Abstract
The accurate segregation or partition of replicated DNA is essential for ensuring stable genome transmission. Partition of bacterial plasmids requires only three elements: a centromere-like DNA site and two proteins, a partition NTPase, and a centromere-binding protein, CBP. Due to this simplicity, partition systems have served as tractable model systems to study the fundamental molecular mechanisms required for DNA segregation at an atomic level. In the last few years, great progress has been made in this endeavor. Surprisingly, these studies have revealed that although the basic partition components are functionally conserved between three types of plasmid partition systems, these systems employ distinct mechanisms of DNA segregation. This review summarizes the molecular insights into plasmid segregation that have been achieved through these recent structural studies.
Introduction
In order to ensure genome stability, DNA must be evenly distributed to daughter cells after replication. This process is termed DNA segregation or partition. Due to its simplicity, the segregation of bacterial plasmids has served as a model for understanding the minimal molecular requirements for DNA segregation. Whereas high copy number bacterial plasmids rely on passive diffusion for plasmid maintenance, low copy number plasmids require so-called partition (par) systems, which are carried on the plasmid DNA, for their retention. The majority of par operons or cassettes contain two genes; one encoding a nucleotide triphosphatase (NTPase) and the second encoding a centromere-binding protein (CBP). In addition, the centromere-like site bound by the centromere-binding protein is located near the par cassette. Only a few plasmid centromeres (Fig. 1a) have been mapped and most consist of multiple repeats. An exception is the P1 centromere, which contains two different repeats that are recognized by the CBP, ParB, as well as a centrally located DNA binding site for the E. coli host integration factor, IHF [1–3].
Figure 1.
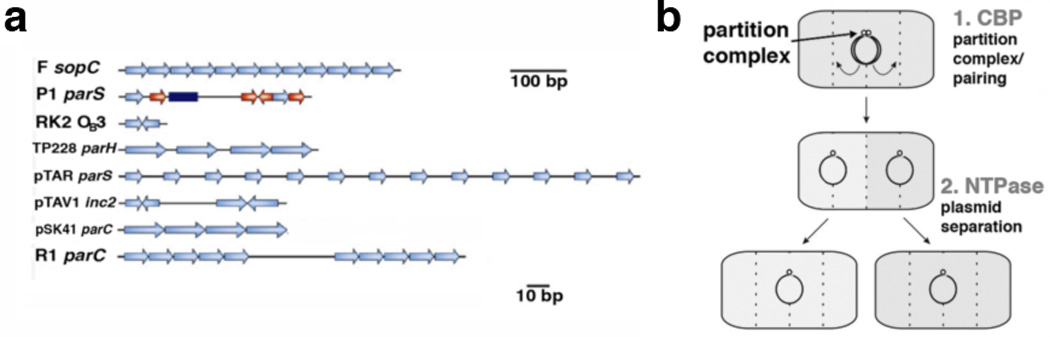
(a) Organization of well characterized centromeres. Repeat sequences are denoted by arrows. The central blue box in the P1 centromere is recognized by IHF. The atypical P1 centromere also has two different repeats bound by ParB [11]. (b) Schematic diagram showing the general steps of plasmid segregation. Plasmids are represented as circles. The first step is formation of the partition complex by CBP binding to the centromere. This is followed by plasmid pairing (interacting circles). The NTPase molecules then interact with partition complexes and drive plasmid separation before cell division.
Because these three components are all that is required to direct the segregation reaction, these bacterial partition (par) systems represent “minimalist” DNA segregation machines [1, 3–4]. The general mechanism involved in bacterial plasmid partition includes three key steps (Fig. 1b). The first step involves the binding of multiple CBPs to the centromere repeats to form a higher order nucleoprotein complex termed the partition complex. The partition complex then recruits the NTPase in the next step. Once recruited the NTPase then actively mediates plasmid separation to opposite bacterial cell poles (Fig. 1b). Three main types of par systems have been recognized, based primarily on the type of NTPase present [5]. The most common are the type I systems, which encode NTPases called ParA and CBPs called ParB. The type I family can be further divided into type Ia and Ib based on the size and sequence of the CBP and NTPase proteins. Type II systems utilize NTPases and CBPs called ParM and ParR and the more recently characterized type III systems use NTPases and CBPs called TubZ and TubR. Partition operons are autoregulated at the transcriptional level. This function is performed by the CBP proteins in the type Ib, II and III systems, and the NTPase in the type Ia systems.
While a general understanding of the bacterial plasmid partition process was obtained several decades ago, it was not until the availability of key structures at atomic resolution that detailed insight into the process began to emerge. The first structure of an NTPase, the R1 ParM protein, revealed the striking finding that it harbors an actin-like fold. This led to the suggestion, which was later confirmed, that ParM utilizes polymers in mediating segregation [6]. Structural studies on CBP proteins, however, proved more difficult as they are highly flexible, multi-domain proteins. Indeed, only four CBP structures were available by 2005, those of type Ib proteins ParG and ω and domains of the type Ia proteins P1 ParB and RP4 KorB [7–11]. These initial structural studies combined with biochemical data revealed the domain organization of the par proteins and suggested mechanisms involved in CBP DNA recognition and NTPase-mediated DNA separation. Despite these insights, key questions remained, including: what types of structures are adopted by partition complexes; how do nucleotides affect NTPase function; and how do the non-actin based NTPase proteins function to segregate plasmids? As described in this review, structural studies in the last few years have shed significant light on and, in some cases, revealed the answers to these questions. Surprisingly, although we now known that each system uses cytoskeletal-like NTPases, the mechanisms employed by each type of partition system turns out to be markedly different.
Type I partition: complexity in CBP structure and NTPase function
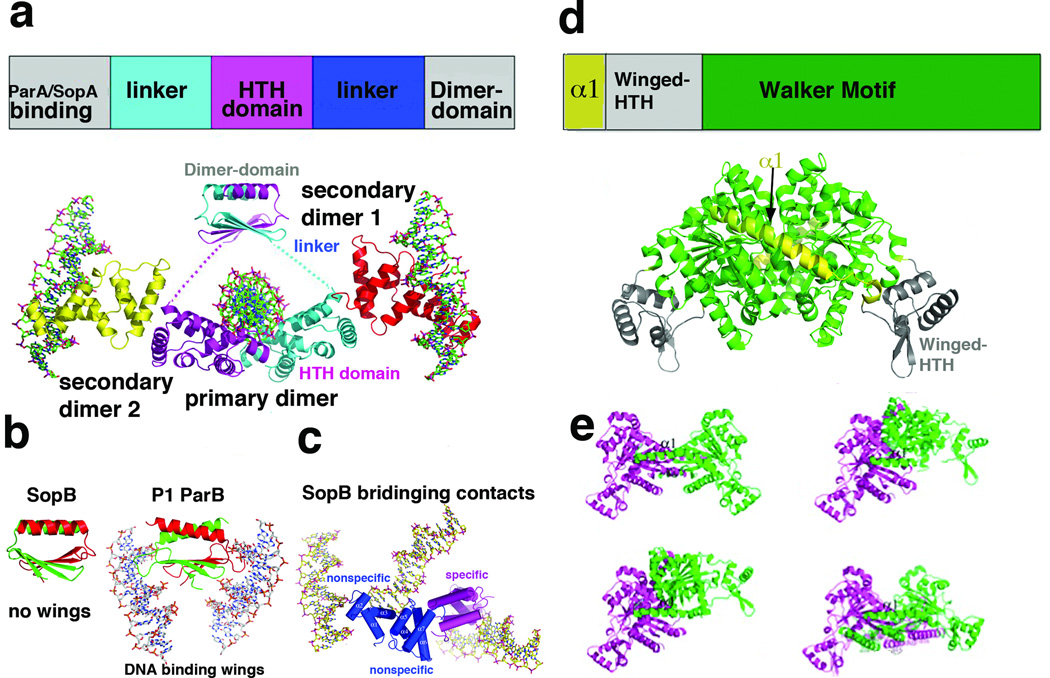
Type I systems share in common the fact that they contain NTPases with Walker box NTPases. However, the type I CBPs show little to no sequence homology. Despite the lack of sequence homology, the structures of type Ib Escherichai coli TP228 ParG and the Streptococcus pyogenes pSM19035 ω revealed that both contain ribbon-helix-helix (RHH) folds [9–10]. Recently, the structure of a third type Ib CBP was solved, that of the Leifsonia xyli subsp. cynodontis pCXC100 ParB [12•]. This structure also contains a RHH, which strongly suggests that all type Ib CBPs contain this DNA binding motif. The only type Ib CBP that has been solved bound to DNA is that of ω. However, because the DNA used in these structural studies contained extra non-centromeric nucleotides, how ω binds multiple repeats was not revealed [13]. Thus, the structure(s) adopted by Ib partition complexes is unknown. In addition to their RHH domains, type Ib CBPs also contain N-terminal arms that interact with their partner NTPases. Importantly, Barilla et al. showed that this arm contains an arginine finger, which appears to be conserved other I CBPs, and which functions in NTPase binding and stimulation of NTP hydrolysis [14]. Type Ia CBPs are structurally distinct from their type Ib counterparts and much more complex. These proteins consist of three domains; an N-terminal NTPase binding domain, a central helix-turn-helix (HTH) domain and a C-terminal dimer-domain (Fig. 2a). Previous structures of separate HTH and dimer-domains of the RP4 KorB and an E. coli P1 ParB fragment that included both domains, revealed their mechanisms of DNA recognition and how P1 ParB recognizes the atypical P1 centromere, which contains two different centromere repeats [7–8, 11, 15]. More recently, structures have become available for a third type Ia CBP, SopB [16••]. SopB is the CBP for the E. coli F plasmid par system, which includes the NTPase, SopA and the sopC centromere [17–24]. Previous studies had shown that SopB can bind DNA both specifically and nonspecifically, the latter leading to DNA coating. The nonspecific coating capability of SopB includes binding between different DNA duplexes or in trans spreading. This DNA coating function was suggested to be critical for stimulating polymerization of SopB’s partner NTPase, SopA, because SopA also binds DNA nonspecifically and SopB DNA binding near the centromere site alleviates the inhibitory affects of nonspecific DNA binding on the polymerization of SopA. According to this model, polymerization of SopA then drives plasmid separation.
Figure 2.
Structure and domain organization of type Ia partition proteins. (a) Top, schematic diagram showing the domain arrangement of type Ia CBPs. Below is the structure of type Ia CBP SopB showing the dimer-domain and HTH domains, which are flexibly attached [16]. Labeled are the primary dimer (that formed on a single palindromic DNA site) and secondary dimers, which mediate bridging between DNA sites. The four SopB subunits are colored differently. (b) Comparison of the SopB dimer-domain with that of P1 ParB. Although the overall folds of the dimer-domains and their dimerization modes are similar, SopB lacks the wings that are present in P1 ParB, which allow the latter protein to bind DNA [15–16]. (c) SopB-DNA structure in which two nonspecific DNA interactions and one specific interaction was captured. These bridging interactions explain how SopB can spread in trans to surrounding DNA once bound to its centromere. (d) Top, schematic diagram of the type Ia NTPase domain organization as revealed by the P1 and P7 ParA structures [29]. Below the diagram is the P1 ParA structure with domains colored as in the schematic. (e) Structures of four different apo ParA structures with the magenta subunit shown in the same orientation to highlight the dramatic conformational flexibility exhibited by the ParA dimer.
Recent structural studies on SopB revealed important insight into these complex DNA binding functions of SopB, but also underscored the difficulties in obtaining structures of the flexible, full length (FL) Ia CBPs. Indeed, while FL SopB bound to an 18 bp centromere site was crystallized and the crystals shown to contain the FL protein, only residues 157–270, which contains the DNA binding HTH domain, were observed. SopB consists of three flexibly linked regions (comprised of residues 1–156, 157–270 and 271–323) and the apparent reason only the HTH domain (residues 157–270) was visible was because it was bound to DNA, which was central to crystal lattice formation. The structure of the SopB dimer-domain, SopB(275–323), was solved separately and revealed a (β2−α)2 dimer arrangement similar to that of P1 ParB [11, 15]. Notably, however, the SopB dimer-domain is missing the “wings” that mediate DNA binding of a second centromere repeat by P1 ParB [11, 15–16] (Fig. 2b). The SopB structures also revealed a “secondary” dimerization motif, which permits SopB to bridge between DNA. In one structure, this secondary dimerization allows one SopB dimer to bridge between three different DNA duplexes, making specific contacts to one DNA site and nonspecific contacts to the other two duplexes. Thus these structures revealed a mechanism for in trans DNA coating and nonspecific DNA binding by SopB (Fig. 2b).
Structures have only recently become available for type I NTPases. The first Ib structure determined was that of S. pyogenes pSM19035 δ, the partner NTPase for ω [25]. As expected, δ contains a deviant Walker motif. ATP binding to Walker box proteins typically leads to their dimerization through the formation of a nucleotide sandwich interaction [26]. However, biochemical data indicate that apo δ is a dimer. The generality of this finding for Ib NTPases remains unknown as δ is the only type Ib NTPase structure yet available. Ia NTPases are also regulated by adenine nucleotide binding, but in a more complex manner. For example, ATP binding by the archetypical type Ia P1 ParA protein activates it for partition, while ADP binding stimulates its operator binding function [27–28]. Recent structures of P1 ParA and its close homolog P7 ParA have started to shed light on Ia NTPase function [29••]. The structures show that the N-terminal 100 residue region, not found in Ib NTPases, harbors a unique structure consisting of a long helix, α1, followed by a winged-HTH motif (Fig. 2d). Unexpectedly, α1 functions in dimerization and four distinct structures of apo ParA showed that P1 (and P7) ParA are dimeric, even in their apo states, and that the apo dimer is flexible (Fig. 2d–e). ParA-ADP structures revealed that ADP binding locks in a specific dimer state and also mediates folding of a C-terminal basic region important for operator binding. The affect of ATP binding on P1 ParA structure remains to be determined.
Although the recent type I NTPase structures have provided insight into nucleotide binding and conformational switching, the question of how the signal of ATP binding is translated to partition is only beginning to become clear from cellular studies. These data indicate that ATP-mediated polymerization and the host nucleoid play central roles in type I partition. In a key study, the Gerdes lab demonstrated that E. coli pB171 ParA polymerized on nucleoid DNA and that the polymers propagated until encountering ParB-parC, which stimulated the ParA ATPase activity [30••]. ATP hydrolysis led to retraction of the polymer, which appeared to somehow attract or carry the ParB-plasmid complex along with it. Such a “pulling” mechanism or ones similar to it, are now supported by data from other type I systems. In particular, P1 ParA-ATP was also shown to polymerize on nucleoid DNA and this polymerization is affected by ParB-parS, which also stimulates ParA ATP hydrolysis [31••]. However, there are likely to be differences in the details of type I mechanisms. For example, as noted although the SopA protein binds nucleoid DNA nonspecifically, the binding appears to prevent its polymerization in this case [19–20].
Type II partition: pushing plasmids apart
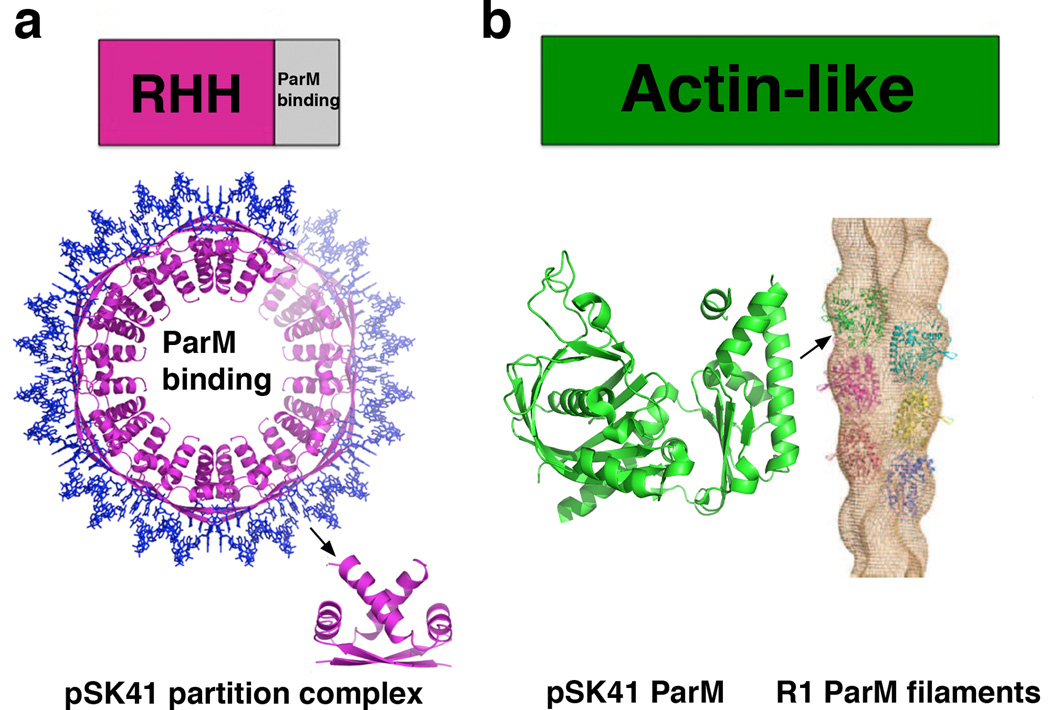
Type II partition, which is the best understood of the plasmid partition processes, is mediated by a so-called insertional polymerization mechanism, in which ParM NTPase filament propagation pushes plasmids apart [32]. Recent structural studies have led to a near atomic level understanding of this process. The CBP-centromere partition complex plays a key role in this process by capping and stabilizing each end of the ParM filament. Insight into the molecular basis of this capture and stabilization mechanism was revealed by the ParR-centromere partition complex structure from the pSK41 multidrug resistance plasmid, which is harbored in Staphylococcus aureus [33••]. pSK41 ParR contains a RHH motif similar to the type Ib CBPs. However, the pSK41 ParR RHH dimer-of-dimers interact cooperatively to generate a continuous protein super-structure that wraps the centromere DNA about itself to create a protein-nucleic acid superhelix (Fig. 3a). Another type II ParR structure, that of pB171 ParR, although solved in the absence of DNA, packed to form a superhelix similar to that of pSK41 ParR, suggesting that all type II partition complexes likely adopt superhelical partition complexes [34]. The pore dimensions of this complex are suitable for ParM filament interaction. Consistent with this idea, the flexible C-terminal regions of ParR that were shown to bind ParM, face the pore [33••]. Moreover, filament growth occurs by addition of ParM-ATP subunits at the ParR-ParM interface.
Figure 3.
Domain organization and structures of type II partition proteins. (a) Top, schematic diagram showing the domain arrangement of type II CBPs. Below is the structure of the pSK41 partition complex superstructure [33]. Shown to the side is one ParR dimer-of-dimer. (b) Domain organization of type II NTPases, which contain actin-like folds. Below is shown the recent structure of the pSK41 ParM protein and to the side is the EM reconstruction of the R1 ParM filament [35–36]. The pSK41 ParM subunit is shown in a similar orientation as the green R1 subunit in the filament to underscore the finding that the regions that make interactions important for filamentation are not conserved between the two structures. This explains why pSK41 ParM forms filaments that are distinct from R1 ParM.
Recent structural and biochemical analyses indicate that there are variations on the insertional polymerization model of type II partition. Specifically, although the structure of the pSK41 NTPase, ParM, revealed a fold similar to R1 ParM, it does not form double stranded filaments like R1 ParM, but rather single stranded polymers that are more stable than R1 filaments (Fig. 3b) [36]. Moreover, although Bacillus subtilis pBET131 AlfA NTPases forms left-handed double stranded filaments, the AlfA polymers are significantly more open, giving them a ribbon-like shape [37–38]. How these differences impact the type II insertional polymerization partition process is not yet known.
Type III partition: A tram model for plasmid transport
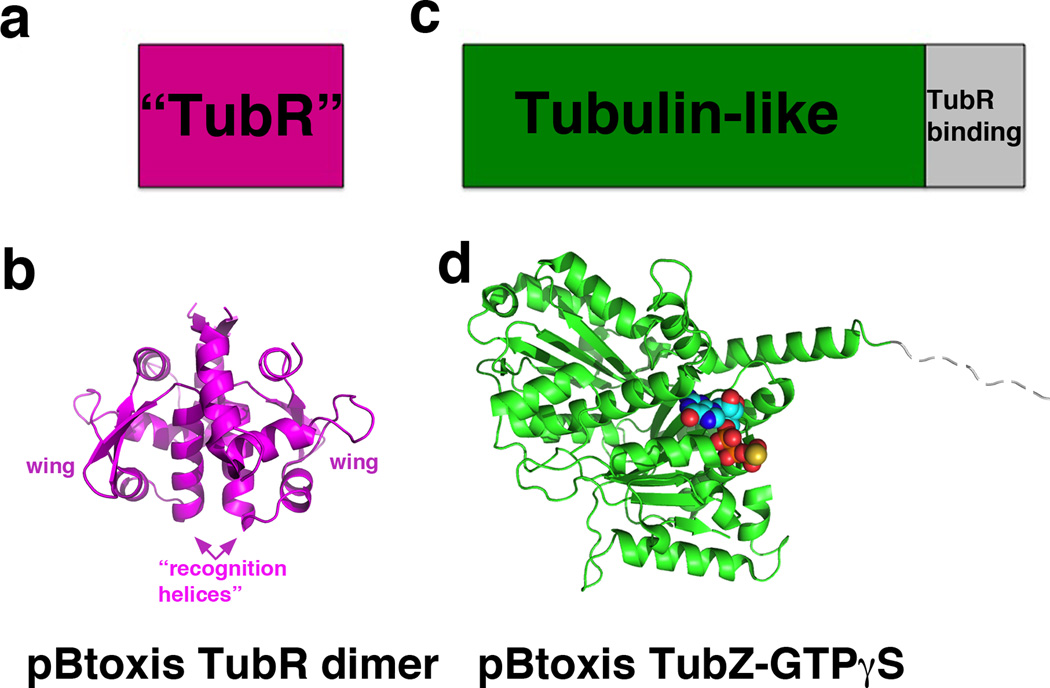
Type III partition systems are the most recently discovered of the plasmid partition systems. The best understood type III system is that encoded on the B. thuringiensis pBtoxis plasmid, which encodes an NTPase, termed TubZ and CBP called TubR [39–42]. Recent studies have revealed the structures of both TubZ and TubR and have suggested a partition process quite different from the pulling or pushing mechanisms employed by the type I and II systems [43••]. TubR forms a highly intertwined dimer with similarity to the ArsR family of winged-HTH transcriptional repressors [44–45]. Notably, all known ArsR proteins bind metals, form distinctive dimers and utilize their recognition helices for DNA recognition. However, TubR does not bind metal and its dimer is dramatically different from the ArsR dimers. Moreover, the TubR recognition helix is utilized in dimerization and hence mostly buried in the dimer interface (Fig. 4a–b). Biochemical studies demonstrated that TubR uses residues its wing and the helix preceding its recognition helix for DNA binding and modeling suggests that the wings could interact with consecutive minor grooves of the DNA, while the N termini of the recognition helices would insert into a single major groove [43••]. Thus, TubR employs a newly described mode of DNA binding.
Figure 4.
Domain organization and recent structures of type III partition proteins. (a) Schematic diagram showing the domain structure of TubR, which consists of a newly described DNA binding fold [43]. (b) The TubR structure is shown below and its DNA binding regions labeled. (c) Domain organization of the type III NTPase TubZ, which harbors a tubulin like fold followed by a flexible TubR binding region. (d) The structure of TubZ-GTPγS is shown below, with the GTPγS shown as cpk.
In addition to TubR, structures of TubZ, in its apo and GTP-γ-S form, were solved (Fig. 4c–d). Prior work had demonstrated that TubZ polymerizes in vivo and in vitro in a GTP dependent manner and undergoes treadmilling [41]. The structures confirmed that TubZ contains a tubulin/FtsZ fold. It also contains a flexible C-terminal tail, not resolved in the structure. Biochemical studies showed that the TubZ C-tail is used to bind TubR. Notably, this TubZ C-tail-TubR interaction is analogous to how FtsZ and tubulin use their C-domains to bind target proteins [43, 46–48]. Subsequent EM structures of TubZ, which showed that it forms double stranded filaments, revealed that its C-terminal tails are indeed solvent exposed [49••]. Thus, the combined data suggest a tram-like mechanism for type III partition. In this model, the TubR-pBtoxis complex becomes attached to TubZ filaments through the interaction of TubR with the surface-exposed C-terminal regions of TubZ. GTP hydrolysis within the TubZ polymer generates treadmilling, elongation at the plus end and retraction at the minus end, which results in translocation of the filament and attached plasmid towards one cell pole. How does the TubR-plasmid complex disengage from the complex? Previous studies revealed that TubZ filaments bend upon reaching the cell pole, which could stimulate TubR-pBtoxis detachment. After unloading the initial TubR-pBtoxis cargo, the filament would continue moving towards the opposite cell pole and could pick up another TubR-pBtoxis complex, which would be transported to the opposite cell pole.
Future directions
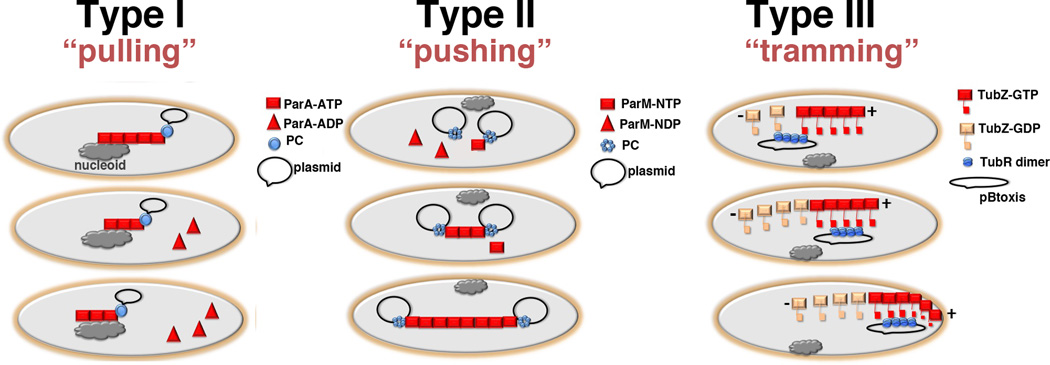
In summary, recent studies have now revealed the structures of the protein components that comprise the partition machinery. These structures combined with biochemical and cellular data have unveiled the general molecular mechanisms utilized by plasmid partition systems (Fig. 5). Remarkably, all these systems use NTPases with distinct types of cytoskeletal folds; actin-like, tubulin-like or Walker box containing. But while all systems use polymer-based separation processes, the specific mechanisms differ. Type I systems appear to “pull”, type II to “push” and type III to “tram/transport” plasmids. Despite the great progress made in establishing these mechanisms and the molecular equipment that drives these processes, an important piece of the puzzle remains unresolved, which is how the CBPs interact with NTPases. These interactions are not only critical for attachment but also, in the case of the type I and II systems, affecting NTPase polymer dynamics. Obtaining a complete molecular description of plasmid partition therefore, will require more biochemical and structural information on CBP-centromere-NTPase-centromere complexes.
Figure 5.
Schematic models for type I, II and III plasmid partition (a) Type I partition utilizes the host cell nucleoid as a “track” for NTPase-ATP binding and polymerization (square). When the NTPase-ATP polymer encounters a ParB-centromere partition complex (shown as a circle), i.e. the ParB attached plasmid, the NTPase activity is activated resulting in dissociation of capping ParA-ADP subunits (triangles) and polymer retraction. The ParB-plasmid is either pulled along in the retreating ParA polymer or is attracted and diffuses toward the moving polymer. The ultimate outcome is the dynamic equi-distribution of ParB-plasmids at opposite ends of the nucleoid. (b) Type II partition uses a pushing or insertional polymerization mode of segregation. In this model, the dynamically unstable ParM filaments are stabilized and propagate only when each end is captured by a ParR-centromere partition complex. The polymer continues to grow upon addition of ParM-ATP or ParM-GTP subunits to the ParR-ParM interface. The outcome is redistribution of replicated plasmids to opposite poles. (c) Type III partition employs a tram mechanism of partition. TubR binds the centromere serving as a high local concentration of binding sites for the C-terminal flexible domains emanating from treadmilling TubZ filaments. Once captured, the TubR-plasmid is transported to the cell pole by the treadmilling TubZ filaments. Upon reaching the membrane the TubZ filament bends, likely dumping its TubR-plasmid cargo, and reverses direction. Now traveling in the opposite direction, the TubZ filament binds another TubR-plasmid cargo and carries it to the opposite pole.
Highlights.
-
►
DNA segregation of low copy number plasmids utilize partition systems.
-
►
Partition systems utilize a centromere, centromere-binding protein and NTPase.
-
►
Three main partition systems, each using a different polymer forming NTPase, drive DNA separation.
Acknowledgements
The author would like to acknowledge support from the Burroughs Wellcome (Burroughs Wellcome Career Development Award 992863), the U.T. M.D. Anderson Cancer Center Trust Fellowship and the National Institutes of Health. I apologize to those whose work was not discussed.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest The author declares no competing conflicts of interest, financial or otherwise.
References
- 1.Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Genet. 2008;9:33–46. doi: 10.1038/nrm2310. [DOI] [PubMed] [Google Scholar]
- 2.Hayes F, Barillà D. The bacterial segrosome: a dynamic nucleoprotein machine for DNA trafficking and segregation. Nat Rev Microbiol. 2006;4:133–143. doi: 10.1038/nrmicro1342. [DOI] [PubMed] [Google Scholar]
- 3.Schumacher MA. Structural biology of plasmid partition proteins. Biochem J. 2008;412:1–18. doi: 10.1042/BJ20080359. [DOI] [PubMed] [Google Scholar]
- 4. Gerdes K, Howard M, Szardenings F. Pushing and pulling in prokaryotic DNA segregation. Cell. 2010;141:927–942. doi: 10.1016/j.cell.2010.05.033. • Excellent review on bacterial DNA segregation, covering both chromosomal and plasmid partition systems. The focus is on the spatiotemporal dynamics of par loci proteins and potential explanations for the observed dynamics using simplified models are discussed.
- 5.Gerdes K, Møller-Jensen J, Bugge Jensen R. Plasmid and chromosome partitioning: surprises from phylogeny. Mol Microbiol. 2000;37:455–466. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- 6.van den Ent F, Møller-Jensen J, Amos LA, Gerdes K, Löwe J. F-actin-like filaments formed by plasmid segregation protein ParM. EMBO J. 2002;21:6935–6943. doi: 10.1093/emboj/cdf672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Delbruck H, Ziegelin G, Lanka E, Heinemann U. An Src Homology 3-like Domain is responsible for Dimerization of the Repressor Protein KorB Encoded by the Promiscuous IncP Plasmid RP4. J Biol Chem. 2002;277:4191–4198. doi: 10.1074/jbc.M110103200. [DOI] [PubMed] [Google Scholar]
- 8.Khare D, Ziegelin G, Lanka E, Heinemann U. Sequence-specific DNA binding determined by contacts outside the helix-turn-helix motif of the ParB homolog KorB. Nat Struct & Mol Biol. 2004;11:656–663. doi: 10.1038/nsmb773. [DOI] [PubMed] [Google Scholar]
- 9.Golovanov AP, Barillà D, Golovanova M, Hayes F, Lian LY. ParG, a protein required for active partition of bacterial plasmids, has a dimeric ribbon-helix-helix structure. Mol Microbiol. 2003;50:1141–1153. doi: 10.1046/j.1365-2958.2003.03750.x. [DOI] [PubMed] [Google Scholar]
- 10.Murayama K, Orth P, de la Hoz AB, Alonso JC, Saenger W. Crystal structure of omega transcriptional repressor encoded by Streptococcus pyogenes plasmid pSM19035 at 1.5 Å resolution. J Mol Biol. 2001;314:789–796. doi: 10.1006/jmbi.2001.5157. [DOI] [PubMed] [Google Scholar]
- 11.Schumacher MA, Funnell BE. Structures of ParB bound to DNA reveal mechanism of partition complex formation. Nature. 2005;438:516–519. doi: 10.1038/nature04149. [DOI] [PubMed] [Google Scholar]
- 12. Huang L, Yin P, Zhu X, Zhang Y, Ye K. Crystal structure and centromere binding of the plasmid segregation protein ParB from pCXC100. Nuc Acids Res. 2011;39:2954–2968. doi: 10.1093/nar/gkq915. •The pCXC100 ParB structure, which is the third structure reported for a type Ib CBP, reveals a RHH fold similar to other Ib CBPs structures. In addition, the pCXC100 centromere is defined via multiple biochemical methods.
- 13.Weihofen WA, Cicek A, Pratto F, Alonso JC, Saenger W. Structures of ω repressors bound to direct and inverted repeats explain modulation of transcription. Nuc Acids Res. 2006;34:1450–1458. doi: 10.1093/nar/gkl015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Barillà D, Carmelo E, Hayes F. The tail of the ParG DNA segregation protein remodels ParF polymers and enhances ATP hydrolysis via and arginine finger-like motif. Proc Natl Acad Sci USA. 2007;104:1811–1816. doi: 10.1073/pnas.0607216104. •• How type I CBPs stimulate the ATPase activities of their partner NTPases has been a central question in partition. This paper describes the important finding that an arginine finger motif in the N-terminal region of ParG is critical for activating the ATPase activity of ParF. Further, comparison of the N-terminal arginine finger motif of ParG with other type I NTPases suggests that this may be a conserved mechanism within type I systems.
- 15.Schumacher MA, Mansoor A, Funnell BE. Structure of a four-way bridged ParB-DNA complex provides insight into P1 segrosome assembly. J Biol Chem. 2007;282:10456–10464. doi: 10.1074/jbc.M610603200. [DOI] [PubMed] [Google Scholar]
- 16. Schumacher MA, Piro KM, Xu W. Insight into F plasmid DNA segregation revealed by structures of SopB and SopB-DNA complexes. Nuc Acids Res. 2010;38:4514–4526. doi: 10.1093/nar/gkq161. •• The paper describes the crystallization and structure determination of the full length type Ia CBP SopB. Unfortunately, due to the flexibility of the protein, only a portion of the structure was ordered. However, the authors utilized the information and constructed domains to obtain high resolution structures of the DNA binding HTH domain in complex with centromere repeats and the C-terminal dimer-domain. Strikingly, the dimer-domain has the same overall fold as the P1 ParB dimer-domain but is missing the DNA binding wings observed in the P1 ParB dimer-domain, explaining why this domain in SopB does not bind DNA. Secondary dimer contacts observed in multiple SopB-DNA structures suggest a mechanism for the in trans DNA spreading capability previously reported for SopB.
- 17.Ogura T, Hiraga S. Partition mechanism of F plasmid: two plasmid gene-encoded products and a cis-acting region are involved in partition. Cell. 1983;32:351–360. doi: 10.1016/0092-8674(83)90454-3. [DOI] [PubMed] [Google Scholar]
- 18.Hatano T, Yamaichi Y, Niki H. Oscillating focus of SopA associated with filamentous structure guides partitioning of F plasmid. Mol Microbiol. 2007;64:1198–1213. doi: 10.1111/j.1365-2958.2007.05728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bouet JY, Ah-Seng Y, Benmeradi N, Lane D. Polymerization of SopA partition ATPase: regulation of DNA binding and SopB. Mol Microbiol. 2007;63:468–481. doi: 10.1111/j.1365-2958.2006.05537.x. [DOI] [PubMed] [Google Scholar]
- 20.Castaing JP, Bouet JY, Lane D. F plasmid partition depends on interaction of SopA with non-specific DNA. Mol Microbiol. 2008;70:1000–1011. doi: 10.1111/j.1365-2958.2008.06465.x. [DOI] [PubMed] [Google Scholar]
- 21.Bouet JY, Lane D. Molecular basis of the supercoil deficit induced by the mini-F plasmid partition complex. J Biol Chem. 2009;284:165–173. doi: 10.1074/jbc.M802752200. [DOI] [PubMed] [Google Scholar]
- 22.Ravin NV, Rech J, Lane D. Mapping of functional domains in F plasmid partition proteins reveals a bipartite SopB-recognition domain in SopA. J Mol Biol. 2003;329:875–889. doi: 10.1016/s0022-2836(03)00525-4. [DOI] [PubMed] [Google Scholar]
- 23.Biek DP, Shi J. A single 43-bp sopC repeat of plasmid mini-F is sufficient to allow assembly of a functional nucleoprotein partition complex. Proc Natl Acad Sci USA. 1994;91:8027–8031. doi: 10.1073/pnas.91.17.8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biek DP, Strings J. Partition functions of mini-F plasmid DNA topology in Escherichia coli. J Mol Biol. 1995;246:388–400. doi: 10.1006/jmbi.1994.0094. [DOI] [PubMed] [Google Scholar]
- 25.Pratto F, Cicek A, Weihofen WA, Lurz R, Saenger W, Alonso JC. Streptococcus pyogenes pSM19035 requires dynamic assembly of ATP-bound ParA and ParB on parS DNA during plasmid segregation. Nucl Acids Res. 2008;36:3676–3689. doi: 10.1093/nar/gkn170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lutkenhaus J, Sundaramoorthy M. MinD and role of the deviant Walker A motif, dimerization and membrane in association. Mol Microbiol. 2003;48:295–303. doi: 10.1046/j.1365-2958.2003.03427.x. [DOI] [PubMed] [Google Scholar]
- 27.Davey MJ, Funnell BE. Modulation of the P1 plasmid partition protein ParA by ATP, ADP, and P1 ParB. J Biol Chem. 1997;272:15286–15292. doi: 10.1074/jbc.272.24.15286. [DOI] [PubMed] [Google Scholar]
- 28.Davis MA, Martin KA, Austin SJ. Biochemical activities of the ParA partition protein of the P1 plasmid. Mol Microbiol. 1992;6:1141–1147. doi: 10.1111/j.1365-2958.1992.tb01552.x. [DOI] [PubMed] [Google Scholar]
- 29. Dunham TD, Xu W, Funnell BE, Schumacher MA. Structural basis for ADP-mediated transcriptional regulation by P1 and P7 ParA. EMBO J. 2009;28:1792–1802. doi: 10.1038/emboj.2009.120. ••The first structure of type Ia NTPases are revealed, those of P1 ParA and its homolog, P7 ParA. The structures shows that the ParA proteins have an extended N-terminal helix, α1, which mediates dimer contacts, followed by a newly described winged-HTH, which is utilized in operator DNA binding. Four different apo structures show that the ParA dimer is flexible. Two different ADP-bound crystal structures show that ADP binding selects a specific dimer state that is important in specific DNA binding. ADP binding also leads to large scale folding of a basic region that, in addition to the winged-HTH, is also essential for operator binding. Mutagenesis and biochemical data are used to support a suggested model for ParA-ADP binding to its operator.
- 30. Ringgaard S, van Zon J, Howard M, Gerdes K. Movement and equipositioning of plasmids by ParA filament disassembly. Proc Natl Acad Sci USA. 2009;106:19369–19374. doi: 10.1073/pnas.0908347106. ••This important study reveals the finding that pB171 ParA, a type Ib NTPase, forms polymers on nucleoid (nonspecific) DNA in an ATP-dependent manner. Further, the studies show that upon encountering a CBP-bound plasmid, which stimulates ParA ATP hydrolysis, the ParA polymer retracts “pulling” or attracting the CBP-bound plasmid with, it, Thus, these data cement the idea that nucleoid DNA is fundamental for type I partition and the authors perform mathematical modeling that support an equidistribution of plasmids by perpetual cycles of ParA assembly and disassembly.
- 31. Vecchuarelli AG, Han YW, Tan X, Mizuuchi M, Ghirlando R, Biertumpfel G, Funnell BE, Mizuucgi K. ATP control of dynamic P1 ParA-DNA interactions: a key role for the nucleoid in plasmid partition. Mol Microbiol. 2010;78:78–91. doi: 10.1111/j.1365-2958.2010.07314.x. ••This paper describes findings similar to the Ringgaard et al. paper regarding a nucleoid requirement for type I partition. Here the authors suggest that a type Ia ParA protein, P1 ParA, undergoes a complex, multi-step structural transition upon ATP binding that ultimately licenses it to bind nucleoid DNA. The authors propose that this kinetically critical time delay switch in conformation combined with ParB-plasmid stimulation of ParA ATPase activity causes an uneven distribution of nucleoid bound ParA and that this unequal distribution is what provides the motive force for ParB-plasmid movement.
- 32.Møller-Jensen J, Borch J, Dam M, Jensen RB, Roepstorff P, Gerdes K. Bacterial mitosis: ParM of plasmid R1 moves plasmid DNA by an actin-like insertional polymerization mechanism. Mol Cell. 2003;12:1477–1487. doi: 10.1016/s1097-2765(03)00451-9. [DOI] [PubMed] [Google Scholar]
- 33. Schumacher MA, Glover TC, Brzoska AJ, Jensen SO, Dunham TD, Skurray RA, Firth N. Segrosome structure revealed by a complex of ParR with centromere DNA. Nature. 2007;450:1268–1271. doi: 10.1038/nature06392. ••Describes the first structure of a FL partition complex, that of ParR-centromere complex from the multidrug resistance plasmid pSK41. The structure reveals a novel higher order protein nucleic acid superhelix with dimensions suitable for capture of ParM filament ends. This study was also the first to show that the C-terminal domains of ParR, which face the pore, bind ParM.
- 34.Møller-Jensen J, Ringgaard S, Mercogliano CP, Gerdes K, Löwe J. Structural analysis of the ParR/parC plasmid partition complex. EMBO J. 2007;26:4413–4422. doi: 10.1038/sj.emboj.7601864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salje J, Löwe J. Bacterial actin: architecture of the ParMRC plasmid DNA partitioning complex. EMBO J. 2008;27:2230–2238. doi: 10.1038/emboj.2008.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popp D, Xu W, Narita A, Brzoska AJ, Skurray RA, Firth N, Ghosdastider U, Maeda Y, Robinson RC, Schumacher MA. Structure and filament dynamics of the pSK41 actin-like ParM protein: implications for plasmid DNA segregation. J Biol Chem. 2010;285:10130–10140. doi: 10.1074/jbc.M109.071613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Polka JK, Kollman JM, Agard DA, Mullins RD. The structure and assembly dynamics of plasmid actin AlfA imply a novel mechanism of DNA segregation. J Bacteriol. 2009;191:6219–6230. doi: 10.1128/JB.00676-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Popp D, Narita A, Ghoshdastider U, Maeda K, Maeda Y, Oda T, Fujisawa T, Onishi H, Ito K, Robinson RC. Polymeric structures and dynamic properties of the bacterial actin AlfA. J Mol Biol. 2010;397:1031–1041. doi: 10.1016/j.jmb.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 39.Larsen RA, Cusumano C, Fujioka A, Lim-Fong G, Patterson P, Pogliano J. Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis. Genes & Dev. 2007;21:1340–1352. doi: 10.1101/gad.1546107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tang M, Bideshi DK, Park H-W, Federici BA. Iteron-binding ORF157 and FtsZ-like ORF156 proteins encoded by pBtoxis play a role in its replication in Bacillus thuringiensis subsp. israelensis. J Bacteriol. 2007;189:8053–8058. doi: 10.1128/JB.00908-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y, Erickson HP. In vitro assembly studies of FtsZ/tubulin-like proteins (TubZ) from Bacillus plasmids- evidence for a capping mechanism. J Biol Chem. 2008;283:8102–8109. doi: 10.1074/jbc.M709163200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tinsley E, Khan SA. A novel FtsZ-like protein is involved in replication of the anthrax toxin-encoding pXO1 plasmid in Bacillus anthracis. J Bacteriol. 2006;188:2829–2835. doi: 10.1128/JB.188.8.2829-2835.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ni L, Xu W, Kumaraswami M, Schumacher MA. Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition. Proc Natl Acad Sci USA. 2010;107:11763–11768. doi: 10.1073/pnas.1003817107. ••The first TubR and TubZ structures are reported as are data demonstrating an interaction between the two proteins. The TubR structure contains an HTH, but utilizes the recognition helices for dimerization and hence they are not accessible for normal HTH-DNA interactions. Biochemical data support a novel mode of DNA binding by TubR. By combining structural and biochemical data, including the knowledge that TubZ undergoes treadmilling, the authors propose a tram model for type III partition.
- 44.Pennella MA, Giedroc DP. Structural determinants of metal selectivity in prokaryotic metal-responsive transcriptional regulators. Biometals. 2005;18:413–428. doi: 10.1007/s10534-005-3716-8. [DOI] [PubMed] [Google Scholar]
- 45.Buesenlehner LS, Pennella MA, Giedroc DP. The SmtB/ArsR family of metalloregulaotry transcriptional repressors: Structure insights into prokaryotic metal resistance. FEMS Microbiol Rev. 2003;27:131–143. doi: 10.1016/S0168-6445(03)00054-8. [DOI] [PubMed] [Google Scholar]
- 46.Errington J, Daniel RA, Scheffers DJ. Cytokinesis in bacteria. Microbiol Mol Biol. 2003;67:52–65. doi: 10.1128/MMBR.67.1.52-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mosyak L, Zhang Y, Glasfeld E, Haney S, Stahl M, Seehra J, Somer WS. The bacterial cell-division ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J. 2000;19:3179–3191. doi: 10.1093/emboj/19.13.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Downing KH. Structural basis for the interaction of tubulin with proteins and drugs that affect microtubule dynamics. Annu Rev Cell Dev Biol. 2000;16:89–111. doi: 10.1146/annurev.cellbio.16.1.89. [DOI] [PubMed] [Google Scholar]
- 49. Aylett CH, Wang Q, Michie KA, Amos LA, Löwe J. Filament structure of bacterial tubulin homologue TubZ. Proc Natl Acad Sci USA. 2010;107:19766–19771. doi: 10.1073/pnas.1010176107. ••Structure of the TubZ polymer is presented and shown to form a double stranded filament. The C-terminal regions of TubZ are located on the surface of the filaments, optimal for interaction with the TubR CBP.
- 50.Delano WL. The PyMOL Molecular Graphics System. San Carlos, California: DeLano Scientific; 2002. [Google Scholar]