Abstract
An important aspect of many malaria molecular epidemiology and transmission studies is RNA-based detection of gametocytes. Ensuring RNA stability represents a challenge in tropical, resource-limited environments, as RNA may quickly degrade when samples are not preserved under adequate conditions. This study investigated the degradation of pfs25 messenger RNA (mRNA), the most widely used Plasmodium falciparum gametocyte marker, in whole blood spiked with cultured P. falciparum gametocytes, exposed to different temperatures for up to 48 hours, and collected with different anticoagulants. The levels of pfs25 mRNA were similar between samples stored at 4°C and 30°C for up to 48 hours before stabilization with RNAprotect (Qiagen, Hilden, Germany). We observed that pfs25 mRNA in heparin-collected blood degraded less than that in ethylenediaminetetraacetic acid (EDTA)–collected blood over the 48-hour period. For field studies aiming for P. falciparum gametocyte detection, immediate stabilization of blood samples is not necessary, as the pfs25 transcript is relatively stable, more so in heparin than EDTA collection tubes.
As malaria transmission decreases, surveillance of the human infectious reservoir, including reliable estimation of gametocyte carriers in endemic areas, is essential.1–3 Molecular epidemiology studies of malaria rely on adequate stabilization and storage of blood samples to ensure the integrity of nucleic acids, particularly RNA, which is prone to rapid degradation.4 Molecular quantification of Plasmodium falciparum gametocytes can be achieved by reverse transcription quantitative polymerase chain reaction (RT-qPCR) targeting the messenger RNA (mRNA) of the pfs25 gene, the most widely used marker of mature, stage V P. falciparum gametocytes.5
When collecting blood samples for gametocyte detection, sample collection and stabilization are important aspects to ensure mRNA integrity.6,7 If the whole blood sample collected in the field is stored at tropical temperatures for extended periods (e.g., the period it takes to reach a field laboratory for processing and freezing), mRNA degradation is thought to occur.8 Therefore, the original gametocyte density may not be deducible from samples that were exposed to temperatures below 37°C for extended periods. Although using appropriate storage media, such as RNAprotect (Qiagen, Hilden, Germany), may stabilize mRNA, the handling of blood samples at the collection site might not be logistically feasible or introduce risks of contamination.
The stability of pfs25 transcripts under field conditions has been investigated previously, but only with respect to dried blood spots.6,7 Our study investigated the stability of pfs25 mRNA in whole blood samples collected with either ethylenediaminetetraacetic acid (EDTA) or heparin as anticoagulants and stored over 48 hours at either 4°C or 30°C to emulate ice pack field conditions or ambient/tropical temperature. We aimed to assess the extent of expected mRNA degradation and inform future studies that collect blood samples in remote field settings for subsequent P. falciparum gametocyte detection. In malaria studies, heparin is sometimes avoided as it interferes with erythrocyte invasion of the parasite and has also been reported to be a potential inhibitor of PCR.9,10 On the other hand, EDTA sample collection and subsequent storage at 4°C and 20°C has been linked to RNA degradation.8
Plasmodium falciparum gametocytes (NF54 strain) were cultured as described previously.11 When outside the incubator, cultures were kept at 37°C on a slide warmer. Of the stage V gametocyte culture, 10 mL was split into two fractions and centrifuged at 500 × g for 5 minutes at 37°C. Gametocytemia was determined as the percentage of gametocyte-infected red blood cells in 5,000 erythrocytes in the resulting pellets. The pellets were then diluted with whole blood collected in tubes containing either 2.6 mg/mL of potassium EDTA (Sarstedt, Germany) or 15–17 USP/mL of lithium heparin (Becton Dickinson, Franklin, Lakes, NH) to 500 gametocytes/μL. Triplicate aliquots of 50 μL were removed from each of the heparin and EDTA preparations and added to 250 μL of commercially available RNA stabilization reagent (RNAprotect). The samples were then placed at −80°C. These samples represent the heparin and EDTA baseline gametocyte samples (time point 0).
Heparin and EDTA gametocyte preparations were further split for incubation at 4°C or 30°C resulting in four experimental treatments (i.e., heparin or EDTA at 4°C or 30°C). All samples were held at these storage conditions over 48 hours. At 2, 4, 6, 8, 12, 24, and 48 hours after time point 0, 50 μL blood from each treatment was added to tubes containing 250 μL of RNAprotect, in triplicate, and the tubes were immediately placed at −80°C.
After 1 week of −80°C storage, the RNAprotect-preserved samples were thawed and RNA extraction was performed using a Qiagen RNeasy 96 Plus Kit as described elsewhere5 with the slight modification to elute the extracted RNA in 60 μL of RNase-Free Water (Qiagen, Hilden, Germany).
TaqMan one-step RT-qPCR targeting the pfs25 transcript was used to detect and quantify the presence of gametocytes in the experimental samples.5,12 The assay and primers have been described previously.5 Each detection experiment included duplicate plasmid controls containing the pfs25 target product (at 104, 103, 102, 10, 5, and 1 copies/μL) to estimate gametocyte densities as pfs25 transcript copies/μL.5
To determine whether patterns of RNA decay in the pfs25 assays were due to total RNA degradation, gametocyte-specific RNA degradation, or technical artefact of the gametocyte-specific assay, TaqMan one-step RT-qPCR assays targeting the human beta-globin transcript, which is the most abundant transcript in erythrocytes, were conducted in parallel. The primers and probe used in this assay have been described previously.13 The assay was performed using the TaqMan RNA-to-Ct™ 1-Step Kit (Applied Biosystems, Carlsbad, CA) in a total volume of 12 μL containing 6 μL of 2× RNA-to-CT™ Mastermix (Applied Biosystems), 670 nM of each of the primers, and 335 nM of the probe. Cycling parameters included reverse transcription for 30 minutes at 48°C, Taq DNA polymerase activation for 10 minutes at 95°C, and 45 cycles of 15 seconds at 95°C denaturation and 1 minute at 58°C annealing, extension, and detection. Positive RNA controls of known concentration, which were serially diluted, were used as standards for quantification, and thus a standard curve of beta-globin (ng/μL) was generated. Both pfs25 and beta-globin assays were run in 384-well plates on a Roche LightCycler480® (Roche, Basel, Switzerland) in triplicate. Spearman rank correlation analysis between replicates indicated minimal intra-assay variation (pfs25 replicate 1 versus 2, r = 0.98 and replicate 1 versus 3, r = 0.95; beta-globin replicate 1 versus 2, r = 0.96 and replicate 1 versus 3, r = 0.95).
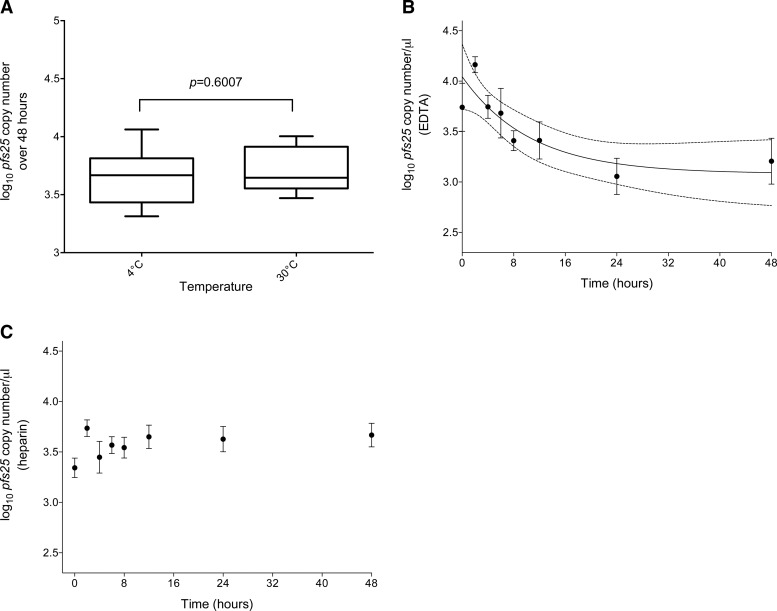
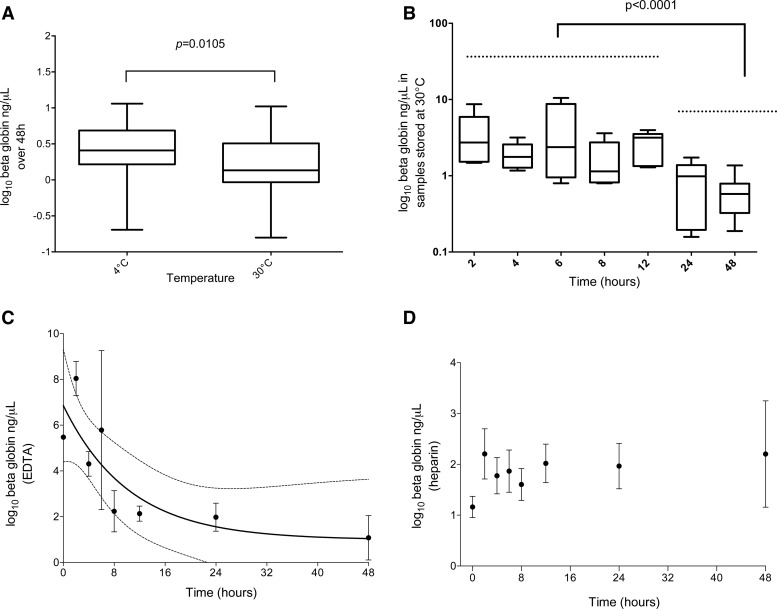
Temperature did not affect the pfs25 mRNA content of samples stabilized over 48 hours significantly (4°C = 3,680 versus 30°C = 3,420 copies/μL, Mann–Whitney P = 0.6007; Figure 1A ). For human beta-globin data, there was moderate evidence that the 4°C storage stabilized human mRNA better than the storage at 30°C (4°C = 2.57 versus 30°C = 1.36 ng/μL Mann–Whitney P = 0.0105; Figure 2A ). The greatest reduction in human mRNA content occurred between 12 and 48 hours (pre-24 hours median ng/μL = 1.97 versus post-24 hours median ng/μL = 0.59, Mann–Whitney P < 0.0001; Figure 2B).
Figure 1.
Messenger RNA levels of Plasmodium falciparum pfs25 transcript (log10 transcript copies/μL) detected by reverse transcription quantitative polymerase chain reaction over 48 hours. (A) Samples stored at 4°C or 30°C (ethylenediaminetetraacetic acid [EDTA] and heparin samples combined). (B) Samples in EDTA (4°C or 30°C samples combined). (C) Samples in heparin (4°C or 30°C samples combined). The error bars represent standard errors of the mean. Pfs25 concentration followed an exponential decay pattern in the samples prepared with blood collected in EDTA (best fit lines in B represent the best fit and 95% confidence interval of an exponential decay function). No degradation was observed in the samples prepared from blood collected in heparin.
Figure 2.
Messenger RNA (mRNA) levels of the human beta-globin transcript (log10 ng/μL) detected by reverse transcription quantitative polymerase chain reaction. (A) Over 48 hours in samples stored at 4°C vs. 30°C, (B) at each time point in samples stored at 30°C, (C) in ethylenediaminetetraacetic acid (EDTA), and (D) in heparin. The error bars represent standard errors of the mean. The greatest decrease in mRNA levels occurred after 24 hours of storage at 30°C. Beta-globin also followed an exponential decay pattern in the samples prepared with blood collected in EDTA (best fit lines in C represent the best fit and 95% confidence interval of an exponential decay function). No degradation was observed in the samples prepared from blood collected in heparin (D).
These data suggest that storing blood in RNAprotect at ambient (tropical) temperature for up to 48 hours does not lead to complete degradation of the pfs25 and beta-globin transcripts, as both are detectable at 48 hours. However, given there was a significant reduction in detectable beta-globin transcript after the 12-hour time point, it may be prudent to stabilize and freeze the sample within 24 hours of collection.
To investigate the effect of anticoagulant, data were stratified by EDTA and heparin collection methods. A significant decrease of pfs25 content over 48 hours was observed in the EDTA samples (EDTA pre-8 hours median pfs25 copies/μL = 7,048 versus EDTA post-8 hours median pfs25 copies/μL = 1,963, Mann–Whitney P = 0.0014) compared with the heparin samples (heparin pre-8 hours median pfs25 copies/μL = 4,280 versus heparin post-8 hours median pfs25 copies/μL = 3,715, Mann–Whitney P = 0.5345). Time degradation profiles of pfs25 EDTA and heparin at each time point are given in Figure 1B and C, respectively.
Human beta-globin mRNA decay analysis resulted in similar profiles as those determined with the P. falciparum gametocyte assays for both EDTA (pre-8 hours median ng/μL = 4.6 versus EDTA post-8 hours median ng/μL = 1.6, Mann–Whitney P = 0.0008; Figure 2C) and heparin samples (pre-8 hours median ng/μL = 1.6 versus post-8 hours median ng/μL = 1.7, Mann–Whitney P = 0.9751; Figure 2D).
As heparin has been shown to inhibit PCR,9,10 heparin concentrations in a representative subset of heparinated RNA samples (N = 16; specifically one replicate from each time point, 0, 2, 4, 6, 8, 12, 24, and 48 hours and from both incubation temperatures 4°C and 30°C) was measured as previously described.14 Heparin was not detected in the mRNA samples (Supplemental Figure 1). It is therefore unlikely that residual heparin influenced the RT-qPCR assays. These findings suggest that both gametocyte and human mRNA degraded to a higher degree in the blood samples collected with EDTA, and thus heparin may be preferable to EDTA when collecting blood samples for RNA-based assays.8
RNA stabilization is a relevant concern for many field studies in remote tropical environments. Transferring samples directly into RNA stabilization reagent at the time of blood collection adds an additional logistic challenge for collection teams and may place samples at risk of contamination. Although short-term ice pack storage conditions are possible for some collection sites, it may be an added logistical challenge for others. This study shows that storage at either 4°C or 30°C did not significantly affect the mRNA stability of P. falciparum gametocyte marker pfs25 over a 48-hour period and of human mRNA up to 24 hours, suggesting that it may be feasible to keep blood samples at tropical ambient conditions for the time required to reach a centralized field laboratory and conduct downstream processing (usually within 24–48 hours). The choice of anticoagulant, however, does seem to affect mRNA stability significantly. In blood samples collected in EDTA, degradation of both pfs25 and human beta-globin transcripts was observed at both temperatures whereas mRNA levels did not decrease significantly in samples collected into heparin.
We observed variability between the starting concentrations of pfs25 and beta-globin mRNA levels in both EDTA and heparin samples. We are confident to rule out inhibition of PCR by heparin, but inherent sources of variability may exist, originating from complex sample preparation (which should also be taken into account when processing field samples for gametocyte quantification) and inhibition during RNA extraction (e.g., through DNase treatment). This may lead to different overall extraction efficiencies between samples. However, the higher instability of mRNA in EDTA samples is clearly significant, as the decay was observed within the first 8 hours in the EDTA samples but not in the heparin samples. The EDTA-related decrease is consistent with a previous study that demonstrated ex vivo instability of mRNA in whole blood collected in EDTA tubes.8
In conclusion, this study shows that sample storage at ambient (tropical) temperature is feasible up to 24 hours and that cooling and/or immediate placement into RNA preservation reagents may not be crucial if samples are collected into heparin. For samples collected into EDTA, transfer into an RNA stabilizing agent should occur within 6–8 hours after collection to avoid excessive RNA degradation.
Supplementary Material
Footnotes
Authors' addresses: Andreea Waltmann, Stephan Karl, Chris Chiu, and Ivo Mueller, Division of Population Health and Immunity, The Walter and Eliza Hall Institute of Medical Research, Victoria, Australia, and Department of Medical Biology, University of Melbourne, Victoria, Australia, E-mails: waltmann@wehi.edu.au, karl@wehi.edu.au, chiu@wehi.edu.au, and ivomueller@fastmail.fm.
References
- 1.Bousema T, Griffin JT, Sauerwein RW, Smith DL, Churcher TS, Takken W, Ghani A, Drakeley C, Gosling R. Hitting hotspots: spatial targeting of malaria for control and elimination. PLoS Med. 2012;9:e1001165. doi: 10.1371/journal.pmed.1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.malERA Consultative Group on Monitoring Evaluation, and Surveillance A research agenda for malaria eradication: monitoring, evaluation, and surveillance. PLoS Med. 2011;8:e1000400. doi: 10.1371/journal.pmed.1000400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bousema T, Drakeley C. Epidemiology and infectivity of Plasmodium falciparum and Plasmodium vivax gametocytes in relation to malaria control and elimination. Clin Microbiol Rev. 2011;24:377–410. doi: 10.1128/CMR.00051-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Houseley J, Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 5.Wampfler R, Mwingira F, Javati S, Robinson L, Betuela I, Siba P, Beck HP, Mueller I, Felger I. Strategies for detection of Plasmodium species gametocytes. PLoS One. 2013;8:e76316. doi: 10.1371/journal.pone.0076316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones S, Sutherland CJ, Hermsen C, Arens T, Teelen K, Hallett R, Corran P, van der Vegte-Bolmer M, Sauerwein R, Drakeley CJ, Bousema T. Filter paper collection of Plasmodium falciparum mRNA for detecting low-density gametocytes. Malar J. 2012;11:266. doi: 10.1186/1475-2875-11-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pritsch M, Wieser A, Soederstroem V, Poluda D, Eshetu T, Hoelscher M, Schubert S, Shock J, Loescher T, Berens-Riha N. Stability of gametocyte-specific Pfs25-mRNA in dried blood spots on filter paper subjected to different storage conditions. Malar J. 2012;11:138. doi: 10.1186/1475-2875-11-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rainen L, Oelmueller U, Jurgensen S, Wyrich R, Ballas C, Schram J, Herdman C, Bankaitis-Davis D, Nicholls N, Trollinger D, Tryon V. Stabilization of mRNA expression in whole blood samples. Clin Chem. 2002;48:1883–1890. [PubMed] [Google Scholar]
- 9.Boyle MJ, Richards JS, Gilson PR, Chai W, Beeson JG. Interactions with heparin-like molecules during erythrocyte invasion by Plasmodium falciparum merozoites. Blood. 2010;115:4559–4568. doi: 10.1182/blood-2009-09-243725. [DOI] [PubMed] [Google Scholar]
- 10.Al-Soud WA, Radstrom P. Purification and characterization of PCR-inhibitory components in blood cells. J Clin Microbiol. 2001;39:485–493. doi: 10.1128/JCM.39.2.485-493.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karl S, Davis TM, St-Pierre TG. A comparison of the sensitivities of detection of Plasmodium falciparum gametocytes by magnetic fractionation, thick blood film microscopy, and RT-PCR. Malar J. 2009;8:98. doi: 10.1186/1475-2875-8-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karl S, Laman M, Koleala T, Ibam C, Kasian B, N'Drewei N, Rosanas-Urgell A, Moore BR, Waltmann A, Koepfli C, Siba PM, Betuela I, Woodward RC, St Pierre TG, Mueller I, Davis TM. Comparison of three methods for detection of gametocytes in Melanesian children treated for uncomplicated malaria. Malar J. 2014;13:319. doi: 10.1186/1475-2875-13-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Irenge LM, Robert A, Gala JL. Quantitative assessment of human beta-globin gene expression in vitro by TaqMan real-time reverse transcription-PCR: comparison with competitive reverse transcription-PCR and application to mutations or deletions in noncoding regions. Clin Chem. 2005;51:2395–2396. doi: 10.1373/clinchem.2005.056630. [DOI] [PubMed] [Google Scholar]
- 14.Klein M, Drongowski R, Linhardt R, Lancer R. A calorimetric assay for chemical heparin in plasma. Anal Biochem. 1982;124:59–64. doi: 10.1016/0003-2697(82)90219-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.