Abstract
Gene loss is the driving force for changes in genome and morphology; however, this particular evolutionary event has been poorly investigated in leguminous plants. Legumes (Fabaceae) have some lineage-specific and diagnostic characteristics that are distinct from other angiosperms. To understand the potential role of gene loss in the evolution of legumes, we compared six genome-sequenced legume species of Papilionoideae, the largest representative clade of Fabaceae, such as Glycine max, with 34 nonlegume plant species, such as Arabidopsis thaliana. The results showed that the putative orthologs of the 34 Arabidopsis genes belonging to 29 gene families were absent in these legume species but these were conserved in the sequenced nonlegume angiosperm lineages. Further evolutionary analyses indicated that the orthologs of these genes were almost completely lost in the Papillionoideae ancestors, thus designated as the legume lost genes (LLGs), and these underwent purifying selection in nonlegume plants. Most LLGs were functionally unknown. In Arabidopsis, two LLGs were well-known genes that played a role in plant immunity such as HARMLESS TO OZONE LAYER 1 and HOPZ-ACTIVATED RESISTANCE 1, and 16 additional LLGs were predicted to participate in plant–pathogen interactions in in silico expression and protein–protein interaction network analyses. Most of these LLGs’ orthologs in various plants were also found to be associated with biotic stress response, indicating the conserved role of these genes in plant defense. The evolutionary implication of LLGs during the development of the ability of symbiotic nitrogen fixation involving plant and bacterial interactions, which is a well-known characteristic of most legumes, is also discussed. Our work sheds light on the evolutionary implication of gene loss events in Papilionoideae evolution, as well as provides new insights into crop design to improve nitrogen fixation capacity.
Keywords: defense response, gene loss, genome evolution, legume, nitrogen fixation
Introduction
Genomic changes such as gene gain and loss events frequently occur during genome evolution (Domazet-Loso and Tautz 2003; Krylov et al. 2003). Genes specifically added to the genome of a species lineage are defined as “taxonomically restricted” genes (TRGs) or orphan genes and these have no significant sequence similarity to genes of other species lineages (Wilson et al. 2005). Comparative genomics has now demonstrated that TRGs are a universal feature of any genome (Khalturin et al. 2009; Tautz and Domazet-Loso 2011; Long et al. 2013; Arendsee et al. 2014) and play essential roles in the species evolution. Drosophila TRGs are involved in the evolution of lineage-specific ecological adaptations (Domazet-Loso and Tautz 2003), and Hydra TRGs play a role in the creation of phylum-specific novelties and in the innate defense system, and are thus also involved in species-specific adaptive processes (Khalturin et al. 2009). Lineage- or species-specific TRGs have also been identified in various plants such as in Arabidopsis thaliana (Lin et al. 2010), Oryza sativa (Campbell et al. 2007), Solanum spp. (Rensink et al. 2005), and legumes (Graham et al. 2004; Schmutz et al. 2010). Some TRGs are preferentially expressed in A. thaliana (Donoghue et al. 2011) and rice (Guo et al. 2007) in reaction to abiotic stresses, whereas O. sativa defense-responsive gene 10 (OsDR10), a rice tribe-specific gene, negatively regulates resistance to a broad spectrum of Xanthomonas oryzae pv. oryzae strains (Xiao et al. 2009). Therefore, TRGs are involved in the response to various stresses, thus contributing to adaptive evolution.
Genes can also be deleted from the genome during evolution. Some members in one gene family are often lost in certain lineages, but, in extreme cases, an entire gene family may be deleted from the genomes of certain lineages (Aravind et al. 2000; Demuth and Hahn 2009), creating lineage-specific lost genes. Gene loss and pseudogenization can lead to immediate loss of gene function, thus severely affecting major physiological processes of organisms. However, it may also open new developmental opportunities, confer a selective advantage, and serve as an engine for evolutionary change in bacterium and animals (Olson 1999; D’Souza et al. 2014; Gladieux et al. 2014). Loss of superfluous genes contributes to bacteria fitness (Koskiniemi et al. 2012) and is the driving force in the adaptation of parasites to eukaryotic cells (Merhej et al. 2009; Cisse et al. 2014; Sharma et al. 2014). The loss of a penicillin-binding protein may contribute to resistance to the cephalosporin drug, ceftazidime, in Burkholderia pseudomallei (Torok et al. 2012). Human-specific loss of a myosin heavy chain isoform expressed in the masticatory muscles has been linked to the weakening of human jaw muscles, which has been suggested to increase cranial capacity in humans (Stedman et al. 2004).
Gene losses are also involved in the evolutionary divergence of floral morphology in plants. The loss of an anthocyanin pathway enzyme is associated with the transition from blue to red floral pigmentation, thus resulting in phenotypic differences among species of the Andean Iochroma of the Solanaceae (Smith and Rausher 2011). The loss of lineage-specific MADS-box genes such as GLOBOSA or DEFICIENS is potentially associated with the evolutionary divergence of floral morphology during the radiation of the Euasterids I within core eudicots (Lee and Irish 2011). Heterotopic expression of a MADS-box gene 2-like from Physalis floridana (MPF2-like) is required for the fruiting calyx inflation trait called “Chinese lantern,” yet is physiologically known as inflated calyx syndrome (ICS) in Physalis (He and Saedler 2005), whereas loss of a copy of MPF2-like genes is involved in the loss of ICS in Tubocapsicum (Khan et al. 2009). Therefore, the evolution of lineage-specific gene loss can result in new morphological traits among species or genera.
The legume (Fabaceae) consisting of three clades, Papilionoideae, Caesalpinioideae, and Mimosaceae, includes important grain, pasture, and agroforestry species and is characterized by unusual flower structure, podded fruit, and the ability of most species to form nodules with rhizobia (De Faria et al. 1989). However, genetic variations that could distinguish legumes from nonlegumes have not been identified. In particular, nodule formation is a developmental process connecting plant and bacterial cell differentiation (Roux et al. 2014). Much of this process remains a mystery although a few symbiosis-related genes have been identified in legumes (Schauser et al. 1999; Catoira et al. 2000; Limpens et al. 2003). Genome sequencing of Glycine max and its comparison with distantly related species such as Populus trichocarpa has revealed specific gene gains in legumes (Schmutz et al. 2010). However, gene loss has not been evaluated in relation to the evolution of legumes. No genome of Caesalpinioideae and Mimosaceae has yet been sequenced, but the whole-genome sequencing of five additional legume species in Papilionoideae, the largest and most widely distributed clade of Fabaceae such as Lotus japonicus (Sato et al. 2008), Medicago truncatula (Young et al. 2011), Cajanus cajan (Varshney et al. 2011), Cicer arietinum (Varshney et al. 2013), and Phaseolus vulgaris (Schmutz et al. 2014) has allowed investigations on lineage-specific losses in Papilionoideae, thereby gaining insights into the adaptive role of gene loss in the entire legume family. In this study, we identified the legume lost genes (LLGs) through genome-wide comparative analyses of legume and nonlegume species. Thirty-four Arabidopsis genes had orthologs in nonlegume species but were not detected in legumes. Eighteen LLGs were directly or indirectly inferred to function in the plant–pathogen interaction in nonlegumes. Therefore, the loss of these genes might have partially contributed to genomic changes that were related to the evolution of symbiotic nitrogen fixation in legumes.
Materials and Methods
Sequence Availability
Whole genome-wide primary transcript sequences of G. max, P. vulgaris, M. truncatula, and 34 nonlegume species were downloaded from Phytozome v10 (http://www.phytozome.net, last accessed March 1, 2015). Sequences of L. japonicus were obtained from Kazusa DNA Research Institute (http://www.kazusa.or.jp/lotus, last accessed March 1, 2015), and those of C. cajan and C. arietinum were downloaded from the International Crops Research Institute for the Semi-Arid Tropics (http://www.icrisat.org, last accessed March 1, 2015). The version of each database is summarized in supplementary table S1, Supplementary Material online.
Identification of LLGs
To identify possible LLGs, we used all coding sequences (CDS) of A. thaliana to conduct Basic Local Alignment Search Tool (BLAST) analysis of sequences of earlier described legume species. The identification steps are presented in supplementary figure S1, Supplementary Material online. Putative LLGs were selected under the BLAST results, with an E-value cutoff of 1 × 10−5. To rule out Arabidopsis-specific genes, the putative LLGs were searched in three selected genomes of Vitis vinifera, Prunus persica, and P. trichocarpa (E-value = 1 × 10−10). Arabidopsis genes without any hits in six legume species but showing homologous sequences in all three nonlegume species were further verified at the protein level in the aforementioned nine species using bidirectional BLASTP (E-value = 1 × 10−4), and bidirectional best hits were defined as putative orthologs. When the best hit in a species of one putative LLG in Arabidopsis was also the best match for another putative LLG in Arabidopsis, the putative LLG’s ortholog (bidirectional best hit) was not considered to be lost from this species. When proteins with bidirectional best hits in three nonlegume species had hits in legumes with an E-value of <1 × 10−4 but did not show bidirectional best hits, these were defined as Group 1 LLGs, indicating that the legume species had lost orthologs of the LLGs. Protein sequences without any hits in the legume species but with bidirectional best hits in the three nonlegume species were defined as Group 2 LLGs. These BLAST results were further verified by orthoMCL v1.4 analysis (Li et al. 2003). Phylogenetic analyses were performed whenever a conflicting signal was observed among bidirectional BLAST and orthoMCL. Nucleotide sequences were aligned using the Clustal X v2.1 program with default parameters (Larkin et al. 2007). Alignments were optimized via manual adjustment, and partial sequences with poor alignment were excluded. Substitution saturation was tested using DAMBE v6.0.1 before phylogenetic analysis (Xia 2013). Unrooted maximum-likelihood trees were constructed using the PhyML v3.1 program using a generalized time-reversible model with 100 bootstrap resamplings (Guindon et al. 2010).
The LLGs’ orthologs were further characterized in 29 other angiosperm species and Selaginella moellendorffii by bidirectional BLASTP using protein sequences (E-value = 1 × 10−4). The phylogeny of the involved plant species was derived from Cogepedia (http://genomevolution.org/wiki/index.php/Sequenced_plant_genomes, last accessed March 1, 2015) and APG III (Angiosperm Phylogeny Group 2009). Ancestral character state reconstruction was performed with the Markov k-state 1 parameter model (Mk1) in Mesquite 3.03 (http://mesquiteproject.org, last accessed March 1, 2015). Information on the gene families of the LLGs was generated from the Phytozome v10 clusters at the angiosperm node. Gene ontology (GO) annotations of LLGs were derived from Blast2GO (Conesa et al. 2005) using the National Center for Biotechnology Information nonredundant database (September 30, 2015).
Identification of Conserved Genes in Angiosperms
The genes conserved in A. thaliana, V. vinifera, P. trichocarpa, P. persica, and all six legumes were used as controls throughout the work. To identify conserved genes, the primary CDS sequences of A. thaliana were subjected to BLAST analysis using the aforementioned three nonlegume and six legume species (E-value = 1 × 10−5), and then Arabidopsis genes showing homologs in the aforementioned nine species were subjected to BLASTP. The resulting protein-encoding genes showing bidirectional best hits in all nine species (E-value = 1 × 10−4) were designated as conserved genes.
Gene Structure Analysis
CDS length, intron number, and intron length covering the CDS of the identified LLGs in nonlegume species were obtained from gff3 profiles downloaded from Phytozome v10 and TAIR10 (http://www.arabidopsis.org, last accessed March 1, 2015).
In Silico Expression Prediction
The expression data of roots, seedlings, expanding leaves, stems, vegetative shoot meristems, whole inflorescences, flowers, and fruits of Arabidopsis were obtained from a previous work (Laubinger et al. 2008). Relative gene expression levels (Z-scores) in different tissues were calculated as previously described (Benedito et al. 2008). When the Z-score value of a given gene in a tissue was not <1.5, the gene was considered highly expressed in the tissue. Gene expression of the identified LLGs under various hormonal treatments and biotic stresses was based on a previous study (Ma and Bohnert 2007). The expression data of the identified LLGs’ orthologs in response to biotic stresses in tomato were taken from the Tomato Functional Genomics database (http://ted.bti.cornell.edu/, last accessed March 1, 2015), and related data of rice and grape were extracted from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/projects/geo, last accessed March 1, 2015). Heat map of gene expression was performed using MeV 4.9.0 (http://www.tm4.org/mev.html, last accessed March 1, 2015).
Gene Coexpression and Enrichment Analyses
The genes coexpressed with the LLGs in A. thaliana were identified using the MAS5 algorithm in CressExpress v3.2 (http://cressexpress.org, last accessed March 1, 2015). The employed parameters were as follows: cutoff value for Kolmogorov–Smirnov quality-control statistic was 0.15, and R2 threshold for pathway-level coexpression was set as 0.36. Genes coexpressed with each LLG were sorted by correlation index, and the top 50 (if <50, then all genes were used) were used in enrichment analysis. Protein sequences of genes coexpressed with each LLG were submitted to KOBAS 2.0 (http://kobas.cbi.pku.edu.cn, last accessed March 1, 2015) for the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Biocyc enrichment analyses. GO enrichment analysis was performed on AGRIGO v1.2 (http://bioinfo.cau.edu.cn/agriGO, last accessed March 1, 2015), with the reference genome locus obtained from TAIR10. The P value of all enrichment analyses was set as 0.05.
Protein–Protein Interaction Prediction
The identified LLGs in Arabidopsis were submitted to the A. thaliana section of STRING 10 (http://string-db.org, last accessed March 1, 2015) to generate protein–protein interaction (PPI) networks.
Microcolinearity Analysis
To identify a syntenic block harboring a LLG between a legume species and a nonlegume species, we first determined the respectively 10 genes upstream and downstream of each LLG in the genome of Arabidopsis. OrthoMCL v1.4 (Li et al. 2003) was used to construct orthologous groups around LLGs across multiple plant taxa with default parameters. The schematic diagram of local genomic synteny was drawn manually.
Selection Test
Gene selection was evaluated by using the ω (ω = dN/dS; dN, nonsynonymous substitution rates; dS, synonymous substitution rates) that was calculated by the codeml program of PAML v4.8 under Models 0 (Yang 2007). The protein sequences were aligned using Clustal-omega v1.2.0, with default parameters (Sievers et al. 2011), and then the nucleotide sequences were aligned using a Perl module derived from ParaAT (Zhang et al. 2012). Poorly aligned regions were removed from each alignment using Gblocks v0.91 b (Castresana 2000) with the following parameters: type of sequence = codons, maximum number of contiguous nonconserved positions = 8, minimum length of a block = 10, and gap positions excluded from all sequences. After performing Gblocks, matrixes with nucleotide sites < 50 were discarded. The significance in ω difference was evaluated using the Kolmogorov–Smirnov test.
Results
LLGs Survey
To identify LLGs, we selected four nonlegume species and six legume species to initiate genome-wide comparisons (supplementary fig. S2, Supplementary Material online). We first used the Arabidopsis genome to probe the genomes of nine other plant species at the nucleotide level, and determined that 70 Arabidopsis genes had homologous sequences in three other nonlegume species, whereas their homologous sequences were not detected in the six legume species (supplementary table S2, Supplementary Material online), indicating a potential loss of these genes in legumes. We then verified these identified putative LLGs at the protein level. Using BLASTP, 34 of these Arabidopsis genes were found to have putative orthologous proteins in all three nonlegume species (V. vinifera, P. trichocarpa, and P. persica), whereas no putative orthologous proteins were detected in the six legume species (table 1 and supplementary table S2, Supplementary Material online). These genes were designated as LLGs and were further divided into two groups. In the legume species, 26 of the LLGs lost their orthologous genes but had homologous sequences and were thus designated as Group 1 LLGs. On the other hand, Group 2 LLGs included eight genes without any homologous sequences in various legume species (table 1).
Table 1.
LLGs
| Query ID | Vv | Pt |
Pp |
Gm |
Pv |
Cc |
Mt |
Ca |
Lj |
Gene Symbol |
|---|---|---|---|---|---|---|---|---|---|---|
| AT1G09195.2 (1) | y (1) | y (1) | y (1) | N (0) | N (0) | N (0) | N (0) | N (0) | N (0) | |
| AT1G35340.1 (1) | y (1) | y (1) | y (1) | N (0) | N (0) | N (0) | n (0) | n (0) | N (0) | |
| AT1G64385.1 (1) | y (1) | y (2) | y (1) | N (0) | n (0) | N (0) | n (0) | N (0) | N (0) | |
| AT2G43210.1 (1) | y (1) | y (2) | y (1) | n (0) | n (0) | n (0) | N (0) | N (0) | n (0) | |
| AT2G43910.2 (2)a | y (0) | y (0) | y (0) | N (0) | N (0) | N (0) | N (0) | N (0) | N (0) | HOL1 |
| AT2G43920.1 (2)a | y (0) | y (0) | y (0) | N (0) | N (0) | N (0) | N (0) | N (0) | N (0) | HOL2 |
| AT2G43940.1 (1)a | y (2) | y (1) | y (1) | n (0) | n (0) | N (0) | N (0) | n (0) | N (0) | HOL3 |
| AT4G14970.1 (1) | y (1) | y (1) | y (1) | N (0) | N (0) | N (0) | N (0) | N (0) | N (0) | |
| AT4G22160.2 (1) | y (2) | y (3) | y (1) | N (0) | N (0) | N (0) | n (0) | n (0) | N (0) | |
| AT4G29560.1 (1) | y (1) | y (1) | y (1) | N (0) | N (0) | N (0) | N (0) | N (0) | N (0) | |
| AT5G44010.1 (1) | y (1) | y (1) | y (1) | N (0) | N (0) | N (0) | N (0) | N (0) | N (0) | |
| AT5G49110.2 (1) | y (1) | y (1) | y (1) | N (0) | N (0) | N (0) | N (0) | N (0) | N (0) | |
| AT5G65740.2 (1) | y (1) | y (1) | y (1) | N (0) | N (0) | N (0) | N (0) | N (0) | N (0) | |
| AT1G13630.1 (1) | y (1) | y (1) | y (2) | n (0) | n (0) | n (0) | n (0) | n (0) | n (0) | |
| AT1G55580.1 (1) | y (1) | y (3) | y (1) | n (0) | n (0) | n (0) | n (0) | n (0) | n (0) | LAS |
| AT1G55590.1 (1) | y (1) | y (1) | y (1) | n (0) | n (0) | n (0) | n (0) | n (0) | n (0) | |
| AT1G68940.3 (3)d | y (2) | y (4) | y (2) | n (2) | n (1) | n (1) | n (1) | n (1) | n (1) | |
| AT1G71120.1 (1) | y (1) | y (1) | y (1) | n (0) | n (0) | n (0) | n (0) | n (0) | n (0) | |
| AT2G05810.1 (1) | y (1) | y (2) | y (1) | n (0) | n (0) | n (0) | n (0) | n (0) | n (0) | |
| AT2G18520.1 (2)b | y (1) | y (2) | y (1) | n (0) | n (0) | n (0) | n (0) | n (0) | n (0) | |
| AT4G36680.1 (2)b | y (1) | y (2) | y (1) | n (0) | n (0) | n (0) | n (0) | n (0) | n (0) | |
| AT2G39100.1 (1) | y (1) | y (1) | y (1) | n (0) | n (0) | n (0) | n (0) | n (0) | n (0) | |
| AT2G45530.1 (1) | y (1) | y (2) | y (1) | n (0) | n (0) | n (0) | n (0) | n (0) | n (0) | |
| AT3G24515.1 (1) | y (1) | y (2) | y (1) | n (0) | n (0) | n (0) | n (0) | n (0) | n (0) | |
| AT3G50950.2 (1) | y (1) | y (1) | y (2) | n (0) | n (0) | n (0) | n (0) | n (0) | n (0) | ZAR1 |
| AT3G61210.1 (3)e | y (0) | y (0) | y (0) | n (0) | n (0) | n (0) | n (0) | n (0) | n (0) | |
| AT4G11670.1 (1) | y (1) | y (1) | y (1) | n (0) | n (0) | n (0) | n (0) | n (0) | n (0) | |
| AT4G24340.1 (2)c | y (1) | y (3) | y (1) | n (0) | n (0) | n (0) | n (0) | n (0) | n (0) | |
| AT4G24350.1 (2)c | y (1) | y (3) | y (1) | n (0) | n (0) | n (0) | n (0) | n (0) | n (0) | |
| AT5G01015.1 (1) | y (1) | y (2) | y (1) | n (0) | n (0) | n (0) | n (0) | n (0) | n (0) | |
| AT5G04840.1 (1) | y (1) | y (2) | y (1) | n (0) | n (0) | n (0) | n (0) | n (0) | n (0) | |
| AT5G10830.1 (1) | y (2) | y (1) | y (2) | n (0) | n (0) | n (0) | n (0) | n (0) | n (0) | |
| AT5G12460.1 (1) | y (1) | y (2) | y (2) | n (0) | n (0) | n (0) | n (0) | n (0) | n (0) | |
| AT5G66160.1 (1) | y (1) | y (1) | y (1) | n (0) | n (0) | n (0) | n (0) | n (0) | n (0) | RMR1 |
Vv, Vitis vinifera; Pt, Populus trichocarpa; Pp, Prunus persica; Gm, Glycine max; Pv, Phaseolus vulgaris; Cc, Cajanus cajan; Mt, Medicago truncatula; Ca, Cicer arietinum; Lj, Lotus japonicus. The symbol y indicates that the LLG had putative orthologs in this species, and n represents that the LLG had homologous protein sequences but no putative ortholog in this species. N indicates that the LLG had no homologous sequence in this species. Genes in bold indicate the Group 2 LLGs. The numbers in parenthesis represent number of genes in this species within the orthoMCL group containing LLG.
HOL1 and HOL2 are in one orthoMCL group; and HOL1, HOL2, and HOL3 belong to the HOL family.
AT2G18520 and AT4G36680 are in one orthoMCL group.
AT4G24340 and AT4G24350 are in one orthoMCL group.
AT1G68940 and two non-LLGs (AT1G20780 and AT1G76390) belong to one orthoMCL group.
AT3G61210.1 and two non-LLGs (AT1G55450 and AT3G54150) comprise one orthoMCL group.
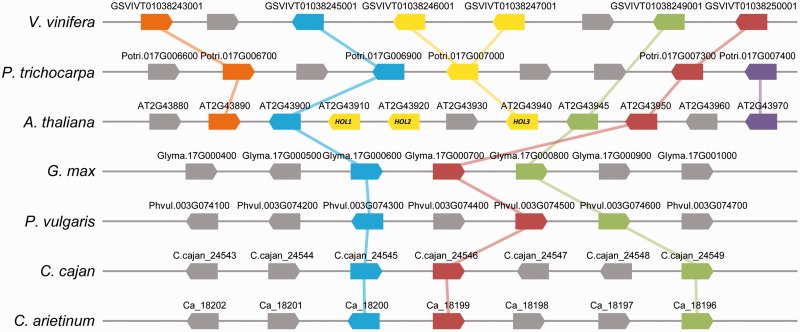
We further performed orthoMCL analyses in the legume species, and 33 orthologs of the aforementioned LLGs were not detected (table 1), thus supporting the BLAST results. Nevertheless, a few inconsistencies were observed. No orthologs of AT1G68940 were detected in the legume species using BLAST, whereas in the orthoMCL analyses, AT1G68940 was determined to have possible orthologs in the legume species, and AT3G61210, AT2G43910, and AT2G43920 seemed to be specific to Arabidopsis (table 1). To assess these inconsistencies, phylogenetic analyses of related genes were performed (supplementary fig. S3, Supplementary Material online), and the results suggested that the orthologs of these genes were present in nonlegumes but absent from legumes. Thus, we ultimately identified 34 LLGs that belonged to 29 gene families in A. thaliana (supplementary table S3, Supplementary Material online). Group 2 LLGs (8 genes) belonged to seven families, and six LLGs in this group formed six single-copy gene family, except for AT2G43910 and AT2G43920 being homologs. In Group 1 LLGs (26 genes), ten were from single-copy gene families in Arabidopsis (supplementary table S3, Supplementary Material online). Multiple-copy gene families could also be lost during evolution such as the HARMLESS TO OZONE LAYER (HOL) family in Arabidopsis, which included AT2G43910 (HOL1), AT2G43920 (HOL2), and AT2G43940 (HOL3), and their orthologs were not detected in legumes (fig. 1 and supplementary table S3, Supplementary Material online).
Fig. 1.—
Local synteny around the HOL genes in legumes and nonlegume species. Species names are provided on the left. The block arrow on the horizontal line represents one open reading frame of a gene and its orientation. The yellow blocks are HOL family genes. The same color block arrows connected with the same color lines indicate the putative orthologs in different species. The gray block arrows are nonhomologous genes. The nomenclature of each putative gene is shown above the corresponding block arrow.
Evidence of Gene Loss from Microsynteny Analyses
To verify LLGs, we assessed for genomic microsynteny around LLGs in nonlegume species in relation to that of legumes. Although these plant species have increasing taxonomic distance and complicated genome structure due to duplications, losses, and segmental reshuffling during evolution, the stretches of syntenic chromosomal segments could be still identified. For example, in nonlegumes (A. thaliana, V. vinifera, P. trichocarpa, and P. persica), 1–3 HOL genes were clustered, and their downstream and upstream regions shared a few conserved genes, which was indicative of synteny (fig. 1). However, no HOL homologous sequences were detected in four legume species (G. max, P. vulgaris, C. cajan, and C. arietinum), although these were conserved in nonlegumes (fig. 1), thus indicating loss of HOL genes in these legumes. Altogether, 20 LLGs were found to maintain a relatively good local synteny among nonlegume species compared with legumes (supplementary table S4, Supplementary Material online). Thus, well maintenance of microsynteny verified the LLGs.
To understand the evolutionary implications of LLGs, we next investigated their evolution history in nonlegume plants.
The Evolution of LLGs in Nonlegume Angiosperms
Selection Pressure
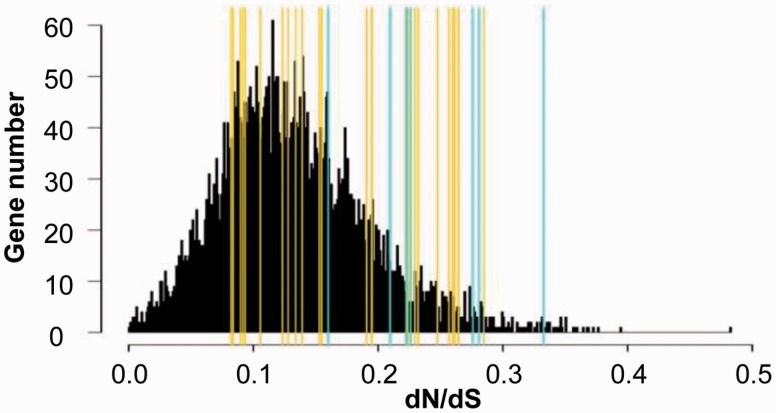
We evaluated the selection pressure of these LLGs in the aforementioned four nonlegume plant species through calculating dN/dS (ω). We found that the ω values of LLGs was <0.35 (0.08 < ω < 0.33) (fig. 2) suggesting that these LLGs might have undergone purifying selection during nonlegume evolution. Moreover, there was no difference in selection pressures between Groups 1 and 2 LLGs (P = 0.15). We further identified 5,935 Arabidopsis genes having reciprocal best hits in the genomes of three nonlegumes and six legumes as conserved genes in angiosperms (supplementary table S5, Supplementary Material online). The ω of these genes in legumes ranged from 0.00010 to 0.65 (supplementary table S5, Supplementary Material online), while it ranged from 0.00064 to 0.48 in the four nonlegume species (indicated by black columns, fig. 2), indicating these conserved genes underwent purifying selection in both nonlegumes and legumes. However, the ω distribution of these conserved genes in nonlegumes was significantly different from that of LLGs (P = 7.81e−07; fig. 2). These results indicated that LLGs were generally conserved during nonlegume evolution, yet might have undergone a relatively relaxed purifying selection compared with conserved genes.
Fig. 2.—
Selection analyses of LLGs in nonlegumes. Selection was evaluated in Arabidopsis thaliana, Prunus persica, Populus trichocarpa, and Vitis vinifera. Yellow lines and cyan lines, respectively, indicate the dN/dS value of the Group 1 LLGs and Group 2 LLGs. The dN/dS distribution of the 5,935 conserved genes in these four plants is presented in black columns.
Gene Loss Patterns
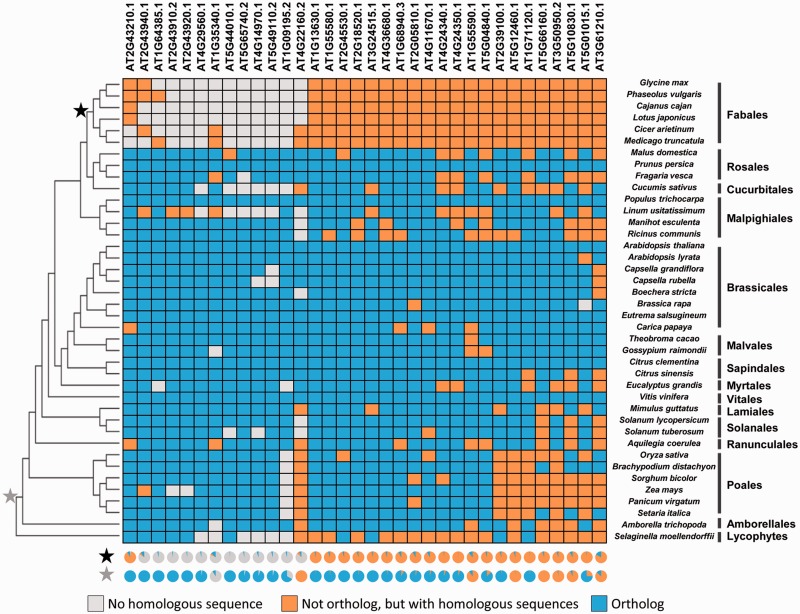
We next investigated the loss pattern of LLGs, including additional 30 genome-sequenced plants that included 29 angiosperm species and S. moellendorffii (supplementary table S1, Supplementary Material online). The orthologs of LLGs in these plant species were identified (supplementary table S6, Supplementary Material online), and the presence–absence pattern of the orthologs of each LLG was mapped to the phylogenetic tree of the involved plants (fig. 3). The presence of the ortholog was indicated in blue. When no ortholog was detected, the presence of putative homologs was indicated in orange. In extreme cases, the absence of homologs was highlighted in gray. The number of species with LLG orthologs in nonlegumes ranged from 17 to 33, and on average, around 28 nonlegume species harbored LLG orthologs. These results again indicated that LLGs were conserved in most plants, whereas these were lost in legumes. Moreover, the absence of LLGs in nonlegumes seemed to be independent of their phylogeny (fig. 3), that is, Cucurbitales and Rosales are closely related to legumes, and the orthologs of 15 LLGs were not detected in C. sativus, whereas these were detected in P. persica, indicating that LLGs might have been randomly lost in a few nonlegume species during evolution. This is consistent with observations made in Poaceae (Poales) and Malpighiales. Poaceae are very different from legumes, and only six LLGs (AT1G09195, AT4G22160, AT2G39100, AT5G12460, AT1G71120, and AT3G50950) were absent from all investigated species of Poaceae (fig. 3), hinting that these might be lost in Poaceae, which was supported by ancestral state reconstruction (supplementary fig. S4, Supplementary Material online). A few LLGs were absent from Euphorbiaceae (Malpighiales) but existed in other species within Malpighiales. Some LLGs, such as AT5G44010, AT4G14970, and AT5G49110, may have undergone multiple, independent gene loss events throughout Angiospermae (fig. 3). Although the systematic loss of a few LLGs in other lineages could not be excluded currently (supplementary fig. S4, Supplementary Material online), the gene loss pattern of these LLGs in nonlegumes was similar to that of the previously identified 5,935 conserved genes in angiosperms (supplementary fig. S5, Supplementary Material online) but different from the evolutionary pattern of legume LLGs that was apparently specific and systematic (fig. 3). Ancestral state reconstruction further showed that the ancestors of Papillionoideae might have lost most of the LLG orthologs, whereas these LLGs’ homologs were found in the ancestors of angiosperms with a few exclusions such as AT1G35340 (fig. 3 and supplementary fig. S4, Supplementary Material online).
Fig. 3.—
LLG evolution in sequenced angiosperms. Gray represents species without any homologous sequence to LLGs. Orange indicates species with LLG homologs but not orthologs. Blue represents species with putative LLG orthologs. The black and gray stars indicate the ancestors of Papillionoideae and Angiospermae, respectively, and their ancestral states are represented at the bottom (for details, see supplementary fig. S4, Supplementary Material online). Forty plant species whose genomes have been sequenced are included, and their phylogeny was deduced from Cogepedia and APG III. Selaginella moellendorffii was used as outgroup.
Comparison of structural features (i.e., protein length, intron number, and average intron length) of conserved genes and LLGs in Arabidopsis (supplementary table S7, Supplementary Material online), as well as assessment of the evolution of LLG structural characteristics in sequenced species (supplementary figs. S6–S8, Supplementary Material online) did not indicate any distinct structural variations.
Functional Clues of the LLGs in Nonlegumes
We further explored the evolutionary implications of these LLGs by investigating the functional roles of these genes in nonlegumes. Literature search revealed that only six LLGs were functionally inferred (table 1), which included HOL1, HOL2, HOL3, LATERAL SUPPRESSOR (LAS), receptor homology-transmembrane-ring H2 domain Protein 1 (RMR1), and HOPZ-ACTIVATED RESISTANCE 1 (ZAR1), and GO annotation revealed that LLGs may involve different biological processes such as those that participate in DNA repair (GO: 0006281) of AT4G14970, AT5G49110, and AT5G65740 (supplementary table S2, Supplementary Material online). Some LLGs have been found to be associated with stress response such as HOL1, HOL2, and ZAR1 (GO: 0006952). AT5G10830 was annotated for respiratory burst involved in defense response (GO: 0002679), and AT3G61210 was annotated for response to ethylene and salt stress (GO: 0009723 and GO: 0009651). The functions of unknown LLGs in nonlegumes were next envisioned through in silico expression and PPI analyses in Arabidopsis because information on these LLGs in other angiosperms is limited.
Expression of LLGs in Arabidopsis
We investigated the expression of LLGs in different tissues of the plant model, Arabidopsis (see Materials and Methods). Besides lacking AT1G09195 expression, the remaining LLGs were differentially expressed in eight tissues, whereas some LLGs showed distinct tissue-specific expression patterns (supplementary fig. S9, Supplementary Material online). Eleven LLGs were highly expressed in roots such as LAS, HOL3, AT4G24340, and AT5G10830, whereas LLGs such as AT1G35340, AT3G50950, HOL2, AT4G24350, and AT1G71120 were highly expressed in expanding leaves, and LLGs AT1G64385, AT2G39100, and AT5G04840 were preferentially expressed in fruits (indicated by the red boxes, supplementary fig. S9, Supplementary Material online). On the other hand, no LLGs were highly expressed in Arabidopsis flowers (Z-score < 0.94).
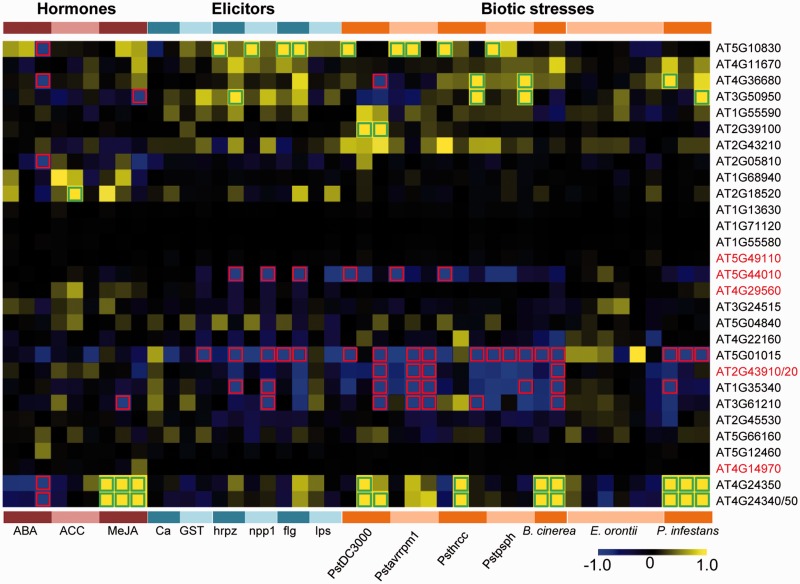
We also investigated LLG expression in response to various biotic stimuli (e.g., hormones, elicitors, and pathogens). The transcript profiles of 30 LLGs were detected, whereas those of four LLGs such as AT1G09195, AT5G65740, AT1G64385, and HOL3 were not detected. In addition, 14 LLGs were involved in response to biotic treatments, including the two function-known genes, HOL1 and ZAR1 (fig. 4). The expression of HOL1 and its close homolog, HOL2, responded to hormone treatments such as abscisic acid (ABA), 1-aminocyclopropane-1-carboxylic acid (ACC), methyl jasmonate (MeJA), as well as to elicitors such as hairpin z (hrpz). In addition, challenging with bacterial pathogens such as Pseudomonas syringae pv. tomato DC3000 (PstDC3000), P. syringae pv. tomato avrRPM1 (Pstavrrpm1), and Botrytis cinerea (B. cinerea) resulted in a significant downregulation in expression of the two HOL genes (fig. 4 and supplementary fig. S10A, Supplementary Material online). ZAR1 also responded to these hormonal treatments and elicitors such as hrpz, and its expression was downregulated during PstDC3000 and Pstavrrpm1 treatments, but upregulated during P. syringae pv. tomato DC3000 hrcC (Psthrcc) and P. syringae pv. phaseolicola (Pstpsph) treatments (fig. 4 and supplementary fig. S10B, Supplementary Material online). Besides these three genes, Arabidopsis orthologs of 11 functionally unknown LLGs also showed differential expression (both upregulation and downregulation) in response to these treatments, which included AT1G35340, AT2G05810, AT2G18520, AT2G39100, AT3G61210, AT4G24340, AT4G24350, AT4G36680, AT5G01015, AT5G10830, and AT5G44010 (fig. 4), indicating that these genes are probably also involved in plant defense response in Arabidopsis. We further investigated the expression of these LLG orthologs in other plants whose transcriptomic variations challenging its own bacterial pathogens are publically available (supplementary table S8, Supplementary Material online), which showed that the orthologs of fourteen LLGs, such as AT1G35340 in rice, tomato, and grape also responded to various biotic stresses (table 2) suggesting that the role of each LLG may be conserved in other nonlegumes.
Fig. 4.—
Heat map of LLG expression under different stresses in Arabidopsis. Hormone treatments include ABA, ACC, and MeJA. Elicitors include CaCl2 (Ca), glutathione S-transferase (GST), hrpz, GST-necrosis-inducing phytophtora Protein 1 (npp1), flagellin (flg), and lipopolysaccharide (lps). Bacterial stresses include Pseudomonas syringae pv. tomato DC3000 (PstDC3000), P. syringae pv. tomato avrRPM1 (Pstavrrpm1), P. syringae pv. tomato DC3000 hrcC (Psthrcc), P. syringae pv. phaseolicola (Pstpsph), Botrytis cinerea (B. cinerea), Erysiphe orontii (E. orontii), and Phytophthora infestans (P. infestans). The column above each treatment represents gene expression of different time points after treatment (for details, see supplementary fig. S10, Supplementary Material online).The color scale indicates the log2 values of expression change (treatments/control). Yellow indicates upregulation under treatments, and blue indicates downregulation under treatments. Green boxes indicate the log2 values of fold changes (treatments/control) > 1.0, and red boxes indicate the log2 values of fold changes (treatments/control) < −1.0. Gene names in red are Group 2 LLGs. AT2G43910/20 represents AT2G43910 (HOL1) and AT2G43920 (HOL2), and AT4G24340/50 represents AT4G24340 and AT4G24350 because one probe ID could detect the two closely related genes.
Table 2.
Summary of LLGs Involved in Plant Defense Response
| LLGs |
Arabidopsis |
Rice | Tomato | Grape | |||
|---|---|---|---|---|---|---|---|
| Function | Expression | PPI | Expression | ||||
| AT1G35340.1 | Y | Y | Y | Y | |||
| AT1G64385.1 | Y | ||||||
| AT2G43910.2 (HOL1) | Y | Y | |||||
| AT2G43920.1 (HOL2) | Y | ||||||
| AT4G14970.1 | Y | ||||||
| AT4G29560.1 | Y | ||||||
| AT5G44010.1 | Y | Y | |||||
| AT5G49110.2 | Y | ||||||
| AT5G65740.2 | Y | ||||||
| AT2G05810.1 | Y | Y | |||||
| AT2G18520.1 | Y | Y | |||||
| AT4G36680.1 | Y | Y | Y | ||||
| AT2G39100.1 | Y | ||||||
| AT3G24515.1 (UBC37) | Y | Y | |||||
| AT3G50950.2 (ZAR1) | Y | Y | Y | Y | |||
| AT3G61210.1 | Y | Y | Y | ||||
| AT4G11670.1 | Y | ||||||
| AT4G24340.1 | Y | Y | Y | ||||
| AT4G24350.1 | Y | Y | Y | ||||
| AT5G01015.1 | Y | ||||||
| AT5G10830.1 | Y | Y | Y | Y | |||
Y indicates that evidence for LLG involvement in defense response was detected. PPI, protein-protein interaction.
Enrichment Analysis of Genes Coexpressed with LLGs
Genes in the same pathway and genes that have related functions often exhibit similar expression patterns, which is why analysis of gene coexpression networks is a useful way of developing functional annotation (Usadel et al. 2009; Lin et al. 2010; Childs et al. 2011). To further explore the function of these LLGs, we performed coexpression analyses. A total of 21 LLG coexpressed gene sets were detected (R2 > 0.36), which in turn were further subjected to functional enrichment analysis. The three coexpressed LLG groups included AT1G35340/AT5G01015, LAS/AT2G05810/AT4G29560, and AT2G18520/AT4G36680 (supplementary table S9, Supplementary Material online). Moreover, the genes coexpressed with LLGs were putatively involved in multiple fundamental biological processes such as DNA replication, ribosome biogenesis, protein processing, and secondary metabolites (supplementary table S10, Supplementary Material online). DNA replication (ath03030) was significantly enriched in the coexpressed genes of AT4G14970 (supplementary table S10, Supplementary Material online). Ribosome biogenesis (ath03008) and pyrimidine ribonucleotide biosynthetic process (GO: 0009220) were significantly enriched in the coexpression genes of AT2G18520 and AT4G36680, whereas protein processing in the endoplasmic reticulum (ath04141) and purine transport (GO: 0006863) were simultaneously enriched in the coexpressed genes of AT2G05810 and LAS (supplementary table S10, Supplementary Material online). The enriched KEGG pathway associated with secondary metabolites such as phenylpropanoid biosynthesis (ath00940) was significantly enriched in coexpression genes of AT5G10830 (supplementary table S10, Supplementary Material online). Furthermore, three LLGs (ZAR1, AT4G24340, and AT4G24350) were associated with plant defense (supplementary table S10, Supplementary Material online). For example, defense-response and incompatible interaction (GO: 0009814) and plant–pathogen interaction (ath04626) were enriched in the gene set that was coexpressed with ZAR1, whereas plant-type hypersensitive response (GO: 0010363) and jasmonic acid biosynthesis (PWY-735) were enriched in the gene set that was coexpressed with AT4G24340/AT4G24350 (supplementary table S10, Supplementary Material online).
PPIs Associated with LLGs
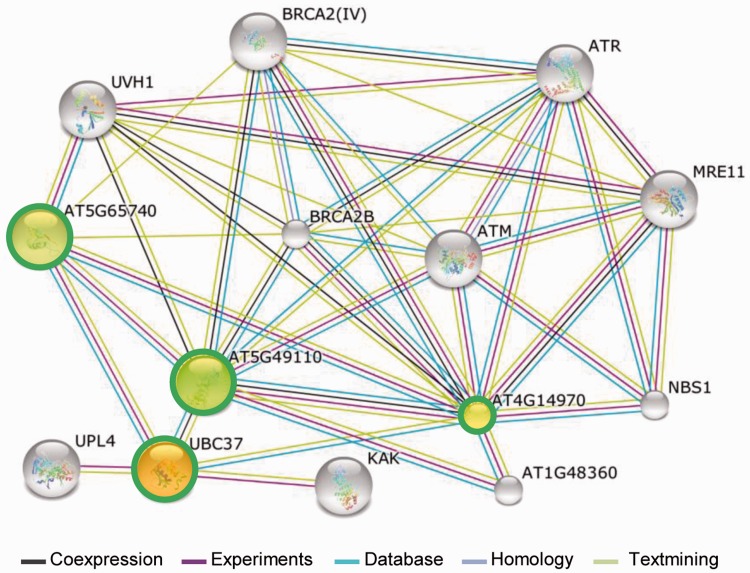
We further predicted PPI networks associated with the identified LLGs. Among all the LLGs examined, four PPI networks were detected (supplementary fig. S11, Supplementary Material online). Three networks involved in each of the two LLGs such as AT2G43210/AT2G05810, AT2G18520/AT4G36680, and AT4G24340/AT4G24350, whereas the largest PPI network was associated with four LLGs, AT3G24515, AT4G14970, AT5G49110, and AT5G65740 (supplementary fig. S11, Supplementary Material online), suggesting that these played a role in protein ubiquitination because AT3G24515 encoded the putative ubiquitin-conjugating enzyme 37 (UBC37).
We also observed that LLGs or LLG PPI networks interacted with various non-LLG proteins that were involved in a wide range of biological processes such as DNA repair, cell division, protein processing, and plant defense (supplementary tables S11 and S12, Supplementary Material online). The LLG (AT2G43210)-interacting proteins included Arabidopsis Cell Division Cycle 48B (AtCDC48B), AtCDC48C, AtCDC48, Radiation-Sensitive 23B (RAD23B), RAD23C, and RAD23D (supplementary table S11, Supplementary Material online), which was suggestive of its involvement in cell division. On the other hand, LLG AT5G10830 appeared to be associated with factors such as BONZAI association Protein 1 (Yang et al. 2007; supplementary table S11, Supplementary Material online), which suggested that it might play a role in plant immunity. Interestingly, the largest PPI network of the four LLGs interacted with ten non-LLG proteins (fig. 5). Eight of these proteins such as ataxia-telangiectasia mutated (ATM), ataxia telangiectasia-mutated and Rad3-related protein (ATR), Nijmegen breakage Syndrome 1 (NBS1), breast cancer 2-like B (BRCA2B), BRCA2 (IV), meiotic recombination 11 (MRE11), ultraviolet-hypersensitive 1 (UVH1), and Fanconi/Fancd2-associated Nuclease I (FAN1) participated in DNA repair, whereas two proteins were from the HECT ubiquitin-protein ligase (UPL) family of proteins such as AT4G38600 (UPL3) and AT5G02880 (UPL4), thus indicating that these LLG proteins is likely involved in ubiquitination (fig. 5 and supplementary table S12, Supplementary Material online).
Fig. 5.—
The largest PPI network associated with LLGs. Each node represents a protein. The four nodes covered by a green circle indicate the four LLGs. The colorful connecting lines represent the types of evidence supporting each association: coexpression (dark brown), experiments (pink), databases (cyan), homology (violet), and text mining (light green).
Other possible developmental roles of these LLGs genes could not be ruled out in plants but extensive data mining suggested that LLGs and their orthologs in nonlegumes are associated primarily with plant defense response (table 2).
Discussion
Gene loss has been investigated in the past decade (Aravind et al. 2000; Moran 2002; Krylov et al. 2003; Wang et al. 2006); however, its importance has only recently attracted attention. Gene loss probably affects organisms to a greater extent than do most amino acid substitutions, thus serving as one of the main drivers in the evolution of gene families, morphological diversity, and adaptation (Lee and Irish 2011; Smith and Rausher 2011; Koskiniemi et al. 2012; De Smet et al. 2013; Dakovic et al. 2014), as well as in organogenesis and speciation (Scannell et al. 2006; Castro et al. 2013). However, gene loss during legume evolution has not been extensively investigated. In this study, we evaluated gene loss events that might have occurred during the evolution of Papilionoideae at the genome-level, and identified 34 LLGs that were lost in a legume-specific manner (fig. 3 and table 1). Altogether 21 LLGs and orthologs in nonlegume species were determined to be associated with plant defense systems (table 2). Therefore, adaptive evolution of Papilionoideae might be implicated in the evolution of these LLGs.
LLGs Are Largely Involved in Plant Defense Response in Nonlegumes
The identified LLGs belonged to multiple gene families, and most of these were not functionally inferred. Based on literature search, gene expression analysis, and PPI prediction, we determined that LLGs might have played diverse roles in nonlegumes. LLG AT5G66160, which encodes the receptor homology region transmembrane domain ring H2 motif Protein 1 (AtRMR1), functions as the sorting receptor of phaseolin, thus facilitating in trafficking protein molecules to its corresponding storage vacuole (Park et al. 2005). LLG AT1G55580, which encodes LAS, plays a key regulatory role in the formation of lateral shoots during the vegetative development of tomato (Schumacher et al. 1999), Arabidopsis (Greb et al. 2003), and cucumber (Yuan et al. 2010). On the other hand, LLG AT2G43210 is possibly involved in cell division because most of its protein partners play essential roles in the cell cycle (supplementary table S11, Supplementary Material online; Park et al. 2008; Farmer et al. 2010). We also determined that a substantial amount of LLGs were involved in biotic stress responses (table 2). HOL1 is involved in the defense response to pathogens in Arabidopsis (Nagatoshi and Nakamura 2009), whereas ZAR1 is responsible for the recognition of the P. syringae Type III secreted effector HopZ1a, which attenuates HopZ1a virulence (Lewis et al. 2010). Similar to the two well-known defense-response genes, 12 LLGs were determined to respond to various biotic stresses in Arabidopsis (fig. 4 and table 2), and ten of the 14 LLGs were also detected in either rice, tomato, or grape (table 2), indicating that these might also participate in the defense response. Moreover, genes sharing a common role or function in a particular pathway were coexpressed. We observed that genes coexpressed with LLGs ZAR1, AT4G24340, and AT4G24350 were enriched in the regulation of plant defense response (supplementary table S10, Supplementary Material online).
The proteins in a PPI network are also probably involved in the same functional pathway (Lin et al. 2015; Zhang et al. 2015). Our PPI network prediction provides substantial informative functional clues for some LLGs. Notably, the largest PPI network that associated four LLGs AT3G24515 (UBC37), AT5G49110, AT5G65740, and AT4G14970 interacted with ATM, ATR, NBS1, BRCA2B, BRCA2(IV), MRE11, UVH1, and FAN1. Arabidopsis ATM, ATR, and NBS1 were involved in double-strand breaks of meiosis (Garcia et al. 2003; Waterworth et al. 2007; Culligan and Britt 2008), and BRCA2B and BRCA2(IV) are important for both DNA break repair and homologous recombination in somatic or meiotic cells (Abe et al. 2009; Seeliger et al. 2012). MRE11 (AT5G54260) plays a role in the early stages of MRE (Bleuyard et al. 2004; Puizina et al. 2004). UVH1 (AT5G41150), also known as AtRAD1, is a homolog of the yeast repair endonuclease RAD1, and is involved in nucleotide excision repair and telomere stability (Fidantsef et al. 2000; Vannier et al. 2009). FAN1 (AT1G48360) is involved in DNA crosslink repair (Herrmann et al. 2015). These observations indicate that these LLGs were presumably involved in DNA repair. Increased somatic recombination was observed in plants subjected to pathogen stress (Lucht et al. 2002). Furthermore, the DNA damage repair proteins, BRCA2 and RAD51, are involved in the regulation of plant defense gene expression (Choi et al. 2001; Durrant et al. 2007; Wang et al. 2010; Song et al. 2011), indicating that the LLGs associated with DNA repair might also be involved in plant responses to microbial pathogens. Furthermore, AT4G38600 (UPL3) and AT5G02880 (UPL4), which were observed in the largest LLGs’ PPI network, exert a role in ubiquitination system (Downes et al. 2003), and ubiquitination is required in plant immunity for the degradation of invading proteins (Trujillo and Shirasu 2010). Therefore, our multiple lines of evidence suggest that around 21 LLGs and orthologs are directly or indirectly involved in plant defense responses.
The Evolution of LLGs in Angiosperms
A gene that is continuously maintained in the genome indicates that it plays an essential role in viability (Krylov et al. 2003). In contrast, a gene without extensive and essential biological functions could be lost during evolution. Therefore, selection could be a significant driving force of gene loss (Koskiniemi et al. 2012). Because there are no whole-genome sequences available for the other two clades of Fabaceae, this study determined that LLGs specifically originated from Papilionoideae during legume evolution. On the other hand, nonlegume angiosperm species apparently had conserved these LLGs in its genome, which subsequently underwent negative selection. Single-copy genes often exhibit higher sequence conservation than nonsingle copy genes (De Smet et al. 2013). In line with this observation, 16 LLGs were single-copy families in Arabidopsis, whereas these maintained a low number of copies in most nonlegume species (supplementary table S3, Supplementary Material online). Therefore, LLGs might have originated from a direct gene deletion from the genome of a legume ancestor, instead of sequence divergence that mainly occurred when a gene is subjected to positive selection. The protein length and exon number of orphan genes, also called TGSs, are significantly different from those of nonorphan genes (Domazet-Loso and Tautz 2003). However, this study determined that the structural evolution of LLGs, lineage-specific lost genes, did not play a role in the emergence of these LLGs during plant evolution. Therefore, the major factors that drove the lineage-specific loss of these LLGs remain unclear. Nevertheless, most LLGs originated from ancestral legumes. Gene loss has been considered as a common and advantageous response during the genome evolution of living organisms (Wang et al. 2006). However, the role of LLGs in legume evolution requires more extensive investigations. Nonetheless, in the light of the putative role of LLGs in response to biotic stresses, we speculate that the loss of these genes plays a beneficial and adaptive role in the evolution of legumes.
Evolutionary Implication of LLGs
Root nodule is specialized organ of legumes, and nodule formation is initiated through the molecular cross-talk between a bacterium and a plant, thus involving a complex and precise interplay between host and symbiont, and shifting the intracellular signaling from defense response to symbiosis (Beck et al. 2008; Nakagawa et al. 2011). Several symbiosis-related genes have been identified in legumes, and their mutants show various defects in the nodule formation (Schauser et al. 1999; Catoira et al. 2000; Limpens et al. 2003). These symbiosis-related genes have orthologs in nonlegumes (Zhu et al. 2006; Zhang et al. 2009). Therefore, the lineage-specific gain or loss of certain genes is likely required in the development of the legume nodulation pathway.
Legume-specific gene families have been identified (Silverstein et al. 2006; Schmutz et al. 2010), and some of these show root- and/or nodule-specific expression (Severin et al. 2010), thus indicating the potential role of lineage-specific gene gain in the formation or maintenance of symbiosis. As compensation, we exploited the possible role of lineage-specific gene loss in the evolution of legumes. Eighteen LLGs in Arabidopsis were determined to participate in defense response (table 2) such as HOL1, HOL2, and ZAR1. In particular, four LLGs in a PPI network (AT3G24515, AT4G14970, AT5G49110, and AT5G65740) were apparently involved in plant–bacterial interactions. Moreover, some LLGs associated with defense responses were highly expressed in roots such as AT2G18520, AT4G36680, AT4G24340, AT3G24515 (UBC37), and AT5G10830 (table 2). LAS was also upregulated in roots but was apparently not associated with nodulation, whereas LAS was predicted to interact with carotenoid cleavage Dioxygenase 7 (supplementary table S11, Supplementary Material online), which is the ortholog in L. japonicus that controls determinate nodulation (Liu et al. 2013), thus suggesting that the root-expressed LAS might also be involved in nodulation. Therefore, LLGs could largely contribute to the improvement of compatibility between legume and rhizobia, thereby facilitating the establishment of reciprocal symbiosis.
In summary, through a genome-wide comparison, we identified a set of LLGs. The mechanisms and driving forces of LLG losses remain elusive; nonetheless, evolutionary loss of certain genes that are involved in plant immunity may provide new insights into elucidating the mechanisms underlying symbiotic nitrogen fixation. This work, for the first time, sheds light on the evolutionary implications of gene loss events in the evolution of Papilionoideae. Whether these findings can be generalized across the entire legume family requires further investigations. Engineering nitrogen-fixed genes in crops is essential for sustainable food production, and the results of this study thus also suggest that knocking out certain LLGs should also be considered in such kind of crop design.
Supplementary Material
Acknowledgments
The authors thank Dr M. Zhao for her valuable suggestions to improve the article. The assistance of Miss Y.Y. Liu in phylogenetic analysis is also acknowledged. This work was supported by the grant (XDA08010105) from the Chinese Academy of Sciences and the CAS/SAFEA International Partner Program for Creative Research Teams of “Systematic and Evolutionary Botany.”
Supplementary Material
Supplementary figures S1–S11 and tables S1–S12 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Literature Cited
- Abe K, et al. 2009. Inefficient double-strand DNA break repair is associated with increased fasciation in Arabidopsis BRCA2 mutants. J Exp Bot 60:2751–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angiosperm Phylogeny Group. 2009. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc 161:105–121. [Google Scholar]
- Aravind L, Watanabe H, Lipman DJ, Koonin EV. 2000. Lineage-specific loss and divergence of functionally linked genes in eukaryotes. Proc Natl Acad Sci U S A. 97:11319–11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arendsee ZW, Li L, Wurtele ES. 2014. Coming of age: orphan genes in plants. Trends Plant Sci. 19:698–708. [DOI] [PubMed] [Google Scholar]
- Beck S, et al. 2008. The Sinorhizobium meliloti MsbA2 protein is essential for the legume symbiosis. Microbiology 154:1258–1270. [DOI] [PubMed] [Google Scholar]
- Benedito VA, et al. 2008. A gene expression atlas of the model legume Medicago truncatula. Plant J. 55:504–513. [DOI] [PubMed] [Google Scholar]
- Bleuyard JY, Gallego ME, White CI. 2004. Meiotic defects in the Arabidopsis rad50 mutant point to conservation of the MRX complex function in early stages of meiotic recombination. Chromosoma 113:197–203. [DOI] [PubMed] [Google Scholar]
- Campbell MA, et al. 2007. Identification and characterization of lineage-specific genes within the Poaceae. Plant Physiol. 145:1311–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552. [DOI] [PubMed] [Google Scholar]
- Castro LF, et al. 2013. Recurrent gene loss correlates with the evolution of stomach phenotypes in gnathostome history. Proc Biol Sci. 281:20132669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catoira R, et al. 2000. Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12:1647–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs KL, Davidson RM, Buell CR. 2011. Gene coexpression network analysis as a source of functional annotation for rice genes. PLoS One 6:e22196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JJ, Klosterman SJ, Hadwiger LA. 2001. A comparison of the effects of DNA-damaging agents and biotic elicitors on the induction of plant defense genes, nuclear distortion, and cell death. Plant Physiol. 125:752–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisse OH, Pagni M, Hauser PM. 2014. Comparative genomics suggests that the human pathogenic fungus Pneumocystis jirovecii acquired obligate biotrophy through gene loss. Genome Biol Evol. 6:1938–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, et al. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. [DOI] [PubMed] [Google Scholar]
- Culligan KM, Britt AB. 2008. Both ATM and ATR promote the efficient and accurate processing of programmed meiotic double-strand breaks. Plant J. 55:629–638. [DOI] [PubMed] [Google Scholar]
- Dakovic N, et al. 2014. The loss of adipokine genes in the chicken genome and implications for insulin metabolism. Mol Biol Evol. 31:2637–2646. [DOI] [PubMed] [Google Scholar]
- De Faria SM, Lewis GP, Sprent JI, Sutherland JM. 1989. Occurrence of nodulation in the leguminosae. New Phytol 111:607–619. [DOI] [PubMed] [Google Scholar]
- De Smet R, et al. 2013. Convergent gene loss following gene and genome duplications creates single-copy families in flowering plants Proc Natl Acad Sci U S A. 110:2898–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuth JP, Hahn MW. 2009. The life and death of gene families. Bioessays 31:29–39. [DOI] [PubMed] [Google Scholar]
- Domazet-Loso T, Tautz D. 2003. An evolutionary analysis of orphan genes in Drosophila. Genome Res. 13:2213–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donoghue MT, Keshavaiah C, Swamidatta SH, Spillane C. 2011. Evolutionary origins of Brassicaceae specific genes in Arabidopsis thaliana. BMC Evol Biol. 11:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes BP, Stupar RM, Gingerich DJ, Vierstra RD. 2003. The HECT ubiquitin-protein ligase (UPL) family in Arabidopsis: UPL3 has a specific role in trichome development. Plant J. 35:729–742. [DOI] [PubMed] [Google Scholar]
- D’Souza G, et al. 2014. Less is more: selective advantages can explain the prevalent loss of biosynthetic genes in bacteria. Evolution 68:2559–2570. [DOI] [PubMed] [Google Scholar]
- Durrant WE, Wang S, Dong X. 2007. Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response. Proc Natl Acad Sci U S A. 104:4223–4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer LM, et al. 2010. The RAD23 family provides an essential connection between the 26S proteasome and ubiquitylated proteins in Arabidopsis. Plant Cell 22:124–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidantsef AL, Mitchell DL, Britt AB. 2000. The Arabidopsis UVH1 gene is a homolog of the yeast repair endonuclease RAD1. Plant Physiol. 124:579–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia V, et al. 2003. AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 15:119–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladieux P, et al. 2014. Fungal evolutionary genomics provides insight into the mechanisms of adaptive divergence in eukaryotes. Mol Ecol 23:753–773. [DOI] [PubMed] [Google Scholar]
- Graham MA, Silverstein KA, Cannon SB, VandenBosch KA. 2004. Computational identification and characterization of novel genes from legumes. Plant Physiol. 135:1179–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greb T, et al. 2003. Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 17:1175–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321. [DOI] [PubMed] [Google Scholar]
- Guo WJ, Li P, Ling J, Ye SP. 2007. Significant comparative characteristics between orphan and nonorphan genes in the rice (Oryza sativa L.) genome. Comp Funct Genomics. 2017: Article ID 21676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C, Saedler H. 2005. Heterotopic expression of MPF2 is the key to the evolution of the Chinese lantern of Physalis, a morphological novelty in Solanaceae Proc Natl Acad Sci U S A. 102:5779–5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann NJ, Knoll A, Puchta H. 2015. The nuclease FAN1 is involved in DNA crosslink repair in Arabidopsis thaliana independently of the nuclease MUS81. Nucleic Acids Res. 43:3653–3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalturin K, Hemmrich G, Fraune S, Augustin R, Bosch TC. 2009. More than just orphans: are taxonomically-restricted genes important in evolution? Trends Genet. 25:404–413. [DOI] [PubMed] [Google Scholar]
- Khan MR, Hu JY, Riss S, He C, Saedler H. 2009. MPF2-like-A MADS-box genes control the inflated calyx syndrome in Withania (Solanaceae): roles of Darwinian selection. Mol Biol Evol. 26:2463–2473. [DOI] [PubMed] [Google Scholar]
- Koskiniemi S, Sun S, Berg OG, Andersson DI. 2012. Selection-driven gene loss in bacteria. PLoS Genet. 8:e1002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krylov DM, Wolf YI, Rogozin IB, Koonin EV. 2003. Gene loss, protein sequence divergence, gene dispensability, expression level, and interactivity are correlated in eukaryotic evolution. Genome Res. 13:2229–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. [DOI] [PubMed] [Google Scholar]
- Laubinger S, et al. 2008. At-TAX: a whole genome tiling array resource for developmental expression analysis and transcript identification in Arabidopsis thaliana. Genome Biol. 9:R112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HL, Irish VF. 2011. Gene duplication and loss in a MADS box gene transcription factor circuit. Mol Biol Evol. 28:3367–3380. [DOI] [PubMed] [Google Scholar]
- Lewis JD, Wu R, Guttman DS, Desveaux D. 2010. Allele-specific virulence attenuation of the Pseudomonas syringae HopZ1a type III effector via the Arabidopsis ZAR1 resistance protein. PLoS Genet. 6:e1000894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13:2178–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpens E, et al. 2003. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302:630–633. [DOI] [PubMed] [Google Scholar]
- Lin H, et al. 2010. Comparative analyses reveal distinct sets of lineage-specific genes within Arabidopsis thaliana. BMC Evol Biol. 10:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, et al. 2015. Comparative genomics reveals new candidate genes involved in selenium metabolism in prokaryotes. Genome Biol Evol. 7:664–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, et al. 2013. CAROTENOID CLEAVAGE DIOXYGENASE 7 modulates plant growth, reproduction, senescence, and determinate nodulation in the model legume Lotus japonicus. J Exp Bot 64:1967–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long M, VanKuren NW, Chen S, Vibranovski MD. 2013. New gene evolution: little did we know. Annu Rev Genet. 47:307–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucht JM, et al. 2002. Pathogen stress increases somatic recombination frequency in Arabidopsis. Nat Genet. 30:311–314. [DOI] [PubMed] [Google Scholar]
- Ma S, Bohnert HJ. 2007. Integration of Arabidopsis thaliana stress-related transcript profiles, promoter structures, and cell-specific expression. Genome Biol. 8:R49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merhej V, Royer-Carenzi M, Pontarotti P, Raoult D. 2009. Massive comparative genomic analysis reveals convergent evolution of specialized bacteria. Biol Direct. 4:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran NA. 2002. Microbial minimalism: genome reduction in bacterial pathogens. Cell 108:583–586. [DOI] [PubMed] [Google Scholar]
- Nagatoshi Y, Nakamura T. 2009. Arabidopsis HARMLESS TO OZONE LAYER protein methylates a glucosinolate breakdown product and functions in resistance to Pseudomonas syringae pv. maculicola. J Biol Chem. 284:19301–19309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, et al. 2011. From defense to symbiosis: limited alterations in the kinase domain of LysM receptor-like kinases are crucial for evolution of legume-rhizobium symbiosis. Plant J. 65:169–180. [DOI] [PubMed] [Google Scholar]
- Olson MV. 1999. When less is more: gene loss as an engine of evolutionary change. Am J Hum Genet. 64:18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Lee D, Lee GJ, Hwang I. 2005. AtRMR1 functions as a cargo receptor for protein trafficking to the protein storage vacuole. J Cell Biol. 170:757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Rancour DM, Bednarek SY. 2008. In planta analysis of the cell cycle-dependent localization of AtCDC48A and its critical roles in cell division, expansion, and differentiation. Plant Physiol. 148:246–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puizina J, Siroky J, Mokros P, Schweizer D, Riha K. 2004. Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell 16:1968–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensink WA, et al. 2005. Comparative analyses of six solanaceous transcriptomes reveal a high degree of sequence conservation and species-specific transcripts. BMC Genomics 6:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux B, et al. 2014. An integrated analysis of plant and bacterial gene expression in symbiotic root nodules using laser-capture microdissection coupled to RNA sequencing. Plant J. 77:817–837. [DOI] [PubMed] [Google Scholar]
- Sato S, et al. 2008. Genome structure of the legume, Lotus japonicus. DNA Res. 15:227–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH. 2006. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature 440:341–345. [DOI] [PubMed] [Google Scholar]
- Schauser L, Roussis A, Stiller J, Stougaard J. 1999. A plant regulator controlling development of symbiotic root nodules. Nature 402:191–195. [DOI] [PubMed] [Google Scholar]
- Schmutz J, et al. 2010. Genome sequence of the palaeopolyploid soybean. Nature 463:178–183. [DOI] [PubMed] [Google Scholar]
- Schmutz J, et al. 2014. A reference genome for common bean and genome-wide analysis of dual domestications. Nat Genet. 46:707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher K, Schmitt T, Rossberg M, Schmitz G, Theres K. 1999. The Lateral suppressor (Ls) gene of tomato encodes a new member of the VHIID protein family. Proc Natl Acad Sci U S A. 96:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeliger K, Dukowic-Schulze S, Wurz-Wildersinn R, Pacher M, Puchta H. 2012. BRCA2 is a mediator of RAD51- and DMC1-facilitated homologous recombination in Arabidopsis thaliana. New Phytol 193:364–375. [DOI] [PubMed] [Google Scholar]
- Severin AJ, et al. 2010. RNA-Seq Atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol. 10:160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R, Mishra B, Runge F, Thines M. 2014. Gene loss rather than gene gain is associated with a host jump from monocots to dicots in the smut fungus Melanopsichium pennsylvanicum. Genome Biol Evol. 6:2034–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein KA, Graham MA, VandenBosch KA. 2006. Novel paralogous gene families with potential function in legume nodules and seeds. Curr Opin Plant Biol. 9:142–146. [DOI] [PubMed] [Google Scholar]
- Smith SD, Rausher MD. 2011. Gene loss and parallel evolution contribute to species difference in flower color. Mol Biol Evol. 28:2799–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, et al. 2011. DNA repair proteins are directly involved in regulation of gene expression during plant immune response. Cell Host Microbe 9:115–124. [DOI] [PubMed] [Google Scholar]
- Stedman HH, et al. 2004. Myosin gene mutation correlates with anatomical changes in the human lineage. Nature 428:415–418. [DOI] [PubMed] [Google Scholar]
- Tautz D, Domazet-Loso T. 2011. The evolutionary origin of orphan genes. Nat Rev Genet. 12:692–702. [DOI] [PubMed] [Google Scholar]
- Torok ME, Chantratita N, Peacock SJ. 2012. Bacterial gene loss as a mechanism for gain of antimicrobial resistance. Curr Opin Microbiol. 15:583–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo M, Shirasu K. 2010. Ubiquitination in plant immunity. Curr Opin Plant Biol. 13:402–408. [DOI] [PubMed] [Google Scholar]
- Usadel B, et al. 2009. Co-expression tools for plant biology: opportunities for hypothesis generation and caveats. Plant Cell Environ 32:1633–1651. [DOI] [PubMed] [Google Scholar]
- Vannier JB, Depeiges A, White C, Gallego ME. 2009. ERCC1/XPF protects short telomeres from homologous recombination in Arabidopsis thaliana. PLoS Genet. 5:e1000380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney RK, et al. 2011. Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat Biotechnol 30:83–89. [DOI] [PubMed] [Google Scholar]
- Varshney RK, et al. 2013. Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat Biotechnol 31:240–246. [DOI] [PubMed] [Google Scholar]
- Wang S, Durrant WE, Song J, Spivey NW, Dong X. 2010. Arabidopsis BRCA2 and RAD51 proteins are specifically involved in defense gene transcription during plant immune responses. Proc Natl Acad Sci U S A. 107:22716–22721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Grus WE, Zhang J. 2006. Gene losses during human origins. PLoS Biol. 4:e52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterworth WM, et al. 2007. NBS1 is involved in DNA repair and plays a synergistic role with ATM in mediating meiotic homologous recombination in plants. Plant J. 52:41–52. [DOI] [PubMed] [Google Scholar]
- Wilson GA, et al. 2005. Orphans as taxonomically restricted and ecologically important genes. Microbiology 151:2499–2501. [DOI] [PubMed] [Google Scholar]
- Xia X. 2013. DAMBE5: a comprehensive software package for data analysis in molecular biology and evolution. Mol Biol Evol. 30:1720–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, et al. 2009. A rice gene of de novo origin negatively regulates pathogen-induced defense response. PLoS One 4:e4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Yang S, Li Y, Hua J. 2007. The Arabidopsis BAP1 and BAP2 genes are general inhibitors of programmed cell death. Plant Physiol. 145:135–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Young ND, et al. 2011. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480:520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan LH, et al. 2010. The Cucumber Lateral Suppressor gene (CLS) is functionally associated with axillary meristem initiation. Plant Mol Biol Rep 28:421–429. [Google Scholar]
- Zhang H, et al. 2015. Novel genes affecting blood pressure detected via gene-based association analysis. G3 5:1035–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XC, Cannon SB, Stacey G. 2009. Evolutionary genomics of LysM genes in land plants. BMC Evol Biol. 9:183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, et al. 2012. ParaAT: a parallel tool for constructing multiple protein-coding DNA alignments. Biochem Biophys Res Commun. 419:779–781. [DOI] [PubMed] [Google Scholar]
- Zhu H, Riely BK, Burns NJ, Ane JM. 2006. Tracing nonlegume orthologs of legume genes required for nodulation and arbuscular mycorrhizal symbioses. Genetics 172:2491–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.