Abstract
Epithelial-to-mesenchymal transition (EMT) is a complex multistep process in which phenotype switches are mediated by a network of transcription factors (TFs). Systematic characterization of all dynamic TFs controlling EMT state transitions, especially for the intermediate partial-EMT state, represents a highly relevant yet largely unexplored task. Here, we performed a computational analysis that integrated time-course EMT transcriptomic data with public cistromic data and identified three synergistic master TFs (ETS2, HNF4A and JUNB) that regulate the transition through the partial-EMT state. Overexpression of these regulators predicted a poor clinical outcome, and their elimination readily abolished TGF-β-induced EMT. Importantly, these factors utilized a clique motif, physically interact and their cumulative binding generally characterized EMT-associated genes. Furthermore, analyses of H3K27ac ChIP-seq data revealed that ETS2, HNF4A and JUNB are associated with super-enhancers and the administration of BRD4 inhibitor readily abolished TGF-β-induced EMT. These findings have implications for systematic discovery of master EMT regulators and super-enhancers as novel targets for controlling metastasis.
INTRODUCTION
Epithelial-to-mesenchymal transition (EMT) is a fundamental developmental process that has been associated with metastasis, in which carcinoma cells lose their epithelial features and acquire invasive properties (1–4). Although metastasis is the main cause of cancer-related death, current EMT markers have not been consistently associated with a poor clinical outcome (5). This disparity is partly because the molecular mechanism of EMT is strongly dependent on the cellular context in which it happens (2,3) and the associated gene regulatory networks (GRNs) are often only partially characterized. For example, TGF-β signaling can induce EMT in numerous cell lines and during the differentiation of multiple tissues and organs, but very different transcriptional responses are observed in different cell types (6,7) and the underlying mechanisms are largely unknown. Recently, it has been shown that cell-type-specific master transcription factors (TFs) are responsible for directing SMADs to gene targets thus can determine cell-type-specific responses of TGF-β signaling (8). This observation is consistent with the extensively characterized transcriptional regulation of the E-cadherin gene during EMT where more than ten TFs have been reported to repress E-cadherin expression in numerous cellular models (2,3), suggesting that additional undiscovered factors could regulate EMT.
Another limitation in our current understanding of EMT is that the established transcriptional regulators are typically studied individually. Hence, the synergistic action among cooperative TFs, which has emerged as a general characteristic of enhancers (9), has not been explored in the transcriptional programs of EMT. Moreover, super-enhancers, which are characterized by unusually high cumulative binding of multiple TFs, show more sensitivity than regular enhancers toward inhibitors of BRD4, a transcriptional coactivator that is generally involved in enhancer function (10). Because master TFs that dictate transcriptional programs in numerous cellular models have been associated with super-enhancers (11,12), it is of particular interest to examine whether master TFs regulating EMT are also associated with super-enhancers and consequently, the utility of BRD4 inhibitors to abolish EMT.
In addition to employing complex molecular mechanisms, EMT also exhibits phenotype dynamics that is characterized by progressive loss of epithelial hallmarks coupled with gradual gain of mesenchymal properties (3,4). Emerging evidence suggests the existence of a partial-EMT (P) state that serves as an intermediate state between the epithelial state (E) to mesenchymal state (M) transition (13–16). The three-state model of EMT suggests that a clear understanding of the partial-EMT state could provide key insights into the metastatic process. Thus far, EMT has been largely studied by analyzing the beginning epithelial state or the ending mesenchymal state, mainly due to the tremendous difficulty in isolating partial-EMT cells (17). However, a number of cellular models, such as TGF-β-induced EMT, have been established (18–20). Thus, a time-course analysis of cells undergoing EMT could reveal the essential characteristics of partial-EMT cells.
In this study, we addressed the hypothesis that the temporal activation of multiple cooperative master TFs mediated by super-enhancers might characterize the partial-EMT state. To this aim we conducted a systematic analysis of the time-course gene expression profile of TGF-β-induced EMT and of public cistromic data. We identified three novel synergistic master TFs that are associated with super-enhancers. The cumulative binding of the master TFs characterizes EMT-associated genes and the synergistic action of the master regulators is essential for the partial-EMT state. Our analysis demonstrated the utility of systematic approaches to identify master regulators of EMT and suggested that super-enhancer-associated master regulators may be valuable for the diagnosis and therapeutic intervention of metastasis.
MATERIALS AND METHODS
Cell culture
A549 cells were maintained in Dulbecco's Modified Eagle's Medium:Nutrient Mixture F-12 (DMEM/F-12) (Corning) supplemented with 10% fetal bovine serum (FBS). To induce EMT, A549 cells were seeded 24 h before induction to reach ∼70% confluence. The induction of EMT was performed following published protocols (21). Specifically, TGF-β1 (Pepro Tech) was added to a final concentration of 4 ng/ml and medium with TGF-β1 was replaced every other day.
RNA isolation and quantitative real-time PCR
Total RNA was isolated from cells using TRIzol reagent (TaKaRa). Reverse transcription was performed using the PrimeScript™ RT Reagent Kit (TaKaRa) and gDNA Eraser (Perfect Real Time) according to the manufacturers’ instructions. Quantitative real-time PCR (qPCR) was performed using the SYBR Premix Ex Taq (Tli RNase H Plus) system on an ABI Step One Plus machine (Applied Biosystems). The experiments were performed in triplicate, and the values were normalized to that of GAPDH. For absolute qPCR, the standard curves were generated using coding regions of SNAI1, SNAI2 and JUNB as described by Denzler et al. (22). The primer sequences are as follows: ETS2 qPCR fwd, CCCCTGTGGCTAACAGTTACA; ETS2 qPCR rev, AGGTAGCTTTTAAGGCTTGACTC; HNF4A qPCR fwd, CACGGGCAAACACTACGGT; HNF4A qPCR rev, TTGACCTTCGAGTGCTGATCC; JUNB qPCR fwd, ACAAACTCCTGAAACCGAGCC; JUNB qPCR rev, CGAGCCCTGACCAGAAAAGTA; FOXP1 qPCR fwd, TGGCATCTCATAAACCATCAGC; FOXP1 qPCR rev, GGTCCACTCATCTTCGTCTCAG; CDH1 qPCR fwd, TCAGGCGTCTGTAGAGGCTT; CDH1 qPCR rev, ATGCACATCCTTCGATAAGACTG; CDH2 qPCR fwd, ACAGTGGCCACCTACAAAGG; CDH2 qPCR rev, CCGAGATGGGGTTGATAATG; SNAI1 qPCR fwd, GGCCCACCTCCAGACCCACT; SNAI1 qPCR rev, GCGGGGACATCCTGAGCAGC; SNAI2 qPCR fwd, TGTGACAAGGAATATGTGAGCC; SNAI2 qPCR rev, TGAGCCCTCAGATTTGACCTG; GAPDH qPCR fwd, GAGTCAACGGATTTGGTCGT; GAPDH qPCR rev, GATCTCGCTCCTGGAAGATG.
Transient siRNA knockdown
Cells were seeded in six-well plates at a density of 1 × 10 5 cells/well and were cultured until they reached 40% confluence. Cells were then transfected with a negative control siRNA (Silencer ®, Life Technologies) or with siRNAs targeting JUNB, ETS2 and HNF4α at a final concentration of 100 nM using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. Twenty-four hours after transfection, TGF-β1 was added to the cell culture to induce EMT. The mRNAs and proteins were extracted for analysis of the effects of the siRNAs at 24 h into EMT. The following are the siRNA sequences: ETS2, sense 5′-GUCUGGUGAACGUGAAUCUTT-3′, antisense 5′-AGAUUCACGUUCACCAGACTT-3′; HNF4A, sense 5′-ACACCACCCUGGAAUUUGATT-3′, antisense 5′-UCAAAUUCCAGGGUGGUGUTT-3′; JUNB, sense 5′-ACAAGGUGAAGACGCUCAATT-3′, antisense 5′-UUGAGCGUCUUCACCUUGUTT-3′.
Transwell migration and invasion assay
The in vitro cell migration assay was performed using Transwell chambers (8-μm pore size; Costar). A549 cells transfected with siRNAs were plated 24 h post-transfection in serum-free medium (5 × 10 4 cells per Transwell). TGF-β1 was added to upper chamber at a final concentration of 5 ng/ml. Medium containing 10% FBS in the lower chamber served as a chemoattractant. In the case of JQ1, 500 nM compound was added to both upper and lower chambers 6 h after the addition of TGF-β1. After 24 h, the non-migrating cells were removed from the upper face of the filters using cotton swabs and the migratory cells located on the lower side of the chamber were stained with crystal violet, air dried, photographed and counted. Images of six random fields at 10× magnification were captured from each membrane, and the number of migratory cells was counted. Similar inserts coated with Matrigel were used to determine cell's invasive potential in the invasion assay.
Fluorescence microscopy and staining for ETS2, HNF4A, JUNB and SMAD3
2×105 A549 cells were seeded in a 30-mm confocal dish. 24 h into TGF-β-induced EMT, cells were fixed in 4% paraformaldehyde, permeabilized in 0.2% Triton X-100 and probed with specific primary antibodies for ETS2 (Santa Cruz, sc-365666), HNF4A (Santa Cruz, sc-6557), JUNB (CST, 3753) or SMAD3 (CST, C67H9). The primary antibodies were then detected using matching fluorescent secondary antibodies. To detect nuclei, cells were co-stained with 4′-6-diamidino-2-phenylindole (DAPI; Invitrogen). Cells were observed on a Leica TCS SP8 Confocal Laser Scanning Microscope. Images were analyzed with Zeiss ZEN software.
Immunoblotting analyses
Protein lysates were prepared in the presence of PIC (protease–inhibitor complex) and PMSF. Twenty-microgram aliquots were separated on 8% SDS-polyacrylamide electrophoresis gels, and the proteins were transferred onto a polyvinylidene difluoride membrane (Merck Millipore). The membrane was incubated for 1 h in blocking buffer (Tris-buffered saline containing 0.1% Tween (TBS-T), and 5% non-fat dry milk) followed by incubation overnight at 4°C with the primary antibodies for ETS2 (Santa Cruz, sc-365666), HNF4A (Santa Cruz, sc-6557), JUNB (CST, 3753), FOXP1 (Abcam, ab16645), CDH1 (CST, 3195S) and CDH2 (CST, 14215S). After washing with TBS-T, the blot was incubated with horseradish peroxidase (HRP)-conjugated secondary antibody and the signals were visualized using an enhanced chemiluminescence system according to the manufacturer's instructions (Kodak).
Immunoprecipitations (IPs)
Twenty-four hours into TGF-β-induced EMT, A549 cells were washed, and whole cell lysates were prepared in lysis buffer containing 20 mM HEPES–KOH (pH 7.9), 150 mM NaCl, 0.5% Nonidet P-40, 1 mM EDTA, and 10% glycerol and freshly supplemented with 10 mM NaF, 1 mM Na3VO4, 1 mM PMSF, and a protease inhibitor mixture (Sigma). Antibodies directed against ETS2 (Santa Cruz, sc-351), HNF4A (Santa Cruz, sc-6557) or JUNB (CST, 3753) were added to 1 mg of a whole cell lysate and this mixture was incubated for 3 h with rotation at 4°C. Prior to adding the antibodies, an aliquot was removed for use as the input sample. Pre-equilibrated Protein A (for rabbit antibody), Protein G (for mouse antibody) or Protein A+G (for goat antibody) Dynabeads in cell-lysis buffer were added to the whole cell lysate/antibody mixtures, and these preparations were incubated overnight with rotation at 4°C. The next day, the Dynabeads were immunomagnetically captured and were washed three times using PBS. The pulled-down proteins were then extracted in sample buffer, separated using SDS-PAGE, and subjected to immunoblotting using the appropriate antibodies followed by detection with Clean-Blot™ IP Detection Kit (HRP) (Thermo Fisher Scientific). All of the co-IPs were repeated two or more times.
ChIP-qPCR
The ChIP assay for BRD4, ETS2, HNF4A and JUNB were performed using HighCell ChIP kit (Diagenode) according to the manufacturer's instructions. A549 cells were harvested at 48 h into TGF-β-induced EMT and 10 7 cells were used per ChIP assay. Chromatin was sonicated to an average fragment size of 800 bp using a Qsonica Q700 sonicator. A fraction (1%) of the sonicated chromatin was used as ‘input’ DNA and the qPCR results were analyzed using the Percent Input Method. The antibodies used for ChIP assay are BRD4 (Bethyl, A301-985A); ETS2 (Santa cruz, sc-365666); HNF4A (Abcam, ab41898) and JUNB (CST, 3753). The primer sequences for qPCR are as follows: EGFR qPCR fwd, GGTTCAAAGAGCAGACGTGG; EGFR qPCR rev, CTGAGAAGCCACGGAAGAGA; ETS2 qPCR fwd, TCTAGGGAGAGGGAACAGCT; ETS2 qPCR rev, CATTCTCAACACAGCAGCCA; HNF4A qPCR fwd, GGTTTTCCCACTACTTCCTGC; HNF4A qPCR rev, GAACCCAGAGCCAGGTGTAT; JUNB qPCR fwd, TTTACAAGGACACGCGCTTC; JUNB qPCR rev, CTTTCCTGGCGTCGTTTCC; VIM qPCR fwd, GTTTCCTCGTTCCCCTTTGG; VIM qPCR rev, GAATTGCTCGTGGGTTGTGT.
RNA sequencing and data analysis
The sequencing libraries were constructed according to the protocol for the Illumina TruSeq Sample preparation kit. Sequencing was performed on the Illumina HiSeq 2500 sequencer. Library construction and sequencing were performed at the Genergy Biotech Co., Ltd. (Shanghai). Paired-end RNA-seq reads with 101 bp at each end were aligned to the ENSEMBL version 75 transcriptome database using the Bowtie program (23). The levels of gene expression were quantified using RSEM software (24). Differential expression analysis was conducted using maSigPro (25), and the significant genes were selected using an FDR cutoff value of 0.05.
Compiling the list of EMT-related genes
We collected the genes with a well-documented role in EMT from several recent reviews (1–3). Those genes were then combined with the four regulators identified in this study (ETS2, HNF4A, JUNB and FOXP1) and genes in the QIAGEN EMT PCR Array to generate a non-redundant list of 120 genes. The gene list is then partitioned into E-markers (overexpressed in epithelial state) and M-markers (overexpressed in partial-EMT or mesenchymal state). The list of E-markers is as follows: AREG, CDH1, COL1A2, CXCL5, ELF3, ELF5, ERBB3, FGFBP1, FOXA1, FOXA2, FOXO3A, FZD7, GCLC, GDF15, GSC, IL1RN, KRT19, MITF, MST1R, OCLN, PTP4A1, RAB27B, SIP1, SMAD3, SNAI3, STEAP1, TM4SF18, TSPAN13. The list of M-markers is as follows: AHNAK, AKT1, BMP7, CALD1, CAMK2N1, CAV2, CD59, CDH2, COL3A1, COL5A2, CTGF, CTNNB1, DSC2, DSP, E47, EGFR, ESR1, ETS2, F11R, FN1, FOXC2, FOXD3, FOXF1, FOXP1, FOXQ1, GATA4, GATA6, GNG11, GSK3B, HMGA2, HNF4A, IGFBP4, ILK, ITGA5, ITGAV, ITGB1, JAG1, JUNB, KLF8, KRT14, KRT7, MAP1B, MMP2, MMP3, MMP9, MSN, NODAL, NOTCH1, NUDT13, PDGFRB, PDK4, PLEK2, PPPDE2, PRDX1, PRX1, PTK2, RAC1, RGS2, SERPINE1, SIX1, SMAD2, SNAI1, SNAI2, SOX10, SOX4, SOX9, SPARC, SPP1, STAT3, TCF3, TCF4, TFPI2, TGFB1, TGFB2, TGFB3, TIMP1, TMEFF1, TMEM132A, TPM1, TWIST1, TWIST2, VCAN, VIM, VPS13A, WNT11, WNT5A, WNT5B, ZEB1, ZEB2, ZNF703.
Motif enrichment analysis
We used the Clover algorithm with the default parameter settings for the motif enrichment analysis (26). Human chromosome 20 sequences were used as the background. The enhancer sequences were extracted from the hg19 genome using the coordinates derived from the ENCODE A549 H3K27ac ChIP-seq data that was also enriched with H3K4me1 but not H3K4me3 signals. Enhancers were classified into three categories: proximal enhancers (located within 10 kb upstream of transcription start sites), distal enhancers (located greater than 10 kb upstream of transcription start sites) and enhancers within gene body and were analyzed separately with Clover.
DREM dynamic regulatory network analysis
We used DREM version 2.0 software (27) to model, analyze and visualize dynamic GRN during EMT, using the following parameters: Filtering Options = Minimum Absolute Expression Change 2 (difference from 0); Search Options = Maximum number of paths out of a split: 2; Model Selection Options = Penalized Likelihood, Node Penalty: 40; Expression Scaling Options = Incorporate expression in regulator data for TF; Minimum TF expression after scaling: 5; Default values were set for other parameters. The key regulators were selected according to the split scores (score threshold < 0.001).
ChIP-seq data from published sources
We used published ChIP-seq data from the Gene Expression Omnibus database for ETS2 (GSE49402), HNF4A (GSE25021 and GSE23436), JUNB (GSE51142), FOXP1 (GSE30992 and GSE51142), SMAD3 (GSE41580 and GSE51510), H3K27ac (GSE36204, GSE29611, GSE17312, GSE26320, GSE52658, GSE16256, GSE27823, GSE40129 and GSE36354) and RNAPII ChIA-PET (GSE33664).
ChIP-seq data analysis and visualization
We downloaded the processed ChIP-seq data from the Cistrome data browser (28) and visualized the ChIP-seq peaks using the UCSC genome browser and the WashU EpiGenome browser. The ChIP-seq peak gene assignment and peak overlap were analyzed using HOMER software (http://homer.salk. edu/homer/). The heatmap matrices were created by counting the tags using a 6-kb window (±3 kb of the ETS2 peak center) and a 25-bp bin size. The tag-density matrix was created by counting the tags using a 3-kb window (±1.5 kb of the ETS2 peak center) and a 25-bp bin size. Super-enhancers were identified as described by Whyte et al. All of the plots were generated using R version 3.1.1 software.
Statistical and survival analysis
Survival analysis was performed using the following public lung cancer data sets: TCGA Lung Adenocarcinoma, GSE50081 and GSE31210. In the case of the TCGA RNA-seq data, samples overexpressing target gene were identified using the procedure outlined by cBioPortal (29,30). Briefly, the gene expression levels in diploid samples (samples harboring no amplification or deletion of the target gene) were used to calculate the mean value and standard deviation for the analyzed gene. In the case of the microarray gene expression values, the R package mclust was used to identify bimodal expression patterns and the lower-expressing cluster was used to calculate the mean value and standard deviation. Samples overexpressing the target gene were then identified using a z -score cutoff of 2. All of the statistical tests and the survival analysis were performed using R version 3.1.1 software.
RESULTS
Time-course gene expression analysis revealed a three-state EMT transcriptome signature
To determine the transcriptomic dynamics over the course of EMT, we used an in vitro model in which the human small cell lung cancer cell line A549 underwent EMT when treated with TGF-β (18). Whereas the control cells maintained the characteristics of an epithelial phenotype, A549 cells treated with TGF-β progressively developed a spindle-like morphology and lost their tight connections (Supplementary Figure S1A). Immunoblotting analysis revealed the progressive loss of an epithelial marker (E-cadherin) and gain of a mesenchymal marker (N-cadherin) (Supplementary Figure S1B), confirming that TGF-β induced EMT in A549 cells.
We then conducted RNA sequencing (RNA-Seq) experiments using TGF-β treated A549 cells with biological replicates at eight time points (Supplementary Figure S2). Global analysis of the EMT transcriptomic data revealed 1,633 differentially expressed genes, which fell into three major clusters (Figure 1A, Supplementary Table S1). The expression of known epithelial-associated genes, such as SMAD3, ELF3 and E-cadherin (also known as CDH1), was, as expected, rapidly downregulated upon TGF-β treatment. In contrast, the expression of mesenchymal-associated genes, such as VIM, FN1 and N-cadherin (also known as CDH2), was induced early and was progressively upregulated throughout the course of EMT. Whereas those two clusters largely recapitulated the classic gene expression changes that occur during EMT, a smaller gene cluster (196 genes) showed transient upregulated expression 12–36 h into EMT. At 12–36 h into EMT, both epithelial and mesenchymal markers were expressed at intermediate levels (Figure 1A), which is consistent with the current definition of partial-EMT. Overall, the transcriptome of A549 cells stimulated with TGF-β to induce EMT exhibited three temporal waves of gene expression, early (epithelial-high), intermediate (partial-EMT-high) and late (mesenchymal-high). This observation is consistent with both the time-course EMT transcriptomic results reported by other groups (31) and the current prevailing regulatory model of EMT, the Cascading Bistable Switches (CBS) model, which predicts three stable states (21).
Figure 1.
Time-course transcriptome analysis reveals dynamics of EMT gene expression program. (A) Heatmap of the time-course gene expression data for A549 cells undergoing TGF-β-induced EMT. The differentially expressed genes are separated into three clusters. Representative EMT genes are indicated. (B) Gene Ontology terms enriched in the three clusters of genes that were differentially expressed during TGF-β-induced EMT.
Next, we analyzed the Gene Ontology (GO) enrichments in each cluster (Figure 1B). The epithelial-high genes were highly enriched with GO terms related to cell cycle. Reduced proliferation is a well-known phenotype associated with TGF-β signaling (6), and the observed enrichment in cell cycle functions for the early-wave genes was consistent with reports demonstrating a reduced proliferation rate in cells undergoing EMT (32). In contrast, the top GO terms enriched in the partial-EMT-high gene cluster were regulation of localization and locomotion, and cell adhesion was the top GO term associated with the mesenchymal-high genes. These results are consistent with the increased cell motility observed upon TGF-β treatment, another hallmark of EMT. Despite these differences, all three clusters were enriched with GO terms related to ‘response to stimulus’, indicating that our clustering captured the key gene expression dynamics of TGF-β-induced EMT.
ETS2, HNF4A and JUNB are putative master TFs of the partial-EMT state
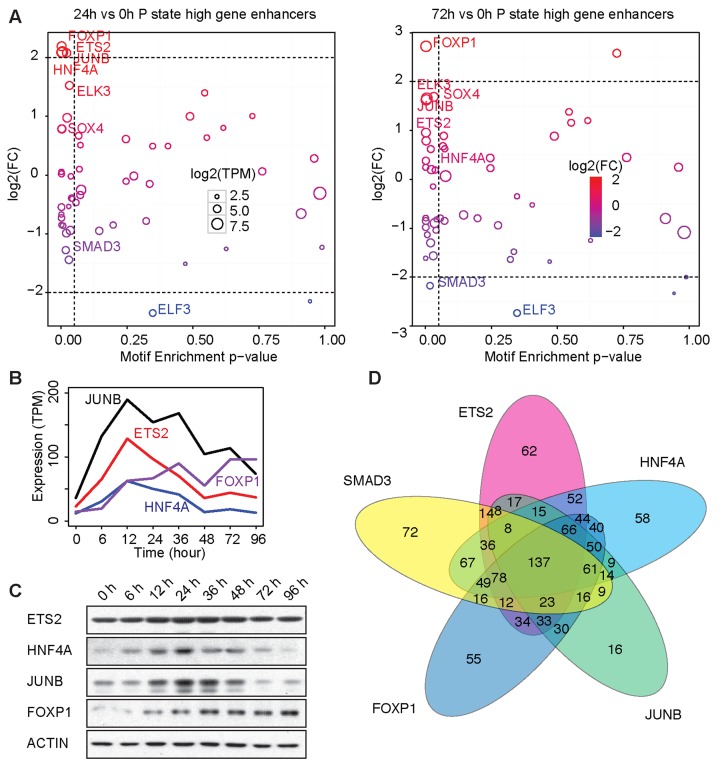
The gene expression dynamics confirmed that A549 cells progressively transited through three distinctive states during TGF-β-induced EMT. These coordinated transcriptomic changes may be the result of differential TF activities. In accordance with the results of previous studies (18), we found that the expression of canonical EMT TFs, such as TWIST1/2 and ZEB1/2, was not significantly altered in A549 cells undergoing TGF-β-induced EMT (Supplementary Figure S3A). Although we observed upregulated SNAI1/2 expression, as reported previously (18), the maximal TPM (transcripts per million) values for SNAI1 (5.58 TPM) and SNAI2 (4.11 TPM), which were confirmed by absolute qPCR (Supplementary Figure S3B), were within the bottom 40% of the values for expressed genes according to the definition of Hebenstreit et al. (33), which suggested that these genes are unlikely to be master TFs for the EMT of A549 cells. Thus additional unknown factors may be involved. We therefore calculated the binding motif enrichment of selected TFs, with the following requirements for each factor: (i) has a known position weight matrix; (ii) with a maximal TPM of >10; (iii) shows differential expression during TGF-β-induced EMT of A549 cells; (iv) is found within enhancers associated with epithelial-high, partial-EMT-high and mesenchymal-high genes. We applied the TPM cutoff to focus on the TFs with relatively high expression levels, an intrinsic characteristic of master TFs that provides advantages in competing for co-factors to dominate the transcriptional landscape (12). To identify potential key regulators responsible for the state transitions, we calculated the fold changes in the expression of each factor during state transition, using their 0-h level of mRNA expression as the reference. We then nominated those TFs with significant binding motif enrichment (P < 0.05) and fold expression change (absolute fold change ≥4) as the potential key regulators (Figure 2A, Supplementary Figure S4). This analysis confirmed the role of known key regulator of EMT, such as SMAD3 and ELF3, in maintaining the epithelial phenotype. Notably, four TFs, ETS2, HNF4A, JUNB and FOXP1, emerged as the top putative regulators of the partial-EMT state in proximal enhancers, distal enhancers and enhancers within gene body (Figure 2A, Supplementary Figure S4). Interestingly, whereas the expression level of FOXP1 increased further upon the transition into the mesenchymal state, the expression levels of ETS2, HNF4A and JUNB fell below the 4-fold cutoff at 72 h into EMT (Figure 2B). We validated the protein levels of the four TFs during EMT (Figure 2C), confirming that ETS2, HNF4A and JUNB were transiently overexpressed 12–36 h into EMT and that FOXP1 displayed monotonic upregulated expression. Consistent with the CBS model of EMT, the E-low, P-high and M-low expression pattern suggested that ETS2, HNF4A and JUNB are likely regulators of the partial-EMT state and that FOXP1 is a candidate driver of the mesenchymal state.
Figure 2.
Integrative analysis identified ETS2, HNF4A, JUNB and FOXP1 as the putative master TFs of EMT. (A) Enrichment P -values of TF binding motifs in the proximal enhancers of partial-EMT high genes (x-axis) were plotted against the fold changes in gene expression during EMT (y-axis; 24 h versus 0 h, left panel; 72 h versus 0 h, right panel). Each circle represents a TF. The dotted lines represent the cutoff for the enrichment (P -value < 0.05) and the expression change (absolute fold change ≥ 4). (B) Levels of ETS2, HNF4A, JUNB and FOXP1 mRNA during EMT as estimated using RNA-seq. (C) Immunoblots showing the levels of ETS2, HNF4A, JUNB and FOXP1 proteins during EMT. (D) Venn diagram showing the overlap of the targets of the five EMT TFs derived from published ChIP-seq data.
To further validate the predicted binding motif enrichment data, we queried the public chromatin immunoprecipitation followed by sequencing (ChIP-seq) data for the known (SMAD3) and putative (ETS2, HNF4A, JUNB and FOXP1) EMT regulators identified in the motif enrichment analysis. Because those ChIP-seq analyses were performed in cells other than A549, we used ENCODE A549 H3K27ac ChIP-seq data as a reference and retained only the TF ChIP-seq peaks that fell within the established A549 cell active enhancers. We then analyzed the ChIP-seq peaks associated with the 1633 genes that were differentially expressed during EMT. Reassuringly, each of the five TFs occupied the enhancers of a substantial number of the 1633 genes, ranging from 512 genes for JUNB to 784 genes for HNF4A (Figure 2D). Furthermore, their target EMT genes that were inferred from ChIP-seq data showed a significant overlap (pair-wise overlap P < 10−16, Fisher's exact test), which was consistent with the binding motif prediction results (Figure 2D).
ETS2, HNF4a and JUNB control the transition through the partial-EMT state in a dynamic GRN model
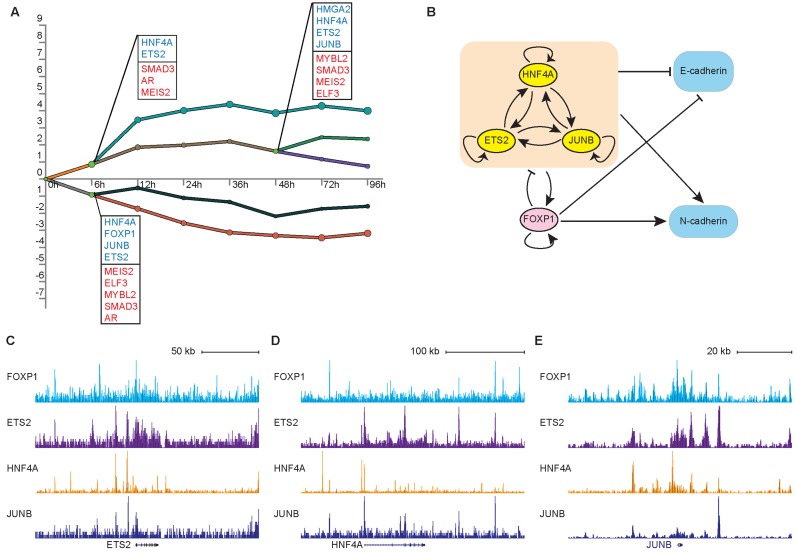
To examine whether the same set of master regulators could be derived using an independent algorithm and to obtain a dynamic, systems-level overview of the roles of the putative EMT regulators, we assembled a dynamic GRN for EMT using the DREM (Dynamic Regulatory Events Miner) software (27). For this analysis, we integrated the time-course gene expression data with the computationally predicted genome-wide binding data for 52 TFs that were differentially expressed during TGF-β-induced EMT. DREM identified a dynamic GRN with prominent splitting points at 6 and 48 h into EMT, which agreed well with the timing of the putative state transitions from E to P and P to M, respectively (Figure 3A). Reassuringly, DREM identified ETS2, HNF4A and JUNB as the top-ranked key regulators at the E to P transition. Similar to the E to P splitting points, ETS2, HNF4A and JUNB were nominated by DREM as the top-ranked regulators at the P to M transition. In addition to ETS2, HNF4a and JUNB, DREM also identified known EMT regulators such as SMAD3, ELF3 and HMGA2 as key TFs operating at the splitting points. Importantly, the expression levels of ETS2, HNF4A and JUNB were upregulated at the splitting points compared to the levels at the 0-h time point, suggesting that they may play more active roles during state transitions than regulators with downregulated expression, such as SMAD3 or ELF3. Overall, the predictions obtained using the two independent computational approaches (DREM and binding motif enrichment) agreed well, further establishing ETS2, HNF4A and JUNB as candidate master regulators of the partial-EMT state.
Figure 3.
ETS2, HNF4A and JUNB are key regulators in a dynamic EMT Gene Regulatory Network model. (A) DREM output showing a dynamic gene regulatory map of EMT. The y-axis shows the levels of gene expression normalized to those at 0 h. Each line represents a path (cluster) of genes with a similar expression pattern, and nodes represent the hidden states of a hidden Markov model. The green nodes represent splitting points. For each splitting point, TFs with a split score < 0.001 were listed in ranked order of scores and were colored according to the expression level changes (blue for upregulation; red for downregulation). (B) Diagram illustrating the putative core regulatory network derived from DREM analysis. (C) Genome browser representation of ETS2, HNF4A, JUNB and FOXP1 binding events near ETS2 gene. (D) Same as (C) for HNF4A gene. (E) Same as (C) for JUNB gene.
To gain further insight into the dynamic GRN associated with EMT, we extracted the core module from the DREM-predicted model, retrieving only the interactions connecting the putative key regulators and the key markers of EMT (CDH1 and CDH2) (Figure 3B). This analysis revealed a highly modular topology, with 22 of 30 possible intra-module interactions implemented (modularity P < 2.2 −16, Fisher's exact test). This module was characterized by feed-forward loops and negative feedback loops, key network motifs to generate bimodality, and was consistent with the canonical EMT regulatory model (21). Specifically, four of four putative EMT regulators had an autoregulatory loop, a common feature of master TFs (12). Finally, the three putative partial-EMT state regulators used a clique motif, the most abundant three-protein interaction motif and one of the most conserved GRN motifs (34,35), suggesting cooperativeness among those regulators.
Validation of the partial-EMT transcriptional regulatory module
According to our dynamic EMT GRN model, the three putative partial-EMT state regulators, ETS2, HNF4A and JUNB, autoregulate and positively regulate each other, repress the expression of the epithelial state markers and upregulate the expression of mesenchymal state regulators/markers. To validate the core partial-EMT regulatory module, we first mined public ChIP-seq data for evidence of direct regulation. Binding events of the putative master TFs on themselves were readily supported by the ChIP-seq data (Figure 3C–E). To validate the ChIP-seq results, we performed ChIP-qPCR on selected sites and confirmed that ETS2, HNF4A and JUNB directly colocalized to their own enhancers in A549 cells (Supplementary Figures S5A and S6). Moreover, gene targets supported by the ChIP-seq data and gene targets supported by the motif enrichment data showed extensive overlap (Supplementary Figure S5B), suggesting that the majority of the predicted regulations were the result of direct TF-DNA interactions.
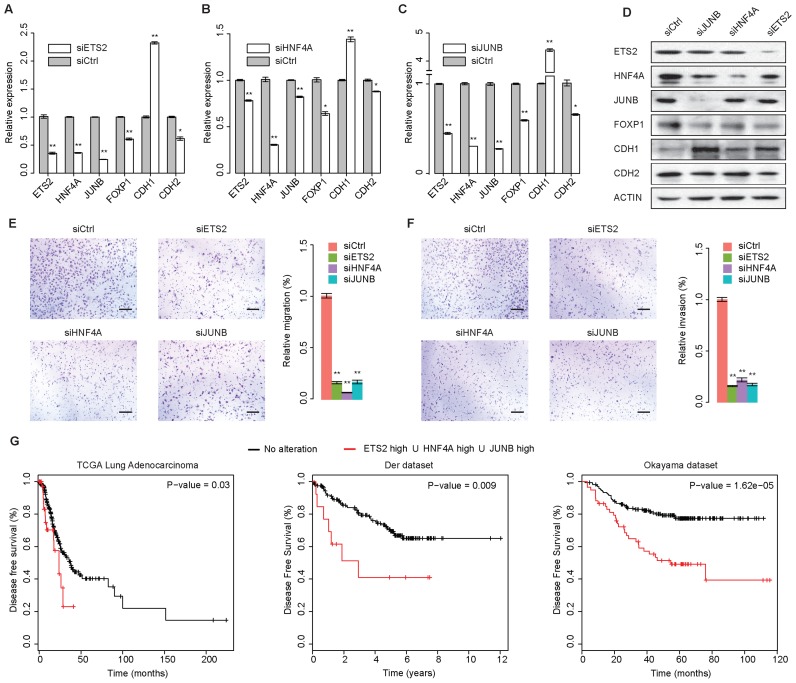
To validate the predicted functional gene regulation by the three putative partial-EMT master regulators, we performed siRNA-mediated silencing of each regulator in A549 cells undergoing TGF-β-induced EMT. To focus on function during the partial-EMT state, siRNAs were introduced into A549 cells 24 h prior to TGF-β treatment, and their effects on mRNA and protein expression were determined 24 h into TGF-β-induced EMT. Compared to the effects of the control siRNAs, siRNAs for ETS2, HNF4A or JUNB significantly reduced the levels of ETS2, HNF4, JUNB, FOXP1 and CDH2 mRNAs and proteins and markedly increased the levels of CDH1 mRNA and protein (Figure 4A–D), which was consistent with the predictions of the putative EMT GRN model. Importantly, knocking down the expression of any one of the ETS2, HNF4A and JUNB trio had significant effects, suggesting that their cooperative activity is critical for EMT. Moreover, treating A549 cells undergoing EMT with siRNAs for ETS2, HNF4A or JUNB substantially reduced their ability to migrate and invade (Figure 4E, F, P < 0.001) without a detectable influence on cell's proliferation or viability (data not shown), supporting the critical roles of these TFs in modulating key phenotypes of EMT. We next tested whether overexpression of the three master TFs was associated with a poor clinical outcome by examining three independent lung adenocarcinoma data sets. We classified the samples into two categories, those that overexpressed at least one of the three master TFs and those in which none of the master TFs was overexpressed. Kaplan-Meier survival analysis revealed that patients with elevated expression of at least one of the three genes had a significantly worse chance of relapse-free survival (Figure 4G), demonstrating that the overexpression of ETS2, HNF4a or JUNB is a robust prognostic indicator.
Figure 4.
ETS2, HNF4A and JUNB play critical roles in partial-EMT. (A) Quantitative reverse transcription polymerase chain reaction results showing the levels of gene expression in A549 cells during TGF-β-induced EMT after silencing ETS2 expression using a specific siRNA. n = 3; error bars indicate mean ± SD. * P < 0.05; ** P < 0.01, determined using the two-tailed Student's t-test. (B) Same as (C) for silencing HNF4A. (C) Same as (A) for silencing JUNB. (D) Immunoblotting analysis of the protein abundance of indicated genes in A549 cells undergoing EMT treated with siRNAs targeting ETS2, HNF4A or JUNB. (E) A549 cells undergoing TGF-β-induced EMT were treated with siRNAs targeting ETS2, HNF4A or JUNB and were subjected to a migration assay. The migratory cells were quantified (bar charts). Scale bars: 100 μm. n = 6; error bars indicate mean ± SD. * P < 0.05; ** P < 0.01, determined using the two-tailed Student's t-test. (F) Same as (E) for the invasion assay. (G) Kaplan–Meier survival analysis based on the ETS2, HNF4A and JUNB expression levels in three independent Lung Adenocarcinoma data sets for disease-free survival.
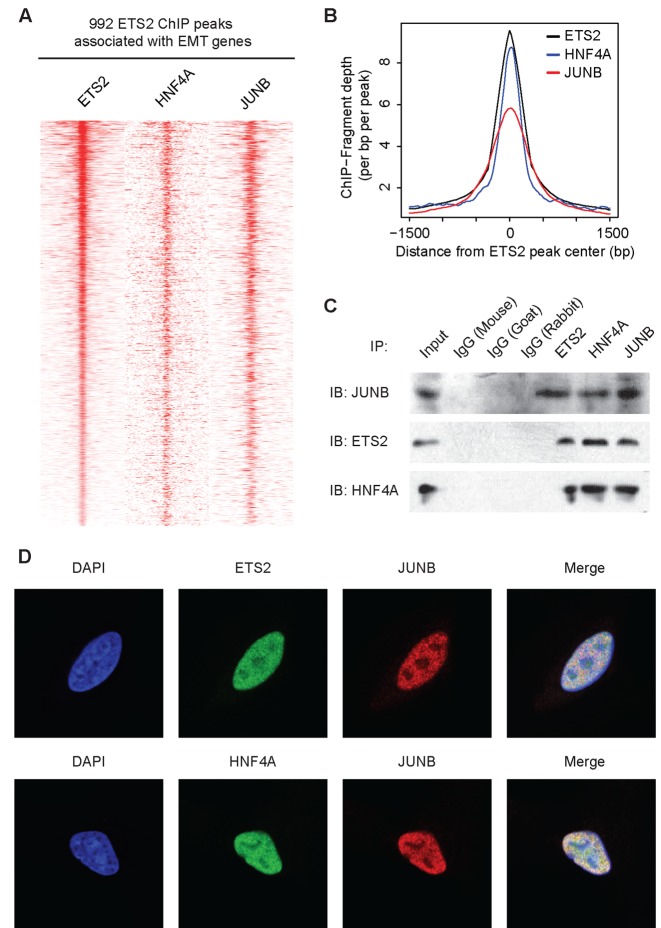
A unique feature of our predicted partial-EMT regulatory circuit was that three master regulators were identified. These master regulators displayed significant co-expression and formed a fully connected module connected by positive feedback loops and autoregulations (a clique). Those features are indicative of cooperative activity. Because a hallmark of enhancer activation is the cooperative assembly of a large trans-factor complex consisting of multiple TFs and their co-factors (9), we proceeded to test the hypothesis that ETS2, HNF4A and JUNB are part of the same trans-activation complex. Consistent with our ChIP-qPCR data, which showed colocalization of ETS2, HNF4A and JUNB on numerous enhancers (Supplementary Figures S5A and S6), examining ChIP-seq data revealed that the binding sites occupied by ETS2 were also highly occupied by HNF4A and JUNB and that the genomic locations of their maximal peak intensities were within 50 bp of each other (Figure 5A, B). We next evaluated the physical interaction of ETS2, HNF4A and JUNB using immunoprecipitation. Compared to the control antibodies, the antibodies specific for each of the three TFs readily pulled down the other two factors (Figure 5C). Moreover, immunofluorescence staining showed that ETS2, HNF4A and JUNB were colocalized in the nucleus of A549 cells 24 h into TGF-β-induced EMT (Figure 5D). Taken together, these data suggested that the three factors indeed are part of the same regulatory complex and cooperatively regulate EMT-associated genes.
Figure 5.
ETS2, HNF4A and JUNB are synergistic master regulators of EMT. (A) Heatmap showing the colocalization of ETS2, HNF4A and JUNB at EMT-associated enhancers. The presence of ETS2, HNF4A and JUNB ChIP-seq peaks are displayed within a 6-kb window centered on the ETS2-bound site. (B) Summary plot for the ETS2, HNF4A and JUNB ChIP-seq peak enrichment across the ETS2-binding site associated with EMT genes. (C) Endogenous association of ETS2, HNF4A and JUNB. At 24 h into TGF-β-induced EMT, A549 cell lysates were prepared and were immunoprecipitated using the indicated antibody. The presence of associated proteins was then analyzed using immunoblotting. (D) Immunofluorescence staining for ETS2, HNF4A or JUNB in A549 cells stimulated with TGF-β for 24 h.
Super-enhancer characterizes master EMT transcriptional regulators
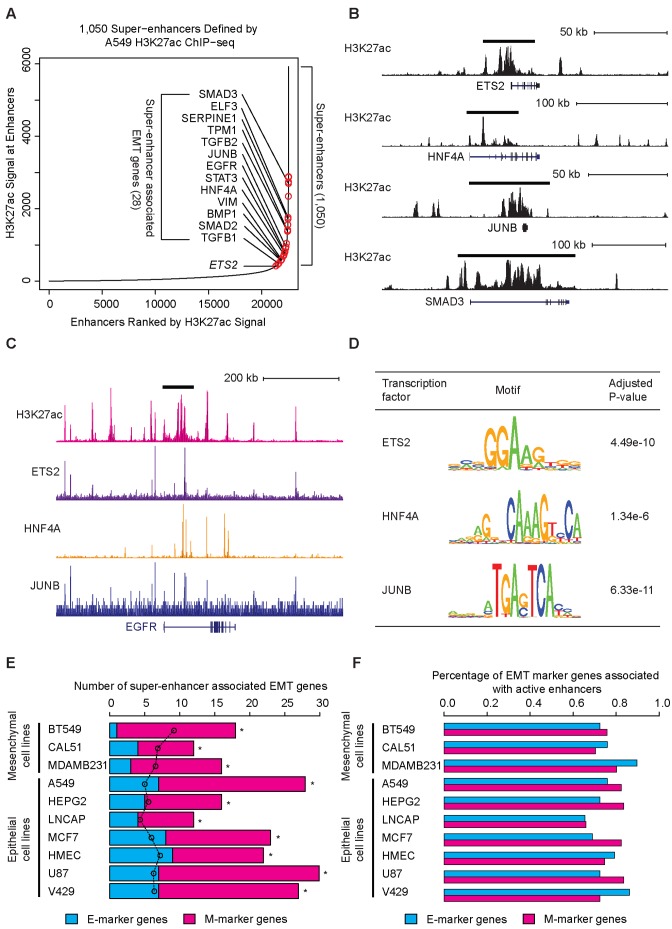
Recent studies suggested that master TFs dominate the transcriptional programs in various cell types and super-enhancers drive the expression of master TFs (10–12,36). To determine whether super-enhancers also characterize the identified EMT master TFs, we analyzed ENCODE A549 H3K27ac ChIP-seq data and identified 1050 super-enhancers (Figure 6A). Whereas the enhancers associated with ETS2 barely missed the cutoff value to be classified as a super-enhancer, HNF4A and JUNB were found to be associated with putative super-enhancers (Figure 6A and B).
Figure 6.
Super-enhancers are associated with master EMT regulators. (A) The distribution of A549 H3K27ac tag intensities revealed 1050 super-enhancers. The red circles indicate EMT genes associated with super-enhancers. (B) Genome browser tracks showing the super-enhancers associated with ETS2, HNF4A, JUNB and SMAD3. The black bars denote super-enhancers. (C) Genome browser tracks showing super-enhancers associated with the EGFR gene and the binding events of ETS2, HNF4A and JUNB around EGFR gene. (D) Table depicting the enrichment of ETS2, HNF4a and JUNB binding motifs in EMT-associated super-enhancers relative to the genomic background. (E) Bar plots showing the enriched association of EMT genes with super-enhancers. Bars represent the observed number of super-enhancer-associated EMT genes and circles represent the expected number of super-enhancer-associated genes assuming no enrichment. * P < 0.001, determined using Fisher's Exact Test. (F) Bar plots showing the universal association of EMT genes with active enhancers. Bars represent the observed percentages of active-enhancer-associated EMT genes.
To test whether other key EMT genes were also associated with super-enhancers, we compiled a list of 120 well-established EMT genes, classified them into epithelial-associated markers (E-markers) and mesenchymal-associated markers (M-markers) and evaluated their intersection with the A549 cell super-enhancer-associated genes. Interestingly, 28 genes from the list of 120 EMT-associated genes were associated with the super-enhancers of A549 cells (Figure 6A, B, Supplementary Figures S7A–C). This enrichment was highly significant (P < 10−14, Fisher's exact test), suggesting that super-enhancers may indeed drive the high expression of numerous key EMT genes. Importantly, binding motifs for ETS2, HNF4A and JUNB were highly enriched in those super-enhancers (P < 10−6), suggesting that those master regulators drive the formation of super-enhancers associated with EMT genes (Figure 6C and D).
We next extended the super-enhancer analyses to 8 additional cell lines. While all examined cell lines demonstrated an enriched association of super-enhancers with EMT markers, neither mesenchymal nor epithelial cell lines displayed biased enrichment toward E-markers or M-markers (Figure 6E, Supplementary Figure S8). The lack of significant association between super-enhancers in mesenchymal cells with M-markers together with the lack of significant association between super-enhancers in epithelial cells with E-markers raised the interesting possibility that EMT marker genes, regardless of their association with epithelial or mesenchymal phenotype, are always marked with active enhancers. To test this hypothesis, we examined the association of all 120 EMT marker genes with active enhancers in the 9 tested cell lines and found that the vast majority of both E-marker and M-marker genes are consistently associated with active enhancers (Figure 6F). This observation is consistent with EMT being a ubiquitous biological process and suggested that the core set of EMT-associated genes are generally active in diverse cell types.
BRD4 inhibitor abolishes TGF-β-induced EMT
BRD4 is a transcriptional coactivator from the bromodomain and extraterminal (BET) subfamily of human bromodomain proteins. Recent studies suggested that BRD4 is generally associated with active enhancers (10). Importantly, JQ1, a recently developed BRD4 inhibitor, preferentially abolishes BRD4 functionality at super-enhancers (10), indicating that JQ1 may abolish TGF-β-induced EMT by inhibiting the master regulators ETS2, HNF4A and JUNB through the disruption of BRD4 functionality. We first analyzed the colocalization of BRD4 to sites occupied by EST2, HNF4A and JUNB by performing ChIP-qPCR assays utilizing the same set of primers used in ETS2, HNF4A and JUNB ChIP-qPCRs. Reassuringly, BRD4 binding is readily detected at tested enhancers near ETS2, HNF4A, JUNB, EGFR and VIM, confirming that BRD4 colocalized to the same sites occupied by ETS2, HNF4A and JUNB (Supplementary Figure S9). Interestingly, while knocking down HNF4A or ETS2 with specific siRNAs significantly reduced BRD4 binding at some of the tested sites, knocking down JUNB consistently reduced BRD4 binding at all the tested sites (Supplementary Figure S9). The result is consistent with a recent proteome-scale protein-protein interaction analysis showed that JUNB makes physical contact with BRD4 (37), suggesting that the master regulator JUNB recruits BRD4 to those sites.
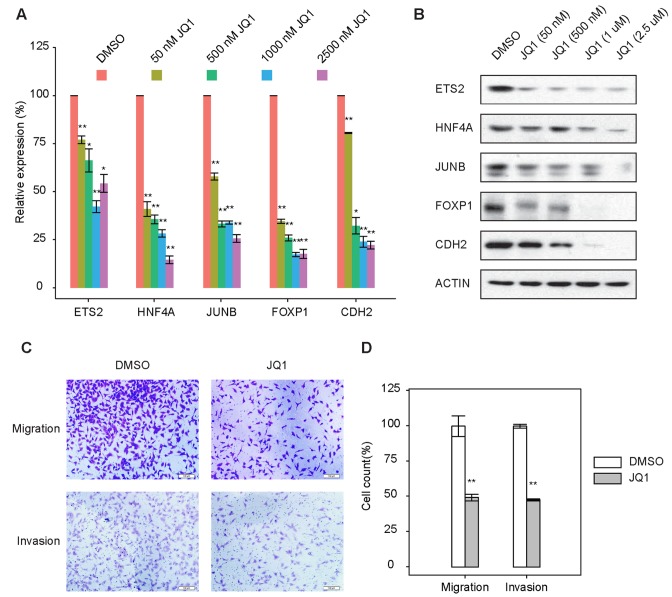
To functionally validate the roles of BRD4 inhibitors in EMT, we treated A549 cells undergoing TGF-β-induced EMT with JQ1, which has been extensively utilized to disrupt super-enhancers (10,36). We tested the effect of JQ1 on the expression of key EMT genes by adding JQ1 to the cell culture medium at 6 h into TGF-β-induced EMT and examined the cells 18 h later. This setup allowed us to specifically target ETS2, HNF4A and JUNB associated super-enhancers at the partial-EMT state without disrupt SMAD3 functionality, which is critical for mediating TGF-β signaling. Indeed, the addition of JQ1 did not interrupt SMAD3 nuclear localization following TGF-β treatment (Supplementary Figure S10). The short JQ1 treatment also excluded molecular and functional impacts from JQ1 induced apoptosis, which occurs after 48 h of JQ1 exposure (38). As expected, the mRNA and protein levels of ETS2, HNF4A and JUNB were significantly reduced compared with those of the DMSO controls (Figure 7A, B), consistent with the association of these genes with super-enhancers. Consequently, the expression of FOXP1 and the mesenchymal marker gene (CDH2) regulated by those master TFs was also downregulated. Furthermore, cells undergoing EMT that were treated with JQ1 displayed significantly reduced abilities to migrate and invade (Figure 7C, D, P < 0.01) and thus were losing key EMT characteristics.
Figure 7.
BRD4 inhibitor abolishes TGF-β-induced EMT. (A) Quantitative reverse transcription polymerase chain reaction results showing the expression levels of indicated genes in A549 cells undergoing EMT that were treated with various concentrations of JQ1. n = 3; error bars indicate mean ± SD. * P < 0.05; ** P < 0.01, as determined using the two-tailed Student's t-test. DMSO, dimethylsulphoxide. (B) Immunoblotting analysis of the expression levels of indicated genes in A549 cells undergoing EMT that were treated with various concentrations of JQ1. (C) A549 cells undergoing TGF-β-induced EMT were treated with 500 nM JQ1 (left panels) or the DMSO control (right panel) and were subjected to a migration assay (top panels) or an invasion assay (bottom panels). (D) The migratory or invasive cells were quantified (bar charts). Scale bars: 100 μm. n = 6; error bars indicate mean ± SD. * P < 0.05; ** P < 0.01, determined using the two-tailed Student's t-test.
DISCUSSION
EMT has been implicated as an integral part of metastasis, and a clear understanding of the underlying molecular mechanism of the latter is crucial for successful clinical therapies. Unfortunately, EMT is a phenomenon that is highly dependent on the cellular context in which it occurs. Despite extensive efforts, the regulatory mechanism of EMT in most cell/tissue types is not fully resolved. Our understanding is particularly poor in the case of partial-EMT, the intermediate state that was recently proposed to hold key insights into metastasis (16). In this study, we employed a systematic approach to identify all potential key regulators and corresponding GRNs of EMT using an integrative analysis of time-course gene expression data and a large repertoire of published ChIP-seq data. We successfully recovered and validated novel partial-EMT regulators using the small cell lung cancer A549 cell line. In principle, our approach can be extended to any system in which time-course gene expression data is accessible and could be valuable for deciphering EMT mechanisms in other cancers.
In sharp contrast to the canonical EMT model, in which a single TF such as SNAI1 of ZEB1 controls the EMT program, we identified multiple master TFs that synergistically control the EMT transcriptional program in A549 cells. The master TFs physically interact and demonstrate cumulative binding at enhancers of EMT-associated genes, suggesting that the synergistic action of multiple master TFs could represent an intrinsic property of the EMT transcriptional program. Similar to the master TFs identified in other cellular models, the master TFs identified in our study are associated with super-enhancers. In addition to A549 cells, JQ1 inhibited migration and invasion of multiple breast and prostate cancer cell lines (data not shown). Moreover, the disruption of BRD4 function by JQ1 effectively inhibits tumor formation and metastasis in numerous mouse models (39,40). Although it is not possible to deconvolute the role of BRD4 inhibitors in EMT using vivo assays owing to the massive apoptosis effects from JQ1 administration (36,38), these observations supported that super-enhancers, the critical role of which in other aspects of tumorigenesis has been demonstrated in several recent studies, are key players in EMT and may together represent a general target for coinhibiting multiple hallmarks of cancers.
Consistent with the mechanism of enhancer formation, we identified 3 synergistic master TFs regulating TGF-β-induced EMT in A549 cells. Interestingly, each of the three factors, ETS2, HNF4A and JUNB, has been previously associated with EMT, albeit in different cellular backgrounds (41–43). Aberrant reactivation of lineage specific TFs is a well-known phenomena during tumorigenesis (44,45). Currently, more than 30 TFs have been implicated in EMT programs underlying a variety of tissue remodeling events (1,2). It is conceivable that many of them could be reactivated during metastasis. Thus, systematic discovery and characterization of their synergistic interactions may offer substantial insights into the molecular mechanisms underlying EMT and may identify novel prognostic markers as well as new therapeutic targets for controlling metastasis.
AVAILABILITY OF SUPPORTING DATA
The data reported in this study have been deposited in GEO under accession number GSE69667.
Supplementary Material
Acknowledgments
The authors would like to thank members of the Laboratory of Systems Biology for participating in helpful discussions and National Center for Protein Science Shanghai for assistance with confocal microscopy.
Authors’ contributions: H.C., Y.L., H.L., S.D. and L.Z. performed the experiments. M.X. conducted the computational analysis. H.C. and H.L. contributed to the computational analysis. P.W. designed and supervised the study, contributed to the computational analysis and wrote the manuscript. All authors have read and approved the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China (NSFC) [31271413]; Science and Technology Commission of Shanghai Municipality [12DZ1910800]; Chinese Academy of Sciences ‘Hundred Talent Program’ (to P.W.). Funding for open access charge: National Natural Science Foundation of China (NSFC) [31271413]; Science and Technology Commission of Shanghai Municipality [12DZ1910800]; Chinese Academy of Sciences ‘Hundred Talent Program’.
Conflict of interest statement. None declared.
REFERENCES
- 1.De Craene B., Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat. Rev. Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 2.Lamouille S., Xu J., Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thiery J.P., Acloque H., Huang R.Y., Nieto M.A. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Yang J., Weinberg R.A. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev. Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 5.Garber K. Epithelial-to-mesenchymal transition is important to metastasis, but questions remain. J. Natl. Cancer Inst. 2008;100:232–233. doi: 10.1093/jnci/djn032. [DOI] [PubMed] [Google Scholar]
- 6.Ikushima H., Miyazono K. TGFbeta signalling: a complex web in cancer progression. Nat. Rev. Cancer. 2010;10:415–424. doi: 10.1038/nrc2853. [DOI] [PubMed] [Google Scholar]
- 7.Massague J., Gomis R.R. The logic of TGFbeta signaling. FEBS Lett. 2006;580:2811–2820. doi: 10.1016/j.febslet.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 8.Mullen A.C., Orlando D.A., Newman J.J., Loven J., Kumar R.M., Bilodeau S., Reddy J., Guenther M.G., DeKoter R.P., Young R.A. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–576. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spitz F., Furlong E.E. Transcription factors: from enhancer binding to developmental control. Nat. Rev. Genet. 2012;13:613–626. doi: 10.1038/nrg3207. [DOI] [PubMed] [Google Scholar]
- 10.Loven J., Hoke H.A., Lin C.Y., Lau A., Orlando D.A., Vakoc C.R., Bradner J.E., Lee T.I., Young R.A. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-Andre V., Sigova A.A., Hoke H.A., Young R.A. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Revenu C., Gilmour D. EMT 2.0: shaping epithelia through collective migration. Curr. Opin. Genet. Dev. 2009;19:338–342. doi: 10.1016/j.gde.2009.04.007. [DOI] [PubMed] [Google Scholar]
- 14.Chao Y., Wu Q., Acquafondata M., Dhir R., Wells A. Partial mesenchymal to epithelial reverting transition in breast and prostate cancer metastases. Cancer Microenviron. 2012;5:19–28. doi: 10.1007/s12307-011-0085-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Theveneau E., Mayor R. Neural crest delamination and migration: from epithelium-to-mesenchyme transition to collective cell migration. Dev. Biol. 2012;366:34–54. doi: 10.1016/j.ydbio.2011.12.041. [DOI] [PubMed] [Google Scholar]
- 16.Jordan N.V., Johnson G.L., Abell A.N. Tracking the intermediate stages of epithelial-mesenchymal transition in epithelial stem cells and cancer. Cell cycle. 2011;10:2865–2873. doi: 10.4161/cc.10.17.17188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu M., Bardia A., Wittner B.S., Stott S.L., Smas M.E., Ting D.T., Isakoff S.J., Ciciliano J.C., Wells M.N., Shah A.M., et al. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science. 2013;339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu J., Hu G., Chen D., Gong A.Y., Soori G.S., Dobleman T.J., Chen X.M. Suppression of SCARA5 by Snail1 is essential for EMT-associated cell migration of A549 cells. Oncogenesis. 2013;2:e73. doi: 10.1038/oncsis.2013.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zavadil J., Bottinger E.P. TGF-beta and epithelial-to-mesenchymal transitions. Oncogene. 2005;24:5764–5774. doi: 10.1038/sj.onc.1208927. [DOI] [PubMed] [Google Scholar]
- 20.Xu J., Lamouille S., Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang J.Y., Tian X.J., Zhang H., Teng Y., Li R.Y., Bai F., Elankumaran S., Xing J.H. TGF-beta-induced epithelial-to-mesenchymal transition proceeds through stepwise activation of multiple feedback loops. Sci. Signal. 2014;7:ra91. doi: 10.1126/scisignal.2005304. [DOI] [PubMed] [Google Scholar]
- 22.Denzler R., Agarwal V., Stefano J., Bartel D.P., Stoffel M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell. 2014;54:766–776. doi: 10.1016/j.molcel.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Conesa A., Nueda M.J., Ferrer A., Talon M. maSigPro: a method to identify significantly differential expression profiles in time-course microarray experiments. Bioinformatics. 2006;22:1096–1102. doi: 10.1093/bioinformatics/btl056. [DOI] [PubMed] [Google Scholar]
- 26.Frith M.C., Fu Y., Yu L., Chen J.F., Hansen U., Weng Z. Detection of functional DNA motifs via statistical over-representation. Nucleic Acids Res. 2004;32:1372–1381. doi: 10.1093/nar/gkh299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schulz M.H., Devanny W.E., Gitter A., Zhong S., Ernst J., Bar-Joseph Z. DREM 2.0: Improved reconstruction of dynamic regulatory networks from time-series expression data. BMC Syst. Biol. 2012;6:104. doi: 10.1186/1752-0509-6-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu T., Ortiz J.A., Taing L., Meyer C.A., Lee B., Zhang Y., Shin H., Wong S.S., Ma J., Lei Y., et al. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol. 2011;12:R83. doi: 10.1186/gb-2011-12-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Javaid S., Zhang J., Anderssen E., Black J.C., Wittner B.S., Tajima K., Ting D.T., Smolen G.A., Zubrowski M., Desai R., et al. Dynamic chromatin modification sustains epithelial-mesenchymal transition following inducible expression of Snail-1. Cell Rep. 2013;5:1679–1689. doi: 10.1016/j.celrep.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evdokimova V., Tognon C., Ng T., Ruzanov P., Melnyk N., Fink D., Sorokin A., Ovchinnikov L.P., Davicioni E., Triche T.J., et al. Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell. 2009;15:402–415. doi: 10.1016/j.ccr.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 33.Hebenstreit D., Fang M., Gu M., Charoensawan V., van Oudenaarden A., Teichmann S.A. RNA sequencing reveals two major classes of gene expression levels in metazoan cells. Mol. Syst. Biol. 2011;7:497. doi: 10.1038/msb.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stergachis A.B., Neph S., Sandstrom R., Haugen E., Reynolds A.P., Zhang M., Byron R., Canfield T., Stelhing-Sun S., Lee K., et al. Conservation of trans-acting circuitry during mammalian regulatory evolution. Nature. 2014;515:365–370. doi: 10.1038/nature13972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeger-Lotem E., Sattath S., Kashtan N., Itzkovitz S., Milo R., Pinter R.Y., Alon U., Margalit H. Network motifs in integrated cellular networks of transcription-regulation and protein-protein interaction. Proc. Natl. Acad. Sci. U.S.A. 2004;101:5934–5939. doi: 10.1073/pnas.0306752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asangani I.A., Dommeti V.L., Wang X., Malik R., Cieslik M., Yang R., Escara-Wilke J., Wilder-Romans K., Dhanireddy S., Engelke C., et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–282. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hein M.Y., Hubner N.C., Poser I., Cox J., Nagaraj N., Toyoda Y., Gak I.A., Weisswange I., Mansfeld J., Buchholz F., et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163:712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 38.Lockwood W.W., Zejnullahu K., Bradner J.E., Varmus H. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc. Natl. Acad. Sci. U.S.A. 2012;109:19408–19413. doi: 10.1073/pnas.1216363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alsarraj J., Hunter K.W. Bromodomain-Containing Protein 4: A Dynamic Regulator of Breast Cancer Metastasis through Modulation of the Extracellular Matrix. Int. J. Breast Cancer. 2012;2012:670632. doi: 10.1155/2012/670632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu Y., Zhou J., Ye F., Xiong H., Peng L., Zheng Z., Xu F., Cui M., Wei C., Wang X., et al. BRD4 inhibitor inhibits colorectal cancer growth and metastasis. Int. J. Mol. Sci. 2015;16:1928–1948. doi: 10.3390/ijms16011928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang M., Li S.N., Anjum K.M., Gui L.X., Zhu S.S., Liu J., Chen J.K., Liu Q.F., Ye G.D., Wang W.J., et al. A double-negative feedback loop between Wnt-beta-catenin signaling and HNF4alpha regulates epithelial-mesenchymal transition in hepatocellular carcinoma. J. Cell Sci. 2013;126:5692–5703. doi: 10.1242/jcs.135053. [DOI] [PubMed] [Google Scholar]
- 42.Gervasi M., Bianchi-Smiraglia A., Cummings M., Zheng Q., Wang D., Liu S., Bakin A.V. JunB contributes to Id2 repression and the epithelial-mesenchymal transition in response to transforming growth factor-beta. J. Cell Biol. 2012;196:589–603. doi: 10.1083/jcb.201109045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Polydorou C., Georgiades P. Ets2-dependent trophoblast signalling is required for gastrulation progression after primitive streak initiation. Nat. Commun. 2013;4:1658. doi: 10.1038/ncomms2646. [DOI] [PubMed] [Google Scholar]
- 44.Darnell J.E., Jr Transcription factors as targets for cancer therapy. Nat. Rev. Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 45.Vaquerizas J.M., Kummerfeld S.K., Teichmann S.A., Luscombe N.M. A census of human transcription factors: function, expression and evolution. Nat. Rev. Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.