Abstract
MicroRNAs (miRNAs) belong to a group of short noncoding RNA molecules with important roles in cellular biology. miRNAs regulate gene expression by repressing translation or degrading the target mRNA. Recently, a growing body of evidence suggests that miRNAs are implicated in many diseases and could be potential biomarkers. Fibrosis and/smooth muscle (SM) dysfunction contributes to the morbidity and mortality associated with several diseases of the gastrointestinal tract (GIT). Currently available therapeutic modalities are unsuccessful in efficiently blocking or reversing fibrosis and/or SM dysfunction. Recent understanding of the role of miRNAs in signaling pathway of fibrogenesis and SM phenotype switch has provided a new insight into translational research. However, much is still unknown about the molecular targets and therapeutic potential of miRNAs in the GIT. This review discusses miRNA biology, pathophysiology of fibrosis, and aging- associated SM dysfunction in relation to the deregulation of miRNAs in the GIT. We also highlight the role of selected miRNAs associated with fibrosis and SM dysfunction-related diseases of the GIT.
Keywords: smooth muscle fibrosis, microRNA, cellular and epigenetic mechanisms, inflammatory mediators, phenotypic plasticity, smooth muscle phenotypic switch, rho kinase
revolution in computational biology led to the identification of ∼2,800 novel human miRNAs, of which more than 1,000 have been validated (data from miRNABase 21; http://www.mirbase.org). miRNAs are a group of endogenous, short (∼22 nucleotides), noncoding RNAs (ncRNAs) with important roles in cellular biology via targeting mRNAs and thereby causing translational repression (2, 6, 44). The first discovered miRNA, lin-4, influenced the development of the nematode Caenorhabditis elegans by repressing the protein expression of its target gene lin-14 (66). Subsequently, miRNAs were found to have a ubiquitous presence among living organisms including human beings; however, their expression profile and biological function could be species or tissue specific (72). miRNAs regulate the expression of over 60% of all mammalian protein coding genes responsible for diverse physiological processes, such as cell growth and differentiation, fibrogenesis, smooth muscle (SM) phenotype switch, and response to stress and aging (7, 39). In addition, miRNA deregulation is being increasingly implicated in several pathophysiological outcomes underlying diseases of various organ systems, such as gastrointestinal tract (GI) dysmotility (75, 91).
In health, GI motility arises from coordinated contractions of smooth muscle cells (SMCs). Although GI SMCs may have spontaneous electrical activity, their contractile function is also regulated by inputs from enteric nervous system including enteric motor neurons and interstitial cells of Cajal (114). Fibrosis, the final outcome in many chronic inflammatory diseases, can result in permanent scarring, organ damage, and even death. In the GIT, fibrosis and/or SM dysfunction contributes to dysmotility, which can lead to intestinal obstruction or rectoanal incontinence (RI). Historically, fibrosis has been viewed as an irreversible process, but recent evidence indicates that it may be reversible (47). Similarly, SM dysfunction is also potentially reversible. The aim of this review is to discuss miRNA biology and its regulation of the pathophysiology of fibrosis and SM dysfunction in the GIT. Regulation of GI SMCs by enteric nervous system is not the focus of our review and is not discussed here. In addition, we highlight the role of selected miRNAs implicated in fibrosis and SM dysfunction underlying diseases of GIT, such as scleroderma and Crohn's disease.
miRNA Biogenesis and Mechanism of Action
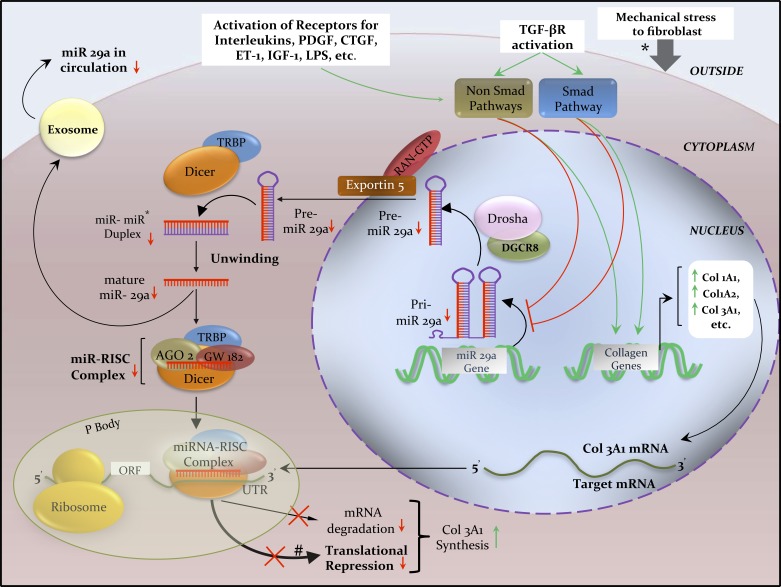
ncRNAs can have a regulatory role over the expression of coding genes. miRNA can be coded by its own gene or produced from introns of other genes. The majority of the characterized miRNA genes are intergenic or oriented antisense to their neighboring genes in the chromosome and are therefore transcribed as independent units. However, in certain situations a miRNA gene can be transcribed along with its host gene; this provides a coregulation of miRNA and the protein-coding mRNA gene. Canonical pathway of miRNA biogenesis (Fig. 1) begins in the nucleus where RNA polymerase II transcribes long molecules called primary miRNAs (pri-miRNAs), which are subsequently processed by the RNase III endonuclease, Drosha, and its cofactor DiGeorge syndrome critical region gene 8 (DGCR8)/Pasha, to yield a shorter (∼70 nucleotides) stem-looped structure called precursor miRNA (pre-miRNA) (48, 68). Subsequently, pre-miRNA is actively transported by exportin 5 to the cytoplasm (137), where a second RNase III enzyme, Dicer (57), and its cofactor transactivating response RNA-binding protein (TRBP) cleave it to form a double-stranded miRNA duplex. Later, the strands are unwound and one of the strands is degraded by Argonaute (Ago) 2 (104), whereas the remaining mature miRNA strand is loaded onto the miRNA-induced silencing complex (RISC). Ago2 and glycine-tryptophan protein (GW182) form the core elements of miRNA-RISC, which are vital for miRNA-mRNA binding. A 6- to 8-nucleotide-long sequence (seed sequence) at the 5′ end of a miRNA binds to complementary sequence in the 3′ untranslated regions (UTR) of target mRNA (102). Finally, translational inhibition or, in some cases, mRNA degradation (5, 87) leads to downregulation of the target protein.
Fig. 1.
Biogenesis of miRNA 29a and its regulation of Col3A1 mRNA inside an activated fibroblast in scleroderma. Schematics illustrate the transcription of pri-miR-29a being regulated through TGF-β, PDGF, CTGF, ET-1, IL-4, and LPS signaling via SMAD and non-SMAD pathways, and by epigenetic silencing. Transcription of Col3A1 mRNA is regulated by the overactive SMAD and non-SMAD signaling pathways. Extracellular exchange of miR 29a via exosomes (small vesicles produced by budding of plasma membrane) is consequently reduced, resulting in low circulating miR 29a levels in the very early stage of scleroderma, which could potentially serve as a disease biomarker (60). In human beings, imperfect complementarity of miR-mRNA binding is more common than perfect complementarity; therefore, miR-induced translational repression (thick black line) is more common that target mRNA degradation (thin black line). *Mechanical stress also activates fibroblasts to synthesize excess collagen (34). Red lines and arrows indicate inhibition or decrease; green lines and arrows indicate stimulation or increase. AGO2, Argonaute2; CTGF, connective tissue growth factor; DGCR8, DiGeorge syndrome critical region gene 8; ET-1, endothelin-1; IGF-1, insulin like growth factor-1; IL-4, interleukin-4; LPS, lipopolysaccharide; ORF, open reading frame; P body, processing body; PDGF-β, platelet-derived growth factor-β; Pre-miR, precursor miR; Pri-miR, primary miR; RAN-GTP, RAS-related nuclear protein-guanosine triphosphate; RISC, RNA-induced silencing complex; TRBP, transactivating response RNA-binding protein; TGF-β, transforming growth factor-β; UTR, untranslated region.
Current evidence points to translational repression occurring mostly at the initiation step of protein synthesis, whereby miRNA-RISC prevents the association of ribosomes with mRNA (56). By contrast, in postinitiation translational repression, ribosomes associate with the mRNAs but are unable to complete translation because of either early dissociation or physical obstruction by associated miRNA. Degradation of mRNA involves deadenylation and decapping by the 5′-3′ exonuclease XRN1 pathway (12, 31).
Although miRNAs bind most frequently to the 3′UTR region of mRNA, other reported sites of binding include the open reading frame (ORF) or the 5′UTR of the target mRNA (81). However, it is seen that binding of miRNA to the 3′UTR of mRNA leads to a greater degree of translational repression compared with binding at the ORF or 5′UTR of target mRNA (4). Interestingly, on rare occasions, miRNAs can also activate mRNA translation (127). Furthermore, a mature miRNA can reenter the nucleus to interact directly with transcription factors or the DNA itself, thereby modulating gene expression (9). Besides the canonical pathway, there are alternative miRNA biogenesis pathways that do not require the ribonuclease Drosha to generate pre-miRNA, such as the splicing-derived miRNAtrons (17, 94).
A single miRNA can bind to multiple mRNAs. Similarly, multiple miRNAs can target a single mRNA (4, 71). However, the role of a miRNA is decided not only by its target specificity but also by its tissue-expression levels. Specific cell types or tissues display a unique miRNA expression profile. Moreover, since one miRNA could regulate a set of mRNAs involved in a specific signaling pathway, the final outcome depends on the complex molecular interactions between the set of miRNAs and their target mRNAs.
Fibrosis Pathophysiology: Cellular and Epigenetic Mechanisms
Fibrogenesis during the physiology of wound healing and tissue repair is well coordinated and tightly regulated. In contrast, pathological fibrosis occurs when extracellular matrix (ECM) synthesis exceeds degradation due to deregulation of the wound healing or tissue repair process (59). Mesenchymal cells, such as fibroblasts and myofibroblasts, are the chief cells responsible for ECM production during fibrosis. As will be discussed later, under certain conditions even nonmesenchymal cells, such as epithelial cells or endothelial cells, can undergo transdifferentiation into ECM-producing fibroblasts or myofibroblasts via epithelial-mesenchymal transition (EMT) (42) or endothelial-mesenchymal transition (EndMT), respectively (138). Fibrosis develops through a set of complex interactions between mesenchymal cells, cytokines and growth factors, and the surrounding ECM. In addition, epigenetic (heritable changes in gene expression or cell phenotype without altering the DNA sequence) modifications within fibroblasts/myofibroblasts are also implicated in the pathobiology of fibrosis (135). As fibrosis progresses, there is increased ECM accumulation and atrophy of neighboring tissues, resulting in loss of normal function and finally organ failure.
Herein, we review the roles of cellular mediators and cellular plasticity and epigenetic regulation in fibrosis of GIT. Specific details of liver fibrosis and molecular mechanisms in fibrosis, such as oxidant stressors, growth factors and cytokines, and immune response, are discussed elsewhere (41, 67, 132).
Cellular players in fibrosis.
Myofibroblast, a phenotypic intermediate cell type between a fibroblast and a SMC, is considered the key cellular player in most fibrotic disorders (14, 40, 52). Myofibroblast arises from transdifferentiation of fibroblasts, fibrocytes, epithelial cells, and endothelial cells. In the gastrointestinal tract, a special group of myofibroblasts called intestinal subepithelial myofibroblasts (SEMFs) are located near the basement membrane of the small and large intestines. Myofibroblasts express their characteristic marker protein α-smooth muscle actin (α-SMA) and synthesize ECM components (mainly collagen) and tissue inhibitors of metalloproteases (14). Myofibroblasts are also a major source of secreted TGF-β during the fibrotic response. In early wound healing physiology, myofibroblasts help in contraction of granulation tissue. Later, myofibroblasts disappear by the process of apoptosis leading to wound resolution. However, prolonged presence of myofibroblasts in pathological conditions contributes to excessive deposition and contraction of ECM as seen in liver cirrhosis (8).
Fibroblasts, once activated, overproduce ECM components: the pathogenetic hallmark of scleroderma. Activated fibroblasts also secrete growth factors, cytokines, and chemokines and express receptors for growth factors, resulting in feed-forward amplification and sustenance of the fibrotic process. In addition, fibroblasts can convert into myofibroblasts (103).
Pericytes are mesenchymal cells located adjacent to endothelial cells of small blood vessels. In fibrotic disorders, activated pericytes undergo hyperplasia and express growth factor receptors (51, 88) in addition to transdifferentiating into fibroblasts and myofibroblasts.
Fibrocytes constitute circulating bone marrow-derived cells with fibroblast-like properties. Roderfeld et al. (112) hypothesized that bone marrow-derived circulating fibrocytes represent key mediators of liver fibrogenesis in mice.
SMCs are also implicated in the pathophysiology of GIT fibrosis in Crohn's disease and ulcerative colitis by producing collagen and matrix metalloproteinases (MMPs) in response to several inflammatory mediators, such as TGF-β and IL-1β. In Crohn's disease, SMC contraction, hyperplasia, and hypertrophy (64, 80) along with increased transmural ECM deposition lead to thickening of bowel wall, stricture formation, and, ultimately, intestinal obstruction. In scleroderma, vascular SMCs can contribute toward fibrosis by transdifferentiating into myofibroblasts.
Hepatic stellate cells (HSCs) are the main effector cells responsible for deposition of ECM during liver fibrosis (67). Stellate cells are also found in the GIT, where they contribute to fibrosis in Crohn's disease and ulcerative colitis (43, 109).
Although mesenchymal stem cells (MSCs) are believed to contribute to fibrosis, their exact role is still controversial (37, 74). A detailed discussion on the role of MSCs and fibrosis can be found elsewhere (37, 74, 77, 126).
Phenotypic plasticity of cells: EMT and EndMT.
Switching of cells from an epithelial to mesenchymal phenotype is important during embryogenesis (65). A similar process, EndMT, is also known to contribute toward fibrosis (111). The role of EMT/EndMT in intestinal fibrosis has been shown in animal models and Crohn's disease in human beings (36, 111).
Epigenetics and fibrosis.
Studies have shown an upregulation of genes in fibrotic myofibroblasts, consistent with activation of signaling pathways involved in fibrosis (63). In addition, collagen genes in scleroderma fibroblasts become autonomous and upregulated (30, 53). Mechanisms of epigenetic regulation in fibrotic disorders include DNA methylation, histone modification, and regulation of miRNAs.
Role of miRNAs in Fibrosis of Gastrointestinal Tract
microRNA databases, validation of databases, and challenges.
MicroRNAs impact gene expression primarily by targeting 3′UTR of the target messenger RNAs. Based on the experimental evidences, thousands of miRNA-target interactions (MTIs) are publically available at http://www.targetscan.org; http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/custom.html; http://www.microrna.org/microrna/home.do; and http://c1.accurascience.com/miRecords/.
To date, miRTarBase provides ∼3,000 MTIs, TarBase provides detailed information on each miRNA-gene interaction, and miRecords includes excellent quality database from experimentally proven miRNA targets obtained from manual literature. In addition, miRWalk database provides human, mouse, and rat validated binding sites on their target genes with different applications for target prediction. These databases are readily available and time saving (11, 24, 33, 58, 70, 73, 119, 136).
Although these databases are being constantly updated by the institutes/organizations, the perspective investigators should be vigilant about the miRs of their interest for the specific mRNA target and use further caution for nonspecific off-targets. It is further challenging considering the milieu of miR targets for mRNAs in different model systems (rodent, human, explant, the organ used, or specific phenotype switching). For example, miR-143 and miR-145 may be highly expressed with an important role in the airway SM contractile dysfunction, which may not be the case in the urinary bladder SM (28). Therefore, there is a dire need for the buildup and regular update of these databases in a model/cell/tissues/organ-specific validation manner.
miRNAs as potential biomarkers.
Being key regulators of cellular survival, proliferation, and SM differentiation (21) and of gain or loss of function under physiological and pathophysiological conditions, miRNAs not surprisingly may also serve as novel biomarkers for a number of diseases involving heart, cancer, kidney, autism, Parkinson's disease, depression, peritoneal fibrosis, autoimmunity, and arthritis (3, 25, 32, 35, 50, 54, 96, 116, 140). Recently, this list has grown to include the role of miRNAs as potential biomarkers in the pathophysiology of different SMs. For example, miR-204 is a major regulator of vascular SMCs (VSMCs) calcification, miR-221, and miRs143/145 are modulators of VSMCs phenotype and contractility (27, 29, 125). Similarly, a large number of miRNAs have been suggested to play important role as biomarkers and therapeutic targets in pulmonary arterial hypertension (16, 115).
Involvement of microRNAs in the remodeling of GI SM in the rodent model suggests a significant impact of microRNAs on phenotypic and functional aspects in the GIT (97, 99). Currently, because of the limited data in the GI SM fibrosis and dysfunction, in this review we have limited our discussion to the following miRNAs.
miRNA-29 family.
The human miRNA-29 family, or simply miRNA-29, includes miRNA-29a, miRNA-29b1, miRNA-29b2, and miRNA-29c, which are encoded by two separate gene clusters. However, all members have consensus seed-sequence that allows them to target the same mRNA. To date, miRNA-29 is the most well-studied regulator of ECM production among all miRNAs. Members of the miRNA-29 family target several genes involved in ECM production (such as collagen, elastin, and fibrillin) and are implicated in multiple fibrosing disorders of the human body, such as scleroderma and liver fibrosis (Table 1). The role of miRNA-29 has been more extensively researched in the hepatology literature compared with the GIT. However, there is mounting evidence to suggest that the pathophysiological role of miRNA-29 in fibrosis of liver and GIT is similar.
Table 1.
Selected miRNAs associated with fibrotic diseases involving the GIT
| Disease Associations | Deregulated Human miRNAs; Chromosome Location | Deregulation in Disease | Validated miR Targets | Affected Pathways and Final Effect in Disease Due to miR Deregulation | Sites Affected and Complications/Clinical Consequences of Fibrosis in Disease | References |
|---|---|---|---|---|---|---|
| Scleroderma (GIT involvement) | 29a and 29b1; chromosome 7 | Downregulated | Col 1A1, Col 1A2 |
PDGF-β and TGF-β, resulting in increased production of type I and type III collagen | Esophagus: Most common site of GIT involvement in SSc. Leads to GERD, stenosis, strictures, and Barrett's esophagus (due to chronic GERD) | 53, 60, 101, 120, 124, 139 |
| Stomach: Watermelon stomach (gastric antral vascular ectasia) and delayed gastric emptying | ||||||
| 29b2 and 29c; chromosome 1 | Col 3A1, ELN, FBN1 |
Small intestine: pseudo-obstruction, malabsorption syndrome (due to bacterial overgrowth), and PCI, constipation, and diarrhea | ||||
| Colon: Pseudo-obstruction, intestinal perforation, constipation, and diarrhea | ||||||
| 92a; chromosome 13 | Upregulated | Integrin αv | Integrin overexpression leading to activation of TGF-β. This results in increased ECM synthesis. | Rectum and anal canal: 2nd most common site of GIT involvement. Constipation (due to impaired relaxation of fibrotic internal anal sphincter). Fecal incontinence and rectal prolapse | ||
| 150; chromosome 19 | Downregulated | Integrinβ3 | Integrin overexpression leading to activation of TGF-β. This results in increased ECM synthesis. | |||
| 196a; chromosomes 12 and 17 | Downregulated | Col1A1, Col1A2 | TGF-β pathway leading to increased collagen production | |||
| 21; chromosome 17 | Upregulated | Smad7 | TGF-β pathway leading to increased collagen production | |||
| Crohn's disease (GIT fibrosis) | 200b; chromosome 1 | Downregulated | ZEB1 and ZEB2 | EMT resulting in increased ECM production | Esophagogastroduodenum: fistulae>strictures Jejunum: strictures > fistulae Ileum: fistulae>strictures Colon: fistulae>>>strictures Anoperineum: fistulae>>>strictures |
20, 69, 92 |
| 19a-3p; chromosome 13 | Downregulated | CTGF and TSP-1 | ||||
| 19b-3p; chromosome13 | Increased collagen production | |||||
| 29b; chromosomes 1 and 7 | Downregulated | Col 1A1, | PDGF-β and TGF-β, resulting in increased production of type I and type III collagen | |||
| Col 1A2, Col 3A1 |
||||||
ECM, extracellular matrix; ELN, elastin; FBN1, fibrillin; GIT, gastrointestinal tract; GERD, gastroesophageal reflux disease; miR, miRNA; PCI, pneumatosis cystoides intestinalis; SSc, scleroderma; ZSB, zinc finger E-box-binding homeobox.
Studies have shown that miRNA-29 is a direct posttranscriptional repressor of collagen gene expression in scleroderma, thereby acting as an antifibrotic miRNA (Fig. 1) (15, 49, 83, 133). Similarly, in the liver, miRNA-29b is antifibrotic by suppressing the genes involved in the activation of HSCs (118). Downregulation of the miRNA-29 family has been reported in patients with advanced hepatic fibrosis. In addition, miRNA-29b overexpression in HSCs resulted in decreased collagen synthesis (112). Studies have shown that TGF-β stimulation produces a reduction in miRNA-29 expression and derepression of collagen synthesis whereas stimulation with hepatocyte growth factor is associated with increased miRNA-29 levels and consequently decreased collagen synthesis (62). Recent studies implicate Toll-like receptors (TLRs) signaling in the pathogenesis of fibrosis in scleroderma by suppressing miRNA-29 expression (13). Although TLR4 polymorphisms are associated with Crohn's disease, their role in fibrosis via interaction with miRNA-29 is unknown (38). In a study by Nijhuis et al. (92), there was significant downregulation of the miR-29 family in the mucosa overlying strictures in Crohn's disease.
miRNA-200 family.
Similar to the miRNA-29 family, members of the human miRNA-200 family are transcribed from two distinct gene clusters. They include miRNA-200a, miRNA-200b, miRNA-200c, miRNA-141, and miRNA-429. On the basis of their seed sequence, the miRNA-200 family is categorized into two functional groups. The first group comprises miRNA-200b, miRNA-200c, and miRNA-429, and the second group includes miRNA-200a and miRNA-141. Because the functional groups have dissimilar seed sequence, they target different mRNAs. The miRNA-200 family participates in a signaling nexus involving the E-cadherin transcriptional repressors zinc finger E-box-binding homeobox (ZEB)1 and ZEB2, and TGF-β, which allows switching of cells between epithelial and mesenchymal phenotypes (45). Expression of the miRNA-200 family inhibits EMT via repressing ZEB1/ZEB2 and TGF-β. Conversely, downregulation of the miRNA-200 family promotes fibrosis by inducing EMT. Studies have established the role of the miRNA-200 family in EMT during fibrosis of several organs, such as lungs and kidneys (128, 135). Recently, deregulation of the miRNA-200 family has been implicated in the pathophysiology of EMT leading to GIT fibrosis (Table 1) (22).
Phenotypic Plasticity of GI Smooth Muscle
In health, GI motility is brought about by a series of coordinated contractions and relaxations of muscles of the gut that enable food to pass from mouth to anus. Except for a part of the esophagus and the external anal sphincter, the rest of the gut musculature is made up of SM. Moreover, not all the SMs of the gut are alike; there are significant differences in their phenotypic makeup. The SMs that form the sphincters (tonic SMs) have a unique contractile mechanism resulting in a higher basal tone compared with phasic SMs. It is well established that the major molecular determinant of basal tone in sphincteric GI SMs, such as internal anal sphincter (IAS), is a RhoA-associated kinase (RhoA)/Rho kinase (ROCK) pathway within the specialized SMCs (105–108).
SMs, unlike cardiac or skeletal muscles, are not terminally differentiated; therefore, they are capable of phenotypic modulation (plasticity). Normally, the GI SMCs exhibit a contractile phenotype characterized by expression of contractile proteins as well as ion channels and signaling molecules involved in contractile function (95). SMC phenotype switch is believed to contribute to the development of fibrosis in Crohn's disease (110). Recently, the role of gut microenvironment in SMC phenotype has received a great deal of attention. It is postulated that gut microbiota can induce phenotypic changes in GI SMCs, leading to GI dysmotility (117). TLRs mediate the interaction between the GIT and microbiota. Moreover, TLRs can regulate miRNAs or mRNAs in humans (82, 123). Changes to the phenotype can affect the “smooth” functioning of SMCs, and the resulting alteration in SM tone may synergize with fibrosis-induced changes to GIT, leading to discordant contractions that are the hallmark of GI dysmotility. Table 2 lists the diseases associated with altered tone due to GI SM phenotype switch.
Table 2.
Implications of altered tone due to smooth muscle phenotype switch in disorders of GIT
| Site of GIT Affected | Disorder |
|---|---|
| Esophageal body | • Diffuse esophageal spasm |
| • Achalasia | |
| • Hypotensive peristalsis | |
| • Hypertensive peristalsis (nutcracker esophagus) | |
| LES | • Achalasia |
| • Hypertensive LES, hypercontracting LES | |
| • Gastroesophageal reflux due to transient LES relaxation | |
| • Hypotensive LES, decreased reflex contraction of LES | |
| Stomach | • Gastric visceral hypersensitivity |
| • Gastric dysrhythmias | |
| • Gastroparesis | |
| • Pylorospasm | |
| Gall bladder | • Biliary dyskinesia |
| Small intestine | • Adynamic ileus |
| • CIPO | |
| Colon and rectosigmoid region | • Colonic inertia, Hirschsprung's disease |
| • CIPO or congenital human MMIHS | |
| Anal canal and internal anal sphincter | • Recurrent anal fissures • Hemorrhoids • Constipation • Rectoanal incontinence |
CIPO, Chronic intestinal pseudo-obstruction; LES, lower esophageal sphincter; MMIHS, megacystic-microcolon-intestinal hypoperistalsis syndrome.
Role of Selected miRNAs in GI Smooth Muscle Phenotype Switch
Phenotypic changes in SMCs occur because of major alteration in gene expression controlled chiefly by serum response factor (SRF). Genes determining SM phenotype are regulated by serum response elements (CArG boxes), to which the transcription factor SRF binds (86). In addition, SRF is regulated by transcriptional coactivators myocardin (MYOCD) and ELK1. RNA-binding protein for multiple spicing 2 (RBPMS2), a marker of SMC precursors, is progressively lost during SMC differentiation. Moreover, it has been found that expression of RBPMS2 in primary cultures of differentiated SMCs changes their phenotype from contractile to proliferative (93). Recently, studies have shown the importance of miRNAs in regulating SM differentiation and proliferation; some of these miRNAs are regulated by SRF via at least one CArG box in promoter regions of miRNAs. Studies have also identified genome-wide miRNAs in human and mouse intestinal SM (97).
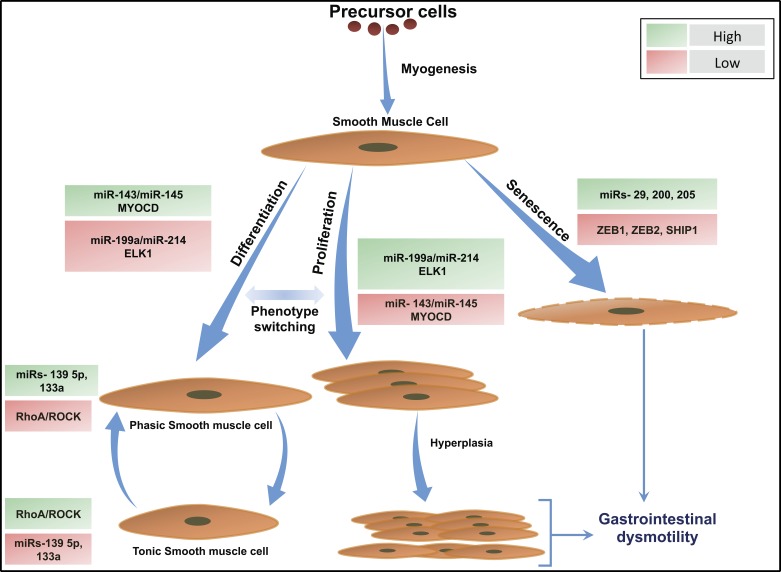
Of all the miRNAs identified in intestinal SM, two miRNA clusters, miRNA-143/miRNA-145 and miRNA-199a/miRNA-214, induced by SRF, regulate SMC differentiation and proliferation, respectively (Fig. 2). miRNA-143 and miRNA-145 are highly expressed in differentiated SMCs but repressed in proliferating SMCs, whereas miRNA-199a and miRNA-214 are highly expressed in proliferating SMCs but repressed in differentiated SMCs. Additionally, gain-of-function or loss-of-function studies with these miRNAs demonstrated that each miRNA could switch SMC phenotype between the proliferating and differentiated state (18, 86, 97).
Fig. 2.
Role of miRNAs in the regulation of GI smooth muscle phenotype switch. Schematics illustrate miRNA-mediated GI smooth muscle switching into proliferative, differentiated, and senescent phenotypes. miRNAs 143/145 control GI SMC phenotype via targeting MYOCD whereas miRNAs 199a/214 target ELK1 and regulate GI SMC phenotype. Upregulation of miRNAs 143/145 and corresponding downregulation of miRNAs 199a/214 lead to differentiation of GI SMCs into a contractile phenotype (97). RhoA/ROCK levels correlate inversely with tissue expression of miRNAs 139 5p/133a in the GI SMCs, resulting in a phasic or tonic GI SMC phenotype (23, 121). Downregulation of miRNAs 143/145 and corresponding upregulation of miRNAs 199a/214 results in GI SMC proliferation that can progress into SMC hypertrophy culminating in GI dysmotility. Senescence of GI SMCs is mediated by miRNAs 29, 200, and 205 via targeting ZEB1, ZEB2, and SHIP1. Green boxes indicate high; red boxes indicate low. GI, gastrointestinal; MYOCD, myocardin; ROCK, rho kinase; SMC, smooth muscle cell; ZEB, zinc finger E-box-binding homeobox.
Phenotype remodeling of SMs occur in various diseases of GIT; however, there is a paucity of literature regarding the exact underlying molecular mechanisms involving SRF signaling and regulation by miRNAs. Deletion of Dicer, the endonuclease responsible for production of mature miRNAs (10), caused developmental failure of the GI SMCs and downregulation of contractile genes and transcription factors (99). Recently, it was shown that loss of SRF-dependent miRNAs resulted apoptosis of GI SMCs (98). Thus miRNAs are essential for the development and establishment of the SMC phenotype, and modulating these molecules might have therapeutic implications in the GIT.
Aging and Oxidative Stress-Related Fibrosis and SM Dysfunction
A prototype of age-related change in the GIT is loss of tone and fibroelastic properties of IAS leading to RI (46, 48). Because of the paucity of literature on age-related changes in the IAS, precise pathogenic mechanisms of aging-associated decrease in the IAS tone are unknown (55, 90, 122). However, data from other SMs, such as vascular SM, have shown an aging-associated increase in oxidative stress and decrease in nitric oxide production (113), which can lead to changes in cellular phenotype and intracellular signaling.
Our earlier studies have shown an oxidative stress-induced decrease in the IAS tone via RhoA/ROCK downregulation (122). Paradoxically, a number of studies have revealed that RhoA/ROCK may be initiators of oxidative stress during disease states based on the findings of ROCK inhibitor-induced halt in the progression of renal fibrosis (89). Recently, it was shown that miRNAs 19b, 20a, and 106a were differentially regulated in aged human cells, such as endothelial cells, renal epithelial cells, and skin fibroblasts (46). In another study, upregulation of miRNA-22 was found to induce senescence in human fibroblasts (134). More recently, Mahavadi et al. (78) have shown that increases in the oxidative stress in gastric SMCs of diabetic rats lead to upregulation of RhoA expression and increase in SMC contraction via decrease in expression of miRNA-133a; however, it is not known whether diabetic status of rats had contributed to their observed findings. Nevertheless, the role of miRNAs in RhoA/ROCK-mediated oxidative stress and/or fibrosis in the GIT awaits further investigation.
Diagnostic and Therapeutic Opportunities
miRNAs as novel diagnostic biomarkers.
In the year 2008, the discovery of miRNAs' presence extracellularly in the serum highlighted the exciting potential of their use as noninvasive disease biomarkers (19). As of today, miRNAs have been found in most body fluids, such as plasma, serum, saliva, tears, breast milk, and urine (130). miRNAs exhibit striking extracellular stability that makes them exciting candidates for use as biomarkers. Although miRNA profiles have been well studied in the cardiovascular, cancer, and neurological fields, there is limited data in the GI literature.
Compared with the GIT, the role of miRNAs in liver fibrosis has been more extensively studied. However, caution should be exercised when extrapolating the roles of miRNAs in liver fibrosis to GIT because of the possibility of tissue-specific differences in the roles of miRNAs. It has been found that serum levels of the miR-29 family were lower in patients with strictures in Crohn's disease compared with those without strictures, suggesting the potential of using miRNAs as biomarkers (92). Similarly, circulating miRNAs in scleroderma have a diagnostic and prognostic value (131) because their expression can be upregulated by using synthetic miRNA mimics. Recent studies have also reported age- and gender-specific influences on miRNA patterns in blood (61, 85). Moreover, the jury is still out on whether miRNAs should be obtained from serum or plasma (84, 129).
miRNA-based therapeutic strategies.
Therapeutic strategies involve the use of miRNA mimics or miRNA antagonists. So far, miRNA-based therapeutic trials in animal models have produced encouraging results with minimal toxicity. Although the miRNA-based therapeutic strategy is promising, certain challenges and issues need to be kept in mind, such as that miR mimics can have off-target effects and miRNA-based drug delivery to the target cells/tissues can pose difficulties because of the presence of RNAses in the blood (100). Novel methods of delivery of miRNAs include use of lenti- or adenovirus vectors, although safety concerns exist. Exosomal delivery of miRNA also appears promising (1).
In a study by Cordes et al. (26), introduction of miRNA-145 via lentiviral-mediated gene transfer into the ligated carotid arteries of mouse led to suppression of VSMC proliferation and promoted differentiation. Conversely, miRNA antagonists, also called antagomirs, can be useful for miRNA silencing. Liu et al. (76) used antagomirs to knock down miRNA-221 and -222 in cultured VSMCs. Recently, Makino et al. (79) described a novel method of miRNA let-7a overexpression in bleomycin-induced fibrosis in mice by intraperitoneal injection of miRNA let-7a. Although miRNA-based therapeutic targeting has yielded promising results in pulmonary and cardiovascular literature, much of the data on the role of miRNAs in GIT is still preliminary. More studies in GIT are warranted.
Concluding Remarks
Fibrosis and SM dysfunction correlate with the morbidity and mortality associated with several diseases of the gastrointestinal tract. However, there is an unmet need of novel markers that reflect the true extent of fibrosis-related gastrointestinal SM dysfunction. In such a scenario, miRNAs have emerged as promising biomarkers for diagnosis and assessment of disease activity and severity. In vitro and in vivo animal models of miRNA-based therapeutic success have given us a ray of hope that miRNA-based therapy for human fibrotic disorders and GI dysmotility could be a reality in the not-so-distant future.
GRANTS
The work was supported by National Institutes of Diabetes and Digestive and Kidney Diseases Grant RO1DK035385 and an institutional grant from Thomas Jefferson University.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
AUTHOR CONTRIBUTIONS
C.V.K., J.S., and S.R. conception and design of research; C.V.K. and J.S. prepared figures; C.V.K., J.S., and S.R. drafted manuscript; C.V.K., J.S., C.T., and S.R. edited and revised manuscript; C.V.K., J.S., C.T., and S.R. approved final version of manuscript.
REFERENCES
- 1.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol 29: 341–345, 2011. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. The functions of animal microRNAs. Nature 431: 350–355, 2004. [DOI] [PubMed] [Google Scholar]
- 3.Anitha A, Thanseem I. microRNA and autism. Adv Exp Med Biol 888: 71–83, 2015. [DOI] [PubMed] [Google Scholar]
- 4.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature 455: 64–71, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagga S, Bracht J, Hunter S, Massirier K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122: 553–563, 2005. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297, 1997. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bataller R, Brenner DA. Liver fibrosis. J Clin Invest 115: 209–218, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Elinat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet 37: 766–770, 2005. [DOI] [PubMed] [Google Scholar]
- 10.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366, 2001. [DOI] [PubMed] [Google Scholar]
- 11.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res 36: D149–D153, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bethune J, Artus-Revel CG, Filipowicz W. Kinetic analysis reveals successive steps leading to miRNA-mediated silencing in mammalian cells. EMBO Rep 13: 716–723, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhattacharyya S, Kelly K, Melichian DS, Tamaki Z, Fang F, Su Y, Feng G, Pope RM, Budinger GR, Mutlu GM, Lafyatis R, Radstake T, Feghali-Bostwick C, Varga J. Toll-like receptor 4 signaling augments transforming growth factor-β responses: a novel mechanism for maintaining and amplifying fibrosis in scleroderma. Am J Pathol 182: 192–205, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhattacharyya S, Varga J. Emerging roles of innate immune signaling and toll-like receptors in fibrosis and systemic sclerosis. Curr Rheumatol Rep 17: 3–9, 2015. [DOI] [PubMed] [Google Scholar]
- 15.Bian EB, Li J, Zhao B. miR-29, a potential therapeutic target for liver fibrosis. Gene 544: 259–260, 2014. [DOI] [PubMed] [Google Scholar]
- 16.Bienertova-Vasku J, Novak J, Vasku A. MicroRNAs in pulmonary arterial hypertension: pathogenesis, diagnosis and treatment. J Am Soc Hypertens 9: 221–234, 2015. [DOI] [PubMed] [Google Scholar]
- 17.Bortolamiol-Becet D, Hu F, Jee D, Wen J, Okamura K, Lin CJ, Ameras SL, Lai EC. Selective suppression of the splicing-mediated microRNA pathway by the terminal uridyltransferase tailor. Mol Cell 59: 217–228, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J, Yin H, Jiang Y, Radhakrishnan SK, Huang ZP, Li J, Shi Z, Kilsodonk EPC, Gui Y, Wang DZ, Zheng XL. Induction of microRNA-1 by myocardin in smooth muscle cells inhibits cell proliferation. Arterioscler Thromb Vasc Biol 31: 368–375, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X, Li Q, Li X, Wang W, Zhang Y, Wang J, Jiang X, Xiang Y, Xu C, Zheng P, Zhang J, Li R, Zhang H, Shang X, Gong T, Ning G, Wang J, Zen K, Zhang J, Zhang CY. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 18: 997–1006, 2008. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y, Ge W, Xu L, Qu C, Zhu M, Zhang W, Xiao Y. miR-200b is involved in intestinal fibrosis of Crohn's disease. Int J Mol Med 29: 601–606, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, Gelfond J, McManus LM, Shireman PK. Temporal microRNA expression during in vitro myogenic progenitor cell proliferation and differentiation: regulation of proliferation by miR-682. Physiol Genomics 43: 621–630, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheny Y, Ge W, Xu L, Qu C, Zhu M, Zhang W, Xiao Y. miR-200b is involved in intestinal fibrosis of Crohn's disease. Int J Mol Med 29: 601–616, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiba Y, Misawa M. MicroRNAs and their therapeutic potential for human disease: MiR-133a and bronchial smooth muscle hyperresponsiveness in asthma. J Pharm Sci 114: 264–268, 2010. [DOI] [PubMed] [Google Scholar]
- 24.Chiu HS, Llobet-Navas D, Yang X, Chung WJ, Ambesi-Impiombato A, Iyer A, Kim HR, Seviour EG, Luo Z, Sehgal V, Moss T, Lu Y, Ram P, Silva J, Mills GB, Califano A, Sumazin P. Cupid: simultaneous reconstruction of microRNA-target and ceRNA networks. Genome Res 25: 257–267, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chung AC. microRNAs in diabetic kidney disease. Adv Exp Med Biol 888: 253–269, 2015. [DOI] [PubMed] [Google Scholar]
- 26.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Sirvastava D. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature 460: 705–710, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cui RR, Li SJ, Liu LJ, Yi L, Liang QH, Zhu X, Liu GY, Liu Y, Wu SS, Liao XB, Yuan LQ, Mao DA, Liao EY. MicroRNA-204 regulates vascular smooth muscle cell calcification in vitro and in vivo. Cardiovasc Res 96: 320–329, 2012. [DOI] [PubMed] [Google Scholar]
- 28.Dahan D, Ekman M, Larsson-Callerfelt AK, Turczynska K, Boettger T, Braun T, Sward K, Albinsson S. Induction of angiotensin-converting enzyme after miR-143/145 deletion is critical for impaired smooth muscle contractility. Am J Physiol Cell Physiol 307: C1093–C1101, 2014. [DOI] [PubMed] [Google Scholar]
- 29.Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Induction of microRNA-221 by platelet-derived growth factor signaling is critical for modulation of vascular smooth muscle phenotype. J Biol Chem 284: 3728–3738, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Derk CT, Jimenez SA. Systemic sclerosis: current views of its pathogenesis. Autoimmun Rev 2: 181–191, 2009. [DOI] [PubMed] [Google Scholar]
- 31.Dijuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science 336: 237–240, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dong H, Wang C, Lu S, Yu C, Huang L, Feng W, Xu H, Chen X, Zen K, Yan Q, Liu W, Zhang C, Zhang CY. A panel of four decreased serum microRNAs as a novel biomarker for early Parkinson's disease. Biomarkers Dec 3: 1–9, 2015. [DOI] [PubMed] [Google Scholar]
- 33.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk—database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform 44: 839–847, 2011. [DOI] [PubMed] [Google Scholar]
- 34.Eckes B, Zweers MC, Zhang ZG, Hallinger R, Mauch C, Aumailley M, Krieg T. Mechanical tension and integrin alpha 2 beta 1 regulate fibroblast functions. J Investig Dermatol Symp Proc 11: 66–72, 2015. [DOI] [PubMed] [Google Scholar]
- 35.Fan L, Qi H, Teng J, Su B, Chen H, Wang C, Xia Q. Identification of serum miRNAs by nano-quantum dots microarray as diagnostic biomarkers for early detection of non-small cell lung cancer. Tumour Biol 2015. December 22 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 36.Flier SN, Tanjore H, Kokkotou EG, Sugimoto H, Zeisberg M, Kalluri R. Identification of epithelial to mesenchymal transition as a novel source of fibroblasts in intestinal fibrosis. J Biol Chem 285: 20202–20212, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Forbes SJ, Russo FP, Rey V, Burra P, Rugge M, Wright NA, Alison MR. A significant proportion of myofibroblasts are of bone marrow origin in human liver fibrosis. Gastroenterology 126: 955–963, 2004. [DOI] [PubMed] [Google Scholar]
- 38.Franchimont D, Vermeire S, Housni H, Pierik M, Van Steen K. Deficient host-bacteria interactions in inflammatory bowel disease? The toll-like receptor (TLR)-4 Asp299gly polymorphism is associated with Crohn's disease and ulcerative colitis. Gut 53: 987–992, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Friedman LM, Avraham KB. MicroRNAs and epigenetic regulation in the mammalian inner ear: implications for deafness. Mamm Genome 20: 581–603, 2009. [DOI] [PubMed] [Google Scholar]
- 40.Gabbiani G. The myofibroblast in wound healing and fibrocontractive diseases. J Pathol 200: 500–503, 2015. [DOI] [PubMed] [Google Scholar]
- 41.Gabrielli A, Avvedimento EV, Krieg T. Mechanisms of disease: Scleroderma. N Engl J Med 360: 1989–2003, 2009. [DOI] [PubMed] [Google Scholar]
- 42.Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal 7: 1–17, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gordon IO, Agrawal N, Goldblum JR, Fiocchi C, Rieder F. Fibrosis in ulcerative colitis: mechanisms, features, and consequences of a neglected problem. Inflamm Bowel Dis 20: 2198–2206, 2014. [DOI] [PubMed] [Google Scholar]
- 44.Green D, Dalmay T, Chapman T. Microguards and micromessengers of the genome. Heredity 116: 125–134, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10: 593–601, 2008. [DOI] [PubMed] [Google Scholar]
- 46.Hackl M, Brunner S, Fortschegger K, Schreiner C, Micutkova L, Muck C. miR-17, miR-19b, miR-20a, and miR-106a are down-regulated in human aging. Aging Cell 9: 291–296, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hammel P, Couvelard A, O'Toole D, Ratouis A, Sauvanet A, Flejou JF, Degott C, Belghiti J, Bernades P, Valla D, Ruszniewski P, Levy P. Regression of liver fibrosis after biliary drainage in patients with chronic pancreatitis and stenosis of the common bile duct. N Engl J Med 344: 418–423, 2001. [DOI] [PubMed] [Google Scholar]
- 48.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev 18: 3016–3027, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He Y, Huang C, Lin X, Li J. MicroRNA-29 family, a crucial therapeutic target for fibrosis disease. Biochimie 95: 1355–1359, 2013. [DOI] [PubMed] [Google Scholar]
- 50.Heegaard NH, Carlsen AL, Skovgaard K, Heegaard PM. Circulating extracellular microRNA in systemic autoimmunity. EXS 106: 171–195, 2015. [DOI] [PubMed] [Google Scholar]
- 51.Helmbold P, Fiedler E, Fischer M, Marsch WC. Hyperplasia of dermal microvascular pericytes in scleroderma. J Cutan Pathol 31: 431–440, 2004. [DOI] [PubMed] [Google Scholar]
- 52.Hinz B, Phan SH, Thannickal VJ, Galli A, Bochaton-Piallat ML, Gabbiani G. The myofibroblast: one function, multiple origins. Am J Pathol 170: 1807–1816, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Honda N, Jinnin M, Kajihara I, Makino T, Makino K, Masuguchi S, Fukushima S, Okamoto Y, Hasegawa M, Fujimoto M, Ihn H. TGF-beta-mediated downregulation of microRNA-196a contributes to the constitutive upregulated type I collagen expression in scleroderma dermal fibroblasts. J Immunol 188: 3323–3331, 2012. [DOI] [PubMed] [Google Scholar]
- 54.Hu B, Song JT, Qu HY, Bi CL, Huang XZ, Liu XX, Zhang M. Mechanical stretch suppresses microRNA-145 expression by activating extracellular signal-regulated kinase 1/2 and upregulating angiotensin-converting enzyme to alter vascular smooth muscle cell phenotype. PLoS One 9: e96338, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huebner M, Margulies RU, Fenner DE, Ashton-Miller JA, Bitar KN, DeLancey JO. Age effects on internal anal sphincter thickness and diameter in nulliparous females. Dis Colon Rectum 50: 1405–1411, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 12: 99–110, 2011. [DOI] [PubMed] [Google Scholar]
- 57.Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science 293: 834–838, 2001. [DOI] [PubMed] [Google Scholar]
- 58.Lee YJ, Kim V, Muth DC, Witwer KW. Validated MicroRNA target databases: an evaluation. Drug Dev Res 76: 389–396, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karsdal MJ, Manon-Jensen T, Genovese F, Kristensen JH. Novel insights into the function and dynamics of extracellular matrix in liver fibrosis. Am J Physiol Gastrointest Liver Physiol 308: G807–G830, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kawashita Y, Jinnin M, Makino T, Kajihara I, Makino K, Honda N, Masuguchi S, Fukushima S, Inoue Y, Ihn H. Circulating miR-29a levels in patients with scleroderma spectrum disorder. J Dermatol Sci 61: 67–69, 2011. [DOI] [PubMed] [Google Scholar]
- 61.Keller A, Meese E. Can circulating miRNAs live up to the promise of being minimal invasive biomarkers in clinical settings? Wiley Interdiscip Rev RNA. 2015 Dec 15. doi: 10.1002/wrna.1320 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 62.Kwiecinski M, Noetel A, Elfimova N, Trebicka J, Schievenbusch S, Strack I, Molnar L, von Brandenstein M, Tox U, Nischt R, Coutelle O, Dienes HP, Odenthal M. Hepatocyte growth factor (HGF) inhibits collagen I and IV synthesis in hepatic stellate cells by miRNA-29 induction. PLoS One 6: e24568, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Larsson O, Diebold D, Fan D, Peterson M, Nho RS, Bitterman PB, Henke CA. Fibrotic myofibroblasts manifest genome-wide derangements of translational control. PLoS One 3: e3220, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lee EY, Stenson WF, DeSchryver-Kecskemeti K. Thickening of muscularis mucosae in Crohn's disease. Mod Pathol 4: 87–90, 1991. [PubMed] [Google Scholar]
- 65.Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: new insights in signaling, development, and disease. J Cell Biol 172: 973–981, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843–854, 1993. [DOI] [PubMed] [Google Scholar]
- 67.Lee UE, Friedman SL. Mechanisms of hepatic fibrogenesis. Best Pract Res Clin Gastroenterol 25: 195–206, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature 425: 415–419, 2003. [DOI] [PubMed] [Google Scholar]
- 69.Lewis A, Mehta S, Hanna LN, Rogalski LA, Jeffery R, Nijhuis A, Kumagai T, Biancheri P, Bundy JG, Bishop CL, Feakins R, Di SA, Lee JC, Lindsay JO, Silver A. Low serum levels of MicroRNA-19 are associated with a stricturing Crohn's disease phenotype. Inflamm Bowel Dis 21: 1926–1934, 2015. [DOI] [PubMed] [Google Scholar]
- 70.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20, 2005. [DOI] [PubMed] [Google Scholar]
- 71.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell 115: 787–798, 2003. [DOI] [PubMed] [Google Scholar]
- 72.Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics 8: 166, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu S, Li JH, Wu J, Zhou KR, Zhou H, Yang JH, Qu LH. StarScan: a web server for scanning small RNA targets from degradome sequencing data. Nucleic Acids Res 43: W480–W486, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu W, Song F, Ren L, Guo W, Wang T, Feng Y, Tang L, Li K. The multiple functional roles of mesenchymal stem cells in participating in treating liver fibrosis. J Cell Mol Med 19: 511–520, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu W, Zhang Q, Li S, Li L, Ding Z, Qian Q, Fan L, Jiang C. The relationship between colonic macrophages and microRNA-128 in the pathogenesis of slow transit constipation. Dig Dis Sci 60: 2304–2315, 2015. [DOI] [PubMed] [Google Scholar]
- 76.Liu X, Cheng Y, Zhang S, Lin Y, Yang J, Zhang C. A necessary role of miR-221 and miR-222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ Res 104: 476–487, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu Y, Yang X, Jing Y, Zhang S, Zong C, Jiang J, Sun K, Li R, Gao L, Zhao X, Wu D, Shi Y, Han Z, Wei L. Contribution and mobilization of mesenchymal stem cells in a mouse model of carbon tetrachloride-induced liver fibrosis. Sci Rep 5: 17762, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mahavadi S, Sriwai W, Kumar DP, Zhou R, Grider JR, Murthy KS. Down-regulation of microRNA-133a due to oxidative stress mediates up-regulation of RHOA expression and increase in rho kinase activity and gastric muscle contraction in diabetes. Gastroenterology 142: S-105, 2012. [Google Scholar]
- 79.Makino K, Jinnin M, Hirano A, Yamane K, Eto M, Kusano T, Honda N, Kajihara I, Makino T, Sakai K, Masuguchi S, Fukushima S, Ihn H. The downregulation of microRNA let-7a contributes to the excessive expression of type I collagen in systemic and localized scleroderma. J Immunol 190: 3905–3915, 2013. [DOI] [PubMed] [Google Scholar]
- 80.Manetti M, Rosa I, Messerini L, Ibba-Manneschi L. Telocytes are reduced during fibrotic remodelling of the colonic wall in ulcerative colitis. J Cell Mol Med 19: 62–73, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martinez-Sanchez A, Murphy CL. MicroRNA target identification—experimental approaches. Biology (Basel) 2: 189–205, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Masotti A. Interplays between gut microbiota and gene expression regulation by miRNAs. Front Cell Infect Microbiol 2: 137, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Maurer B, Stanczyk J, Jungel A, Akhmetshina A, Trenkmann M, Brock M, Kowal-Bielecka O, Gay RE, Michel BA, Distler JH, Gay S, Distler O. MicroRNA-29, a key regulator of collagen expression in systemic sclerosis. Arthritis Rheum 62: 1733–1743, 2010. [DOI] [PubMed] [Google Scholar]
- 84.McDonald JS, Milosevic D, Reddi HV, Grebe SK, Algeciras-Schimnich A. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem 57: 833–840, 2011. [DOI] [PubMed] [Google Scholar]
- 85.Meder B, Backes C, Haas J, Leidinger P, Stahler C, Grossmann T, Vogel B, Frese K, Giannitsis E, Katus HA, Meese E, Keller A. Influence of the confounding factors age and sex on microRNA profiles from peripheral blood. Clin Chem 60: 1200–1208, 2014. [DOI] [PubMed] [Google Scholar]
- 86.Miano JM, Long X, Fujiwara K. Serum response factor: master regulator of the actin cytoskeleton and contractile apparatus. Am J Physiol Cell Physiol 292: C70–C81, 2007. [DOI] [PubMed] [Google Scholar]
- 87.Moser JJ, Fritzler MJ. Cytoplasmic ribonucleoprotein (RNP) bodies and their relationship to GW/P bodies. Int J Biochem Cell Biol 42: 828–843, 2010. [DOI] [PubMed] [Google Scholar]
- 88.Murthy KS. Signaling for contraction and relaxation in smooth muscle of the gut. Annu Rev Physiol 68: 345–374, 2006. [DOI] [PubMed] [Google Scholar]
- 89.Nagatoya K, Moriyama T, Kawada N, Takeji M, Oseto S, Murozono T, Ando A, Imai E, Hori M. Y-27632 prevents tubulointerstitial fibrosis in mouse kidneys with unilateral ureteral obstruction. Kidney Int 61: 1684–1695, 2002. [DOI] [PubMed] [Google Scholar]
- 90.Nakano R, Paran TS, Rolle U, Puri P. Age-related changes in the neuromuscular development of the internal anal sphincter. J Pediatr Surg 43: 1106–1110, 2008. [DOI] [PubMed] [Google Scholar]
- 91.Nezami BG, Mwangi SM, Lee JE, Jeppsson S, Anitha M, Yarandi SS, Farris AB III, Srinivasan S. MicroRNA 375 mediates palmitate-induced enteric neuronal damage and high-fat diet-induced delayed intestinal transit in mice. Gastroenterology 146: 473–483, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nijhuis A, Biancheri P, Lewis A, Bishop CL, Giuffrida P, Chan C, Feakins R, Poulsom R, Di Sabatino A, Corazza GR, MacDonald TT, Lindsay JO, Silver AR. In Crohn's disease fibrosis-reduced expression of the miR-29 family enhances collagen expression in intestinal fibroblasts. Clin Sci (Lond) 127: 341–350, 2014. [DOI] [PubMed] [Google Scholar]
- 93.Notarnicola C, Rouleau C, Le Guen L, Virsolvy A, Richard S, Faure S, De Santa BP. The RNA-binding protein RBPMS2 regulates development of gastrointestinal smooth muscle. Gastroenterology 143: 687–697, 2012. [DOI] [PubMed] [Google Scholar]
- 94.Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130: 89–100, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev 84: 767–801, 2004. [DOI] [PubMed] [Google Scholar]
- 96.Paek SY, Han L, Weiland M, Lu CJ, McKinnon K, Zhou L, Lim HW, Elder JT, Mi QS. Emerging biomarkers in psoriatic arthritis. IUBMB Life 67: 923–927, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Park C, Hennig GW, Sanders KM, Cho JH, Hatton WJ, Redelman D, Park JK, Ward SM, Miano JM, Yan W, Ro S. Serum response factor-dependent MicroRNAs regulate gastrointestinal smooth muscle cell phenotypes. Gastroenterology 141: 164–175, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Park C, Lee MY, Slivano OJ, Park PJ, Ha S, Berent RM, Fuchs R, Collins NC, Yu TJ, Syn H, Park JK, Horiguchi K, Miano JM, Sanders KM, Ro S. Loss of serum response factor induces microRNA-mediated apoptosis in intestinal smooth muscle cells. Cell Death Dis 6: e2011, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Park C, Yan W, Ward SM, Hwang SJ, Wu Q, Hatton WJ, Park JK, Sanders KM, Ro S. MicroRNAs dynamically remodel gastrointestinal smooth muscle cells. PLoS One 6: e18628, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Pecot CV, Calin GA, Coleman RL, Lopez-Berestein G, Sood AK. RNA interference in the clinic: challenges and future directions. Nat Rev Cancer 11: 59–67, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Peng WJ, Tao JH, Mei B, Chen B, Li BZ, Yang GJ, Zhang Q, Yao H, Wang BX, He Q, Wang J. MicroRNA-29: a potential therapeutic target for systemic sclerosis. Expert Opin Ther Targets 16: 875–879, 2012. [DOI] [PubMed] [Google Scholar]
- 102.Pillai RS. MicroRNA function: multiple mechanisms for a tiny RNA? RNA 11: 1753–1761, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rajkumar VS, Howell K, Csiszar K, Denton CP, Black CM, Abraham DJ. Shared expression of phenotypic markers in systemic sclerosis indicates a convergence of pericytes and fibroblasts to a myofibroblast lineage in fibrosis. Arthritis Res Ther 7: R1113–R1123, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123: 621–629, 2005. [DOI] [PubMed] [Google Scholar]
- 105.Rattan S, Benjamin P, Maxwell PJ 4th. RhoA/ROCK-kinase: pathophysiologic and therapeutic implications in gastrointestinal smooth muscle tone and relaxation. Gastroenterology 138: 13–18, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rattan S, De Godoy MAF, Patel CA. Rho kinase as a novel molecular therapeutic target for hypertensive internal anal sphincter. Gastroenterology 131: 108–116, 2006. [DOI] [PubMed] [Google Scholar]
- 107.Rattan S, Singh J. RhoA/ROCK pathway is the major molecular determinant of basal tone in intact human internal anal sphincter. Am J Physiol Gastrointest Liver Physiol 302: G664–G675, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rattan S, Singh J, Kumar S, Phillips B. Nature of extracellular signal that triggers RhoA/ROCK activation for the basal internal anal sphincter tone in humans. Am J Physiol Gastrointest Liver Physiol 308: G924–G933, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Rieder F, Fiocchi C. Intestinal fibrosis in inflammatory bowel disease: progress in basic and clinical science. Curr Opin Gastroenterol 24: 462–468, 2008. [DOI] [PubMed] [Google Scholar]
- 110.Rieder F, Fiocchi C. Intestinal fibrosis in IBD a dynamic, multifactorial process. Nat Rev Gastroenterol Hepatol 6: 228–235, 2009. [DOI] [PubMed] [Google Scholar]
- 111.Rieder F, Kessler SP, West GA, Bhilocha S, de la Motte C, Sadler TM, Gopalan B, Stylianou E, Fiocchi C. Inflammation-induced endothelial-to-mesenchymal transition: a novel mechanism of intestinal fibrosis. Am J Pathol 179: 2660–2673, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Roderfeld M, Rath T, Voswinckel R, Dierkes C, Dietrich H, Zahner D, Graf J, Roeb E. Bone marrow transplantation demonstrates medullar origin of CD34+ fibrocytes and ameliorates hepatic fibrosis in Abcb4−/− mice. Hepatology 51: 267–276, 2010. [DOI] [PubMed] [Google Scholar]
- 113.Rubio-Ruiz ME, Perez-Torres I, Soto ME, Pastelin G, Guarner-Lans V. Aging in blood vessels. Medicinal agents for systemic arterial hypertension in the elderly. Ageing Res Rev 18: 132–147, 2014. [DOI] [PubMed] [Google Scholar]
- 114.Sanders KM, Koh SD, Ro S, Ward SM. Regulation of gastrointestinal motility: insights from smooth muscle biology. Nat Rev Gastroenterol Hepatol 9: 633–645, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sarrion I, Milian L, Juan G, Ramon M, Furest I, Carda C, Cortijo GJ, Mata RM. Role of circulating miRNAs as biomarkers in idiopathic pulmonary arterial hypertension: possible relevance of miR-23a. Oxidat Med Cell Longev 2015: 792846, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schulte C, Molz S, Appelbaum S, Karakas M, Ojeda F, Lau DM, Hartmann T, Lackner KJ, Westermann D, Schnabel RB, Blankenberg S, Zeller T. miRNA-197 and miRNA-223 predict cardiovascular death in a cohort of patients with symptomatic coronary artery disease. PLoS One 10: e0145930, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Scirocco A, Matarrese P, Carabotti M, Ascione B, Malorni W, Severi C. Cellular and molecular mechanisms of phenotypic switch in gastrointestinal smooth muscle. J Cell Physiol 231: 295–302, 2016. [DOI] [PubMed] [Google Scholar]
- 118.Sekiya Y, Ogawa T, Yoshizato K, Ikeda K, Kawada N. Suppression of hepatic stellate cell activation by microRNA-29b. Biochem Biophys Res Commun 412: 74–79, 2011. [DOI] [PubMed] [Google Scholar]
- 119.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: a comprehensive database of experimentally supported animal microRNA targets. RNA 12: 192–197, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sing T, Jinnin M, Yamane K, Honda N, Makino K, Kajihara I, Makino T, Sakai K, Masuguchi S, Fukushima S, Ihn H. microRNA-92a expression in the sera and dermal fibroblasts increases in patients with scleroderma. Rheumatology (Oxford) 51: 1550–1556, 2012. [DOI] [PubMed] [Google Scholar]
- 121.Singh J, Addya S, Fortina P, Rattan S. Role of microRNA-139-5p (miR-139-5p) in the myogenic basal tone of internal anal sphincter (IAS) vs. the phasic rectal smooth muscle (RSM): studies in purified smooth muscle cells (SMCs). Gastroenterology 144: S-364, 2013. [Google Scholar]
- 122.Singh J, Kumar S, Krishna CV, Rattan S. Aging-associated oxidative stress leads to decrease in IAS tone via RhoA/ROCK downregulation. Am J Physiol Gastrointest Liver Physiol 306: G983–G991, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Singh N, Shirdel EA, Waldron L, Zhang RH, Jurisica I, Comelli EM. The murine caecal microRNA signature depends on the presence of the endogenous microbiota. Int J Biol Sci 8: 171–186, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Steen SO, Iversen LV, Carlsen AL, Burton M, Nielsen CT, Jacobsen S, Heegaard NH. The circulating cell-free microRNA profile in systemic sclerosis is distinct from both healthy controls and systemic lupus erythematosus. J Rheumatol 42: 214–221, 2015. [DOI] [PubMed] [Google Scholar]
- 125.Thi Hien T, Turczynska KM, Dahan D, Ekman M, Grossi M, Sjogren J, Nilsson J, Braun T, Boettger T, Garcia Vaz E, Stenkula K, Sward K, Gomez MF, Albinsson S. Elevated glucose levels promote contractile and cytoskeletal gene expression in vascular smooth muscle via Rho/protein kinase C and actin polymerization. J Biol Chem. 2015 Dec 18. pii: jbc.M115.654384. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Usunier B, Benderitter M, Tamarat R, Chapel A. Management of fibrosis: the mesenchymal stromal cells breakthrough. Stem Cells Int 2014: 340257, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science 318: 1931–1934, 2007. [DOI] [PubMed] [Google Scholar]
- 128.Wang B, Koh P, Winbanks C, Coughlan MT, McClelland A, Watson A, Jandeleit-Dahm K, Burns WC, Thomas MC, Cooper ME, Kantharidis P. miR-200a prevents renal fibrogenesis through repression of TGF-beta2 expression. Diabetes 60: 280–287, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang GK, Zhu JQ, Zhang JT, Li Q, Li Y, He J, Qin YW, Jing Q. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J 31: 659–666, 2010. [DOI] [PubMed] [Google Scholar]
- 130.Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K. The microRNA spectrum in 12 body fluids. Clin Chem 56: 1733–1741, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wuttge DM, Carlsen AL, Teku G, Steen SO, Wildt M, Vihinen M, Hesselstrand R, Heegaard NH. Specific autoantibody profiles and disease subgroups correlate with circulating micro-RNA in systemic sclerosis. Rheumatology (Oxford) 54: 2100–2107, 2015. [DOI] [PubMed] [Google Scholar]
- 132.Wynn TA. Cellular and molecular mechanisms of fibrosis. J Pathol 214: 199–210, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Xiao J, Meng XM, Huang XR, Chung AC, Feng YL, Hui DS, Yu CM, Sung JJ, Lan HY. miR-29 inhibits bleomycin-induced pulmonary fibrosis in mice. Mol Ther 20: 1251–1260, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Xu D, Takeshita F, Hino Y, Fukunaga S, Kudo Y, Tamaki A, Matsunaga J, Takahashi RU, Takata T, Shimamoto A, Ochiya T, Tahara H. miR-22 represses cancer progression by inducing cellular senescence. J Cell Biol 193: 409–424, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yang IV, Pedersen BS, Rabinovich E, Hennessy CE, Davidson EJ. Relationship of DNA methylation and gene expression in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 190: 1263–1272, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yang JH, Li JH, Shao P, Zhou H, Chen YQ, Qu LH. starBase: a database for exploring microRNA-mRNA interaction maps from Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res 39: D202–D209, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhou H, Chen X, Chen L, Zhou X, Zheng G, Zhang H, Huang W, Cai J. Anti-fibrosis effect of scutellarin via inhibition of endothelial-mesenchymal transition on isoproterenol-induced myocardial fibrosis in rats. Molecules 19: 15611–15623, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Zhu H, Li Y, Qu S, Luo H, Zhou Y, Wang Y, Zhao H, You Y, Xiao X, Zuo X. MicroRNA expression abnormalities in limited cutaneous scleroderma and diffuse cutaneous scleroderma. J Clin Immunol 32: 514–522, 2012. [DOI] [PubMed] [Google Scholar]
- 140.Zurawek D, Kusmider M, Faron-Gorecka A, Gruca P, Pabian P, Kolasa M, Solich J, Szafran-Pilch K, Papp M, Dziedzicka-Wasylewska M. Time-dependent miR-16 serum fluctuations together with reciprocal changes in the expression level of miR-16 in mesocortical circuit contribute to stress resilient phenotype in chronic mild stress — An animal model of depression. Eur Neuropsychopharmacol 26: 23–36, 2016. [DOI] [PubMed] [Google Scholar]