Abstract
We present the results of a laboratory-based surveillance of dengue in Taiwan in 2014. A total of 240 imported dengue cases were identified. The patients had arrived from 16 countries, and Malaysia, Indonesia, the Philippines, and China were the most frequent importing countries. Phylogenetic analyses showed that genotype I of dengue virus type 1 (DENV-1) and the cosmopolitan genotype of DENV-2 were the predominant DENV strains circulating in southeast Asia. The 2014 dengue epidemic was the largest ever to occur in Taiwan since World War II, and there were 15,492 laboratory-confirmed indigenous dengue cases. Phylogenetic analysis showed that the explosive dengue epidemic in southern Taiwan was caused by a DENV-1 strain of genotype I imported from Indonesia. There were several possible causes of this outbreak, including delayed notification of the outbreak, limited staff and resources for control measures, abnormal weather conditions, and a serious gas pipeline explosion in the dengue hot spot areas in Kaohsiung City. However, the results of this surveillance indicated that both active and passive surveillance systems should be strengthened so appropriate public health measures can be taken promptly to prevent large-scale dengue outbreaks.
Introduction
Dengue virus (DENV), a mosquito-borne flavivirus, is the most prevalent arbovirus in tropical and subtropical regions in the world.1 The DENV genome consists of a single-stranded, positive sense RNA, which is approximately 10,700 nucleotides and contains a long open reading frame that encodes three structural proteins (capsid [C], premembrane/membrane [M], and envelope [E] proteins) and seven nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5).2,3 There are four serotypes of DENV (DENV-1 to DENV-4) that cause dengue. They produce a wide clinical spectrum of illness ranging from inapparent infection to moderate febrile illness, dengue fever, and, finally, severe and fatal hemorrhagic disease. Infection induces a lifelong protective immunity to the homologous serotype but only provides brief protection against heterologous serotypes.4–6
In recent decades, the number of dengue cases has rapidly expanded worldwide due to the growing habitats of the mosquito vectors Aedes aegypti and Aedes albopictus resulting from global climate change, a growing number of susceptible human hosts, and more frequent DENV exposure caused by increased international trade and travel.7,8 An estimated 3.97 billion people living in 128 countries are now at risk of dengue, and annually, an estimated 390 million dengue infections occur worldwide.9,10 Dengue is endemic to most countries in the Asia-Pacific region, with nearly 75% of the global population exposed to dengue living in this region.1,11
Taiwan is located in the western Pacific region; Ae. albopictus is found throughout Taiwan, whereas Ae. aegypti is only distributed in southern Taiwan.12 Over the last decade, multiple DENV strains have caused dengue outbreaks in Taiwan every year. Most of the epidemic strains were introduced from neighboring Asia-Pacific countries through air travel.13–16 To reduce the introduction of DENV strains into Taiwan and prevent local outbreaks, both passive (hospital-based reporting systems) and active surveillance systems (such as fever screening at airports and seaports, self-reports, and expanded screening for contacts of confirmed cases) have been implemented by central and local health departments. Samples of suspected dengue cases were submitted for laboratory diagnosis and confirmation. In this study, we present the results of the laboratory-based dengue surveillance in Taiwan in 2014.
Materials and Methods
Human serum samples.
Dengue is a notifiable infectious disease in Taiwan, and suspected cases must be reported within 24 hours of clinical diagnosis.17 Human serum samples of suspected dengue cases must be submitted to the Centers for Disease Control, Taiwan (Taiwan CDC) or a dengue laboratory in Kaohsiung Medicine University Hospital (approved by Taiwan CDC) to confirm DENV infection. The human serum samples used in this study were derived from confirmed dengue cases. Serum samples collected 1–7 days after the onset of symptoms were considered acute-phase samples, and those collected 8–30 days after the onset of disease were considered convalescent-phase samples. An imported dengue case was defined as an infected patient who had traveled abroad more than 2 weeks before the onset of illness, because in that scenario, it is likely that the patient was infected abroad. An indigenous case was defined as an infected patient who had not traveled overseas.
Laboratory diagnosis.
DENV infection was defined as a febrile illness associated with the detection of DENV RNA by reverse transcription-polymerase chain reaction (RT-PCR), isolation of DENV by cell culture, detection of DENV NS1 antigen (Ag), or detection of DENV-specific immunoglobulin M (IgM) or IgG antibodies.18–21 Isolation of DENV was performed using a mosquito cell line (clone C6/36 of Ae. albopictus cells). For each acute-phase serum sample, 50 μL of the samples diluted at ratios of 1:20, 1:40, 1:80, and 1:160 with Roswell Park Memorial Institute 1640 medium (Gibco/BRL, Life Technologies, Auckland, New Zealand) containing 1% fetal calf serum was added to a 96-well microtiter plate. Then, 1 × 105 cells/100 μL/well of C6/36 were added to the microtiter plate and incubated for 7 days at 28°C. Cells were harvested, and infection was confirmed by immunofluorescence assay using DENV serotype-specific monoclonal antibodies. The viruses were subcultured in C6/36 cells and harvested for nucleotide sequencing after the first or second passage. Isolated viruses were identified using the nomenclature of serotype/country of origin/strain/year of isolation. To detect and differentiate DENV serotypes in acute-phase samples, we performed one-step, SYBR Green I–based, real-time RT-PCR (QuantiTect SYBR Green RT-PCR Kit; Qiagen, Hilden, Germany) using the Mx3000P quantitative PCR system (Stratagene, La Jolla, CA).18 Real-time RT-PCR was performed using two sets of consensus primers: one primer set targeted a region of the NS5 gene to detect all flaviviruses and the other primer set targeted a region of the C gene to detect all DENV serotypes. The DENV serotypes of the positive samples were then confirmed by DENV serotyping using four sets of serotype-specific primers targeting the C gene.13 A commercial DENV NS1 Ag strip rapid test kit (Bio-Rad Laboratories, Marnes La Coquette, France)21 and SD Dengue NS1 Ag test (Standard Diagnostics, Inc., Kyonggi-do, Korea) were used to detect DENV NS1 Ag in acute-phase and convalescent-phase serum samples. E/M-specific capture IgM and IgG enzyme-linked immunosorbent assays were used to detect DENV-specific IgM and IgG antibodies, as previously described.19
Virus isolates selected for complete E gene sequencing.
For phylogenetic analyses, nucleotide sequences of complete E genes of DENV isolates from all imported cases and representative indigenous cases were determined. Representative indigenous cases were selected based on their place of residence, date of onset of illness, and recovered DENV serotype. From all of the imported dengue cases identified in Taiwan in 2014, 139 strains (65 DENV-1, 42 DENV-2, 20 DENV-3, and 12 DENV-4) were isolated. For indigenous cases, a total of 158 isolates (156 DENV-1 and 2 DENV-2) were obtained.
Preparation of viral RNA, RT-PCR amplification, and nucleotide sequencing.
Viral RNA was extracted from either acute-phase serum samples or culture supernatant of C6/36 cells infected with each of the isolated DENV strains using the QIAamp Viral RNA Mini Kit (Qiagen). Primers used for amplification and sequencing of E gene sequences of DENV were described previously.13,14 RT-PCR was performed using the SuperScript III One-Step RT-PCR System with Platinum Taq High Fidelity (Invitrogen, Life Technologies, Carlsbad, CA). The complementary DNA synthesis step was performed at 55°C for 30 minutes. Then, PCR was performed with the following parameters: 94°C for 2 minutes, 40 cycles of 94°C for 15 seconds, 50°C for 30 seconds, and 68°C for 1 minute, and a prolonged elongation step at 68°C for 5 minutes. The PCR products were purified using the Qiagen QIAquick Gel Extraction Kit (Qiagen). Nucleotide sequences were determined using ABI Prism automated DNA sequencing (Applied Biosystems, Foster City, CA). Overlapping nucleotide sequences were combined for analysis and edited using the Lasergene software package (DNASTAR Inc., Madison, WI).
Phylogenetic analysis.
A total of 37 DENV-1 and 32 DENV-2 sequences of Taiwan isolates selected from representative strains of imported cases and indigenous cases from major local DENV-1 and DENV-2 outbreaks combined with global reference sequences of different genotypes available in GenBank were used for the phylogenetic analyses. The nucleotide sequences were aligned using the Clustal W software.22 Phylogenetic and molecular analyses were conducted using MEGA version 6 (http://www.megasoftware.net/).23 Phylogenetic trees were generated using the maximal likelihood method based on the general time-reversible model. The reliability of the analyses was calculated using 1,000 bootstrap replications. The nucleotide sequences of DENV isolates from imported and indigenous cases were submitted to GenBank with accession numbers KT175076–KT175140.
Results
Imported dengue cases in Taiwan, 2014.
A total of 240 laboratory-confirmed imported dengue cases were identified in Taiwan in 2014 (Table 1). Among them, 117 cases (48.8%) were identified by fever screening at airports. Most imported cases arrived from Southeast and East Asia, with Malaysia, Indonesia, the Philippines, and China being the most frequent importing countries. In addition, cases were imported from the Indian subcontinent (India and Bangladesh), the South Pacific region (Nauru, Tuvalu, and French Polynesia), and the Middle East (Saudi Arabia).
Table 1.
Serotype distributions of DENV strains from imported dengue cases in Taiwan, 2014
| Country of origin | Case | Fever screening | DENV-1 | DENV-2 | DENV-3 | DENV-4 | Unknown |
|---|---|---|---|---|---|---|---|
| Malaysia | 71 | 41 | 30 | 24 | 2 | 1 | 14 |
| Indonesia | 58 | 35 | 12 | 10 | 20 | 2 | 14 |
| Philippines | 33 | 12 | 7 | 6 | 1 | 5 | 14 |
| China | 23 | 7 | 14 | 2 | – | – | 7 |
| Singapore | 12 | 9 | 7 | 2 | – | 1 | 2 |
| Myanmar | 10 | 3 | 2 | 3 | – | 1 | 4 |
| Thailand | 10 | 3 | 2 | 3 | 1 | 1 | 3 |
| Vietnam | 9 | 3 | 3 | 1 | – | 1 | 4 |
| Cambodia | 3 | 1 | 1 | – | 1 | – | 1 |
| India | 3 | 2 | 1 | – | – | – | 2 |
| Bangladesh | 2 | – | 1 | – | – | – | 1 |
| Nauru | 2 | – | – | – | – | – | 2 |
| French Polynesia | 1 | – | – | – | – | – | 1 |
| Tuvalu | 1 | 1 | – | 1 | – | – | 0 |
| Saudi Arabia | 1 | – | – | 1 | – | – | 0 |
| Japan | 1 | – | – | – | – | – | 1 |
| Total | 240 | 117 | 80 | 53 | 25 | 12 | 70 |
DENV = dengue virus.
Dengue epidemics in Taiwan, 2014.
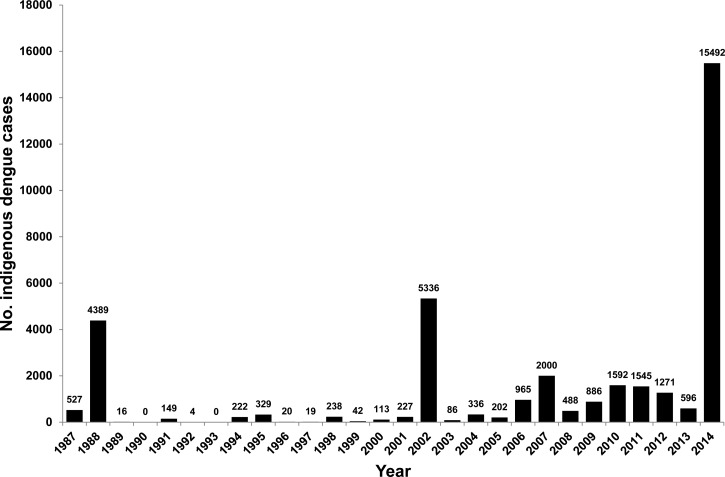
A total of 15,492 laboratory-confirmed indigenous dengue cases were recorded in 2014 (Figure 1 ). Among them, 14 cases were infected with DENV-2 from January to February 2014. These cases represented the last wave of the 2013 outbreak in southern Taiwan. The remaining 15,478 cases were infected with either DENV-1 or DENV-2 from May to December 2014. There were 136 severe hemorrhagic fever cases and 20 deaths.
Figure 1.
Number of indigenous dengue cases in Taiwan between 1987 and 2014.
Transmission dynamics of epidemic DENV strains.
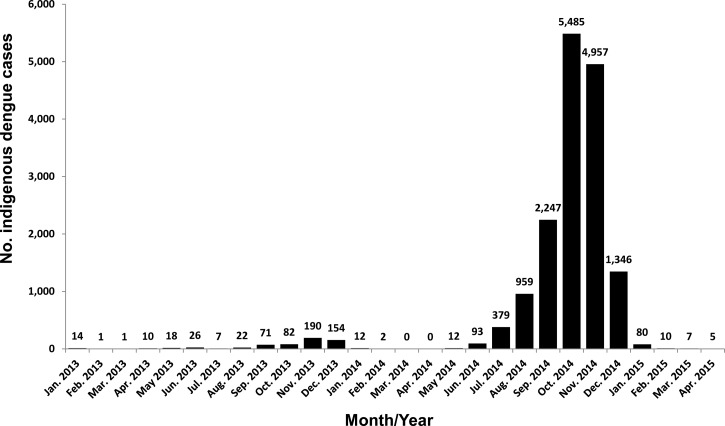
Table 2 summarizes the transmission dynamics, areas infected, and total cases estimated for each of the three DENV strains. The major dengue outbreak was caused by a strain of DENV-1, D1/Taiwan/806KH1405a/2014, and the outbreak began in the Cianjhen District, Kaohsiung City, in May and later spread to Tainan City, Pingtung County, and then throughout Taiwan. The epidemic virus continued to be transmitted in southern Taiwan in 2015, and approximately 100 dengue cases were identified from January to April 2015 (Figure 2 ). Another DENV-1 strain, D1/Taiwan/820KH1409a/2014, was isolated from a dengue patient who lived in Gangshan District, Kaohsiung City. In addition, a DENV-2 strain, D2/Taiwan/807KH1412a/2014, was isolated from a patient who lived in Sanmin District, Kaohsiung City. This DENV-2 strain caused a small outbreak in Samnin District, Kaohsiung City, with two cases identified in December 2014 and five additional cases identified from January to March 2015. Figure 2 shows the monthly distribution of indigenous dengue cases: the epidemic began in May 2014, peaked in October and November 2014, and then declined in January 2015.
Table 2.
Summary of the dengue epidemics in Taiwan from May to December 2014
| DENV strain | Serotype/genotype | Epidemic area infected | First case | Last case | Total cases |
|---|---|---|---|---|---|
| D1/Taiwan/806KH1405a/2014 | DENV-1 genotype I | Kaohsiung City | May 20 | – | 14,995 |
| Chiayi City | June 14 | November 21 | 6 | ||
| Penghu County | July 7 | December 4 | 16 | ||
| Tainan City | July 19 | – | 152 | ||
| New Taipei City | July 19 | December 6 | 16 | ||
| Taoyuan City | August 18 | November 30 | 5 | ||
| Taichung City | August 29 | December 13 | 15 | ||
| Changhua County | September 6 | December 18 | 4 | ||
| Taitung County | September 7 | – | 16 | ||
| Pingtung County | September 11 | – | 214 | ||
| Hsinchu County | September 11 | September 11 | 2 | ||
| Taipei City | September 12 | – | 13 | ||
| Hsinchu City | September 12 | October 27 | 4 | ||
| Yilan County | September 20 | September 20 | 1 | ||
| Miaoli County | September 24 | October 18 | 2 | ||
| Chiayi County | October 3 | November 27 | 4 | ||
| Nantou County | October 14 | November 15 | 2 | ||
| Yunlin County | October 15 | November 17 | 7 | ||
| Hualien County | November 21 | November 21 | 1 | ||
| D1/Taiwan/820KH1409a/2014 | DENV-1 genotype II | Gangshan District, | September 1, 2015 | September 1, 2014 | 1 |
| Kaohsiung City | |||||
| D2/Taiwan/807KH1412a/2014 | DENV-2 Asian genotype I | Sanmin District, | December 3, 2015 | – | 2 |
| Kaohsiung City |
DENV = dengue virus.
“–” Indicates the epidemic continued to occur in 2015.
Figure 2.
Monthly distribution of indigenous dengue cases from January 2013 to April 2015.
Phylogenetic analysis of DENV-1 and DENV-2 strains isolated from indigenous and imported cases in Taiwan, 2014.
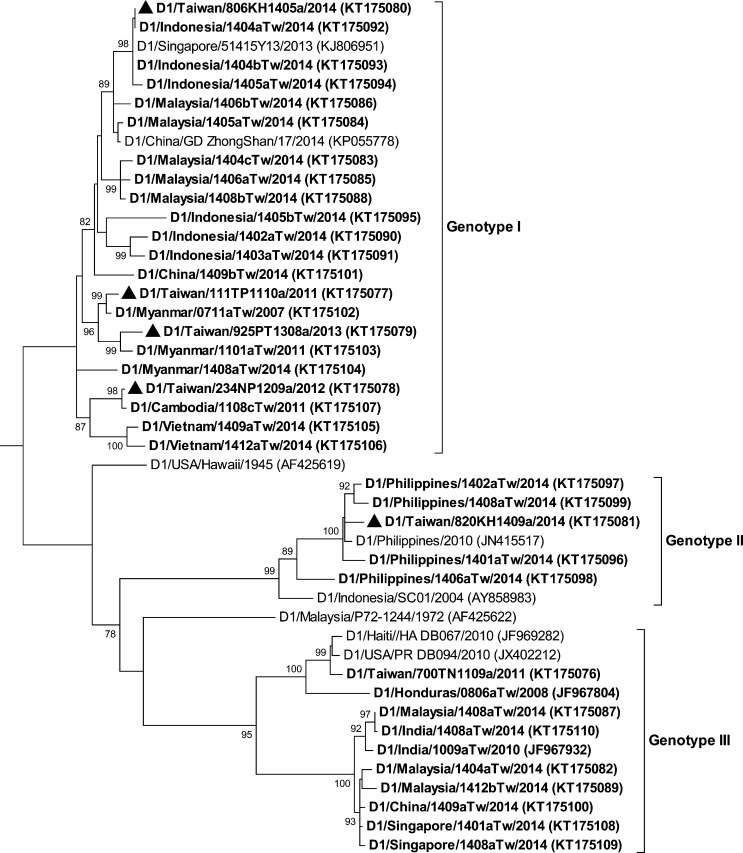
The complete E gene sequences of three distinct DENV isolates (D1/Taiwan/806KH1405a/2014, D1/Taiwan/820KH1409a/2014, and D2/Taiwan/807KH1412a/2014) from indigenous index cases and E gene sequences of viruses isolated from imported dengue cases were determined and compared with the sequences available in GenBank and the Taiwan CDC DENV sequence database. The DENV genotype designations were based on the classification of A-Nuegoonpipat and others24 and Twiddy and others25 for the DENV-1 and DENV-2 strains, respectively. Figure 3 shows the phylogenetic tree of the complete E gene sequences of the DENV-1 strains. Genotype I of DENV-1 was the most prevalent genotype of DENV strains isolated from dengue cases arriving from Southeast Asian countries in 2014. DENV strains of genotype I of DENV-1 were isolated from cases imported from Indonesia, Malaysia, China, Myanmar, and Vietnam. The epidemic strain, D1/Taiwan/806KH1405a/2014, which caused a large outbreak in 2014, belongs to this genotype and is most closely related to virus strains from Indonesia. DENV strains of genotype II of DENV-1 were isolated from cases imported from the Philippines. Another local strain, D1/Taiwan/820KH1409a/2014, belonged to this genotype and is most closely related to strains from the Philippines. DENV strains of genotype III of DENV-1 were isolated from cases imported from Malaysia, India, China, and Singapore. Figure 4 shows the phylogenetic tree of DENV-2. DENV strains of the Asian genotype I of DENV-2 were isolated from cases imported from Malaysia, Myanmar, and Thailand. The local strain, D2/Taiwan/807KH1412a/2014, belongs to this genotype and is most closely related to virus strains isolated from Myanmar. DENV strains of the cosmopolitan genotype of DENV-2 have a wide geographic distribution and were isolated from cases imported from Malaysia, Indonesia, the Philippines, China, Singapore, Thailand, Vietnam, Tuvalu, and Saudi Arabia.
Figure 3.
Phylogenetic tree of dengue virus type 1 (DENV-1). The phylogenetic tree is based on the complete E gene sequences of 46 strains of DENV-1, including 37 Taiwan isolates obtained from imported and indigenous dengue cases. The tree was constructed using the maximum likelihood method and the general time reversible model. Bootstrap support values greater than 75 are shown. The Taiwan isolates from imported dengue cases are designated in boldface. The epidemic strains isolated from major dengue outbreaks in Taiwan are designated in boldface and with a solid triangle (▴). Viruses were identified by using the nomenclature of serotype/country of origin/strain/year of isolation. GenBank accession numbers are shown in parentheses.
Figure 4.
Phylogenetic tree of dengue virus type 2 (DENV-2). The phylogenetic tree is based on the complete E gene sequences of 43 strains of DENV-2, including 32 Taiwan isolates obtained from imported and indigenous dengue cases. See the legend of Figure 3 for further details.
Amino acid mutations in the E protein of DENV-1 isolates during a major epidemic.
The complete E gene sequences from 155 DENV-1 isolates of the major DENV-1 epidemic were determined, and sequence substitutions relative to the major epidemic index strain D1/Taiwan/806KH1405a/2014 were examined. E gene sequences of 122 isolates were identical to D1/Taiwan/806KH1405a/2014. In addition, 25 synonymous substitutions were identified in 21 isolates (data not shown), 11 nonsynonymous mutations were identified in nine isolates, and both synonymous and nonsynonymous mutations were identified in two isolates. Table 3 summarizes the mutations resulting in amino acid changes in the E gene relative to the D1/Taiwan/806KH1405a/2014 strain. Mutations were variable between isolates and were not located in sequences involved in receptor binding or membrane fusion activities.26
Table 3.
Summary of mutations resulting in amino acid changes in envelope (E) protein gene of DENV-1 isolates relative to the strain D1/Taiwan/806KH1405a in 2014
| Isolates | Date of sample collection | Amino acid position in the E protein | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 47 | 95 | 114 | 157 | 159 | 161 | 171 | 203 | 205 | 227 | 339 | 400 | 492 | ||
| D1/Taiwan/806KH1405a | May 24 | K | T | I | E | G | T | T | E | S | S | T | K | M |
| D1/Taiwan/831KH1406b | June 20 | – | – | T | – | – | – | – | – | – | – | – | – | – |
| D1/Taiwan/710TN1407a | July 19 | – | – | – | – | E | – | – | – | – | – | – | – | – |
| D1/Taiwan/241NP1408a | August 15 | – | – | – | – | – | – | – | – | – | – | – | – | T |
| D1/Taiwan/814KH1408a | August 17 | R | – | – | – | – | – | – | – | – | – | – | – | – |
| D1/Taiwan/844KH1409a | September 3 | – | – | – | – | – | – | – | – | – | P | – | – | – |
| D1/Taiwan/845KH1409a* | September 14 | – | – | – | – | – | I | – | – | L | – | – | – | – |
| D1/Taiwan/830KH1410a | October 1 | R | – | – | – | – | – | – | – | – | – | – | – | – |
| D1/Taiwan/604CY1410a | October 3 | – | A | – | – | – | – | – | – | – | – | N | – | – |
| D1/Taiwan/807KH1411f | November 5 | – | – | – | – | – | – | A | – | – | – | – | – | – |
| D1/Taiwan/710TN1411a | November 6 | – | – | – | K | – | – | – | G | – | – | – | – | – |
| D1/Taiwan/807KH1411i* | November 10 | – | – | – | – | – | – | – | – | – | – | – | R | – |
DENV = dengue virus.
Indicates both synonymous and nonsynonymous mutations were identified. K = lysine; T = threonine; I = isoleucine; E = glutamic acid; G = glycine; S = serine; M = methionine; R = arginine; L = leucine; P = proline; N = asparagine.
Discussion
A total of 240 imported dengue cases and 15,492 indigenous dengue cases were identified in Taiwan in 2014. Malaysia, Indonesia, the Philippines, and China were the most frequent importing countries of DENV, which reflects the frequency of air travel between Taiwan and these nations and the high intensity of massive dengue outbreaks during the same period in these countries. Indeed, the number of dengue cases in Malaysia and China in 2014 had significantly increased compared with previous years.27 Genotype I of DENV-1 and the cosmopolitan genotype of DENV-2 were the most prevalent DENV strains isolated from imported dengue cases arriving from Southeast Asian countries in 2014, which corresponds to the results of studies conducted over the last decade.14,15 Interestingly, a DENV-2 strain isolated from a case imported from Tuvalu had the cosmopolitan genotype and is closely related to viral strains from Fiji, which suggests that these viruses were co-circulated between these two nations. In addition, a DENV-2 strain isolated from a case imported from Saudi Arabia had the cosmopolitan genotype and is closely related to viral strains from South and Southeast Asian countries (Figure 4). Because air travel has become increasingly popular and convenient, DENVs can spread quickly throughout the world via infected passengers.
The dengue outbreak of 2014 was the largest ever to occur in Taiwan since World War II (Figure 1). There are several possible causes of this large epidemic. First, notification of locally acquired dengue cases reported to the health authority in the early phases of the epidemic may have been delayed. In late May 2014, several laboratory-confirmed indigenous dengue cases were identified in multiple districts in Kaohsiung City (Cianjhen, Siaogan, Linyuan, and Cijin Districts), thus, indicating that dengue may have transmitted throughout the community for some time without notification to health authorities. The epidemic had initially affected the Cianjhen fishing port, which is a tourist attraction where many local residents also resided, this situation may have accelerated the spread of the virus. In Taiwan, patients with dengue fever often sought medical care on average of two to three times before being diagnosed and reported to the health authorities. Primary care physicians usually do not report the suspected dengue cases because they either do not recognize the disease or they consider the reporting procedure too sophisticated and inconvenient.28 Because most of the dengue patients who initially sought medical attention were in the acute viremic stage of illness, delay in reporting dengue disease could have increased the risk of exposure of patients to mosquito bites, subsequently increasing the risk of DENV transmission. To increase the physician's willingness to report suspected dengue cases, a simplified reporting procedure must be developed, furthermore, a reward/penalty system for reporting communicable diseases should be established to improve the disease reporting compliance in primary care doctors. Moreover, in recent years, dengue outbreaks have shifted toward outbreaks occurring in the earlier months of the year. Before 2012, dengue outbreaks typically started between June and August; however, during 2012 to 2014, dengue outbreaks began in April or May. Because the dengue epidemic season overlapped with the viral respiratory diseases season, physicians may have had a lower level of awareness about dengue fever. With unrecognized dengue patients and a high mosquito density between April and December in Taiwan (http://www.cdc.gov.tw/), a human-to-mosquito-to-human transmission cycle of DENV could have successfully established, and thus increased the risk of a dengue epidemic.
Secondly, dengue control measures may have been insufficient and ineffective in the early phase of the epidemic, which allowed the number of dengue cases to continue to increase in later months. In Taiwan, dengue control measures were conducted according to the guidelines for dengue/chikungunya control, Taiwan CDC (http://www.cdc.gov.tw). These control measures contained vector density surveillance, mosquito breeding source elimination, community mobilization for environment management, and health education. Additional control efforts in dengue outbreak and gas explosion area included dengue case investigation, providing dengue patients with insect repellent, emergency indoor and outdoor insecticide spring for adult mosquitoes. However, due to ignoring the possibility of a large outbreak, vector control measures have not been fully conducted in Kaohsiung City. Vector breeding sources have not been completely eliminated and some local residents were against the continuous insecticide spraying in their communities, therefore, the dengue epidemic was still ongoing in some districts in Kaohsiung City.
The third possible cause of the dengue outbreak is the result of a serious gas pipeline explosion that occurred in dengue epidemic hot spot areas (Cianjhen and Lingya Districts) in Kaohsiung City on 31 July 2014. After the incident, all traffic and personnel access had been controlled or limited during August 1–8. Vector control personnel were thus unable to enter the disaster area to carry out emergency control measures, while the collapsed infrastructure spawned many mosquito breeding sites that likely caused an increase in mosquito density. After August 8, despite carrying out reduction and elimination of mosquito breeding sites and chemical control for adult mosquitoes, an abnormal rainfall pattern with a large amount of concentrated precipitation within a short period of time was observed during August 8–14. Consequently, 2 weeks after the gas explosion, a significant increase in the number of dengue cases in the disaster area was realized.29 In response to the pipeline explosion, a great deal of man power and material resources was used for aiding the disaster relief. This led to financial and labor resource limitations to address the unprecedented outbreak of dengue. Under the circumstances, insufficient resources developed a much more serious case of a dengue epidemic in Kaohsiung City. After the gas explosion incident, the abnormal rainfall pattern with a large amount of rainfall was observed in early August and was followed by an unusually dry October in southern Taiwan in 2014 (http://www.cwb.gov.tw/eng/index.htm). Whether these abnormal weather conditions affected the growth cycle of mosquito vectors and made the dengue outbreak even worse requires further study.
The phylogenetic study showed that a DENV-1 strain, D1/Taiwan/806KH1405a/2014, was responsible for the large outbreak in Taiwan in 2014. The E gene sequence of this epidemic strain is identical to that of a DENV-1 strain (D1/Indonesia/1404aTw/2014) isolated from a case imported from Indonesia in 2014. This epidemic strain is different from those that previously circulated in Taiwan (Figure 3), which indicates that constant importations of DENV strains from international travelers are responsible for the outbreaks that occur each year. Because of increased global trade and travel, DENV strains are expected to be introduced into Taiwan more frequently. The geographic location of Taiwan (an island in Southeast Asia), its climate conditions (tropical and subtropical), and its vector distributions (Ae. albopictus throughout the island and Ae. aegypti in southern Taiwan) provide an ideal environment for the transmission of DENV.30,31 Therefore, without successful disease surveillance systems, dengue control measures and community participation, a large dengue outbreak is likely to occur in southern Taiwan every year.
There have been several DENV outbreaks in southern Taiwan since 1987; however, except for the DENV-1 epidemic in 1988, which had 4,389 confirmed cases, and the DENV-2 epidemic in 2002, which had 5,336 confirmed cases, all local outbreaks have been small in scale, with dengue cases of less than 2,000 (Figure 1).16 During 2005, we have conducted a seroepidemiological study of dengue in Kaohsiung City, which had been the worst dengue epidemic city in Taiwan. The seroprevalence rates of dengue ranged from 8.0% to 64.6% in different age groups (mean = 25.4%) with higher prevalence rates among older adults (Supplemental Table 1). Most people are susceptible to DENV infections; therefore, DENV can easily spread in Taiwan.
Because of the constant importation of DENV strains by international travelers and the environmental conditions that favor the transmission of DENV, dengue is a continuing threat in Taiwan. To reduce population exposure to DENV infections and minimize the magnitude of local epidemics, public health authorities should strengthen passive and active surveillance systems; in particular, they should encourage clinicians to notify health authorities of patients who have contracted DENV abroad. In addition, various vector control methods, such as chemical control, biological control, genetically engineered mosquito vectors, and vaccine development, should be implemented to reduce the risk of dengue.
Supplementary Material
ACKNOWLEDGMENTS
We thank Mei-Chun Chang and Huai-Chin Hu for their expert technical assistance. We also thank Eric Shu and Charles Shu for their great editing job on the manuscript.
Footnotes
Financial support: This work was supported in part by grants MOHW103-CDC-C-315-000205 and MOHW104-CDC-C-315-000101 from Centers for Disease Control, Ministry of Health and Welfare, Taiwan, Republic of China.
Authors' addresses: Shu-Fen Chang, Tung-Chien Hsu, Chien-Ling Su, Chien-Chou Lin, and Pei-Yun Shu, Center for Research, Diagnostics, and Vaccine Development, Centers for Disease Control, Ministry of Health and Welfare, Taipei, Taiwan, Republic of China, E-mails: vivi@cdc.gov.tw, tchsu@cdc.gov.tw, sue@cdc.gov.tw, jjlin@cdc.gov.tw, and pyshu@cdc.gov.tw. Cheng-Fen Yang, Center for Research, Diagnostics, and Vaccine Development, Centers for Disease Control, Ministry of Health and Welfare, Taipei, Taiwan, Republic of China, and Department of Biotechnology and Laboratory Science in Medicine, National Yang Ming University, Taiwan, Republic of China, E-mail: joyceyyang@cdc.gov.tw.
References
- 1.WHO . Global Strategy for Dengue Prevention and Control, 2012–2020. Geneva, Switzerland: World Health Organization; 2012. [Google Scholar]
- 2.Calisher CH, Karabatsos N, Dalrymple JM, Shope RE, Porterfield JS, Westaway EG, Brandt WE. Antigenic relationships between flaviviruses as determined by cross-neutralization tests with polyclonal antisera. J Gen Virol. 1989;70:37–43. doi: 10.1099/0022-1317-70-1-37. [DOI] [PubMed] [Google Scholar]
- 3.Lindenbach BD, Rice CM. Molecular biology of flaviviruses. Adv Virus Res. 2003;59:23–61. doi: 10.1016/s0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- 4.Burke DS, Nisalak A, Johnson DE, Scott RM. A prospective study of dengue infections in Bangkok. Am J Trop Med Hyg. 1988;38:172–180. doi: 10.4269/ajtmh.1988.38.172. [DOI] [PubMed] [Google Scholar]
- 5.Nimmannitya S. Clinical spectrum and management of dengue haemorrhagic fever. Southeast Asian J Trop Med Public Health. 1987;18:392–397. [PubMed] [Google Scholar]
- 6.WHO . Dengue Guideline for Diagnosis, Treatment, Prevention and Control. Geneva, Switzerland: World Health Organization; 2009. [PubMed] [Google Scholar]
- 7.Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11:480–496. doi: 10.1128/cmr.11.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibbons RV, Vaughn DW. Dengue: an escalating problem. BMJ. 2002;324:1563–1566. doi: 10.1136/bmj.324.7353.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brady OJ, Gething PW, Bhatt S, Messina JP, Brownstein JS, Hoen AG, Moyes CL, Farlow AW, Scott TW, Hay SI. Refining the global spatial limits of dengue virus transmission by evidence-based consensus. PLoS Negl Trop Dis. 2012;6:e1760. doi: 10.1371/journal.pntd.0001760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. The global distribution and burden of dengue. Nature. 2013;496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murray NE, Quam MB, Wilder-Smith A. Epidemiology of dengue: past, present and future prospects. Clin Epidemiol. 2013;5:299–309. doi: 10.2147/CLEP.S34440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang LH, Hsu EL, Teng HJ, Ho CM. Differential survival of Aedes aegypti and Aedes albopictus (Diptera: Culicidae) larvae exposed to low temperatures in Taiwan. J Med Entomol. 2007;44:205–210. doi: 10.1603/0022-2585(2007)44[205:dsoaaa]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 13.Huang JH, Liao TL, Chang SF, Su CL, Chien LJ, Kuo YC, Yang CF, Lin CC, Shu PY. Laboratory-based dengue surveillance in Taiwan, 2005: a molecular epidemiologic study. Am J Trop Med Hyg. 2007;77:903–909. [PubMed] [Google Scholar]
- 14.Shu PY, Su CL, Liao TL, Yang CF, Chang SF, Lin CC, Chang MC, Hu HC, Huang JH. Molecular characterization of dengue viruses imported into Taiwan during 2003–2007: geographic distribution and genotype shift. Am J Trop Med Hyg. 2009;80:1039–1046. [PubMed] [Google Scholar]
- 15.Huang JH, Su CL, Yang CF, Liao TL, Hsu TC, Chang SF, Lin CC, Shu PY. Molecular characterization and phylogenetic analysis of dengue viruses imported into Taiwan during 2008–2010. Am J Trop Med Hyg. 2012;87:349–358. doi: 10.4269/ajtmh.2012.11-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chang SF, Huang JH, Shu PY. Characteristics of dengue epidemics in Taiwan. J Formos Med Assoc. 2012;111:297–299. doi: 10.1016/j.jfma.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 17.Shu PY, Chien LJ, Chang SF, Su CL, Kuo YC, Liao TL, Ho MS, Lin TH, Huang JH. Fever screening at airports and imported dengue. Emerg Infect Dis. 2005;11:460–462. doi: 10.3201/eid1103.040420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shu PY, Chang SF, Kuo YC, Yueh YY, Chien LJ, Sue CL, Lin TH, Huang JH. Development of group- and serotype-specific one-step SYBR green I-based real-time reverse transcription-PCR assay for dengue virus. J Clin Microbiol. 2003;41:2408–2416. doi: 10.1128/JCM.41.6.2408-2416.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shu PY, Chen LK, Chang SF, Yueh YY, Chow L, Chien LJ, Chin C, Lin TH, Huang JH. Comparison of capture immunoglobulin M (IgM) and IgG enzyme-linked immunosorbent assay (ELISA) and nonstructural protein NS1 serotype-specific IgG ELISA for differentiation of primary and secondary dengue virus infections. Clin Diagn Lab Immunol. 2003;10:622–630. doi: 10.1128/CDLI.10.4.622-630.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shu PY, Huang JH. Current advances in dengue diagnosis. Clin Diagn Lab Immunol. 2004;11:642–650. doi: 10.1128/CDLI.11.4.642-650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shu PY, Yang CF, Kao JF, Su CL, Chang SF, Lin CC, Yang WC, Shih H, Yang SY, Wu PF, Wu HS, Huang JH. Application of the dengue virus NS1 antigen rapid test for on-site detection of imported dengue cases at airports. Clin Vaccine Immunol. 2009;16:589–591. doi: 10.1128/CVI.00475-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.A-Nuegoonpipat A, Berlioz-Arthaud A, Chow V, Endy T, Lowry K, Mai le Q, Ninh TU, Pyke A, Reid M, Reynes JM, Su Yun ST, Thu HM, Wong SS, Holmes EC, Aaskov J. Sustained transmission of dengue virus type 1 in the Pacific due to repeated introductions of different Asian strains. Virology. 2004;329:505–512. doi: 10.1016/j.virol.2004.08.029. [DOI] [PubMed] [Google Scholar]
- 25.Twiddy SS, Farrar JJ, Vinh Chau N, Wills B, Gould EA, Gritsun T, Lloyd G, Holmes EC. Phylogenetic relationships and differential selection pressures among genotypes of dengue-2 virus. Virology. 2002;298:63–72. doi: 10.1006/viro.2002.1447. [DOI] [PubMed] [Google Scholar]
- 26.Nayak V, Dessau M, Kucera K, Anthony K, Ledizet M, Modis Y. Crystal structure of dengue virus type 1 envelope protein in the postfusion conformation and its implications for membrane fusion. J Virol. 2009;83:4338–4344. doi: 10.1128/JVI.02574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.WHO Update on the Dengue Situation in the Western Pacific Region. Northern Hemisphere. 2015. http://www.wpro.who.int/emerging_diseases/dengue_biweekly_20150519.pdf Available at. Accessed June 1, 2015.
- 28.Tan HF, Yeh CY, Chang HW, Chang CK, Tseng HF. Private doctors' practices, knowledge, and attitude to reporting of communicable diseases: a national survey in Taiwan. MBC Infect Dis. 2009;9:e11. doi: 10.1186/1471-2334-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen CM, Kou HW, Liu DP. Impact assessment of petrochemical gas explosions on dengue fever epidemic in Kaohsiung City [in Chinese] Taiwan Public Health Assoc. 2014;33:563–567. [Google Scholar]
- 30.Weaver SC, Barrett AD. Transmission cycles, host range, evolution and emergence of arboviral disease. Nat Rev Microbiol. 2004;2:789–801. doi: 10.1038/nrmicro1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naish S, Dale P, Mackenzie JS, McBride J, Mengersen K, Tong S. Climate change and dengue: a critical and systematic review of quantitative modelling approaches. BMC Infect Dis. 2014;14:e167. doi: 10.1186/1471-2334-14-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.