Abstract
CRISPR-Cas9 is widely applied for genome engineering in various organisms. The assembly of single guide RNA (sgRNA) with the Cas9 protein may limit the Cas9/sgRNA effector complex function. We developed a FRET-based assay for detection of CRISPR–Cas9 complex binding to its targets and used this assay to investigate the kinetics of Cas9 assembly with a set of structurally distinct sgRNAs. We find that Cas9 and isolated sgRNAs form the effector complex efficiently and rapidly. Yet, the assembly process is sensitive to the presence of moderate concentrations of non-specific RNA competitors, which considerably delay the Cas9/sgRNA complex formation, while not significantly affecting already formed complexes. This observation suggests that the rate of sgRNA loading into Cas9 in cells can be determined by competition between sgRNA and intracellular RNA molecules for the binding to Cas9. Non-specific RNAs exerted particularly large inhibitory effects on formation of Cas9 complexes with sgRNAs bearing shortened 3′-terminal segments. This result implies that the 3′-terminal segment confers sgRNA the ability to withstand competition from non-specific RNA and at least in part may explain the fact that use of sgRNAs truncated for the 3′-terminal stem loops leads to reduced activity during genomic editing.
INTRODUCTION
CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)–Cas (CRISPR-associated genes) systems provide prokaryotes with adaptive hereditable immunity by acquiring fragments of foreign DNA (spacers) and upon subsequent encounters destroying DNA molecules harboring sequences matching acquired spacers (1–3). In DNA-targeting CRISPR–Cas nucleoprotein complexes, the CRISPR RNA (crRNA) forms a hybrid with the complementary strand of foreign DNA, displacing the non-complementary strand to form an R-loop (4,5). The Type II CRISPR–Cas systems require only one protein, Cas9 and two partially complementary RNAs, the crRNA containing spacer sequence and trans-activating tracrRNA, to bind to and cleave target DNA (6–8). The crRNA and tracrRNA pair (dual guide RNA, dgRNA) can be replaced with a single guide RNA (sgRNA) made up of fused crRNA and tracrRNA molecules (8). The Cas9/sgRNA system has been successfully used for genome editing in various eukaryotic organisms (9–11). A Cas9 derivative that lacks endonuclease activity but can bind target DNA when programmed with sgRNA (dCas9) (8) is used for transcriptome modulation (12) and visualization of genomic loci in live cells (13). Considerable efforts have been made to improve the targeting efficiency and specificity of Cas9/sgRNA systems by rational choice of targeted sequences and through protein and sgRNA engineering (13–25).
The crystal structure of Cas9 in complex with sgRNA and target DNA shows that the sgRNA assumes a T-shaped architecture comprising a guide:target heteroduplex, a repeat:anti-repeat duplex and 3′-terminal stem loops 1–3 (26). A sgRNA with shortened tracrRNA 3′-tail containing only stem loop 1 (sgRNA(+48)) is the minimal RNA that efficiently programs popular Streptococcus pyogenes Cas9 (SpCas9) to recognize and cleave its targets in vitro (8). However, genome engineering studies showed that sgRNA(+48) generates mutations in mammalian and plant cells much less efficiently than sgRNAs containing all three tracrRNA stem loops (9,14,27–29). Activity of sgRNA truncated for stem loop 3, sgRNA(+67), is ∼2-fold lower than that of the full length sgRNA (26,27). In vitro measurements of equilibrium dissociation constant (Kd) for Cas9 complexes with the full-length sgRNA and an sgRNA truncated for the stem loops 2 and 3 indicate that the stem loops significantly contribute into the overall sgRNA–Cas9 binding energy, yet the truncated sgRNA version still retains a high affinity (Kd is about 1 nM) for Cas9 (24). Northern blot analyses revealed decreased levels of sgRNA(+48) and sgRNA(+67) expression in human cells, leading to a model that sgRNA abundance determines the efficiency of CRISPR–Cas9 targeting (14). Expression of sgRNAs was found to be increased in the presence of Cas9, implying that sgRNA loading into Cas9 can protect it from degradation (28,30). It was proposed that stem loops 2 and 3 support the Cas9/sgRNA complex formation thus enhancing the stability of sgRNA and improving its in vivo activity (26). Analysis of factors that influence Cas9-mediated mutagenesis in zebrafish embryos supports the suggestion that sgRNA stability is an important determinant of its activity (18,31). These data have led to a proposal that assembly of sgRNA into Cas9 and/or sgRNA expression levels may limit the efficiency of Cas9-mediated gene targeting, particularly in the case of less abundant and/or relatively short-lived sgRNAs (14,18,28,32). We surmised that kinetic and mechanistic characterization of sgRNA loading into Cas9 in vitro may help to better understand mechanisms of the in vivo assembly process. We developed a fluorometric molecular beacon-like assay for detection of CRISPR–Cas9 complex binding to its targets and applied this assay to study the assembly of Cas9/sgRNA complexes. Our data reveal that the binding of isolated Cas9 to sgRNAs is a quite rapid and efficient process that, therefore, is unlikely to limit the rate of Cas9-mediated gene targeting. However, we found that the Cas9/sgRNA complex formation rate considerably decreases in the presence of moderate concentrations of non-specific RNA. This effect becomes particularly strong with sgRNAs truncated for the 3′-terminal stem loops 2 and 3. These data suggest that the rate of Cas9/sgRNA assembly in cells is determined by a competition between sgRNA and intracellular RNA molecules for the binding to Cas9 and that the 3′-terminal stem loops 2 and 3 confer guide RNAs the ability to more effectively withstand the inhibitory effect of non-specific RNAs.
MATERIALS AND METHODS
Proteins
Streptococcus pyogenes proteins Cas9 (SpCas9) wild-type (wt) and catalytically inactive dCas9 harboring point mutations D10A/H840A were expressed in Escherichia coli BL21(DE3) using the expression plasmids pMJ806 and pMJ841, respectively, (Addgene, https://www.addgene.org/) and purified essentially as previously described (8).
RNAs
All guide RNAs, including crRNA, tracrRNA and sgRNAs used in this study were made in vitro with T7 RNA polymerase (New England Biolabs) transcribing from polymerase chain reaction-generated dsDNA templates according to manufacturer recommendations. The sequences of the templates are listed in Supplementary Table S1. Transcribed RNAs were purified using 10% denaturing polyacrylamide gelelectrophoresis. Dual guide RNA was formed by mixing equimolar amounts of crRNA and tracrRNA in a buffer containing 40 mM Tris, pH 7.9, 100 mM NaCl; heating for 30 s at 90°C and slowly cooling the reactions. Yeast tRNA and total RNA from human lung tissue were purchased from Thermo Fisher Scientific.
DNA probes
DNA probes were prepared from unmodified and chromophore-labeled DNA oligonucleotides synthesized by Integrated DNA Technologies. Target double-stranded (ds) DNA and Cas9 beacon DNA constructs shown in Figure 1A were formed by mixing equimolar amounts of synthetic complementary strands (final concentrations were within low μM range) in a buffer containing 40 mM Tris, pH 7.9, 100 mM NaCl; heating for 2 min at 90°C and slowly cooling the reactions to 20°C. The protospacer adjacent motif (PAM)-distal ends of the beacon target and non-target strands are labeled with fluorescein and Iowa Black® FQ, respectively. pUC19 plasmid vector was purchased from New England Biolabs.
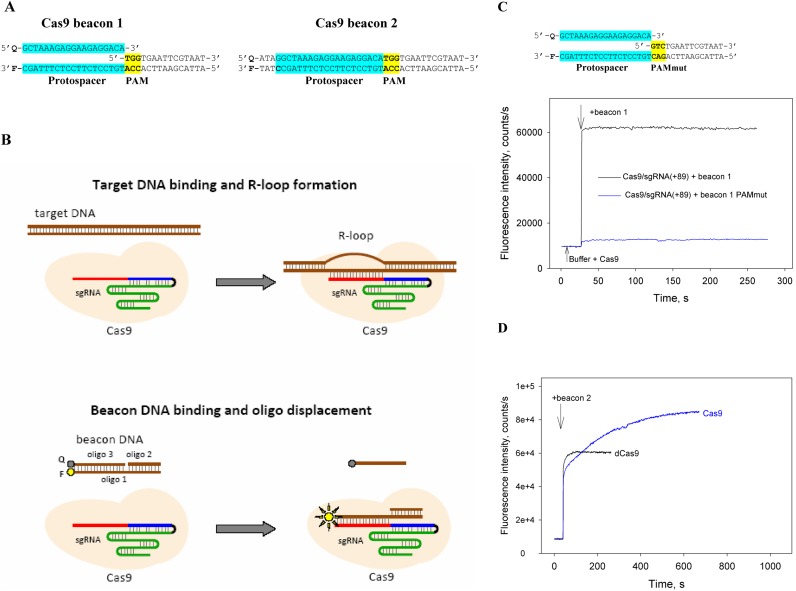
Figure 1.
A Cas beacon assay to detect Cas9/guide RNA complexes. (A) Structures of Cas9 beacon DNA constructs. The PAM and protospacer sequences are highlighted in yellow and blue. (B) Schematic representation of target DNA and Cas9 beacon 1 binding. The circles labeled F and Q indicate the fluorophore and quencher. (C) Measuring of Cas9/sgRNA(+89) complex binding to beacon 1. Time dependence of the increase in fluorescence upon the addition of 1 nM beacon 1 or a control beacon 1 derivative bearing mutated PAM sequence to 3 nM Cas9 pre-incubated with 5 nM sgRNA(+89) for 30 min. (D) Measuring of beacon 2 interaction with Cas9/sgRNA(+89) and dCas9/sgRNA(+89) complexes. Time dependence of the increase in fluorescence upon the addition of 1 nM beacon 2 to 3 nM Cas9 or dCas9 pre-incubated with 5 nM sgRNA(+89) for 30 min.
Fluorometric measurements
If not otherwise stated, fluorescence measurements were performed using a QuantaMaster QM4 spectrofluorometer (PTI) in binding buffer [20 mM Tris HCl (pH 7.9), 120 mM NaCl, 5% glycerol, 0.1 mM DTT and 1 mM MgCl2] containing 0.02% Tween 20 at 25°C. The measurements were performed in a buffer containing 1 mM MgCl2 since this concentration of Mg2+ is close to estimates of intracellular concentration of free Mg2+ (33). Final assay mixtures (800 μl) contained 3 nM of a Cas protein (Cas9 or dCas9), 1 nM labeled Cas beacon construct and guide RNAs at various concentrations. The fluorescein fluorescence intensities were recorded with an excitation wavelength of 498 nm and an emission wavelength of 520 nm. Time-dependent fluorescence changes were monitored after addition of negligible volume of Cas beacon or guide RNA to a cuvette followed by manual mixing; the mixing dead-time was 15 s. The Cas9 beacon binding half-time (t0.5) values presented are averages obtained from at least three individual experiments, the error is ∼20%.
RESULTS
Design and validation of Cas9 beacon oligonucleotide displacement assay
To detect formation of specific Cas9-guide RNA complexes, we developed a fluorometric assay based on measuring emission from fluorescently labeled derivatives of target DNA. The target DNA derivatives are prepared by annealing two or three oligonucleotides to generate a ∼ 40-nt fragment of DNA comprising a protospacer, a functional PAM and ∼15-bp downstream of PAM. We named these DNA constructs ‘Cas9 beacons’. The PAM-distal ends of the beacon target and non-target strands are labeled with a fluorescent label and fluorescence quencher, respectively. We tested two Cas9 beacons shown in Figure 1A. Beacon 1 is shorter than beacon 2 by 4 nt and consists of three oligonucleotides: its non-target strand contains a discontinuity between PAM and the protospacer segment (Figure 1A). The longer beacon 2 consists of just two fully complementary oligonucleotides. The baseline fluorescence intensity of both beacons is expected to be low due to quenching of the fluorescence label by the nearby quencher via FRET mechanism. We expected that both beacons would mimic native target DNA and would bind Cas9 complexes with sgRNA containing an appropriate spacer segment (Figure 1B). Cas9 binding to beacon 1 and formation of a DNA–RNA heteroduplex should result in dissociation of oligo 3, leading to increase in fluorescence intensity due to disappearance of the quenching effect (Figure 1B, bottom). Indeed, upon the addition of 3 nM of SpCas9 pre-incubated with 5 nM sgRNA(+89) (shown in Figure 2) to 1 nM beacon 1, fluorescence intensity rapidly increased by ∼50-fold, reaching peak intensity during the mixing time (Figure 1C). We performed several control experiments to verify that the increase in fluorescence intensity was due to specific interaction between Cas9/sgRNA(+89) and the beacon. First, no change in fluorescence intensity was observed upon the addition of 1 nM beacon 1 to 10 nM sgRNA(+89) without Cas9 (Supplementary Figure S1A). Second, fluorescence signal of beacon 1 with a PAM mutation known to severely hinder the interaction of target DNA with the effector complex (4,5,34,35) did not increase in the presence of Cas9/sgRNA(+89) complex (Figure 1C). Third, a double-stranded DNA fragment containing SpCas9 PAM and a protospacer targeted by sgRNA(+89) spacer segment competed with the beacon for the binding to the Cas9/sgRNA(+89) complex (Supplementary Figure S1B). The very slow signal increase observed upon the beacon 1 addition to Cas9/sgRNA(+89) pre-incubated with the target DNA fragment (Supplementary Figure S1B) is consistent with reported data indicating that Cas9 associates tightly with both ends of cut target DNA duplex (35). A catalytically inactive Cas9 mutant dCas9, when programmed with sgRNA, binds target DNA with affinities similar to those of a wt effector complex (8). Consistently, Cas9 and dCas9 complexes with sgRNA(+89) generated similar fluorescence signals upon the addition of the beacon 1 in the absence or presence of competitor target dsDNA (Supplementary Figure S1B and C).
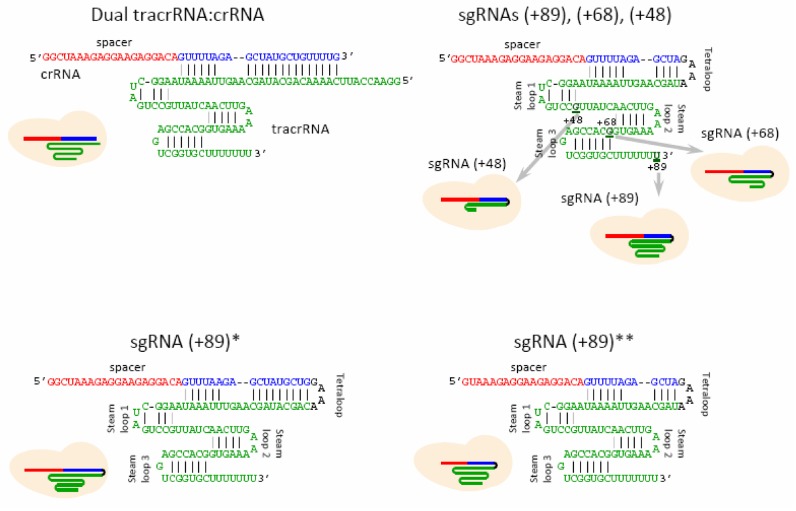
Figure 2.
Structures of guide RNAs.
The addition of Cas9/sgRNA(+89) complex to beacon 2 resulted in biphasic increase in fluorescence intensity (Figure 1D). We surmise that these kinetic phases represented the initial separation of the fluorescent label and quencher molecules caused by beacon 2 binding, followed by a slower cleavage reaction that should lead to dissociation of quencher-labeled fragment of the non-target strand. This explanation is supported by kinetic curve obtained with the dCas9/sgRNA(+89) complex, which showed no slow kinetic component (Figure 1D).
Overall, we conclude, that the data above show that interaction of the Cas9 beacons 1 and 2 with the effector complex is specific and mimics the binding of native target DNA and can be used as a tool to study Cas9 complex interactions with their targets and target cleavage. In what follows we will use only the Cas9 beacon 1 construct to analyze target binding by effector complexes assembled with various sgRNAs.
Cas9/sgRNA complexes bind beacon rapidly
Structures of sgRNAs and dgRNA used in this work are shown in Figure 2. The sgRNA(+89) molecule used to develop the beacon assay (above) contains stem loops 1–3 of tracrRNA 3′ tail (nucleotides 23 to 89 of the native tracrRNA sequence), and is thus similar to sgRNAs most widely used for CRISPR-Cas9-mediated mutagenesis and editing. sgRNA(+68) and sgRNA(+48) lack stem loop 3 or stem loops 2 and 3, respectively. sgRNA(+89)* is a derivative of sgRNA(+89) with a modified stem of the tetraloop structure (13). dCas9 programmed with such sgRNAs showed increased target DNA binding in human cells and it was suggested that this effect may be in part due to improvement of the assembly step (13). We also tested a derivative of sgRNA(+89) truncated by 2 nt at the 5′-end, i.e. in the spacer segment ((sgRNA(+89)**). Such truncated sgRNAs lead to a considerable improvement in Cas9 cleavage fidelity in vivo (15). The level of sgRNA expression or delivery to cells frequently needs to be optimized to lower off-target editing (14,20,36–38). In this work, we used 5 and 50 nM guide RNA concentrations that correspond to a typical concentration range of sgRNAs injected into zebrafish embryos (25–300 pg injected sgRNA (18,31,37) per ∼0.2 μl embryo (39)).
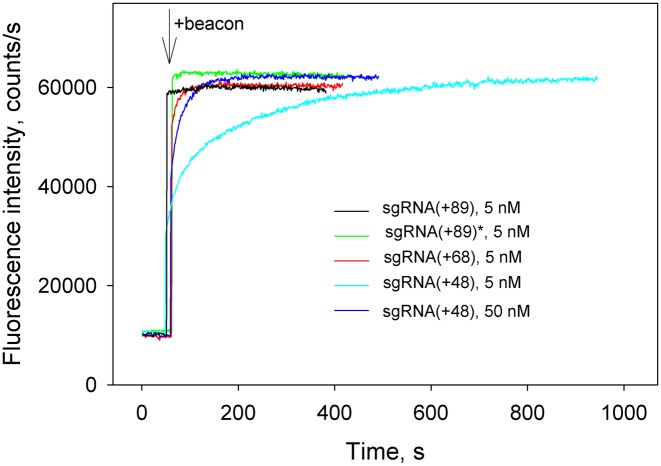
The kinetic traces for changes in fluorescence upon the binding of beacon to Cas9/sgRNA complexes obtained by incubation of 3 nM Cas9 with 5 nM of sgRNAs for 30 min are shown in Figure 3. Almost identical kinetic traces were obtained when dCas9 was used instead of Cas9 in similar experiments (data not shown). The beacon very rapidly bound to Cas9 complexes with sgRNA(+89) and sgRNA(+89)* (Figure 3). Rapid mixing kinetic measurement with Cas9/sgRNA(+89) showed that the binding half-time was about 4 s (Supplementary Figure S2). The binding of the beacon to Cas9 pre-incubated with sgRNA(+68) was slower and was complete within 40 s (Figure 3). The kinetics of beacon binding to Cas9 pre-incubated with sgRNA(+48) was biphasic with distinct fast and slow components (Figure 3). Increasing the pre-incubation time did not cause any change in the kinetics (data not shown), evidencing that equilibrium binding of Cas9 to sgRNA(+48) has been reached. Increasing sgRNA(+48) concentration to 50 nM decreased the amount of binding attributable to the slow component (Figure 3). The kinetic data obtained with sgRNA(+48) can be explained by its lower affinity for Cas9 compared to other sgRNAs used (24). Thus, a noticeable fraction of Cas9 remains unbound at 5 nM sgRNA(+48) concentration. The fast kinetic component of the beacon binding observed at 5 nM sgRNA(+48) corresponds to beacon binding by Cas9/sgRNA(+48) complexes that were formed before the beacon addition. The slow kinetic component is due to additional ongoing assembly that is stimulated by the coupled binding of Cas9/sgRNA(+48) complex to the beacon.
Figure 3.
Measuring of Cas beacon binding to Cas9 complexes with sgRNAs bearing 3′-tails of different lengths. Time dependence of the increase in fluorescence upon the addition of 1 nM beacon to 3 nM Cas9 pre-incubated with 5 nM of sgRNA(+89), sgRNA(+89)*, sgRNA(+68) and 5 or 50 nM sgRNA(+48) for 30 min.
Using the Cas9 beacon to monitor Cas9/sgRNA complex assembly
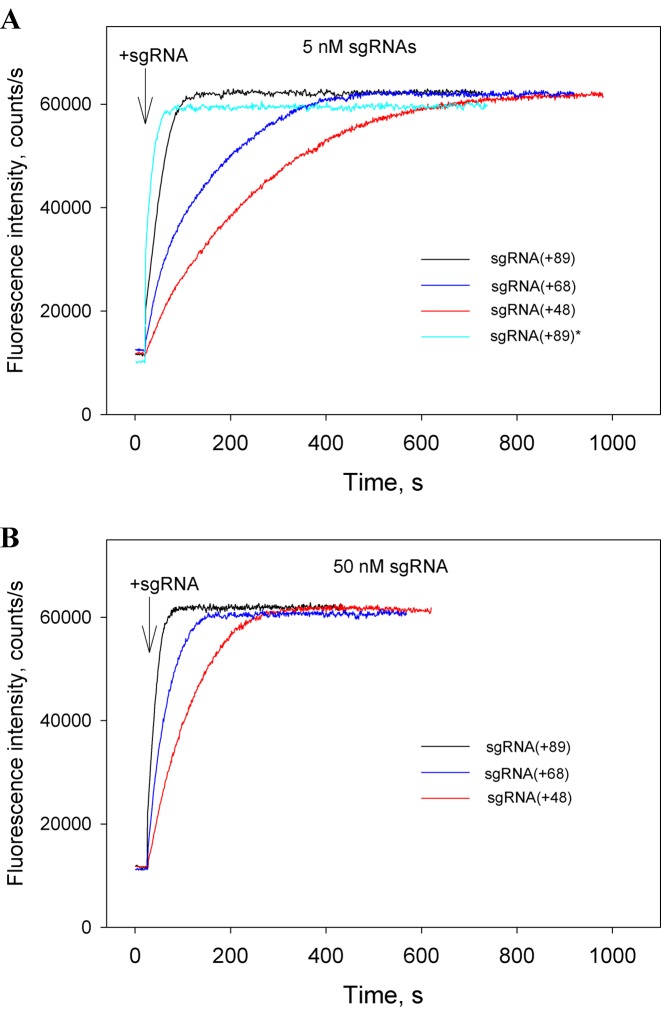
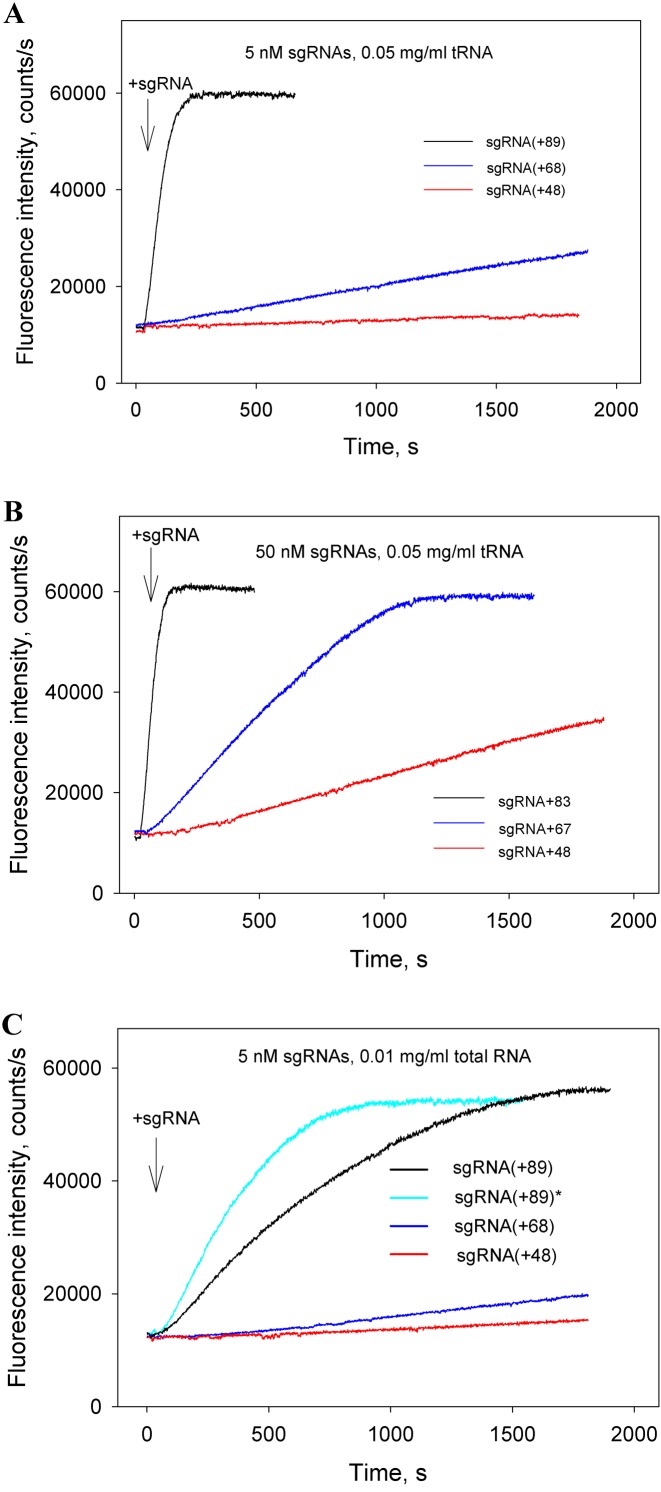
In the following series of experiments, we excluded the Cas9/sgRNA pre-incubation step and measured the kinetics of beacon binding immediately upon the addition of sgRNAs to samples containing Cas9 and the beacon. The kinetic traces obtained in this way (Figure 4A and B) are considerably slower than corresponding traces measured with preformed Cas9/sgRNA complexes (Figure 3). The difference could be attributed to limitation of the rate of beacon binding by the relatively slow assembly of the Cas9/sgRNA complexes. In other words, traces in Figure 4A and B closely represent the kinetics of formation of Cas9/sgRNA complexes capable of target recognition. As seen from data in Figure 4A, Cas9 most rapidly forms complexes with sgRNA(+89) and sgRNA(+89)*, whereas complexes with sgRNA(+68) and sgRNA(+48) form ∼5- and 10-fold slower, respectively. Thus, the 3′-terminal stem loops 2 and 3 of sgRNA significantly affect the rate of sgRNA binding to Cas9.
Figure 4.
Cas beacon assay for the assembly of sgRNAs into Cas9. Time dependence of the increase in fluorescence upon the addition of 5 nM (A) or 50 nM (B) sgRNAs to samples containing Cas9 and beacon.
Effect of non-specific RNA and DNA on formation of Cas9/sgRNA complexes
Most proteins binding nucleic acids are capable of both specific and non-specific binding. We examined whether formation of Cas9/sgRNA complexes can be affected by non-specific RNA and DNA competitors using yeast tRNA, total RNA from human lung tissue and the pUC19 plasmid. Inside the cells, only free RNA molecules are expected to be involved in the competition with sgRNA for the binding to Cas9. Most cellular RNA, with a notable exception of tRNA, is recruited into nucleoprotein complexes that are not expected to compete with sgRNA. While intracellular concentrations of unbound RNAs are unknown, a pool of free RNA molecules must exist due to degradation of nucleoprotein complexes and other processes. With these considerations in mind, we used total human lung RNA at a concentration (0.01 mg/ml) which is much lower than the typical total cellular RNA concentration (∼10 mg/ml, 40). The concentrations of yeast tRNA and pUC19 DNA were 0.05 and 0.1 mg/ml, respectively.
Incubation of preformed Cas9/sgRNA(+89) and Cas9/sgRNA(+68) complexes with RNA competitors for 2 h had no effect on beacon binding (Supplementary Figure S3A). In contrast, a signal generated upon the beacon addition to the Cas9/sgRNA(+48) complex decreased 2- to 3-fold after incubation with total human lung RNA (Supplementary Figure S3B). We conclude that sgRNA(+89) and sgRNA(+68) remain tightly bound to Cas9 once the complex is formed, whereas the Cas9/sgRNA(+48) complex is unstable in the presence of non-specific RNA.
When added to Cas9 before the addition of sgRNAs, the RNA competitors significantly delayed beacon binding (compare data in Figures 5A–C and 4A and B). These data indicate that non-specific RNAs readily bind to Cas9 and affect the Cas9/sgRNA assembly step. Total RNA produced larger effects than tRNA in these experiments (Figure 5A and C), possibly because longer or differently structured non-specific RNAs have a higher affinity for Cas9. As expected, formation of the sgRNA(+89) complex with dCas9 was also considerably delayed in the presence of total RNA (Supplementary Figure S4). The loading of sgRNA(+89)* into Cas9 in the presence of total RNA was slightly faster than that of sgRNA(+89) (Figure 5C). Importantly, data in Figure 5A–C show that formation of Cas9 complexes with sgRNA(+68) and, most strikingly, with sgRNA(+48), is affected by non-specific RNA much more stronger than formation of Cas9/sgRNA(+89) and Cas9/sgRNA(+89)* complexes. We suggest that the low cleavage efficiency observed with sgRNA(+48) in eukaryotic cells is in part a consequence of its inability to compete with intracellular RNA for binding to Cas9.
Figure 5.
Effect of non-specific RNA competitors on formation of Cas9/sgRNA complexes. (A and B) Time dependencies of the increase in fluorescence upon the addition of 5 nM (A) or 50 nM (B) sgRNAs to samples containing Cas9, beacon and 0.05 mg/ml yeast tRNA; (C) Time dependence of the increase in fluorescence upon the addition of 5 nM sgRNAs to samples containing Cas9, beacon and 0.01 mg/ml total RNA from human lung.
We also tested sgRNA(+89)**, a derivative of sgRNA(+89) truncated by two 5′-nt, in experiments similar to those described above. Since sgRNA(+89)** showed behavior indistinguishable from that of sgRNA(+89), we conclude that this spacer segment truncation has no significant influence on the kinetics of Cas9/sgRNA complex formation.
In contrast to RNA competitors, the pUC19 DNA noticeably decreased the rate of beacon binding to preformed Cas9/sgRNA(+89) complex (Supplementary Figure S5A). This result is in agreement with reported Cas9/sgRNA binding to non-specific DNA sites (35). When added to Cas9 before the addition of sgRNA(+89), pUC19 moderately delayed formation of the Cas9/sgRNA(+89) complex (Supplementary Figure S5B), similarly to the effect produced by tRNA.
Cas9 interactions with dgRNA
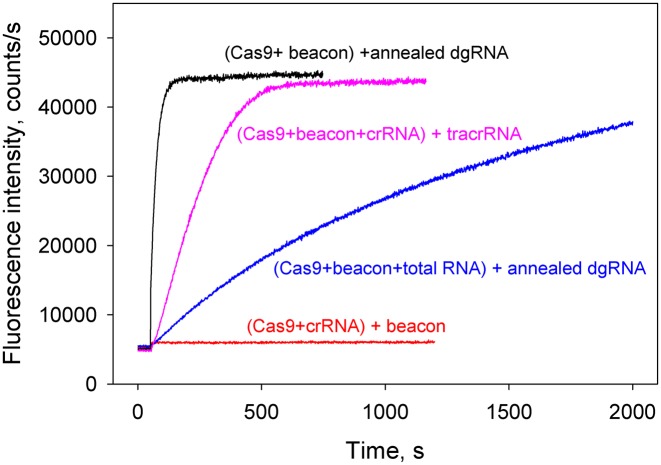
Cas9 interactions with dgRNA were analyzed next. The Cas9 beacon rapidly bound to preformed complex of Cas9 with 5 nM annealed crRNA and tracrRNA (Supplementary Figure S6). No increase in fluorescence intensity was observed upon addition of the beacon to Cas9 pre-incubated with 50 nM crRNA alone (Figure 6), consistent with reported inability of SpCas9 crRNA to function without tracrRNA (6,8). The rate of beacon binding measured upon the addition of 5 nM annealed crRNA and tracrRNA to Cas9 and beacon was fast and similar to that observed in experiment with sgRNA(+89) (Figures 4A and 6) indicating rapid Cas9–dgRNA complex formation. In the presence of 0.01 mg/ml total RNA, the kinetics of dgRNA assembly with Cas9 was delayed (t0.5 values are 25 s and 14 min, respectively) (Figure 6). The relatively moderate effect of total RNA on the Cas9/dgRNA assembly as compared with that exerted on the Cas9/sgRNA(+48) complex formation (Figures 5C and 6) is consistent with a suggestion that stem loops 2 and 3 determine the ability of guide RNAs to withstand competition from non-specific RNA. We also performed an order of addition Cas9/dgRNA assembly experiment in which beacon binding was measured upon the addition of tracrRNA to sample containing Cas9, beacon and crRNA (Figure 6). The kinetic trace observed in this experiment was 9-fold slower (t0.5 = 220 s) than the trace obtained with pre-annealed dgRNA, suggesting that the effector complex formation was limited by the crRNA/tracrRNA annealing step.
Figure 6.
Cas9 assembly with dgRNA. Time dependencies of the increase in fluorescence upon the addition of annealed crRNA and tracrRNA to samples containing Cas9 and beacon in the absence or presence of 0.01 mg/ml total human lung RNA (black and blue curves, respectively). Pink curve shows time dependence of the increase in fluorescence upon the addition of tracrRNA to sample containing Cas9, beacon and crRNA. The crRNA and tracrRNA concentrations were 5 nM in these experiments. No increase in fluorescence intensity was observed upon beacon addition to Cas9 pre-incubated with 50 nM crRNA (red curve).
Effect of salt composition on rate of the Cas9/sgRNA(+89) complex formation
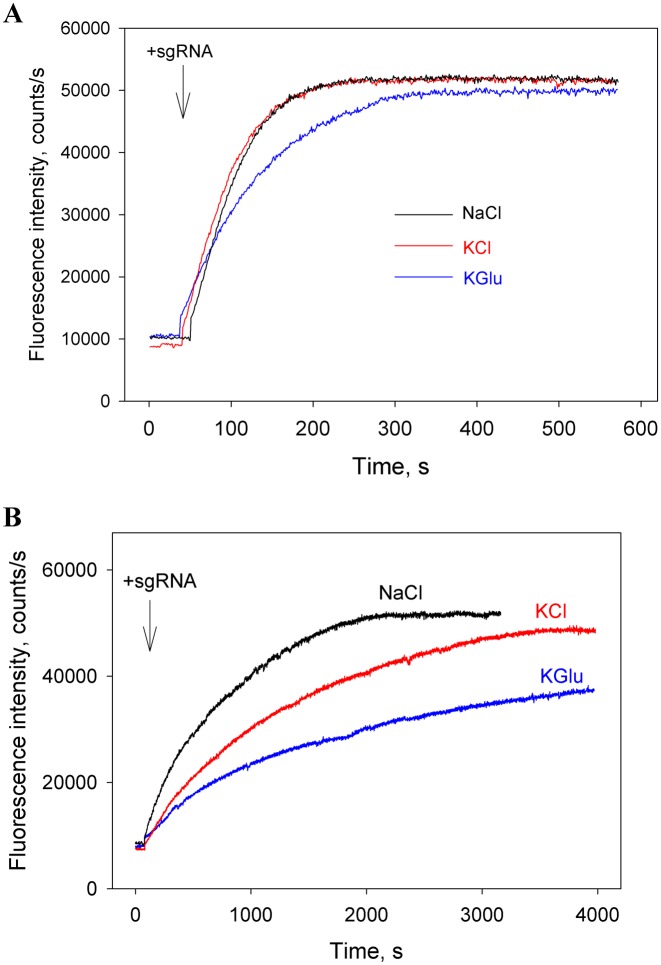
Protein–nucleic acid interactions are sensitive to the concentrations and nature of electrolyte ions in solution (41,42). We evaluated the influence of ion composition on beacon binding to the preformed Cas9/sgRNA(+89) complex and on the Cas9/sgRNA(+89) assembly in assay mixtures containing 120 mM of either NaCl, KCl or KGlu. The kinetics of beacon bindings to the preformed Cas9/sgRNA(+89) complex were similar to that shown in Figure 1C in all assay mixtures (data not shown). The effect of salt composition on kinetics of the Cas9/sgRNA(+89) assembly was measured in the absence or presence of total human RNA similarly to the experiments shown in Figures 4A and 5C. In the presence of total human lung RNA, the assembly halftimes in the samples containing NaCl, KCl or KGlu were about 10, 20 and 40 min, respectively (Figure 7B). On the other hand, only small differences between the kinetic traces obtained in the absence of non-specific RNA were observed (Figure 7A). This result suggests that the difference in the assembly rates observed in the presence of total RNA might be due to a more tight binding of non-specific RNA to Cas9 in the samples containing KCl and KGlu. Since mammalian cytosol typically contain higher concentration of K+ than Na+ and chloride is present only at low concentration (43), we reason that the KGlu containing assay mixture may more closely mimic the intracellular ionic conditions.
Figure 7.
Effect of salt composition on the Cas9 assembly with sgRNA(+89). Time dependencies of the increase in fluorescence upon the addition of sgRNA(+89) to samples containing Cas9 and beacon in the absence (A) or presence (B) of 0.01 mg/ml total human lung RNA. The measurements were carried out in the assay mixtures containing 120 mM of either NaCl, KCl or KGlu.
DISCUSSION
To elucidate factors determining efficiency of the Cas9-guide RNA assembly process and their potential significance for Cas9 function, we measured the kinetics of Cas9 assembly with a set of distinct sgRNA versions and correlated the data with reported efficiencies of in vivo gene targeting by these sgRNAs. In order to mimic the intracellular conditions, we also performed measurements in the presence of non-specific RNA. The Cas9/sgRNA assembly was detected using a novel fluorometric Cas9 beacon assay. The advantages of this assay are that it reports only on sgRNA–Cas9 complexes capable of specific binding to target DNA and allows measuring both fast and slow kinetics. Similar assays may also be instrumental in studies of assembly of other CRISPR–Cas surveillance complexes and their interaction with target DNAs.
Our study provides several new insights into the assembly of Cas9 effector complex. The experiments with sgRNA(+89) and its truncated derivatives sgRNA(+68) and sgRNA(+48) reveal that the 3′-terminal stem loops 2 and 3 accelerate the sgRNA loading process. In the absence of non-specific RNA, Cas9 assembly with these sgRNAs occurred on a time scale of few minutes (Figure 4A and B). The assembly rates considerably decreased in the presence of moderate concentrations of non-specific RNAs. For example, the sgRNA(+89) loading into Cas9 in KGlu containing buffer occurred on a ∼1 h time scale in the presence of 0.01 mg/ml total human lung RNA, which is about 30-fold slower than the loading without non-specific RNA competitors (Figure 7A and B). These data suggest that the rate of Cas9/sgRNA effector complex formation in cells is determined by competition between sgRNA and free cellular RNAs for binding to Cas9. We speculate that the assembly of Cas9/sgRNA complexes in cells might be even slower as the ratio of concentrations of non-specific RNA competitors and sgRNA in cells may be higher than that in our experiments. Taking into account that stability of sgRNAs increases upon the binding to Cas9 (28,30), such slow assembly may be a cause of significant loss of sgRNAs due to degradation before the loading takes place. This consideration is consistent with a report on high efficiency and accelerated rate of genome editing upon delivery of pre-assembled Cas9/sgRNA complexes in cells (44). We did not observe a noticeable destabilization of preformed Cas9/sgRNA complexes by non-specific RNA, except for the Cas9/sgRNA(+48) complex. This observation is in agreement with time course analyses of sgRNA abundance in zebrafish embryos that suggests that sgRNAs are stably retained in cells once they are loaded into Cas9 (18).
Wright et al. reported that a split Cas9 enzyme in which the nuclease lobe and α-helical lobe are expressed as separate polypeptides efficiently cleaves target DNA in vitro but has low in-cell genome-editing activity compared to that of the wt Cas9 enzyme (24). It seems possible that this difference may be due to destabilization of sgRNA complex with the split Cas9 version by cellular non-specific RNA.
The formation of Cas9/sgRNA(+68) and, in particular, Cas9/sgRNA(+48) complexes was significantly more susceptible to the presence of non-specific RNA competitors compared to sgRNA(+89) and dgRNA (Figures 5A–C and 6). These data imply that the 3′-terminal stem loops 2 and 3 confer guide RNAs the ability to more effectively overcome the inhibition effect of non-specific RNAs. The differences in the rates of the Cas9 assembly with sgRNA(+89), sgRNA(+68), and sgRNA(+48) in the presence of non-specific RNA correlate well with reported frequencies of indels generated by Cas9 complexes with these sgRNAs during genomic editing experiments (9,14,26–28) and with levels of their apparent expression in human cells (14). Thus, the very low abundance of sgRNA(+48) (14) may at least in part be a consequence of its particularly slow loading into Cas9.
The rate of Cas9 assembly with sgRNA(+89) was only about 2-fold slower than with sgRNA(+89)* containing a modified tetraloop segment, either in the absence or presence of RNA competitors. This result seems to suggest that observed superior properties of sgRNA(+89)* compared to sgRNA(+89) (13,45) are mainly due to other reasons than improvement of assembly such as, for example, a higher expression level of sgRNA(+89)* (13).
Supplementary Material
Acknowledgments
We thank Dr. E. Peter Geiduschek for careful reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by NIH grant National Institutes of Health [GM10407 to K.S.]; Russian Science Foundation [14-14-00988]; and Ministry of Education and Science of Russian Federation project [14.B25.31.0004]. Funding for open access charge: NIH and Russian Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Brouns S.J., Jore M.M., Lundgren M., Westra E.R., Slijkhuis R.J., Snijders A.P., Dickman M.J., Makarova K.S., Koonin E.V., van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garneau J.E., Dupuis M.E., Villion M., Romero D.A., Barrangou R., Boyaval P., Fremaux C., Horvath P., Magadán A.H., Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2011;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 4.Jore M.M., Lundgren M., van Duijn E., Bultema J.B., Westra E.R., Waghmare S.P., Wiedenheft B., Pul U., Wurm R., Wagner R., et al. Structural basis for CRISPR RNA-guided DNA recognition by Cascade. Nat. Struct. Mol. Biol. 2011;18:529–536. doi: 10.1038/nsmb.2019. [DOI] [PubMed] [Google Scholar]
- 5.Szczelkun M.D., Tikhomirova M.S., Sinkunas T., Gasiunas G., Karvelis T., Pschera P., Siksnys V., Seidel R. Direct observation of R-loop formation by single RNA-guided Cas9 and Cascade effector complexes. Proc. Natl. Acad. Sci. U.S.A. 2014;111:9798–9803. doi: 10.1073/pnas.1402597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A., Eckert M.R., Vogel J., Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sapranauskas R., Gasiunas G., Fremaux C., Barrangou R., Horvath P., Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang W.Y., Fu Y., Reyon D., Maeder M.L., Tsai S.Q., Sander J.D., Peterson R.T, Yeh J.R., Joung J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sander J.D., Joung J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014;32:347–55. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E., Stern-Ginossar N., Brandman O., Whitehead E.H., Doudna J.A., et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen B., Gilbert L.A., Cimini B.A., Schnitzbauer J., Zhang W., Li G.W., Park J., Blackburn E.H., Weissman J.S., Qi L.S., et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O., et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doench J.G., Hartenian E., Graham D.B., Tothova Z., Hegde M., Smith I., Sullender M., Ebert B.L., Xavier R.J., Root D.E. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Josephs E.A., Kocak D.D., Fitzgibbon C.J., McMenemy J., Gersbach C.A., Marszalek P.E. Structure and specificity of the RNA-guided endonuclease Cas9 during DNA interrogation, target binding and cleavage. Nucleic Acids Res. 2015;43:8924–8941. doi: 10.1093/nar/gkv892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreno-Mateos M.A., Vejnar C.E., Beaudoin J.D., Fernandez J.P., Mis E.K., Khokha M.K., Giraldez A.J. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods. 2015;12:982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ran F., Hsu P.D., Lin C.-Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., Church G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsai S.Q., Wyvekens N., Khayter C., Foden J.A., Thapar V., Reyon D., Goodwin M. J., Aryee M.J., Joung J.K. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guilinger J.P., Thompson D.B., Liu D.R. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davis K.M, Pattanayak V., Thompson D.B., Zuris J.A., Liu D.R. Small molecule-triggered Cas9 protein with improved genome-editing specificity. Nat. Chem. Biol. 2015;11:316–318. doi: 10.1038/nchembio.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright A.V., Sternberg S.H., Taylor D.W., Staahl B.T., Bardales J.A., Kornfeld J.E., Doudna J.A. Rational design of a split-Cas9 enzyme complex. Proc. Natl. Acad. Sci. U.S.A. 2015;112:2984–2989. doi: 10.1073/pnas.1501698112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishimasu H., Ran F.A., Hsu P.D., Konermann S., Shehata S.I., Dohmae N., Ishitani R., Zhang F., Nureki O. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Briner A., Donohoue P., Gomaa A., Selle K., Slorach E., Nye C., Haurwitz R., Beisel C., May A., Barrangou R. Guide RNA functional modules direct Cas9 activity and orthogonality. Mol. Cell. 2014;56:333–339. doi: 10.1016/j.molcel.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 28.Jinek M., East A., Cheng A., Lin S., Ma E., Doudna J. RNA-programmed genome editing in human cells. Elife. 2013;2:1–9. doi: 10.7554/eLife.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou H., Liu B., Weeks D.P., Spalding M.H., Yang B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014;42:10903–10914. doi: 10.1093/nar/gku806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu X., Scott D.A., Kriz A.J., Chiu A.C., Hsu P.D., Dadon D.B., Cheng A.W., Trevino A.E., Konermann S., Chen S., et al. Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat. Biotechnol. 2014;32:670–676. doi: 10.1038/nbt.2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gagnon J.A., Valen E., Thyme S.B., Huang P., Akhmetova L., Pauli A., Montague T.G., Zimmerman S., Richter C., Schier A.F. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang T., Wei J.J., Sabatini D.M., Lander E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343:80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rink T.J., Tsien R.Y., Pozzan T. Cytoplasmic pH and free Mg2+ in lymphocytes. J. Cell Biol. 1982;95:189–196. doi: 10.1083/jcb.95.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Semenova E., Jore M.M., Datsenko K.A., Semenova A., Westra E.R., Wanner B., van der Oost J., Brouns S.J., Severinov K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sternberg S.H., Redding S., Jinek M., Greene E.C., Doudna J.A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hwang W.Y., Fu Y., Reyon D., Maeder M.L., Tsai S.Q., Sander J.D., Peterson R.T., Yeh J.R., Joung J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pattanayak V., Lin S., Guilinger J.P., Ma E., Doudna J.A., Liu D.R. High-throughput profiling of off-target DNA cleavage reveals RNAprogrammed Cas9 nuclease specificity. Nat. Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hagedorn M., Kleinhans F.W., Artemov D., Pilatus U. Characterization of a major permeability barrier in the zebrafish embryo. Biol. Reprod. 1998;59:1240–1250. doi: 10.1095/biolreprod59.5.1240. [DOI] [PubMed] [Google Scholar]
- 40.Priami C., Morine M.J. Analysis of Biological Systems. London: Imperial College Press; 2015. [Google Scholar]
- 41.Record M.T., Mazur S. J., Melaqon P., Roe J.H., Shaner S.L., Unger L. Double helical DNA: conformations, physical properties, and interactions with ligands. Annu. Rev. Biochem. 1981;50:997–1024. doi: 10.1146/annurev.bi.50.070181.005025. [DOI] [PubMed] [Google Scholar]
- 42.Leirmo S., Harrison C., Cayley D.S., Burgess R.R., Record M.T. Replacement of potassium chloride by potassium glutamate dramatically enhances protein–DNA interactions in vitro. Biochemistry. 1987;26:2095–2101. doi: 10.1021/bi00382a006. [DOI] [PubMed] [Google Scholar]
- 43.Lodish H., Berk A., Zipursky S., Matsudaira P., Baltimore D., Darnell J. Molecular Cell Biology. NY: W.H. Freeman; 2000. [Google Scholar]
- 44.Kim S., Kim D., Cho S.W., Kim J., Kim J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014;24:1012–1019. doi: 10.1101/gr.171322.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dang Y., Jia G., Choi J., Ma H., Anaya E., Ye C., Shankar P., Wu H. Optimizing sgRNA structure to improve CRISPR-Cas9 knockout efficiency. Genome Biol. 2015;16:280. doi: 10.1186/s13059-015-0846-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.