Abstract
African trypanosomes (Trypanosoma brucei spp.) cause devastating diseases in sub-Saharan Africa. Trypanosomes differentiate repeatedly during development in tsetse flies before gaining mammalian infectivity in fly salivary glands. Lipid phosphate phosphatases (LPPs) are involved in diverse biological processes, such as cell differentiation and cell migration. Gene sequences encoding two putative T. brucei LPP proteins were used to search the T. brucei genome, revealing two additional putative family members. Putative structural features and transcript abundance during parasite development in tsetse fly were characterized. Three of the four LPP proteins are predicted to have six transmembrane domains, while the fourth shows only one. Semiquantitative gene expression revealed differential regulation of LPPs during parasite development. Transcript abundance for three of the four putative LPP genes was elevated in parasites infecting salivary glands, but not mammalian-infective metacyclic cells in fly saliva, indicating a potential role of this family in parasite establishment in tsetse salivary glands.
Trypanosoma brucei spp. cause human African trypanosomiasis and animal African trypanosomiasis in sub-Saharan Africa.1 Transmitted by the bite of an infected tsetse fly (Glossina spp.), trypanosomes undergo several distinct developmental stages during their life cycle.2 Once taken up by tsetse fly during blood feeding, mammalian bloodstream form (BSF) trypanosomes differentiate into the procyclic form (PF) in the midgut (MG). PF cells proliferate and migrate to the cardia, differentiating into epimastigotes. In some flies, parasites continue to invade the salivary glands where they ultimately develop into mammalian-infective metacyclic form (MCF) trypomastigotes.2 A tight control of gene expression occurs during tsetse colonization by T. brucei, where unique transcripts from parasites infecting midgut, cardia, and salivary glands have been reported.3,4
Lipid phosphate phosphatases (LPPs) are members of the phosphatidic acid phosphatase superfamily.5 LPP enzymes catalyze the dephosphorylation of phosphorous lipids, important in a broad range of physiological functions including cell migration, proliferation, and differentiation.5,6 Eukaryotic LPP proteins span the lipid bilayer six times and have three conserved domains constituting the catalytic site (named C1, C2, and C3).6 The function of LPP proteins in parasitic protozoa remains unknown. The genome of Trypanosoma cruzi codes for at least three LPPs, which might be involved in cell differentiation.7 In this study, four T. brucei genes encoding putative LPP proteins (TbLPP) were identified, and their tsetse tissue–specific transcription were profiled. In addition, we evaluated the transcriptional regulation of these genes both during parasite development in vivo and in vitro.
An RNA sequencing–based transcriptomic analysis of T. brucei brucei infecting tsetse midgut, proventriculus, and salivary glands revealed differential abundance of transcripts associated with two genes (Tb927.10.13700 and Tb927.10.13930), whose products were predicted to be members of the LPP family (National Center for Biotechnology Information Sequence Read Archive under accession nos. SRP002243 and SRA059559). Using the tool BLAST-N, we searched paralogous sequences at the publically available T. brucei reference genome database.8 Two additional genes (Tb927.8.480 and Tb927.10.13400) coding for other potential members of the LPP family of proteins were identified. We named these genes TbLPP1 (Tb927.8.480), TbLPP2 (Tb927.10.13400), and TbLPP3 (Tb927.10.13930), following the established nomenclature for this category of proteins. Because of its similarity to TbLPP2, the fourth gene was named TbLPP2-related protein (Tb927.10.13700).
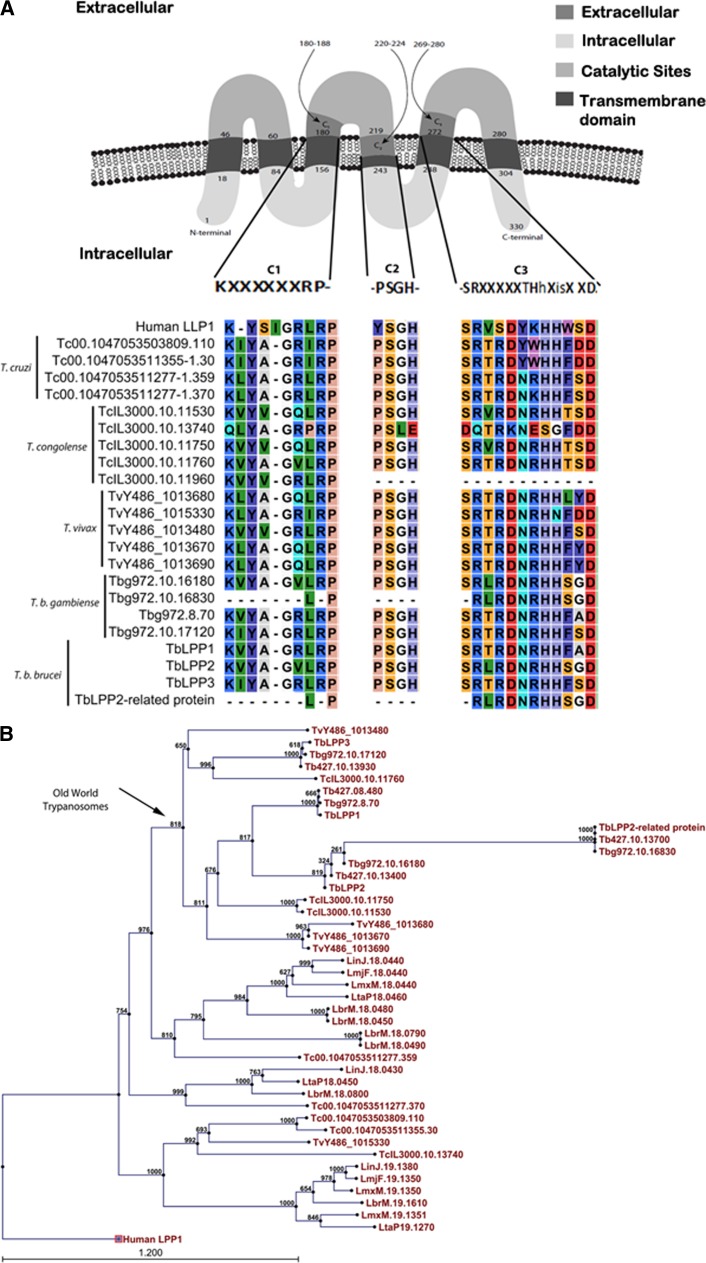
We used the TMHMM server 2.0 to predict transmembrane helices.9 TbLPP1–3 genes were predicted to code for proteins with six putative membrane-spanning domains that localize on the cell surface, with catalytic sites located between the second and third loop facing the extracellular milieu (Figure 1A ). Alignment of all four proteins confirmed that the putative TbLPP1–3 proteins contain the three conserved catalytic domains (C) characteristic of the LPP family: C1, C2, and C3. The TbLPP2-related protein however putatively codes for a truncated protein, which contains only a single transmembrane domain and the conserved domain C3 (Figure 1A). The TbLPP2-related protein was identified in a T. brucei mitochondrial proteome analysis.14 A phylogenetic analysis using available sequences for Trypanosomatidae revealed that the TbLPPs from Old World trypanosomes (T. b. brucei, Trypanosoma vivax, and Trypanosoma congolense) are evolutionarily conserved (Figure 1B). Moreover, TbLPP2-related protein has been found in the genomes of the T. brucei complex (T. b. brucei strain 927, T. b. brucei strain 427, and Trypanosoma brucei gambiense) and also in the genome of Trypanosoma evansi strain STIB 805, in agreement with the topology of the consensus phylogenic tree proposed for this group.
Figure 1.
Bioinformatic analyses of the Trypanosoma brucei lipid phosphate phosphatase (TbLPP) sequences. (A) The alignment of TbLPPs with LPP homologous sequences was performed using the software CLC Workbench using the ClustalW algorithm. The three catalytic motifs that define the LPP family were identified by searches on National Center for Biotechnology Information Conserved Domain database,10 Pfam,11 and Prosite. The tool TargetP 4.112 was used to predict the subcellular localization of TbLPPs. Tb = Trypanosoma brucei brucei strain 927; Tb427 = T. b. brucei strain 427; Tbg = Trypanosoma brucei gambiense; TcIL = Trypanosoma congolense; TvY = Trypanosoma vivax. (B) Phylogenetic inference of LPPs from Trypanosomatidae. The phylogenetic tree was constructed by ClustalW alignment using the neighbor-joining algorithm13 with a 1000-bootstrap analysis with the commercial program CLC DNA Workbench 6.2.
To evaluate the level of divergence among the TbLPPs, a pairwise protein comparison was performed using EMBOSS Matcher-Alignment program. This analysis revealed that TbLPPs possess a high level of divergence, suggesting they might have different preference for substrates (Table 1). Interestingly, despite the putative structural differences observed between TbLPP2 and TbLPP2-related protein—TbLPP2 containing ∼330 amino acids and TbLPP2-related protein around ∼70 amino acids—these proteins showed high identity (97%), suggesting that these sequences may have recently diverged, possibly after a gene duplication event (Table 1). In further support of this hypothesis, both genes are located on chromosome 10, and locus mapping, using the chromosome map available at TritryDB,8 revealed that downstream coding sequences flanking these two genes are composed of homologous proteins (Supplemental Figure 1).
Table 1.
Comparison of identity and similarity of TbLPPs among themselves
| Genes | TbLPP2 | TbLPP3 | TbLPP-related protein | |||
|---|---|---|---|---|---|---|
| Identity (%) | Similarity (%) | Identity (%) | Similarity (%) | Identity (%) | Similarity (%) | |
| TbLPP1 | 56.6 | 68.4 | 39.9 | 57.7 | 61.9 | 81 |
| TbLPP2 | – | – | 39.2 | 57.4 | 96.7 | 100 |
| TbLPP3 | – | – | – | – | 54.7 | 71.7 |
The tool EMBOSS Matcher-Alignment was used to identify local similarities between two sequences using an algorithm based on LALIGN application.15
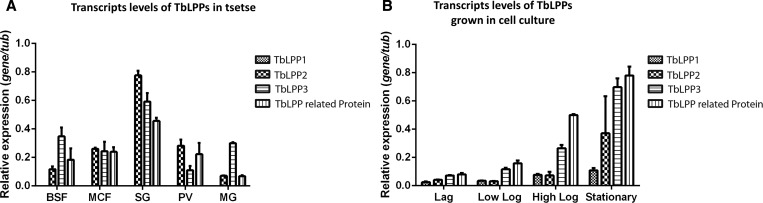
We then evaluated the tissue and developmental stage–regulated transcriptional abundance of the TbLPP-encoding genes using semiquantitative reverse transcriptase polymerase chain reaction. BSF T. b. brucei (RUMP 503) was expanded in rats, harvested at peak parasitemia, and used to infect Glossina morsitans morsitans (Westwood). Transcripts were analyzed using complementary DNA templates normalized against T. brucei α-tubulin prepared from trypanosome-infected tsetse midgut, cardia, and salivary glands, as well as the MCF parasites isolated from tsetse saliva, and BSF trypanosomes were experimentally raised in rodent blood using previously described methods.4 Transcripts corresponding to TbLPP2, TbLPP3, and TbLPP2-related protein were the most abundant in salivary glands, followed by BSF and cardia samples (Figure 2A and Supplemental Figure 2). We could not detect transcripts of the TbLPP1 gene in any of the tissue samples tested using our assay conditions (Figure 2A). The transcript levels of TbLPP3 were seven times higher in the midgut samples than that of TbLPP2 and TbLPP2-related protein (Figure 2A). Although we could detect low level of expression for the TbLPP genes from the BSF samples, transcript levels for these genes were too low for reproducible amplification from MCF parasites.
Figure 2.
Differential transcript abundance of TbLPPs. (A) Reverse transcriptase polymerase chain reaction (RT-PCR) using gene-specific primers in a 35-cycle PCR. The TbLPPs were normalized to tubulin (tub) (mean ± standard error [SE], from three independent biological samples; Supplemental Table 1). BSF, MCF, SG, cardia, and MG from 42-day-old infected tsetse flies. (B) Differential transcript abundance of TbLPPs in procyclic parasites from different phases of cellular growth. RT-PCR using gene specific primers in a 35-cycle PCR from parasites from lag, low log, high log, and stationary phases (mean ± SE, from three independent biological samples). BSF, MCF, SG, cardia, and MG from 42-day-old infected tsetse flies. (A and B) Statistical analyses are shown in Supplemental Tables 2 and 3. BSF = bloodstream form; MCF = metacyclic form; SG = salivary glands; MG = midgut from 42-day-old infected tsetse flies.
TbLPP2, TbLPP3, and TbLPP2-related protein were highly expressed in salivary glands. Within the salivary glands, three important steps of T. brucei development take place: the differentiation of free epimastigotes into epithelia-attached epimastigotes, the differentiation of attached epimastigotes to attached trypomastigotes, and ultimately, the differentiation of the attached trypomastigotes to the free MCFs.16 All these processes involve the attachment of the parasite to the luminal surface of the salivary glands, putting the parasite membrane—where the TbLPPs are predicted to be located—in an intimate contact with the host phospholipids (from epithelial cells). We speculate that the high levels of the TbLPPs observed in salivary gland stages might be important for the uptake of phospholipids from salivary gland epithelium, supporting parasite growth.
We analyzed the transcriptional activity of the TbLPP genes at different procyclic culture growth stages: the lag phase, the low and high log phases, and the stationary phase, respectively. Transcript abundance of all TbLPP genes differed as a function of the phase of cellular growth at which the parasites were collected (Figure 2B and Supplemental Figure 3). TbLPP3 and TbLPP2-related protein showed the highest abundance of transcripts in parasites from the stationary phase. Interestingly, higher levels of TbLPP1 transcripts were detected from stationary stage cultures maintained in vitro than PF cells obtained from the fly midgut in vivo. Typically, microbial cultures in the stationary phase show shortage of nutrients and accumulation of microbial by-products of metabolism. It is possible that the level of nutrients available for parasite growth contribute for the transcriptional control of TbLPPs, since they are important for lipid uptake.17
In conclusion, we infer that the abundance of transcripts encoding T. brucei LPPs change during the colonization of tsetse fly. The wide range of functions associated with LPPs make these proteins an interesting model for studying basic aspects of cell biology—cell differentiation of T. brucei in different organs of tsetse—and host cell colonization. Moreover, their preferential expression in the metacyclic stages in the salivary glands of the fly open up studies into their functional biology with inferences for control of parasite infections in the fly.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Yineng Wu for assistance with sample normalization and Geoffrey Attardo for assistance with enzyme structure illustration.
Footnotes
Financial support: This work was supported by the Ambrose Monell Foundation to Serap Aksoy and the National Institute of Allergy and Infectious Diseases award no. AI028798 to Elisabetta Ullu and ChristianTschudi. Thiago L. Alves e Silva's doctorate internship was sponsored by the Brazilian funding agency Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq).
Authors' addresses: Thiago Luiz Alves e Silva, Department of Molecular Microbiology and Immunology, Johns Hopkins University, Baltimore, MD, E-mail: tlasbio@gmail.com. Amy F. Savage, Department of Biology, Bard College, Annandale-on-Hudson, NY, E-mail: asavage@bard.edu. Serap Aksoy, Department of Epidemiology of Microbial Diseases, Yale School of Public Health, New Haven, CT, E-mail: serap.aksoy@yale.edu.
References
- 1.Courtin F, Jamonneau V, Duvallet G, Garcia A, Coulibaly B, Doumenge JP, Cuny G, Solano P. Sleeping sickness in west Africa (1906–2006): changes in spatial repartition and lessons from the past. Trop Med Int Health. 2008;13:334–344. doi: 10.1111/j.1365-3156.2008.02007.x. [DOI] [PubMed] [Google Scholar]
- 2.Sharma R, Gluenz E, Peacock L, Gibson W, Gull K, Carrington M. The heart of darkness: growth and form of Trypanosoma brucei in the tsetse fly. Trends Parasitol. 2009;25:517–524. doi: 10.1016/j.pt.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Telleria EL, Benoit JB, Zhao X, Savage AF, Regmi S, Alves e Silva TL, O'Neill M, Aksoy S. Insights into the trypanosome-host interactions revealed through transcriptomic analysis of parasitized tsetse fly salivary glands. PLoS Negl Trop Dis. 2014;8:e2649. doi: 10.1371/journal.pntd.0002649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Savage AF, Cerqueira GC, Regmi S, Wu Y, El Sayed NM, Aksoy S. Transcript expression analysis of putative Trypanosoma brucei GPI-anchored surface proteins during development in the tsetse and mammalian hosts. PLoS Negl Trop Dis. 2012;6:e1708. doi: 10.1371/journal.pntd.0001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brindley DN, Pilquil C, Sariahmetoglu M, Reue K. Phosphatidate degradation: phosphatidate phosphatases (lipins) and lipid phosphate phosphatases. Biochim Biophys Acta. 2009;1791:956–961. doi: 10.1016/j.bbalip.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pascual F, Carman GM. Phosphatidate phosphatase, a key regulator of lipid homeostasis. Biochim Biophys Acta. 2013;1831:514–522. doi: 10.1016/j.bbalip.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gimenez AM, Santander VS, Villasuso AL, Pasquare SJ, Giusto NM, Machado EE. Regulation of phosphatidic acid levels in Trypanosoma cruzi. Lipids. 2011;46:969–979. doi: 10.1007/s11745-011-3577-6. [DOI] [PubMed] [Google Scholar]
- 8.Kissinger JC. A tale of three genomes: the kinetoplastids have arrived. Trends Parasitol. 2006;22:240–243. doi: 10.1016/j.pt.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 10.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, Bateman A, Bernard T, Binns D, Bork P, Burge S, de Castro E, Coggill P, Corbett M, Das U, Daugherty L, Duquenne L, Finn RD, Fraser M, Gough J, Haft D, Hulo N, Kahn D, Kelly E, Letunic I, Lonsdale D, Lopez R, Madera M, Maslen J, McAnulla C, McDowall J, McMenamin C, Mi H, Mutowo-Muellenet P, Mulder N, Natale D, Orengo C, Pesseat S, Punta M, Quinn AF, Rivoire C, Sangrador-Vegas A, Selengut JD, Sigrist CJ, Scheremetjew M, Tate J, Thimmajanarthanan M, Thomas PD, Wu CH, Yeats C, Yong SY. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, Heger A, Holm L, Sonnhammer EL, Eddy SR, Bateman A, Finn RD. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 13.Mailund T, Brodal GS, Fagerberg R, Pedersen CN, Phillips D. Recrafting the neighbor-joining method. BMC Bioinformatics. 2006;7:29. doi: 10.1186/1471-2105-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Cui J, Nilsson D, Gunasekera K, Chanfon A, Song X, Wang H, Xu Y, Ochsenreiter T. The Trypanosoma brucei MitoCarta and its regulation and splicing pattern during development. Nucleic Acids Res. 2010;38:7378–7387. doi: 10.1093/nar/gkq618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Homer N, Merriman B, Nelson SF. Local alignment of two-base encoded DNA sequence. BMC Bioinformatics. 2009;10:175. doi: 10.1186/1471-2105-10-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rotureau B, Subota I, Buisson J, Bastin P. A new asymmetric division contributes to the continuous production of infective trypanosomes in the tsetse fly. Development. 2012;139:1842–1850. doi: 10.1242/dev.072611. [DOI] [PubMed] [Google Scholar]
- 17.Roberts RZ, Morris AJ. Role of phosphatidic acid phosphatase 2a in uptake of extracellular lipid phosphate mediators. Biochim Biophys Acta. 2000;1487:33–49. doi: 10.1016/s1388-1981(00)00081-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.