Abstract
Reef-building corals depend on symbiotic mutualisms with photosynthetic dinoflagellates in the genus Symbiodinium. This large microalgal group comprises many highly divergent lineages (“Clades A–I”) and hundreds of undescribed species. Given their ecological importance, efforts have turned to genomic approaches to characterize the functional ecology of Symbiodinium. To date, investigators have only compared gene expression between representatives from separate clades—the equivalent of contrasting genera or families in other dinoflagellate groups—making it impossible to distinguish between clade-level and species-level functional differences. Here, we examined the transcriptomes of four species within one Symbiodinium clade (Clade B) at ∼20,000 orthologous genes, as well as multiple isoclonal cell lines within species (i.e., cultured strains). These species span two major adaptive radiations within Clade B, each encompassing both host-specialized and ecologically cryptic taxa. Species-specific expression differences were consistently enriched for photosynthesis-related genes, likely reflecting selection pressures driving niche diversification. Transcriptional variation among strains involved fatty acid metabolism and biosynthesis pathways. Such differences among individuals are potentially a major source of physiological variation, contributing to the functional diversity of coral holobionts composed of unique host–symbiont genotype pairings. Our findings expand the genomic resources available for this important symbiont group and emphasize the power of comparative transcriptomics as a method for studying speciation processes and interindividual variation in nonmodel organisms.
Keywords: dinoflagellates, phylogenetics, RNAseq, symbiosis, transcriptome, zooxanthellae
Introduction
The concept that adaptation and speciation are driven largely by natural selection on variant individuals of a population is central to evolutionary biology. Much like other types of genetic diversity, gene expression variation is extensive, highly heritable, and subject to selection (Ranz and Machado 2006; Voolstra et al. 2007; Wittkopp et al. 2008). The role of differential gene expression in ecological speciation has received renewed interest in the genomics era because the molecular biology of nonmodel organisms with unique evolutionary histories can now be studied in great detail at relatively low cost (Wolf et al. 2010). For example, among two recently diverged species of cordgrass, only one is successful at invading environments perturbed by climate change, and it exhibits unique expression patterns at growth- and stress-related genes (Chelaifa et al. 2010). A similar study in daisies illustrated that a comparative transcriptomic framework can be used to identify selective processes affecting ecological speciation (Chapman et al. 2013). Additionally, transcription-based assays of microbial metagenomes have revealed unique niche diversification (e.g., specialization for certain substrates, metabolic pathways, or environments) that is otherwise hidden due to functional redundancy in the genomes of many bacteria (Gifford et al. 2013). Thus, comparative genomics can also provide a means to recognize important functional variation in organisms that are difficult to probe phenotypically, such as corals and their symbionts.
Coral reef ecosystems support tremendous marine biodiversity and ecological goods and services (Moberg and Folke 1999). Coral productivity and growth depend on a mutualism with endosymbiotic dinoflagellates known as Symbiodinium (Muscatine and Porter 1977; Muscatine 1990; Yellowlees et al. 2008). This microalgal “genus” is incredibly diverse, encompassing at least nine major lineages that show ribosomal divergence equivalent to that found among different genera, families, or even orders of other dinoflagellates (Rowan and Powers 1992). Likewise, Symbiodinium exhibit many unique ecologies, ranging from “host-specialized” taxa commonly found as symbiotic partners of corals (Parkinson, Coffroth, et al. 2015), to “ecologically cryptic” taxa with alternate nonsymbiotic lifestyles (LaJeunesse et al. 2015), to completely “free-living” taxa that thrive independently in the water column (Jeong et al. 2014). Unlike their mostly obligate coral hosts, many Symbiodinium can survive ex hospite and are maintained in culture. In the natural environment, stressful conditions can cause the association between corals and host-specialized symbionts to break down in a process called coral bleaching, which can lead to colony mortality (Fitt et al. 2001). Climate change is predicted to drive more frequent and intense bleaching events (Hoegh-Guldberg 1999), prompting a major research focus on how climate-related stressors might affect coral–dinoflagellate symbioses in the future. Accordingly, the last decade has generated many studies describing coral host transcription in various contexts (Meyer and Weis 2012), but comparable studies in Symbiodinium are still in their early stages (Leggat et al. 2007; Leggat, Yellowlees, et al. 2011; Lin 2011).
With the incorporation of next-generation sequencing technology, genomic resources for Symbiodinium have expanded greatly despite their status as a nonmodel organism. The first draft genome was released in 2013 (Shoguchi et al. 2013), with the complete chloroplast genome following shortly thereafter (Barbrook et al. 2014). Multiple mRNA transcriptomes are available (Bayer et al. 2012; Ladner et al. 2012; Baumgarten et al. 2013; Rosic et al. 2014; Xiang et al. 2015), representing the four major clades known to associate with scleractinian corals (Clades A, B, C, and D). Recent efforts have expanded in important new directions, such as the description of Symbiodinium microRNAs (Baumgarten et al. 2013), the comparison of orthologous genes among clades (Voolstra et al. 2009; Ladner et al. 2012; Barshis et al. 2014; Rosic et al. 2014), the completion of another draft genome (Lin et al. 2015), and the development of the Aiptasia–Symbiodinium system for in-depth cellular and physiological research (Weis et al. 2008; Sunagawa et al. 2009; Lehnert et al. 2012, 2014; Xiang et al. 2013; Baumgarten et al. 2015).
Dinoflagellate genomes are unique among eukaryotes for multiple reasons (Leggat, Yellowlees, et al. 2011). Of particular note, dinoflagellates including Symbiodinium modulate nuclear-encoded protein levels predominantly by posttranscriptional processes (Morse et al. 1989; Leggat, Seneca, et al. 2011). It is now understood that dinoflagellates also exhibit some measure of transcriptional regulation, albeit changes in expression profiles are minimal when exposed to different environmental conditions (Erdner and Anderson 2006; Moustafa et al. 2010). For example, the number and magnitude of expression changes among Symbiodinium exposed to thermal stress are relatively small compared with their animal hosts (Leggat, Seneca, et al. 2011). Barshis et al. (2014) found that two Symbiodinium spp. in Clades C and D did not alter gene expression when exposed to temperature stress in hospite, even though the host response involved the modulation of hundreds of genes (Barshis et al. 2013, 2014). Interestingly, a large number of transcriptional differences were maintained, or fixed, for the two species from different clades regardless of temperature treatment (Barshis et al. 2014). This suggests that fixed expression differences are likely to be evident in strains cultured ex hospite under identical controlled environmental conditions. Differences in these “stable-state” expression profiles among lineages may strongly reflect evolutionary divergence, some of which may be adaptive. These expression patterns may also correspond to functional differences among distantly related species. If lineage-specific expression extends to the subcladal level—that is, between species within clades or among individual strains within species—it will be critical to recognize this source of variation when interpreting Symbiodinium genomic data and account for it in future experimental designs.
By comparing different isoclonal cell lines (strains), it is possible to reveal intraspecific variation in genomic features that underlie ecological and physiological phenotypes. For example, unique genes distinguish strains of nitrogen-fixing rhizobial bacteria with different symbiotic efficiencies and host specificities (Galardini et al. 2011; Österman et al. 2015). At the level of transcription, toxic and nontoxic strains of the dinoflagellate Alexandrium minutum maintain fixed expression differences at shared genes (Yang et al. 2010). We may expect similar patterns among Symbiodinium strains, but this idea has never been tested. Symbiodinium belonging to Clade B are ideal candidates for further genomic characterization because several ecologically distinct species within this group were recently described (LaJeunesse et al. 2012; Parkinson, Coffroth, et al. 2015), a draft genome exists for the member species Symbiodinium minutum (Shoguchi et al. 2013), and multiple genetically distinct cultures are available for several species.
Currently, the extent of variation among species within a single Symbiodinium clade and among individual strains within a single species is mostly unknown (Parkinson and Baums 2014). To address this knowledge gap, we analyzed stable-state gene expression among four species representing the two major evolutionary radiations within Clade B: The Pleistocene (B1) radiation and the Pliocene (B19) radiation (sensu LaJeunesse 2005). For each radiation, two species with different ecologies were studied: Either host-specialized taxa or ecologically cryptic taxa. Although these latter species were originally cultured from coral tissues, they have never been detected as the numerically dominant symbionts in cnidarian mutualisms, and therefore were probably commensals or free-living contaminants isolated from the mucus or gastrovascular cavity (Parkinson, Coffroth, et al. 2015). Where available, we incorporated biological replication in the form of distinct isoclonal cell cultures. The genomic resources developed here should assist in the design and interpretation of future comparative transcriptional analyses among Symbiodinium strains, species, and clades, as well as broaden our understanding of speciation among microeukaryotes.
Materials and Methods
Culturing
Isoclonal cultures (strains) of Clade B Symbiodinium were maintained at the Pennsylvania State University. They were originally acquired from the Robert K. Trench and Buffalo Undersea Reef Research collections. This study included one strain of Symbiodinium aenigmaticum (mac04-487), four strains of S. minutum (mac703, Mf1.05b, rt002, and rt351), one strain of Symbiodinium pseudominutum (rt146), and four strains of Symbiodinium psygmophilum (HIAp, Mf10.14b.02, PurPFlex, and rt141), for the analysis of ten individual transcriptomes. Most strains are available from the Provasoli-Guillard National Center for Marine Algae and Microbiota at Bigelow Laboratory for Ocean Sciences, East Boothbay, Maine (LaJeunesse et al. 2012; Parkinson, Coffroth, et al. 2015) or by request. Within S. minutum and S. psygmophilum, strains were confirmed to represent distinct genotypes based on repeat length variation at the microsatellite locus Sym15 (Pettay and LaJeunesse 2007) and haplotype differences in the psbA noncoding region (LaJeunesse and Thornhill 2011).
Single cells were originally isolated from host tissues by Schoenberg and Trench (1980) using modified methods of McLaughlin and Zahl (1959) or by Mary Alice Coffroth following the methods of Santos et al. (2001). To establish initial crude cultures, several drops of a heavy suspension of symbiont cells were transferred into nutrient-enriched filtered seawater (Provasoli 1968) and then spread onto semisolid agar (0.8%) containing the same seawater. Vegetative cells from viable colonies on agar were then transferred to liquid medium ASP-8A (Ahles 1967). To generate isoclonal lines, only individual motile cells were transferred to fresh media. For this experiment, an additional transfer to new media was made to synchronize all cultures. Final cultures were grown in 50 ml volumes in Erlenmeyer flasks for 2 weeks up to concentrations ∼1 × 106 cells · ml− 1. Cultures were maintained in incubators at 26 °C under Philips fluorescent tubes (Koninklijke Philips Electronics, Amsterdam, The Netherlands) delivering 80–120 µmol · m−2 · s−1 photosynthetically active radiation on a 12:12 (light:dark) photoperiod. All cultures grew together under identical conditions until processed simultaneously for RNA extraction.
RNA Isolation and Sequencing
At the sixth hour of the light photoperiod on the last day of the second week of growth postsynchronization, all target cultures were transferred to 50 ml tubes and centrifuged at 3000 RCF (relative centrifugal force). The media was decanted and the pellets were flash frozen in liquid nitrogen. Pellets were ground with a prechilled mortar and pestle and transferred into 1.5 ml tubes. Nucleic acids were extracted with TriReagent (Thermo Fisher Scientific, Waltham, MA) and RNA was isolated and cleaned with the RNeasy Kit (Qiagen, Venlo, The Netherlands) according to the manufacturer’s protocols.
Total RNA isolations were shipped on dry ice to the KAUST Red Sea Research Center, where they were quality checked using a Bioanalyzer 2000 (Agilent, Santa Clara, CA) prior to library preparation. For Illumina 2 × 100 bp paired-end sequencing, 180 bp libraries were generated from oligo(dT)-enriched mRNA using the NEBNext Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA) according to the manufacturer’s protocols. Each read pair ideally yielded a partially overlapping 180 bp contiguous sequence, allowing for additional quality control. mRNA sequencing libraries for each of the ten samples were multiplexed in equimolar concentrations and run on one lane on the Illumina HiSeq 2000 platform, producing a total of 142 million paired-end reads. All raw RNAseq data are available in the NCBI Sequence Read Archive database under accession numbers http://www.ncbi.nlm.nih.gov/bioproject/PRJNA274856/ (S. aenigmaticum), http://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA274852 (S. minutum), http://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA274855 (S. pseudominutum), and http://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA274854 (S. psygmophilum).
Transcriptome Assemblies and Annotation
Adapters and low quality nucleotides (<20 Phred score in ASCII 33 format) were removed from raw reads with Trimmomatic (Bolger et al. 2014). Reads were error corrected with the error correction module of the AllPATHS-LG pipeline (Gnerre et al. 2011; Ribeiro et al. 2012). Quality-controlled reads combined from all samples on a per-species basis were assembled using the Trinity package (Grabherr et al. 2011) with minimum k-mer coverage of 2 and minimum contig length of 250 bp to generate one reference assembly per species (four total). For mapping purposes, we further reduced each reference assembly to include only the longest transcript for a related set of splice variants of a gene. For each sample, reads were mapped back to the reduced assembly for the appropriate species with Bowtie2 (Langmead and Salzberg 2012), and quantified by summing counts of all transcripts per gene with the program eXpress (Roberts and Pachter 2013), producing effective read counts and FPKM values (Fragments Per Kilobase of transcript per Million mapped reads).
Each reference assembly was annotated by iterative searches of the longest transcript per gene against SwissProt, TrEMBL, and NCBI nr sequence databases (UniProt Consortium 2013; Pruitt et al. 2014) using BLASTX (Altschul et al. 1990) and the October 2013 releases. Only hits with e-values < 1 × 10−5 were retained. All genes remaining unannotated after BLASTX against the first database were passed onto the next sequentially. Gene Ontology (GO) categories were assigned through the BLASTX hit to SwissProt or TrEMBL databases, and subsequent mapping to the UniProt-GOA database (Dimmer et al. 2012). The assembled and annotated transcriptome sequences for each species are available at http://reefgenomics.org.
The recently published S. minutum draft genome (Shoguchi et al. 2013) was derived from strain Mf1.05b, which was included in this study. To compare our sequencing results to this resource, we aligned our Mf1.05b reads to the exome of the draft genome to estimate the proportion of mappable reads using Bowtie 2; 73% of the raw reads aligned.
To assess how comparable our Clade B species assemblies were in terms of gene content independent of expression, complete assemblies were uploaded into Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, www.ingenuity.com, last accessed January 2014). IPA compares user-provided gene lists with reference canonical pathways in the manually curated Ingenuity Knowledge Base. It generates a ratio of genes present versus total genes belonging to a pathway and tests for the probability of significant enrichment for that pathway in the Symbiodinium transcriptome with Fisher’s exact tests. The Ingenuity Knowledge Base is designed mainly for model organism data, so results should be interpreted in the context of pathways that are well annotated and highly conserved across eukaryotes. We were less concerned about pathway identity and more interested in whether representation within a pathway was similar across Clade B species. Thus, we compared ratio values for all transcriptomes at all pathways determined to be significantly enriched in the dataset at Padj < 0.05. As an additional metric of comparison across Clade B species, we mined all transcriptomes for repeats and flanking regions using the software SciRoKo (Kofler et al. 2007) and considered repeat motifs between 2 and 6 bp.
Ortholog Identification and Differential Expression between Species, Lineages, and Ecologies
To test for differential expression between the four species, it was necessary to identify a set of comparable orthologous genes. Open reading frames (ORFs) were predicted for each transcriptome with TransDecoder (Haas et al. 2013). Orthologous genes were identified via reciprocal BLASTP of ORFs pairwise for each species within the program InParanoid (Remm et al. 2001), retaining hits with bitscores >300 (supplementary table S1, Supplementary Material online). Multiparanoid (Alexeyenko et al. 2006) was then used to identify orthologs that occurred exactly four times (once in each species). Additional domain-based functional annotations were assigned using the Pfam database v27 (Finn et al. 2014) and are provided in supplementary table S2, Supplementary Material online.
Most current software designed to analyze differential expression for RNAseq data assumes raw read counts among samples mapped to one common reference transcriptome, and therefore only accepts integer values as input. To compare expression at orthologous genes across species, it was necessary to normalize read counts by transcript length using FPKM to account for species-specific sequence length differences. This normalization produced many decimal expression values that were still informative. For this reason, we scaled FPKM by a common factor such that the lowest expressed gene’s value equaled 1 and then rounded values to the nearest integer. Thus, a scaled FPKM of 50 means the gene is expressed 50 times higher than the lowest expressed gene retained in the data set. This way, all orthologs could be compared in the scaled FPKM space.
Scaled FPKM data were then used as input for the R package edgeR (Robinson et al. 2010), which accommodates data sets with unequal replication when performing comparisons among treatments (in this case, species). No additional normalization procedures were carried out within the program. Significant differential expression was determined by pairwise comparisons among species with a false discovery rate–adjusted P value (FDR) of < 0.1. To additionally test by lineage, all S. minutum and S. pseudominutum samples were grouped as “Pleistocene” and all S. aenigmaticum and S. psygmophilum samples were grouped as “Pliocene.” To test by ecology, all S. minutum and S. psygmophilum samples were grouped as host-specialized, and all S. aenigmaticum and S. pseudominutum samples were grouped as ecologically cryptic.
Multidimensional scaling (MDS) plots were generated in edgeR using the plotMDS function. The distances between pairs of RNA samples correspond to the leading log2-fold-changes, which is the average (root-mean-square) of the largest absolute log2-fold-changes (Robinson et al. 2010). In all three comparisons of the MDS plots (non-DEG [differentially expressed gene] only, non-DEG + DEG, and DEG only), similar clustering was observed among the four replicates of the two species with replicates. It was therefore reasonable to assume that (hypothetical) replicates of the other two species would show a similar distribution. Because variation between replicates was consistent, we assumed that the distances between any of the species (which contributes to the number of DEGs identified) was not affected by the number of replicates within a species. This was incorporated into the between-species comparison by calculating the distribution with edgeR once over all samples/replicates before comparing species. One distribution value was used for all subsequent comparisons, including those with species that had only one replicate.
Differential Expression within Species
For both S. minutum and S. psygmophilum, isoclonal cultures of four individual strains each were available, providing two opportunities to test for differential expression within a species. Each species was analyzed separately in edgeR (Robinson et al. 2010). Raw read counts were normalized with the geometric mean method.
Unlike hybridization-based techniques, the level of technical variation from sequencing is predictable and can therefore be distinguished from biological variation (Chen et al. 2014). In this case, the technical coefficient of variation describes the measurement error derived from the uncertainty with which the abundance of every gene is estimated from the sequencing platform, which decreases with increasing total counts for each gene in an RNA sample. In contrast, the biological coefficient of variation (BCV) describes the variation of the unknown, true abundance of each gene among replicates of RNA samples that will remain, even if sequencing depth could be increased indefinitely. Thus, the BCV represents the most important and main source of variation in expression studies using a high-throughput deep-sequencing approach (McCarthy et al. 2012). In RNA expression studies, the BCV is usually determined from the biological replicates of RNA samples so the total variation of gene abundances can be calculated by considering the following equation: Total CV2 = Technical CV2 + Biological CV2 (McCarthy et al. 2012).
Due to the lack of replicates for the comparison of the four genotypes of S. minutum and S. psygmophilum, we set the BCV to a fixed value a priori under the assumption that a majority of genes were not differentially expressed, which we considered appropriate for Symbiodinium of the same species under stable-state conditions. Although a value of ∼1% is suggested for technical replicates and a value of ∼10% is suggested for unique samples from separate but genetically identical model organisms, a value of ∼40% is appropriate for independent biological samples (Chen et al. 2014) and was chosen for our expression analysis among the four genotypes of both Symbiodinium species.
Significance of DEGs was determined by pairwise comparisons among individuals based on the negative binomial distribution with FDR < 0.1. A pairwise Euclidian distance matrix for all strain comparisons within and between S. minutum and S. psygmophilum was computed based on scaled FPKM values using PRIMER v6 software (Clarke and Gorley 2006).
Visualization and Functional Analyses
DEGs between and within species were visualized as heatmaps by plotting scaled FPKM expression values with Gene-E (Gould 2015). Lists were tested for GO term functional enrichment with the R/Bioconductor package topGO (Alexa and Rahnenfuhrer 2010), using the default “weight01” Alexa algorithm with the recommended cutoff of P < 0.05.
Results
Transcriptome Assemblies
We targeted isoclonal strains from four Clade B Symbiodinium species: S. aenigmaticum (n = 1 strain: mac04-487); S. minutum (n = 4 strains: mac703, Mf1.05b, rt002, and rt351); S. pseudominutum (n = 1 strain: rt146); and S. psygmophilum (n = 4 strains: HIAp, Mf10.14b.02, PurPFlex, and rt141). We reared the ten cultures under identical conditions in one incubator to assess stable-state conditions in the absence of environmental variability. Symbiodinium minutum and S. pseudominutum belong to the Pleistocene (B1) radiation. The former is a host-specialized mutualist because it commonly associates with the anemone Aiptasia sp. The latter is ecologically cryptic—although it has been isolated from the background symbiont population of four cnidarians, it has never been identified as a dominant symbiont. Symbiodinium psygmophilum and S. aenigmaticum belong to the Pliocene (B19) radiation; the former is host-specialized, the latter is ecologically cryptic. Note the uneven distribution of strains within species. This limitation was based on which cultures were available in the collection and meant that certain contrasts (e.g., 4 S. minutum strains vs. 4 S. psygmophilum strains) potentially had more power to detect differential expression than others (e.g., 1 S. aenigmaticum strain vs. 1 S. pseudominutum strain). However, for these data it was unlikely that the number of replicates affected power given that the same variance distribution was used for all comparisons (see Materials and Methods).
From these cultures, we generated ten high-quality short-read RNAseq libraries (table 1). Across species, sequencing statistics were quite similar. Total reads per sample averaged 14.3 million, while on average 88.5% of reads per sample passed quality control. For each of the four species, we generated a single reference assembly from either a combination of all strains of a given species (in the cases of S. minutum and S. psygmophilum) or from the single representative strain (in the cases of S. aenigmaticum and S. pseudominutum). The number of assembled genes per transcriptome averaged 48,700, the number of predicted ORFs averaged 41,300, the contig N50 statistic averaged 1,515 bp, mean transcript length per transcriptome averaged 1,078 bp, and annotation success averaged 46.5%. These values are in agreement with previously published Clade B Symbiodinium transcriptomes (Bayer et al. 2012; Baumgarten et al. 2013; Shoguchi et al. 2013).
Table 1.
Sequencing Statistics for the Ten Strains (A) and Transcriptome Assembly Statistics for the Four Species (B) of Clade B Symbiodinium
| A | ||||||||
|---|---|---|---|---|---|---|---|---|
| Species | Strain | Radiation | Ecology | Total Read Count (M) | Remaining After QC (%) | |||
| Symbiodinium minutum | mac703 | Pleistocene (B1) | Host-specialized | 10.9 | 89.03 | |||
| S. minutum | Mf1.05b | Pleistocene (B1) | Host-specialized | 19.3 | 88.40 | |||
| S. minutum | rt002 | Pleistocene (B1) | Host-specialized | 12.4 | 88.00 | |||
| S. minutum | rt351 | Pleistocene (B1) | Host-specialized | 8.7 | 88.68 | |||
| Symbiodinium psygmophilum | HIAp | Pliocene (B19) | Host-specialized | 13.4 | 88.14 | |||
| S. psygmophilum | Mf10.14b.02 | Pliocene (B19) | Host-specialized | 11.1 | 88.63 | |||
| S. psygmophilum | PurPflex | Pliocene (B19) | Host-specialized | 11.7 | 88.55 | |||
| S. psygmophilum | rt141 | Pliocene (B19) | Host-specialized | 19.5 | 88.47 | |||
| Symbiodinium pseudominutum | rt146 | Pleistocene (B1) | Ecologically cryptic | 23.7 | 88.83 | |||
| Symbiodinium aenigmaticum | mac04-487 | Pliocene (B19) | Ecologically cryptic | 11.9 | 88.10 | |||
| B | ||||||||
| Species | Assembly Length (Mbp) | Gene Count | Predicted ORF Count | Genes Annotated (%) | Longest Gene Length (bp) | Mean Gene Length (bp) | N50 (bp) | GC Content (%) |
| S. minutum | 57.2 | 51,199 | 42,929 | 47.3 | 37,483 | 1,118 | 1,579 | 51.33 |
| S. psygmophilum | 57.2 | 50,745 | 42,740 | 47.7 | 31,367 | 1,128 | 1,618 | 51.37 |
| S. pseudominutum | 51.3 | 47,411 | 40,716 | 46 | 31,393 | 1,081 | 1,508 | 51.51 |
| S. aenigmaticum | 44.6 | 45,343 | 38,923 | 44.9 | 24,202 | 984 | 1,355 | 51.39 |
Note.—The S. minutum and S. psygmophilum assemblies in (B) are composited from the reads of all respective strains listed in (A).
After uploading reference assemblies into IPA software, we identified 19 relevant canonical pathways with significant gene set representation in all species, including fatty acid beta-oxidation, nitric oxide signaling, oxidative stress response, cell cycle control, RNA processes, and protein ubiquitination (supplementary fig. S1, Supplementary Material online). We compared the ratio of genes observed to total associated genes per pathway among transcriptomes; each pathway was evenly represented in each Clade B Symbiodinium species. The four species were also roughly equivalent in terms of their proportions of microsatellite repeat motifs (supplementary fig. S2, Supplementary Material online).
Between-Species Expression Differences
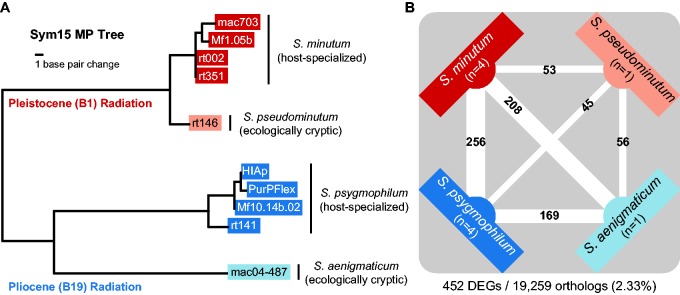
In order to compare gene expression between Symbiodinium species, we identified orthologs via a reciprocal BLAST approach on predicted ORFs. All species shared a total of 19,359 orthologs after filtering out paralogs and low quality matches (average pairwise ortholog count: 27,784; supplementary table S1, Supplementary Material online). We then scaled expression values relative to the lowest expressed gene in the data set (scaled FPKM) to allow for statistical comparison with the R package edgeR. We detected a total of 452 significant DEGs in pairwise species comparisons using expression dispersion estimates derived from all samples across Clade B (fig. 1a). Of these DEGs, 184 could be annotated. The distribution ranged from a low of 45 DEGs between S. pseudominutum and S. psygmophilum to a high of 256 DEGs between S. minutum and S. psygmophilum (fig. 1b). We visualized any annotated gene that was differentially expressed in at least one species with a heatmap to show relative expression patterns for all samples (fig. 2). A full list of annotations, expression values, and DEG list memberships can be found in supplementary table S3, Supplementary Material online.
Fig. 1.—
(A) Phylogenetic relationships and ecologies for the Clade B Symbiodinium species used in this experiment. The maximum parsimony tree was generated based on microsatellite Sym15 flanker region data from Parkinson, Coffroth, et al. (2015) and the methodologies described therein. (B) The numbers of differentially expressed genes (DEGs) between Clade B Symbiodinium species. Counts are placed on the lines connecting the two species being contrasted. Line thickness is scaled by the number of DEGs. Also depicted are the numbers (n) of cultured strains (clonal cell lines) included per species. Counts below the diagram show the total number of genes differentially expressed in at least one species and the total number of comparable orthologs across all species.
Fig. 2.—
Expression heatmaps of annotated DEGs among species. Colors are scaled to the minimum (purple) and maximum (orange) expression value per gene. Any gene that is differentially expressed in at least one species is included.
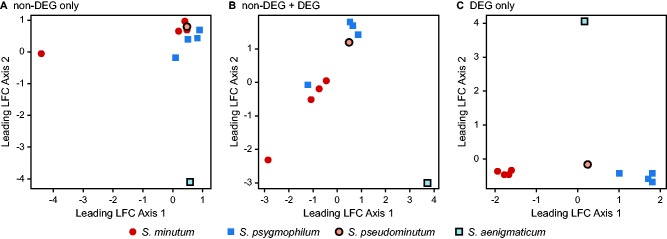
We also visualized the ten samples in an MDS plot, which shows spatial clustering based on similarity in gene expression values (fig. 3). When we clustered samples based on the expression of 18,907 genes that were not significantly differentially expressed in species-level contrasts, 8 of the 10 samples spanning three species grouped closely, with only the S. minutum strain mac703 and S. aenigmaticum strain mac04-487 separating from the others (fig. 3a). When we also included the 452 DEGs, species began to resolve, with one cluster consisting mostly of S. minutum, one of mostly S. psygmophilum, and the single S. aenigmaticum remaining unique (fig. 3b). All species segregated well when only the DEGs were considered (fig. 3c).
Fig. 3.—
Multidimensional scaling plots depicting sample clustering along the primary and secondary leading log2-fold change (LFC) axes using the following expression values: (A) Only non-DEGs (18,907), (B) non-DEGs and DEGs together (19,359), and (C) only DEGs (452). No border = host-specialized; border = ecologically cryptic; circles = Pleistocene (B1) radiation, squares = Pliocene (B19) radiation.
We subsequently conducted a GO term enrichment analysis on DEGs in order to assess which pathways were differentially represented (supplementary table S2, Supplementary Material online). The S. psygmophilum–S. pseudominutum contrast was enriched for processes including photosynthesis, response to cold, and transmembrane transport. The S. pseudominutum–S. minutum contrast was enriched for photosynthesis and apoptosis. The S. aenigmaticum–S. pseudominutum contrast was enriched for photosynthesis and heat acclimation. The S. psygmophilum–S. aenigmaticum contrast was enriched for photosynthesis and mitosis. The S. aenigmaticum–S. minutum contrast was enriched for stress response. The S. psygmophilum–S. minutum contrast was enriched for photosynthesis, phagocytosis, and cell signaling.
Within-Species Expression Differences
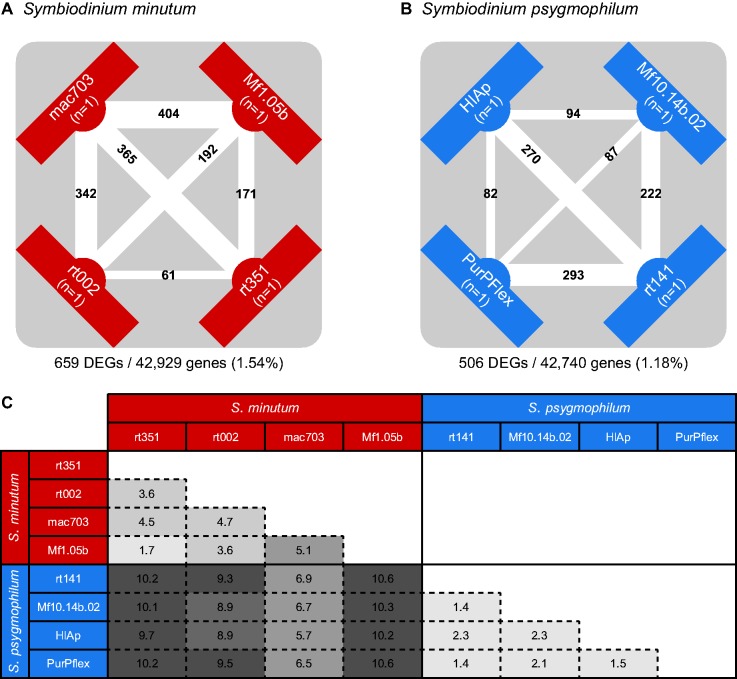
To understand the extent of gene expression differences among individuals within species, we first quantified expression variability by estimating the BCV and subsequently performed pairwise comparisons of the four distinct strains of S. minutum or S. psygmophilum using a fixed BCV that was more conservative (lower) than the original BCV estimate (Chen et al. 2014). We identified many pairwise expression differences among strains, ranging from 61 to 404 DEGs for S. minutum (fig. 4a) and 82 to 293 DEGs for S. psygmophilum (fig. 4b).
Fig. 4.—
The numbers of DEGs among strains within (A) Symbiodinium minutum and (B) Symbiodinium psygmophilum. Counts are placed on the lines connecting the two strains being contrasted. Line thickness is scaled by the number of DEGs. Counts below the diagram show the total number of genes differentially expressed in at least one strain and the total number of comparable genes (ORFs) across all strains. (C) Distances between pairs of strains based on Euclidean distance (×103) of expression estimates for all pairwise comparisons of S. minutum and S. psygmophilum strains. Boxes are shaded such that light coloration reflects relatively small values and dark coloration reflects relatively large values.
To further illustrate differences between pairs of strains for S. minutum and S. psygmophilum, we calculated an expression–strain distance matrix based on Euclidean distances between all pairwise strain comparisons using scaled FPKM values to assess variance (fig. 4c). Technical variation would be expected to be equally distributed across all samples. The distance matrix revealed a nonrandom distribution of variation in gene expression between pairs of strains for a given species. Also, the distance matrix showed that pairwise distances between strains from different species exceeded any within-species variation, and that both species exhibited distinct variance distributions among member strains.
Finally, we generated heatmaps to show the subset of all annotated genes differentially expressed in at least one individual among the four S. minutum strains (fig. 5a) and among the four S. psygmophilum strains (fig. 5b). In S. minutum, DEGs between strains were most highly enriched for the processes of malonyl-CoA biosynthesis, protein polymerization, long-chain fatty acid biosynthesis and metabolism, microtubule and nuclear envelope organization, GTP catabolism, and mitosis regulation (supplementary table S2, Supplementary Material online). In S. psygmophilum, DEGs between strains were most highly enriched for the processes of DNA replication and biosynthesis, sulfate assimilation and hydrogen sulfide biosynthesis, and microtubule organization (supplementary table S2, Supplementary Material online). A full list of annotations, expression values, and DEG list memberships can be found in supplementary table S3, Supplementary Material online.
Fig. 5.—
Expression heatmaps of annotated DEGs among individual strains (clonal cell lines) within (A) Symbiodinium minutum and (B) Symbiodinium psygmophilum. Colors are scaled to the minimum (purple) and maximum (orange) expression value per gene. Any gene that is differentially expressed in at least one strain is included.
Discussion
Fixed differences in gene expression ultimately influence the phenotypic variation available for selection to act upon. We anticipated that a comparative analysis of Symbiodinium spp. transcription would improve our understanding of adaptation and speciation among microeukaryotes. Indeed, we found that despite an overall similarity in gene content and expression among Clade B species with distinct ecologies, all cultures exhibited lineage-specific expression differences diagnostic for each species. Overrepresentation of photosynthesis-related gene expression variation among species likely reflects adaptation to unique light regimes over evolutionary time. Extensive disparity in the expression of fatty acid metabolism genes among strains within species may translate into differences in membrane composition, thermal tolerance, energy reserves, and growth rates. These differences may play a crucial role in coral–dinoflagellate symbiosis ecology and evolution. By examining the stable-state transcriptomes of cultures reared independently of their hosts under identical environmental conditions, we infer that these differences stem from genotypic rather than environmental factors. Our efforts reinforce the utility of comparative transcriptomics for studying speciation and functional variation in dinoflagellates and other nonmodel organisms (Chelaifa et al. 2010; Chapman et al. 2013; Gifford et al. 2013).
Partitioning the Variation in Gene Expression
When comparing multiple species, expression differences can be attributed to 1) technical variation, 2) within-species variation, and 3) among-species variation, with the proportion of variable genes expected to increase from 1) to 2) to 3) (Whitehead and Crawford 2006). Our results matched this general trend. Technical variation was inferred to be low based on the agreement between our data and transcriptome statistics from other studies that included the same S. minutum Mf1.05b strain, the high mapping success achieved between our Mf1.05b reads and the draft genome derived from the same strain (73%), and the nonrandom distribution of expression differences among species (fig. 4c). The percentage of orthologous genes differentially expressed within species (1.54% for S. minutum and 1.18% for S. psygmophilum; fig. 4) was roughly half that found between species (2.33%; fig. 2b). Overall, DEGs make up a small proportion of the entire transcriptome, as has been found before for Symbiodinium and other dinoflagellates (Baumgarten et al. 2013; Barshis et al. 2014; Xiang et al. 2015).
Between-Species Variation
By comparing closely related species within a clade, we greatly expanded our comparative power to determine what genetic changes underlie speciation among Symbiodinium. We were able to identify at least four times as many orthologs shared between Clade B species as has been possible using similar methods to compare species across separate clades (Ladner et al. 2012; Barshis et al. 2014; Rosic et al. 2014). Genetic divergence between clades is massive (Rowan and Powers 1992), and thus comparisons among species within clades reveal finer-scale differences likely to be important in physiological and ecological processes. Overall, stable-state gene expression was similar among Clade B Symbiodinium. Of the nearly 20,000 orthologs shared by S. aenigmaticum, S. minutum, S. pseudominutum, and S. psygmophilum, only 452 (2.3%) were differentially expressed between species. Thus a substantial portion of the transcriptome maintains relatively constant expression levels across members of Clade B. This result mirrors similar studies in other systems such as flowering plants where only a small proportion of interspecific orthologs were differentially expressed (Chapman et al. 2013).
The species comparison with the greatest number of DEGs was S. minutum versus S. psygmophilum (fig. 1b), which fit expectations for several reasons. First, our replication scheme (four strains per species) may have enhanced our ability to detect fixed differences between these species’ transcriptomes (though this is unlikely; see Materials and Methods). Second, both species associate with different hosts and likely diverged in part due to coevolutionary constraints imposed by those hosts, whereas ecologically cryptic species may not have faced the same constraints. Third, they are from phylogenetically divergent lineages. Finally, S. minutum is warm water adapted, while S. psygmophilum is cold water adapted (Thornhill et al. 2008), likely contributing to expression differences.
Interestingly, the contrasts with the second- and third-most abundant DEG counts both involved S. aenigmaticum (fig. 1b), a very distinct species from the Pliocene radiation and one that appears to have undergone rapid evolution (fig. 1a; LaJeunesse 2005; Parkinson, Coffroth, et al. 2015). The three species pair comparisons with the least number of DEGs all involved S. pseudominutum (fig. 1b). In fact, this species was roughly equidistant from all other species based on DEG number and MDS position (fig. 3c). Its position might be explained on the one hand by its close evolutionary history with S. minutum, and on the other by its cryptic ecology shared with S. aenigmaticum. Based on these results, fixed differences in gene expression may not always correspond to phylogenetic similarity.
Multidimensional scaling offered a complementary analysis for visualizing the similarities in expression among all strains without a priori knowledge of species membership (fig. 3). By restricting the data set to only non-DEGs, almost all replicates from all species (8 of 10) clustered together (fig. 3a), matching the expectation that at stable-state these Clade B Symbiodinium generally maintain similar expression profiles. When both non-DEGs and DEGs were included in the analysis, each species was mostly resolved, showing that non-DEGs contributed little to either species-level signal or noise (fig. 3b). As expected, when only the DEGs were considered, all species resolved well (fig. 3c). Note however that the distant positioning of S. aenigmaticum in all three MDS plots indicates that a large proportion of expression variation for this species is unique.
In addition to pairwise comparisons, we also contrasted groups of replicate species by lineage (2 species from the Pleistocene radiation vs. 2 Pliocene radiation species) or by ecology (2 host-specialized species vs. 2 ecologically cryptic species). The Pleistocene—Pliocene contrast was equivalent to the S. minutum–S. psygmophilum comparison in terms of identity of DEGs, meaning that the species contrast either captured all the differences between major lineages, or that adding just one more strain to each group did not affect expression variation sufficiently to alter our detection of DEGs, even though the strain belonged to a different species. Similarly, the ‘host-specialized’—‘ecologically cryptic’ contrast only recovered four unique genes that had not been identified in any of the species contrasts. The probable identity of only one of these DEGs was determined (a general mRNA splicing factor). These results indicate that differential expression of a particular set of genes does not necessarily explain shared ecological attributes of phylogenetically distinct species.
Photosynthesis Gene Expression Differences between Species
Expression differences among closely related species were consistently enriched for photosynthesis genes (supplementary table S2, Supplementary Material online). Here, overrepresentation of plastid genes cannot be attributed to light intensity differences because all cultures were reared under identical light conditions. In fact, although we might expect these genes to be regulated by light intensity in Symbiodinium as they are in other photosynthetic organisms (Escoubas et al. 1995; Pfannschmidt 2003), only minor (or no) changes in photosynthesis-related gene expression have been detected in cultures exposed to varying light levels (McGinley et al. 2013; Xiang et al. 2015). Thus, we conclude that different species evolved unique expression levels among photosynthesis-related genes. These differences may relate to inherent variation in the circadian rhythm among species (Van Dolah et al. 2007; Sorek and Levy 2012) or, more likely, to functional variation in photosynthesis biochemistry. For example, during heat stress, thermally sensitive Symbiodinium taxa suffer physiological disruption of PSII photochemistry (Warner et al. 1999; Robison and Warner 2006) and associated downregulation of core photosynthesis genes (McGinley et al. 2012), whereas thermally tolerant species do not. The maintenance of distinct expression patterns at key genes may underlie the capacity for certain Symbiodinium species to occupy distinct niches, as has been demonstrated for three diatom species in the genus Pseudonitzschia (Di Dato et al. 2015).
Evolutionary Significance of Gene Expression Variation
In biogeographic surveys of marine mutualisms, depth and latitude (correlates of light availability) are often primary factors explaining the distribution of Symbiodinium diversity (Rowan and Knowlton 1995; LaJeunesse et al. 2004, 2014; Frade et al. 2008; Finney et al. 2010; Sanders and Palumbi 2011). Thus, light availability represents a main axis of niche differentiation for this group. Symbiodinium possess a diverse array of light-harvesting proteins (Boldt et al. 2012), which may be both the cause and consequence of ecological specialization. Many such genes have been transferred to the nuclear genome (Bachvaroff et al. 2004), while others are encoded on plastid minicircles (Zhang et al. 1999; Moore et al. 2003; Barbrook et al. 2014). Minicircles are subject to different transcriptional mechanisms than nuclear encoded genes (Dang and Green 2010), which may also facilitate specialization to different light regimes. Given that a majority of expression variation between divergent species is expected to accumulate neutrally over time (Khaitovich et al. 2005), it is intriguing that expression differences between Symbiodinium species are consistently enriched for photosynthesis genes (Baumgarten et al. 2013; Barshis et al. 2014; Rosic et al. 2014; this study). This evidence suggests that species-specific differences in gene expression are functionally important and influenced by natural selection tied to niche diversification.
Within-Species Variation
Within each of the two species with four isoclonal cultures, we detected hundreds of DEGs: 659 unique genes among S. minutum strains (fig. 4a) and 506 unique genes among S. psygmophilum strains (fig. 4b). Interestingly, only four annotated genes differentially expressed among S. minutum overlapped with those among S. psygmophilum, and enriched categories only overlapped for housekeeping genes which regulate biochemical processes like nucleic acid synthesis and microtubule organization (supplementary table S2, Supplementary Material online). Thus, transcriptional variation among strains differs from species to species (fig. 5). Furthermore, nonrandom gene expression differences among strains of a given species exist even under identical rearing conditions (fig. 4c), emphasizing that a degree of expression variation among Symbiodinium strains is genetically determined and potentially subject to natural selection. Thus, the extent of variation among isoclonal strains may be much greater than previously assumed. Although inter-individual differences are known to play a significant role in symbiosis ecology and evolution in terrestrial systems (Shuster et al. 2006; Whitham et al. 2006; Hughes et al. 2008), such evidence has been lacking for coral–dinoflagellate associations (Parkinson and Baums 2014). Although ∼500 of the ∼40,000 genes represents a small fraction of the transcriptome, such differences may be important, especially because overall differential expression of genes within a Symbiodinium species responding to stress seems low (Barshis et al. 2014; but see Baumgarten et al. 2013).
For example, putative “symbiosis genes” have been identified by comparing symbiotic versus aposymbiotic cnidarian hosts (Meyer and Weis 2012). The expression levels of similar genes in the symbiont may also play a role in maintaining functional associations. Two such genes varied among S. minutum strains: An ABC transporter (up to 4.2-fold) and a glutathione reductase (up to 9.5-fold). There were also clear differences in the expression of long chain fatty acid CoA ligase (up to 12.2-fold), long chain acyl-CoA synthetase (up to 8.8-fold), and six acetyl-CoA carboxylases (up to 12.5-fold), indicating that certain strains regulate fatty acid metabolism differently. These genes may be related to cell membrane composition, which in turn can affect thermal sensitivity (Tchernov et al. 2004; Diaz-Almeyda et al. 2011). They may also relate to energy storage and nutrient availability, perhaps contributing to different growth rates observed among some of these strains ex hospite (Parkinson and Baums 2014). Under environmental change, these functional differences may impact stress tolerance among genotypic host–symbiont combinations in a population (Parkinson and Baums 2014; Parkinson, Banaszak, et al. 2015), partly explaining why some coral colonies of a given species bleach while others do not, even when sharing the same symbiont species (Goulet et al. 2008; LaJeunesse et al. 2010). Similar fine-scale variation has been observed among maize strains with distinct flavonoid content (Casati and Walbot 2003) and among dinoflagellate strains with distinct toxicities (Yang et al. 2010).
Conclusions
Comparisons among deeply sequenced transcriptomes can reveal the extent and function of molecular variation that is critical to speciation in nonmodel organisms. Such work provides important baselines against which experimentally manipulated samples might be compared and more accurately interpreted. Our data reveal the extent of expression variation that occurs among strains of Symbiodinium and emphasizes how natural selection on existing populations may play a critical role in the response of coral–dinoflagellate symbioses to climate change. The genomic resources described here should improve functional investigations into marine symbiosis biology, particularly as model systems continue to be developed (Baumgarten et al. 2015). Future studies should examine the same strains exposed to different stressors (thermal, osmotic, and/or light) in order to characterize the relationship between physiological and gene expression phenotypes. Each strain should also be brought into an experimental host (e.g., the model Aiptasia [= Exaiptasia]) and observed in symbiosis, which would provide insight into how changes in gene expression work to maintain stable cnidarian–dinoflagellate mutualisms. Our findings underscore that important transcriptional differences exist at different taxonomic ranks among dinoflagellates, from clades to species to strains. Future Symbiodinium genomics experiments should be designed such that clade-level questions incorporate different species to serve as a representative sampling of the clade under study, while species-level questions should incorporate distinct strains to serve as a representative sampling of the species under study. Such designs will improve our understanding of Symbiodinium genetic, functional, and phylogenetic diversity.
Supplementary Material
Supplementary tables S1–S3 and figures S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
Mary Alice Coffroth provided cultures, and Maren Ziegler assisted with PRIMER software. This work was supported by the National Science Foundation (DEG-0750756 to J.E.P., OCE-0928764 to T.C.L. and I.B.B.), the Pennsylvania State University, and baseline research funds to CRV by King Abdullah University of Science and Technology (KAUST).
Literature Cited
- Ahles MD. 1967. Some aspects of the morphology and physiology of Symbiodinium microadriaticum [phd dissertation]. [New York]: Fordham University. [Google Scholar]
- Alexa A, Rahnenfuhrer J. 2010. topGO: enrichment analysis for gene ontology. R package version 2.2.0. http://bioconductor.uib.no/2.7/bioc/html/topGO.html Last accessed: January 2014.
- Alexeyenko A, Tamas I, Liu G, Sonnhammer ELL. 2006. Automatic clustering of orthologs and inparalogs shared by multiple proteomes. Bioinformatics 22:E9–E15. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215:403–410. [DOI] [PubMed] [Google Scholar]
- Bachvaroff TR, Concepcion GT, Rogers CR, Herman EM, Delwiche CF. 2004. Dinoflagellate expressed sequence tag data indicate massive transfer of chloroplast genes to the nuclear genome. Protist 155:65–78. [DOI] [PubMed] [Google Scholar]
- Barbrook AC, Voolstra CR, Howe CJ. 2014. The chloroplast genome of a Symbiodinium sp. clade C3 isolate. Protist 165:1–13. [DOI] [PubMed] [Google Scholar]
- Barshis DJ, Ladner JT, Oliver J, Palumbi SR. 2014. Lineage specific transcriptional profiles of Symbiodinium spp. unaltered by heat stress in a coral host. Mol Biol Evol. 31:1343–1352. [DOI] [PubMed] [Google Scholar]
- Barshis DJ, et al. 2013. Genomic basis for coral resilience to climate change. Proc Natl Acad Sci U S A. 110:1387–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarten S, et al. 2013. Integrating microRNA and mRNA expression profiling in Symbiodinium microadriaticum, a dinoflagellate symbiont of reef-building corals. BMC Genomics 14:704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarten S, et al. 2015. The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc Natl Acad Sci U S A. 112:11893–11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer T, et al. 2012. Symbiodinium transcriptomes: genome insights into the dinoflagellate symbionts of reef-building corals. PLoS One 7:e5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldt L, Yellowlees D, Leggat W. 2012. Hyperdiversity of genes encoding integral light-harvesting proteins in the dinoflagellate Symbiodinium sp. PLoS One 7:e47456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casati P, Walbot V. 2003. Gene expression profiling in response to ultraviolet radiation in maize genotypes with varying flavonoid content. Plant Physiol. 132:1739–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MA, Hiscock SJ, Filatov DA. 2013. Genomic divergence during speciation driven by adaptation to altitude. Mol Biol Evol. 30:2553–2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelaifa H, Mahe F, Ainouche M. 2010. Transcriptome divergence between the hexaploid salt-marsh sister species Spartina maritima and Spartina alterniflora (Poaceae). Mol Ecol. 19:2050–2063. [DOI] [PubMed] [Google Scholar]
- Chen Y, Lun AT, Smyth GK. 2014. Differential expression analysis of complex RNA-seq experiments using edgeR In: Nettleton D, Datta S, editors. Statistical analysis of next generation sequencing data. Springer International Publishing: Switzerland. p. 51–74. [Google Scholar]
- Clarke K, Gorley R. 2006. PRIMER v6: user manual/tutorial. Plymouth (UK): PRIMER-E; 192pp. [Google Scholar]
- Dang YK, Green BR. 2010. Long transcripts from dinoflagellate chloroplast minicircles suggest “rolling circle” transcription. J Biol Chem. 285:5196–5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz-Almeyda E, Thome PE, El Hafidi M, Iglesias-Prieto R. 2011. Differential stability of photosynthetic membranes and fatty acid composition at elevated temperature in Symbiodinium. Coral Reefs 30:217–225. [Google Scholar]
- Di Dato V, et al. 2015. Transcriptome sequencing of three Pseudo-nitzschia species reveals comparable gene sets and the presence of Nitric Oxide Synthase genes in diatoms. Sci Rep. 5:12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmer EC, et al. 2012. The UniProt-GO Annotation database in 2011. Nucleic Acids Res. 40:D565–D570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdner DL, Anderson DM. 2006. Global transcriptional profiling of the toxic dinoflagellate Alexandrium fundyense using massively parallel signature sequencing. BMC Genomics 7:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escoubas JM, Lomas M, Laroche J, Falkowski PG. 1995. Light-intensity regulation of cab gene: transcription is signaled by the redox state of the plastoquinone pool. Proc Natl Acad Sci U S A. 92:10237–10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. 2014. Pfam: the protein families database. Nucleic Acids Res. 42:D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finney JC, et al. 2010. The relative significance of host-habitat, depth, and geography on the ecology, endemism, and speciation of coral endosymbionts in the genus Symbiodinium. Microb Ecol. 60:250–263. [DOI] [PubMed] [Google Scholar]
- Fitt WK, Brown BE, Warner ME, Dunne RP. 2001. Coral bleaching: interpretation of thermal tolerance limits and thermal thresholds in tropical corals. Coral Reefs 20:51–65. [Google Scholar]
- Frade PR, De Jongh F, Vermeulen F, Van Bleijswijk J, Bak RPM. 2008. Variation in symbiont distribution between closely related coral species over large depth ranges. Mol Ecol. 17:691–703. [DOI] [PubMed] [Google Scholar]
- Galardini M, et al. 2011. Exploring the symbiotic pangenome of the nitrogen-fixing bacterium Sinorhizobium meliloti. BMC Genomics 12:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford SM, Sharma S, Booth M, Moran MA. 2013. Expression patterns reveal niche diversification in a marine microbial assemblage. ISME J. 7:281–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnerre S, et al. 2011. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A. 108:1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould J. 2015. Gene-E. Available from: http://www.broadinstitute.org/cancer/software/GENE-E/index.html. Last accessed: June 2015.
- Goulet TL, LaJeunesse TC, Fabricius KE. 2008. Symbiont specificity and bleaching susceptibility among soft corals in the 1998 Great Barrier Reef mass coral bleaching event. Mar Biol. 154:795–804. [Google Scholar]
- Grabherr MG, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 8:1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegh-Guldberg O. 1999. Climate change, coral bleaching and the future of the world's coral reefs. Mar Freshwater Res. 50:839–866. [Google Scholar]
- Hughes AR, Inouye BD, Johnson MTJ, Underwood N, Vellend M. 2008. Ecological consequences of genetic diversity. Ecol Lett. 11:609–623. [DOI] [PubMed] [Google Scholar]
- Jeong HJ, et al. 2014. Genetics and morphology characterize the dinoflagellate Symbiodinium voratum, n. sp., (Dinophyceae) as the sole representative of Symbiodinium clade E. J Eukaryot Microbiol. 61:75–94. [DOI] [PubMed] [Google Scholar]
- Khaitovich P, Paabo S, Weiss G. 2005. Toward a neutral evolutionary model of gene expression. Genetics 170:929–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Schlotterer C, Lelley T. 2007. SciRoKo: a new tool for whole genome microsatellite search and investigation. Bioinformatics 23:1683–1685. [DOI] [PubMed] [Google Scholar]
- Ladner JT, Barshis DJ, Palumbi SR. 2012. Protein evolution in two co-occurring types of Symbiodinium: an exploration into the genetic basis of thermal tolerance in Symbiodinium clade D. BMC Evol Biol. 12:217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaJeunesse TC. 2005. “Species” radiations of symbiotic dinoflagellates in the Atlantic and Indo-Pacific since the Miocene-Pliocene transition. Mol Biol Evol. 22:570–581. [DOI] [PubMed] [Google Scholar]
- LaJeunesse TC, Lee SY, Gil-Agudelo DL, Knowlton N, Jeong HJ. 2015. Symbiodinium necroappetens sp. nov. (Dinophyceae): an opportunist ‘zooxanthella’ found in bleached and diseased tissues of Caribbean reef corals. Eur J Phycol. 50:223–238. [Google Scholar]
- LaJeunesse TC, Parkinson JE, Reimer JD. 2012. A genetics-based description of Symbiodinium minutum sp. nov. and S. psygmophilum sp. nov. (Dinophyceae), two dinoflagellates symbiotic with cnidaria. J Phycol. 48:1380–1391. [DOI] [PubMed] [Google Scholar]
- LaJeunesse TC, Thornhill DJ. 2011. Improved resolution of reef-coral endosymbiont (Symbiodinium) species diversity, ecology, and evolution through psbA non-coding region genotyping. PLoS One 6:e29013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaJeunesse TC, et al. 2004. Closely related Symbiodinium spp. differ in relative dominance in coral reef host communities across environmental, latitudinal and biogeographic gradients. Mar Ecol Prog Ser. 284:147–161. [Google Scholar]
- LaJeunesse TC, et al. 2010. Host-symbiont recombination versus natural selection in the response of coral-dinoflagellate symbioses to environmental disturbance. Proc Biol Sci. 277:2925–2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaJeunesse TC, et al. 2014. Ecologically differentiated stress tolerant endosymbionts in the dinoflagellate genus Symbiodinium Clade D are different species. Phycologia 53:305–319. [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggat W, Hoegh-Guldberg O, Dove S, Yellowlees D. 2007. Analysis of an EST library from the dinoflagellate (Symbiodinium sp.) symbiont of reef-building corals. J Phycol 43:1010–1021. [Google Scholar]
- Leggat W, Seneca F, et al. 2011. Differential responses of the coral host and their algal symbiont to thermal stress. PLoS One 6:e26687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggat W, Yellowlees D, Medina M. 2011. Recent progress in Symbiodinium transcriptomics. J Exp Mar Biol Ecol. 408:120–125. [Google Scholar]
- Lehnert EM, Burriesci MS, Pringle JR. 2012. Developing the anemone Aiptasia as a tractable model for cnidarian-dinoflagellate symbiosis: the transcriptome of aposymbiotic A. pallida. BMC Genomics 13:271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnert EM, et al. 2014. Extensive differences in gene expression between symbiotic and aposymbiotic cnidarians. G3 (Bethesda) 4:277–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ. 2011. Genomic understanding of dinoflagellates. Res Microbiol. 162:551–569. [DOI] [PubMed] [Google Scholar]
- Lin SJ, et al. 2015. The Symbiodinium kawagutii genome illuminates dinoflagellate gene expression and coral symbiosis. Science 350:691–694. [DOI] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40:4288–4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinley MP, Suggett DJ, Warner ME. 2013. Transcript patterns of chloroplast-encoded genes in cultured Symbiodinium spp. (Dinophyceae): testing the influence of a light shift and diel periodicity. J Phycol. 49:709–718. [DOI] [PubMed] [Google Scholar]
- McGinley MP, et al. 2012. Transcriptional response of two core photosystem genes in Symbiodinium spp. exposed to thermal stress. PLoS One 7:e50439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin JJA, Zahl PA. 1959. Axenic zooxanthellae from various invertebrate hosts. Ann N Y Acad Sci. 77:55–72. [Google Scholar]
- Meyer E, Weis VM. 2012. Study of cnidarian-algal symbiosis in the “omics” age. Biol Bull. 223:44–65. [DOI] [PubMed] [Google Scholar]
- Moberg F, Folke C. 1999. Ecological goods and services of coral reef ecosystems. Ecol Econ. 29:215–233. [Google Scholar]
- Moore RB, Ferguson KM, Loh WKW, Hoegh-Guldberg C, Carter DA. 2003. Highly organized structure in the non-coding region of the psbA minicircle from clade C Symbiodinium. Int J Syst Evol Microbiol. 53:1725–1734. [DOI] [PubMed] [Google Scholar]
- Morse D, Milos PM, Roux E, Hastings JW. 1989. Circadian regulation of bioluminescence in Gonyaulax involves translational control. Proc Natl Acad Sci U S A. 86:172–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa A, et al. 2010. Transcriptome profiling of a toxic dinoflagellate reveals a gene-rich protist and a potential impact on gene expression due to bacterial presence. PLoS One 5:e9688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscatine L. 1990. The role of symbiotic algae in carbon and energy flux in reef corals In: Dubinsky, editor. Ecosystems of the world 25: Coral reefs. New York: Elsevier. p. 75–87. [Google Scholar]
- Muscatine L, Porter JW. 1977. Reef corals—mutualistic symbioses adapted to nutrient-poor environments. Bioscience 27:454–460. [Google Scholar]
- Österman J, Mousavi SA, Koskinen P, Paulin L, Lindström K. 2015. Genomic features separating ten strains of Neorhizobium galegae with different symbiotic phenotypes. BMC Genomics 16:348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JE, Banaszak AT, Altman NS, LaJeunesse TC, Baums IB. 2015. Intraspecific diversity among partners drives functional variation in coral symbioses. Sci Rep. 5:15667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JE, Baums IB. 2014. The extended phenotypes of marine symbioses: ecological and evolutionary consequences of intraspecific genetic diversity in coral-algal associations. Front Microbiol. 5:445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson JE, Coffroth MA, LaJeunesse TC. 2015. New species of Clade B Symbiodinium (Dinophyceae) from the greater Caribbean belong to different functional guilds: S. aenigmaticum sp. nov., S. antillogorgium sp. nov., S. endomadracis sp. nov., and S. pseudominutum sp. nov. J Phycol. 51:850–858. [DOI] [PubMed] [Google Scholar]
- Pettay DT, LaJeunesse TC. 2007. Microsatellites from clade B Symbiodinium spp. specialized for Caribbean corals in the genus Madracis. Mol Ecol Notes. 7:1271–1274. [Google Scholar]
- Pfannschmidt T. 2003. Chloroplast redox signals: how photosynthesis controls its own genes. Trends Plant Sci. 8:33–41. [DOI] [PubMed] [Google Scholar]
- Provasoli L. 1968. Media and prospects for the cultivation of marine algae. In: Watanabe H, Hattori A, editors. Culture and Collection of Algae. Proceedings of the US-Japan Conference. Japanese Society for Plant Physiology; Hakone, Japan. p. 63–75.
- Pruitt KD, et al. 2014. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 42:D756–D763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranz JM, Machado CA. 2006. Uncovering evolutionary patterns of gene expression using microarrays. Trends Ecol Evol. 21:29–37. [DOI] [PubMed] [Google Scholar]
- Remm M, Storm CEV, Sonnhammer ELL. 2001. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol. 314:1041–1052. [DOI] [PubMed] [Google Scholar]
- Ribeiro FJ, et al. 2012. Finished bacterial genomes from shotgun sequence data. Genome Res. 22:2270–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pachter L. 2013. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods. 10:71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison JD, Warner ME. 2006. Differential impacts of photoacclimation and thermal stress on the photobiology of four different phylotypes of Symbiodinium (Pyrrhophyta). J Phycol. 42:568–579. [Google Scholar]
- Rosic N, et al. 2014. Unfolding the secrets of coral–algal symbiosis. ISME J. 9:844–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan R, Knowlton N. 1995. Intraspecific diversity and ecological zonation in coral algal symbiosis. Proc Natl Acad Sci U S A. 92:2850–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan R, Powers DA. 1992. Ribosomal RNA sequences and the diversity of symbiotic dinoflagellates (Zooxanthellae). Proc Natl Acad Sci U S A. 89:3639–3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders JG, Palumbi SR. 2011. Populations of Symbiodinium muscatinei show strong biogeographic structuring in the intertidal anemone Anthopleura elegantissima. Biol Bull. 220:199–208. [DOI] [PubMed] [Google Scholar]
- Santos SR, Taylor DJ, Coffroth MA. 2001. Genetic comparisons of freshly isolated versus cultured symbiotic dinoflagellates: implications for extrapolating to the intact symbiosis. J Phycol. 37:900–912. [Google Scholar]
- Schoenberg DA, Trench RK. 1980. Genetic variation in Symbiodinium (=Gymnodinium) microadriaticum Freudenthal, and specificity in its symbiosis with marine invertebrates. 1. Isoenzyme and soluble-protein patterns of axenic cultures of Symbiodinium microadriaticum. Proc Biol Sci. 207:405–427. [Google Scholar]
- Shoguchi E, et al. 2013. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr Biol. 23:1399–1408. [DOI] [PubMed] [Google Scholar]
- Shuster SM, Lonsdorf EV, Wimp GM, Bailey JK, Whitham TG. 2006. Community heritability measures the evolutionary consequences of indirect genetic effects on community structure. Evolution 60:991–1003. [PubMed] [Google Scholar]
- Sorek M, Levy O. 2012. Influence of the quantity and quality of light on photosynthetic periodicity in coral endosymbiotic algae. PLoS One 7:e43264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunagawa S, et al. 2009. Generation and analysis of transcriptomic resources for a model system on the rise: the sea anemone Aiptasia pallida and its dinoflagellate endosymbiont. BMC Genomics 10:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchernov D, et al. 2004. Membrane lipids of symbiotic algae are diagnostic of sensitivity to thermal bleaching in corals. Proc Natl Acad Sci U S A. 101:13531–13535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornhill DJ, Kemp DW, Bruns BU, Fitt WK, Schmidt GW. 2008. Correspondence between cold tolerance and temperate biogeography in a Western Atlantic Symbiodinium (Dinophyta) lineage. J Phycol. 44:1126–1135. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium 2013. Update on activities at the Universal Protein Resource (UniProt) in 2013. Nucleic Acids Res. 41:D43–D47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dolah FM, et al. 2007. Microarray analysis of diurnal- and circadian-regulated genes in the Florida red-tide dinoflagellate Karenia brevis (Dinophyceae). J Phycol. 43:741–752. [Google Scholar]
- Voolstra C, Tautz D, Farbrother P, Eichinger L, Harr B. 2007. Contrasting evolution of expression differences in the testis between species and subspecies of the house mouse. Genome Res. 17:42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voolstra CR, et al. 2009. The host transcriptome remains unaltered during the establishment of coral-algal symbioses. Mol Ecol. 18:1823–1833. [DOI] [PubMed] [Google Scholar]
- Warner ME, Fitt WK, Schmidt GW. 1999. Damage to photosystem II in symbiotic dinoflagellates: a determinant of coral bleaching. Proc Natl Acad Sci U S A. 96:8007–8012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis VM, Davy SK, Hoegh-Guldberg O, Rodriguez-Lanetty M, Pringle JR. 2008. Cell biology in model systems as the key to understanding corals. Trends Ecol Evol. 23:369–376. [DOI] [PubMed] [Google Scholar]
- Whitehead A, Crawford DL. 2006. Variation within and among species in gene expression: raw material for evolution. Mol Ecol. 15:1197–1211. [DOI] [PubMed] [Google Scholar]
- Whitham TG, et al. 2006. A framework for community and ecosystem genetics: from genes to ecosystems. Nat Rev Genet. 7:510–523. [DOI] [PubMed] [Google Scholar]
- Wittkopp PJ, Haerum BK, Clark AG. 2008. Regulatory changes underlying expression differences within and between Drosophila species. Nat Genet. 40:346–350. [DOI] [PubMed] [Google Scholar]
- Wolf JBW, Lindell J, Backstrom N. 2010. Speciation genetics: current status and evolving approaches. Philos Trans R Soc B Biol Sci. 365:1717–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang T, Nelson W, Rodriguez J, Tolleter D, Grossman AR. 2015. Symbiodinium transcriptome and global responses of cells to immediate changes in light intensity when grown under autotrophic or mixotrophic conditions. Plant J. 82:67–80. [DOI] [PubMed] [Google Scholar]
- Xiang TT, Hambleton EA, DeNofrio JC, Pringle JR, Grossman AR. 2013. Isolation of clonal axenic strains of the symbiotic dinoflagellate Symbiodinium and their growth and host specificity. J Phycol. 49:447–458. [DOI] [PubMed] [Google Scholar]
- Yang I, et al. 2010. Comparative gene expression in toxic versus non-toxic strains of the marine dinoflagellate Alexandrium minutum. BMC Genomics 11:248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellowlees D, Rees TAV, Leggat W. 2008. Metabolic interactions between algal symbionts and invertebrate hosts. Plant Cell Environ. 31:679–694. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Green B, Cavalier-Smith T. 1999. Single gene circles in dinoflagellate chloroplast genomes. Nature 400:155–159 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.