increased by the siRNA-mediatd knockdown of QKI and PAPD4 (Supplementary Figure S3A). On the other hand, the knockdown of QKI and PAPD4 had no significant effect on the deandeylation rate of BGG-2×QRE complement mRNA (Supplementary Figure S3B). These results strengthen our conclusion that QKI-7 and PAPD4 regulate the length of the hnRNPA1 poly(A) tail in a QRE-dependent manner.
QKI and PAPD4 regulate translation of hnRNPA1 mRNA in a QRE-dependent manner
To determine if the observed change in the hnRNPA1 poly(A) tail length affects translation of the mRNA, we examined the effect of QKI-7 (1–295) on the amount of protein produced from BGG hnRNPA1 3′ UTR mRNA. HEK293T cells were co-transfected with a reporter plasmid expressing either 5×Flag-tagged BGG hnRNPA1 3′ UTR or BGG hnRNPA1 3′ UTR ΔQRE mRNA, a reference plasmid expressing EGFP as a transfection/loading control, and a plasmid expressing either 5×Myc-tagged GST, QKI-7 or QKI-7 (1–295), which cannot interact with PAPD4. The level of 5×Flag-BGG protein produced from the BGG hnRNPA1 3′ UTR mRNA was decreased to ∼50% by overexpression of QKI-7 (1–295) in a dominant negative manner (Figure 6H). Conversely, the level of 5×Flag-BGG protein produced from the BGG hnRNPA1 3′ UTR ΔQRE mRNA showed an ∼60% reduction as compared to that from BGG hnRNPA1 3′ UTR mRNA, while overexpression of both QKI-7 and QKI-7 (1–295) had no significant effect (Figure 6H). To assess translation efficiency of the BGG mRNA, BGG hnRNPA1 3′ UTR mRNA was detected by Northern blot analysis (Supplementary Figure S4A) and the amount of BGG protein was normalized to the mRNA level (Supplementary Figure S4B). The translation efficiency of BGG hnRNPA1 3′ UTR mRNA was decreased to ∼60% by overexpression of QKI-7 (1–295), whereas the translation efficiency of BGG hnRNPA1 3′ UTR ΔQRE showed ∼50% reduction as compared to BGG hnRNPA1 3′ UTR. Overexpression of QKI-7 and QKI-7 (1–295) had no significant effect (Supplementary Figure S4B). These results indicate that QKI-7 and PAPD4 regulate the efficiency of hnRNPA1 mRNA translation in a QRE-dependent manner.
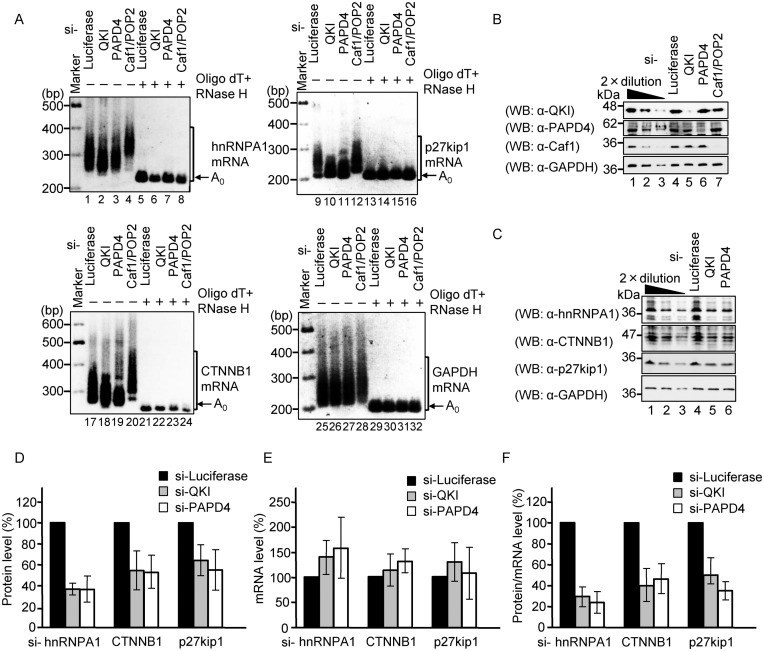
QKI and PAPD4 also regulate polyadenylation and translation of endogenous hnRNPA1, p27kip1 and CTNNB1 mRNAs
Our next series of experiments examined the effect of knockdown of QKI and PAPD4 on poly(A) tail length of endogenous hnRNPA1 mRNA and its protein product. HEK293T cells were transfected with siRNA against either QKI, PAPD4, Caf1/Pop2 or luciferase (control), and the poly(A) tail length of hnRNPA1 mRNA was analyzed using an RL-PAT assay. A previous study had shown that knockdown of Caf1/Pop2 generally increases mRNA poly(A) tail length, and we confirmed that, under our assay conditions, knockdown of Caf1/Pop2 increases the length of the poly(A) tail on hnRNPA1 mRNA (Figure 7A, lane 4). In sharp contrast, the knockdown of QKI and PAPD4 decreased the length of the hnRNPA1 mRNA poly(A) tail (Figure 7A). The expression of QKI, PAPD4 and Caf1 proteins were confirmed by Western blotting (Figure 7B). Similarly, for two other QKI target mRNAs (p27kip1 and CTNNB1), the length of the poly(A) tail was also decreased as a consequence of knocking down either QKI or PAPD4, whereas the poly(A) tail length of GAPDH mRNA, which does not contain a QRE, was not affected (Figure 7A).
Figure 7.
QKI and PAPD4 regulate the poly(A) tail length of endogenous QRE-containing mRNA. (A) HEK293T cells were transfected with Luciferase siRNA, QKI siRNA, PAPD4 siRNA or Caf1/Pop2 siRNA. The poly(A) tail length of the endogenous hnRNPA1 mRNA, p27kip1 mRNA, CTNNB1 mRNA and GAPDH mRNA was analyzed using an RL-PAT assay. (B) Protein expression was analyzed by Western blotting with the indicated antibodies. (C) HEK293T cells were transfected with Luciferase siRNA, QKI siRNA or PAPD4 siRNA. hnRNPA1, p27kip1, CTNNB1 and GAPDH protein expression was analyzed by Western blotting. (D) The amount of hnRNPA1, p27kip1 and CTNNB1 proteins was measured and normalized to the GAPDH protein. The protein level in Luciferase siRNA transfected cells was defined as 100%. The results were derived from three independent experiments and shown as the means ± SD. (E) The amount of hnRNPA1, p27kip1 and CTNNB1 mRNAs was measured by Real-Time PCR (RT-PCR). hnRNPA1, p27kip1 and CTNNB1 mRNA levels were normalized to GAPDH mRNA. The mRNA level in Luciferase siRNA-transfected cells was defined as 100%. The results were derived from three independent experiments and shown as the means ± SD. (F) The translation efficiency was calculated by normalization of protein level to the corresponding mRNA level.
To test the impact of QKI and PAD4 knockdown on translation of the hnRNPA1, CTNNB1 and p27kip1 mRNAs, we analyzed the proteins by Western blotting. The results indicated that the hnRNPA1, CTNNB1 and p27kip1 protein levels were decreased to 36%, 54% and 64%, respectively, by knockdown of QKI, and were decreased to a similar degree (36%, 53% and 55%, respectively), by knockdown of PAPD4 (Figure 7C and D). Finally, the levels of endogenous hnRNPA1, CTNNB1 and p27kip1 mRNAs were analyzed by RT-PCR (Figure 7E) and the amount of each protein was normalized to the corresponding mRNA level to estimate translation efficiency. The translation efficiency of hnRNPA1, CTNNB1 and p27kip1 was decreased to 29%, 39% and 50%, respectively, by knockdown of QKI, and was also decreased to 24%, 46% and 35%, respectively, by knockdown of PAPD4, (Figure 7F). These results are consistent with a model in which QKI, in a complex with PAPD4, induces polyadenylation of its target mRNAs to positively regulate its gene expression.
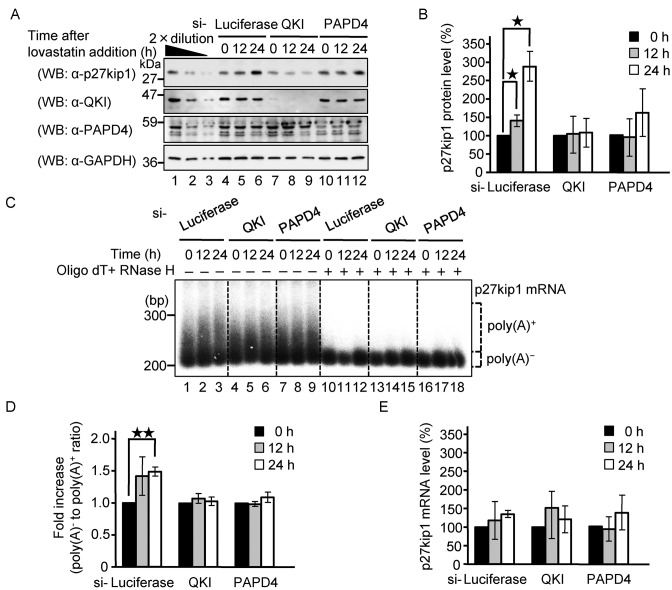
Lovastatin increases poly(A) tail length of p27kip1 mRNA and increases production of the encoded protein via QKI and PAPD4
A previous study showed that high cell density or the anti-mitogenic agent lovastatin activates translation of p27kip1 without affecting its mRNA level to induce cell cycle arrest at the G1 phase in HeLa cells (37). In conjunction with the findings reported here, this led us to speculate that the lovastatin-induced increase in the level of p27kip1 protein might be mediated by QKI-induced polyadenylation of its mRNA. To test this possibility, we examined the effect of knocking down QKI and PAPD4 on the lovastatin-induced increase of p27kip1 protein. As shown in Figure 8A and B, lovastatin increased the p27kip1 protein level in a time-dependent manner, whereas knockdown of QKI and PAPD4 repressed the increase in p27kip1 protein levels. Therefore, we also analyzed the poly(A) tail length of p27kip1 mRNA under these conditions. Lovastatin increased the length of the poly(A) tail of p27kip1 mRNA in a time dependent manner (Figure 8C and D), whereas knockdown of QKI and PAPD4 suppressed the increase in poly(A) tail length. The p27kip1 mRNA levels were not significantly affected by these treatments (Figure 8E). We conclude that lovastatin increases the length of the p27kip1 mRNA poly(A) tail and the levels of its protein product via QKI and PAPD4.
Figure 8.
Lovastatin increases the poly(A) tail length of p27kip1 mRNA and its protein output via QKI and PAPD4. HeLa cells were transfected with siRNA against either Luciferase, QKI or PAPD4. Forty-eight hours later, HeLa cells were harvested at the specified times after the addition of lovastatin. (A) The total cell lysate was analyzed by western blotting with the indicated antibodies. (B) The amount of p27kip1 protein was measured and normalized to the GAPDH protein. The protein level at the 0 h time point was defined as 100%. The results were derived from three independent experiments and shown as the means ± SD. *P < 0.05. (C) The poly(A) tail length of the endogenous p27kip1 mRNA was analyzed using an RL-PAT assay and the amplified cDNA was detected by Southern blotting. To quantitatively estimate distribution of the poly(A) tail length, the mRNAs were digested with RNase H in the presence of oligo(dT) to localize deadenylated (A0) mRNA and the corresponding region of the mRNA signal was designated as poly(A)−; the upper portion was designated as poly(A)+ as indicated. (D) The amount of poly(A)+ portion was measured and normalized to the poly(A)− portion. The poly(A) tail level at 0 h time point was defined as 100%. The results were derived from three independent experiments and shown as the means ± SD. **P < 0.01. (E) The amount of p27kip1 mRNA was measured by RT-PCR and was normalized to that of GAPDH mRNA. The mRNA level at the 0 h time point was defined as 100%. The results were derived from three independent experiments and shown as the means ± SD.
DISCUSSION
To date, cytoplasmic polyadenylation has been most extensively studied in the context of germ line and early embryonic development. In oocytes and early stage embryos, where transcription is silenced, it is straightforward to determine whether poly(A) tail elongation is due to cytoplasmic polyadenylation. In somatic cells, however, the steady-state length of mRNA poly(A) tails reflects the balance among three processes: co-transcriptional polyadenlyation, post-transcriptonal deadenylation and cytoplasmic polyadenylation. Therefore, it is critical to distinguish whether any observed increase in the length of the poly(A) tail is due to the synthesis of new mRNA, to reduced deadenylation or to cytoplasmic polyadenylation. In the current study, we have utilized a transcriptional pulse-chase in combination with suppression of deadenylase activity to overcome this barrier. This strategy allowed us to substantially advance our understanding of cytoplasmic polyadenylation by identifying QKI-7 as a novel cytoplasmic polyadenylation specificity factor that functions in somatic cells. Our data demonstrate that QKI-7 induces poly(A) tail elongation in the absence of transcription and deadenylation. We showed that (i) QKI-7 but not other QKI isoforms specifically induced poly(A) tail lengthening, (ii) QKI-7 induced poly(A) tail lengthening even when transcription was shut off and the two major deadenylases, Caf1-Ccr4 and Pan2-Pan3, were rendered inactive by dominant negative mutants, (iii) the unique C-terminal region of QKI-7 was necessary for poly(A) tail lengthening activity as well as for the physical interaction with the poly(A) polymerase PAPD4 (Gld-2), (iv) QKI-7-induced poly(A) tail lengthening was abrogated by the siRNA-mediated knockdown of PAPD4, (v) the poly(A) tail of a reporter mRNA appended with the hnRNPA1 3′ UTR was destabilized by deletion of the QRE or knockdown of either QKI or PAPD4, (vi) the poly(A) tail length of endogenous QKI target mRNA was decreased by knockdown of either QKI or PAPD4, (vii) the steady-state level of endogenous hnRNPA1 protein was reduced by knockdown of either QKI or PAPD4, and (viii) anti-mitogenic agent lovastatin, which is known to arrest cells in the G1 phase of the cell cycle, increased the poly(A) tail length of p27kip1 mRNA and its protein product through the collaborative action of QKI and PAPD4. From these results, we conclude that QKI-7 recruits PAPD4 to target mRNAs (i.e. hnRNPA1 and p27kip1 mRNAs) so as to induce cytoplasmic polyadenylation of the mRNA and to increase its protein output.
Importantly, QKI was identified as the gene responsible for the characteristic phenotype of body tremor and severe dysmyelination observed in the quaking viable mouse (qkv). Reduced expression of QKI in the oligodendrocyte lineage has been shown to cause the dysmyelination phenotype. Previous studies demonstrated that QKI controls myelination in the CNS by regulating mRNA expression of genes involved in oligodendrocyte differentiation and maturation, which include MBP, p27kip1, and MAP1b mRNAs. Moreover, both QKI and hnRNPA1 regulate an overlapping set of transcripts related to oligodendrocyte differentiation, and QKI positively regulates hnRNPA1 expression (36,38). These results led to the hypothesis that hnRNPA1 is a primary target of QKI, which indirectly regulates oligodendrocyte precursor genes by increasing the intracellular concentration of hnRNPA1 (36). However, how QKI enhances hnRNPA1 expression remained unclear. The results presented here showing that QKI-7 induces the polyadenylation of its target mRNA provide an answer to this question. The mechanism involves PAPD4, also known as Gld-2, which had been previously established as an enzyme responsible for cytoplasmic polyadenylation in the germline and the brain (39,40). Although we did not directly determine whether QKI-7-induced polyadenylation occurred in the cytoplasm or nucleus, it is likely that QKI-7 in complex with PAPD4 induces cytoplasmic polyadenylation of target mRNAs because only QKI-7, which is localized exclusively in the cytoplasm (41), and not other QKI isoforms, specifically induces the reaction.
Interestingly, QKI has been identified as a candidate gene for schizophrenia. A susceptibility locus was mapped to chromosome 6q25–26, in which a 0.5 Mb haplotype containing QKI gene segregated with schizophrenia in a large Swedish pedigree (27). Several genes related to myelin formation, such as myelin basic protein (MBP), proteolipid protein 1 (PLP1) and myelin-associated glycoprotein (MAG) as well as QKI, were shown to be down-regulated in schizophrenic patients (42). Among the QKI isoforms, QKI-7 was severely reduced (27), suggesting that down-regulation of QKI-7 might be the primary cause of decreased expression of myelin-related genes, thereby significantly contributing to the pathology of schizophrenia. As described above, QKI regulates myelin formation by stabilizing MBP, p27kip1 and MAP1b mRNAs (43–46) and also by regulating the alternative splicing of MBP, PLP and MAG mRNAs (47). Ryder and colleagues suggested that QKI-induced up-regulation of hnRNPA1 may alter the regulation of alternative splicing of many genes including MAG by hnRNPA1 (36). Based on our work, it is tempting to speculate that QKI-7 in complex with PAPD4/Gld2 plays a pivotal role in myelination by promoting elongation of the hnRNPA1 mRNA poly(A) tail, disorders of which contribute to the onset of schizophrenia.
In the present study, we also showed that QKI-7, in a complex with PAPD4, induces polyadenylation and translation of p27kip1 mRNA as well as hnRNPA1 mRNA. The cdk inhibitor p27kip1 binds to various cyclin/CDK complexes and typically causes cells to arrest at the G1 phase of the cell cycle. Previous work had shown that both the anti-mitogenic agent lovastatin and density-mediated growth arrest induce p27kip1 by activating p27kip1 translation without affecting its mRNA level (37). Here, we showed that lovastatin induces cytoplasmic polyadenylation and translation activation of p27kip1 mRNA via QKI and PAPD4. We have not determined precisely how QKI and PAPD4 function in lovastatin-induced cytoplasmic polyadenylation of p27kip1; however, previous studies had demonstrated that QKI is a downstream target of the Src protein tyrosine kinases (Src-PTKs). QKI has several proline-rich SH3-binding motifs and C-terminal tyrosine cluster (P motif and Y motif in Figure 1A, respectively), which are important for Src-dependent phosphorylation of QKI. Src-PTKs act transiently to phosphorylate QKI, which negatively regulates the binding of QKI to its target mRNAs. Therefore, it is conceivable that a mitogenic signal inactivates QKI through activation of Src-PTKs that mediate its phosphorylation. On the other hand, since statin was reported to inhibit Src-PTKs, it is also conceivable that anti-mitogenic signals (e.g. statin, contact inhibition) induce activation of QKI through inhibition of Src-PTKs, thereby leading to the cytoplasmic polyadenylation and translational activation of p27kip1 mRNA. These possibilities are now under investigation in our laboratory.
Cytoplasmic polyadenylation by the cytoplasmic polyadenylation element binding protein CPEB has been extensively investigated in the early development of Xenopus oocytes (48). CPEB forms a complex with the cleavage and polyadenylation specificity factor CPSF as well as with the scaffold protein symplekin, which recruits the non-canonical poly(A) polymerase Gld2/PAPD4 to form the cytoplasmic polyadenylation machinery (18). In the present study, we showed that QKI-7 also recruits Gld2/PAPD4 to target mRNAs in somatic cells. Therefore, as with negative regulation by deadenylation in which specific RNA binding proteins recruit deadenylases (i.e. Caf1-Ccr4), recruitment of poly(A) polymerase by specific RNA binding proteins may be a general mechanism for the positive regulation of gene expression. Additional work will be required to elucidate the detailed molecular mechanism for QKI-induced polyadenylation and translational regulation to determine whether other factors such as CPSF and symplekin are also involved.
Recently, dynamic changes in mRNA poly(A) tail length have been reported to play key roles in various biological processes. In some cases, CPEBs have been implicated in these processes. However, in most cases, it has not been determined whether cytoplasmic polyadenylation is involved in the regulatiory mechanism. We believe that the methodology developed in our study will be useful for distinguishing whether the observed poly(A) tail elongation is due to cytoplasmic polyadenylation, which will then pave the way to uncover novel polyadenylation specificity factors in somatic cells in addition to QKI-7 and CPEBs.
Supplementary Material
Acknowledgments
We thank Tadashi Baba, Shin-ichi Kashiwabara and Ann-Bin Shyu for providing antibodies.
Author Contributions: R.Y., S.H. designed the experiments; R.Y., T.T., H.M., T.M. performed the experiments; R.Y., S.H. analyzed the data; R.Y., S.H. wrote the paper; S.H. conceived and directed the study.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministry of Education, Culture, Sports, Science and Technology of Japan Grant-in-Aid for Scientific Research on Innovative Areas ‘RNA regulation’ [No. 20112006, to S.H. in part]; Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research (B) [No. 25291004 to S.H.]; Ministry of Health, Labour and Welfare of Japan Grant-in-Aid [H24-Bsou-kanen-ippan-011 to S.H.]. Funding for open access charge: Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research (B) [No. 25291004 to S.H.]; Ministry of Health, Labour and Welfare of Japan Grant-in-Aid [H27-Bsou-kanen-ippan-011 to S.H.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Wells S.E., Hillner P.E., Vale R.D., Sachs A.B. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 2.Gallie D.R. A tale of two termini: a functional interaction between the termini of an mRNA is a prerequisite for efficient translation initiation. Gene. 1998;216:1–11. doi: 10.1016/s0378-1119(98)00318-7. [DOI] [PubMed] [Google Scholar]
- 3.Gallie D.R. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 4.Coller J., Parker R. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- 5.Shyu A.B., Belasco J.G., Greenberg M.E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 6.Decker C.J., Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 7.Lai W.S., Carballo E., Strum J.R., Kennington E.A., Phillips R.S., Blackshear P.J. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol. Cell. Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sandler H., Kreth J., Timmers H.T., Stoecklin G. Not1 mediates recruitment of the deadenylase Caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic Acids Res. 2011;39:4373–4386. doi: 10.1093/nar/gkr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leppek K., Schott J., Reitter S., Poetz F., Hammond M.C., Stoecklin G. Roquin promotes constitutive mRNA decay via a conserved class of stem-loop recognition motifs. Cell. 2013;153:869–881. doi: 10.1016/j.cell.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 10.Hosoda N., Funakoshi Y., Hirasawa M., Yamagishi R., Asano Y., Miyagawa R., Ogami K., Tsujimoto M., Hoshino S. Anti-proliferative protein Tob negatively regulates CPEB3 target by recruiting Caf1 deadenylase. EMBO J. 2011;30:1311–1323. doi: 10.1038/emboj.2011.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogami K., Hosoda N., Funakoshi Y., Hoshino S. Antiproliferative protein Tob directly regulates c-myc proto-oncogene expression through cytoplasmic polyadenylation element-binding protein CPEB. Oncogene. 2014;33:55–64. doi: 10.1038/onc.2012.548. [DOI] [PubMed] [Google Scholar]
- 12.Chen C.Y., Zheng D., Xia Z., Shyu A.B. Ago-TNRC6 triggers microRNA-mediated decay by promoting two deadenylation steps. Nat. Struct. Mol. Biol. 2009;16:1160–1166. doi: 10.1038/nsmb.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fabian M.R., Mathonnet G., Sundermeier T., Mathys H., Zipprich J.T., Svitkin Y.V., Rivas F., Jinek M., Wohlschlegel J., Doudna J.A., et al. Mammalian miRNA RISC recruits CAF1 and PABP to affect PABP-dependent deadenylation. Mol. Cell. 2009;35:868–880. doi: 10.1016/j.molcel.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zekri L., Huntzinger E., Heimstadt S., Izaurralde E. The silencing domain of GW182 interacts with PABPC1 to promote translational repression and degradation of microRNA targets and is required for target release. Mol. Cell. Biol. 2009;29:6220–6231. doi: 10.1128/MCB.01081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J.I., Reed R.B., Grabowski P.J., Artzt K. Function of quaking in myelination: regulation of alternative splicing. Proc. Natl. Acad. Sci. U.S.A. 2002;99:4233–4238. doi: 10.1073/pnas.072090399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novoa I., Gallego J., Ferreira P.G., Mendez R. Mitotic cell-cycle progression is regulated by CPEB1 and CPEB4-dependent translational control. Nat. Cell Biol. 2010;12:447–456. doi: 10.1038/ncb2046. [DOI] [PubMed] [Google Scholar]
- 17.Burns D.M., D'Ambrogio A., Nottrott S., Richter J.D. CPEB and two poly(A) polymerases control miR-122 stability and p53 mRNA translation. Nature. 2011;473:105–108. doi: 10.1038/nature09908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barnard D.C., Ryan K., Manley J.L., Richter J.D. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 19.Charlesworth A., Wilczynska A., Thampi P., Cox L.L., MacNicol A.M. Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation. EMBO J. 2006;25:2792–2801. doi: 10.1038/sj.emboj.7601159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu L., Good P.J., Richter J.D. The 36-kilodalton embryonic-type cytoplasmic polyadenylation element-binding protein in Xenopus laevis is ElrA, a member of the ELAV family of RNA-binding proteins. Mol. Cell. Biol. 1997;17:6402–6409. doi: 10.1128/mcb.17.11.6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vishnu M.R., Sumaroka M., Klein P.S., Liebhaber S.A. The poly(rC)-binding protein alphaCP2 is a noncanonical factor in X. laevis cytoplasmic polyadenylation. RNA. 2011;17:944–956. doi: 10.1261/rna.2587411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmid M., Kuchler B., Eckmann C.R. Two conserved regulatory cytoplasmic poly(A) polymerases, GLD-4 and GLD-2, regulate meiotic progression in C. elegans. Genes Dev. 2009;23:824–836. doi: 10.1101/gad.494009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kondo T., Furuta T., Mitsunaga K., Ebersole T.A., Shichiri M., Wu J., Artzt K., Yamamura K., Abe K. Genomic organization and expression analysis of the mouse qkI locus. Mamm. Genome. 1999;10:662–669. doi: 10.1007/s003359901068. [DOI] [PubMed] [Google Scholar]
- 24.Chenard C.A., Richard S. New implications for the QUAKING RNA binding protein in human disease. J. Neurosci. Res. 2008;86:233–242. doi: 10.1002/jnr.21485. [DOI] [PubMed] [Google Scholar]
- 25.Ebersole T.A., Chen Q., Justice M.J., Artzt K. The quaking gene product necessary in embryogenesis and myelination combines features of RNA binding and signal transduction proteins. Nat. Genet. 1996;12:260–265. doi: 10.1038/ng0396-260. [DOI] [PubMed] [Google Scholar]
- 26.Wu J., Zhou L., Tonissen K., Tee R., Artzt K. The quaking I-5 protein (QKI-5) has a novel nuclear localization signal and shuttles between the nucleus and the cytoplasm. J. Biol. Chem. 1999;274:29202–29210. doi: 10.1074/jbc.274.41.29202. [DOI] [PubMed] [Google Scholar]
- 27.Aberg K., Saetre P., Jareborg N., Jazin E. Human QKI, a potential regulator of mRNA expression of human oligodendrocyte-related genes involved in schizophrenia. Proc. Natl. Acad. Sci. U.S.A. 2006;103:7482–7487. doi: 10.1073/pnas.0601213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruan L., Osawa M., Hosoda N., Imai S., Machiyama A., Katada T., Hoshino S., Shimada I. Quantitative characterization of Tob interactions provides the thermodynamic basis for translation termination-coupled deadenylase regulation. J. Biol. Chem. 2010;285:27624–27631. doi: 10.1074/jbc.M110.138867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Funakoshi Y., Doi Y., Hosoda N., Uchida N., Osawa M., Shimada I., Tsujimoto M., Suzuki T., Katada T., Hoshino S. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes Dev. 2007;21:3135–3148. doi: 10.1101/gad.1597707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ogami K., Cho R., Hoshino S. Molecular cloning and characterization of a novel isoform of the non-canonical poly(A) polymerase PAPD7. Biochem. Biophys. Res. Commun. 2013;432:135–140. doi: 10.1016/j.bbrc.2013.01.072. [DOI] [PubMed] [Google Scholar]
- 31.Saito S., Hosoda N., Hoshino S. The Hbs1-Dom34 protein complex functions in non-stop mRNA decay in mammalian cells. J. Biol. Chem. 2013;288:17832–17843. doi: 10.1074/jbc.M112.448977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakanishi T., Kubota H., Ishibashi N., Kumagai S., Watanabe H., Yamashita M., Kashiwabara S., Miyado K., Baba T. Possible role of mouse poly(A) polymerase mGLD-2 during oocyte maturation. Dev. Biol. 2006;289:115–126. doi: 10.1016/j.ydbio.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 33.Ezzeddine N., Chang T.C., Zhu W., Yamashita A., Chen C.Y., Zhong Z., Yamashita Y., Zheng D., Shyu A.B. Human TOB, an antiproliferative transcription factor, is a poly(A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation. Mol. Cell. Biol. 2007;27:7791–7801. doi: 10.1128/MCB.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Proudfoot N.J. Ending the message: poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.D'Ambrogio A., Nagaoka K., Richter J.D. Translational control of cell growth and malignancy by the CPEBs. Nat. Rev. Cancer. 2013;13:283–290. doi: 10.1038/nrc3485. [DOI] [PubMed] [Google Scholar]
- 36.Zearfoss N.R., Clingman C.C., Farley B.M., McCoig L.M., Ryder S.P. Quaking regulates Hnrnpa1 expression through its 3′ UTR in oligodendrocyte precursor cells. PLoS Genet. 2011;7:e1001269. doi: 10.1371/journal.pgen.1001269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hengst L., Reed S.I. Translational control of p27Kip1 accumulation during the cell cycle. Science. 1996;271:1861–1864. doi: 10.1126/science.271.5257.1861. [DOI] [PubMed] [Google Scholar]
- 38.Zhao L., Mandler M.D., Yi H., Feng Y. Quaking I controls a unique cytoplasmic pathway that regulates alternative splicing of myelin-associated glycoprotein. Proc. Natl. Acad. Sci. U.S.A. 2010;107:19061–19066. doi: 10.1073/pnas.1007487107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L., Eckmann C.R., Kadyk L.C., Wickens M., Kimble J. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans. Nature. 2002;419:312–316. doi: 10.1038/nature01039. [DOI] [PubMed] [Google Scholar]
- 40.Rouhana L., Wang L., Buter N., Kwak J.E., Schiltz C.A., Gonzalez T., Kelley A.E., Landry C.F., Wickens M. Vertebrate GLD2 poly(A) polymerases in the germline and the brain. RNA. 2005;11:1117–1130. doi: 10.1261/rna.2630205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hardy R.J., Loushin C.L., Friedrich V.L., Jr, Chen Q., Ebersole T.A., Lazzarini R.A., Artzt K. Neural cell type-specific expression of QKI proteins is altered in quakingviable mutant mice. J. Neurosci. 1996;16:7941–7949. doi: 10.1523/JNEUROSCI.16-24-07941.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aberg K., Saetre P., Lindholm E., Ekholm B., Pettersson U., Adolfsson R., Jazin E. Human QKI, a new candidate gene for schizophrenia involved in myelination. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2006;141:84–90. doi: 10.1002/ajmg.b.30243. [DOI] [PubMed] [Google Scholar]
- 43.Li Z., Zhang Y., Li D., Feng Y. Destabilization and mislocalization of myelin basic protein mRNAs in quaking dysmyelination lacking the QKI RNA-binding proteins. J. Neurosci. 2000;20:4944–4953. doi: 10.1523/JNEUROSCI.20-13-04944.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Larocque D., Galarneau A., Liu H.N., Scott M., Almazan G., Richard S. Protection of p27(Kip1) mRNA by quaking RNA binding proteins promotes oligodendrocyte differentiation. Nat. Neurosci. 2005;8:27–33. doi: 10.1038/nn1359. [DOI] [PubMed] [Google Scholar]
- 45.Zhao L., Ku L., Chen Y., Xia M., LoPresti P., Feng Y. QKI binds MAP1B mRNA and enhances MAP1B expression during oligodendrocyte development. Mol. Biol. Cell. 2006;17:4179–4186. doi: 10.1091/mbc.E06-04-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang G., Fu H., Zhang J., Lu X., Yu F., Jin L., Bai L., Huang B., Shen L., Feng Y. RNA-binding protein quaking, a critical regulator of colon epithelial differentiation and a suppressor of colon cancer. Gastroenterology. 2010;138:231–240. doi: 10.1053/j.gastro.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu J.I., Reed R.B., Grabowski P.J., Artzt K. Function of quaking in myelination: regulation of alternative splicing. Proc. Natl. Acad. Sci. U.S.A. 2002;99:4233–4238. doi: 10.1073/pnas.072090399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hake L.E., Richter J.D. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell. 1994;79:617–627. doi: 10.1016/0092-8674(94)90547-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.