Abstract
Microscopic evaluation of skin biopsies is the monitoring and evaluation (M and E) method currently used by multiple onchocerciasis elimination programs in Africa. However, as repeated mass drug administration suppresses microfilarial loads, the sensitivity and programmatic utility of skin snip microscopy is expected to decrease. Using a pan-filarial real-time polymerase chain reaction with melt curve analysis (qPCR-MCA), we evaluated 1) the use of a single-step molecular assay for detecting and identifying Onchocerca volvulus microfilariae in residual skin snips and 2) the sensitivity of skin snip microscopy relative to qPCR-MCA. Skin snips were collected and examined with routine microscopy in hyperendemic regions of Uganda and Ethiopia (N = 500 each) and “residual” skin snips (tissue remaining after induced microfilarial emergence) were tested with qPCR-MCA. qPCR-MCA detected Onchocerca DNA in 223 residual snips: 139 of 147 microscopy(+) and 84 among microscopy(−) snips, suggesting overall sensitivity of microscopy was 62.3% (139/223) relative to qPCR-MCA (75.6% in Uganda and 28.6% in Ethiopia). These findings demonstrate the insufficient sensitivity of skin snip microscopy for reliable programmatic monitoring. Molecular tools such as qPCR-MCA can augment sensitivity and provide diagnostic confirmation of skin biopsies and will be useful for evaluation or validation of new onchocerciasis M and E tools.
Introduction
Onchocerciasis, or river blindness, is a vector-borne filarial neglected tropical disease caused by the parasitic worm Onchocerca volvulus. Second only to trachoma as an infectious cause of blindness worldwide, onchocerciasis has been targeted for elimination in several African countries by 2025.1 Community-directed, once- or twice-per-year mass drug administration (MDA) with ivermectin (IVM; Mectizan®, Merck and Co., Kenilworth, NJ) is the intervention of choice for controlling both morbidity and parasite transmission.2 As a microfilaricide that also temporarily suppresses production of microfilariae (MF), IVM MDA is an effective strategy to interrupt transmission. However, long-lived (10–15 years) reproductive adult filariae necessitate sustained long-term programs of annual or twice-per-year treatment for complete elimination of transmission risk.3,4 The availability of effective tools to determine infection status will be of critical importance to elimination programs, both for monitoring and evaluation (M and E) of program success and for determining when to stop treatment.
The current World Health Organization guidelines that define the epidemiologic criteria for determining interruption of transmission require a 5-year cumulative incidence of infection of < 0.1% in children under five and untreated immigrants.5 Within the endemic areas of the Americas, the evaluation whether a program has met these criteria has been based on serologic detection of IgG4 antibody response to the OV-16 antigen.6 Within the African context, however, several countries continue to measure infection status and prevalence through visual, microscopy-based detection of MF in skin snip biopsies.7 In hyperendemic areas, untreated areas, and areas with heavy burdens of disease morbidity, this is a satisfactory tool for identifying patent infections and transmission potential. However, reduced microfiladermia is accompanied by reduced sensitivity of skin snip microscopy.8 Thus, IVM MDA and the associated sustained suppression of microfiladermia are expected to further reduce the sensitivity of skin snip microscopy in later stages of program M and E.7,9 Moreover, sensitivity of skin snip microscopy is also known to be limited in hypoendemic areas, further calling into question its utility for mapping as elimination programs expand to include regions of reduced endemicity.10
Here we describe the design and evaluation of a single-step DNA-based method to detect patent O. volvulus infection in skin snips as an alternative to microscopic evaluation. As previously described, length polymorphisms of the first internal transcribed spacer (ITS1) of the ribosomal DNA (rDNA) repeat can be used for the detection and differentiation of filarial parasite species.11–13 Highly conserved regions within the ITS1 and in the flanking 18S and 5.8S tracts allow for development of “generalist” primers that can be used across related species. Thus, with a single set of primers, DNA from an array of filarial species can be amplified and then differentiated on the basis of amplicon length or sequence polymorphism. We adapted this approach to a real-time polymerase chain reaction with melt curve analysis (qPCR-MCA), such that the amplification, detection, and differentiation of differently sized PCR products occur within a single tube and within the time frame of a single PCR. Reducing the assay to a single preparation eliminates the need for downstream processing and diminishes the risk of subsequent transfer or cross-contamination error. The ability of this qPCR-MCA to reliably detect and identify O. volvulus was compared with that of other commonly used molecular methods such as conventional PCR with agarose gel visualization and PCR–enzyme linked immunosorbent assay (PCR-ELISA) detection of the O-150 repeat. In addition, we evaluated the sensitivity of field skin snip microscopy relative to DNA-based methods.
Methods
Skin snip collection.
Skin snips, which are superficial skin biopsies taken from the skin around the iliac crest, were collected as part of a study to evaluate diagnostic tests for onchocerciasis that could be used in the context of elimination programs in Africa. Study sites were selected in Ethiopia and Uganda where IVM distribution had occurred for less 3 three years; the last IVM distribution occurred 5 months before the study. A convenience sample of villagers was taken, as the objective was to find infected individuals, not define prevalence of infection. Five hundred participants ≥ 5 years of age and having resided within the sample sites for ≥ 10 years or since birth for children under 10 were enrolled from each site. A basic epidemiological questionnaire was administered, two skin snips were taken, and blood samples were collected from each participant. The protocol was approved by the institutional review boards of the U.S. Centers for Disease Control and Prevention, Ethiopian Health and Nutrition Research Institute of the Ministry of Health, and the Uganda National Council for Science and Technology of the Ministry of Health.
Microscopic evaluation of skin snips.
Immediately after collection, each snip was individually placed into the well of a round-bottom 96-well microtiter plate with approximately 40 μL physiological saline in each well and incubated for 12–24 hours at room temperature to induce microfilarial emergence.14 After incubation, the two snips from each participant (residual skin snips) were transferred to a single 2-mL cryovial containing 400 μL of RNAlater® preservative (Life Technologies; Grand Island, NY). The remaining incubation media was examined microscopically at 40–400× magnification to determine identity and number of MF per snip. After microscopic evaluation, transfer of the remaining media and MF to the corresponding cryovial was attempted during field processing in Uganda. However, MF adhered to the surfaces of the examination slides and, given the uncertain efficacy of the transfer, this procedure was eliminated during field processing in Ethiopia. Examined specimens were maintained at −80 to −20°C until tested at the Centers for Disease Control and Prevention laboratories in Atlanta, GA.
DNA extraction.
Before extraction of genomic DNA from the residual skin snip specimens, snips and free-floating MF were isolated from RNAlater preservative. To facilitate efficient transfer and centrifugal separation of MF, 400 μL of a 1× phosphate-buffered saline solution containing 0.05% Tween-20 (PBS-Tween) were added to each cryovial (i.e., 1:1 [v/v] of RNAlater and PBS-Tween). Contents were briefly vortexed to ensure complete mixing, centrifuged briefly to remove liquids from the cryovial top, and then transferred to a sterile 2-mL microcentrifuge tube. To ensure complete transfer of specimens, each cryovial was rinsed with an additional 400 μL volume of PBS-Tween, vortexed, briefly centrifuged, and the contents were transferred to the corresponding 2-mL microcentrifuge tube. Microcentrifuge tubes and their contents were then centrifuged at 20,000 × g for 10 minutes in a tabletop fixed-rotor microcentrifuge. Supernatant was removed via pipette and returned to the corresponding cryovial for backup storage at −80°C. During each round of isolation and extraction, an extraction negative control (sham extraction) was created by performing the above procedures on an unused cryovial containing 400 μL RNAlater. Sham extractions were carried forward throughout the entire process of extraction and molecular evaluation to confirm lack of cross-contamination during processing of sample batches.
Whole genomic DNA was extracted from isolated specimens using the QIAamp DNA Investigator Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions for tissue extraction, using a 6–12 hour digest at 56°C with mixing to ensure complete lysis. Purified DNA was eluted from QIAamp MinElute columns with 50 μL Buffer ATE after a 20-minute incubation at room temperature and quantified with a NanoDrop ND-2000 spectrophotometer (ThermoScientific, Wilmington, DE).
qPCR-MCA detection of O. volvulus.
Conserved tracts within the ITS1 and 5.8S regions of the rDNA gene and flanking a polymorphic segment of the ITS1were targeted for primer design (ITS1-UNIF: 5′-GGTGwTATTCGTTGGTGTCTAT-3′ and 5.8S-EXTR: 5′-AGCTAGCTGCGTTCTTCAT-3′, the latter primer being a modified version of the UNI-1R primer), resulting in a 246–270 bp amplicon corresponding to the 3′ end of the ITS1.13 Known larval and/or adult isolates of Dirofilaria immitis, Brugia malayi, and O. volvulus were used to characterize specific melt curve profiles of the amplicon and establish the ability of the assay to differentiate species on the basis of melt temperature (Tm). Although neither D. immitis nor B. malayi are expected to have epidemiologic relevance to human onchocerciasis, availability of well-characterized biological and genetic materials through the Filariasis Research Reagent Resource Center makes these species well-suited for inclusion as technical controls for a pan-filarial assay. The qPCR-MCA was performed on a Stratagene Mx30005P system (Agilent Technologies, Santa Clara, CA) in 10 μL volumes containing 1× EvaGreen Supermix with Low ROX (BioRad, Inc., Hercules, CA), 0.4 μM of each primer, and 0.5 μL genomic DNA, with the platform's standard EvaGreen melt curve analysis protocol (10 minutes at 95°C; 40 cycles of 10 seconds at 95°C, 15 seconds at 60°C, and 20 seconds at 72°C; complete denaturation of double-stranded PCR product for 1 minute at 95°C; and dissociation curve data collection over a 0.01°C/second ramp from 60°C to 95°C). Each 96-well assay included a positive control panel of D. immitis, B. malayi, and O. volvulus genomic DNA from the known larval or adult isolates, as well as a negative control panel consisting of nonendemic Homo sapiens genomic DNA, the sham extraction corresponding to the batch of samples being tested, and a no-template negative control. All samples and controls were assayed in duplicate and the individual performing the PCR testing was blind to the results from the field microscopy.
Assays were considered valid if all positive controls yielded amplicons of the appropriate size and Tm as determined during standardization of the protocol (D. immitis: 78.4 ± 0.2°C; B. malayi: 77.4 ± 0.2°C; O. volvulus: 79.4 ± 0.19°C) and negative controls were free of product. For skin snip samples to be considered positive, the Tm was required to be within the range specified for O. volvulus and have a cycle threshold (CT) value < 36—the value below which amplification artifacts were not seen during protocol standardization. Samples with the appropriate melt temperature and CT values of 36–38 were considered indeterminate and re-assayed, while samples with a CT > 38 were considered negative.
Conventional molecular detection.
Conventional PCR with agarose gel product visualization was performed with the primers described above as a visual verification of product during qPCR-MCA protocol standardization and then to compare the performance of qPCR-MCA and conventional PCR during snip processing. Reactions were performed in 25 μL volumes containing 1× buffer, 0.2 mM dNTP, 0.4 μM each primer, 0.63 U Taq, and 1 μL genomic DNA, with a thermocycling protocol of 5 minutes at 94°C; 35 cycles of 30 seconds at 94°C, 30 seconds at 60°C, and 30 seconds at 72°C; and a final extension of 72°C for 7 minutes. As above, all assays included a panel of D. immitis, B. malayi, and O. volvulus positive controls and a negative control panel of nonendemic human genomic DNA, the sham extraction, and a no-template well. Amplicons were separated on a 1.2% agarose gel and visualized with EZ-Vision®Two fluorescent dye (Amresco, Solon, OH) and an ultraviolet transilluminator. Assays were considered valid if all positive control wells contained visible product of the appropriate size and negative control wells did not. Likewise, skin snip samples were considered positive if wells contained visible product of the same size as the positive controls.
For the purposes of external quality control and methodological comparison, a random subset of approximately 25% of the skin snip samples were also assayed via the standard O-150 PCR-ELISA protocol regularly used to detect O. volvulus in Simulium flies in the Americas.15 Amplification of the O-150 repeat was performed in duplicate for all samples, including two high- and low-concentration aliquots of adult O. volvulus genomic DNA, two nonendemic human genomic DNA negative controls, and one no-template negative control. ELISA reactions to detect O-150 product were performed in duplicate for each PCR replicate and scored as previously described.15
Finally, for quality control of qPCR-MCA specificity, a randomly selected subset of 24 qPCR-MCA-positive skin snip samples were also subjected to standard Sanger sequencing of the entire ITS1 (external primers 18S-EXTF: 5′-TGCTGTAACCATTACCGAAAG-3′ modified from and 5.8S-EXTR as above; internal primers 18S-INTF: 5′-TGAGCCGTTTCGAGAAAAGC-3′, ITS1-UNIR: 5′-TyAGGCGATAAyTTGGATAAAG-3′, and ITS1-UNIF as above).13 In brief, externally amplified PCR product was purified (StrataPrep PCR Purification kit; Agilent Technologies), sequenced bidirectionally with internal primers and BigDye® Terminator v3.1 chemistry (LifeTechnologies, Inc.), and analyzed on an ABI 3130xl genetic analyzer (LifeTechnologies, Inc.). Electropherograms were evaluated and trimmed manually and assembled in ChromasPro v1.7.6 (Technelysium, South Brisbane, Australia) before interrogation of the National Center for Biotechnology Information's nucleotide database via the Basic Local Alignment Search Tool using the megablast algorithm.
Results
Skin snip microscopy versus PCR-based detection.
Skin snip biopsies from a total of 1,000 participants (500 each from the Ugandan and Ethiopian sites) were examined for the presence of O. volvulus MF via standard field microscopy and a qPCR-MCA approach to molecular detection. Skin snips were incubated in physiological saline for an average of 23.3 hours (±1.3 hours) before microscopic examination. A total of 147 participants (14.7%) were positive for O. volvulus microfiladermia by microscopic examination—127 in Uganda (25.4%) and 20 in Ethiopia (4%). Onchocerca volvulus DNA was detected in skin snips from a total of 223 participants (22.3%) by qPCR-MCA—160 in Uganda (32%) and 63 in Ethiopia (12.6%). Relative to PCR-based detection of MF in skin snips, standard field microscopy demonstrated an overall sensitivity of 62.3% (Uganda: 75.6%; Ethiopia: 28.6%; Table 1). Among individuals with low microfiladermia (≤ 2 MF total), sensitivity of microscopy decreased to 40.4% overall and 56.2% and 13.5% within the Ugandan and Ethiopian groups, respectively (Table 1).
Table 1.
Performance of skin snip microscopy relative to qPCR-MCA DNA-based detection
| All samples | qPCR-MCA positive | ||
|---|---|---|---|
| Overall | Uganda | Ethiopia | |
| Microscopy positive | 139 | 121 | 18 |
| Microscopy negative | 84 | 39 | 45 |
| Microscopy sensitivity (%) | 62.3 | 75.6 | 28.6 |
| Samples with ≤ 2 MF total | qPCR-MCA positive | ||
| Overall | Uganda | Ethiopia | |
| Microscopy positive | 57 | 50 | 7 |
| Microscopy negative | 84 | 39 | 45 |
| Microscopy sensitivity (%) | 40.4 | 56.2 | 13.5 |
MF = microfilariae; qPCR-MCA = real-time polymerase chain reaction with melt curve analysis.
Overall agreement between field microscopy of incubated skin snips and qPCR-MCA was 90.8% (908/1,000), with the vast majority (91.3%; 84/92) of disagreement comprising samples negative by microscopy and positive by qPCR-MCA. Of the eight samples positive by microscopy and negative by qPCR-MCA, average microfilarial load was 1.63 MF/person (median = 1, range: 1–5 MF); only one of these eight samples was positive by the other molecular methods (i.e., conventional PCR and O-150 PCR-ELISA).
qPCR-MCA review and molecular method comparison.
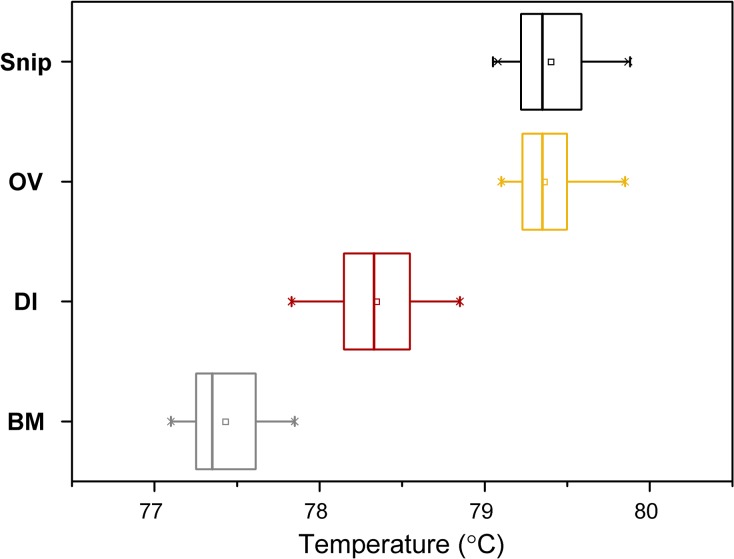
The species-specific Tm for positive controls showed minimal variation among assays and Tm for positive skin snip reactions was identical to the known adult worm control (Figure 1 ). In addition to Tm, the melt curve profiles of each control species and the positive skin snip reactions consistently comprised single peaks indicating lack of secondary or nonspecific product (Figure 2 ). Likewise, determination of infection status was highly consistent, with 96.2% of samples having 100% agreement both within assays (i.e., between technical replicates) and among repeated assays.
Figure 1.
Box plot of melt curve analysis–derived melting temperatures observed in individual real-time polymerase chain reaction with melt curve analysis reactions illustrating limited variation in Tm observed in reactions containing Brugia malayi (BM, N = 156), Dirofilaria immitis (DI, N = 79), and Onchocerca volvulus (OV, N = 82) positive controls and MF-positive skin snips (Snip, N = 509) from Uganda and Ethiopia. Box boundaries indicate one standard deviation and whiskers delineate minimum and maximum observed Tm.
Figure 2.
Overlay of all melt curves generated for positive control specimens (Brugia malayi, Dirofilaria immitis, and Onchocerca volvulus) and positive skin snips, demonstrating the technical reproducibility of the melt curve as an indicator of sample identity.
Determinations by qPCR-MCA and conventional PCR were in agreement for 98.5% of the samples, with over 93% of disagreements (14/15) comprising samples positive by qPCR-MCA and negative by conventional PCR. Eight of the 14 qPCR-MCA positive, conventional PCR negative samples were determined to be positive with high confidence; two were positive by microscopy (1–2 MF/person); and six produced consistently positive qPCR-MCA results (Tm 79.3–79.8°C and CT ≤ 36). The remaining six qPCR-MCA-positive/PCR-negative samples had indeterminate initial results (i.e., replicate disagreement or appropriate Tm but the CT fell in the indeterminate range of 36–38) followed by positive results. It is unclear whether these are true positives at the limit of assay detection or false positives. Conventional PCR identified one positive sample missed by qPCR-MCA (MF load = 1).
Within the subset of samples evaluated by both qPCR-MCA and O-150 PCR-ELISA, there was 89.5% (222/248) agreement between assays. The majority of discordant results were positive by qPCR-MCA and negative by O-150 PCR-ELISA (88.5%; 23/26), with microscopy and/or conventional PCR supporting positive qPCR-MCA outcomes for 17 samples. Six of the samples with conflicting qPCR-MCA and O-150 PCR-ELISA results were positive by microscopy, with five samples falsely negative by O-150 PCR-ELISA (average = 88.6 MF/person; range = 2–360 MF) and one qPCR-MCA false negative (1 MF).
Sanger sequencing of the ITS1 region of the rDNA was attempted for 24 qPCR-MCA positive samples, nine of which were O-150-PCR-ELISA negative samples. Overall, 21 specimens returned ITS1 sequences (656–751 bp) with ≥ 99% homology to previously published O. volvulus ITS1 sequences, including seven of the nine specimens positive by qPCR-MCA and negative by O-150 PCR-ELISA. The remaining three samples did not produce sufficient product for sequencing. One was positive by all three molecular assays and two were positive only by qPCR-MCA.
Discussion
Herein, we have demonstrated that single-step reaction of qPCR-MCA reliably detects and identifies O. volvulus MF in skin snip specimens. The qPCR-MCA results were well-supported by the other molecular methods evaluated, with over 92% of qPCR-MCA-positive, microscopy-negative samples being positive by at least one other molecular method. Likewise, qPCR-MCA exhibited a sensitivity equal to or greater than the multistep processes of conventional PCR and O-150 PCR-ELISA.
More importantly, our results clearly illustrate the limited sensitivity of skin snip microscopy relative to qPCR-MCA, particularly in scenarios of suppressed microfiladermia. The substantial difference between the Ugandan and Ethiopian field sites in the sensitivity of microscopy relative to qPCR-MCA may be a prime example of how reliance on skin snip microscopy can result in inconsistent and unreliable program assessment. A full explanation regarding the differences in snip performance between the two sites requires additional analysis, although there could be site-specific difference in force of transmission.
Comparison among molecular assays tested during this investigation indicated that amplification of the ITS1 target had a greater overall sensitivity relative to O-150 PCR-ELISA and this sensitivity appears to be maximized in the real-time amplification and detection of qPCR-MCA. Both the O-150 locus and the rDNA gene are multicopy elements within the genome, improving the likelihood of detecting even a single MF. The primary limitation of using O-150 PCR-ELISA in this comparison may be the additional post-amplification manipulation required for detection of PCR product. Real-time amplification and detection of the O-150 repeat have generally exhibited greater sensitivity relative to methods requiring post-amplification manipulation for product detection.16,17 Therefore, it is possible that differences observed here are largely due to the constraints of the detection method, rather than genomic properties of the molecular target being amplified. This highlights that in selecting a method for implementation, the added cost of a real-time platform may be somewhat offset by the combined benefits of time saved, reduced crossover contamination risk, and improved assay sensitivity.
The application of PCR-based tools for the detection of parasite DNA in host or vector tissues is not new. For O. volvulus and other filariae, a variety of molecular techniques and targets have been demonstrated to be more sensitive for the detection of parasitic infections than standard parasitological evaluation of biological specimens. These methods include the O-150 PCR-ELISA used here, qPCR-MCA of the O-150 repeat, TaqMan-based qPCR, loop-mediated isothermal amplification, specific oligonucleotide capture with magnetic beads, nested PCR combining general and species-specific primers, and PCR with general filarial primers followed by restriction fragment length polymorphism analysis to determine taxonomic identity.13,15–23 However, with few exceptions, these procedures require at least one post-amplification step for final determination, are specific to only one genus, or require multiple specific reaction components.16,17 The procedure used here allows for single-tube assessment of both presence and identity in a single 90-minute reaction. Moreover, it capitalizes on the dual existence of highly conserved and highly variable regions within the ITS1–5.8S region of rDNA to allow for general filarial detection and identification without increasingly specific and complicated reagent or processing needs. Exact costs of establishing any molecular protocol in reference or regional laboratories will vary among laboratories and countries depending on the availability of equipment through other well-supported control programs (e.g., malaria and human immunodeficiency virus/acquired immunodeficiency syndrome) and reagent suppliers. But the flexibility and simplified needs of qPCR-MCA—fewer specific probes and reaction components and the ability to use less specialized and costly qPCR platforms—may provide an economical and practical alternative relative to other previously reported methods. Preliminary evaluations with other relevant filarial species (e.g., Loa loa, Mansonella ozzardi, and Mansonella perstans; see Supplemental Figure 1) and sample types (e.g., filtered blood, dried blood spots, residual test strip blood), indicate that this method could be applied for supporting mapping or M and E activities for a broad array of filarial genera, principally as a tool for confirmation of infection. The data presented here suggest robustness of the assay for O. volvulus detection, but further evaluations including samples from additional geographical origins are needed for complete methodological evaluation and to determine if the assay allows for identification of mixed infections.
Our results corroborate previous observations regarding the relative performance of microscopy and molecular detection of O. volvulus.10,16,24,25 It is clear that microscopy-based methods will miss patent infections, which is not ideal within the elimination context. Although microscopic evaluation of skin snips was a critical component of early onchocerciasis control activities, the preponderance of evidence indicates its limited utility outside of baseline mapping and initial monitoring of meso- and hyperendemic areas. This underscores the need to adapt skin snip and other methodologies to support the needs of programs as they move toward elimination. In the context of elimination, highly sensitive tools that identify patent infections are of great importance. This is particularly true during the determination of whether a program has interrupted transmission and during post-treatment surveillance. An assay must be sufficiently sensitive to detect low-level patent infections and specific enough to minimize false-positive results. In the low prevalence settings of the elimination context, concern for false-positive results is of paramount concern. Molecular tests, such as qPCR-MCA, offer the most sensitive and specific test for confirmation of results from other less-invasive testing modalities (e.g., serologic tests) when stopping decisions are being made and during post-treatment surveillance, where distinguishing residual antibody reactivity from a patent and potentially recrudescent infection is important. This may also be important as strategies to map hypoendemic areas evolve, again as a method to help interpret results from other testing modalities. In light of the results presented here and in the aforementioned comparisons, it is clear that molecular detection of parasites in skin snips will provide a specific and much more sensitive confirmatory assay.
Supplementary Material
ACKNOWLEDGMENTS
We extend sincere thanks to the Ugandan and Ethiopian communities and field teams for their generous participation, Mark Eberhard for O. volvulus macrofilariae, and the Filariasis Research Reagent Resource Center (FR3) for provision of Brugia and Dirofilaria specimens.
Footnotes
Financial support: This work was supported by a grant from the Bill & Melinda Gates Foundation. Elizabeth A. Thiele was supported by a Postdoctoral Fellowship through the Oak Ridge Institute for Science and Education (ORISE).
Authors' addresses: Elizabeth A. Thiele, Department of Biology, Vassar College, Poughkeepsie, NY, E-mail: elthiele@vassar.edu. Vitaliano A. Cama, Francisca Abanyie, and Paul T. Cantey, Parasitic Diseases Branch, Division of Parasitic Diseases and Malaria, Centers for Disease Control and Prevention, Atlanta, GA, E-mails: vcama@cdc.gov, why6@cdc.gov, and pcantey@cdc.gov. Thomson Lakwo, Vector Control Division, Ministry of Health, Kampala, Uganda, E-mail: tlakwo@gmail.com. Sindeaw Mekasha, Markos Sleshi, and Amha Kebede, Ethiopian Public Health Institute, Addis Ababa, Ethiopia, E-mails: mekashasindeaw@yahoo.com, markossleshi@yahoo.com, and amha.kebede@gmail.com.
References
- 1.World Health Organization African programme for onchocerciasis control: meeting of National Task Forces, September, 2011. Wkly Epidemiol Rec. 2011;86:541–549. [PubMed] [Google Scholar]
- 2.Richards FO, Miri E, Meredith S, Guderian R, Sauerbrey M, Remme H, Packard R, Ndiaye JM. Onchocerciasis. Bull World Health Organ. 1998;76:147–149. [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts JMD, Neumann E, Göckel CW, Highton RB. Onchocerciasis in Kenya 9, 11 and 18 years after elimination of the vector. Bull World Health Organ. 1967;37:195–212. [PMC free article] [PubMed] [Google Scholar]
- 4.Turner HC, Walker M, Churcher TS, Basáñez MG. Modelling the impact of ivermectin on River Blindness and its burden of morbidity and mortality in African Savannah: EpiOncho projections. Parasit Vectors. 2014;7:241. doi: 10.1186/1756-3305-7-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization . Criteria for Certification of Interruption of Transmission/Elimination of Human Onchocerciasis. Geneva, Switzerland: WHO; 2001. Document WHO/CDS/CEE/2001.18a. [Google Scholar]
- 6.Lindblade KA, Arana B, Zea-Flores G, Rizzo N, Porter CH, Dominguez A, Cruz-Ortiz N, Unnasch TR, Punkosdy GA, Richards J, Sauerbrey M, Castro J, Catú E, Oliva O, Richards FO., Jr Elimination of Onchocercia volvulus transmission in the Santa Rosa focus of Guatemala. Am J Trop Med Hyg. 2007;77:334–341. [PubMed] [Google Scholar]
- 7.Diawara L, Traoré MO, Badji A, Bissan Y, Doumbia K, Goita SF, Konaté L, Mounkoro K, Sarr MD, Seck AF, Toé L, Tourée S, Remme JH. Feasibility of onchocerciasis elimination with ivermectin treatment in endemic foci in Africa: first evidence from studies in Mali and Senegal. PLoS Negl Trop Dis. 2009;3:e497. doi: 10.1371/journal.pntd.0000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor HR, Munoz B, Keyvan-Larijani E, Greene BM. Reliability of detection of microfilariae in skin snips in the diagnosis of onchocerciasis. Am J Trop Med Hyg. 1989;41:467–471. doi: 10.4269/ajtmh.1989.41.467. [DOI] [PubMed] [Google Scholar]
- 9.Basáñez M-G, Pion SDS, Boakes E, Filipe JAN, Churcher TS, Boussinesq M. Effect of single-dose ivermectin on Onchocerca volvulus: a systematic review and meta-analysis. Lancet Infect Dis. 2008;8:310–322. doi: 10.1016/S1473-3099(08)70099-9. [DOI] [PubMed] [Google Scholar]
- 10.Boatin BA, Toe L, Alley ES, Nagelkerke NJ, Borsboom G, Habbema JD. Detection of Onchocerca volvulus infection in low prevalence areas: a comparison of three diagnostic methods. Parasitology. 2002;125:545–552. [PubMed] [Google Scholar]
- 11.Morales-Hojas R, Post RJ, Shelley AJ, Maia-Herzog M, Coscaron S, Cheke RA. Characterisation of nuclear ribosomal DNA sequences from Onchocerca volvulus and Mansonella ozzardi (Nematoda: Filarioidea) and development of a PCR-based method for their detection in skin biopsies. Int J Parasitol. 2001;31:169–177. doi: 10.1016/s0020-7519(00)00156-9. [DOI] [PubMed] [Google Scholar]
- 12.Rishniw M, Barr SC, Simpson KW, Frongillo MF, Franz M, Dominguez Alpizar JL. Discrimination between six species of canine microfilariae by a single polymerase chain reaction. Vet Parasitol. 2006;135:303–314. doi: 10.1016/j.vetpar.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 13.Tang TH, Lopez-Velez R, Lanza M, Shelley AJ, Rubio JM, Luz SL. Nested PCR to detect and distinguish the sympatric filarial species Onchocerca volvulus, Mansonella ozzardi and Mansonella perstans in the Amazon Region. Mem Inst Oswaldo Cruz. 2010;105:823–828. doi: 10.1590/s0074-02762010000600016. [DOI] [PubMed] [Google Scholar]
- 14.Collins RC, Brandling-Bennett AD, Holliman RB, Campbell CC, Darsie RF. Parasitological diagnosis of onchocerciasis: comparisons of incubation media and incubation times for skin snips. Am J Trop Med Hyg. 1980;29:35–41. doi: 10.4269/ajtmh.1980.29.35. [DOI] [PubMed] [Google Scholar]
- 15.Unnasch TR, Meredith SE. The use of degenerate primers in conjunction with strain and species oligonucleotides to classify Onchocerca volvulus. In: Clapp JP, editor. Methods in Molecular Biology. Vol. 50. Totowa, NJ: Humana Press Inc.; 1996. pp. 293–303. [DOI] [PubMed] [Google Scholar]
- 16.Fink DL, Fahle GA, Fischer S, Fedorko DF, Nutman TB. Toward molecular parasitologic diagnosis: enhanced diagnostic sensitivity for filarial infections in mobile populations. J Clin Microbiol. 2011;49:42–47. doi: 10.1128/JCM.01697-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lloyd MM, Gilbert R, Taha NT, Weil GJ, Meite A, Kouakou IMM, Fischer PU. Conventional parasitology and DNA-based diagnostic methods for onchocerciasis elimination programmes. Acta Trop. 2015;146:114–118. doi: 10.1016/j.actatropica.2015.03.019. [DOI] [PubMed] [Google Scholar]
- 18.Fischer P, Buttner DW, Bamuhiiga J, Williams SA. Detection of the filarial parasite Mansonella streptocerca in skin biopsies by a nested polymerase chain reaction-based assay. Am J Trop Med Hyg. 1998;58:816–820. doi: 10.4269/ajtmh.1998.58.816. [DOI] [PubMed] [Google Scholar]
- 19.Nuchprayoon S, Junpee A, Poovorawan Y, Scott AL. Detection and differentiation of filarial parasites by universal primers and polymerase chain reaction-restriction fragment length polymorphism analysis. Am J Trop Med Hyg. 2005;73:895–900. [PubMed] [Google Scholar]
- 20.Jiménez M, González LM, Carranza C, Bailo B, Pérez-Ayala A, Muro A, Pérez-Arellano JL, Gárate T. Detection and discrimination of Loa loa, Mansonella perstans and Wuchereria bancrofti by PCR-RFLP and nested-PCR of ribosomal DNA ITS1 region. Exp Parasitol. 2011;127:282–286. doi: 10.1016/j.exppara.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 21.Poole CB, Tanner NA, Zhang Y, Evans TC, Jr, Carlow CK. Diagnosis of brugian filariasis by loop-mediated isothermal amplification. PLoS Negl Trop Dis. 2012;6:e1948. doi: 10.1371/journal.pntd.0001948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gopal H, Hassan HK, Rodriguez-Perez MA, Toe LD, Lustigman S, Unnasch TR. Oligonucleotide based magnetic bead capture of Onchocerca volvulus DNA for PCR pool screening of vector black flies. PLoS Negl Trop Dis. 2012;6:e1712. doi: 10.1371/journal.pntd.0001712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pilotte N, Torres M, Tomaino FR, Laney SJ, Williams SA. A TaqMan-based multiplex real-time PCR assay for the simultaneous detection of Wuchereria bancrofti and Brugia malayi. Mol Biochem Parasitol. 2013;189:33–37. doi: 10.1016/j.molbiopara.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 24.Zimmerman PA, Guderian RH, Aruajo E, Elson L, Phadke P, Kubofcik J, Nutman TB. Polymerase chain reaction-based diagnosis of Onchocerca volvulus infection: improved detection of patients with onchocerciasis. J Infect Dis. 1994;169:686–689. doi: 10.1093/infdis/169.3.686. [DOI] [PubMed] [Google Scholar]
- 25.Toé L, Boatin BA, Adjami A, Back C, Merriweather A, Unnasch TR. Detection of Onchocerca volvulus infection by O-150 polymerase chain reaction analysis of skin scratches. J Infect Dis. 1998;178:282–285. doi: 10.1086/517454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.