Abstract
DREB1/CBF genes, known as major regulators of plant stress responses, are rapidly and transiently induced by low temperatures. Using a Yeast one Hybrid screening, we identified a putative Phytochrome-Interacting bHLH Factor (OsPIF14), as binding to the OsDREB1B promoter. bHLH proteins are able to bind to hexameric E-box (CANNTG) or N-box (CACG(A/C)G) motifs, depending on transcriptional activity. We have shown that OsPIF14 binds to the OsDREB1B promoter through two N-boxes and that the flanking regions of the hexameric core are essential for protein-DNA interaction and stability. We also showed that OsPIF14 down-regulates OsDREB1B gene expression in rice protoplasts, corroborating the OsPIF14 repressor activity observed in the transactivation assays using Arabidopsis protoplasts. In addition, we showed that OsPIF14 is indeed a Phytochrome Interacting Factor, which preferentially binds to the active form (Pfr) of rice phytochrome B. This raises the possibility that OsPIF14 activity might be modulated by light. However, we did not observe any regulation of the OsDREB1B gene expression by light under control conditions. Moreover, OsPIF14 gene expression was shown to be modulated by different treatments, such as drought, salt, cold and ABA. Interestingly, OsPIF14 showed also a specific cold-induced alternative splicing. All together, these results suggest the possibility that OsPIF14 is involved in cross-talk between light and stress signaling through interaction with the OsDREB1B promoter. Although in the absence of stress, OsDREB1B gene expression was not regulated by light, given previous reports, it remains possible that OsPIF14 has a role in light modulation of stress responses.
Keywords: Rice phytochrome-interacting factor 14 (OsPIF14), OsDREB1B, protein-DNA interaction, alternative splicing, light regulation, cold stress
1. Introduction
Plant growth and development are extremely influenced by environmental conditions. Abiotic stresses such as cold, drought and salinity are responsible for major losses in crop yield worldwide. In response to these environmental factors, plants have evolved mechanisms in order to cope with extreme conditions, such as the production of osmoprotectants and regulatory proteins involved in signaling pathways1,2. Among the latter, transcription factors (TFs) play a very important role in response to these stresses, since they can regulate the expression of many genes by binding to specific cis-acting elements in the promoter regions. A single TF can therefore have a major effect in the response to a specific stimulus.
The Dehydration-Responsive Element-Binding 1/C-Repeat-Binding Factor (DREB1/CBF) TFs belong to the AP2/ERF family and have been described as being rapidly and transiently induced by low temperature3–5. When present in the cell nucleus, DREB1 proteins bind to a conserved cis-motif, the Dehydration-Responsive Element/C-Repeat (DRE/CRT), present in the promoter region of stress-inducible genes, thus regulating their transcription4,5. DREB1 proteins were initially identified in Arabidopsis5,6, but homologues have been identified in several other plants, such as rice3, maize7, Eucalyptus8, grape9, cotton10 and almond11, which is illustrative of their relevance in plant stress responses. In rice, OsDREB1A to OsDREB1G have been identified as homologues of the Arabidopsis DREB1/CBFs3,12,13. Our work focuses on the regulation of OsDREB1B, that was initially described as highly and specifically induced in response to cold3. More recently, this gene has been shown to respond to other stresses, such as drought, oxidative and mechanical stress as well as to ABA and salicylic acid14,15. The over-expression of this gene in rice provided increased tolerance to cold, drought and salinity16, whereas in tobacco it led to increased tolerance to oxidative and freezing stress, as well as to infection by tobacco streak virus14.
Temperature and light signaling have been previously described to cross-talk17, and phytochrome signaling in particular was described as a regulator of DREB1/CBF expression in Arabidopsis18–20. Phytochromes are photosensitive chromoproteins that can reversibly interconvert between two different forms: the inactive red-light absorbing Pr and the active far-red light absorbing Pfr21. In Arabidopsis there are five genes that code for phytochromes (phyA to phyE), whereas in rice there are three members (phyA to phyC), which function as the only photoreceptors to perceive red and far-red light22. Upon activation by red light, the Pfr active form of phytochromes migrates into the nucleus, where it interacts with TFs of the basic helix-loop-helix family (bHLH), referred to as Phytochrome Interacting Factors (PIFs21). This interaction usually results in a proteasome-dependent degradation of the PIFs, modulating the expression of genes regulated by PIFs. This regulatory mechanism was observed for example for PIF123, PIF324 and PIF525, but it does not seem to be the case of the more recently identified PIF7, because even though it co-localizes with phyB in nuclear speckles after a red light pulse, this protein appears to be light-stable26. Additionally, a set of putative PIFs has also been described in rice27, but so far the interaction between these proteins and the rice phytochromes is yet to be shown, as well as their stability under light/dark conditions.
The animal bHLH proteins are typically classified into six major groups (group A to group F) depending on its basic domain and consequent DNA cis-element binding. Animal group A is associated with bHLH activators, which recognize E-box sequences (CAGCTG or CACCTG). Animal group B seems to be the closest to the bHLH ancestor and is also found in plants and yeast. The basic domain of group B bHLHs are characterized for having an arginine at position 13 (R13), which is crucial to bind to hexameric DNA sequences with a guanine at position 4 (e.g. G-box - CACGTG). Additionally, a leucine zipper can be found only in this group, however it is not present in all group B bHLH proteins. Animal group C represents the bHLH-PAS proteins, which are characterized by a PAS domain and binding to ACGTG or GCGTG core motifs, but they are not found in plants. Group D animal bHLHs are characterized by a lack of the basic domain, thus being unable to bind DNA. Group E animal bHLHs bind to N-boxes (e.g. CACGCG or CACGAG), which are cis-elements associated with bHLHs with repressor activity. Additionally, these bHLHs have a proline in their basic domain and a WRPW sequence in their C-termini. Moreover, since group E factors also have R13 in their basic domain these proteins can bind to group B cis-responsive elements and the other way around, however with less affinity. Animal group F bHLHs are called COE-bHLH and are not found in plants. Additionally, in 2010 plant bHLH proteins were clustered in 26 subfamilies based on phylogenetic analyses. For better characterization of bHLH proteins read28–32.
In our work, we focused on the transcriptional regulation of OsDREB1B. We have previously identified seven Zn Finger TFs as binding to the promoter of OsDREB1B, in a Yeast One-Hybrid (Y1H) screening15. Here, we report the identification and characterization of another TF binding to the promoter of OsDREB1B. This new TF was previously reported as Phytochrome Interacting factor 3 – Like 14 (OsPIL14)27, which belongs to the bHLH protein family and is predicted to be a putative PIF. Here, we have shown that OsPIL14 interacts with the photoactivated form of phyB and therefore named it OsPIF14. We have also studied OsPIF14 gene response to different abiotic stresses, its transcriptional activity, and characterized the OsPIF14 interaction with the respective cis-element present in the OsDREB1B promoter.
2. Materials and Methods
2.1 Cold–induced cDNA expression library
The cDNA expression library was prepared as previously described15. Briefly, eight-day-old rice seedlings (Oryza sativa L. cv. Nipponbare) grown at 28°C and 12h/12h photoperiod, were subjected to a 5°C treatment. Whole seedlings were sampled after 2h, 5h, and 24h of cold treatment and the RNA extracted was used to construct the cDNA library following the manufacturer instructions (HybriZAP-2.1 XR Library Construction Kit (Stratagene)).
2.2 Yeast One-Hybrid
The OsDREB1B promoter fragment used as bait for the Yeast One-Hybrid screening ranged from −488bp to −3bp counting from the ATG start codon and was isolated by PCR using the primers 5′-ATGCGGCCGCTCGGAGTAACACTCGTGCAG-3′ and 5′-GGACTAGTTGACTCTCTCTGGTTCACTTCG-3′ (underlined sequences represent adaptors with restriction enzyme sites). This fragment was cloned as a SpeI-NotI fragment in the pHIS3/pINT1 vector system33,34 and integrated into yeast strain Y187 (Clontech). This bait strain was then transformed with the rice cold-induced cDNA expression library. Over one million yeast colonies were screened in CM-His− medium supplemented with 5mM 3-amino-1,2,4-triazole, as described33. The plasmids from the yeast clones that actively grew on selective medium were extracted and the cDNA insert sequenced. These sequences were used to search for homology in the rice genome, using the BLAST algorithm. Plasmids containing genes encoding transcription factors were re-transformed into the respective bait strain, to confirm activation of the HIS3 reporter gene.
To divide OsDREB1B promoter (−488 to −3bp from ATG) in two different baits we isolated both promoter sequences by PCR combining the primers described below and the new pair of primers 5′-GGACTAGTTGCTGCTGCTACTCCAGCTT-3′ and 5′-ATGCGGCCGCCCAAAAACCCAACAGAAACC-3′. Fragments were cloned as described below.
For the direct Y1H, we used the identified Y1H clone or the full coding sequence of the OsPIF14 gene, depending on the situation. The full coding sequence of the OsPIF14 gene was cloned into vector pAD-WT (Stratagene), by replacement of the coding region of the wild-type lambda cI, fragment C, downstream of the GAL4 activation domain, using EcoRI and PstI. The yeast bait strains harboring the OsDREB1B promoter fragments ranging from −1945 to −388bp have been described elsewhere15.
2.3 Abiotic Stress Treatments
Rice seedlings were grown hydroponically in nutritive medium35 at 28°C, 700μmol fotons.m−2.s−1, 70% humidity and 12h/12h photoperiod for 14 days. The seedlings were then transferred to stress conditions 4h after the start of the light period. At the same time, control seedlings were transferred to fresh nutritive medium (mock control). Temperature treatments were performed by transferring the seedlings to growth chambers at the desired temperature in pre-cooled, or pre-warmed, medium. For salt and ABA treatments, seedlings were transferred to nutritive media supplemented with 200mM NaCl or 100μM ABA, respectively. Drought treatment was performed by maintaining the seedlings over dry absorbent paper. All other conditions were maintained throughout the assays. For semi-quantitative RT-PCR analysis, time points consisting of ten plants were sampled (roots and shoots separately). Arabidopsis thaliana ecotype Col-0 seeds were vernalized for 4 days at 4°C in the dark and then germinated at 22°C on MS plates (MS basal salts, 0.05% MES buffer, 1% sucrose, pH 5.7, 0.6% agar) in a 16h/8h photoperiod. For the cold treatment assay, 10-day-old seedlings were transferred to 5°C 4h after the start of the light period, and kept there for 1 or 4 hours. Ten whole seedlings were collected at each time point.
2.4 Semi-quantitative RT-PCR and RT-qPCR
Total RNA from both rice and Arabidopsis seedlings was extracted using the RNeasy Plant Mini kit (Qiagen). For semi-quantitative RT-PCR analysis, first strand cDNA was synthesized from 1μg total RNA, using an oligo-dT primer and the SuperscriptII reverse transcriptase (Invitrogen) following the manufacturer’s instructions. The cDNA was then amplified by PCR using gene-specific primers (See Table S1). For rice, ACTIN1 (Os03g50885) was used as an internal control for all experiments, except for shoots in the drought assay and roots at 10°C, where EUKARYOTIC ELONGATION FACTOR 1-α (Os03g08060) and UBIQUITIN-CONJUGATING ENZYME E2 (UBC2; Os02g42314) were used, respectively. Besides a biological replicate, measurements were performed in triplicate for each time point and efficiency curves were prepared in duplicate.
For quantitative PCR analysis, RNA extraction was conducted as described above. First strand cDNA synthesis was performed using 4μg total RNA with an oligo-dT primer using Transcriptor High Fidelity cDNA Synthesis Kit (Roche), according to the manufacturer’s instructions. The cDNA was then amplified by PCR using gene-specific primers (See Table S2). Ubiquitin-conjugating enzyme E2 was used as internal control. Real Time PCR was done in a Lightcycler 480 (Roche), using Lightcycler 480 Master I Mix (Roche). Determinations were performed in triplicate for each time point and efficiency curves in duplicate. Relative expression levels were determined by Roche’s E-method using the Lightcycler 480 software.
Total RNA from rice protoplasts was extracted using the Direct-zol kit from Zymo. 500ng of RNA were used to synthesize cDNA using oligo-dT primer and the Superscript III reverse transcriptase (Invitrogen) following the manufacturer’s instructions. The quantitative PCR analysis was performed as described above.
2.5 Rice protoplasts transformation
Rice protoplasts were prepared from etiolated wild-type seedlings (7 DAG), using an protocol previously described by our group36. The protoplasts were transformed with 10ug of OsPIF14::HA, cloned into the pHBT95 plasmid to construct the 35S::OsPIF14::HA cassette. Water transformed protoplasts were used as control. After transformation protoplasts were incubated in 24 well plate under dark, at room temperature for 18h.
2.6 Transactivation assay
The effector plasmid was built by recombining the full coding sequences of gene OsPIF14 into vector pH7WG2 (VIB-Ghent University), to be under the control of the full CaMV35S promoter. The reporter plasmid used was the pLUCm35GUS15. The OsDREB1B promoter region used in the Y1H was cloned upstream of the minimal 35S promoter, as a SalI fragment, to drive the expression of the GUS gene. The reporter plasmid was confirmed by restriction analysis and sequencing.
Arabidopsis protoplasts were prepared as described37. For each independent transformation, 5μg of reporter plasmid and 10μg of effector plasmid were used. Each transformation was performed in triplicate. Cells were incubated for 24h at 22°C in the dark and then collected at 450g for 1min in a swing-out rotor. Cell lysis and determination of GUS and Luciferase levels was performed as described15. Readings were performed in triplicate. Activation of gene expression was calculated as a GUS/LUC ratio.
2.7 Yeast-two hybrid assay
The full coding sequence of OsPIF14 was cloned into vector pAD-WT, as described in the Y1H section. The sequences coding for the C-terminal non-photoactive regions of rice phytochromes A, B and C were cloned in vector pDONR221 (Invitrogen), according to the manufacturer’s instructions. The cDNA sequences that were used encoded a.a. 620–1129 for OsphyA (Os03g0719800), 654–1172 for OsphyB (Os03g0309200) and 620–1138 for OsphyC (Os03g0752100). These sequences were then recombined into vector pBD-GW. This vector was obtained by cloning a Gateway (GW) cassette into plasmid pBD-GAL4 Cam (Stratagene). Sense orientation and translational fusion between GAL4-BD and PHY encoding genes were confirmed by restriction digestion and sequencing, respectively.
The bait and prey plasmids were transformed into yeast strain AH109 (Stratagene), as described33. Yeast colonies were plated on CM-leu-trp-his media and growing colonies were confirmed by PCR.
2.8 Bimolecular Fluorescence Complementation
The OsPIF14 coding region was cloned into the vector YFPN43, to be in fusion with the N-terminal portion of the Yellow Fluorescent Protein (YFP). The non-photoactive C-terminal portions of the three rice phytochromes were cloned in vector YFPC43, to be in fusion with the C-terminal portion of the YFP. Cloning in YFNC43 and YFCN43 vectors was done according to Gateway technology (Invitrogen). To use as negative control in the interaction assays with OsPIF14, the Arabidopsis SNF1 kinase homolog 10 (Akin10) was tested in fusion with the N-terminal portion of the YFP. These plasmids, together with a construct harboring the silencing suppressor HcPro38, were transformed into Agrobacterium thumefaciens strain LBA4404. Agro-infiltration of tobacco leaves was performed as described by Wydro et al. (2006), with modifications. Briefly, Agrobacterium cultures harbouring the constructs were grown overnight in LB medium supplemented with 150μM acetosyringone. The bacteria were centrifuged and the pellets ressuspended in 10mM MgCl2 150μM acetosyringone and incubated for 2h at RT. The Agrobacterium strains were diluted and combined to a maximum total OD600 of 0.5 (0.2 OD from each of the YFP reporter vectors and 0.1 OD from HcPro) to infiltrate Nicotiana benthamiana leaves. After incubation in the dark for 2 days, the abaxial epidermis of the leaves was detached after gluing on a microscope slide with medical adhesive (Hollister). The samples were observed with a confocal microscope (Leica SP5).
2.9 In vitro co-immunoprecipitation assay
The vectors for this assay were prepared as following: the OsPIF14:GAD was constructed by removing the Arabidopsis PIF7 coding region from plasmid PIF7:GAD26 and replacing it with the OsPIF14 coding region, using NdeI and BamHI. Vectors for in vitro expression of the rice phytochromes were obtained by recombining their full coding sequences from pDONR221 into pDEST17 (Invitrogen). Control constructs GAD:PIF3 and GAD were previously described39. Recombinant proteins were produced in vitro using the TNT Quick Coupled Transcription/Translation System (Promega) in the presence of [35S]Methionine. Preparation of bait and prey was performed as described by Khanna et al. (2004). Light treatments were performed by exposing the samples to 4min of red light (660nm) or 4min of red light followed by 4min of far-red light (750nm). Binding and washes were carried out as described 40, and samples run on 10% SDS-PAGE gels. Gels were dried and signals obtained using a STORM 860 PhosphorImager (Molecular Dynamics).
2.10 Expression and purification of recombinant thioredoxin-tagged OsPIF14
OsPIF14 coding sequence was recombined into pET32a (Novagen, USA), in fusion with a thioredoxin tag. pET32a/OsPIF14 was used to transform Rosetta pLysS for protein production. Cells were grown to an OD600 0.6 and protein expression was induced with 100 μM IPTG and allowed to occur for 4h at 28°C. Cells were harvested by centrifugation (3.500xg, 20 min, 4°C). The bacterial pellet was then resuspended in 20 mM sodium phosphate pH 7.5, 500 mM NaCl, 10 mM imidazole, 250 μM MgCl2, 1.25 mM PMSF, 1x Complete Protease Inhibitor (Roche) and 8 μg/ml DNase. Cells were lysed by enzymatic digestion (Lysozyme 50mg/ml) for 1hour at 4°C with agitation. The lysate was centrifuged for 1 hour at 18.000xg and 4°C. The supernatant was filtered with a 0.45 μm filter and soluble thioredoxin-OsPIF14 protein was purified using the HiTrap IMAC FF system (GE Healthcare). Column-bond protein was eluted with 20 mM sodium phosphate pH 7.5, 500 mM NaCl and 250 mM imidazole. Eluted protein was further purified by size exclusion in 20 mM sodium phosphate pH 7.5, 200 mM NaCl, 1 mM EDTA, 1 mM DTT and 5% glycerol using a HiLoad 16/600 Superdex 200 pg column (GE Healthcare) and samples were stored at −80°C. The thioredoxin tag was produced similarly as described above.
2.11 Electrophoretic Mobility Shift Assay (EMSA)
DNA probes were generated by annealing oligonucleotide pairs, in a PCR machine as described in 41 and the annealing temperatures (Table S3). The OsPIF14-DNA binding reaction was performed in a volume of 10 μL, which contained 10 mM HEPES pH 7.9, 40 mM KCl, 1 mM EDTA pH8, 50 fmol of labeled probe, 1 mM DTT, 50 ng herring sperm DNA, 15 μg BSA and 500 ng of purified thioredoxin::OsPIF14 or 500 ng of purified thioredoxin. The reactions were incubated for 1 hour on ice and loaded onto native 5% polyacrylamide gel (37.5:1). Electrophoresis was run as described by41.
3. Results
3.1 OsPIF14 is a novel bHLH TF that binds to the promoter of OsDREB1B
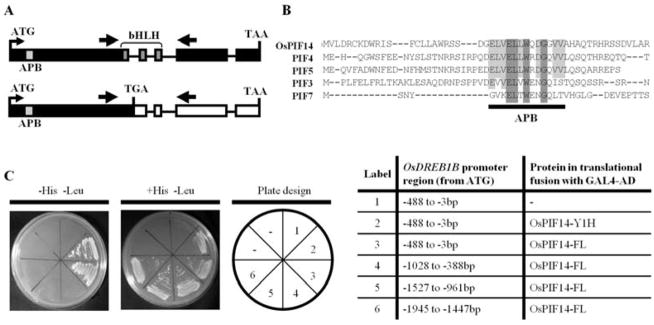
In order to identify TFs that regulate the expression of OsDREB1B, we screened a cold-induced rice cDNA expression library, using as bait four overlapping fragments covering 1945bp upstream the OsDREB1B start codon. In addition to the seven Zinc Finger TFs previously identified as binding to OsDREB1B15, we identified a rice putative Phytochrome Interacting Factor (PIF) binding to the fragment −488bp to −3bp before ATG. Figure 1C shows that PIF binds specifically to this fragment of the OsDREB1B promoter. This TF belongs to the bHLH family and had been previously named Phytochrome Interacting factor 3-Like 14 (OsPIL14) by Nakamura et al. (2007). In 2010, this rice PIL was clustered within the plant bHLH subfamily VII (a+b). These proteins show a phytochrome interaction region at the N-terminal and a domain of unknown function on C-terminal32. The sequence of this gene was differently predicted in two public databases (GenBank reference Os07g0143200 and Rice Genome Annotation Project 2013, reference Os07g05010) and also different from the sequence we isolated. We identified a 1,245bp coding sequence encoding a 414 amino acid-long protein (JN400276), with a predicted molecular weight of 44.49 kDa and a pI of 4.714. This protein has a putative phytochrome B interacting region (active phytochrome binding; APB) at the N-terminal of the protein and a conserved bHLH DNA binding domain, extending from amino acid 222 to 275 (Fig. 1A and 1B). This gene was found to have four introns, two of which within the bHLH coding region. When comparing its predicted APB domain with that of some of the Arabidopsis PIFs (Fig. 1B), several amino acid residues are conserved between all the proteins, but the rice PIL seems to have the highest degree of identity to Arabidopsis PIF4 and PIF5 (85% APB homology; Fig.1B). Since, in this manuscript (Figure 8), we show that OsPIL14 indeed interacts with the photoactivated form of phytochrome B, from now on we will call it OsPIF14. This protein clustered with both AtPIF4 and AtPIF5, together with a rice putative PIF (PIL13)27,42.
Figure 1.
Characterization of the rice phytochrome-interacting factor 14. A, Schematic representation of the OsPIF14 transcript present under control conditions (top) and the alternative splice form that appears under cold stress (bottom). APB - Active Phytochrome Binding motif (light grey box); bHLH - basic Helix Loop Helix domain (dark grey). Thick arrows represent RT-PCR primer locations. Black boxes represent translated exons, white boxes represent non-translated exons and lines represent introns. B, Alignment of the N-terminal a.a. sequence of OsPIF14 with the same region of the Arabidopsis PIF3, PIF4, PIF5, and PIF7, showing the conserved APB domain. Dark shaded boxes show a.a. residues conserved in all the protein sequences, whereas light shaded boxes show residues conserved between OsPIF14 and some of the Arabidopsis PIFs within the APB domain. C, Yeast One-Hybrid assay testing the interaction of OsPIF14 with the promoter of OsDREB1B. Protein coded by the cDNA clone that we identified in the Y1H screen (OsPIF14-Y1H), was tested as well as the full length protein (OsPIF14-FL). The OsPIF14-FL protein was then tested for interactions against several fragments of the OsDREB1B promoter. On the left is the selection plate, showing positive protein-DNA interactions. In the middle panel the yeast growth in medium supplemented with histidine is shown. On the right panel the plate design is shown. The table to the right details the yeast bait strains and the constructs used in each portion of the plates.
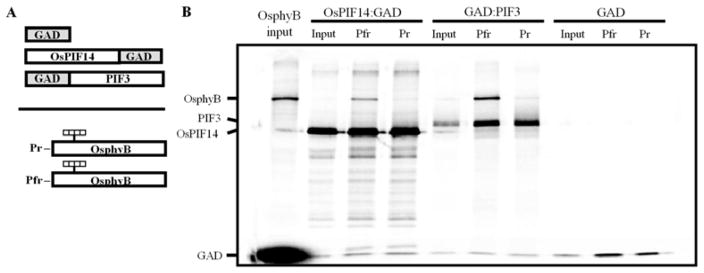
Figure 8.
Analysis of the interaction between OsPIF14 and the Pr and Pfr forms of OsphyB, using a co-immunoprecipitation assay. A, Schematic representation of the protein constructs used for the assay. OsPIF14 was used as bait, in a translational fusion with the GAL4 AD. The GAL4 AD vector was used as a negative control for the interaction, whereas a fusion of GAD with the Arabidopsis PIF3 was used as the positive control. The Pr and Pfr forms of OsphyB were used as prey. B, SDS-PAGE separation of pellet fractions for each interaction and inputs for the proteins used in the assay. Data from a representative experiment is shown.
In order to confirm the binding of OsPIF14 to the promoter of OsDREB1B, we re-transformed the yeast bait strain with the plasmid containing the cDNA clone that we identified in the Y1H screen (Fig. 1C). Moreover, the full length transcript was isolated and cloned in a plasmid in order to be in a translational fusion with the GAL4-Activation Domain (GAD). This construct was also transformed into the yeast bait strain. In both cases, we could see an activation of the HIS3 reporter. We also tested the interaction of the full length OsPIF14 with other regions of the OsDREB1B promoter. As seen in Fig. 1C, the protein only interacted with the promoter region ranging from −488 to −3bp, counting from the ATG start codon. Using direct Y1H, we further tested whether OsPIF14 could also bind to the promoter of OsDREB1A, the other cold-responsive DREB1/CBF gene in rice. In this case, we could not observe an activation of the reporter HIS3 for any of the bait strains (Fig. S1) indicating that the interaction between OsPIF14 and the −488 to −3bp OsDREB1B promoter is specific.
3.2 OsPIF14 binds to OsDREB1B promoter through an extended version of N-BOX
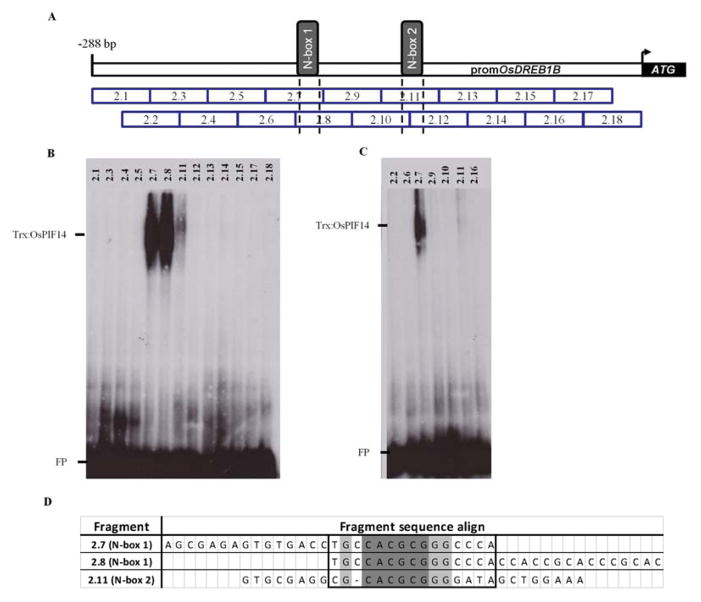
It is described that bHLH TFs can bind to E-box (CANNTG) or N-box (CACG(C/A)G) depending on their transcriptional activity29. In order to identify the specific DNA sequence to which OsPIF14 binds, we analyzed the OsDREB1B promoter bait where OsPIF14 was identified, searching for E-box or N-box cis-elements using the Geneious program. In silico results revealed the presence of two N-boxes (CACGCG), within the 288bp upstream of the start codon of OsDREB1B that we called N-box 1 and N-box 2 (Fig. 2A). To confirm that OsPIF14 binds to these cis-elements, we divided the bait fragment (−488bp to −3bp from ATG) into two fragments (fragment 1: −488bp to −222bp and fragment 2: −288bp to −3bp from ATG) and constructed two yeast baits for direct Y1H. Both yeast bait strains were re-transformed with the plasmid containing the OsPIF14 cDNA clone that we identified in the Y1H screen. The results of direct Y1H, showed that OsPIF14 only binds to fragment 2 (Fig. S2). In addition, we designed 18 overlapping small fragments (double-stranded oligonucleotides with 30bp each) spanning fragment 2 (Fig. 2A) and used them as probes in Electrophoretic mobility shift assays (EMSA) carried out with the recombinant Trx::OsPIF14 protein. We identified three small fragments (2.7, 2.8 and 2.11) in which Trx::OsPIF14 binds (Fig. 2B and 2C). Fragments 2.7 and 2.8 share the same N-box domain (N-box 1; N1) and Trx::OsPIF14 is able to bind to both sequences. The Trx alone do not bind to any of these probes (Fig. S3A). Hence, we can conclude that Trx::OsPIF14 binds within the 2.7/2.8 overlapping DNA sequence (Fig. 2D, square box). Trx::OsPIF14 also binds to N-box 2 (N2) in fragment 2.11, but the interaction seems to be much weaker than that observed for N1. In order to test whether OsPIF14 actually binds to the N-box, we designed a mutated probe for N-box 1 (N1_m2), in which the middle Cytosine and Adenine were changed to an Adenine and Thymine, respectively (Fig. 3C). This double point mutation completely abolished the protein-DNA interaction, showing that Trx::OsPIF14 in fact binds to the N-box and that both nucleotides are crucial for DNA binding (Fig 3A).
Figure 2.
OsPIF14 binds to both OsDREB1B promoter N-box (N-box 1 and 2). A, Schematic representation of the eighteen probes used in Electrophoretic Mobility Shift Assay (EMSA) to screen OsDREB1B promoter. Each probe has 30 bases length in which 15 bases overlap with next probe. N-box 1 and 2 squares represent the sequence (CACGCG). Probes 2.7, 2.8 and 2.11 have intact N-box, while 2.12 lacks the first base of N-box, as represented by dotted lines. Probe 2.7 and 2.8 share the same N-box. B and C, EMSA results of OsDREB1B promoter probes screen showing that OsPIF14 binds to both N-box of OsDREB1B promoter with different intensities. FP, free probe. D, alignment of the three probes to which OsPIF14 binds in EMSA. Square represents the important bases for OsPIF14-DNA binding. Dark shaded bases represent the N-box. Light shaded bases represents the common bases between the three probes.
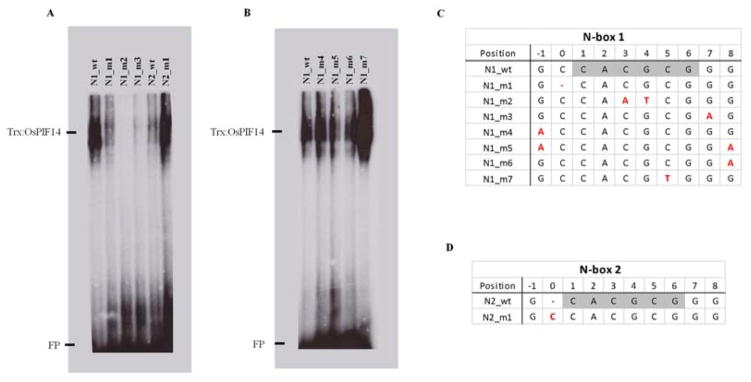
Figure 3.
Flanking region of N-box plays an important role in OsPIF14-DNA binding. A, point mutation on N-box (N1_m2) and first flanking base (N1_m1, N1_m3, N2_m1). B, single or double point mutation on second flanking base (N1_m4, N1_m6, N1_m5, respectively) and point mutation to change N-box to G-box (N1_m7). C and D, represents N-box 1 and 2 (light shaded bases) and the two flanking bases of the wild type (wt) and mutated (m) probes used in EMSA. Red marked bases represents the point mutations (substitutions), (−) represents base deletion or absence (for N1_m1 and N2_wt, respectively). Full probe sequence is represented in Supplemental Table S3. FP, Free Probe
Despite N-box 1 and N-box 2 sharing the same six core nucleotides CACGCG (Fig. 2D), EMSA results showed different signal intensities (Fig. 3A), which could be due to differences in the flanking region (Fig 2D). Comparing the two bases from 5′ and 3′ flanking regions of N-box 1 and N-box 2, they only differ in one Cytosine in the 5′ region (Fig. 2D). In order to investigate the importance of this Cytosine for protein-DNA interaction we did point mutations in both probes (Fig. 3C and D). We first added the lacking 5′ Cytosine to N-box 2 (N2_m1) and removed the corresponding Cytosine from N-box 1 (N1_m1). When the 5′ Cytosine is added (N2_m1) the signal intensity of the mutated probe is similar to the wild type version of N-box 1 (Fig. 3A). On the other hand, when the 5′ Cytosine is removed from N-box 1 (N1_m1) the signal intensity decreases reaching the level of N-box 2 wild type (Fig. 3A). These results show that this Cytosine is not essential, but plays an important role in Trx::OsPIF14-DNA binding strength. In general, cis-elements are composed of an even number of base-pairs. In order to evaluate whether the corresponding nucleotide in the 3′ region of the cis-element is also important, we made a point mutation (N1_m3, Fig. 3C). When we changed this nucleotide from a Guanine to an Adenine, the EMSA signal is much weaker compared to that obtained with the WT probe (N1_wt), thus showing the importance of this nucleotide for the protein-DNA interaction. In order to evaluate whether the subsequent nucleotides were also important for this interaction, we mutated them in the 5′ and 3′ region, individually and simultaneously (N1_m4, N1_m5, N1_m6). The results revealed that these nucleotides are not important for OsPIF14-DNA interaction (Fig. 3B). We can then conclude that OsPIF14 binds to both N-box 1 and N-box 2 of OsDREB1B promoter, though it prefers the extended version (CCACGCGG).
3.3 OsPIF14 has properties of a bHLH Group B protein
In order to further characterize the OsPIF14 bHLH domain, we aligned the basic domain of bHLH proteins belonging to group A, B and E (Fig. 4), as those three groups are the most similar to OsPIF14. Group B is characterized by an arginine at position 13 (R13), which is also present in OsPIF14 (Fig. 4). This R13 confers the capability of binding to cis-elements that have a Guanine at position 4 (G4; Fig 3C/D), a feature shared between N-box and G-box31. As we showed before, OsPIF14 is able to bind to the N-box motif (CACGCG), however we do not know if OsPIF14 is able to bind to the G-box cis-element (CACGTG). In order to test that, we changed the N-box to a G-box in which a Cytosine at position 5 was changed to a Thymine in N-box 1 (Fig. 3C, N1_m7). The EMSA results clearly showed that OsPIF14 is able to bind to G-box (Fig. 3B) and the EMSA signal was stronger for the G-box than for the N-box. In addition, it is reported that bHLH proteins from group B are not able to bind to cis-elements of group A because they lack the G4 (e.g. CACCTG)31. To test this, we used a mutated G-box probe, changing the G4 to a Cytosine (G-box MUT). In this case we observed that OsPIF14 loses its ability to bind to DNA confirming that Guanine at position 4 is important for DNA binding (Fig. S3B).
Figure 4.
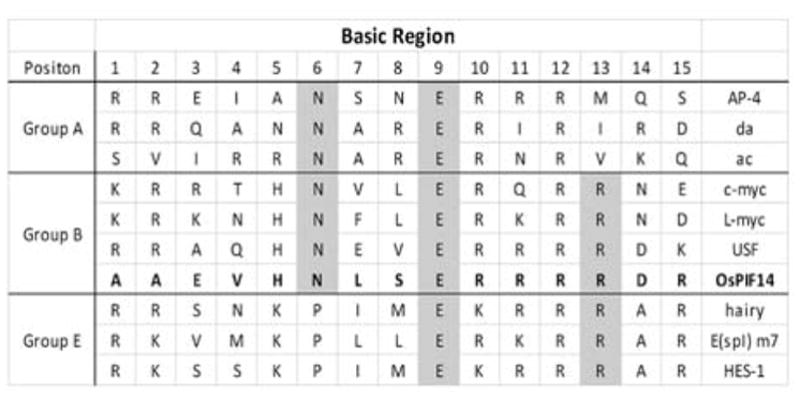
OsPIF14 belongs to Group B bHLH. Alignment of the basic domain of the most related bHLH groups (Group A, B and E). Shaded bases represents the similarities between OsPIF14 and other bHLH groups. Adapted from Ohsako et. al. 1994.
3.4 OsPIF14 is a repressor of OsDREB1B
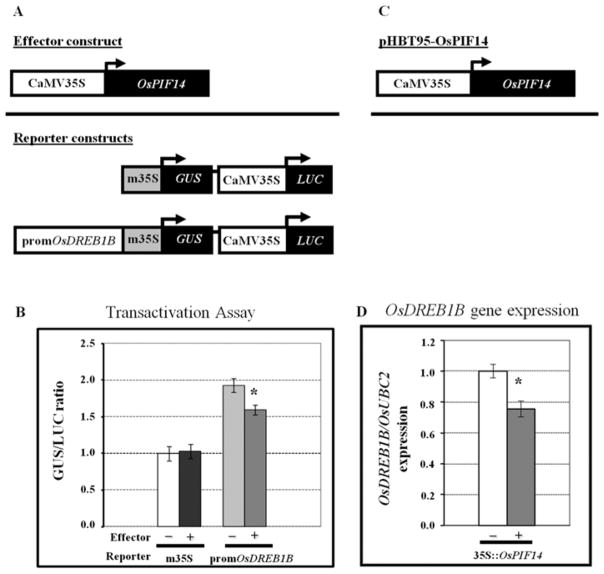
Drosophila bHLH TFs that bind to the N-box motif have been characterized as repressors29. To investigate whether OsPIF14 is a transcriptional repressor or activator, we have performed a transactivation assay in Arabidopsis protoplasts. The full length OsPIF14 cDNA clone was used as the effector, while the GUS gene driven by the same OsDREB1B promoter fragment, used as bait in the Y1H screening, was used as reporter (Fig. 5A). Fig. 5B shows that co-transformation of OsPIF14 with the reporter vector resulted in a statistically significant, albeit modest, decrease in GUS activity, as compared to the reporter vector alone. This decrease was not observed when the reporter did not contain the promoter fragment of OsDREB1B, indicating that the presence of this sequence is necessary for the binding of OsPIF14 and consequent repression of transcription. The OsPIF14 transcriptional repression activity and its binding to OsDREB1B promoter was also tested in vivo in rice protoplasts. The rice protoplasts were transformed with the full OsPIF14 CDS sequence driven by the 35S and the expression of endogenous OsDREB1B was analyzed after 18h of dark incubation (Fig. 5D). The results showed that OsPIF14 over-expression leads to an OsDREB1B down regulation (as compared to non-transformed protoplasts), indicating that OsPIF14 binds to OsDREB1B promoter, thus repressing its gene expression. These results confirmed that OsPIF14 is indeed a repressor and provides further evidence for in vivo binding to the OsDREB1B promoter.
Figure 5.
Analysis of the OsPIF14 transcriptional activity. A, Constructs used for Arabidopsis protoplast transformation (pCambia 1391z). Effector construct used corresponds to the OsPIF14 coding region under the control of the full CaMV 35S promoter. Reporter constructs contain the GUS gene driven by the minimal CaMV 35S promoter (control vector) or under the minimal CaMV 35S promoter plus the fragment of the OsDREB1B promoter used as bait in the Y1H screening. The LUC gene under the control of the full CaMV 35S promoter was used to normalize GUS expression levels. B, Transactivation analysis of OsPIF14 as a GUS/LUC activity ratio. Data from a representative experiment is shown. Values shown are multiples of the GUS/LUC ratio obtained with the control vector without effector. Data represents mean +/− SD (N=3). C, Construct used for rice protoplast transformation (pHBT95). OsPIF14 corresponds to the full OsPIF14 CDS sequence. D, expression of endogenous OsDREB1B after 18h of dark incubation. Results were normalized to 1 and represent the mean of two independent experiments +/− SD (N=5). * - Differences statistically significant (t-test, p<0.05).
3.5 OsPIF14 transcript is regulated by alternative splicing under cold stress
To understand how OsPIF14 influences the plant responses to cold, and whether it is also involved in the responses to other stresses, we have investigated OsPIF14 expression under different abiotic stress conditions. Two week-old rice seedlings were subjected to cold, salt, drought and ABA treatments over a period of 24h (Fig. 6A). The cold-induced gene expression of DREB1/CBFs in Arabidopsis was previously shown to be gated by the circadian clock, and to be more pronounced when stress is applied 4h after the start of the light period43. We decided to start our assays at that time of the day since we expect to observe higher differences in OsDREB1B regulation after treatment imposition.
Figure 6.
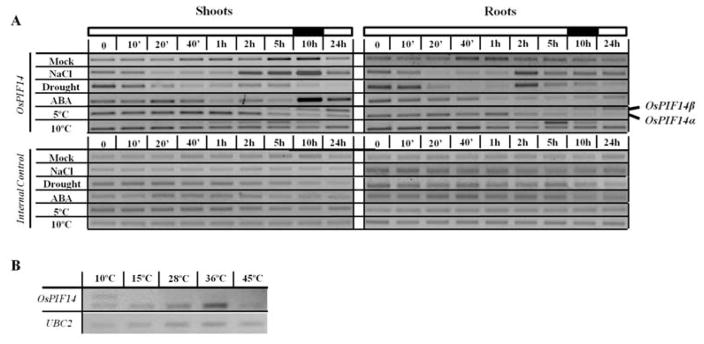
Expression profile of OsPIF14 in two week-old rice seedlings under different treatments. The plants were grown in a 12h:12h photoperiod and all the treatments started 4h after the beginning of the light period. A, OsPIF14 gene expression pattern under different abiotic stress conditions. The black and white bars on top indicate the time of the day at which the time-points were taken (light or dark). Roots and shoots were analyzed separately. Two alternative splice forms identified under cold stress are indicated on the right. B, Analysis of the alternative splicing of the OsPIF14 transcript after exposing the plants to different temperatures for 4h. Whole plants were assayed.
The gene expression studies showed that under control conditions the OsPIF14 transcript level slightly increases in shoots during late afternoon and the night period (5h and 10h in Fig. 6A, respectively), indicating a light/dark responsive behavior, as previously described27. In rice seedlings subjected to different treatments, the OsPIF14 gene expression was repressed after 20min under salt and drought and 1h after ABA treatment, in both roots and shoots. The transcript was then up-regulated some hours afterwards: under salt stress this up-regulation occurred 2h after stress, both in roots and shoots; in the case of drought, the same pattern was observed for roots, but was not so pronounced in shoots; and, in the case of ABA treatment, the up-regulation only occurred in shoots and only 10h after treatment imposition.
The transcriptional regulation of OsPIF14 under cold was unique, in the sense that it was the only stress inducing the appearance of an upper mRNA band, both in shoots and roots (Fig. 6A). After cloning and sequencing, we found that the second band was an alternative splicing form of the transcript (Fig. 1A). Under cold conditions, the alternative splicing of the OsPIF14 transcript leads to the retention of the first intron, and consequent formation of a premature stop codon. Since the stop codon is positioned more than 50 nucleotides upstream of the next exon-exon junction, we predict that this splicing form would be down-regulated by nonsense-mediated mRNA decay (NMD)44. On top of that, the putative protein product lacks a complete bHLH domain, thus it is expected to be transcriptionally inactive if produced. We named this splicing form OsPIF14β, whereas the splicing form under control conditions will be referred to as OsPIF14α. Interestingly, the gene expression pattern of both splicing forms seems to be slightly different when rice seedlings were subjected to different temperatures (Fig. 6A). At 5°C, transcripts of the βform were detected 40min after stress onset, increasing along the treatment, whereas the α-form started to decline after 1 to 2h. In seedlings subjected to 10°C, the transcript level of the β-form appeared 20min after stress onset, reaching a peak at 5h and declining to non-detectable levels afterwards. The α-form did not show significant alterations along the 10°C treatment. The expression pattern of both forms was similar in both roots and shoots. To determine whether this alternative splicing event was specific to low temperature, or if it occurred in temperature shifts in general, we analyzed the OsPIF14 gene expression after transferring rice plants to different temperatures for 5h (Fig. 6B). We could only detect the presence of the alternative splicing form OsPIF14β, when the plants were transferred to 10°C and not to 15°, 28°C, 36°C or 45°C. This indicates that this splicing event is specific to low temperatures.
Given that PIF7 was described as a regulator of DREB1/CBF expression in Arabidopsis20, we wanted to know whether, under cold conditions, any of the Arabidopsis PIF transcripts would also undergo alternative splicing. Thus, 10-day-old Arabidopsis seedlings were subjected to 5°C for 4h and the gene expression of PIF1, 3, 4, 5, 6 and 7 was analyzed by semi-quantitative RT-PCR, using intron-spanning primers (Fig. S4A). Interestingly, this analysis identified alternative splicing forms for PIF3, 6 and 7 both under control and cold conditions. The PIF6 splicing forms observed had been previously reported45. For PIF3 and PIF7, the splicing events corresponded to the retention of introns in the vicinity of the bHLH coding region (Fig. S4B), leading to the formation of premature stop codons, similarly to what happens with OsPIF14.
3.6 OsPIF14 interacts with the active form of OsphyB
In Arabidopsis it is known that phytochromes, upon activation by red light, migrate into the nucleus, where they interact with PIFs. This interaction usually results in a proteasome-dependent degradation of the PIFs, modulating the expression of genes regulated by PIFs. The interaction between rice phytochromes (A, B, and C) and putative PIFs had not yet been shown in rice. Thus, we have analyzed whether OsPIF14 interacted with any of the rice phytochromes. Initially, we performed a Yeast Two-Hybrid assay (Y2H), using the coding region of OsPIF14 as prey, and as baits the C-terminal non-photoactive coding regions of the three rice phytochromes, respectively (A, B and C; Fig. 7A). From the yeast growth rate in histidine-lacking medium (Fig. 7B), our results indicated that OsPIF14 interacts with OsPHYB C-terminal domain in a stronger manner than it does with the comparable OsPHYA or OsPHYC domains. To further confirm these results, a Bimolecular Fluorescence Complementation assay (BiFC) was performed (Fig. S5). YFP fluorescence signals could be detected in the nucleus of Nicotiana benthamiana leaf cells co-infiltrated with the OsPIF14::YFPN and OsPHYB::YFPC fusion proteins (Fig. S5B). No fluorescence was detected when leaves were co-infiltrated with OsPIF14::YFPN plus either OsPHYA::YFPC or OsPHYC::YFPC.
Figure 7.
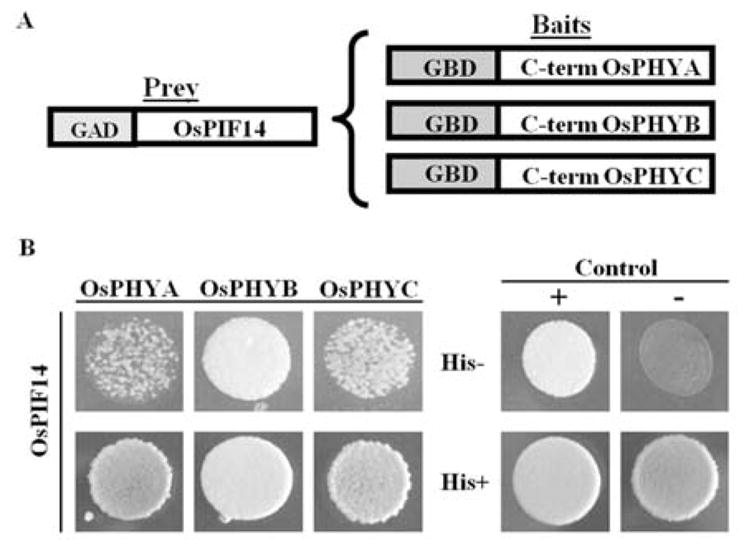
Analysis of the interaction between OsPIF14 and the three rice phytochromes, using a Yeast Two-Hybrid assay. A, Protein constructs used in the Y2H assay. OsPIF14 was used as prey, in a translational fusion with the GAL4 AD. The C-terminal non-photoactive regions of rice PHYA, B and C were fused with the GAL4 BD to be used as baits. B, Analysis of protein-protein interactions in yeast growing in histidine-lacking medium. Positive control used was the interaction between pAD-WT and pBD-WT, and negative control was the absence of interaction between GAD:OsPIF14 and pBD-WT. Bottom panel shows yeast growth in histidine-supplemented medium.
The Y2H and BiFC results support the hypothesis that OsPIF14 can bind to the C-terminal domain of OsPHYB. In order to test whether this binding was dependent on the active or inactive form of the full-length phytochrome, we expressed OsPIF14 and the full-length OsphyB in vitro and analyzed their interaction using a co-immunoprecipitation assay. As shown in Fig. 8B, OsPIF14 interacts preferably with the active Pfr form of OsphyB. The positive interaction between OsphyB-Pfr and Arabidopsis PIF3 was used as a positive control for the interaction39. Although we observed that OsPIF14 binds to the active form of OsphyB and that OsPIF14 represses OsDREB1B expression, we could not observe a significant regulation of OsDREB1B expression by light under normal, non-stress conditions (Fig. S6).
4. Discussion
The DREB1/CBF regulon has long been known to play an important role in the cold stress response in plants. Furthermore, it has already been reported that, in Arabidopsis, DREB1 genes are light regulated, having their expression dependent on light quality (red/far-red ratio)18,19. PIF7 was described as a possible link between light and cold signaling in Arabidopsis, since it binds to the promoter of DREB1C, and the pif7 null mutant was shown to have altered transcript levels of DREB1B and DREB1C20. In our work, using a Y1H system, we have identified OsPIF14 as binding to the promoter of OsDREB1B. We assessed the role of OsPIF14 in the regulation of OsDREB1B and its possible involvement in the link between light and cold signaling in rice. Even though PIF7 was described as a regulator of DREB1C and DREB1B in Arabidopsis20, the OsPIF14 APB domain shows the least degree of identity to PIF7, whereas PIF4 and PIF5 show the highest identity (85%), in agreement with what was previously reported by Nakamura, 2007. Our Y1H assays show that OsPIF14 binds specifically to the OsDREB1B promoter region ranging from −288 to −3bp, from the ATG start codon and it does not bind to the promoter of OsDREB1A. This means that OsPIF14 does not bind to the promoter of all members of DREB1/CBFs. The same was observed in Arabidopsis for PIF720. In order to better understand OsDREB1B regulation by OsPIF14, we have analyzed in detail the interaction of OsPIF14 with the cis-element present in the OsDREB1B promoter. Previous studies have reported that the flanking region of cis-elements play an important role in the activity of TFs, specially the bases closest to the cis-element46. Here, we showed that OsPIF14 has higher affinity to an extended version of the N-box (CCACGCGG). These results illustrate the complexity of bHLH-DNA binding which acts as a mechanism of specificity. Since OsPIF14 binds to the N-boxes present in the OsDREB1B promoter, and given that these boxes are associated with repressors, we examined whether OsPIF14 acts as a repressor29. We observed that OsPIF14 represses the expression of the reporter gene driven by the OsDREB1B promoter in arabidopsis protoplasts and also the endogenous OsDREB1B in rice protoplasts. These results are in agreement with those describing Hairy, a bHLH TF from Drosophila that binds to an N-box, acts as a repressor, and also prefers to have a Cytosine at 5′ flanking region of cis-element31. Actually, it was previously reported that a rice PIL15 acts as a repressor of genes involved in auxin pathway and cell wall organization or biogenesis47. Arabidopsis PIF7 was also described as a repressor of CBF2/DREB1C20. However, it is interesting to note that several Arabidopsis PIFs, including PIF7, have also been described as activators of transcription26,48,49. This may indicate that transcriptional activity of these proteins depends on promoter context, as recently suggested50, and/or additional interacting proteins. Our transactivation studies in Arabidopsis protoplasts showed an activation of the reporter gene even without the effector (35S::OsPIF14). This induction is most probably due to the fact that OsDREB1B promoter is induced by mechanical stress15, and the process of preparing Arabidopsis protoplasts surely triggers a stress response.
The fact that OsDREB1B promoter has two N-boxes and OsPIF14 recognizes them with different binding strength suggests a complex regulation of OsDREB1B repression. In order to further characterize the interaction OsPIF14-DNA, we have analyzed the basic region of OsPIF14. It has been reported that Arginine at position 13 (R13) within basic DNA binding region is crucial for binding to cis-elements with Guanine at position 4 (G4) (e.g. N-box (CACGCG) or G-box (CACGTG)), which is a mechanism to ensure that repressors do not bind to activator cis-elements (e.g. CACCTG)31. Here, we show the presence of R13 within the OsPIF14 basic domain and observe that OsPIF14 binds to cis-elements with G4, whose interaction is lost when the Guanine is changed to a Cytosine. All together these results suggest that G4 is important for OsPIF14-DNA binding affinity. Moreover, our EMSA results show that OsPIF14 is able to bind both N-box and G-box sequences. In this context, these results allowed us to characterize OsPIF14 as a bHLH belonging to group B. OsPIF14, showed higher binding affinity to G-box than to N-box, as described for group B proteins. We also showed that the presence of a 5′Cytosine and a 3′Guanine flanking the N-box increases OsPIF14 ability to bind to the N-box core motif. This suggests that OsPIF14 may bind to a large variety of gene promoters with different binding strengths according not only to cis-element core motif but also to the flanking regions.
OsPIF14 was shown to be regulated at the transcriptional level under several abiotic stress conditions. In response to salt, drought, and ABA treatments, the transcript levels are down-regulated within 20min to 1h and, in most cases, the levels are recovered after some hours. This suggests that OsPIF14 could have a potential role in rice responses to different abiotic stresses, through the regulation of OsDREB1B and/or possibly other downstream genes. In fact, it was already demonstrated that over-expression of maize PIF3 (ZmPIF3) confers more tolerance to dehydration and salt to rice plants51. The increased tolerance is due to activation of stress responsive genes, like OsDREB2A. In addition, phytochrome signaling has already been implicated in the response to salt stress in Mesembryanthemum crystallinum52, to drought in tomato plants53, and to ABA metabolism in Nicotiana plumbaginifolia54.
In contrast to the above-mentioned treatments, the effect of phytochrome signaling in general, and of PIFs in particular, in the Arabidopsis responses to cold has been well described17–20. Previously, we showed that under mild cold conditions (10°C) OsDREB1B stays up-regulated during the first 10h, reaching control levels after 24h, while under severe cold conditions (5°C) OsDREB1B stays induced even after 24h of stress imposition15. Those results indicate that at 5°C the down-regulation of OsDREB1B over time is compromised. Here, we observed that low temperatures induce the formation of an alternative splice form of the OsPIF14 transcript. The α–form, which is transcriptional active and the β–form, which corresponds to the transcriptionally inactive form since it lacks the complete bHLH domain. In addition, we predict that this β–form is down-regulated by NMD due to its premature stop codon. Interestingly, the expression patterns of both splice forms, α and β, were different when rice seedlings were subjected to 10°C or 5°C. In response to a mild stress (10°C), the expression of the α–form is not significantly altered, whereas the β–form is only expressed during the first 10hours of cold. After 24 hours of cold (10°C) only the α–form is present, which correlates with the low OsDREB1B levels due to the repressor activity of OsPIF14. When rice seedlings were subjected to a more severe stress (5°C), the constitutive α–form seems to be replaced by the β–form along the treatment, especially at 24h. We hypothesize that after 24h at 5°C the low levels of α–form are not enough to down regulate OsDREB1B. These results suggest a differential response of OsPIF14 to low temperatures, depending on how severe the stress is. Moreover, we observed that this splicing event only occurs when the plants are transferred to cold conditions, and not to warmer temperatures, which indicates that OsPIF14β must play a role in the plant responses specific to cold.
We also demonstrated that the Arabidopsis PIF3, 6 and 7 also show alternative splice forms both under control and cold conditions, while the OsPIF14 splice forms are cold specific. PIF6 had already been shown to have two alternative splice forms with different roles in Arabidopsis seed dormancy45. These results, together with ours, indicate that alternative splicing in PIFs may be a common mechanism for the regulation of PIF protein levels in the cells, in response to environmental conditions or certain developmental stages. We cannot rule out other putative splice forms for the genes tested, eventually not detected in our assays. It is yet to be shown whether these PIF alternative splice forms, so far identified, code for proteins that maintain the ability to bind phytochromes, since they have an intact APB domain. If so, this mechanism of regulation may be more than a simple way to modulate PIF protein levels, it might also regulate phytochrome signaling itself. Arabidopsis PIF3 was previously described as controlling hypocotyl cell elongation through its binding to phyB, modulating the abundance of this photoreceptor, independently of being able to bind DNA49. PIF4, PIF5 and PIF7 have also been reported to modulate phyB levels, in a process involving the proteasome pathway26,55. This was further confirmed by the finding that PIF3 modulate phyB levels by enhancing its in vivo poly-ubiquitination through the recruitment of LRB [Light-Response Bric-a-Brack/Tramtrack/Broad (BTB)] E3 ubiquitin ligases to the PIF3-phyB complex56. Moreover, for PIF4 and 5 it was already demonstrated that COP1 is also important in this in vitro degradation process57,58. A question thus arises on whether the OsPIF14β transcript, that has a longer 3′UTR when compared to the α-form, is eliminated by nonsense-mediated mRNA decay59 or results in a shorter, truncated protein. If the OsPIF14β transcript is translated into a functional protein, it is possible that it has a role in the regulation of phytochrome protein levels in rice, modulating the light signaling pathway, for example under cold conditions.
In Arabidopsis, several PIFs have been shown to interact with the active forms of the phytochromes25,26,39,40. However, to the best of our knowledge, no reports have yet been published on the interaction between PIFs and phytochromes in other species, including rice. Our co-immunoprecipitation results show that OsPIF14 preferably binds to the active form of OsphyB. Nevertheless, the Y2H assay, performed using the C-terminal non-photoactive forms of the rice phytochromes, also indicated that this terminal portion of the phytochrome B appears to be sufficient for the binding. Similar results had already been published for PIF3 in Arabidopsis39,60,61. Interestingly, the Y2H assays also show that OsPIF14 can bind weakly to the C-terminal domains of the rice phytochromes A and C. However, we were not able to validate this interaction either by BiFC or co-immunoprecipitation assays. It is also interesting to note that the heterologous Arabidopsis PIF3 binds to OsphyB more efficiently than OsPIF14 does. The OsPIF14-OsphyB interaction might result in the sequestration of OsPIF14 or its targeting for degradation. In Arabidopsis, PIF7, which was implicated in cold stress responses20, was described as light stable even though it interacts with the active form of phyB26. It is possible, therefore, that OsPIF14 is functionally more similar to PIF7, even though its APB sequence is more similar to PIF4 and 5.
In our work, we have shown that the OsPIF14 transcript is regulated by low temperature and, in addition, it has been reported that phyB levels are dependent on PIFs in Arabidopsis26,55–58. Thus, it may be that the regulation of PIFs by temperature has an effect on phytochrome levels, modulating their downstream signaling at different temperatures.
Overall our data provide evidence that both light and cold-temperature signaling pathways may converge on the OsDREB1B regulon of rice, likely via different mechanisms. The action of OsPIF14 may be regulated through its preferential interaction with the red light active form of OsphyB, which in turn could target OsPIF14 for degradation or, alternatively, sequester it and abrogate its function (as a repressor of OsDREB1B gene expression). It will be important to determine whether OsPIF14-OsphyB interaction prevents OsPIF14 from binding the OsDREB1B promoter. OsPIF14 binds to the OsDREB1B promoter N-boxes thereby acting as a repressor, but the regulation of OsDREB1B may be also modulated through alternative splicing of the OsPIF14 transcript. Moreover, we cannot rule out that other TFs repress the OsDREB1B expression, which under different light and temperature conditions is certainly regulated by additional TFs that are yet to be identified.
Supplementary Material
Figure S1. Y1H testing interaction of OsPIF14 with the promoter of OsDREB1A.
Figure S2. Direct Y1H of OsPIF14 against both halves of OsDREB1B promoter bait (−488 to −3bp from ATG)
Figure S3. EMSA controls for interaction OsPIF14 and N-boxes and interaction between OsPIF14 and G-box WT (CACGTG) or G-box MUT (CACCTG).
Figure S4. Expression pattern and alternative splice forms of the PIF genes in Arabidopsis seedlings.
Figure S5. Analysis of the interaction between OsPIF14 and different rice phytochromes, using a BiFC system.
Figure S6. OsDREB1B gene expression pattern determined by qRT-PCR.
Table S1. Primers used for semi-quantitative RT-PCR.
Table S2. Primers used for quantitative RT-qPCR.
Table S3. OsDREB1B promoter probes used for EMSA.
Acknowledgments
Funding: A.M.C., D.D.F. and T.L. were financed by Fundação para a Ciência e Tecnologia (FCT) through the fellowship SFRH/BD/29258/2006, SFRH/BD/74946/2010 and SFRH/BPD/102872/2014, respectively. I.A.A. was supported by SFRH/BPD/78314/2011 and FCT Investigator, financed by POPH (QREN). NS was supported by Programa Ciência 2007 and FCT Investigator, financed by POPH (QREN). This work was funded by National Institutes of Health Grant GM-R01-47475, Department of Energy Grant DE-FG03-87ER13742, and USDA Agricultural Research Service Current Research Information System Grant 5335-21000-032-00D to P.H.Q, by Research unit GREEN-it “Bioresources for Sustainability” (UID/Multi/04551/2013) and by FCT projects POCI/BIA-BCM/56063/2004 and PTDC/BIA_BCM/099836/2008.
BiFC vectors were kindly provided by Alejandro Ferrando, Universidad de Valencia, Spain (www.ibmcp.upv.es/FerrandoLabVectors.php).
Literature Cited
- 1.Hirayama T, Shinozaki K. Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 2010;61:1041–52. doi: 10.1111/j.1365-313X.2010.04124.x. [DOI] [PubMed] [Google Scholar]
- 2.Saibo NJM, Lourenço T, Oliveira MM. Transcription factors and regulation of photosynthetic and related metabolism under environmental stresses. Ann Bot. 2009;103:609–23. doi: 10.1093/aob/mcn227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubouzet JG, et al. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003;33:751–63. doi: 10.1046/j.1365-313x.2003.01661.x. [DOI] [PubMed] [Google Scholar]
- 4.Gilmour SJ, et al. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 1998;16:433–42. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- 5.Liu Q, et al. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci U S A. 1997;94:1035–40. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin F, et al. Cloning and functional analysis of a novel DREB1/CBF transcription factor involved in cold-responsive gene expression in Zea mays L. Plant Cell Physiol. 2004;45:1042–52. doi: 10.1093/pcp/pch118. [DOI] [PubMed] [Google Scholar]
- 8.El Kayal W, et al. Expression profile of CBF-like transcriptional factor genes from Eucalyptus in response to cold. J Exp Bot. 2006;57:2455–69. doi: 10.1093/jxb/erl019. [DOI] [PubMed] [Google Scholar]
- 9.Xiao H, Siddiqua M, Braybrook S, Nassuth A. Three grape CBF/DREB1 genes respond to low temperature, drought and abscisic acid. Plant Cell Environ. 2006;29:1410–21. doi: 10.1111/j.1365-3040.2006.01524.x. [DOI] [PubMed] [Google Scholar]
- 10.Shan DP, et al. Cotton GhDREB1 increases plant tolerance to low temperature and is negatively regulated by gibberellic acid. New Phytol. 2007;176:70–81. doi: 10.1111/j.1469-8137.2007.02160.x. [DOI] [PubMed] [Google Scholar]
- 11.Barros PM, Gonçalves N, Saibo NJM, Oliveira MM. Functional characterization of two almond C-repeat-binding factors involved in cold response. Tree Physiol. 2012;32:1113–28. doi: 10.1093/treephys/tps067. [DOI] [PubMed] [Google Scholar]
- 12.Chen JQ, Meng XP, Zhang Y, Xia M, Wang XP. Over-expression of OsDREB genes lead to enhanced drought tolerance in rice. Biotechnol Lett. 2008;30:2191–8. doi: 10.1007/s10529-008-9811-5. [DOI] [PubMed] [Google Scholar]
- 13.Wang Q, et al. Overexpression of a rice OsDREB1F gene increases salt, drought, and low temperature tolerance in both Arabidopsis and rice. Plant Mol Biol. 2008;67:589–602. doi: 10.1007/s11103-008-9340-6. [DOI] [PubMed] [Google Scholar]
- 14.Gutha LR, Reddy AR. Rice DREB1B promoter shows distinct stress-specific responses, and the overexpression of cDNA in tobacco confers improved abiotic and biotic stress tolerance. Plant Mol Biol. 2008;68:533–55. doi: 10.1007/s11103-008-9391-8. [DOI] [PubMed] [Google Scholar]
- 15.Figueiredo DD, et al. Seven zinc-finger transcription factors are novel regulators of the stress responsive gene OsDREB1B. J Exp Bot. 2012;63:3643–56. doi: 10.1093/jxb/ers035. [DOI] [PubMed] [Google Scholar]
- 16.Ito Y, et al. Functional analysis of rice DREB1/CBF-type transcription factors involved in cold-responsive gene expression in transgenic rice. Plant Cell Physiol. 2006;47:141–53. doi: 10.1093/pcp/pci230. [DOI] [PubMed] [Google Scholar]
- 17.Franklin KA. Light and temperature signal crosstalk in plant development. Curr Opin Plant Biol. 2009;12:63–8. doi: 10.1016/j.pbi.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 18.Franklin KA, Whitelam GC. Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat Genet. 2007;39:1410–3. doi: 10.1038/ng.2007.3. [DOI] [PubMed] [Google Scholar]
- 19.Kim HJ, Kim YK, Park JY, Kim J. Light signalling mediated by phytochrome plays an important role in cold-induced gene expression through the C-repeat/dehydration responsive element (C/DRE) in Arabidopsis thaliana. Plant J. 2002;29:693–704. doi: 10.1046/j.1365-313x.2002.01249.x. [DOI] [PubMed] [Google Scholar]
- 20.Kidokoro S, et al. The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol. 2009;151:2046–57. doi: 10.1104/pp.109.147033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Franklin K, Quail PH. Phytochrome functions in Arabidopsis development. J Exp Bot. 2010;61:11–24. doi: 10.1093/jxb/erp304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takano M, et al. Phytochromes are the sole photoreceptors for perceiving red/far-red light in rice. Proc Natl Acad Sci U S A. 2009;106:14705–10. doi: 10.1073/pnas.0907378106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen H, Moon J, Huq E. PIF1 is regulated by light-mediated degradation through the ubiquitin-26S proteasome pathway to optimize photomorphogenesis of seedlings in Arabidopsis. Plant J. 2005;44:1023–35. doi: 10.1111/j.1365-313X.2005.02606.x. [DOI] [PubMed] [Google Scholar]
- 24.Al-Sady B, Ni W, Kircher S, Schäfer E, Quail PH. Photoactivated phytochrome induces rapid PIF3 phosphorylation prior to proteasome-mediated degradation. Mol Cell. 2006;23:439–46. doi: 10.1016/j.molcel.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 25.Shen Y, Khanna R, Carle CM, Quail PH. Phytochrome induces rapid PIF5 phosphorylation and degradation in response to red-light activation. Plant Physiol. 2007;145:1043–51. doi: 10.1104/pp.107.105601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leivar P, et al. The Arabidopsis phytochrome-interacting factor PIF7, together with PIF3 and PIF4, regulates responses to prolonged red light by modulating phyB levels. Plant Cell. 2008;20:337–52. doi: 10.1105/tpc.107.052142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakamura Y, Kato T, Yamashino T, Murakami M, Mizuno T. Characterization of a set of phytochrome-interacting factor-like bHLH proteins in Oryza sativa. Biosci Biotechnol Biochem. 2007;71:1183–91. doi: 10.1271/bbb.60643. [DOI] [PubMed] [Google Scholar]
- 28.Atchley WR, Fitch WM. A natural classification of the basic helix-loop-helix class of transcription factors. Proc Natl Acad Sci. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fisher A, Caudy M. The function of hairy-related bHLH repressor proteins in cell fate decisions. BioEssays. 1998;20:298–306. doi: 10.1002/(SICI)1521-1878(199804)20:4<298::AID-BIES6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 30.Ledent V, Vervoort M. The basic helix-loop-helix protein family: comparative genomics and phylogenetic analysis. Genome Res. 2001;11:754–70. doi: 10.1101/gr.177001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohsako S, Hyer J, Panganiban G, Oliver I, Caudy M. Hairy function as a DNA-binding helix-loop-helix repressor of Drosophila sensory organ formation. Genes Dev. 1994;8:2743–2755. doi: 10.1101/gad.8.22.2743. [DOI] [PubMed] [Google Scholar]
- 32.Pires N, Dolan L. Origin and diversification of basic-helix-loop-helix proteins in plants. Mol Biol Evol. 2010;27:862–74. doi: 10.1093/molbev/msp288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ouwerkerk PB, Meijer AH. Yeast one-hybrid screening for DNA-protein interactions. Curr Protoc Mol Biol. 2001;Chapter 12(Unit 12.12) doi: 10.1002/0471142727.mb1212s55. [DOI] [PubMed] [Google Scholar]
- 34.Meijer AH, Ouwerkerk PB, Hoge JH. Vectors for transcription factor cloning and target site identification by means of genetic selection in yeast. Yeast. 1998;14:1407–15. doi: 10.1002/(SICI)1097-0061(199811)14:15<1407::AID-YEA325>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 35.Yoshida S, Forno DA, Cock JH, Gomez KAG. Laboratory manual for physiological studies of rice. 1976 [Google Scholar]
- 36.Lourenço T, et al. Isolation and characterization of rice (Oryza sativa L.) E3-ubiquitin ligase OsHOS1 gene in the modulation of cold stress response. Plant Mol Biol. 2013;83:351–63. doi: 10.1007/s11103-013-0092-6. [DOI] [PubMed] [Google Scholar]
- 37.Anthony RG, et al. A protein kinase target of a PDK1 signalling pathway is involved in root hair growth in Arabidopsis. EMBO J. 2004;23:572–81. doi: 10.1038/sj.emboj.7600068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wydro M, Kozubek E, Lehmann P. Optimization of transient Agrobacterium-mediated gene expression system in leaves of Nicotiana benthamiana. Acta Biochim Pol. 2006;53:289–98. [PubMed] [Google Scholar]
- 39.Ni M, Tepperman JM, Quail PH. PIF3, a Phytochrome-Interacting Factor Necessary for Normal Photoinduced Signal Transduction, Is a Novel Basic Helix-Loop-Helix Protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- 40.Khanna R, et al. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell. 2004;16:3033–44. doi: 10.1105/tpc.104.025643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Serra TS, et al. OsRMC, a negative regulator of salt stress response in rice, is regulated by two AP2/ERF transcription factors. Plant Mol Biol. 2013;82:439–55. doi: 10.1007/s11103-013-0073-9. [DOI] [PubMed] [Google Scholar]
- 42.Carretero-Paulet L, et al. Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 2010;153:1398–412. doi: 10.1104/pp.110.153593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fowler SG, Cook D, Thomashow MF. Low temperature induction of Arabidopsis CBF1, 2, and 3 is gated by the circadian clock. Plant Physiol. 2005;137:961–8. doi: 10.1104/pp.104.058354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci U S A. 2003;100:189–92. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Penfield S, Josse EM, Halliday KJ. A role for an alternative splice variant of PIF6 in the control of Arabidopsis primary seed dormancy. Plant Mol Biol. 2010;73:89–95. doi: 10.1007/s11103-009-9571-1. [DOI] [PubMed] [Google Scholar]
- 46.Fisher F, Goding CR. Single amino acid substitutions alter helix-loop-helix protein specificity for bases flanking the core CANNTG motif. EMBO J. 1992;11:4103–4109. doi: 10.1002/j.1460-2075.1992.tb05503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou J, et al. Overexpression of OsPIL15, a phytochrome-interacting factor-like protein gene, represses etiolated seedling growth in rice. J Integr Plant Biol. 2014;56:373–87. doi: 10.1111/jipb.12137. [DOI] [PubMed] [Google Scholar]
- 48.Huq E, et al. Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science. 2004;305:1937–41. doi: 10.1126/science.1099728. [DOI] [PubMed] [Google Scholar]
- 49.Al-Sady B, Kikis EA, Monte E, Quail PH. Mechanistic duality of transcription factor function in phytochrome signaling. Proc Natl Acad Sci U S A. 2008;105:2232–7. doi: 10.1073/pnas.0711675105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leivar P, Quail PH. PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 2011;16:19–28. doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gao Y, et al. A maize phytochrome-interacting factor 3 improves drought and salt stress tolerance in rice. Plant Mol Biol. 2015;87:413–28. doi: 10.1007/s11103-015-0288-z. [DOI] [PubMed] [Google Scholar]
- 52.Cockburn W, Whitelam GC, Broad A, Smith J. The participation of phytochrome in the signal transduction pathway of salt stress responses in Mesembryanthemum crystallinum L. J Exp Bot. 1996;47:647–653. [Google Scholar]
- 53.Biehler K, Haupt S, Beckmann J, Fock H, Becker TW. Simultaneous CO 2 - and 16 O 2 / 18 O 2 -gas exchange and fluorescence measurements indicate differences in light energy dissipation between the wild type and the phytochrome-deficient aurea mutant of tomato during water stress. J Exp Bot. 1997;48:1439–1449. [Google Scholar]
- 54.Kraepiel Y, et al. Analysis of phytochrome- and ABA-deficient mutants suggests that ABA degradation is controlled by light in Nicotiana plumbaginifolia. Plant J. 1994;6:665–672. [Google Scholar]
- 55.Khanna R, et al. The basic helix-loop-helix transcription factor PIF5 acts on ethylene biosynthesis and phytochrome signaling by distinct mechanisms. Plant Cell. 2007;19:3915–29. doi: 10.1105/tpc.107.051508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ni W, et al. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science. 2014;344:1160–4. doi: 10.1126/science.1250778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jang IC, Henriques R, Seo HS, Nagatani A, Chua NH. Arabidopsis PHYTOCHROME INTERACTING FACTOR proteins promote phytochrome B polyubiquitination by COP1 E3 ligase in the nucleus. Plant Cell. 2010;22:2370–83. doi: 10.1105/tpc.109.072520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ni W, et al. Multisite light-induced phosphorylation of the transcription factor PIF3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyB levels in Arabidopsis. Plant Cell. 2013;25:2679–98. doi: 10.1105/tpc.113.112342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kertész S, et al. Both introns and long 3′-UTRs operate as cis-acting elements to trigger nonsense-mediated decay in plants. Nucleic Acids Res. 2006;34:6147–57. doi: 10.1093/nar/gkl737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu Y, Tepperman JM, Fairchild CD, Quail PH. Phytochrome B binds with greater apparent affinity than phytochrome A to the basic helix-loop-helix factor PIF3 in a reaction requiring the PAS domain of PIF3. Proc Natl Acad Sci U S A. 2000;97:13419–24. doi: 10.1073/pnas.230433797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ni M, Tepperman JM, Quail PH. Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature. 1999;400:781–4. doi: 10.1038/23500. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Y1H testing interaction of OsPIF14 with the promoter of OsDREB1A.
Figure S2. Direct Y1H of OsPIF14 against both halves of OsDREB1B promoter bait (−488 to −3bp from ATG)
Figure S3. EMSA controls for interaction OsPIF14 and N-boxes and interaction between OsPIF14 and G-box WT (CACGTG) or G-box MUT (CACCTG).
Figure S4. Expression pattern and alternative splice forms of the PIF genes in Arabidopsis seedlings.
Figure S5. Analysis of the interaction between OsPIF14 and different rice phytochromes, using a BiFC system.
Figure S6. OsDREB1B gene expression pattern determined by qRT-PCR.
Table S1. Primers used for semi-quantitative RT-PCR.
Table S2. Primers used for quantitative RT-qPCR.
Table S3. OsDREB1B promoter probes used for EMSA.