Abstract
Facilitating the evolution of new gene functions, gene duplication is a major mechanism driving evolutionary innovation. Gene family expansions relevant to host/symbiont interactions are increasingly being discovered in eukaryotes that host endosymbiotic microbes. Such discoveries entice speculation that gene duplication facilitates the evolution of novel, endosymbiotic relationships. Here, using a comparative transcriptomic approach combined with differential gene expression analysis, we investigate the importance of endosymbiosis in retention of amino acid transporter paralogs in aphid genomes. To pinpoint the timing of amino acid transporter duplications we inferred gene phylogenies for five aphid species and three outgroups. We found that while some duplications arose in the aphid common ancestor concurrent with endosymbiont acquisition, others predate aphid divergence from related insects without intracellular symbionts, and still others appeared during aphid diversification. Interestingly, several aphid-specific paralogs have conserved enriched expression in bacteriocytes, the insect cells that host primary symbionts. Conserved bacteriocyte enrichment suggests that the transporters were recruited to the aphid/endosymbiont interface in the aphid common ancestor, consistent with a role for gene duplication in facilitating the evolution of endosymbiosis in aphids. In contrast, the temporal variability of amino acid transporter duplication indicates that endosymbiosis is not the only trait driving selection for retention of amino acid transporter paralogs in sap-feeding insects. This study cautions against simplistic interpretations of the role of gene family expansion in the evolution of novel host/symbiont interactions by further highlighting that multiple complex factors maintain gene family paralogs in the genomes of eukaryotes that host endosymbiotic microbes.
Keywords: aphid, buchnera, amino acid transporter, gene duplication, symbiosis
Introduction
Gene family expansion is a key player in the evolution of innovation (Ohno 1970; Arnegard et al. 2010; Deng et al. 2010; Kondrashov 2012; Voordeckers et al. 2012), but elucidating the factors maintaining paralogs in a genome can be tricky (Innan and Kondrashov 2010). The vast majority of paralogs that arise in a population are lost through stochastic birth and death processes (Lynch and Conery 2000), presenting an evolutionary conundrum when ancient paralogs are found. A complete understanding of paralog retention requires detailed knowledge of a number of traits, such as paralog expression and function, and the ecological and molecular mechanisms driving paralog diversification. While expression, function, ecology, and molecular traits facilitate explaining the maintenance of anciently acquired paralogs, obtaining data about all these traits is often intractable. In these cases, a combination of natural history and phylogeny of the taxa of interest can provide a framework for proposing the ecological and biological factors that underlie paralog retention.
One trait that may drive paralog maintenance is the evolution of endosymbiosis between multicellular eukaryotes and microorganisms. In endosymbiosis, microbial symbionts reside intracellularly within hosts, commonly providing a nutritional benefit. New gene paralogs may facilitate the establishment and maintenance of a symbiotic partnership through expression diversification (e.g., from a nonsymbiotic tissue to a symbiotic tissue) or possibly through functional diversification (e.g., toward a function specialized for endosymbiosis). Indeed, genomic studies suggest a role for gene duplication in endosymbiosis—these studies have found lineage-specific duplication in genes functionally relevant to symbiotic interactions across systems as divergent as the legume/Rhizobia endosymbiosis (Young et al. 2011), the endosymbiosis between corals or anemones and Symbiodinium (Shinzato et al. 2011; Baumgarten et al. 2015), and the endosymbiosis between the pea aphid Acyrthosiphon pisum and its endosymbiont Buchnera aphidicola (Huerta-Cepas et al. 2010; Price et al. 2011). A. pisum, a sap-feeding insect, has undergone expansions in over 2000 gene families (Huerta-Cepas et al. 2010; International Aphid Genomics Consortium 2010), some of which may be mechanistically important for its relationship with Buchnera. Particularly intriguing in A. pisum are lineage-specific expansions in amino acid transporter genes (Price et al. 2011; Duncan et al. 2014; Dahan et al. 2015; Wilson and Duncan 2015)—genes whose membrane-bound protein products are crucial for nutritional exchange between A. pisum and Buchnera (Price et al. 2014, 2015). Indeed, amino acid transporters are over-represented among A. pisum gene families that underwent large expansions resulting in more than ten paralogs (Huerta-Cepas et al. 2010). Similarly, we recently discovered that several sap-feeding insects (also with endosymbionts) experienced lineage-specific expansions in amino acid transporters. Interestingly, independent expansion of amino acid transporters in multiple sap-feeding insect lineages is a pattern that parallels other independently evolved signatures of host/endosymbiont genome co-evolution (Wilson and Duncan 2015).
The mechanistic importance of gene duplication in amino acid transporters for endosymbiosis in these insects is further supported by the observation that some lineage-specific paralogs in A. pisum and the citrus mealybug Planococcus citri are enriched in bacteriocytes (Price et al. 2011; Duncan et al. 2014), the specialized insect cells where symbionts reside. Bacteriocyte enrichment of some paralogs implies paralog evolution to operate in a symbiotic context because bacteriocytes represent the interface between the insect host and the endosymbiont. This host/endosymbiont interface is made up of three membrane barriers separating host tissues from endosymbionts: (1) The plasma membrane surrounding the bacteriocyte, (2) the insect-derived membrane surrounding individual symbiont cells (symbiosomal membrane), and (3) the inner and outer bacterial membranes of each symbiont cell. While the symbiosomal membrane is the most immediate interface between host (bacteriocyte cytoplasm) and endosymbiont, the bacteriocyte plasma membrane is also an important part of the host/endosymbiont interface because of the role it plays in regulating metabolic output of the symbiont (Price et al. 2014). In addition to independent recruitment of amino acid transporter paralogs to the host/symbiont interface of both A. pisum and P. citri, tests for signatures of selection in the pea aphid found, in one expansion, an elevated rate of evolution in the transition from high gut expression to enriched bacteriocyte expression. The elevated rate of evolution suggests functional evolution corresponding to a shift toward symbiotic expression (Price et al. 2011). Lastly, expansions in sap-feeding insects are significantly associated with increased gene duplication rates and decreased gene loss rates (Dahan et al. 2015), supporting an adaptive explanation for the retention of duplicate amino acid transporters—an explanation that may relate to shared traits among these insects, such as endosymbiosis with nutrient-provisioning bacteria.
Despite the evidence supporting endosymbiosis as a factor influencing the retention of amino acid transporter paralogs in sap-feeding insects, transcriptomic and expression data indicate that other biological factors are also at play. For example, in aphids, some paralogs show biased expression in males (Duncan et al. 2011), where symbionts are less abundant than in females (Douglas 1989). In fact, accelerated rates of evolution also correlate with the evolution of male-biased expression in one male-biased paralog, suggesting a derived sex-biased function. Further, despite bacteriocyte enrichment in some citrus mealybug and pea aphid paralogs, most paralogs are not enriched in bacteriocytes (Price et al. 2011; Duncan et al. 2014), suggesting that other features of these insects influence paralog retention. Lastly, while we found lineage-specific duplications in amino acid transporters of four sap-feeding insects—pea aphids (Price et al. 2011), citrus mealybugs, potato psyllids, and whiteflies (Duncan et al. 2014)—we found no evidence for duplication in the sap-feeding cicada (Duncan et al. 2014), indicating that gene duplication in amino acid transporters is not necessary for the evolution of endosymbiosis between sap-feeding insects and bacteria. These data do not rule out the possibility that gene duplication has played an important role in the evolution and maintenance of endosymbiosis in some sap-feeding insects. Even so, evidence that other factors influence paralog retention makes it unclear if endosymbiosis plays a primary or secondary role in selection for the maintenance of amino acid transporter paralogs.
Here, using a comparative transcriptomic approach, we leverage the phylogeny and natural history of aphids and their close relative, the grape phylloxera Daktulosphaira vitifoliae (see table 1 and fig. 1 for taxonomic classifications and relationships among taxa in this study), to investigate life history traits underlying the retention of amino acid transporter paralogs. Aphids and phylloxera belong to different families of the hemipteran group Aphidomorpha—Aphididae and Phylloxeridae, respectively. The common ancestor of Aphididae established an endosymbiotic relationship with the bacterium B. aphidicola around 160–280 MYA (Moran et al. 1993), and since that initial infection, Buchnera has been vertically inherited by nearly all extant aphids. In contrast, Phylloxeridae, including D. vitifoliae, lacks Buchnera or other intracellular symbionts (Vorwerk et al. 2007; Medina et al. 2011). If endosymbiosis between aphids and Buchnera initially drove paralog retention in amino acid transporters, we expect to find that duplication took place, at least initially, in the aphid common ancestor. If, however, duplication predates the aphid/phylloxera split or postdates aphid diversification, then the initial trait influencing paralog maintenance is more likely not endosymbiosis with Buchnera, but another trait that evolved concurrently with gene duplication.
Table 1.
Taxon Sampling Within Aphidomorpha
| Family |
|---|
| Subfamily |
| Tribe |
| Genus species |
| Phylloxeridae |
| D. vitifoliae |
| Aphididae |
| Eriosomatinae |
| Pemphigini |
| P. obesinymphae |
| Tamaliinae |
| T. coweni |
| Aphidinae |
| Aphidini |
| A. nerii |
| Macrosiphini |
| M. persicae |
| A. pisum |
Fig. 1.—
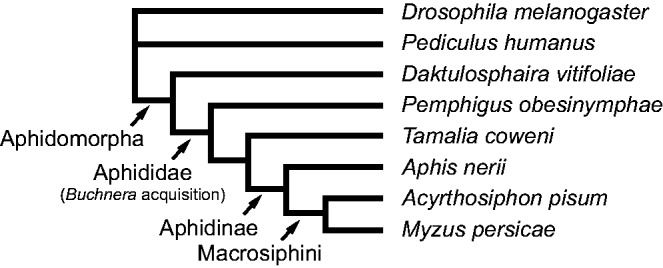
Phylogenetic relationships among sampled taxa. Acquisition of the aphid endosymbiont Buchnera and relevant higher taxonomic classifications are mapped. Tree structure is based on phylogenetic analyses reported in Nováková et al. (2013), Misof et al. (2014) and Dahan et al. (2015).
Materials and Methods
Taxon Sampling, Specimen Collection, Identification, and Vouchering
We sampled amino acid transporters from several aphid genera representing three subfamilies and multiple tribes across the Aphididae (table 1 and fig. 1). Aphid species included A. pisum, Myzus persicae, Aphis nerii, Tamalia coweni, and Pemphigus obesinymphae. We consider these taxa as representative of aphids because they include tribes at a range of positions across the aphid phylogeny, including members of Pemphigini, which are usually supported as sister to the rest of aphids (Nováková et al. 2013). In addition, we sampled amino acid transporters from an outgroup of aphids, the grape phylloxera D. vitifoliae, and two Aphidomorpha outgroups (Pediculus humanus and Drosophila melanogaster; fig. 1).
Myzus persicae and A. pisum data were generated from established isofemale laboratory lines. M. persicae RNAseq and differential expression data, generated in this study, came from laboratory clones G006, G002, and BTI Red (also known as USDA) (Ramsey et al. 2007). Data for A. pisum, generated in previously published studies using RNAseq and qRT-PCR (Hansen and Moran 2011; Price et al. 2011; Macdonald et al. 2012; Duncan et al. 2014), came from laboratory clones LSR1 (Caillaud et al. 2002), 9-2-1 (Russell and Moran 2006), 5A (Sandström and Moran 2001), and CWR09/18 (Macdonald et al. 2012). A. nerii were collected from Asclepias spp. in Miami, FL (Miami-Dade county), Atlanta, GA (Dekalb county), and Minnesota. The Atlanta and Miami populations are maintained as isofemale clones in the laboratory of Patrick Abbot at Vanderbilt University. A. nerii were identified based on host plant and distinctive morphology. T. coweni were collected from various sites and host plants in Arizona, Nevada, and California, as reported by Miller et al. (2015). P. obesinymphae were collected in the vicinity of Nashville, TN (Davidson county) on Populus deltoides subsp. deltoides and were identified by Patrick Abbot based on distinctive gall morphology. D. vitifoliae were collected at the vineyards of Château Couhins in Bordeaux, France on Vitis vinifera cv. Cabernet franc and were identified by aphid biologists at the French National Institute for Agricultural Research (INRA). An isofemale clone of D. vitifoliae (INRA-Pcf7) is maintained at INRA. Voucher specimens for T. coweni are deposited in the Smithsonian Institution Department of Entomology, the Canadian National Collection of Insects, and collections at Washington State University and California State University, Chico (Miller et al. 2015). In addition, we annotated transcripts corresponding to cytochrome c oxidase subunit 1 (CO1) for A. nerii, T. coweni, P. obesinymphae, and D. vitifoliae to serve as identity vouchers. In brief, we used a protein sequence for A. pisum CO1 (Genbank ID YP_002323931.1) as a query in local TBLASTN searches against transcriptomes for the other aphids and D. vitifoliae. Top hits (all with e-value = 0.0) were used as queries in reciprocal BLASTX searches against the NCBI refseq protein database to confirm homology with A. pisum CO1. Reciprocal BLAST searches returned one CO1 sequence for each species except A. nerii, which had two CO1 sequences. Sequences are provided in supplementary file 1, Supplementary Material online.
Transcriptome Sequencing and Assembly
Transcriptomes were sequenced for A. nerii and M. persicae. Total RNA was extracted from whole adult, asexual female A. nerii bodies and a combination of whole bodies, bacteriocyte, and gut for adult, asexual female M. persicae. Total RNA was sent to the Hussman Institute for Human Genomics (University of Miami Miller School of Medicine) for library preparation and paired end sequencing on the Illumina HiSeq platform. Raw RNAseq reads for A. nerii and M. persicae were deposited in the NCBI Sequence Read Archive (SRA) under BioProject PRJNA296778. Raw reads for T. coweni (a combination of paired end and single end reads) and P. obesinymphae (single end reads) were provided by Patrick Abbot, and are available on NCBI in the SRA (T. coweni accession numbers: SRX1305377, SRX1305445, SRX1305282, SRX1304838 [Miller et al. 2015]; P. obeiynymphae reads are stored under BioProject PRJNA301746). Reference transcriptomes were assembled for T. coweni, P. obesinymphae, A. nerii, and the M. persicae G006 clone. Reads for these taxa were filtered to a minimum quality score of 30 over 95% of the read, resulting in a combination of paired end and single end reads for A. nerii and G006. All reads from each taxon kept after the filtering process were assembled into a single reference transcriptome for each species in Trinity (7/17/14 release) (Haas et al. 2013) using the Blacklight system at the Pittsburgh Supercomputing Center. A fully assembled transcriptome for D. vitifoliae was provided by colleagues at INRA, for which raw sequencing reads are accessible via NCBI (BioProject PRJNA294954). The other three taxa in our dataset—A. pisum, P. humanus, and D. melanogaster—have fully sequenced genomes, from which we used amino acid transporter sequences that were annotated in a previous study (Price et al. 2011).
Amino Acid Transporter Annotation
Using assembled transcriptomes, we annotated amino acid transporters in the Amino Acid-Polyamine-Organocation (APC) (TC # 2.A.3) and Amino Acid-Auxin-Permease (AAAP) (TC # 2.A.18) families from T. coweni, P. obesinymphae, M. persicae, A. nerii, and D. vitifoliaea as previously described (Price et al. 2011; Duncan et al. 2014). In brief, we used a stand-alone PERL script underlying the ORF prediction available at http://bioinformatics.ysu.edu/tools/OrfPredictor.html, last accessed February 29, 2016 where transcripts were translated into the six reading frames. The translated transcripts were searched for functional domains that significantly matched (e ≤ 0.001) known APC, and AAAP families in HMMER v3.0 (Eddy 2009; Finn et al. 2011). HMMER hits were verified through BLAST searches to the NCBI refseq protein database. Transcripts with significant similarity (e ≤ 0.001) to APC or AAAP sequences from D. melanogaster or A. pisum were selected for further computational processing.
Transcriptomes generated many unique but similar transcripts identified as APC or AAAP members through HMMER and BLAST. We collapsed amino acid transporter transcripts into conservative sets of representative loci for each taxon using methods we previously developed (Duncan et al. 2014). For M. persicae, which has a draft genome sequence, we collapsed all transcripts that mapped to the same location in the genome. For remaining taxa, we followed a series of steps. First, we collapsed all transcripts with the same Trinity component number into the longest representative transcript. Next, we followed the methods we previously described (Duncan et al. 2014). In brief, we estimated the pairwise rate of synonymous substitutions (Ks) among transcripts that clustered together in preliminary phylogenetic analyses using the Goldman Yang method (Goldman and Yang 1994) in KaKs_Calculator v1.2 (Zhang et al. 2006). We collapsed all transcripts with a pairwise Ks value < 0.25, keeping the longest sequence to represent the locus. If two similar transcripts met the cutoff Ks of ≥0.25, but overlapped <50 bp, we collapsed the shorter transcript into the longer transcript to validate a conservative estimation of locus number. We chose the threshold Ks value (0.25) because we previously found that it slightly underestimates the number of true amino acid transporter paralogs in A. pisum, collapsing only three very recently duplicated APC paralogs (Duncan et al. 2014). Thus, this threshold is appropriate for conservative estimation of amino acid transporter paralogs in related species.
Differential Expression Analysis
The differential expression of M. persicae amino acid transporters between bacteriocyte and whole body tissues was quantified using the transcriptome data from this study. M. persicae clones G006, G002, and BTI Red were treated as replicates. Differential expression analysis was conducted with the RSEM package (v.1.2.22) (Li and Dewey 2011) and edgeR (v.3.10.2) from the Bioconductor package (Robinson et al. 2010). In brief, the processed forward RNAseq reads for all three M. persicae clones were mapped to reference transcripts in a strand-specific manner using bowtie2 (v.2.2.4) (Langmead and Salzberg 2012) and mapped reads were counted using PERL script rsem-calculate-expression.pl from the RSEM package. The counts from RSEM were scaled to the whole transcriptomes and normalized by relative log expression. The significantly differentially expressed amino acid transporters between bacteriocyte and whole insect were identified using edgeR negative binomial models. Four amino acid transporter sequences comprised several truncated, partial transcripts that supported full-length gene models in the M. persicae genome (Mper-APC09, Mper-APC12, Mper-AAAP06, and Mper-AAAP20; the M. persicae draft genome assembly is available at www.aphidbase.com, last accessed February 29, 2016). In these four cases, the differential expression analysis was performed by mapping raw RNAseq reads to the gene models instead of the transcripts.
Phylogenetic Analysis
Transcript sequences were translated to protein using Seaview (v.4.5.2) (Gouy et al. 2010), and protein sequences were aligned using MAFFT (v.7.158b) (Katoh et al. 2002) using default parameters. Alignments were trimmed in TRIMAL (v.1.4) (Capella-Gutiérrez et al. 2009) using a gap threshold of 25%. Prottest (v.3.4) (Abascal et al. 2005) determined the best-fit model of protein evolution to be either LG + G (APC family) or LG + I + G (AAAP) based on the Akaike Information Criterion. Maximum likelihood (ML) phylogenies were inferred for the APC and AAAP families in RAxML (v.8.0.26) (Random Axelerated Maximum Likelihood) (Stamatakis 2006; Ott et al. 2007) using the best-fit model of protein evolution and the fast bootstrap option. Bootstrap replicate number was chosen by the bootstrap convergence criterion “autofc”.
We further inferred Bayesian phylogenies in MrBayes (v.3.2) using WAG + G (APC family) or WAG + I + G (AAAP), as MrBayes does not implement the LG amino acid substitution model. Two independent runs, each with four chains, were run for one to five million generations, until the standard deviation of split frequencies between runs converged to <0.01. Appropriate parameter sampling and convergence were determined by visually inspecting trace files in Tracer (v.1.6) (Rambaut et al. 2014). Tracer was also used to determine burn-in values of each dataset (10% of generations), which we discarded when constructing Bayesian consensus trees. In the figures presented here, we mapped ML bootstrap support onto Bayesian consensus trees using SumTrees (v.3.0) from the DendroPy package (Sukumaran and Holder 2010).
Although we annotated all amino acid transporters in the AAAP family, because of large divergence in that family (Duncan et al. 2014), we inferred the relationships among a reduced set of sequences corresponding to the “arthropod expanded clade”. The arthropod expanded clade consists of arthropod orthologs to the mammalian SLC36 family of proton-coupled amino acid transporters (Price et al. 2011; Thwaites and Anderson 2011). Two human SLC36 sequences (SLC36A1 and SLC36A2), previously shown to belong to the sister clade of the arthropod expanded clade (Price et al. 2011) were used as outgroups. Outgroup sequences for the APC family are members of the sister clade of Na-K-Cl transporters (ACYPI001649, ACYPI007138) (Price et al. 2011). Untrimmed transcript sequences translated into protein as well as trimmed Bayesian and ML alignments of the APC family and reduced AAAP family (“arthropod expanded clade”) are provided as supplementary files 2–5, Supplementary Material online, and are also available by request from RPD.
Results
The slimfast Expansion Predates Aphid–Phylloxera Divergence
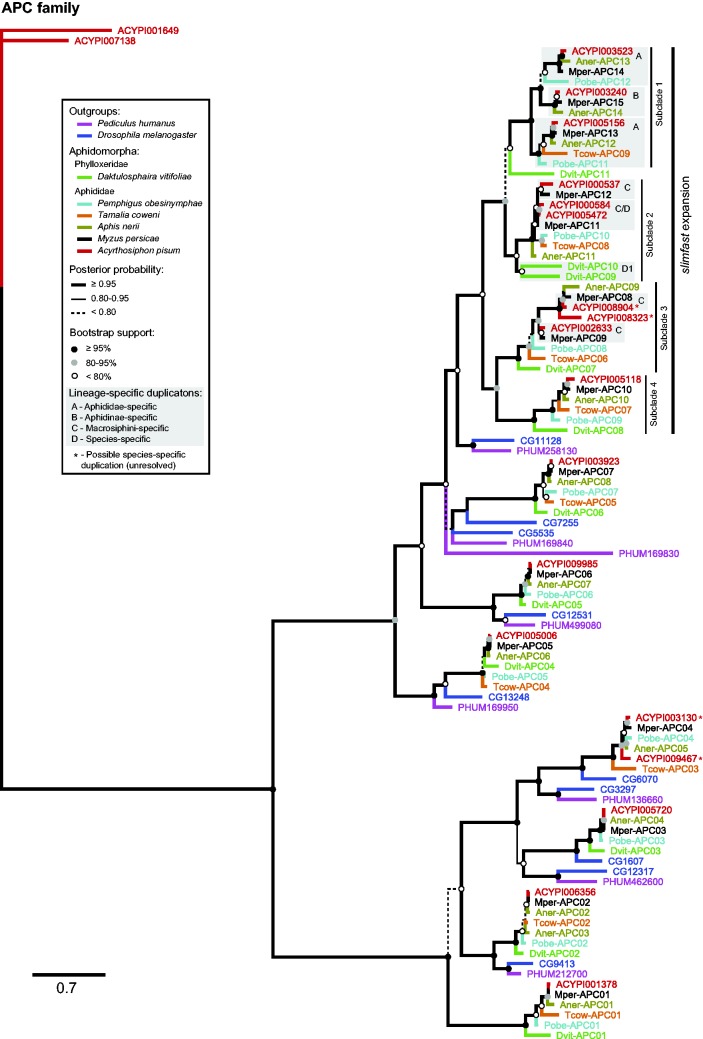
Previously, we identified an aphid-specific expansion in the APC transporter slimfast (named for the D. melanogaster ortholog, CG11128; Colombani et al. 2003) (Price et al. 2011). Here, we find APC members of different Aphidomorpha (table 1 and fig. 1) interleaved within four subclades of the slimfast expansion. These data imply that slimfast expanded into minimally four paralogs before the divergence of aphids and the grape phylloxera, D. vitifoliae (fig. 2).
Fig. 2.—
Bayesian phylogeny of amino acid transporters in the APC family. Branches are color coded by taxon, as indicated in the key. Node support is shown both as branch weight (Bayesian posterior probability) and circles on nodes (ML bootstrap support). The slimfast expansion is marked in the upper right, along with the four subclades resulting from duplication events that predate aphid/phylloxera divergence. Duplication events occurring at different time scales in aphid evolution gave rise to lineage-specific clades highlighted by gray boxes. Letters in gray boxes refer to (A) clades resulting from duplication events predating Aphididae divergence, (B) clades resulting from duplication events predating Aphidinae divergence, (C) clades resulting from duplication events predating Macrosiphini divergence, and (D) species-specific duplication events.
Amino Acid Transporters Duplicated at Multiple Time Scales in Aphid Evolution
APC Family
Following the aphid/phylloxera split, additional gene duplication events were inferred in the slimfast expansion at four different taxonomic levels within aphids: (1) predating aphid diversification (fig. 2, gray boxes labeled A), (2) predating diversification of the subfamily Aphidinae (fig. 2, gray box labeled B), (3) predating diversification of the tribe Macrosiphini (fig. 2, gray boxes labeled C), as well as (4) at least one duplication event specific to A. pisum (fig. 2, gray box labeled D). In addition, our phylogenetic analysis supports one D. vitifoliae-specific gene duplication following aphid/phylloxera divergence (fig. 2, gray box labeled D1). Species varied in the number of slimfast paralogs they encoded, ranging from four in T. coweni to 10 in A. pisum. Variation in number of slimfast paralogs resulted at least in part from differences in additional gene duplications during aphid diversification, but may also indicate gene losses in some species or under-sampling.
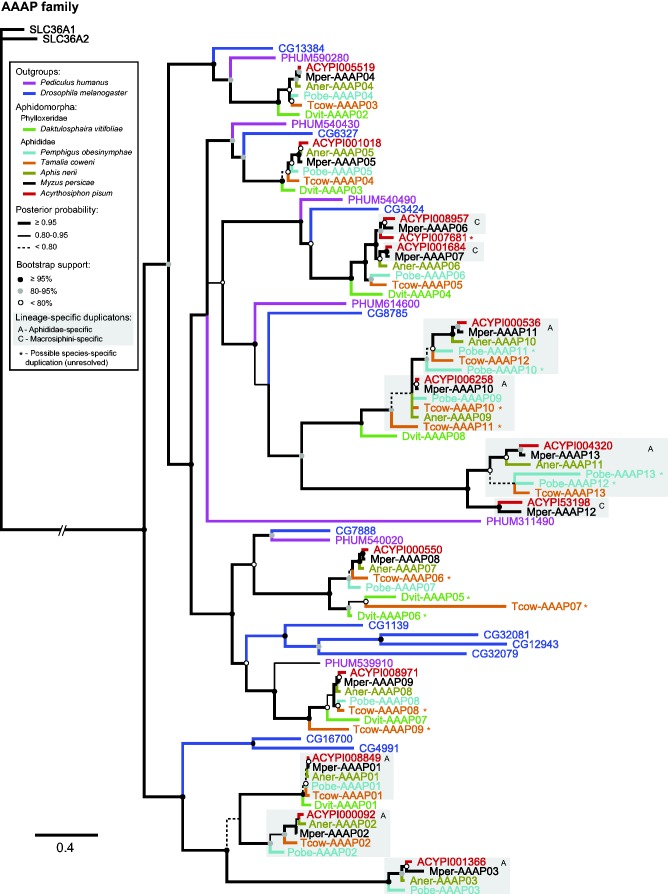
AAAP Family
In contrast to the APC slimfast expansion, no AAAP duplications predated the phylloxera/aphid divergence. However, mirroring other duplication patterns we found in the APC slimfast expansion, duplication in the AAAP family happened at different time scales during aphid evolution (fig. 3). We found strong support for several duplications predating aphid diversification (fig. 3, gray boxes labeled A), as well as support for duplications specific to the tribe Macrosiphini (fig. 3, gray boxes labeled C). Asterisks beside gene IDs in figures 2 and 3 mark genes that may be products of species-specific gene duplication. However, given our tree topology, we are unable to confidently infer that these sequences result from species-specific duplication.
Fig. 3.—
Bayesian phylogeny of amino acid transporters in the AAAP family. Branches are color coded by taxon, as indicated in the key. Node support is shown both as branch weight (Bayesian posterior probability) and circles on nodes (ML bootstrap support). Duplication events occurring at different time scales in aphid evolution gave rise to lineage-specific clades that are highlighted by gray boxes. Letters in gray boxes refer to (A) clades resulting from duplication events predating Aphididae divergence, and (C) clades resulting from duplication events predating Macrosiphini divergence. Outgroup sequences (SLC36A1 and SLC36A2) are from humans.
Bacteriocyte Expression Is Dynamic Among Aphids
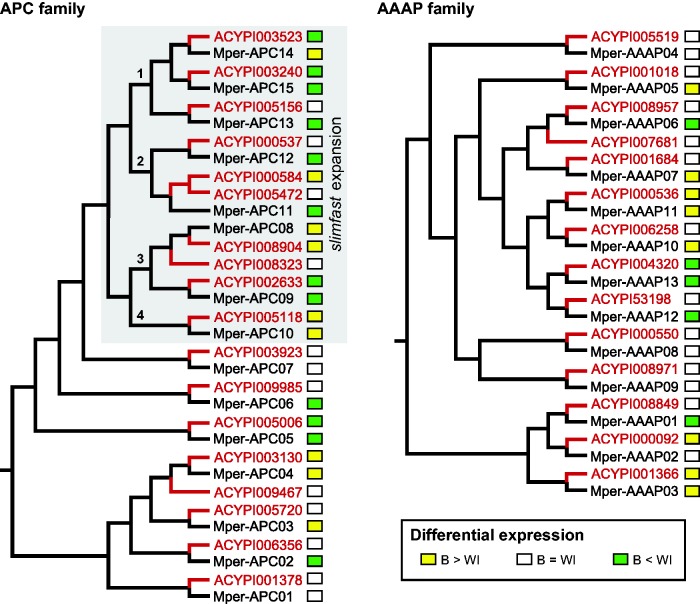
Our differential expression analysis identified 25 significantly differentially expressed amino acid transporters in the M. persicae transcriptome (based on a 2-fold difference in either direction and P ≤ 0.05, FDR ≤ 0.05; supplementary table S1, Supplementary Material online). Differentially expressed amino acid transporters included 11 bacteriocyte-enriched transcripts and 14 bacteriocyte-depleted transcripts. Of these differentially expressed amino acid transporters, ten bacteriocyte-enriched and 12 bacteriocyte-depleted transcripts were members of the APC family or the reduced AAAP family that appear in our phylogenetic analyses (fig. 4).
Fig. 4.—
Differential expression of amino acid transporters in A. pisum and M. persicae. Phylogenies depict relationships between A. pisum and M. persicae amino acid transporters in the APC and AAAP families (based on figs. 2 and 3). The slimfast expansion in the APC family is highlighted with a gray box, and numbers are mapped onto nodes to indicate the four slimfast subclades resulting from gene duplications predating aphid/phylloxera divergence. Two-fold or greater differential expression between bacteriocyte (B) and whole insect (WI) is mapped onto trees. Expression data for M. persicae amino acid transporters are reported in supplementary file 6. Differential expression for A. pisum transporters is based on consistent differential expression in the same direction found across four studies: Hansen and Moran (2011), Price et al. (2011), Macdonald et al. (2012), and Duncan et al. (2014).
Differential expression results for amino acid transporters in M. persicae were qualitatively compared with previously published expression results for orthologous amino acid transporters in A. pisum (Hansen and Moran 2011; Price et al. 2011; Macdonald et al. 2012; Duncan et al. 2014). Bacteriocyte expression was not necessarily consistent between A. pisum and M. persicae, evident by three patterns displayed by orthologous pairs: (1) Both have the same relative bacteriocyte expression (enriched, depleted or not significantly different from whole insect), (2) one species has enriched or depleted bacteriocyte expression while its ortholog is not significantly differentially expressed between whole insect and bacteriocyte, or (3) one species has bacteriocyte-enriched expression while the other has bacteriocyte-depleted expression (fig. 4). In the APC family, three pairs of A. pisum/M. persicae orthologs showed conserved bacteriocyte enrichment: ACYPI008904/Mper-APC08, ACYPI005118/Mper-APC10, and ACYPI003130/Mper-APC04. Notably, the first two pairs are members of the slimfast expansion (figs. 2 and 4). In the AAAP family, two orthologous pairs show conserved bacteriocyte enrichment: ACYPI000536/Mper-AAAP11 and ACYPI001366/Mper-AAAP03, both of which are members of aphid-specific expansions (figs. 3 and 4).
Discussion
Recent studies on symbiotic organisms support a role for gene duplication in the evolution of endosymbiosis (Price et al. 2011; Shinzato et al. 2011; Young et al. 2011; Duncan et al. 2014; Baumgarten et al. 2015; Dahan et al. 2015). However, most studies do not address the possibility of a gene duplication/endosymbiosis connection in more than one species and, as we have found in sap-feeding insects, nonsymbiotic traits may influence the retention of duplicate genes (Duncan et al. 2011, 2014). To gain insight into the role of endosymbiosis in retaining aphid-specific amino acid transporter paralogs, we used a comparative transcriptomic approach to pinpoint the timing of amino acid transporter duplications in the Aphidomorpha (table 1 and fig. 1). Our results support a complex and dynamic evolutionary history of amino acid transporters in these insects—a history that was likely shaped by multiple biological and ecological factors. In support of a role for gene duplication in the evolution of endosymbiosis, we inferred several duplication events in the aphid common ancestor, corresponding to the acquisition of the aphid endosymbiont, Buchnera (figs. 1–3). However, we also inferred duplication events both earlier and later than the evolution of endosymbiosis in aphids. Duplication at these earlier and later time scales implies that gene duplication and retention has also been driven by factors other than endosymbiosis.
The slimfast Expansion Was Not Driven by the Evolution of Endosymbiosis
We posit that the aphid/phylloxera slimfast expansion, at least initially, was not driven by endosymbiosis. Phylloxera lack an endosymbiont, and assuming that the shared ancestor of aphids and phylloxera also lacked an endosymbiont, the expansion predated the evolution of endosymbiosis. However, the relationship of the aphid/phylloxera lineage, derived from within Sternorrhyncha, suggests that endosymbiosis originated in the common ancestor of Sternorrhyncha and was secondarily lost in phylloxera. Even if endosymbiosis originated in the sternorrhynchan common ancestor, the slimfast expansion likely was not driven primarily by primary endosymbiosis because the expansion postdates aphid divergence from other major sternorrhynchan lineages (Duncan et al. 2014). Furthermore, despite their shared ancestry, sternorrhynchan lineages may have evolved endosymbiosis independently, supported by ongoing discoveries of convergent patterns of host/symbiont genome coevolution (reviewed by Wilson and Duncan 2015). Given the timing of the slimfast expansion, its origin is more likely influenced by a trait shared by aphids and phylloxera, such as their complex life cycle that involves both sexual and asexual reproduction (Blackman and Eastop 2000; Forneck and Huber 2009). Indeed, we previously reported on male-biased and asexual female-biased slimfast paralogs in aphids (Duncan et al. 2011), supporting the notion that slimfast paralogs were retained as a result of selection for divergence to fulfill sex-specific roles.
Dynamic Evolution of Amino Acid Transporters in Aphids
APC Family
Despite evidence that the slimfast expansion was driven by a shared trait between aphids and phylloxera, retention of slimfast paralogs in aphids was most likely influenced by more complex and dynamic factors and processes. For example, gene duplication within the slimfast expansion continued after aphids and phylloxera diverged (fig. 2), implying that (1) additional selective pressures, perhaps shifting from ancestral selective pressures, influenced the retention of additional slimfast paralogs as they emerged, (2) additional slimfast paralogs emerged by chance and were retained through nonadaptive processes (e.g., the classic model of subfunctionalization known as Duplication, Degeneration, Complementation [Force et al. 1999]), or (3) aphids are particularly prone to gene duplication. Notably, these three possibilities are not mutually exclusive and could all be operating.
Another important aspect of paralog evolution in the slimfast expansion is highlighted by two pairs of A. pisum/M. persicae paralogs that have conserved bacteriocyte enrichment: ACYPI008904/Mper-APC08 and ACYPI005118/Mper-APC10. A change in expression in these two orthologous pairs from the typical gut expression of slimfast (Price et al. 2011) toward bacteriocyte enrichment implies that these slimfast paralogs were recruited to the aphid/Buchnera symbiotic interface and retained for a role in endosymbiosis. In addition to being recruited to bacteriocytes, ACYPI008904 and Mper-APC08 may have diverged functionally from other members of their subclade (fig. 2). Subclade 3 of the slimfast expansion experienced gene duplication in the common ancestor of Macrosiphini, resulting in multiple orthologous pairs for A. pisum and M. persicae while the other three aphid species each have only one gene. Gene duplication in subclade 3 provides an opportunity for functional divergence among paralogs while still fulfilling their ancestral function in two ways: (1) by evolving novel function and/or expression (neofunctionalization) or (2) by partitioning, and possibly optimizing or specializing, ancestral functions in sister paralogs (subfunctionalization). Indeed, supporting divergence following gene duplication in subclade 3, while ACYPI008904 and Mper-APC08 are both highly enriched in bacteriocytes, the closely related paralogs ACYPI002633 and Mper-APC09 are both bacteriocyte-depleted (Price et al. 2011) (fig. 3). In fact, our previous work in A. pisum revealed that ACYPI008904 and ACYPI002633 have very different expression profiles—while ACYPI008904 is enriched in bacteriocytes (Price et al. 2011) as well as asexual adult females (Duncan et al. 2011), ACYPI002633 is enriched in both gut (Price et al. 2011) and adult male A. pisum (Duncan et al. 2011). In addition, the branches leading to both ACYPI002633 and the clade containing ACYPI008904 and ACYPI008323 experienced accelerated rates of evolution (Duncan et al. 2011; Price et al. 2011), supporting the possibility that gene duplication was followed by both expression and functional divergence.
Without expression data for the other three aphid species, we cannot infer if the different expression profiles of the A. pisum and M. persicae APC paralogs result from neofunctionalization or subfunctionalization, both of which have implications for the role of endosymbiosis in the evolution of amino acid transporters. If bacteriocyte expression is novel in ACYPI008904 and Mper-APC08, then amino acid transporter recruitment to bacteriocytes is an ongoing process in aphid evolution. In contrast, if single-copy aphid orthologs are also expressed in bacteriocytes, then we could infer that Buchnera infection coincided with recruitment of the single-copy, ancestral gene of subclade 3 to aphid bacteriocytes. In light of expression data for M. persicae (supplementary file S6, Supplementary Material online) and A. pisum (Hansen and Moran 2011; Price et al. 2011; Macdonald et al. 2012), we predict that the ancestral aphid gene of subclade 3 operated at the symbiotic interface, coincident with Buchnera infection. After all, the sister clade, subclade 4, contains the only additional slimfast paralogs with conserved bacteriocyte enrichment between A. pisum (ACYPI005118) and M. persicae (Mper-APC10). Thus, parsimony would predict that aphid slimfast members in both subclades 3 and 4 inherited bacteriocyte expression from their common ancestor—or, since subclades 3 and 4 appeared before the aphid/phylloxera split, their ancestral gene may have been expressed in the cells that gave rise developmentally to aphid bacteriocytes after the aphids and phylloxera diverged from a common ancestor (Wilson and Duncan 2015).
A scenario in which bacteriocyte expression evolved in single-copy aphid orthologs would imply that the ancestral aphid gene of subclade 3 operated in endosymbiosis in addition to other, previously defined functions in place prior to aphid/phylloxera divergence. Importantly, there is a precedent for genes with broad expression to play important roles in endosymbiosis. Indeed, the A. pisum AAAP member ACYPI001018 (also known as ApGLNT1) is both globally highly expressed and also a key regulator of Buchnera metabolic output (Price et al. 2014, 2015)—a role that is very possibly conserved in M. persicae, given the conserved high bacteriocyte expression of the ortholog Mper-AAAP05.
AAAP Family
Amino acid transporters in the AAAP family, like the slimfast paralogs, expanded at different time scales during aphid evolution. AAAP members resulting from duplication in the aphid common ancestor, including ACYPI000536/Mper-AAAP11 and ACYPI001366/Mper-AAAP03, are prime candidates for facilitating transporter divergence toward a role in endosymbiosis. The fact that both these pairs of orthologs have conserved bacteriocyte enrichment while related paralogs have different expression profiles (fig. 4, supplementary file 6, Supplementary Material online) suggests that gene duplications in the aphid common ancestor were followed by functional divergence to symbiotic and nonsymbiotic roles. Our phylogeny also supports three gene duplications predating the diversification of the tribe Macrosiphini (fig. 3). Retention of these sets of orthologs is unlikely to have been driven by acquisition of the primary endosymbiont Buchnera, given that they lack conservation of differential expression between A. pisum and M. persicae.
Gene Duplication and Endosymbiosis in Aphids
The factors driving gene duplication in the amino acid transporters of Aphidomorpha are complex—a pattern that may also influence our understanding of gene duplications in other host genomes and their role in endosymbiosis. Our data continue to point toward the importance of nonsymbiotic traits in driving selection for paralog retention. At the same time, our data support a role for endosymbiosis in maintaining duplicated amino acid transporters in aphids. Importantly, we find that the most parsimonious explanation for conserved bacteriocyte-enrichment in orthologous pairs is that amino acid transporters were recruited to bacteriocytes in the aphid common ancestor—coinciding with acquisition of the primary, obligate endosymbiont, Buchnera.
Given the repeated support we have found for both symbiotic and nonsymbiotic roles for gene duplication in sap-feeding insects (Duncan et al. 2011, 2014; Price et al. 2011; Dahan et al. 2015), we caution against drawing strong conclusions about the role endosymbiosis plays in gene duplications found in the genomes of other symbiotic systems. An understanding of the natural history of a focal taxon can indeed help with making predictions about the evolutionary significance of interesting genomic patterns like gene duplication. However, by using a comparative approach, we have found that multiple complex factors maintain paralogs in a group of genes that are functionally critical to host/symbiont interactions. Moving forward, we advocate using a comparative framework with as much information as possible about the expression and function of duplicated genes. Differential expression analysis of symbiotic and nonsymbiotic tissues using RNAseq or qRT-PCR provides a valuable layer of information that contributes to our ability to infer the role of duplicated genes and the factors contributing to their retention in a genome. These genomic and transcriptomic approaches pave the way for the next phase in understanding why genomes evolved their particular architecture through application of functional genomic approaches.
Supplementary Material
Supplementary table S1 and supplementary files S1–S6 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Patrick Abbot for the raw Illumina reads for P. obesinymphae and T. coweni. Claude Rispe and François Delmotte generously provided the assembled D. vitifoliae transcriptome, and Nicole Gerardo and Stephanie Chiang provided total RNA for the A. nerii transcriptome. Transcriptomes were assembled and analyzed on the Blacklight system at the Pittsburgh Supercomputing Center. Blacklight is part of the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation (NSF) grant number OCI-1053575. This work was supported by NSF Doctoral Dissertation Improvement Grant DEB-1406631 (R.P.D.), NSF IOS-1121847 (A.C.C.W.), NSF IOS-1354154 (A.C.C.W.), NSF Graduate Research Fellowship DG1E-0951782 (R.P.D.), an REU supplement to IOS-1121847 (A.C.C.W.) for support of undergraduate researcher DMN and USDA-NIFA award 2010-65105-2055 (A.C.C.W.).
Literature Cited
- Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105. [DOI] [PubMed] [Google Scholar]
- Arnegard ME, Zwickl DJ, Lu Y, Zakon HH. 2010. Old gene duplication facilitates origin and diversification of an innovative communication system—twice. Proc Natl Acad Sci U S A. 107:22172–22177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarten S, et al. 2015. The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc Natl Acad Sci U S A. 112:11893–11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman R, Eastop VF. 2000. Aphids on the World’s Crops. 2nd ed New York: John Wiley & Sons, LTD. [Google Scholar]
- Caillaud M, Boutin M, Braendle C, Simon J-C. 2002. A sex-linked locus controls wing polymorphism in males of the pea aphid, Acyrthosiphon pisum (Harris). Heredity 89:346–352. [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombani J, et al. 2003. A nutrient sensor mechanism controls Drosophila growth. Cell 114:739–749. [DOI] [PubMed] [Google Scholar]
- Dahan RA, Duncan RP, Wilson ACC, Dávalos LM. 2015. Amino acid transporter expansions associated with the evolution of obligate endosymbiosis in sap-feeding insects (Hemiptera: sternorrhyncha). BMC Evol Biol. 15:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C, Cheng C-HC, Ye H, He X, Chen L. 2010. Evolution of an antifreeze protein by neofunctionalization under escape from adaptive conflict. Proc Natl Acad Sci U S A. 107:21593–21598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AE. 1989. Mycetocyte symbiosis in insects. Biol Rev Camb Philos Soc. 64:409–434. [DOI] [PubMed] [Google Scholar]
- Duncan RP, et al. 2014. Dynamic recruitment of amino acid transporters to the insect/symbiont interface. Mol Ecol. 23:1608–1623. [DOI] [PubMed] [Google Scholar]
- Duncan RP, Nathanson L, Wilson AC. 2011. Novel male-biased expression in paralogs of the aphid slimfast nutrient amino acid transporter expansion. BMC Evol Biol. 11:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 2009. A new generation of homology search tools based on probabilistic inference. Genome Inf. 23:205–211. [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39:W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A, et al. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151:1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forneck A, Huber L. 2009. (A)sexual reproduction - a review of life cycles of grape phylloxera, Daktulosphaira vitifoliae. Entomol Exp Appl. 131:1–10. [Google Scholar]
- Goldman N, Yang Z. 1994. A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol Biol Evol. 11:725–736. [DOI] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. 2010. SeaView Version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 27:221–224. [DOI] [PubMed] [Google Scholar]
- Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 8:1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen AK, Moran NA. 2011. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc Natl Acad Sci U S A. 108:2849–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, Marcet-Houben M, Pignatelli M, Moya A, Gabaldon T. 2010. The pea aphid phylome: a complete catalogue of evolutionary histories and arthropod orthology and paralogy relationships for Acyrthosiphon pisum genes. Insect Mol Biol. 19:13–21. [DOI] [PubMed] [Google Scholar]
- Innan H, Kondrashov F. 2010. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 11:97–108. [DOI] [PubMed] [Google Scholar]
- International Aphid Genomics Consortium 2010. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 8:e1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov FA. 2012. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc R Soc B. 279:5048–5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Conery J. 2000. The evolutionary fate and consequences of duplicate genes. Science 290:1151–1155. [DOI] [PubMed] [Google Scholar]
- Macdonald SJ, Lin GG, Russell CW, Thomas GH, Douglas AE. 2012. The central role of the host cell in symbiotic nitrogen metabolism. Proc R Soc B. 279:2965–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina RF, Nachappa P, Tamborindeguy C. 2011. Differences in bacterial diversity of host-associated populations of Phylloxera notabilis Pergande (Hemiptera: Phylloxeridae) in pecan and water hickory. J Evol Biol. 24:761–771. [DOI] [PubMed] [Google Scholar]
- Miller DG, Lawson SP, Rinker DC, Estby H, Abbot P. 2015. The origin and genetic differentiation of the socially parasitic aphid Tamalia inquilinus. Mol Ecol. 24:5751–5766. [DOI] [PubMed] [Google Scholar]
- Misof B, et al. 2014. Phylogenomics resolves the timing and pattern of insect evolution. Science 346:763–767. [DOI] [PubMed] [Google Scholar]
- Moran NA, Munson MA, Baumann P, Ishikawa H. 1993. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc R Soc B. 253:167–171. [Google Scholar]
- Nováková E, et al. 2013. Reconstructing the phylogeny of aphids (Hemiptera: Aphididae) using DNA of the obligate symbiont Buchnera aphidicola. Mol Phylogenet Evol. 68:42–54. [DOI] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by Gene Duplication. New York: Springer. [Google Scholar]
- Ott M, Zola J, Stamatakis A, Aluru S. 2007. Large-scale maximum likelihood-based phylogenetic analysis on the IBM BlueGene/L. New York: ACM Press; p. 1. [Google Scholar]
- Price DRG, et al. 2014. Aphid amino acid transporter regulates glutamine supply to intracellular bacterial symbionts. Proc Natl Acad Sci U S A. 111:320–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price DRG, Duncan RP, Shigenobu S, Wilson ACC. 2011. Genome expansion and differential expression of amino acid transporters at the aphid/Buchnera symbiotic interface. Mol Biol Evol. 28:3113–3126. [DOI] [PubMed] [Google Scholar]
- Price DRG, Wilson ACC, Luetje CW. 2015. Proton-dependent glutamine uptake by aphid bacteriocyte amino acid transporter ApGLNT1. Biochim Biophys Acta. 1848:2085–2091. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Suchard MA, Xie D, Drummond AJ. 2014. Tracer v.1.6. http://beast.bio.ed.ac.uk/Tracer.
- Ramsey JS, et al. 2007. Genomic resources for Myzus persicae: EST sequencing, SNP identification, and microarray design. BMC Genomics 8:423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JA, Moran NA. 2006. Costs and benefits of symbiont infection in aphids: variation among symbionts and across temperatures. Proc R Soc B. 273:603–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandström JP, Moran NA. 2001. Amino acid budgets in three aphid species using the same host plant. Physiol Entomol. 26:202–211. [Google Scholar]
- Shinzato C, et al. 2011. Using the Acropora digitifera genome to understand coral responses to environmental change. Nature 476:320–323. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. [DOI] [PubMed] [Google Scholar]
- Sukumaran J, Holder MT. 2010. DendroPy: a Python library for phylogenetic computing. Bioinformatics 26:1569–1571. [DOI] [PubMed] [Google Scholar]
- Thwaites DT, Anderson CMH. 2011. The SLC36 family of proton-coupled amino acid transporters and their potential role in drug transport. Br J Pharmacol. 164:1802–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voordeckers K, et al. 2012. Reconstruction of ancestral metabolic enzymes reveals molecular mechanisms underlying evolutionary innovation through gene duplication. PLoS Biol. 10:e1001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorwerk S, Martinez-Torres D, Forneck A. 2007. Pantoea agglomerans-associated bacteria in grape phylloxera (Daktulosphaira vitifoliae, Fitch). Agric Entomol. 9:57–64. [Google Scholar]
- Wilson ACC, Duncan RP. 2015. Signatures of host/symbiont genome coevolution in insect nutritional endosymbioses. Proc Natl Acad Sci U S A. 112:10255–10261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young ND, et al. 2011. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480:520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, et al. 2006. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinformatics 4:259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.