Abstract
Chemotherapeutics such as doxorubicin (DOX) and paclitaxel (PXL) have dose limiting systemic toxicities including cardiotoxicity and peripheral neuropathy. Delivery strategies to minimize these undesirable effects are needed and could improve efficacy, while reducing patient morbidity. Here DOX and PXL were conjugated to a nanodendron (ND) through an MMP9-cleavable peptide linker, producing two new therapies, ND2DOX and ND2PXL designed to improve delivery specificity to the tumor microenvironment and reduce systemic toxicity. Comparative cytotoxicity assays were performed between intact ND-drug conjugates and the MMP9 released drug in cell lines with and without MMP9 expression. While ND2DOX was found to loose cytotoxicity due to the modification of DOX for conjugation to the ND; ND2PXL was determined to have the desired properties for a prodrug delivery system. ND2PXL was found to be cytotoxic in MMP9-expressing mouse mammary carcinoma (R221-Aluc) (53%) and human breast carcinoma (MDA-MB-231) (66%) at a concentration of 50 nM (in PXL) after 48 hours. Treating ND2PXL with MMP9 prior to the cytotoxicity assay resulted in a faster response; however, both cleaved and intact versions of the drug reached the same efficacy as the unmodified drug by 96 hours in the R221A-luc and MDA-MB-231 cell lines. Further studies in modified Lewis lung carcinoma cells that either do (LLCMMP9) or do not express (LLCRSV) MMP9 demonstrate the selectivity of ND2PXL for MMP9. LLCMMP9 cells were only 20% viable after 48 hours of treatment while LLCRSV were not affected. Inclusion of an MMP inhibitor, GM6001, when treating the LLCMMP9 cells with ND2PXL eliminated the response of the MMP9 expressing cells (LLCMMP9). The data presented here suggests that these NDs, specifically ND2PXL, are non-toxic until activated by MMP9, a protease common in the microenvironment of tumors, indicating that incorporation of chemotherapeutic or cytostatic agents onto the ND platform have potential for tumor-targeted efficacy with reduced in vivo systemic toxicities.
Keywords: nanomaterials, prodrug, MMP9, paclitaxel, dendrons, drug delivery
Introduction
Chemotherapeutic drugs are generally administered at the maximum tolerated dose so as to achieve efficacy while avoiding severe systemic toxicities. Nonetheless, patients treated with systemic chemotherapy are prone to debilitating side effects such as neurotoxicity, myelotoxicity and cardiotoxicity. Thus, new strategies are necessary to minimize damage of chemotherapeutics to healthy cells and tissues [1–3]. These strategies include the delivery of therapy through nanomaterials to the target site, particularly tumors, thereby reducing systemic, off-target effects of chemotherapeutic drugs. A related and somewhat more sophisticated approach involves the incorporation of the drug into a delivery vehicle that can be activated in the tumor microenvironment, leading to the localized release of the medication as has been successful in imaging [4–15].
The development of drugs with relatively low off-site toxicity and the desired delivery properties has been approached by coupling the chemotherapeutic drug to the carrier through an enzymatically cleaved linker designed to be cleaved by a protease present principally in diseased tissues. Tumor microenvironments are enriched with a variety of extracellular proteases including matrix metalloproteinases (MMPs), a family of extracellular zinc-dependent enzymes capable of degrading all components of the extra-cellular matrix and that have been implicated in cancer progression and establishment of metastatic disease [16–18]. In the clinical setting, MMP9 has been implicated in tumor growth and metastasis and found to be an indicator of poor prognosis, particularly in breast cancer patients [19, 20]. In addition, it has been shown that MMP (including MMP9) expression can be used to distinguish benign from malignant tumors and identify aggressive tumors associated with poor outcome [17, 20]. These clinical observations have been evaluated in preclinical models of cancer using genetic manipulation of specific MMP family members in both breast cancer cells and the mouse host microenvironment to examine the roles of MMPs in tumor progression in lung, bone and liver organ-specific metastasis [21–24]. Active MMP9 has been found to be present in high concentrations in both the primary tumor as well as at the earliest stage of metastatic disease. Targeting MMP9 expression has the potential to identify and target micrometastatic lesions that may be present in breast cancer patients but are undetectable by current imaging technology. The gene promoter of MMP9 is highly regulated and has strong oncogene-inducible response elements, such that MMP9 protein expression is limited under normal conditions and often up-regulated in cancer. By contrast, MMP2 has a “housekeeping” gene promoter region; that results in widespread constitutive and responsive to general changes in cellular metabolism but is only modestly inducible by growth factors or cytokines [25]. Taken collectively, MMP-cleavable linkers are attractive components to use in activatable drugs that target the earliest stages of breast cancer metastasis. Since our long-term goal was to generate a chemotherapeutic agent with selectivity for tumor over normal tissues, we chose to target MMP9 when designing the peptide linker. In the prototype, NDDrug formulations described here, the drug is linked to the ND through a peptide linker that includes the sequence AVRWLLTA. This peptide sequence was reported to be a good substrate for MMP9 [26, 27] which was confirmed in our laboratory by incorporating this sequence into an analogous proteolytic beacon and confirming that this sequence is cleaved by MMPs with significant selectivity for MMP9.
Taxane and anthracycline based regimens, such as paclitaxel (PXL) and doxorubicin (DOX) respectively, have been used extensively to treat of a variety of cancers including early and advanced breast cancer. As non-targeted formulations, these as well as other chemotherapeutics present dose-limiting systemic toxicities. For DOX, cardiomyopathy, the major dose-limiting side effect, limits the cumulative dose even in patients with healthy heart function [28]. While liposomal formulations of DOX, such as DOXIL®, exhibit reduced cardiotoxicity compared with standard anthracycline therapy [29], significant cardiotoxicity remains when DOXIL is given in combination therapy, e.g. with trastuzumab in HER2-positive early-stage breast cancer patients [30]. Likewise, PXL is considered standard and preferred chemotherapy for early high risk and metastatic breast cancer but can cause severe toxicities, such as hypersensitivity reaction, myelosuppression and neurotoxicity [31]. Although hypersensitivity and neutropenia are significantly attenuated by the albumin-based nanoparticle formulation of PXL, Abraxane® (Abx), patients treated with Abx remain prone to neuropathy, with an incidence reported to be higher than with other formulations of PXL [31]. It is anticipated that targeted formulations of DOX or PXL have the potential to reduce cardiomyopathy or neuropathy, respectively, thereby permitting higher doses to enhance efficacy versus early stage and/or advanced disease.
Here we describe a new class of nanodendrons (ND) that have been specially designed to act as delivery vehicles for chemotherapeutics and in vitro results with either an anthracycline (DOX), or a taxane (PXL) incorporated. In particular, we have synthesized these agents to not only carry the drug, but to function as a prodrug, releasing the therapy only when activated by a physiological factor that is present within the tumor microenvironment. This report focuses on the design, synthesis and assessment of in vitro cytotoxicity of novel chemotherapeutic agents formulated as MMP9-activatable NDs that deliver DOX or PXL to the protease-rich microenvironment of tumors. Our MMP9-activatable drugs are desirable in that: a) they can be synthesized with high purity, b) are not toxic to cells that do not express MMP9 and c) the design allows modifications to optimize solubility and pharmacokinetics. The ND system described here is amendable allowing targeting of other proteases, use of other drugs and the incorporation of other functionalities (e.g. imaging agents, multiple drugs and solubilizing agents).
These NDs have been designed for activation by MMP9, an MMP present in a number of cancers. The therapeutic activatable nanoparticle consists of a dendron with multiple MMP9 selective peptides that when enzymatically cleaved releases DOX or PXL from the ND delivery vehicle. Since this activation occurs primarily in the presence of MMP9, selective delivery is expected, minimizing systemic toxicities. Our ND platform is based on a polyester dendron backbone [32] modified for rapid incorporation of peptides (and subsequently drugs) with high yielding methods that reliably functionalize all four terminal groups on each dendron. Both DOX and PXL have been incorporated into this novel MMP9-activatable ND delivery platform to obtain ND2DOX and ND2PXL, respectively. Both compounds exhibit MMP9-dependent cleavage; while ND2DOX exhibits no detectable cytotoxicity, ND2PXL is highly cytotoxic upon MMP9 activation and has been assessed versus four cancer cell lines. ND2PXL is sensitive against three cell lines that express MMP9 while not affecting the viability of a cell line that expresses no detectable amount of MMP9. Cytotoxicity of ND2PXL is comparable to unmodified PXL, albeit with a delayed response time. In addition, cytotoxicity was significantly attenuated by co-administration with GM6001, a broad spectrum MMP inhibitor, in MMP9 expressing cell lines. Overall, these results demonstrate the construction and potential of our new protease-activated therapeutics to deliver active drug selectively to diseased cells.
Results and Discussion
All of the NDs reported here utilize the same MMP9-cleavable peptide sequence, AVRWLLTA (supplemental, Figure S4). The nomenclature for the NDs include a subscript 2 (i.e. ND2) to indicate this is a second generation dendron while the drug is abbreviated and superscripted to indicate which therapeutic is incorporated. ND2DOX did not produce the desired cytotoxicity upon protease treatment because the DOX molecule activity is lost when the drug is modified for conjugation. It is noteworthy that modification at the carbohydrate amine with glutaric anhydride had been reported previously to be an effective approach in the development of prodrugs based on DOX [33, 34]. In our hands this was not the case, therefore we suggest that it is imperative to test new formulations of prodrugs at all stages of development to ensure that the drug will be both inactivated (on scaffold) and re-activated (upon cleavage).
Four MMP9-selective peptides were linked to the ND scaffold after minor modifications to the dendron backbone to obtain a pure product
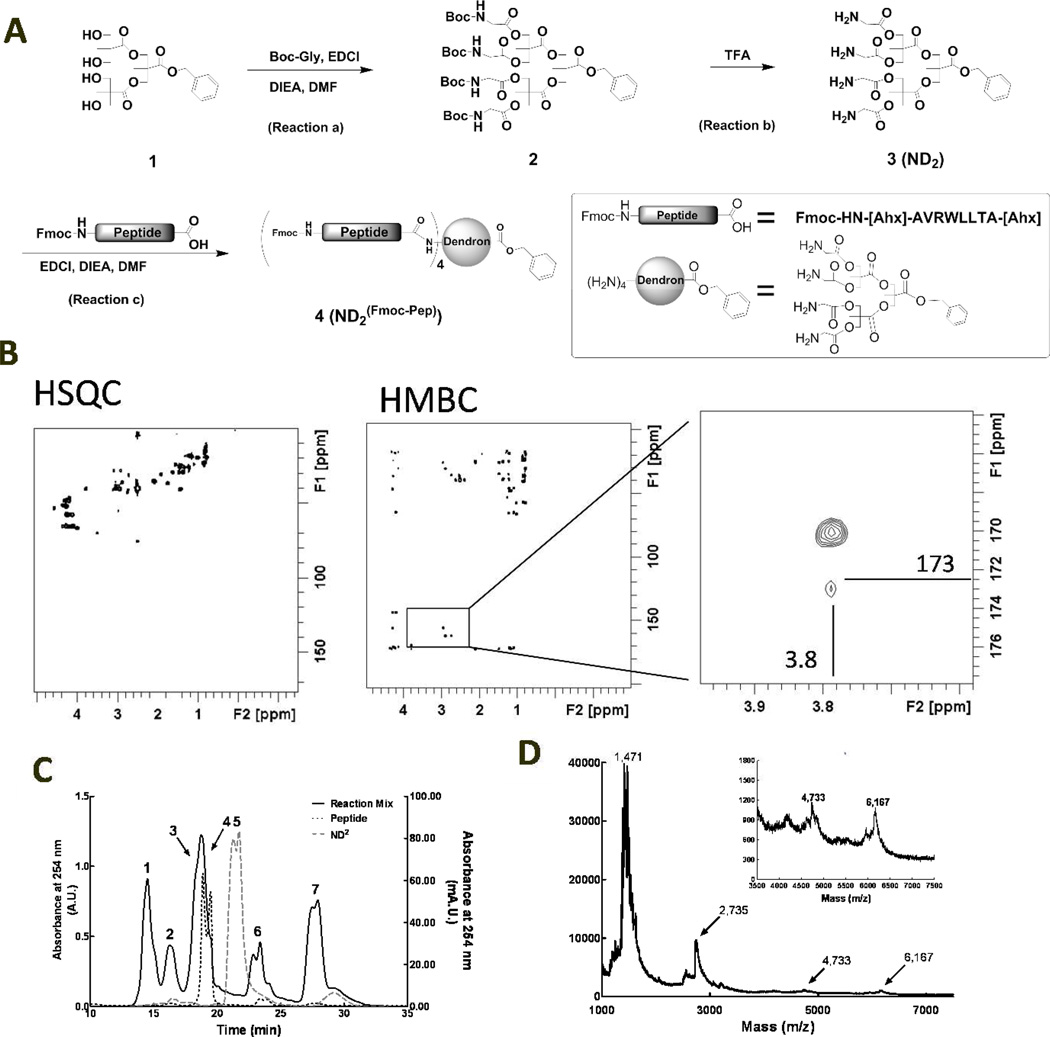
The ND platform was built by modifying the peripheral alcohols of a polyester dendron [32] to glycine residues (ND2) then incorporating an MMP9-cleavable peptide and drug (DOX or PXL) onto the nanomaterial structure. Characterization of ND2Pep was initially complicated when a product ion peak could not be detected in the matrix assisted laser desorption ionization time of flight mass spectrometer (MADLI-TOF MS). To combat this issue, new methods were developed to characterize the NDs; specifically, 1) to show that the peptide covalently attached to the dendron, 2) the number of peptides attached and 3) the crude material was pure after preparatory HPLC. A second generation polyester dendron (1) [32] was modified via two synthetic steps to provide glycines on the surface. The resulting surface amines were then used to rapidly and reliable attach the MMP9-selective peptide via standard coupling procedures. To do this, first Boc-glycine was attached to the hydroxyls on the surface of dendron 1 using EDCI (or DCC) with excess Boc-Gly (reaction a, Figure 1A) to modify all the hydroxyls (2, ND2Boc). Next, ND2Boc was treated with TFA (reaction b) to remove the Boc protecting groups, yielding ND2 (3). For the glycine incorporation (reaction a), comparable results, though with longer reaction times, were obtained using DCC instead of EDCI. Both ND2Boc (2) and ND2 (3) produced the expected nuclear magnetic resonance (NMR) spectra (see Figure 1B and supplementary data for the spectra and proton assignments) and molecular weights via electrospray ionization mass spectrometry (ESI+ MS) of 1,107.5 and 685.4 Da. for 2 and 3 respectively (see supplemental Figures S1 and S11).
Figure 1.
Conjugation and subsequent characterization of an MMP9-cleavable peptide to ND2. A: Schematic representation of the synthetic pathway to ND2(Fmoc-Pep) (product 4). B: HSQC and HMBC NMR spectra of ND2(Fmoc-Pep). The expanded image (right panel) of the HMBC (center panel) highlights the peak at 3.8 and 173 ppm that is indicative of coupling between the carboxy terminus of the peptide and the methylene protons on the surface Gly residues of ND2. C: SEC traces of Fmoc-[Ahx]-AVRWLLTA-[Ahx], treated with EDCI (black dotted), ND2 (gray dashed) and the coupling reaction mix of these two components (black solid) indicating progress of the reaction after 24 hours. Peaks 1–7 were identified as: desired product, ND2PXL, with 4 peptides attached to the dendron(Peak 1); 3 peptides attached to the dendron (Peak 2); a combination of ND2 with either 1 or 2 peptides attached (Peaks 3 and 4); ND2 starting material (Peak 5) (not found after reaction for 24 hours); and small molecule coupling agents and side products (peaks 6 and 7). D: MALDI-TOF MS of ND2(Fmoc-Pep), product 4; the inset expands the 3000 to 7500 Da. region including the molecular weight ion peak at 6,167 Da.
Following modification of the polyester dendron, incorporation of the protease-cleavable peptide was readily obtained; however, novel methods of characterization were required to ensure purity and the desired number of peptides attached to the ND. ND2 was reacted with 4.4 equivalents of MMP9 substrate peptide [26, 27], Fmoc-[Ahx]-AVRWLLTA-[Ahx], to functionalize all four surface amines. Although this coupling reaction was successfully completed with other coupling agents, including HATU and BOP, EDCI was found to give the highest overall yields and be the easiest to purify with these NDs. Characterization of ND2(Fmoc-Pep) (4) and the subsequent side products containing less than 4 peptides per ND, (reaction c, Figure 1A) included NMR, size exclusion chromatography (SEC), MALDI-TOF-MS) analysis and a microscale ninhydrin assay to measure primary amines (see supplemental data and figure S15) [6]. This extensive characterization was completed to ensure that there was a pure product with precisely 4 peptides per ND prior to moving forward with drug attachment.
To assess attachment, 2-dimensional NMR techniques (HMBC and HSQC) were utilized to determine the connectivity of the organic ND structures. HSQC produces signals from protons coupled to other protons separated by 3 bonds; while HMBC produces signals between a proton and a carbon that are 3 or 4 bonds apart. Analysis of the starting materials and the product by NMR indicated that ND2 covalently attaches to the carboxy terminal of Fmoc-[Ahx]-AVRWLLTA-[Ahx]. As seen in the 2-D NMR (Figure 1B), a proton at 3.8 ppm is coupled to a carbon at 173 ppm. From analysis of ND2 and Fmoc-[Ahx]-AVRWLLTA-[Ahx] starting materials, these peaks were assigned to the methylene on the glycine portion of ND2 and the carboxylic acid carbonyl carbon from the peptide (now an amide) (see supplemental Figures S3, S7, S8, S9 and S13). For these molecules, the HMBC was particularly useful in that it produced a correlation between the methylene on the glycine portion of the dendron and the carbonyl carbon from the peptide. The only circumstance in which that carbon could couple to those protons in the HMBC NMR is if they are covalently attached. This type of coupling requires characterizing the starting materials and is limited due to the potential for signal overlap from other materials, particularly when working with large, complex molecules. In this system signal overlap was not an issue as the carbon and proton signals in question were resolvable from the other signals in the molecules (Figure 1B). Knowing that the peptide is covalently attached to the dendron was the first step in determining the overall structure and purity.
SEC was used to characterize the relative size of the products obtained in reaction c (Figure 1A). Calibration using standards for SEC were attempted, but did not correspond with known molecular weights of the starting materials, likely due to the structural diversity in the dendron starting materials. The calibration standards consist of proteins of increasing molecular weight while our samples are a mix of small peptides and synthetic materials. Although they have similar molecular weights, the NDs and their starting materials have different chemical properties such as solubility, shape (particularly in solution) and molecular interactions with the column packing material. Thus, the molecular weights of the starting materials, characterized by mass spectrometry, did not correlate with the MW standards kit; the order of elution (i.e. larger molecules eluted earlier) was preserved under the conditions the chromatograms were acquired for these ND-drug conjugates. The reaction mix produced four peaks that eluted earlier than either the peptide or the dendron starting material on the SEC column; indicating that four compounds with molecular weights larger than the starting materials were formed (Figure 1C peaks 1–4). The SEC peaks were collected and concentrated on the analytical scale for further analysis.
A ninhydrin assay reported from our laboratory [6] was used to determine the concentration, and thus the number, of primary amines on the surface of each dendron fraction isolated from the SEC spectra. Briefly, a solution containing the sample to be tested was boiled with ninhydrin reagent and the color change was quantified by reading the absorbance at 560 nm. The concentration of primary amine in each sample was calculated based on a standard curve acquired with known concentrations of leucine. Based on the ninhydrin response to the peptide and the dendron (with and without the amines available for reaction) alone, the number of amines on the dendron (and thus the number of peptides) was deduced (see supplemental for further details). The first two peaks in the SEC chromatogram were found by the ninhydrin assay, to have 4 (peak 1) and 3 (peak 2) peptides attached per dendron. The remaining two peaks (peaks 3 & 4), isolated together from SEC were found to have an average of 1.7 peptides per dendron (ninhydrin assay method), indicating a mixture of two products with either 1 or 2 peptides attached, reflecting incomplete reaction. Following extensive characterization of peak 1 that yielded only fragments in the MS and not the expected MW, a MALDI-TOF MS was obtained serendipitously from a non-protease treated control of compound 4 when looking for cleavage products after protease treatment. These samples had been spotted onto the MALDI plate with buffer in the solution, which typically inhibits ionization of the parent ion in the MALDI; however in this case, the before protease control produced the desired, single charged ion that had been absent in previous analyses. The presence of four peptides on the product isolated as peak 1 (Figure 1C) was confirmed by MALDI-TOF MS, yielding the molecular weight 6,167 Da. (Figure 1D), identifying this product as compound 4, ND2Fmoc-Pep (Figure 1A). Upon synthesizing repeated batches of 4, the bulk of the product (72–85% yield) was isolated as pure ND2(Fmoc-Pep) containing four peptides per molecule.
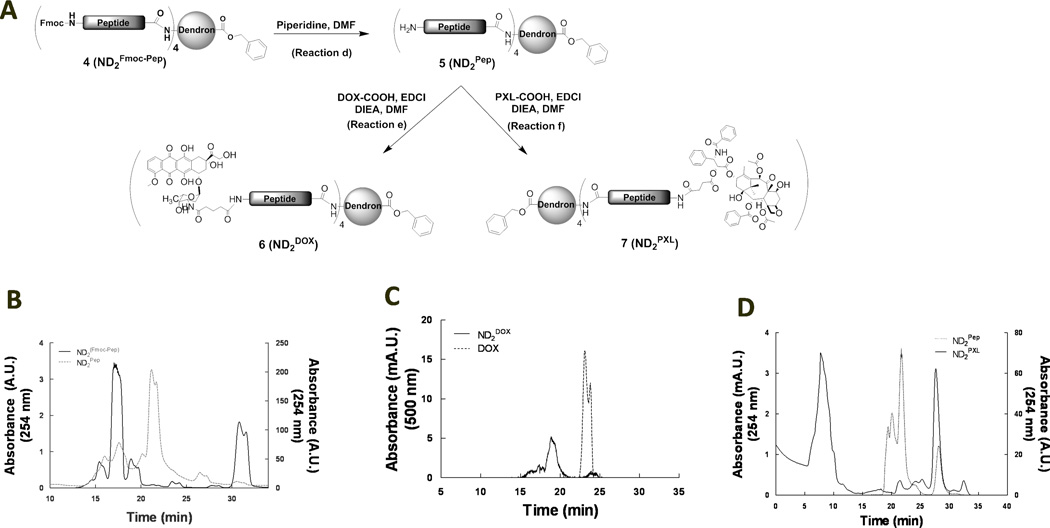
Doxorubicin (DOX) was attached to ND2(Fmoc-Pep) with minimal steps (3 total) in excellent yield
Glutaric anhydride was reacted with (DOX) as previously described to modify the amine on the carbohydrate moiety and to obtain a carboxylic acid for conjugation, DOX-COOH [34]. ND2Fmoc-Pep was deprotected (reaction d, Figure 2A) yielding compound 5, ND2Pep, that eluted from SEC at 22 minutes, several minutes after the peak elution time for the protected ND2Fmoc-Pep starting compound (Figure 2B). DOX-COOH was then attached to ND2Pep using standard peptide coupling methods, primarily EDCI (reaction e, Figure 2A). A new molecule, as evaluated by SEC, with the characteristic DOX absorbance at 500 nm was obtained and purified by preparatory HPLC. While DOX elutes from the SEC column at 23–24 min, the main reaction product with an absorbance at 500 nm eluted at 19 min (Figure 2C), indicating an increase in size. In addition, ninhydrin assay confirmed the absence of primary amines in the purified product consistent with the incorporation of four DOX per ND2Pep in the final product ND2DOX, 6 (Figure 2A).
Figure 2.
Drug conjugation and characterization of activatable DOX (ND2DOX) and PXL (ND2PXL). A: Schematic representation of the synthetic pathway from ND2(Fmoc-Pep) (4) to either ND2DOX (6) or ND2PXL (7). B: SEC of ND2(Fmoc-Pep) (solid, left y-axis) and ND2Pep (dotted, right y-axis)). C: SEC of DOX (dotted) and ND2DOX (solid). D: SEC of ND2Pep (dotted, left y-axis) and of the reaction mix to link PXL to ND2Pep (solid, right y-axis).
Paclitaxel (PXL) was attached to ND2(Fmoc-Pep) with minimal steps (3 total) in excellent yield (85 %)
PXL was modified as previously described to provide a carboxylic acid conjugation handle (PXL-COOH) while maintaining therapeutic efficacy [35]. Analogous to the synthesis of ND2DOX, PXLCOOH was attached to ND2Pep, (compound 5, Figure 2A), using EDCI coupling (reaction f, Figure 2A). To obtain ND2PXL (7), excess PXL-COOH (4.4 equivalents) was used to ensure conjugation to the N-terminal on all of the four peptides linked to ND2. ND2PXL was purified and characterized by SEC, collecting the first peak that eluted (Figure 2D). This peak was further characterized using MALDI-TOF MS, which gave several fragments consistent with ND2PXL. Reaction with ninhydrin indicated that no primary amines were present in the final product.
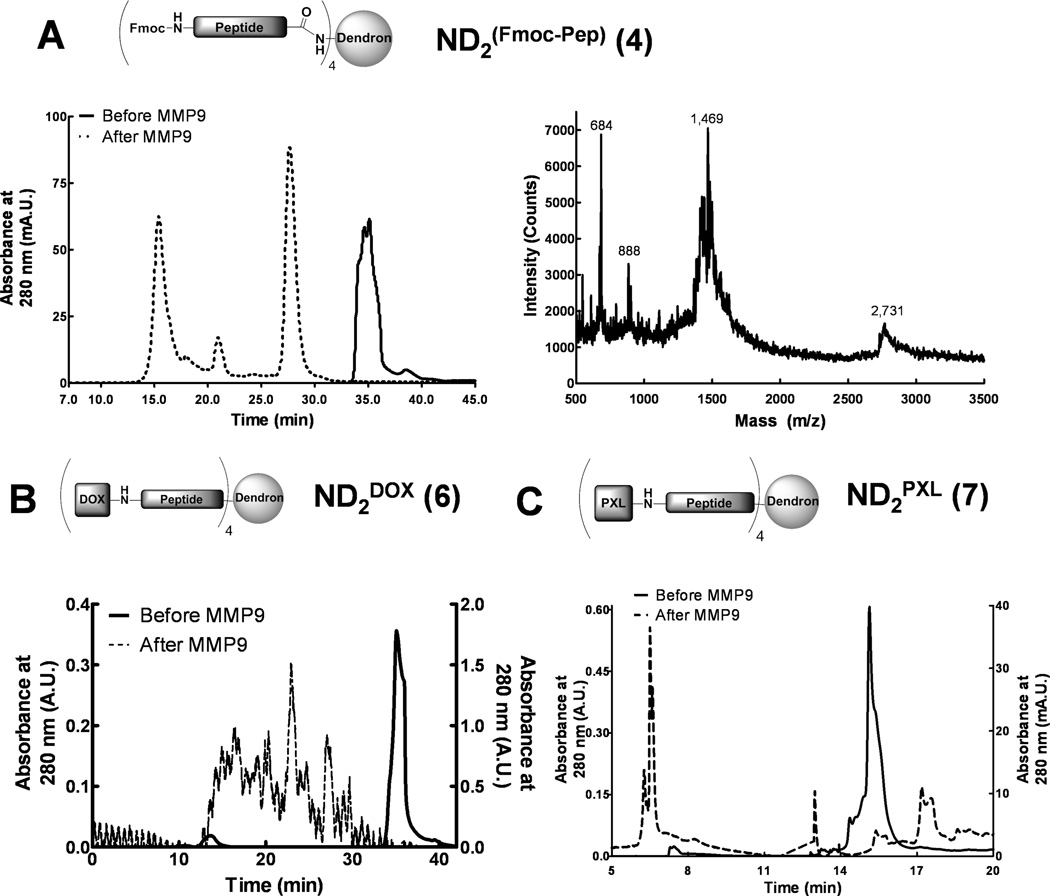
Peptide-dendron molecules are cleaved by MMP9
The three peptide-dendron conjugates (ND2(Fmoc-Pep), ND2DOX and ND2PXL) were treated with MMP9, trypsin (a positive control) and buffer (negative control) to test for proteolytic cleavage. For these experiments an excess of enzyme was used to ensure complete cleavage for subsequent in vitro toxicity testing; however, we have demonstrated that cleavage occurs at physiologically relevant concentrations in MMP-cleavable beacons [5, 6]. HPLC analysis before and after treatment with MMP9 shows that ND2(Fmoc-Pep), ND2PXL and ND2DOX are all cleaved by MMP9 (Figure 3). The HPLC of ND2(Fmoc-Pep), monitoring the peptide component at 280 nm, provided a single peak that elutes at 35 minutes. Following treatment with MMP9, ND2(Fmoc-Pep) elutes as two major peaks at 27.5 and 15.3 minutes and a minor peak at 20.8 minutes (Figure 3A). Mass spectrometry (MALDI-TOF) confirmed the proteolytic cleavage of ND2(Fmoc-Pep) yielding the expected cleavage products at 2,731 Da., corresponding with LLTA-[Ahx])4-ND, and 888 Da. for Fmoc-[Ahx]-AVRW (Figure 3A, right panel). In addition, a peak at 1,469 was observed in the MALDI-TOF spectrum, which correlates with further degradation of the dendron fragment to A-[Ahx]4-ND. Intact ND2DOX (6) also shows a single peak that eluted at 35 minutes in reverse phase HPLC. This peak is completely lost upon treatment with MMP9 (Figure 3B). MMP9-treated ND2DOX exhibits multiple peaks of low intensity that are not well-resolved (Figure 3B) suggesting that the expected DOX-[Ahx]-AVRW fragment(s) obtained after MMP9 cleavage may be susceptible to additional cleavage and/or may be poorly soluble. A similar, heterogeneous, poorly-resolved mixture of products was observed in HPLC analyses of trypsin-treated ND2DOX (not shown) whereas control (buffer treated) samples had no change from the original HPLC. HPLC analysis of ND2PXL (Figure 3C) showed a single peak at 15.5 minutes that disappears while several new peaks (6.3, 6.5, 13 and 17 minutes) appear after treatment with MMP9, indicative of protease digestion. For all three compounds, ND2(Fmoc-Pep), ND2DOX and ND2PXL, controls (buffer-treated) showed no degradation whereas trypsin-treated samples were cleaved yielding HPLC traces with multiple peaks as observed in the MMP9-treated counterparts.
Figure 3.
MMP9 treatment of ND2(Fmoc-Pep), ND2DOX and ND2PXL. A: Reverse phase C18 HPLC of ND2(Fmoc-Pep) before and after treatment with MMP9 (left panel) and the MALDI-TOF MS spectrum of the MMP9-treated reaction mix showing the cleavage products at 2,731 (LLTA-[Ahx]4-ND2) and 1,469 (A-[Ahx]4-ND2) Da. (right). B: Reveres phase C18 HPLC of ND2DOX before and after MMP9 treatment. C: Reverse phase C18 HPLC of ND2PXL before and after MMP9 treatment. HPLC conditions as per methods. In the chromatograms shown in A, B, and C, the solid line represents samples before MMP9 treatment while the dotted line is the same sample after overnight incubation with excess MMP9.
Cleavage of ND2PXL or ND2DOX by MMP9 confirms that further interrogation through functionalizing both ends of the peptide sequence does not change the ability of MMP9 to cleave the MMP9-selective sequence AVRWLLTA. HPLC traces before and after treatment with MMP9 of all the ND2-peptide compounds leads to the formation of at least two new components in solution. NDs treated with buffer (no protease) remained intact while those treated with trypsin also formed multiple components overnight. Digestion into more than two components is likely due to further degradation of the initial digestion products. Upon isolation, the cleaved products of ND2PXL treated with MMP9 provided the molecular weight for one of the expected cleavage peaks PXL-[Ahx]-AVRW and further fragmentation products when analyzed by MALDI-TOF MS. Similar cleavage patterns were seen with ND2PXL cleaved by trypsin, the positive control. Although, cleavage by trypsin may cause concern that in vivo delivery my not be feasible, he digestion of the peptide by trypsin is not of biological concern in the design of a chemotherapeutic agent since trypsin secretion is limited to the exocrine pancreas and the enzyme is delivered directly to the gut through specific ducts. An IV-delivered chemotherapeutic agent would not come in contact with trypsin. These results are consistent with the MMP-selective beacons developed in our laboratory [5, 6, 12] and suggest that in addition to DOX and PXL, further drugs and imaging moieties can be coupled to the scaffold without loss of MMP9 ability to cleave this peptide sequence.
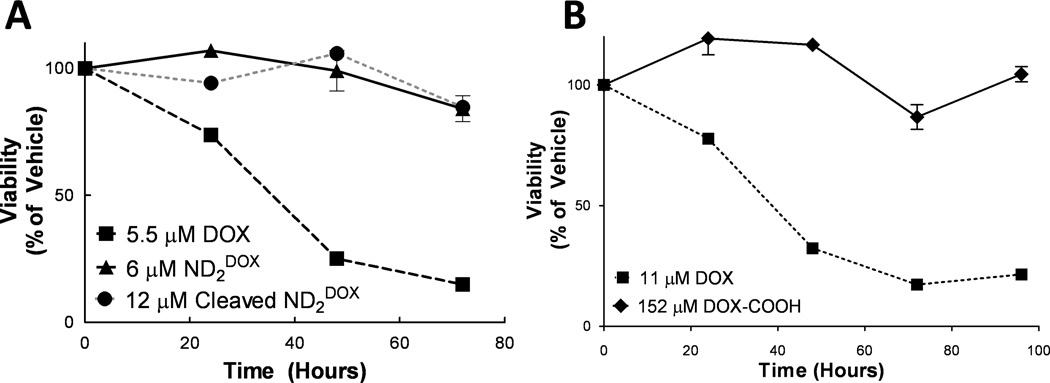
ND2DOX is ineffective at killing R221A-luc cells
The toxicity of the NDs was tested to ensure that ND2DOX and ND2PXL are not toxic until cleaved with MMP9. Cytotoxicity of ND2DOX was assessed in R221A-luc cells using a trypan blue exclusion assay for viability and measuring at 24, 48, 72 and 96 hours following the first dose. R221A-luc cells have a < 25 % viability after 48 hours as assessed using a trypan blue exclusion assay of cytotoxicity (Figure 4A); however, ND2DOX at 6 µM showed no appreciable cytotoxicity (Figure 4A). The R221A-luc cells were shown to be sensitive to DOX at concentrations as low as 184 nM via the MTS assay (see supplemental Figure S17). Treatment of ND2DOX with MMP9 to obtain the cleavage product also did not induce cell death. In vitro testing of R221A-luc cells treated with ND2DOX resulted in no observable cytotoxicity of either the intact or MMP9-treated ND2DOX at concentrations as high as 6 µM in DOX (Figure 4). In contrast, the growth of cells treated with unmodified DOX was inhibited at concentrations as low as 20 nM. Further investigation concluded that even the simplest modification of DOX tested in these studies, DOX-COOH, at concentrations as high as 152 µM (25-fold higher concentration than unmodified DOX), for up to 96 hours (4 doses) produced no detectable cytotoxicity (Figure 4B). This most basic modification of doxorubicin, DOX-COOH, gave viability comparable to vehicle-treated cells, indicating modification of DOX results in a molecule that is not toxic in vitro. Thus, modification of DOX at the carbohydrate amine to generate a carboxylic acid blocks DOX cytotoxicity against R221A-luc cells. Although some literature suggested that this modification was acceptable [33, 34], others have reported that this specific modification eliminates the toxicity of DOX [36]. Modifications to other functional groups on DOX have been reported for conjugation to nanomaterials [37, 38] which have the potential to work with our ND system; we opted to incorporate a different drug, PXL.
Figure 4.
Assessment of cytotoxicity of DOX modified compounds ND2DOX (with and without MMP9 treatment) and DOX-COOH compared to DOX. A: viability of 6 µM (in DOX) ND2DOX (triangle) and 12 µM (in DOX) cleaved ND2DOX (circle) compared to 5.5 µM DOX (square). B: viability of 152 µM DOX-COOH (triangle) comared to 11 µM DOX (rhombus).
ND2PXL’s cytotoxicity versus two breast cancer cell lines is time and dose dependent
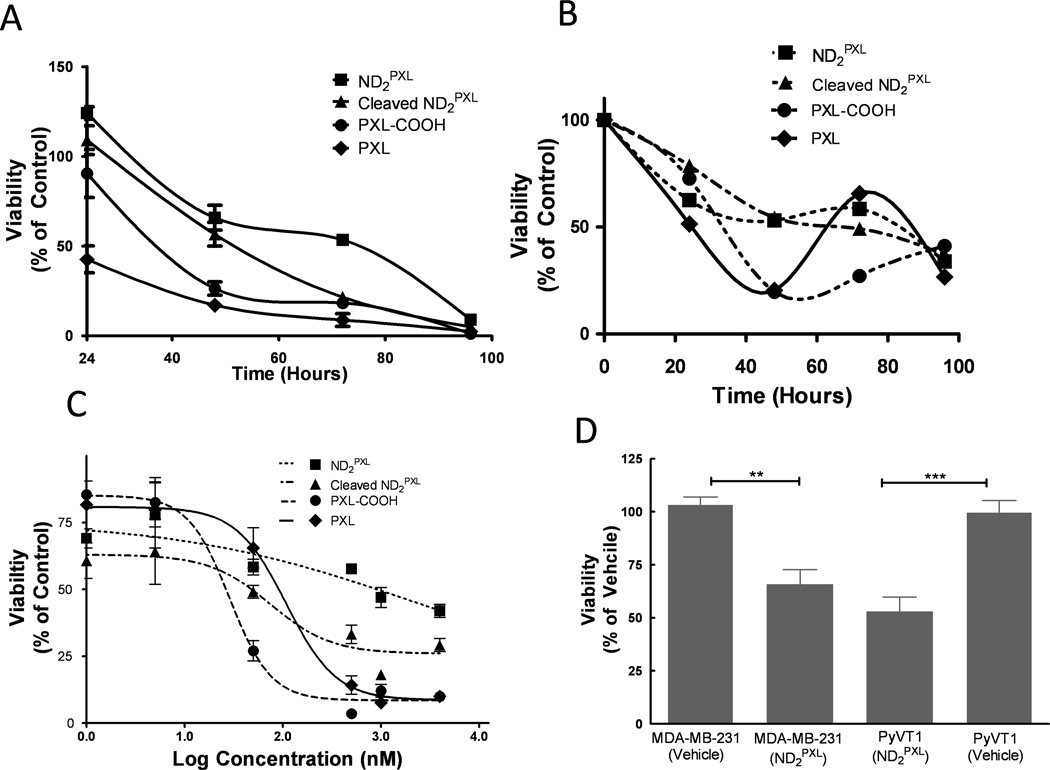
ND2PXL was tested, with and without pretreatment of MMP9, for cytotoxicity as compared to both PXL and PXL-COOH versus MDA-MB-231 (human) and R221A-luc (mouse) breast cancer cell lines, both of which are known to express MMP9 ([39] and [24] respectively) (Figure 5). These two breast cancer cell lines were tested for sensitivity to ND2PXL in anticipation for in vivo efficacy and systemic toxicity studies. In MDA-MB-231 cells, PXL and PXL-COOH are more potent over the first 48 hours of treatment than the same dose of PXL delivered using ND2PXL with (cleaved) and without (intact) pretreatment with MMP9. However, by 48 hours both cleaved and intact ND2PXL are significantly more cytotoxic compared to vehicle-treated cells (Figure 5A&D) and at even longer times (72 and 96 hours), MMP9-cleaved ND2PXL has cytotoxicity similar to PXL and PXL-COOH. For intact ND2PXL, a longer treatment time (96 hours) is required to achieve cytotoxicity (<10% of viable MDA-MB-231 cells remaining) comparable to the PXL and PXL-COOH molecules. Thus, by 96 hours, ND2PXL has cytotoxicity versus MDA-MB-231 cells comparable to PXL, PXL-COOH or pre-cleaved ND2PXL (Figure 5A). Although cytotoxicity of ND2PXL is time dependent and directly related to the activation of the ND-prodrug, this human cell line is sensitive to ND2PXL. Given that MMP9-treated ND2PXL produces a toxic effect faster than untreated ND2PXL, MMP9-cleavage is implicated in the activation mechanism.
Figure 5.
Assessment of ND2PXL cytotoxicity. A & B: Cell viability (A, MDA-MB-231 and B, R221A-luc) was measured over time (as indicated) following addition of either PXL, PXL-COOH, ND2PXL or ND2PXL following pretreatment with MMP9, each at a comparable concentration of PXL (50 nM). C: Dose-response plot of R221A-luc cell viability after 72 hours of treatment (compounds each 50 nM in PXL). D: Viability of MDA-MB231 and R221A-luc cells following 48 hours of ND2PXL treatment (50 nM in PXL) as compared with vehicle-treated cells.
The R221A-luc cell line is derived from a spontaneous primary mammary tumor in an MMTV-PyVT mouse (FVB background) and provides a fully immunocompetent mouse model of breast cancer for preclinical efficacy testing of novel therapeutic agents. In this cell line, a similar pattern is observed as seen in the MDA-MB-231 cell line. Intact ND2PXL and MMP9-treated ND2PXL involve a slower evolution of cytotoxicity as compared with PXL or PXL-COOH (Figure 5B); however after 96 hours all dosing regimens produce similar toxicity. R221A-luc cells treated with ND2PXL or MMP9-treated ND2PXL retain 52 (+/−5) % viability after 48 hours of treatment (Figure 5B) whereas cells treated with either PXL or PXL-COOH are <25% viable. However, after 96 hours of treatment, the four PXL-containing compounds including ND2PXL and MMP9-treated ND2PXL have comparable cytotoxicity versus R221A-luc cells with viability ranging from 27 to 41%. All forms of PXL tested are dose-dependent versus the R221A-luc cells (Figure 5C). PXL and PXL-COOH exhibit typical curves of dose dependency with marked increases in toxicity above 100 nM and calculated EC50 values in the range of 20–50 nM for both PXL and PXL-COOH after 72 hours of treatment (Figure 5C). As compared with PXL and PXL-COOH, the ND2PXL forms (intact or MMP9-treated) of the drug show less concentration dependence against R221A-luc cells. The intact and MMP9-treated forms of ND2PXL have estimated EC50 values in the range of 1 QM and 100 nM (in drug concentration) respectively. The cytotoxicity of untreated ND2PXL is similar in the two cell lines; with the viability of the MDA-MB-231 and R221A-luc cells lines following treatment with ND2PXL (50 nM in PXL for 48 hours) being uh66 and 52%, respectively, compared to vehicle-treated cells. The slower response to and reduced cytotoxicity of intact ND2PXL as compared with PXL and PXL-COOH, observed with both the MDA-MB-231 and R221A-luc cell lines, suggest that the cytotoxicity of ND2PXL is dependent on cleavage of the MMP9 substrate peptide. These observations indicate that structural differences between PXL/PXL-COOH and PXL-[Ahx]-AVRW could account for this difference, making cellular internalization or microtubular binding slower with the bulkier PXL-[Ahx]-AVRW. Alternatively, further enzymatic degradation of PXL-[Ahx]-AVRW may be necessary prior to the onset of cytotoxic effects. Regardless of the complete mechanism of activation, the in vitro data presented here provides evidence of the NDs potential to deliver PXL to MMP9-rich tumor microenvironments.
Cytotoxicity of ND2PXL is enhanced by expression of MMP9 and attenuated by treatment with an MMP inhibitor
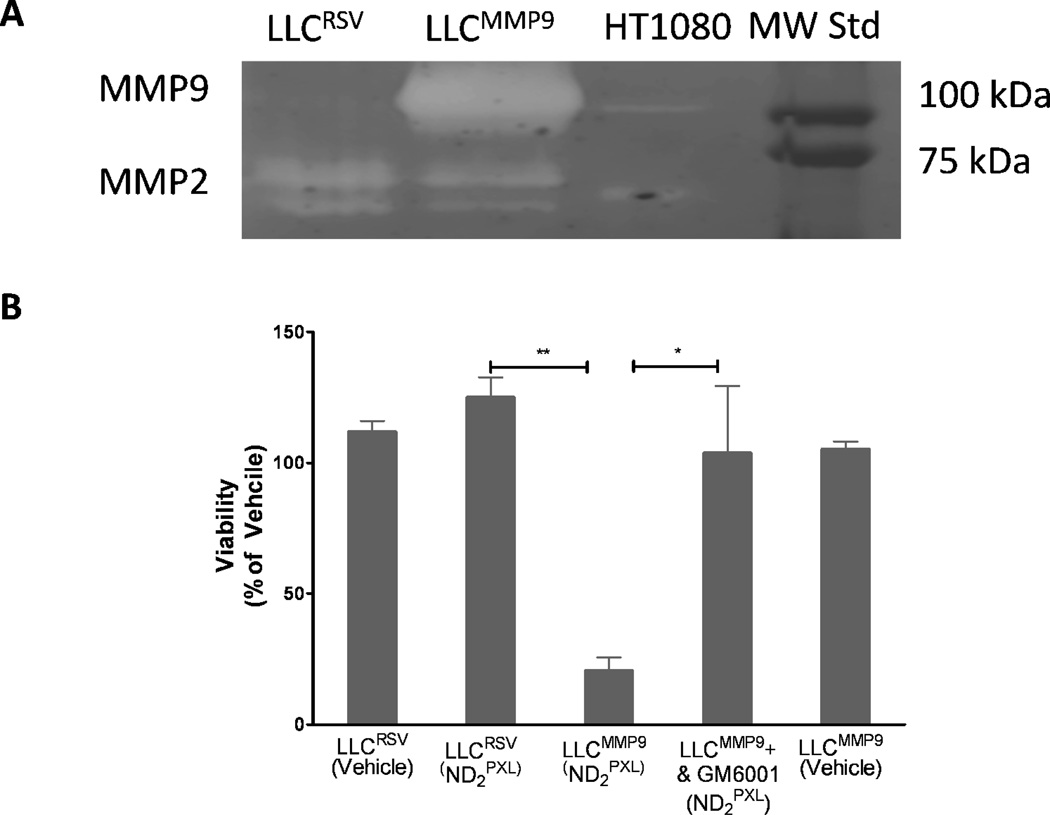
The efficacy of ND2PXL was also tested against two cell lines derived from LLC cells: LLCRSV, that like the parental cell line express very low levels of MMP9, and LLCMMP9, which were transfected to express relatively high levels of MMP9. Since these two cell lines are essentially identical except for the over-expression of MMP9 in the LLCMMP9 line, they were used to assess the effect of MMP9 expression on the in vitro efficacy of ND2PXL. Gelatin zymography (Figure 6A) confirmed that although both LLCRSV and LLCMMP9 express both the pro and active forms of MMP2, significant expression of MMP9 is detected only in conditioned media from LLCMMP9 cells. Both LLCRSV and LLCMMP9 cells were treated with 50 nM ND2PXL; after 48 hours of treatment the LLCMMP9 cells were only 20% viable while the LLCRSV cells were not affected by ND2PXL (Figure 6B) with viability comparable to vehicle-treated cells. LLCRSV cells were sensitive to PXL, other formulations of PXL, including PXL-COOH and MMP9-treated ND2PXL. While PXL, PXL-COOH and MMP9-treated ND2PXL are cytotoxic to LLCRSV cells (50 nM for 48 hours); ND2PXL (not protease treated) exhibit no significant cytotoxicity versus LLCRSV cells (supplemental Figure S20). Thus, expression of MMP9 is required for cytotoxicity of ND2PXL in this cell line.
Figure 6.
ND2PXL cytotoxicity in vitro is dependent on MMP9 activity. A: Zymogram of conditioned media from LLCRSV and LLCMMP9 cells. B: Viability of LLCRSV and LLCMMP9 cells, with low and high MMP9 expression, respectively, and LLCMMP9 cells in the, presence of 10 QM GM6001 and vehicle-treated cells, each after 48 hours of treatment with 50 nM ND2PXL.
The cytotoxic effect of 50 nM ND2PXL on LLCMMP9 cells was abrogated by inclusion, in the culture medium, of GM6001, a broad spectrum MMP inhibitor, with cells remaining viable under the same conditions (Figure 6B). Thus, the cytotoxic effect of ND2PXL is mediated, by proteases that are inhibited by GM6001. When the LLCMMP9 cells are treated with GM6001, the ND2PXL toxicity seen in cells treated with ND2PXL alone disappears, suggesting that the toxicity is due to MMP activity. Since both LLCRSV and LLCMMP9 cell lines express MMP9 (Figure 6A), but there is minimal cytotoxicity of ND2PXL in LLCRSV cells, the activation and cytotoxicity of ND2PXL in LLCMMP9 cells can be attributed to the expression of MMP9 in these cells. LLCRSV with no detectable MMP9 expression remain viable when treated for up to 48 hours with 5 µM of ND2PXL (supplemental Figure S20). The results of the cytotoxicity studies with the four cell lines, three that express MMP9 (MDA-MB-231, R221A-luc and LLCMMP9) and one that does not have detectable MMP9 (LLCRSV) are consistent with ND2PXL being non-toxic until cleaved by MMP9. Therefore, the prodrug formulation, ND2PXL, should be activated in vivo preferentially in MMP9-expressing tissues such as in tumors rather than in healthy tissues that generally express low levels of MMP9. All indications are consistent with ND2PXL being non-toxic until cleaved by MMP9 and therefore, should be activated preferentially in MMP9-expressing tissues, such as tumors rather than healthy tissue, which generally express low levels of MMP9.
Interestingly, cytotoxicity in response to ND2PXL, pretreated with MMP9 to release PXL-[Ahx]-AVRW (see Figure 3C) also evolves more slowly than with PXL of PXL-COOH in both MDA-MB-231 and R221A-luc cells (figures 5A & 5B respectively); which is consistent with either slower uptake and/or additional metabolism of PXL-[Ahx]-AVRW. Regardless of the detailed mechanism for activation of ND2PXL following cleavage with MMP9, the in vitro data presented here provides evidence for the potential of this ND platform to deliver PXL to MMP9-rich tumor microenvironments. Likewise, although the design of the linkage to DOX in ND2DOX appears to preclude the generation of cytotoxic DOX from the compound, the platform has potential to deliver chemotherapeutic compounds selectively to MMP9-enriched microenvironments such as in tumors. In addition, the peptide linker in the ND platform can be readily modified so as to target the drug to be activated by other MMPs or proteases (e.g. cathepsins) not only in tumors, but potentially in other disease settings (e.g. inflammatory disease).
A few protease-activated prodrugs have been reported to successfully reduce tumor volume in vivo and killed cells in vitro [40–42]. Such prodrugs consist of three main components: 1) a protease-cleavable peptide sequence, 2) a chemotherapeutic and 3) a delivery platform. The peptide sequences are specific for the protease being targeted and a number of sequences have been published [26, 27]. Although the sequence AVRWLLTA has been reported as a good substrate for MMP9 with selectivity versus MMP2, no protease-activated prodrug formulations have been described using this peptide linker. MMP9-targeted protease-activatable prodrugs have used either DOX or methotrexate with a somewhat improved efficacy in vitro and in vivo compared to their unconjugated counterparts [40, 43, 44]; however, these are the first reports of protease-activated drugs containing PXL to our knowledge. The drug carrier systems utilized by others include a polymer or another large nanoparticle, all of which are polydisperse in size and functionalization and not the defined molecules reported here. The ND systems presented here, are designed with an MMP9-sequence not previously reported in any prodrugs and utilizing PXL rather than chemotherapeutics that have been reported elsewhere. Limiting the specificity of the peptide to MMP9 was based on the general drug discovery principle of trade-offs between selectivity and efficacy; more selective agents have fewer off-target side effects but may sacrifice some degree of efficacy. In the ND platform both the peptide sequence and the drug can be exchanged with others to optimize the selectivity of activation and therapy for the specific need such as efficacy of delivery depending on the specific therapeutic target. The NDs are unique among drug carriers in that they provide a platform that is size tunable, can be constructed and purified as a well-defined molecule and have the ability to be attached to further moieties, making the ND multi-functional.
The MMP9-activatable NDs reported here were synthesized by bridging an MMP9-selective peptide between PXL (or DOX) and the ND scaffold. Using ND2PXL (or ND2DOX) and MMP9-treated ND2PXL or ND2DOX releasing PXL-[Ahx]-AVRW or DOX-[Ahx]-AVRW respectively, cellular toxicity studies were completed and demonstrate the utility of the NDs for drug delivery. Treatment with intact ND2PXL elicited toxicity only in cell lines that express MMP9 with minimal toxicity in a cell line that has negligible MMP9 expression. ND2PXL was significantly less effective versus LLCRSV as compared with LLCMMP9 cells (i.e. blocking the MMP-activity in R221A-luc and LLCMMP9 cells significantly reduced the toxicity of the ND2PXL). The broad spectrum MMP inhibitor, GM6001, significantly diminished the cytotoxicity of ND2PXL versus the MMP9-expressing cell lines R221A-luc and LLCMMP9. These ND2PXL results highlight the potential of our NDs as activatable drug delivery agents. In addition to producing cytotoxic affects against two breast cancer cell lines R221A-luc and MDA-MB-231), the MMP9 specificity of ND2PXL was confirmed in cells expressing negligible (LLCRSV) and high (LLCMMP9) levels of MMP9 and with the use of an MMP inhibitor to demonstrate the MMP9 is necessary for the toxicity of ND2PXL.
Experimental Procedures
General Methods
MDA-MB-231 cells were purchased from ATCC and cultured in DMEM, 10% FBS and gentamycin with 5% CO2. R221A-Luc cells were [24] cultured in DMEM without L-glutamine, 10% FBS, gentamycin (50 µg/mL) and puromycin (10 µg/mL) with 5% CO2. LLCRSV and LLCMMP9 cells [45] were cultured in DMEM, 10% FBS and gentamycin with 5% CO2. The custom ordered peptides were purchased from Genscript™ (Piscataway, NJ). The enzymes were purchased from Calbiochem (Darmstadt, Germany). All other chemicals were reagent grade and purchased from Fisher Scientific (Pittsburgh, PA) or Aldrich Chemical (St. Louis, MO) companies and used as is unless otherwise indicated.
NDPXL Synthesis with peptide coupling
ND2 was synthesized and characterized according to published methods [32]. Gly residues were attached to all surface hydroxyls by reaction with excess Boc-Gly, 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDCI), diisopropyl ethyl amine (DIEA) in dimethyl formamide (DMF) (Reaction A, Figure 1A). Product, 2, was purified on silica gel with a hexane:ethyl acetate gradient on the Biotage SP1 system. The Boc protecting groups were removed just prior to conjugation to provide primary amines. The ND2 (3) was reacted with Fmoc-[Ahx]-AVRWLLTA-[Ahx]-COOH that had been activated with EDCI (1.2 mole equivalents) and DIEA (3.6 mole equivalents) in DMF for 30 min. The reaction was complete after overnight stirring, yielding compound 4. The Fmoc group was removed with piperidine followed by conjugation of PXL-COOH [35] using the same EDCI coupling methodology. Experimental details can be found in the supplemental information.
ND2Pep(5), ND2DOX (6) and ND2PXL (7)
These compounds were synthesized by standard peptide coupling methods, purified using preparatory HPLC (C18) and characterized with SEC, MS and NMR. See supplemental for experimental details of each compound.
Cleavage of Peptide compounds with Proteases
The compound to be tested for cleavage by protease was dissolved in methanol at 10 mg/mL; aliquots, usually 100 ul were diluted to 1 mg/mL with tricine buffer. A 10 µL aliquot of solution was injected into the HPLC (C18 silica with 55% 0.05% TFA in Acetonitrile/ 45% 0.065% TFA in water) for a zero time point. Following HPLC, 100 ng of enzyme (MMP9 or Trypsin) was added to the reagent solution and the reaction mixture was incubated at 37 °C overnight. Again, analytical HPLC was used to assess the percentage of the sample had been cleaved. For samples which produced a solid after protease treatment, the solid was collected, dissolved in 100% methanol and injected into the HPLC. The samples were then characterized with ESI and MALDI-TOF MS to determine the cleavage products. Precipitated solids, when present, contained the cleaved product.
Cytotoxicity studies
Cells were plated at 40,000 – 60,000 cells/mL into 24 well plates (0.5 mL/well) in appropriate culture medium. A separate plate was used for each time point measured in the assay (i.e. four plates were used to measure 4 time points). The cells were incubated overnight at 37 °C. After examination under the microscope to ensure the cells were attached and healthy, the media was aspirated from each well and replaced with media containing the drug, ND or vehicle; solutions were prepared in media immediately prior to use. The cells were incubated for up to 96 hours, replacing the drug-media solutions every 24 hours with samples being collected/and completed using the trypan blue exclusion assay. For typsinization, the media was removed by aspiration and each well was washed with 0.5 mL of Dulbecco's phosphate-buffered saline (DPBS). The DPBS was removed and each well was treated with 200 µl of trypsin. The plate was incubated for 10 - 15 minutes and then agitated, either with a shaker or by hand. Finally, 40 µl of trypan blue (100 µg/mL) was added to each well (typically 4 wells at a time) and the cells were counted with a hemocytometer (total cells and live cells). The dosing and counting was continued for the remaining time points (up to 96 hours) with the cells given fresh media (with or without drug) every 24 hours until counted.
Zymography
Conditioned media was collected from plates of confluent cells after incubation for 48 hours with serum free media. Equal protein concentrations were determined using the BCA assay (Pierce). Equal amounts of protein were loaded on 10% SDS-polyacrylamide gels containing 0.1 mg/ml gelatin and run at 100 volts for 4 hours in non-reducing conditions. After electrophoresis, gels were washed with 2.5 % Triton-X twice for 15 minutes and incubated at 37°C in substrate buffer (50mM Tris-HCL, pH 7.5, 10 mM CaCl2) overnight. A control gel was incubated in substrate buffer that contained EDTA. Gels were stained with 0.5% Coomassie blue, 50% methanol, 10% acetic acid and destained in 50% methanol, 10% acetic acid.
Supplementary Material
Acknowledgments
This work was supported in part by Susan G. Komen for the Cure® Grant KG090434, NSF IGERT, 0333392, DOD BCRP, BC073450 and NCI 2R25CA092043.
Footnotes
Supporting Information: Detailed synthetic procedures, characterization data (NMRs, MS spectra, etc.) and additional cytotoxicity data is available in the supporting information. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Kratz F, Elsadek B. Clinical impact of serum proteins on drug delivery. J Control Release. 2012;161(2):429–445. doi: 10.1016/j.jconrel.2011.11.028. [DOI] [PubMed] [Google Scholar]
- 2.Elsadek B, Kratz F. Impact of albumin on drug delivery--new applications on the horizon. J Control Release. 2012;157(1):4–28. doi: 10.1016/j.jconrel.2011.09.069. [DOI] [PubMed] [Google Scholar]
- 3.Wolinsky JB, Colson YL, Grinstaff MW. Local drug delivery strategies for cancer treatment: gels, nanoparticles, polymeric films, rods, and wafers. J Control Release. 2012;159(1):14–26. doi: 10.1016/j.jconrel.2011.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bremer C, Tung CH, Weissleder R. Molecular imaging of MMP expression and therapeutic MMP inhibition. Acad Radiol. 2002;9(Suppl 2):S314–S315. doi: 10.1016/s1076-6332(03)80214-3. [DOI] [PubMed] [Google Scholar]
- 5.McIntyre JO, Matrisian LM. Molecular imaging of proteolytic activity in cancer. Journal of Cellular Biochemistry. 2003;90(6):1087–1097. doi: 10.1002/jcb.10713. [DOI] [PubMed] [Google Scholar]
- 6.McIntyre JO, et al. Development of a novel fluorogenic proteolytic beacon for in vivo detection and imaging of tumour-associated matrix metalloproteinase-7 activity. Biochem J. 2004;377(Pt 3):617–628. doi: 10.1042/BJ20030582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lebel R, et al. Novel Solubility-Switchable MRI Agent Allows the Noninvasive Detection of Matrix Metalloproteinase-2 Activity In Vivo in a Mouse Model. Magnetic Resonance in Medicine. 2008;60(5):1056–1065. doi: 10.1002/mrm.21741. [DOI] [PubMed] [Google Scholar]
- 8.Scherer RL, McIntyre JO, Matrisian LM. Imaging matrix metalloproteinases in cancer. Cancer Metastasis Rev. 2008;27(4):679–90. doi: 10.1007/s10555-008-9152-9. [DOI] [PubMed] [Google Scholar]
- 9.Scherer RL, et al. Optical imaging of matrix metalloproteinase-7 activity in vivo using a proteolytic nanobeacon. Mol Imaging. 2008;7(3):118–131. [PMC free article] [PubMed] [Google Scholar]
- 10.Jastrzebska B, et al. New Enzyme-Activated Solubility-Switchable Contrast Agent for Magnetic Resonance Imaging: From Synthesis to in Vivo Imaging. Journal of Medicinal Chemistry. 2009;52(6):1576–1581. doi: 10.1021/jm801411h. [DOI] [PubMed] [Google Scholar]
- 11.Jastrzebska B, et al. Monitoring of MMPs activity in vivo, non-invasively, using solubility switchable MRI contrast agent. Peptides for Youth. 2009;611:453–454. doi: 10.1007/978-0-387-73657-0_196. [DOI] [PubMed] [Google Scholar]
- 12.McIntyre JO, Scherer RL, Matrisian LM. Near-infrared optical proteolytic beacons for in vivo imaging of matrix metalloproteinase activity. Methods Mol Biol. 2010;622:279–304. doi: 10.1007/978-1-60327-299-5_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Achilefu S. Rapid response activatable molecular probes for intraoperative optical image-guided tumor resection. Hepatology. 2012;56(3):1170–1173. doi: 10.1002/hep.25807. [DOI] [PubMed] [Google Scholar]
- 14.Akers WJ, et al. Detection of MMP-2 and MMP-9 activity in vivo with a triple-helical peptide optical probe. Bioconjug Chem. 2012;23(3):656–663. doi: 10.1021/bc300027y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, et al. NIR fluorophore-hollow gold nanosphere complex for cancer enzyme-triggered detection and hyperthermia. Peptides for Youth. 2013;765:323–328. doi: 10.1007/978-1-4614-4989-8_45. [DOI] [PubMed] [Google Scholar]
- 16.Coussens LM, Fingleton B, Matrisian LM. Matrix metalloproteinase inhibitors and cancer: trials and tribulations. Science. 2002;295(5564):2387–2392. doi: 10.1126/science.1067100. [DOI] [PubMed] [Google Scholar]
- 17.Liotta LA, et al. Metastatic potential correlates with enzymatic degradation of basement membrane collagen. Nature. 1980;284(5751):67–68. doi: 10.1038/284067a0. [DOI] [PubMed] [Google Scholar]
- 18.McCawley LJ, Matrisian LM. Tumor progression: defining the soil round the tumor seed. Curr Biol. 2001;11(1):R25–R27. doi: 10.1016/s0960-9822(00)00038-5. [DOI] [PubMed] [Google Scholar]
- 19.Martin MD, Matrisian LM. Matrix metalloproteinases as prognostic factors for cancer. Clinical Lab. Invest. 2005;28:16–18. [Google Scholar]
- 20.Martin MD, Matrisian LM. The other side of MMPs: protective roles in tumor progression. Cancer Metastasis Rev. 2007;26(3–4):717–724. doi: 10.1007/s10555-007-9089-4. [DOI] [PubMed] [Google Scholar]
- 21.Acuff HB, et al. Matrix metalloproteinase-9 from bone marrow-derived cells contributes to survival but not growth of tumor cells in the lung microenvironment 1. Cancer Research. 2006;66(1):259–266. doi: 10.1158/0008-5472.CAN-05-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorden DL, et al. Resident stromal cell-derived MMP-9 promotes the growth of colorectal metastases in the liver microenvironment. Int J Cancer. 2007;121(3):495–500. doi: 10.1002/ijc.22594. [DOI] [PubMed] [Google Scholar]
- 23.Martin M, et al. Effect of ablation or inhibition of stromal matrix metalloproteinase-9 on lung metastasis in a breast cancer model is dependent on genetic background. Cancer Res. 2008;68(15) doi: 10.1158/0008-5472.CAN-08-0537. E-Pub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin MD, et al. Effect of ablation or inhibition of stromal matrix metalloproteinase-9 on lung metastasis in a breast cancer model is dependent on genetic background. Cancer Res. 2008;68(15):6251–6259. doi: 10.1158/0008-5472.CAN-08-0537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan CH, Boyd DD. Regulation of matrix metalloproteinase gene expression. Journal of Cellular Physiology. 2007;211(1):19–26. doi: 10.1002/jcp.20948. [DOI] [PubMed] [Google Scholar]
- 26.Kridel SJ. Substrate hydrolysis by matrix metalloproteinase-9. J Biol Chem. 2001;276(23):20572–20578. doi: 10.1074/jbc.M100900200. [DOI] [PubMed] [Google Scholar]
- 27.Chen EI, et al. A residue in the S2 subsite controls substrate selectivity of matrix metalloproteinase-2 and matrix metalloproteinase-9. J Biol Chem. 2003;278(19):17158–17163. doi: 10.1074/jbc.M210324200. [DOI] [PubMed] [Google Scholar]
- 28.Singal PK, Iliskovic N. Doxorubicin-induced cardiomyopathy. N Engl J Med. 1998;339(13):900–905. doi: 10.1056/NEJM199809243391307. [DOI] [PubMed] [Google Scholar]
- 29.Airoldi M, et al. Clinical activity and cardiac tolerability of non-pegylated liposomal doxorubicin in breast cancer: a synthetic review. Tumori. 2011;97(6):690–692. doi: 10.1177/030089161109700602. [DOI] [PubMed] [Google Scholar]
- 30.Slamon D, et al. Adjuvant trastuzumab in HER2-positive breast cancer. N Engl J Med. 2011;365(14):1273–1283. doi: 10.1056/NEJMoa0910383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gradishar WJ, et al. Phase III trial of nanoparticle albumin-bound paclitaxel compared with polyethylated castor oil-based paclitaxel in women with breast cancer. Journal of Clinical Oncology. 2005;23(31):7794–7803. doi: 10.1200/JCO.2005.04.937. [DOI] [PubMed] [Google Scholar]
- 32.Malkoch ME, Malmstrom E, Hult A. Rapid and efficient synthesis of aliphatic ester dendrons and dendrimers. Macromolecules. 2002;35(22):8307–8314. [Google Scholar]
- 33.Horton DPW, Varela O. Synthesis and antitumor activity of 3'-deamino-3'-hydroxydoxorubicin. A facile procedure for the preparation of doxorubicin analogs. The Journal of Antibiotics. 1984;37(8):853–858. doi: 10.7164/antibiotics.37.853. [DOI] [PubMed] [Google Scholar]
- 34.Gabathuler QCDASJCR. Efficient One-Pot Synthesis of Doxorubicin Conjugates Through Its Amino Group to Melanotransferrin P97. Synthetic Communications. 2003;33(14):2401–2421. [Google Scholar]
- 35.Majoros IJ, et al. PAMAM dendrimer-based multifunctional conjugate for cancer therapy: synthesis, characterization, and functionality. Biomacromolecules. 2006;7(2):572–579. doi: 10.1021/bm0506142. [DOI] [PubMed] [Google Scholar]
- 36.Arcomone FCA, Cassinellie G, Di Marco A, Penco S. Doxorubicin and related compounds. II. Structure-activity consideration. Developments in oncology. 1982;10:179–189. [Google Scholar]
- 37.Albright CF, et al. Matrix metalloproteinase-activated doxorubicin prodrugs inhibit HT1080 xenograft growth better than doxorubicin with less toxicity. Mol Cancer Ther. 2005;4(5):751–760. doi: 10.1158/1535-7163.MCT-05-0006. [DOI] [PubMed] [Google Scholar]
- 38.Nasrolahi Shirazi A, et al. Design and biological evaluation of cell-penetrating Peptide-Doxorubicin conjugates as prodrugs. Mol Pharm. 2013;10(2):488–499. doi: 10.1021/mp3004034. [DOI] [PubMed] [Google Scholar]
- 39.Rudolph-Owen LA, et al. The matrix metalloproteinase matrilysin influences early-stage mammary tumorigenesis. Cancer Res. 1998;58(23):5500–5506. [PubMed] [Google Scholar]
- 40.Kline T, et al. Novel antitumor prodrugs designed for activation by matrix metalloproteinases-2 and-9. Mol Pharm. 2004;1(1):9–22. doi: 10.1021/mp0340183. [DOI] [PubMed] [Google Scholar]
- 41.Law B, Tung CH. Proteolysis: a biological process adapted in drug delivery, therapy, and imaging. Bioconjug Chem. 2009;20(9):1683–1695. doi: 10.1021/bc800500a. [DOI] [PubMed] [Google Scholar]
- 42.Choi KY, et al. Protease-activated drug development. Theranostics. 2012;2(2):156–178. doi: 10.7150/thno.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chau Y, Tan FE, Langer R. Synthesis and characterization of dextran-peptide-methotrexate conjugates for tumor targeting via mediation by matrix metalloproteinase II and matrix metalloproteinase IX. Bioconjug Chem. 2004;15(4):931–941. doi: 10.1021/bc0499174. [DOI] [PubMed] [Google Scholar]
- 44.Chau Y, et al. Investigation of targeting mechanism of new dextran-peptide-methotrexate conjugates using biodistribution study in matrix-metalloproteinase-overexpressing tumor xenograft model. J Pharm Sci. 2006;95(3):542–551. doi: 10.1002/jps.20548. [DOI] [PubMed] [Google Scholar]
- 45.Acuff HB, et al. Matrix metalloproteinase-9 from bone marrow-derived cells contributes to survival but not growth of tumor cells in the lung microenvironment. Cancer Res. 2006;66(1):259–266. doi: 10.1158/0008-5472.CAN-05-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.