Abstract
Region and cell-type specific differences in the molecular make up of colon epithelial cells have been reported. Those differences may underlie the region-specific characteristics of common colon epithelial diseases such as colorectal cancer and inflammatory bowel disease. DNA methylation is a cell-type specific epigenetic mark, essential for transcriptional regulation, silencing of repetitive DNA and genomic imprinting. Little is known about any region-specific variations in methylation patterns in human colon epithelial cells. Using purified epithelial cells and whole biopsies (n = 19) from human subjects, we generated epigenome-wide DNA methylation data (using the HELP-tagging assay), comparing the methylation signatures of the proximal and distal colon. We identified a total of 125 differentially methylated sites (DMS) mapping to transcription start sites of protein-coding genes, most notably several members of the homeobox (HOX) family of genes. Patterns of differential methylation were validated with MassArray EpiTYPER. We also examined DNA methylation in whole biopsies, applying a computational technique to deconvolve variation in methylation within cell types and variation in cell-type composition across biopsies. Including inferred epithelial proportions as a covariate in differential methylation analysis applied to the whole biopsies resulted in greater overlap with the results obtained from purified epithelial cells compared with when the covariate was not included. Results obtained from both approaches highlight region-specific methylation patterns of HOX genes in colonic epithelium. Regional variation in methylation patterns has implications for the study of diseases that exhibit regional expression patterns in the human colon, such as inflammatory bowel disease and colorectal cancer.
Keywords: DNA methylation, epigenome-wide sequencing, deconvolution, region-specific colonic variation
the human colon is a conduit for the excretion of the byproducts of digestion. Normal colonic homeostasis depends on the maintenance of a barrier between the colonic lumen and the host, consisting primarily of a single layer of intestinal epithelial cells (IECs). IECs in the colon play essential roles both in the absorption of fecal water (57) and as innate immune cells, controlling the interface between a potentially hostile colonic luminal environment and the host (42).
The colon is subject to several common diseases including cancer of colon epithelial cells (colorectal carcinoma) and inflammatory bowel diseases, among others. Researchers investigating those diseases often utilize biopsies, obtained from the colon epithelium during colonoscopy, for molecular and cellular analyses. Those biopsy samples consist of a mixture of different cell types including epithelial cells, stromal cells such as fibroblasts, immune cells such as macrophages and lymphocytes, and endothelial cells. Thus, the results of analyses of homogenates of those heterogeneous biopsies represent the aggregate values of the different cell types present in the biopsies. Researchers who need to study single homogenous cell types have used laboratory techniques to disaggregate cell types from one another in the biopsies, although those procedures have some drawbacks (29). Therefore, computational techniques to deconvolute data obtained from homogenates of heterogeneous biopsies could be of value to researchers seeking to assess cell-type specific molecular factors from such samples.
Another important consideration in the use of colon epithelial biopsies in disease pathogenesis research is the choice of appropriate control material. Researchers often compare biopsies obtained from different colon regions, but little is known about whether there are region-specific differences in the molecular make up of IECs in the colon. The importance of region-specific differences in human colon epithelium is highlighted by the fact that some common diseases of the colon, such as those mentioned above, exhibit regional variation in their localization in the colon. For these reasons, a better understanding of any molecular differences between epithelial cells of different colon regions would be important for the correct interpretation of experimental data based on biopsies of those regions.
The methylation of cytosine nucleotides at the 5′-position of DNA is a crucial epigenetic mechanism for the control of gene expression. As well as playing important roles in embryonic development and maintenance of normal tissue differentiation (reviewed in Ref. 31), DNA methylation abnormalities have been identified as key contributors to the neoplastic transformation of IECs in the colon (36). For example, variable DNA methylation patterns have been observed in ulcerative colitis-associated cancer (14, 53) and have contributed to aberrant epigenetic gene silencing in sporadic colorectal cancer (10, 20). This indicates that DNA methylation differences play roles in disease pathogenesis, but there have been no prior studies exploring whether any epithelial cell-specific regional variations in DNA methylation patterns are present between distinct colonic regions. Knowledge of any such differences in DNA methylation is necessary to correctly interpret the results of DNA methylation profiling datasets that are commonly generated and that may underlie regional expression of specific genes and disease states in the human colon.
As a first step toward assessing this possibility, we evaluated DNA methylation, comparing proximal and distal colonic regions by two distinct approaches. First, because it has been documented that individual cell types can have a specific DNA methylation identity (47, 48, 63) including even closely related cell types (21), we utilized laboratory techniques to isolate and characterize a purified epithelial cell population from proximal and distal colonic regions. Second, we used whole unpurified biopsies from the same subjects, examining whether DNA methylation patterns in the epithelial component could be inferred in the heterogeneous cell mixtures from computational deconvolution techniques.
MATERIALS AND METHODS
Patient recruitment and collection of biopsies.
All patient recruitment and sample collection were performed under human subjects protocol approval from the Galway University Hospitals Research Ethics Committee. Patients enrolled in the study were at University Hospital Galway undergoing colonoscopy for the evaluation of symptoms. Biopsies were collected from five different individuals where both the colonoscopic appearance and histological evaluation of biopsies were normal. No patients were taking any medication known to alter the DNA methylome (e.g., folic acid, sulfasalazine or valproic acid).
Isolation of epithelial cells from pinch biopsies.
Previous techniques for isolating purified IECs relied on isolation of cells at room temperature or 37°C (4, 62). We developed modifications of those techniques that allowed for epithelia to be obtained at 4°C to limit the detrimental effects to membrane integrity, cellular viability, and molecular degradation that can occur at higher temperatures (13).
Ten colonoscopic pinch biopsies were taken from the proximal and distal areas of the colon from healthy patients and stored in ice-chilled PBS. Biopsies then underwent washing (3×) with 5 ml of ice-chilled PBS and centrifuged at 250 g for 5 min at 4°C. After the third wash, the ice-chilled PBS was replaced with 25 ml of chelation buffer (1 mM EDTA, 1 mM EGTA, 0.5 M DTT, 55 mM d-sorbitol, 44 mM sucrose with distilled H20 at pH 7.3) and stored for 2 h at 4°C on a rocker. After chelation, the samples were then shaken by hand for 30 s. The cell suspension consisting mostly of intact colon crypts was transferred to a new centrifuge tube, and this step was repeated until no more visible cells were liberated. Finally, the cell suspension was centrifuged at 250 g for 10 min at 4°C, the supernatant was discarded, and the pellet of cells was resuspended in 2 ml of 0.5% BSA in PBS. We aliquoted 200 μl of the sample for cell staining. The remainder of the cells was centrifuged at 250 g at 4°C, and the resulting cell pellet was used for genomic DNA extraction. All reagents were purchased from Sigma-Aldrich (http://www.sigmaaldrich.com).
Cell staining and flow cytometry.
Each sample was incubated with 5 μl of blocking IgG goat serum (Sigma-Aldrich) and stored for 20 min at 4°C. Samples were then double stained with an epithelial-specific marker [FITC Anti-Human CD326 (EpCAM), Biolegend], an immune cell-specific maker (APC Anti-Human CD45, Biolegend), or each marker's isotype control (FITC Mouse IgG2b, κ, APC Mouse IgG, κ, both Biolegend). Samples were then stored for a further 20 min in the dark at 4°C, centrifuged at 320 g at 4°C, and washed twice with 200 μl of 0.5% BSA in PBS. Disaggregation of crypts into a single cell suspension was achieved through pipetting of the cell isolate. Subsequently, cells were examined by flow cytometry with FACSCanto, and data analysis was performed using WinMDI (version 2.8) software.
HELP-tagging assay library preparation.
Extraction of genomic DNA was performed by the method derived from the Albert Einstein University online database (WASP system). All steps outlined in the protocol were followed exactly. Please refer to supplementary materials and methods for a detailed protocol description with no modifications.1
Genomic DNA (3 μg) was digested overnight in 200 μl reactions containing methyl-sensitive restriction enzyme HpaII, thus enriching the hypomethylated portion of the genome. We ran 5 μl of the digest on a 1% agarose gel and used the remainder for library preparation. We added 300 μl of Tris/EDTA (TE) buffer (pH 8.0) to the rest of the digest as well as 500 μl of phenol-chloroform and mixed well. The sample was then centrifuged at top speed for 20 min. The aqueous phase of the sample was then transferred to a new tube and precipitated with 1 μl of glycogen and 50 μl of 3 M sodium acetate. We added 800 μl of isopropanol, and the sample was incubated at −20°C for 2 h and then spun at top speed for 20 min. The supernatant was then removed, and the pellet of DNA was washed with 70% ethanol. The sample was then air-dried and resuspended in 20 μl of TE buffer. Adapter EcoP15I side (AE adapter) ligation was performed in a 50 μl reaction containing 2× Quick ligase buffer, 0.5 μl of 0.1 μM AE adapter, digested DNA, deionized water, and 3 μl of Quick Ligase for 15 min at room temperature. All subsequent steps up to polymerase chain reaction (PCR) amplification were performed using the protocol developed by Suzuki et al. (61). Please refer to supplementary materials and methods for the exact protocol description and Table 1 for a full list of adapters and primers used during library preparation.
Table 1.
List of adapters and primers used during HELP-tagging library preparation
| Name | Sequence (5′-3′) |
|---|---|
| AE | 5′-AcagtaatacgactcactatagggagaaggctCAAGCAGAAGACGGCATACGACAGCAG (AE_1) |
| 5′-p-CGCTGCTGTCGTATGCCGTCTTCTGCTTGagccttctccctatagtgagtcgtattactg*T (*T=Inverted-dT) (AE_2_Int_dT) | |
| AS1 | HT Adapter F 5′- ACACTCTTTCCCTACACGACGCTCTTCCGATCTTATGACG*T |
| HT Adapter R 5′- pCGTCATAAGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT | |
| AS2 | HT Adapter F 5′- ACACTCTTTCCCTACACGACGCTCTTCCGATCTACATCTC*T |
| HT Adapter R 5′- pGAGATGTAGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT | |
| AS3 | HT Adapter F 5′- ACACTCTTTCCCTACACGACGCTCTTCCGATCTCGGTACA*T |
| HT Adapter R 5′- pTGTACCGAGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT | |
| AS4 | HT Adapter F 5′- ACACTCTTTCCCTACACGACGCTCTTCCGATCTGTCATGA*T |
| HT Adapter R 5′- pTCATGACAGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT | |
| PS | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCT |
| PE | CAAGCAGAAGACGGCATACGACAGCAG |
The PCR product was extracted from a 3.5% low molecular weight agarose gel electrophoresis and purified by Mini-Elute gel extraction kit (Qiagen). Purified products were analyzed by Bioanalyzer to ensure integrity and purity followed by Illumina sequencing. All enzymes utilized for the HELP assay were purchased from New England Biosciences (http://www.neb.com). All adapters and primers were purchased from the WASP system at Albert Einstein University.
Processing of sequence data.
Illumina sequencing was performed on an Illumina HiSeq 2000, at the Epigenomics Shared Facility of Albert Einstein University. For this assay, single-end 36–50 base pair (bp) sequencing was required. The images generated by the Illumina sequencer were analyzed with Illumina pipeline software (version 1.4). Default read length of 36 bp was used for initial data preprocessing. Post sequence isolation, in which adapter sequence on the 3′-end was found; adapter sequence was replaced with poly (N) sequence of equal length. The ELAND alignment algorithm was performed on these sequences with the sequence length set to 27 bp. Data generated by the ELAND pipeline were used to count the number of aligned sequences overlapping each CCGG site in the hg19 build of the human genome.
During the alignment process, a maximum of two mismatches in each sequence was accepted. Statistics produced from alignment included the quantity of sequence tags that were excluded because of too many matches to the reference genome (a maximum of 10 matches) or for which there was no match at all. For all nonunique alignments, sequences were assigned a partial count for each alignment location amounting to 1/n, where n represented the total number of alignments. The number of sequences associated with each HpaII site was then divided by the total number of sequences (including partial counts) aligning to all HpaII sites in the same sample to normalize the data between experiments (61).
To measure the level of DNA methylation at each CCGG site, the normalized accumulative proportion (NAP) count for the HpaII digested sample was compared with the reference NAP count for the MspI digest. If the site is hypermethylated, the HpaII NAP count for this site should be less than the MspI NAP count. This is because the HpaII restriction enzyme is not capable of cutting DNA when the internal cytosine is methylated, whereas the MspI enzyme is able to cut regardless of the cytosine methylation state (61). The DNA methylation angle score was calculated using the arctangent of the ratio of HpaII NAP count and MspI NAP count as described previously (30). This allows normalization of HpaII counts in terms of variability of the MspI representation. DNA methylation levels reported here were calculated as one minus the DNA methylation angle score, for ease of interpretability, and range from 0 (no DNA methylation) to 100 (complete DNA methylation).
Data analysis.
CCGG sites with fewer than five MspI reads were excluded from all analyses to improve DNA methylation estimation accuracy. Quantile normalization was then carried out to account for possible variation in total CCGG methylation among samples. Two R packages were used for DNA methylation analysis. First, limma was used to identify individual differentially methylated CCGG sites [i.e., differentially methylated sites (DMS)] (59). Second, Bumphunter was used to identify differentially methylated clusters of CCGG sites i.e., differentially methylated regions (DMRs) (27). A false discovery rate analysis [Benjamini-Hochberg (BH)] was used to correct for multiple testing. We used a false discovery threshold of 0.05 to decide statistical significance.
We tested for enrichment of gene ontology categories among genes for which at least one DMS was found close (± 2 kb) to the transcription start site (TSS). The HELP-tagging assay profiles the DNA methylation status at CCGG sites. However, different genes may be associated with very different numbers of such sites, with genes associated with larger numbers of CCGG sites having a greater chance of being associated with at least one DMS. This can result in severe bias in gene set analysis (17). The R package Goseq was utilized to correct this bias. Goseq calculates a probability weighting function for a list of genes based on a given bias. In this case, the bias was based on the total number of CCGG sites mapping to each gene. The Wallenius approximation was used to calculate the over and under representation of Gene Ontology (GO) categories among differentially methylated genes. A false discovery rate analysis (BH) was used to correct for multiple testing. We used a false discovery threshold of 0.05 to decide statistical significance.
The R package CellMix (16) was utilized to estimate the proportion of epithelial cells in each whole biopsy sample. This was carried out by the semisupervised nonnegative matrix factorization (ssNMF) method, implementing the ssKL algorithm (15). ssNMF takes into account prior knowledge of marker CCGG sites for each cell type in a given sample. Marker sites were used to map each basis component (W) to one of the real cell type signatures (H), with the objective of estimating more stable and meaningful cell population estimates (15). In this case we selected a set of marker sites for epithelial and nonepithelial cell types based on differences in DNA methylation patterns observed between pure and whole biopsy (mixed cell) samples. The binary nature of DNA methylation was utilized to select markers, i.e., sites that were both nonmethylated (methylation score ranging from 0 to 10) across pure samples and intermediately methylated (methylation score ranging from 30 to 70) across mixed cell samples were selected as epithelial marker sites (n = 84). Sites that were intermediately methylated across both pure and mixed samples were chosen as nonepithelial marker sites (n = 12).
The UCSC table browser (32) was used to obtain coordinates of genomic regions including CpG Islands, gene bodies, and intergenic and intragenic regions. The mammalian expression atlas and enhancer peaks were obtained from the FANTOM consortium (1, 12). The epigenomics roadmap consortium (8) was used to obtain coordinates for genomic regions for ChromHMM states in colonic mucosa. In this case all CCGG sites as well as DMS were mapped to each state (n = 18). Enrichment of differential methylation was conditioned on the CCGG density at each state. Significant enrichment was measured with a Fisher exact test. These states, as well as states for other cell lines and tissues can be downloaded from http://egg2.wustl.edu/roadmap/data/byFileType/chromhmmSegmentations/ChmmModels/core_K27ac/jointModel/final/.
Mapping of all CCGG sties, DMS and DMRs to candidate genomic locations such as CpG Islands, gene body, intergenic, intragenic, enhancers, and TSS regions was carried out using customized python scripts.
Validation of differentially methylated sites/regions.
Targeted DNA methylation analysis was performed using MassArray EpiTYPER (Agena Bioscience) technology as previously described (49, 50). In brief, 1 μg of genomic DNA was bisulphite-treated, where nonmethylated cytosine is converted to uracil while methylated cytosine remains unchanged. Using mass spectrometry, we performed semiquantitative methylation analysis by comparing signals (the area under the peak) representing methylated and nonmethylated DNA template. A T-specific cleavage reaction was performed to discover methylation sites and to determine methylation ratios within a given target region. Methylation calls were performed by the EpiTYPER software v1.2 (Agena Bioscience) and written to an Oracle 8i database. Statistical analysis of individual CpGs was performed using the R package limma (59) to identify patterns of differential methylation. A false discovery rate analysis (BH) was used to correct for multiple testing. We used a false discovery threshold of 0.05 to decide statistical significance.
For both HELP-tagging and MassArray EpiTYPER data, differential methylation is expressed as delta methylation (Δ Methylation). Δ Methylation reflects the geometric mean methylation score of one sample group relative to another i.e., a Δ Methylation value of > 0 reflects hypermethylation in the distal colon, and a Δ Methylation value of < 0 reflects hypermethylation in the proximal colon.
RESULTS
We report DNA methylation analysis comparing proximal and distal colonic regions in purified epithelial cell populations and the results of a novel deconvolution technique in heterogeneous whole biopsy samples from healthy individuals.
Establishment of a colonic cell suspension enriched in epithelial cells.
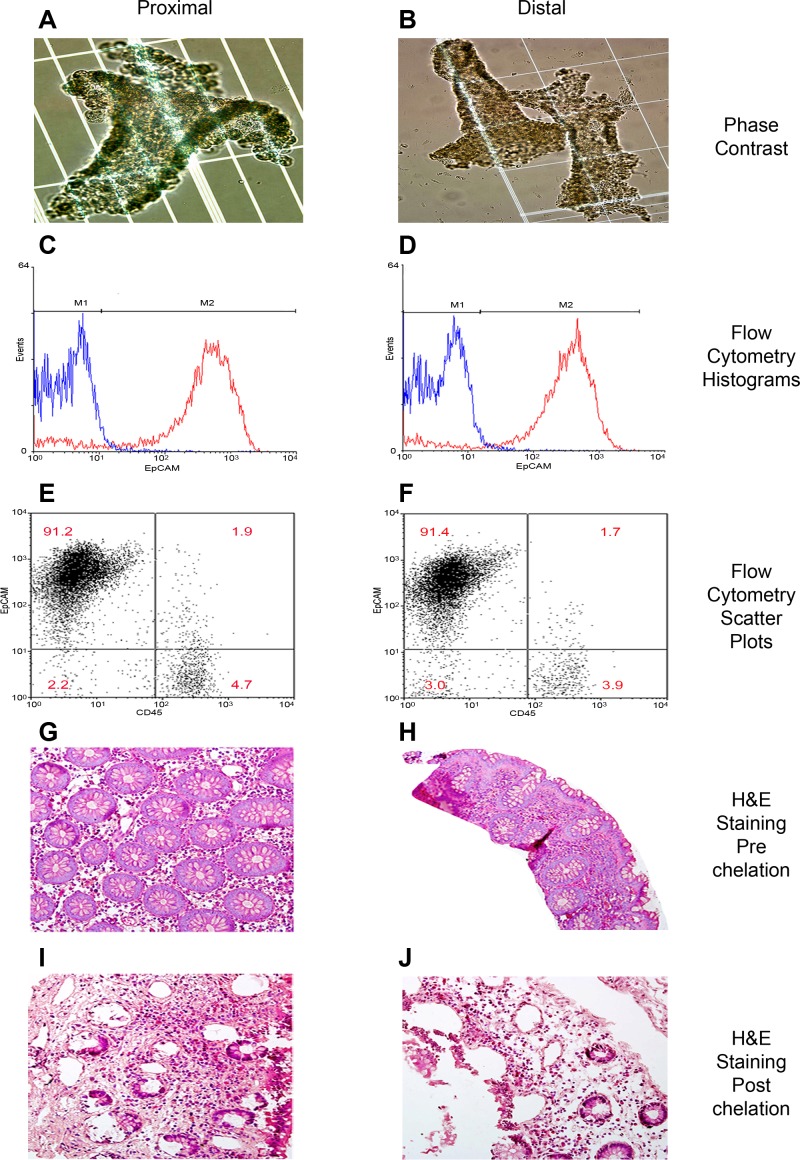
Our method is a modification of two previously developed techniques (11, 64) that allows the detachment of whole epithelial crypts from mucosal biopsies of the colon. Histologic examination of the biopsies postdetachment of epithelial crypts revealed that whole crypt structures could be removed from the biopsies leaving cells of the lamina propria behind (Fig. 1, A and B). We used flow cytometry with markers specific for IECs and bone marrow-derived cells to assess the cellular make-up of the suspensions resulting from the chelation procedure. We found that > 90% of the cell suspension comprised EpCAM-positive cells, indicative of epithelial cells (Fig. 1, C–F, Table 2). Approximately 5% of the suspension cells were stained with the CD45 antibody, indicating bone marrow origin. The double negative cells in this analysis could be stromal cells such as fibroblasts or endothelial cells, which are neither epithelial nor of bone marrow origin. Compared with whole colon biopsies, which display high proportions of nonepithelial cells by morphological appearance (Fig. 1, G–J), these data indicate a successful enrichment of epithelial cells in the suspension.
Fig. 1.
Classification of cellular proportions in proximal and distal colonic regions. Intestinal epithelial cells (IECs) isolated from mucosal pinch biopsies are illustrated (A–F). Phase contrast microscopy of intestinal crypts post-IEC isolation of proximal (A) and distal (B) regions. IECs were labeled with fluorescent antibodies EpCAM and CD45 to distinguish cell populations. Representative histograms of EpCAM-labeled cells (red) and its isotype control (blue) are illustrated in proximal (C) and distal (D) regions. Quantification of the percentage of epithelial cells in the IEC isolate was then performed. Representative scatter plots of EpCAM-positive cells (top left), CD45-positive cells (bottom right) and double negative cells (bottom left and top right) are illustrated in proximal (E) and distal (F) regions. Representative examples of hematoxylin and eosin (H&E) staining of colonic mucosal tissue are illustrated before (G and I) and after (H and J) chelation in proximal and distal regions.
Table 2.
Estimated proportions of epithelial cell isolate postchelation in proximal and distal samples
| Sample | EpCAM Positive, % | CD45 Positive, % | Double Negative, % |
|---|---|---|---|
| Proximal 1 | 90.1 | 3.7 | 6.2 |
| Proximal 2 | 89.6 | 4.4 | 6.0 |
| Proximal 3 | 91.0 | 5.6 | 3.4 |
| Proximal 4 | 91.2 | 4.7 | 4.1 |
| Proximal 5 | 89.1 | 5.9 | 5.0 |
| Distal 1 | 92.8 | 5.2 | 2.0 |
| Distal 2 | 93.0 | 4.8 | 2.2 |
| Distal 3 | 90.7 | 3.8 | 5.5 |
| Distal 4 | 92.1 | 5.1 | 2.8 |
| Distal 5 | 91.4 | 3.9 | 4.7 |
Sequencing data and coverage.
DNA methylation was assayed in both pure epithelial cell samples and whole biopsies using the HELP-tagging assay (materials and methods). Multiplexing of HELP-tagging libraries for both pure and whole biopsy samples was done with four libraries per lane. For each sample, DNA methylation was measured at ∼1.9 million CCGG sites; however, only ∼1.6 million sites remained when sites with fewer than 5 MspI reads were removed. DNA methylation levels were measured by a modified version of the angle methylation score; this ranged from 0 (no DNA methylation) to 100 (complete DNA methylation). The average number of reads for all samples varied from 10.9 to 20.2 million HpaII reads per sample with an average depth of coverage between 9.9X and 18.2X (Table 3).
Table 3.
HELP-tagging read count for each lane
| Lane Number | Average Read Number per Lane, millions | Average Aligned Hits,% | Average Coverage |
|---|---|---|---|
| Lane 1 | 15.58 | 86.25 | 14.3 X |
| Lane 2 | 20.18 | 83.75 | 18.16 X |
| Lane 3 | 16.9 | 83.5 | 15.22 X |
| Lane 4 | 10.93 | 84 | 9.89 X |
| Lane 5 | 15.83 | 81.75 | 13.95 X |
Genome-wide patterns of DNA methylation.
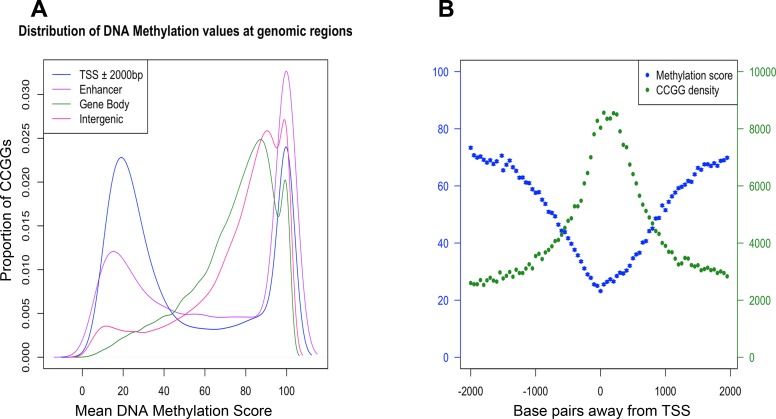
The distribution of DNA methylation values shows that the majority of CCGG sites in the genome were methylated (i.e., DNA methylation score ≥ 70) (Fig. 2A). Consistent with previous reports (19, 24, 46), a higher proportion of nonmethylated CCGG sites (i.e., DNA methylation score ≤ 30) fell within the vicinity of the TSS of genes (± 2 kb). The distribution in this region was bimodal, whereas other genomic locations such as the gene body and intergenic regions were predominantly methylated (Fig. 2A).
Fig. 2.
Global DNA methylation patterns relative to protein-coding genes. Distribution of all DNA methylation values across the genome partitioned by position relative to transcription start site (TSS) peaks, enhancer peaks, gene body, and intergenic regions (A). Mean DNA methylation score (blue), standard error (blue bars), and density (green) of CCGG sites with respect to their position relative to TSS peaks (B).
Gene regulation by epigenetic modification takes place at promoters and distally located regulatory elements (6). To characterize the DNA methylation patterns at promoter regions, we utilized the mammalian expression atlas generated by the FANTOM consortium (12) to map CCGG sites that fall within the vicinity of TSS peaks (± 2 kb). Results illustrated the relationship between CCGG rich and depleted regions and the relative DNA methylation state at these regions. As expected, the proportion of CCGG sites was higher at the TSS, becoming relatively depleted up and downstream of the TSS. However, DNA methylation decreased in close proximity to TSSs (mean score = 21 at the TSS) and then increased both upstream and downstream from each TSS peak (mean score = 74.1 ± 2 kb from TSS) (Fig. 2B). This characterized the typically unbalanced nature of DNA methylation in a normal state, with 70–80% of the genome being methylated, whereas nonmethylated loci generally tended to cluster in groups around the TSS of protein-coding genes (5).
Identification of differentially methylated sites between proximal and distal colonic regions in purified epithelial cell samples.
The R package limma was used to identify site-specific DNA methylation differences between proximal and distal colonic regions. We identified 125 DMS between proximal and distal samples. Of these DMS, 93 (74.4%) showed higher methylation in the distal colon compared with the proximal colon. A small portion of the DMS (n = 17, i.e., 14%) mapped to multiple locations within the same gene. Of the 125 DMS, 78 DMS mapped to 61 unique genes (Supplementary Table S1). Generally, those DMS that mapped to multiple locations within the same gene had similar DNA methylation patterns, as has been documented in previous studies (34). This was the case, for example for HOXB3, HOXB4, HOXB6, HOXB7, and HOXC4 were protein-coding genes that showed similar patterns of differential methylation at multiple CCGG sites (Supplementary Table S1).
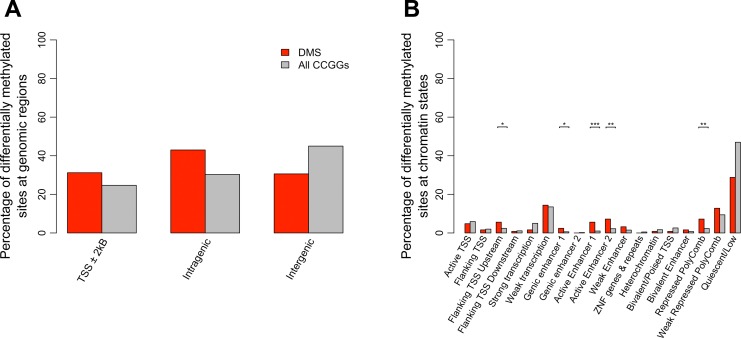
The results also indicated an enrichment of DMS at TSS peaks (∼31% of CCGG sites that were located near TSSs were differentially methylated compared with ∼21% of all CCGG sites, P value = 0.03, Fisher exact test) (Table 4). However, the majority, 64% of DMS were located at noncoding intragenic or intergenic regions (Fig. 3A). Analysis identified 125 DMS mapping to TSS peaks associated with 33 genes that were differentially methylated between proximal and distal regions of colonic epithelial cells (Table 4).
Table 4.
List of significantly DMS that map to candidate TSS peaks (± 2 kb)
| DMS | Gene Name | Description | Δ Methylation | FDR |
|---|---|---|---|---|
| chr17-46683559 | HOXB7 | homeobox B7 | 92.47932 | 6.64E-07 |
| chr17-46683644 | HOXB7 | homeobox B7 | 86.18336 | 6.80E-06 |
| chr17-46674011 | HOXB6 | homeobox B6 | 86.86708 | 0.000403693 |
| chr20-48324489 | B4GALT5 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 | 25.50876 | 0.002045373 |
| chr3-119785950 | GSK3B | glycogen synthase kinase 3 beta | 76.18214 | 0.002045373 |
| chr4-6578275 | MAN2B2 | mannosidase, alpha, class 2B, member 2 | 67.02742 | 0.002045373 |
| chr17-27053049 | TLCD1 | TLC domain containing 1 | −82.34036 | 0.002498093 |
| chr17-46683607 | HOXB7 | homeobox B7 | 81.83062 | 0.002597815 |
| chr16-89688808 | DPEP1 | dipeptidase 1 (renal) | −81.10938 | 0.002825292 |
| chr3-72939150 | GXYLT2 | glucoside xylosyltransferase 2 | 81.08082 | 0.002937034 |
| chr12-54428544 | HOXC4/HOXC5 | homeobox C5 | −70.1646 | 0.003048136 |
| chr17-75112449 | SEC14L1 | SEC14-like 1 | −55.19028 | 0.003048136 |
| chr20-22562585 | FOXA2 | forkhead box A2 | −74.31288 | 0.003048136 |
| chr17-46629480 | HOXB3 | homeobox 3 | 67.2539 | 0.003352509 |
| chr17-9054773 | NTN1 | netrin 1 | 81.72508 | 0.003352509 |
| chr20-22562511 | FOXA2 | forkhead box A2 | −78.6755 | 0.00431715 |
| chr1-201982462 | ELF3 | E74-like factor 3 (ets domain transcription factor, epithelial-specific) | −78.33404 | 0.005491125 |
| chr8-99961645 | OSR2 | odd-skipped related 2 (Drosophila) | −71.71648 | 0.005873741 |
| chr17-46683048 | HOXB7 | homeobox B7 | 62.81628 | 0.005873741 |
| chr17-46671931 | HOXB6 | homeobox B6 | 76.75316 | 0.005873741 |
| chr20-22562467 | FOXA2 | forkhead box A2 | −65.65948 | 0.005873741 |
| chr12-54423547 | HOXC6 | homeobox C6 | −60.96322 | 0.007053416 |
| chr19-46022568 | VASP | vasodilator-stimulated phosphoprotein | 80.23486 | 0.008287636 |
| chr10-63852409 | ARID5B | AT rich interactive domain 5B (MRF1-like) | 56.37224 | 0.012281356 |
| chr17-46654129 | HOXB4 | homeobox B4 | 82.07232 | 0.012281356 |
| chr13-99461626 | DOCK9 | dedicator of cytokinesis 9 | 22.2441 | 0.013171602 |
| chr16-2013144 | RPS2 | ribosomal protein S2 | 65.14832 | 0.016932408 |
| chr1-45002565 | RNF220 | ring finger protein 220 | 42.79432 | 0.017525011 |
| chr4-154143656 | TRIM2 | tripartite motif-containing 2 | 21.11232 | 0.017525011 |
| chr7-45148667 | TBRG4 | transforming growth factor beta regulator 4 | 43.28968 | 0.017525011 |
| chr17-77807502 | CBX4 | chromobox homolog 4 (Pc class homolog, Drosophila) | 45.12846 | 0.026487069 |
| chr21-43371580 | C2CD2 | C2 calcium-dependent domain containing 2 | 22.36632 | 0.029436478 |
| chr10-86005070 | RGR | retinal G protein-coupled receptor | 79.64252 | 0.032571793 |
| chr11-93476463 | C11orf54 | chromosome 11 open reading frame 54 | 50.5306 | 0.032571793 |
| chr17-46673605 | HOXB6 | homeobox 6 | 84.34082 | 0.036240058 |
| chr19-18812199 | CRTC1 | CREB regulated transcription coactivator 1 | 77.80982 | 0.036240058 |
| chr11-45948422 | GYLTL1B | glycosyltransferase-like 1B | 34.04874 | 0.036802373 |
| chr9-137002691 | WDR5 | WD repeat domain 5 | 74.93544 | 0.03721202 |
| chr13-21597674 | LATS2 | large tumor suppressor, homolog 2 (Drosophila) | 76.966 | 0.043869031 |
Benjamini-Hochberg adjusted P value cut-off of 0.05.
TSS, transcription start site; DMS, differentially methylated site; FDR, false discovery rate.
Fig. 3.
Annotation of CCGG sites to genomic regions and ChromHMM states. The percentage of CCGG sites (gray bars) and differentially methylated sites (DMS) (red bars) annotating to TSSs, intragenic and intergenic regions is shown (A). Results indicated enrichment of DMS at TSS peaks (P value = 0.03, Fisher exact test). However, the majority of DMS were located at noncoding intragenic or intergenic regions. The total percentage of probes is greater than 100% as several probes were classified as belonging in more than one class of genomic region. The percentage of CCGG sites (gray bars) and DMS (red bars) mapping to 18 active and repressed genomic states in colonic mucosa using the ChromHMM model is shown (B). Using the genomic states defined by the ChromHMM model, we tested for significant enrichment of DMS, conditioned on the CCGG density at each genomic state. P values were determined using a fisher exact test (*<0.05, **<0.005, ***<0.005). Results indicate significant enrichment at enhancers, TSS regions as well as repressed polycomb regions.
GO analysis of differential DNA methylation events at promoter regions was then carried out using the R package Goseq. Following bias correction, we identified genes that were assigned to seven GO categories for biological processes (GO:BP) that were significantly enriched (FDR <0.05). Notably, genes were enriched for GO terms associated with embryological development (GO:0048706, BH adjusted P = 8.8 × 10−3) and anterior-posterior formation (GO:0009952, BH adjusted P = 0.02). These included a considerable number of genes from the HOX clusters (HOXB3, HOXB4, HOXB6, HOXB7, HOXC4, HOXC5, and HOXC6) (Table 4).
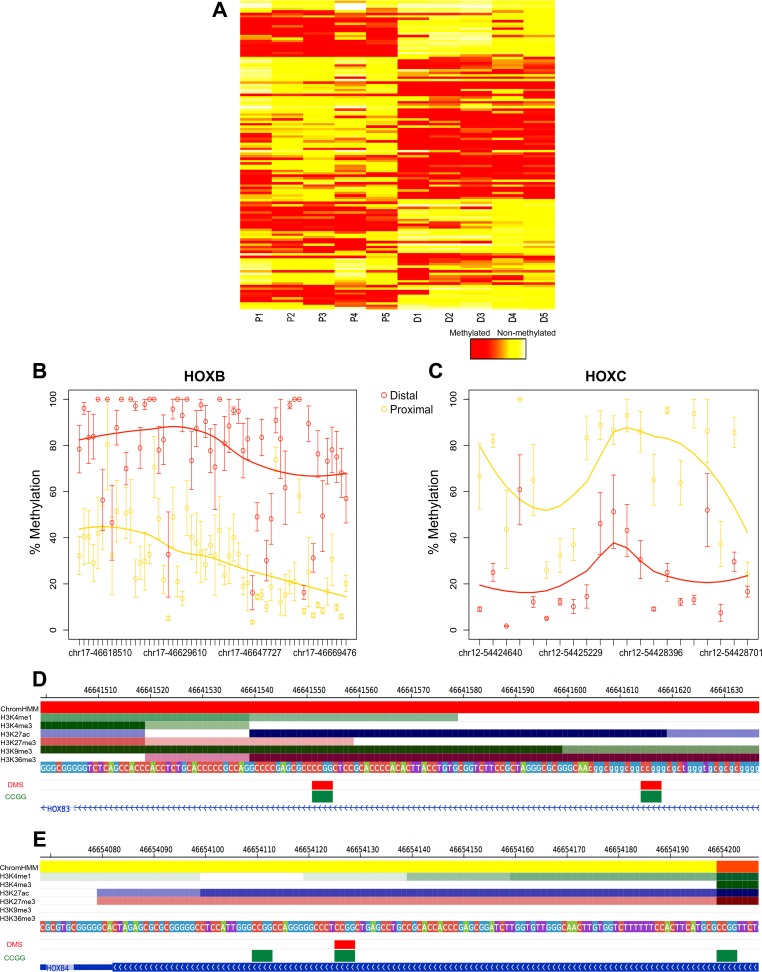
To give these differential methylation patterns identified further epigenetic context, we utilized publically available CHIP-Seq data of active and repressed histone marks (n = 6, H3K4me3, H3K4me1, H3K36me3, H3K27me3, H3K9me3, and H3K27ac). These data were employed to describe the chromatin arrangements present in colonic mucosa using the ChromHMM model (9). The roadmap epigenomics consortium expanded on a model developed by Ernst and Kellis (9), generating a tissue and cell-type specific 18 state model that encapsulated all key interactions between these chromatin marks at various genomic elements. We utilized this model to calculate enrichment of differential methylation at a number of genomic states including promoter, enhancer, transcribed, and repressed regions. We found DMS as well as all CCGGs were most heavily distributed at a quiescent/low activity state (Table 5), as was documented in previous studies (8, 39). We also tested for significant enrichment of DMS, conditioned on the CCGG density at each genomic state (Fig. 3B). Enrichment was identified upstream from the active TSS (P value = 0.03, Fisher exact test), at enhancer states (genic enhancer: P value = 0.04, active enhancer 1: P value = 3.4e-04, active enhancer 2: P value = 2e-03, Fisher exact test), as well as transcriptionally repressed polycomb regions (P value = 2e-03, Fisher exact test) (Fig. 3B). DMS at the active TSS region of HOXB3, active enhancer region of HOXB4, as well as the CHIP-seq profiles of the set of six chromatin marks assayed in colonic mucosa, are shown in Fig. 4, D and E, respectively. The figure illustrates elevated levels of H3K27ac at these regions, a histone mark that highlights active regulatory elements and differentiates active enhancers and promoters from their inactive complements.
Table 5.
Total number of CCGG loci mapping the each ChromHMM state (n = 18)
| ChromHMM State | Mapped DMS, n | Mapped CCGG Sites, n |
|---|---|---|
| 1) Active TSS | 6 | 98793 |
| 2) Flanking TSS | 2 | 34168 |
| 3) Flanking TSS upstream | 7 | 40614 |
| 4) Flanking TSS downstream | 1 | 18671 |
| 5) Strong transcription | 2 | 83295 |
| 6) Weak transcription | 18 | 225469 |
| 7) Genic enhancer 1 | 3 | 10395 |
| 8) Genic enhancer 2 | 0 | 3365 |
| 9) Active enhancer 1 | 7 | 17187 |
| 10) Active enhancer 2 | 9 | 37577 |
| 11) Weak enhancer | 4 | 25863 |
| 12) ZNF genes and repeats | 0 | 7926 |
| 13) Heterochromatin | 1 | 28139 |
| 14) Bivalent/poised TSS | 1 | 42915 |
| 15) Bivalent enhancer | 2 | 12297 |
| 16) Repressed polycomb | 9 | 37756 |
| 17) Weak repressed polycomb | 16 | 156986 |
| 18) Quiescent/low | 37 | 782010 |
Fig. 4.
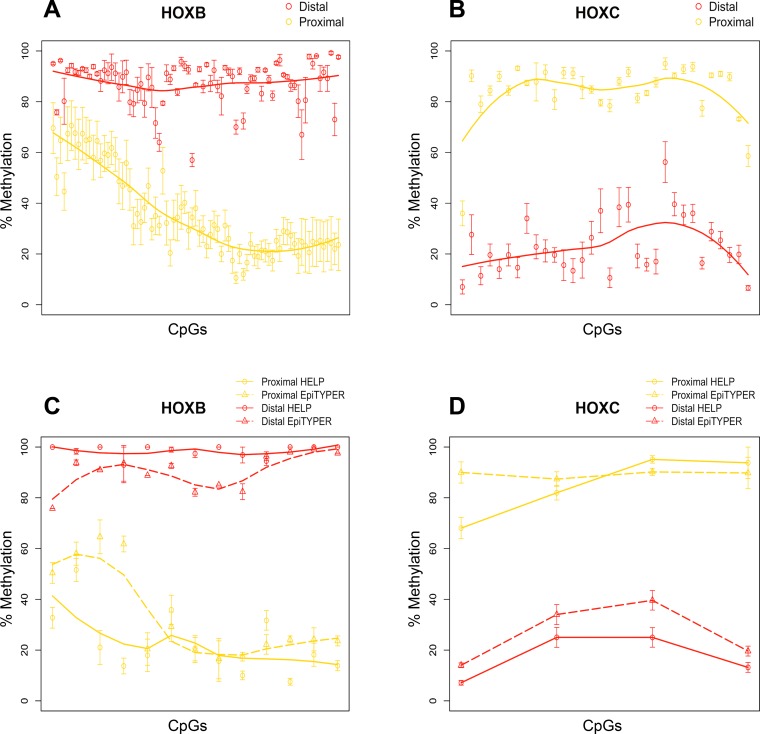
Site and region-specific differential methylation between proximal and distal regions: HELP-tagging assay. DNA methylation values of DMRs using a color scale from red (high DNA methylation) to yellow (low DNA methylation). Columns represent samples (P, proximal; D, distal), and rows represent all CCGG sites that fall within each DMR (A). Scatter plot illustrating individual CCGG loci (circles) contained within DMRs mapping to HOXB genes (B) and HOXC genes (C) between proximal (yellow) and distal (red) samples. Data points are presented as means ± SE. DMS mapping to the active TSS region (red) of HOXB3 (D), active enhancer region (yellow) of HOXB4 (E), as well as the CHIP-seq profiles of the set of 6 histone marks assayed in colonic mucosa are shown. Below the histone marks, DMS (red) and all CCGG sites (green) mapping to each gene segment are also illustrated.
Functionally relevant sites of DNA methylation are often associated with associated with individual CpGs (44), as well as genomic regions such as at CpG islands (26), CpG island shores, and CpG island shelves (25). We used Bumphunter (27) to identify differentially methylated regions (DMRs). The Bumphunter method has the benefit of combining surrogate variable analysis (40), a method used for modeling unexplained heterogeneity (e.g., batch effects) in genomic measurements, regression modeling, smoothing techniques, and multiple comparison methods to provide reliable lists of DMRs.
This analysis identified 11 DMRs between proximal and distal colonic regions. Consistent with the results from limma, Bumphunter identified several differentially methylated genomic regions close to homeobox (HOX) genes, including HOXB2, HOXB3, HOXB5, HOXB6, HOXC4, and HHEX (Fig. 4, A–C; Table 6). A high proportion of these peaks were identified at loci associated with functionally relevant genomic regions, i.e., ∼55% were located at CpG islands (two peaks in the gene body of HOXB3 and HOXC4, the TSS of FOXA2 and HOXB5, and an intragenic region of HHEX), ∼27% at CpG island shores (± 1 kb from CpG island; gene body of HOXB2, an intergenic region between HOXB5 and HOXB6 and an intragenic region of HOXB2), and 9% at CpG island shelves (± 2 kb from CpG island; the gene body region of OSR2).
Table 6.
List of significantly DMRs located by Bumphunter
| Chr Number | Start Position | End Position | CpG Context | Gene Symbol | Δ Methylation | FDR |
|---|---|---|---|---|---|---|
| Chr 17 | 46641476 | 46642110 | CpGI | HOXB3 | 54.881204 | 0.0032 |
| Chr 17 | 46647727 | 46648208 | Open Sea | HOXB3 | 53.093463 | 0.0088 |
| Chr 20 | 22562221 | 22563809 | CpGI | FOXA2 | −49.289813 | 0.0104 |
| Chr 17 | 46671413 | 46672379 | CpGIShore | HOXB5/HOXB6 | 50.047402 | 0.0128 |
| Chr 17 | 46629027 | 46629972 | CpGIShore | HOXB3 | 49.601861 | 0.0184 |
| Chr 10 | 94455582 | 94456148 | CpGI | HHEX | 42.943158 | 0.0208 |
| Chr 12 | 54428314 | 54428935 | CpGI | HOXC4 | −47.370940 | 0.0208 |
| Chr 17 | 46669476 | 46670126 | CpGI | HOXB5 | 45.448684 | 0.0208 |
| Chr 12 | 54424612 | 54425447 | CpGI | HOXC4 | −42.839777 | 0.0259 |
| Chr 17 | 46618510 | 46619969 | CpGIShore | HOXB2 | 38.690672 | 0.0292 |
| Chr 8 | 99961123 | 99962167 | CpGIShelf | OSR2 | −45.342486 | 0.0424 |
Benjamini-Hochberg adjusted P value cut-off of 0.05.
DMR, differentially methylated region; Chr, chromosome; CpGI, CpG island; CpGIShore, <2 kb from CpG Island; CpGIShelf, <4 kb from CpG Island; Open Sea, >4kb from CpG Island.
We next performed validation for the defined DMRs/DMS in our purified epithelial cell samples. Targeted CpGs contained within 17 amplicons that mapped to HOXB and HOXC genes were chosen to validate the patterns of differential methylation observed from the HELP-tagging data. These amplicons contained 17 CpGs assessed by the HELP-tagging assay, as well as neighboring CpGs (due to amplicons design) where we also measured differential methylation (n = 116 CpGs with informative methylation percentages). The R package limma was used to identify site-specific DNA methylation differences between proximal and distal colonic regions. Of the 116 CpGs assessed, we identified 112 DMS mapping to HOXB (n = 79 CpGs) and HOXC (n = 33 CpGs) genes (Fig. 5, A and B). On a site-specific level, the Δ Methylation of specific CpG sites (n = 17) mapping to HOXB and HOXC genes assessed in both the HELP-tagging assay and EpiTYPER MassArray is shown in Table 7 and Fig. 5, C and D. All of these sites were differentially methylated in the same direction according to both methylation assays.
Fig. 5.
Validation of differentially methylated regions and sites by MassArray EpiTYPER. Scatter plot of differentially methylated CpGs mapping to HOXB (n = 79 CpGs) (A) and HOXC (n = 33 CpGs) (B) genes between proximal (yellow) and distal (red) samples measured by MassArray EpiTYPER. Comparison of DNA methylation patterns of individual CpGs mapping to HOXB (C) and HOXC (D) genes assessed by HELP-tagging (circles) and MassArray EpiTYPER (triangles) in proximal (yellow) and distal (red) samples. All data points are presented as means ± SE. Results indicate a consistent pattern of differential methylation from HELP-tagging and MassArray EpiTYPER, 2 independent DNA methylation assays.
Table 7.
Δ Methylation of specific CpG sites (n = 17) mapping to HOXB and HOXC genes assessed in both the HELP-tagging assay and EpiTYPER MassArray
| DMS | Δ Methylation (HELP) | FDR | Δ Methylation (EpiTYPER) | FDR |
|---|---|---|---|---|
| chr12-54423547 | −65.076 | 0.00059452 | −76.0 | 9.91E-09 |
| chr12-54424689 | −56.983 | 0.01946927 | −53.4 | 1.87E-06 |
| chr12-54428544 | −55.726 | 0.000536599 | −50.6 | 6.23E-07 |
| chr12-54428644 | −71.916 | 0.001873619 | −70.2 | 7.77E-09 |
| chr17-46629480 | 67.253 | 0.003352509 | 25.4 | 0.00383449 |
| chr17-46639805 | 46.71816 | 0.046413584 | 35.8 | 0.000730088 |
| chr17-46641555 | 78.9683 | 0.019383318 | 26.4 | 0.002913226 |
| chr17-46641618 | 79.24532 | 0.040733787 | 31.8 | 0.001595676 |
| chr17-46654129 | 82.07232 | 0.012281356 | 68.4 | 1.04E-07 |
| chr17-46660594 | 41.57789491 | 0.0325717925 | 63.4 | 2.08E-06 |
| chr17-46671931 | 76.75316 | 0.005873741 | 62.2 | 4.52E-06 |
| chr17-46673605 | 84.340 | 0.036240058 | 68.4 | 1.04E-07 |
| chr17-46674011 | 86.86708 | 0.000403693 | 65.0 | 5.10E-08 |
| chr17-46683048 | 62.816 | 0.005873741 | 74.2 | 5.91E-08 |
| chr17-46683559 | 92.47932 | 6.64E-07 | 73.8 | 2.68E-06 |
| chr17-46683607 | 81.83062 | 0.002597815 | 75.0 | 7.14E-06 |
| chr17-46683644 | 86.18336 | 6.80E-06 | 74.0 | 6.08E-06 |
Benjamini-Hochberg adjusted P value cut-off of 0.05.
To determine how differential methylation patterns regulate gene expression, we utilized two previously published datasets comparing genome-wide transcriptome profiles of proximal and distal colonic mucosa (18, 37). Although the number intersecting differentially methylated and differentially expressed genes between both datasets was quite low [n = 4 Glebov et al. (18), n = 3 LaPointe et al. (37)], the most variable differentially methylated genes containing DMRs at CpG island shores and CpG island shelves were also differentially expressed in both studies. These included genes HOXB2, HOXB6, and FOXA2. In fact genes HOXB2 [Δ Methylation + 38.69 (Table 6), LogFC = −0.73, Glebov et al.] and HOXB6 [Δ Methylation + 50.04 (Table 6), LogFC −1.49, LaPointe et al.; LogFC −0.29, Glebov et al.] were both hypermethylated and downregulated in the distal colon. FOXA2 showed the opposite pattern, as it was hypermethylated and downregulated in the proximal colon [Δ Methylation + 49.29 (Table 6), LogFC −1.35, La Pointe et al.]. These results provide some evidence that DNA methylation may regulate the expression patterns of given protein-coding genes in different colonic regions.
Computational deconvolution of DNA methylation patterns in epithelial cells from whole biopsies.
Tissues obtained from human subjects consist of mixtures of different types of cells that can occur in varying proportions. We sought to develop reliable computational deconvolution techniques that can infer the DNA methylation patterns in individual cell types within the heterogeneous mixtures. Such techniques have been applied extensively to gene expression data (54) and more recently also to DNA methylation data (22). In this study the availability of DNA from whole biopsies and experimentally purified epithelial cells from the same subjects allowed us to investigate the accuracy of computational deconvolution techniques applied to DNA methylation data.
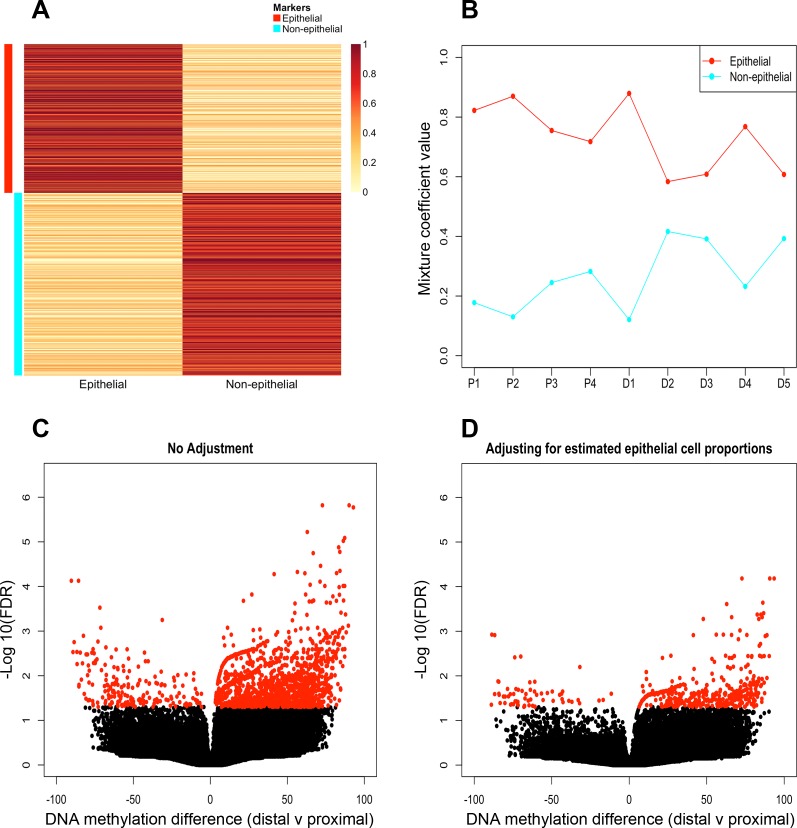
We observed a higher proportion of sites at intermediate DNA methylation levels (i.e., defined as sites with DNA methylation scores between 30 and 70) in the whole biopsies compared with the purified epithelial cell samples (P = 0.028, Table 8). This suggested that sites at intermediate DNA methylation levels may result from mixtures of different cell types with distinct DNA methylation patterns. We therefore assessed whether the epithelial cell component of whole colon biopsies could be accurately inferred by computational methods. We used CellMix for this purpose, a computational tool that we developed previously to deconvolve gene expression data from heterogeneous samples (16), although it has also been recently used for DNA methylation data (65). CellMix provides a unified framework for gene expression deconvolution, enabling access to several different computational methods that have been applied to this problem. We chose the ssNMF method (15). We restricted the analysis to sites found at intermediate DNA methylation levels in the whole biopsy samples and therefore corresponding to positions that may be differentially methylated between the cell types that constitute the whole biopsy samples. ssNMF makes use of a set of signature sites that are, in this application, nonmethylated in the cell type of interest and intermediately methylated in the mixed samples. These are putatively marker sites for the epithelium. They enable the ssNMF method to determine the component of the variation in DNA methylation across samples that is attributable to variation in the proportion of epithelial cells. This in turn allows CellMix to estimate the epithelium proportion in the mixed samples. The mean methylation of the epithelium and nonepithelium cell-type marker sites is shown in Fig. 6A and the estimated proportions of epithelial cells in each sample are shown in Fig. 6B. Results indicate that epithelial cells are the most abundant cell type in whole biopsies, with the epithelial proportion being relatively higher in proximal samples compared with distal samples and intersample variability being relatively lower in proximal samples (mean epithelial proportion ∼79% ± 6.8) relative to distal samples (mean epithelial proportion ∼68% ± 12.9).
Table 8.
Number of intermediate sites in proximal and distal samples in pure and whole biopsy samples
| Pure Samples | Number of CCGG Sites (30-70 score) | Whole Biopsy | Number of CCGG sites (30-70 score) |
|---|---|---|---|
| P1 | 185345 | P1 | 184066 |
| P2 | 229650 | P2 | 240504 |
| P3 | 149158 | P3 | 191809 |
| P5 | 138091 | P5 | 189068 |
| D1 | 115113 | D1 | 190231 |
| D2 | 193359 | D2 | 227548 |
| D3 | 173641 | D3 | 185728 |
| D4 | 142948 | D4 | 192485 |
| D5 | 166793 | D5 | 198909 |
P, proximal; D, distal.
Fig. 6.
Identification of DNA methylation differences in proximal and distal colonic mucosa following deconvolution. A marker map illustrating the mean methylation of the epithelium (red) and nonepithelium (cyan) cell-type marker sites are annotated by colored bands (A). Profile plot illustrating cell proportion estimates of epithelial (red) and nonepithelial (cyan) populations in whole biopsy samples (B). Volcano plots of differential DNA methylation analysis pre- (C) and postdeconvolution (D). x-Axis, DNA methylation difference between distal and proximal samples; y-axis, −Log10 for each nonsignificant (black) and significant (red) (Benjamini-Hochberg adjusted P value of 0.05) site of whole biopsy samples.
We used the estimated epithelial proportions as a covariate in an analysis of differential DNA methylation in the whole biopsy samples. The objective was to reduce the impact of variation in the epithelial component (the main constituent cell type of the whole biopsy samples) on the differential DNA methylation analysis of those samples. The number of DMS identified between proximal and distal regions in whole biopsies was reduced from 1,820 to 537 sites (Fig. 6, C and D) when this covariate was included (Supplementary Table S2). These 537 sites mapped to TSS peaks associated with 124 genes. Although the number of DMS identified in the deconvolution corrected whole biopsy samples was far greater than those identified in purified epithelium, there were DMS from 15 genes that were consistently differentially methylated in both datasets, including HOXB5, HOXB6, HOXB7, GXYLT2, HOXC4, HOXC5, FOXA2, OSR2, LATS2, and RGR (Table 9). Consistent with results from pure epithelial cell types, post-CCGG bias correction, differential DNA methylation analysis identified the enrichment of genes with GO terms associated with embryological development (GO:0048706, BH adjusted P = 0.036) between proximal and distal colonic regions. These included a considerable number of genes from the HOX clusters [HOXB5, HOXB6, HOXB7, HOXB9, and HOXC5 (Supplementary Table S2)].
Table 9.
List of significantly DMS shared between pure and whole biopsy samples (following deconvolution)
| DMS | Gene Symbol | Genomic Region | Δ Methylation (Pure) | Δ Methylation (WB) | FDR (Pure) | FDR (WB) |
|---|---|---|---|---|---|---|
| chr17-46683559 | HOXB7 | TSS | 92.47932 | 93.52100253 | 6.64E-07 | 6.57E-05 |
| chr20-12021310 | N/A | Intergenic | 88.88558 | 78.45390423 | 2.01E-06 | 0.01962828 |
| chr 3-64490712 | N/A | Intergenic | 84.62064 | 76.62662015 | 0.000717469 | 0.005626721 |
| chr20-12021361 | N/A | Intergenic | 80.57726 | 77.78940502 | 0.002498093 | 0.016091634 |
| chr14-64071629 | WDR89 | GB | 86.37394 | 81.14626421 | 0.002683528 | 0.026326852 |
| chr 4-41158891 | AFB2 | GB | 85.76930 | 87.80469167 | 0.002825292 | 0.001284722 |
| chr 3-72939150 | GXYLT2 | TSS | 81.08082 | 66.10503178 | 0.002937034 | 0.000484394 |
| chr12-54428544 | HOXC4/HOXC5 | TSS | −70.16460 | −71.89992964 | 0.003048136 | 0.043214188 |
| chr20-22562585 | FOXA2 | TSS | −74.31288 | −67.94694421 | 0.003048136 | 0.049633641 |
| chr6-156919819 | N/A | Intergenic | −67.64680 | 65.41223713 | 0.003048136 | 0.040605349 |
| chr20-22562511 | FOXA2 | TSS | −78.67550 | −67.53913942 | 0.00431715 | 0.048195907 |
| chr 8-41454062 | AGPAT6 | N/A | 85.91622 | 87.3631527 | 0.004677339 | 0.024438054 |
| chr17-46671931 | HOXB5/HOXB6 | TSS | 76.75316 | 64.30906393 | 0.005873741 | 0.016979544 |
| chr 8-99961645 | OSR2 | TSS | −71.71648 | −74.55163091 | 0.005873741 | 0.025501327 |
| chr13-112786724 | N/A | Intergenic | 68.93572 | 75.33322323 | 0.012997517 | 0.016091634 |
| chr12-54428644 | HOXC5 | TSS | −80.64078 | −88.94722457 | 0.015139795 | 0.04438151 |
| chr5-134662970 | N/A | Intergenic | −69.45856 | −80.19381577 | 0.017525011 | 0.040783135 |
| chr13-21597264 | LATS2 | TSS | 78.17346 | 82.16007176 | 0.01946927 | 0.045614529 |
| chr10-86005070 | RGR | TSS | 79.64252 | 81.91855932 | 0.032571793 | 0.016979544 |
| chr13-21597674 | LATS2 | GB | 76.96600 | 75.08835236 | 0.04386903 | 0.033262593 |
| chr 7-47544612 | TNS3 | GB | 61.95122 | 43.49234966 | 0.04571766 | 0.014225618 |
Benjamini-Hochberg adjusted P value cut-off of 0.05.
WB, whole biopsy; GB, gene body.
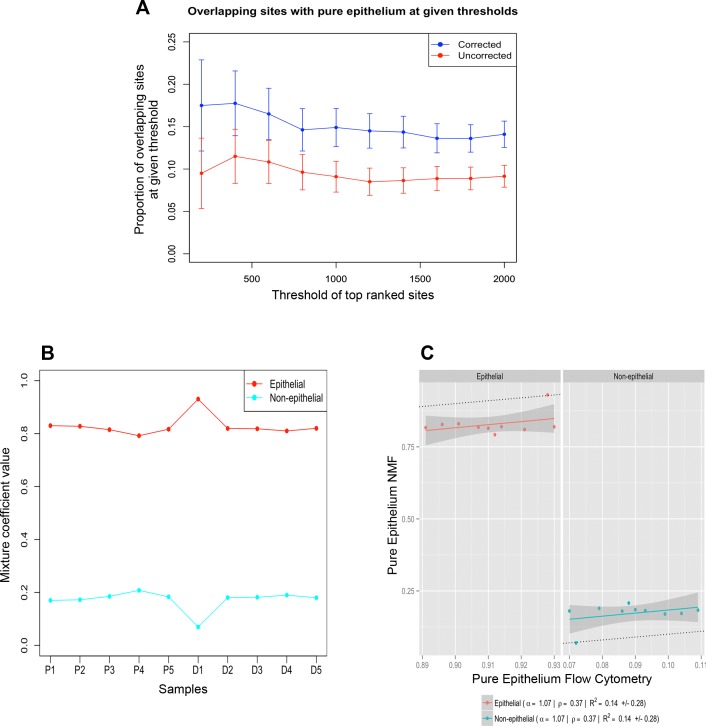
The purpose of incorporating the estimated abundance of epithelial cells as a covariate into the regression models used to identify differential DNA methylation based on the whole biopsies was to reduce the impact of variation in cellular composition on the detection of differential DNA methylation. We wished to test whether differences that we find in the pure epithelial cells can be identified from whole pinch biopsies without the need, in future studies, to purify the epithelial component. Our results indicate that incorporating this covariate does result in somewhat greater similarity between the results based on the whole biopsies and purified epithelium (Fig. 7A). However, the specificity of the deconvolution assay was not optimal as the proportion of overlapping DMS between the deconvolution corrected dataset with pure epithelium was still relatively low (mean proportion of overlapping sites = 0.151).
Fig. 7.
Validation of deconvolution accuracy. Overlapping differentially methylated sites (DMS). The proportions of overlapping DMS between whole biopsies (red) and deconvolution corrected whole biopsies (blue) with pure epithelial samples. Lists of DMS identified in each of the 3 cases were ranked according to the mean difference in DNA methylation. For each rank threshold, t, shown on the x-axis, the number of sites in common between the top t sites in the corrected or uncorrected lists and the top t sites identified in the purified epithelium is shown (A). The SEs were constructed based on the upper and lower limits of the estimated confidence interval of the overlap proportions. Profile plot illustrating cell proportion estimates of epithelial (red) and nonepithelial (cyan) populations in whole purified cell samples (B). Scatter plot illustrating the correlation (Pearson = ρ) between measured cell signature estimates of the purified cell samples using flow cytometry (x-axis) and cell signature estimates of the purified samples generated by deconvolution (y-axis) (C).
We also applied the same deconvolution method to the purified epithelial cell samples. These samples still consist of mixtures of cell types (they are composed of roughly 90% epithelium with the remaining 10% corresponding to other cell types). Given that estimates of the epithelial proportions for these samples were obtained from flow cytometry, they provide a basis to estimate the accuracy of cell-type proportion estimates produced by computational deconvolution methods (Table 2). Although the estimates of the proportion of the dominant cell type were more stable in the pure samples (similar to the pattern seen using flow cytometry) relative to whole biopsy samples (Figs. 6B and 7B, respectively), the correlation between measured cell signature estimates of the purified cell samples from flow cytometry and cell signature estimates of the purified samples according to deconvolution was weak (Pearson correlation = 0.37, Fig. 7C). Low correlation is partly the result of low variance in the epithelial proportion across samples. The observation of a correlation in this case is also potentially biased, as the marker sites used were identified on the basis of the purified samples to identify the epithelium marker sites.
Although we have shown that computational deconvolution does partially correct for the effects of differences in cell-type composition of the whole biopsies, substantial differences in methylation patterns remained between the deconvolved and purified cell samples. Therefore, we suggest that experimental purification is likely to remain the gold standard method to identify changes in DNA methylation within specific cellular components of biological samples.
DISCUSSION
In this work, we have developed and verified a method to isolate highly purified colonic epithelial cells from endoscopic pinch biopsies. We compared the DNA methylation profiles of colonic epithelial cells from proximal and distal colon regions. Of the 1.6 million CCGG loci assessed, we identified 125 DMS between proximal and distal colonic regions. Although the number of sites where we observed significant variation was quite few, we identified a targeted dichotomy of methylation patterns in colonic regions specific to HOX genes, namely hypermethylation of HOXB genes in the distal colon and hypermethylation of HOXC genes in the proximal colon. Furthermore, we adapted a computational deconvolution approach previously developed for analysis of gene expression profiles to study DNA methylation patterns in whole colon endoscopic biopsies. This work provides novel insights into regional variation in colonic epithelial epigenomics, heightening the influence of DNA methylation in the position-specific expression of HOX genes along the gastrointestinal tract, as previously documented (66).
During embryonic development, the proximal colon is established by the embryonic midgut, supplied by the superior mesenteric artery, whereas the distal colon is established from the hindgut and supplied by the inferior mesenteric artery (30a). These embryologically divergent origins are reflected at the histological level where the proximal colon has a multilayered capillary network, whereas the distal colon is single layered (3, 58). The distal colon also has a longer average crypt length compared with the proximal colon (2). At the level of gene expression, prior work has documented some gene and protein expression variation along the proximal-distal gradient (37, 52, 66). The molecular mechanisms that underlie regional variation in gene expression patterns in the colon are not understood, but it is possible that epigenetic factors, such as DNA methylation, might control such regional variation.
Using statistical algorithms, we identified sets of genes differentially methylated between proximal and distal colon regions, specifically around transcription start sites. We identified 10 different HOX genes (HOXB2, HOXB3, HOXB4, HOXB5, HOXB6, HOXB7, HOXC4, HOXC5, HOXC6, and HHEX) that were differentially methylated between proximal and distal areas. In particular, we observed a defined enrichment of region-specific DNA methylation differences in HOXB genes (n = 6), with loci showing hypermethylation in distal samples as well as DNA methylation differences in HOXC genes (n = 3), with loci showing hypermethylation in proximal samples (Fig. 4). These results also showed strong reproducibility, with methylation levels validating well using MassArray EpiTYPER (Fig. 5).
HOX genes are a highly conserved family that encodes transcription factors, incorporating a HOX sequence that regulates segmentation and pattern formation (38). In total, there are 39 HOX genes in the human genome, located in four distinct clusters (A–D) (7). These are a gene family that specifies segment identity, playing an essential role in the formation of the skeletal system and central nervous system (23, 67). However, other evidence suggests that they also play a role in regulating pattern formation in organs, for instance in the gastrointestinal tract. Region-specific differential gene expression of homeotic genes has been reported in the small intestine in mice (28) and the large intestine in humans (37). In fact, Yahagi et al. (66) established a position-specific expression pattern of HOX genes along the entire anteroposterior axis of the adult gastrointestinal tract. Other work has shown that alterations to the expression pattern of HOXC8 caused malformation of gastric epithelium (51). Furthermore, upregulation of HOXC6 expression is associated with poor survival in gastric cancer patients (68). Therefore, some evidence suggests that HOX genes might contribute to the progression of gastric carcinogenesis and may be a significant predictor of poor survival in patients.
Our results further demonstrate that HOX genes are differentially regulated at the level of DNA methylation in proximal and distal colon segments of healthy human colon, which implies a functional role for epigenetic regulation of their expression. Therefore, building this model to explain the underlying differences observed in region-specific genomic and epigenomic patterns of HOX genes represents a valuable tool for interpreting experimental data on diseases that exhibit region-specific expression in the colon such as inflammatory bowel disease and colorectal cancer. For example, one form of this inflammatory bowel disease, ulcerative colitis, most commonly affects the distal colon (35), while another form, Crohn's disease, more commonly affects the proximal colon (60). The more common chromosomal instability pathway of colorectal carcinogenesis results in tumors developing predominantly in the distal colon, while the microsatellite instability pathway is more common in the proximal colon (43).
The application of DNA methylation analysis to human samples is an increasingly common technique in disease pathogenesis research. However, because DNA methylation patterns are cell-type specific, the precision of DNA methylation data can be adversely affected by the heterogeneous cellular make-up of clinically obtained samples such as endoscopic biopsies of the gastrointestinal mucosa. We therefore employed a deconvolution approach to account for this heterogeneity and to test whether this could be used as way to reduce the impact of variation in cell-type composition of heterogeneous tissue samples in differential DNA methylation analysis. The number of DMS identified in whole biopsies (n = 1,820) decreased significantly when the inferred epithelial cell proportion was included as a covariate in differential DNA methylation analysis (n = 537) (Fig. 6). Moreover, the overlap in DMS detected from purified epithelial cells and whole biopsy samples increased, suggesting that this approach can correct, at least partially, for the effects of differences in cell-type composition of the whole biopsies. Nonetheless, the specificity of the assay was not optimal, as the number of DMS in whole biopsy samples remained far greater than the number identified from the pure epithelial cell isolates (n = 125). Furthermore, the correlation between estimates of the purified cell samples generated by deconvolution and the measured flow cytometry estimates was also weak. This suggests that, as it was applied here, computational deconvolution cannot fully substitute for laboratory-based purification of epithelial cells. This may be because our analysis did not account for variation in the less abundant cell types between the whole biopsies.
Prior work by other research groups has highlighted variation in DNA methylation patterns of colonic mucosa in both normal (45) and diseased states (55). In particular, Luo et al. (45) demonstrated distinct differential methylation between normal colon mucosa from subjects with no history of colon neoplasms and the normal colon mucosa from subjects with concurrent colorectal cancer. However, as the subject matter as well as the source material utilized in these studies were not directly comparable to ours, we decided not to incorporate their findings into the results that we obtained.
To the best of our knowledge, this is the first methylome map modeling epithelial-specific epigenetic modifications between proximal and distal regions in normal colonic mucosa. Our data indicate that the differences in DNA methylation patterns between proximal and distal regions are quite specific, predominantly targeted at HOX genes. We hypothesize that this significant variation could be related to embryonic development and anterior-posterior formation of the gastrointestinal tract. These insights should be used to obtain a better understanding of how biological processes of the colonic epithelium vary between colonic regions. Furthermore, our data, combined with previous DNA methylation and gene expression studies specific to colonic mucosa, will contribute to a greater understanding of the colon epithelial epigenome, potentially implicating inflammatory related diseases of epithelial origin.
GRANTS
A. Barnicle received a scholarship from the Irish Research Council, and further support was provided by AbbVie.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
Author contributions: A.B., C.S., A.G., and L.J.E. conception and design of research; A.B. performed experiments; A.B. analyzed data; A.B., C.S., A.G., J.M.G., and L.J.E. interpreted results of experiments; A.B. prepared figures; A.B. drafted manuscript; A.B., C.S., A.G., J.M.G., and L.J.E. edited and revised manuscript; C.S. and L.J.E. approved final version of manuscript.
Supplementary Material
ACKNOWLEDGMENTS
We thank Coralie Mureau for assistance with hematoxylin and eosin staining and microscopy. We also thank staff from Einstein's Center of Epigenomics including the Epigenomics Shared Facility and computational Epigenomics group. Finally we acknowledge the staff at the Genomics Shared Resource supported by Roswell Park Cancer Institute and National Cancer Institute. Sample processing for DNA methylation validation was performed by this group under the grant number P30CA-016056.
Footnotes
The online version of this article contains supplemental material.
REFERENCES
- 1.Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, Ntini E, Arner E, Valen E, Li K, Schwarzfischer L, Glatz D, Raithel J, Lilje B, Rapin N, Bagger FO, Jørgensen M, Andersen PR, Bertin N, Rackham O, Burroughs a M, Baillie JK, Ishizu Y, Shimizu Y, Furuhata E, Maeda S, Negishi Y, Mungall CJ, Meehan TF, Lassmann T, Itoh M, Kawaji H, Kondo N, Kawai J, Lennartsson A, Daub CO, Heutink P, Hume a D, Jensen TH, Suzuki H, Hayashizaki Y, Müller F, Forrest ARR, Carninci P, Rehli M, Sandelin A. An atlas of active enhancers across human cell types and tissues. Nature 507: 455–61, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arai T, Kino I. Morphometrical and cell kinetic studies of normal human colorectal mucosa. Comparison between the proximal and the distal large intestine. Acta Pathol Jpn 39: 725–730, 1989. [DOI] [PubMed] [Google Scholar]
- 3.Araki K, Furuya Y, Kobayashi M, Matsuura K, Ogata T, Isozaki H. Comparison of mucosal microvasculature between the proximal and distal human colon. J Electron Microsc (Tokyo) 45: 202–206, 1996. [DOI] [PubMed] [Google Scholar]
- 4.Barnard JA, Beauchamp RD, Coffey RJ, Moses HL. Regulation of intestinal epithelial cell growth by transforming growth factor type beta. Proc Natl Acad Sci USA 86: 1578–1582, 1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21, 2002. [DOI] [PubMed] [Google Scholar]
- 6.Blattler A, Yao L, Witt H, Guo Y, Nicolet CM, Berman BP, Farnham PJ. Global loss of DNA methylation uncovers intronic enhancers in genes showing expression changes. Genome Biol 15: 469, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boncinelli E, Simeone A, Acampora D, Mavilio F. HOX gene activation by retinoic acid. Trends Genet 7: 329–334, 1991. [DOI] [PubMed] [Google Scholar]
- 8.Consortium Roadmap Epigenomics, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu YC, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh KH, Feizi S, Karlic R, Kim AR, Kulkarni A, Li D, Lowdon R, Elliott G, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJM, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai LH, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Meth 9: 215–216, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esteller M, Garcia-Foncillas J, Andion E, Goodman SN, Hidalgo OF, Vanaclocha V, Baylin SB, Herman JG. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med 343: 1350–1354, 2000. [DOI] [PubMed] [Google Scholar]
- 11.Flint N, Cove FL, Evans GS. A low-temperature method for the isolation of small-intestinal epithelium along the crypt-villus axis. Biochem J 280: 331–334, 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Forrest ARR, Kawaji H, Rehli M, Baillie JK, de Hoon MJL, Lassmann T, Itoh M, Summers KM, Suzuki H, Daub CO, Kawai J, Heutink P, Hide W, Freeman TC, Lenhard B, Bajic VB, Taylor MS, Makeev VJ, Sandelin A, Hume a D., Carninci P, Hayashizaki Y. A promoter-level mammalian expression atlas. Nature 507: 462–70, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gallo-Payet N, Hugon JS. Insulin receptors in isolated adult mouse intestinal cells: studies in vivo and in organ culture. Endocrinology 114: 1885–1901, 1984. [DOI] [PubMed] [Google Scholar]
- 14.Garrity-Park MM, Loftus EV, Sandborn WJ, Bryant SC, Smyrk TC. Methylation status of genes in non-neoplastic mucosa from patients with ulcerative colitis-associated colorectal cancer. Am J Gastroenterol 105: 1610–1619, 2010. [DOI] [PubMed] [Google Scholar]
- 15.Gaujoux R, Seoighe C. Semi-supervised nonnegative matrix factorization for gene expression deconvolution: a case study. Infect Genet Evol 12: 913–921, 2012. [DOI] [PubMed] [Google Scholar]
- 16.Gaujoux R, Seoighe C. CellMix: a comprehensive toolbox for gene expression deconvolution. Bioinformatics 29: 2211–2212, 2013. [DOI] [PubMed] [Google Scholar]
- 17.Geeleher P, Hartnett L, Egan LJ, Golden A, Raja Ali RA, Seoighe C. Gene-set analysis is severely biased when applied to genome-wide methylation data. Bioinformatics 29: 1851–1857, 2013. [DOI] [PubMed] [Google Scholar]
- 18.Glebov OK, Rodriguez LM, Nakahara K, Jenkins J, Cliatt J, Humbyrd CJ, DeNobile J, Soballe P, Simon R, Wright G, Lynch P, Patterson S, Lynch H, Gallinger S, Buchbinder A, Gordon G, Hawk E, Kirsch IR. Distinguishing right from left colon by the pattern of gene expression. Cancer Epidemiol Biomarkers Prev 12: 755–762, 2003. [PubMed] [Google Scholar]
- 19.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130: 77–88, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, Kane MF, Kolodner RD, Vogelstein B, Kunkel TA, Baylin SB. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA 95: 6870–6875, 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hodges E, Molaro A, Dos Santos CO, Thekkat P, Song Q, Uren PJ, Park J, Butler J, Rafii S, McCombie WR, Smith AD, Hannon GJ. Directional DNA methylation changes and complex intermediate states accompany lineage specificity in the adult hematopoietic compartment. Mol Cell 44: 17–28, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformat 13: 86, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hunt P, Whiting J, Nonchev S, Sham MH, Marshall H, Graham A, Cook M, Allemann R, Rigby PW, Gulisano M. The branchial Hox code and its implications for gene regulation, patterning of the nervous system and head evolution. Dev Suppl 2: 63–77, 1991. [PubMed] [Google Scholar]
- 24.Illingworth RS, Bird AP. CpG islands–'a rough guide'. FEBS Lett 583: 1713–1720, 2009. [DOI] [PubMed] [Google Scholar]
- 25.Irizarry a R, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, Ji H, Potash JB, Sabunciyan S, Feinberg AP. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet 41: 178–86, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 33, Suppl: 245–254, 2003. [DOI] [PubMed] [Google Scholar]
- 27.Jaffe AE, Murakami P, Lee H, Leek JT, Fallin MD, Feinberg AP, Irizarry RA. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol 41: 200–209, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.James R, Kazenwadel J. Homeobox gene expression in the intestinal epithelium of adult mice. J Biol Chem 266: 3246–3251, 1991. [PubMed] [Google Scholar]
- 29.Jenke AC, Postberg J, Raine T, Nayak KM, Molitor M, Wirth S, Kaser A, Parkes M, Heuschkel RB, Orth V, Zilbauer M. DNA methylation analysis in the intestinal epithelium-effect of cell separation on gene expression and methylation profile. PLoS One 8: e55636, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jing Q, McLellan A, Greally JM, Suzuki M. Automated computational analysis of genome-wide DNA methylation profiling data from HELP-tagging assays. Methods Mol Biol 815: 79–87, 2012. [DOI] [PubMed] [Google Scholar]
- 30a.Johnson LR, Barrett KE, Merchant JL, Ghishan FK, Said HM, Wood JD. Physiology of the Gastrointestinal Tract (4th Ed.). Burlington, MA: Elsevier Academic, 2006. (accessed online 17 Nov 2014, http://books.google.com/books?id=CNwLlih2C60C&pgis=1). [Google Scholar]
- 31.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13: 484–492, 2012. [DOI] [PubMed] [Google Scholar]
- 32.Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. The UCSC Table Browser data retrieval tool. Nucleic Acids Res 32: D493–D496, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaufman TC, Seeger MA, Olsen G. Molecular and genetic organization of the antennapedia gene complex of Drosophila melanogaster. Adv Genet 27: 309–62, 1990. http://www.ncbi.nlm.nih.gov/pubmed/1971986 (28 Mar. 2015). [DOI] [PubMed] [Google Scholar]
- 34.Kaz AM, Wong CJ, Luo Y, Virgin JB, Kay Washington M, Willis JE, Leidner RS, Chak A, Grady WM. DNA methylation profiling in Barrett's esophagus and esophageal adenocarcinoma reveals unique methylation signatures and molecular subclasses. Epigenetics 6: 1403–1412, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koutroubakis IE. Recent advances in the management of distal ulcerative colitis. World J Gastrointest Pharmacol Ther 1: 43–50, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lao VV, Grady WM. Epigenetics and colorectal cancer. Nat Rev Gastroenterol Hepatol 8: 686–700, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.LaPointe LC, Dunne R, Brown GS, Worthley DL, Molloy PL, Wattchow D, Young GP. Map of differential transcript expression in the normal human large intestine. Physiol Genomics 33: 50–64, 2008. [DOI] [PubMed] [Google Scholar]
- 38.Lawrence PA, Morata G. Homeobox genes: their function in Drosophila segmentation and pattern formation. Cell 78: 181–189, 1994. http://www.ncbi.nlm.nih.gov/pubmed/7913879 (22 Mar. 2015). [DOI] [PubMed] [Google Scholar]
- 39.Lay FD, Triche TJ, Tsai YC, Su SF, Martin SE, Daneshmand S, Skinner EC, Liang G, Chihara Y, Jones PA. Reprogramming of the human intestinal epigenome by surgical tissue transposition. Genome Res 24: 545–53, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet 3: 1724–1735, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis EB. A gene complex controlling segmentation in Drosophila. Nature 276: 565–570, 1978. [DOI] [PubMed] [Google Scholar]
- 42.Liévin-Le Moal V, Servin AL. The front line of enteric host defense against unwelcome intrusion of harmful microorganisms: mucins, antimicrobial peptides, and Microbiota. Clin Microbiol Rev 19: 315–337, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lindblom A. Different mechanisms in the tumorigenesis of proximal and distal colon cancers. Curr Opin Oncol 13: 63–69, 2001. [DOI] [PubMed] [Google Scholar]
- 44.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462: 315–322, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo Y, Wong CJ, Kaz AM, Dzieciatkowski S, Carter KT, Morris SM, Wang J, Willis JE, Makar KW, Ulrich CM, Lutterbaugh JD, Shrubsole MJ, Zheng W, Markowitz SD, Grady WM. Differences in DNA methylation signatures reveal multiple pathways of progression from adenoma to colorectal cancer. Gastroenterology 147: 418–429, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mohn F, Schübeler D. Genetics and epigenetics: stability and plasticity during cellular differentiation. Trends Genet 25: 129–136, 2009. [DOI] [PubMed] [Google Scholar]
- 48.Montaño CM, Irizarry RA, Kaufmann WE, Talbot K, Gur RE, Feinberg AP, Taub MA. Measuring cell-type specific differential methylation in human brain tissue. Genome Biol 14: R94, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Novakovic B, Yuen RK, Gordon L, Penaherrera MS, Sharkey A, Moffett A, Craig JM, Robinson WP, Saffery R. Evidence for widespread changes in promoter methylation profile in human placenta in response to increasing gestational age and environmental/stochastic factors. BMC Genomics 12: 529, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ollikainen M, Smith KR, Joo EJH, Ng HK, Andronikos R, Novakovic B, Aziz NKA, Carlin JB, Morley R, Saffery R, Craig JM. DNA methylation analysis of multiple tissues from newborn twins reveals both genetic and intrauterine components to variation in the human neonatal epigenome. Hum Mol Genet 19: 4176–4188, 2010. [DOI] [PubMed] [Google Scholar]
- 51.Pollock RA, Jay G, Bieberich CJ. Altering the boundaries of Hox3.1 expression: evidence for antipodal gene regulation. Cell 71: 911–923, 1992. [DOI] [PubMed] [Google Scholar]
- 52.Van der Post S, Hansson GC. Membrane protein profiling of human colon reveals distinct regional differences. Mol Cell 13: 2277–2287, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saito S, Kato J, Hiraoka S, Horii J, Suzuki H, Higashi R, Kaji E, Kondo Y, Yamamoto K. DNA methylation of colon mucosa in ulcerative colitis patients: Correlation with inflammatory status. Inflamm Bowel Dis 17: 1955–1965, 2011. [DOI] [PubMed] [Google Scholar]
- 54.Shen-Orr SS, Gaujoux R. Computational deconvolution: extracting cell type-specific information from heterogeneous samples. Curr Opin Immunol 25: 571–578 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Simmer F, Brinkman AB, Assenov Y, Matarese F, Kaan A, Sabatino L, Villanueva A, Huertas D, Esteller M, Lengauer T, Bock C, Colantuoni V, Altucci L, Stunnenberg HG. Comparative genome-wide DNA methylation analysis of colorectal tumor and matched normal tissues. Epigenetics 7: 1355–1367, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Singh SK, Binder HJ, Boron WF, Geibel JP. Fluid absorption in isolated perfused colonic crypts. J Clin Invest 96: 2373–2379, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Skinner SA, O'Brien PE. The microvascular structure of the normal colon in rats and humans. J Surg Res 61: 482–490, 1996. [DOI] [PubMed] [Google Scholar]
- 59.Smyth G. limma: Linear Models for Microarray Data. In: Bioinformatics and Computational Biology Solutions Using R and Bioconductor, edited by Gentleman R, Carey VJ, Huber W, Irizarry RA, Dudoit S. Springer, 2005, p. 397–420. [Google Scholar]
- 60.Soucy G, Wang HH, Farraye FA, Schmidt JF, Farris AB, Lauwers GY, Cerda SR, Dendrinos KG, Odze RD. Clinical and pathological analysis of colonic Crohn's disease, including a subgroup with ulcerative colitis-like features. Mod Pathol 25: 295–307, 2012. [DOI] [PubMed] [Google Scholar]
- 61.Suzuki M, Jing Q, Lia D, Pascual M, McLellan A, Greally JM. Optimized design and data analysis of tag-based cytosine methylation assays. Genome Biol 11: R36, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Traber PG, Gumucio DL, Wang W. Isolation of intestinal epithelial cells for the study of differential gene expression along the crypt-villus axis. Am J Physiol Gastrointest Liver Physiol 260: G895–G903, 1991. [DOI] [PubMed] [Google Scholar]
- 63.Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, Pauli-Behn F, Cross MK, Williams BA, Stamatoyannopoulos JA, Crawford GE, Absher DM, Wold BJ, Myers RM. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res 23: 555–567, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Whitehead RH, Demmler K, Rockman SP, Watson NK. Clonogenic growth of epithelial cells from normal colonic mucosa from both mice and humans. Gastroenterology 117: 858–865, 1999. [DOI] [PubMed] [Google Scholar]
- 65.Wijetunga NA, Delahaye F, Zhao YM, Golden A, Mar JC, Einstein FH, Greally JM. The meta-epigenomic structure of purified human stem cell populations is defined at cis-regulatory sequences. Nat Commun 5: 5195, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yahagi N, Kosaki R, Ito T, Mitsuhashi T, Shimada H, Tomita M, Takahashi T, Kosaki K. Position-specific expression of Hox genes along the gastrointestinal tract. Congenit Anom (Kyoto) 44: 18–26, 2004. [DOI] [PubMed] [Google Scholar]
- 67.Yokouchi Y, Sasaki H, Kuroiwa A. Homeobox gene expression correlated with the bifurcation process of limb cartilage development. Nature 353: 443–445, 1991. [DOI] [PubMed] [Google Scholar]
- 68.Zhang Q, Jin XS, Yang ZY, Wei M, Liu BY, Gu QL. Upregulated Hoxc6 expression is associated with poor survival in gastric cancer patients. Neoplasma 60: 439–445, 2013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.