Abstract
Bromodomain and extra terminal domain (BET) proteins are important epigenetic regulators facilitating the transcription of genes in chromatin areas linked to acetylated histones. JQ1, a BET protein inhibitor, has antiproliferative activity against many cancers, mainly through inhibition of c-MYC and upregulation of p21. In this research, we investigated the use of JQ1 for human osteosarcoma (OS) treatment. JQ1 significantly inhibited the proliferation and survival of OS cells inducing G1 cell cycle arrest, premature senescence, but little effect on apoptosis. Interestingly, c-MYC protein levels in JQ1-treated cells remained unchanged, whereas the upregulation of p21 protein was still observable. Although effective in vitro, JQ1 alone failed to reduce the size of the MNNG/HOS xenografts in immunocompromised mice. To overcome the resistance of OS cells to JQ1 treatment, we combined JQ1 with rapamycin, an mammalian target of rapamycin (mTOR) inhibitor. JQ1 and rapamycin synergistically inhibited the growth and survival of OS cells in vitro and in vivo. We also identified that RUNX2 is a direct target of bromodomain-containing protein 4 (BRD4) inhibition by JQ1 in OS cells. Chromatin immunoprecipitation (ChIP) showed that enrichment of BRD4 protein around RUNX2 transcription start sites diminished with JQ1 treatment in MNNG/HOS cells. Overexpression of RUNX2 protected JQ1-sensitive OS cells from the effect of JQ1, and siRNA-mediated inhibition of RUNX2 sensitized the same cells to JQ1. In conclusion, our findings suggest that JQ1, in combination with rapamycin, is an effective chemotherapeutic option for OS treatment. We also show that inhibition of RUNX2 expression by JQ1 partly explains the antiproliferative activity of JQ1 in OS cells.
Keywords: osteosarcoma, JQ1, rapamycin, synergism, RUNX2
Histone acetylation and deacetylation are important epigenetic processes for regulating gene expression through chromatin remodeling. Addition of acetyl groups to lysine residues of histone proteins by histone acetyltransferases weakens polar interaction between histone proteins and DNA, loosens chromatin structure, and makes genes more accessible to transcription.1 Removal of acetyl groups by histone deacetylases (HDACs) reverses the effect.1 “Reader” proteins such as bromodomain and extra terminal domain (BET) family of proteins recognize these acetylation “marks”, recruit important transcription activators at specific promoter regions, and help transcriptional initiation and elongation of genes.2 Deregulation of these proteins is often found in several diseases including cancer.1 Therefore, they are attractive potential therapeutic targets for new anticancer drug development.
While the majority of anticancer drug studies targets HDACs, a growing number of studies focus on targeting the reader proteins. To date, I-BET762 and JQ1 are the two most potent small molecule inhibitors which target the BET family of proteins.1 JQ1 is an inhibitor of bromodomain-containing protein 4 (BRD4), which recruits positive transcription elongation factor b (P-TEFb) complex to the chromatin and activates transcription by phosphorylation of RNA polymerase II.3 JQ1 competitively binds to the acetyl-lysine recognition pocket of BRD4, displaces BRD4 from chromatin, and represses transcription of genes in the affected chromatin region.4 JQ1 has potent chemotherapeutic activity against many hematological malignancies, including acute myeloid leukemia,5 multiple myeloma6 and acute lymphoblastic leukemia (ALL).7 In these cancers, JQ1 shows its antiproliferative activity mainly through the inhibition of c-MYC expression, a well-known oncogene whose constitutive overexpression is responsible for several cancer types.8 JQ1 also has activity against several solid tumors, including NUT midline carcinoma,4 lung adenocarcinoma (LAC),2 neuroblastoma9 and medulloblastoma.10 Interestingly, increasing number of studies show that inhibition of c-MYC is not always the main mediator of JQ1 activity. JQ1 can achieve its activity through the inhibition of other genes such as interleukin 7 receptor (IL7R) in ALL7 and FOS-like antigen in LAC.2 Considering drastic difference in histone acetylation patterns of different cancer types, it is reasonable to assume that antiproliferative activity of JQ1 is achieved by the inhibition of actively transcribed genes whose expression is dependent on histone acetylation and is crucial for the proliferation and survival of the particular cell type.2 Thus, JQ1 can show activity across a wide range of cancers and can be a potential therapeutic agent for cancers with limited chemotherapeutic options such as human osteosarcomas (OS).
In this research, we explore the potential use of JQ1 for OS treatment. Majority of OS cells show various levels of copy number (CN) gains and overexpression of a region in chromosome 8 q-arm (8q23.3-qter), in which c-MYC resides.11 Therefore, we initially hypothesized that JQ1 might mediate its activity through inhibition of c-MYC in OS cells. Rather, we discovered that the Runt-related transcription factor (RUNX2) and its downstream targets are suppressed by JQ1 treatment which partly explains JQ1 activity in OS cells. We also found that rapamycin synergistically potentiates JQ1. Rapamycin is a well-known inhibitor of mTOR which binds FK506 binding protein 12 (FKBP12) and blocks the formation of mTOR complexes.12 Taken together, we find that JQ1 has its antiprolfierative activity in OS cells, partly through the inhibition of RUNX2 and its downstream targets. We also show that JQ1 in combination with rapamycin is a good chemotherapeutic option for OS treatment.
Materials and Methods
OS cell culture
Seven human OS cell lines were used in the study (U2OS, G292, MG-63, HT-161, MNNG/HOS, SAOS-2 and SJSA). Each cell line except HT-161 was obtained from American Tissue Type Culture Collection (ATCC, Rockville, MD). HT-161 was received from Dr. Emil Bogenmann.13 All cell lines except HT-161 were authenticated by short tandem repeat profiling, and confirmed that all DNA profiles for the cell lines matched the DNA profiles provided by DSMZ. OS cell lines were maintained in DMEM medium (Mediatech, Herndon, VA) supplemented with 10% fetal bovine serum (Thermo Scientific HyClone, Logen, UT) in a humidified incubator at 37°C supplied with 5% CO2. Only the cells in exponential growth phase were used in the study.
Chemical compounds
JQ1 was generously provided by Dr. James E. Bradner.4 Rapamycin was purchased from Biotang (Waltham, MA). Both JQ1 and rapamycin were solubilized with DMSO to a stock concentration of 10 mM. All chemical compounds were freshly dissolved on the day of experiment.
Measurement of cell growth and survival
For dose-response tests, cytotoxicity LDH detection kit was used to measure cell survival and determine IC50 and according to the manufacturer’s protocol (Clontech, Mountain View, CA). For pulse-exposure experiments, 3-(4,5-dimethylthiazol-2-yl)–2,5-diphenyltetrazolium bromide (MTT) assay was used as previously described.14 Clonogenic ability of OS cells under JQ1 exposure was monitored by soft-agar colony formation assay as previously described.15
Measurement of cell cycle, apoptosis and cellular senescence
Analyses of cell cycle and apoptosis were performed as previously described.14 Senescence staining was done using a cellular senescence assay kit according to the manufacturer’s protocol (Cell Biolabs, San Diego, CA).
Drug interaction analysis
Synergism between JQ1 and rapamycin was determined quantitatively by isobologram and combination index (CI) analysis adapted from the median-principle methods of Chou and Talalay.16,17 CompuSyn 1.0 software was available free and used for CI analysis (ComboSyn, Paramus, NJ).
Growth of MNNG/HOS xenografts in anthymic nude mice with in vivo treatment and tumor histochemistry
All animal experiments strictly followed the guidelines of Cedars-Sinai Medical Center and the National Institute of Health (NIH). Female nu/nu athymic nude mice (5–6 weeks old; average weight 21 g; specific pathogen-free) from Harlan laboratories (Indianapolis, IN) were maintained in a pathogen-free condition with sterilized chow and water. 5 × 106 cells of MNNG/HOS cells were mixed with 200 µL of Matrigel solution (BD Biosciences, San Jose, CA) per injection, and the mixture was injected subcutaneously on the upper flanks of nude mice. After 72 hr, tumor size was measured, and any outliers were ruled out by one-way analysis of variance (ANOVA) test. Five mice were randomly assigned to each experimental group: (i) PBS (diluent-specific control), (ii) JQ1 (50 mg/kg body weight), (iii) rapamycin (1.5 mg/kg body weight) and (iv) JQ1 with rapamycin. Intraperitoneal (i.p.) injections of either PBS or JQ1 were made daily, and rapamycin was administered every other day. Body weights were monitored every day. Tumor size was measured every 3 days, and the tumor volumes were measured using the following formula: A (length) × B (width) × C (height) × 0.5236.18 The experiment was stopped at Day 24, and all mice were sacrificed. At autopsy, tumors were excised, weighed and fixed in 10% PBS-buffered formalin and maintained in 70% ethanol. Fixed tumors were embedded in paraplast (Oxford Labware, St. Louis, MO), sliced and stained with hematoxylin and eosin (H&E) staining for histopathological examination as described previously.14
Quantitative reverse-transcription real-time PCR (qRT-PCR)
RNA purification, cDNA construction and qRT-PCR were performed as previously described.15 Primer sets for the detection of genes are listed in Supporting Information Table S1.
Western blotting
Western blotting was performed as previously described.14 Antibodies used for Western blotting are listed in Supporting Information Table S2. ImageJ was used to process blot images and analyze band intensity.19
Microarray analysis
MNNG/HOS cells were treated with DMSO (control), 7.5 µM JQ1, 12.5 nM rapamycin or both for 6 hr and total RNA was harvested using RNeasy mini kit (Qiagen, Valencia, CA). Prior to hybridization, quality check of the samples was done using Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA). Samples were hybridized onto HumanHT-12 v4 Expression BeadChip Kit from Illumina (Santa Clara, CA), washed and scanned according to the manufacturer’s protocol. All scanned raw data were obtained from the GenomeStudio software with the subtraction of the background. Normalization of raw data was then performed across all samples based on the Cross Correlation Method,20 and normalized data were further log2-transformed. Differentially expressed genes (DEGs) were identified based on the fold change cutoff of 2. Gene set enrichment analysis (GSEA) was performed based on the normalized data using GSEA v2.0 tool (http://www.broad.mit.edu/gsea).21 In order to demonstrate that the enrichment of a gene set is mainly due to the DEGs, the GSEA was run twice using the expression data of either the whole transcriptome or only the DEGs.
Chromatin Immunoprecipitation
Binding of BRD4 protein on RUNX2 promoter regions was determined by ChIP using MAGnify chromatin immunoprecipitation system according to the manufacturer’s protocol (Life Technologies, Grand Island, NY). BRD4 antibody for ChIP was purchased from Bethyl Laboratories (Montgomery, TX). ChIP primer sets for RUNX2 promoter regions are listed in Supporting Information Table S1.
Modulation of RUNX2 expression level in OS cell lines
For overexpression of RUNX2, pEF-BOS-RUNX2 expression vector was obtained from Dr. Yoshiaki Ito.22 G292 and MNNG/HOS cells were transfected with either RUNX2 expression vector or empty vector (EV) control using jet-PRIME DNA transfection reagent according to the manufacturer’s protocol (BIOPARC, Illkirch, France). Subsequent JQ1 treatment was performed after 48 hr of incubation for 12 hr. Expression of exogenous RUNX2 was validated by qRT-PCR.
For inhibition of RUNX2, Trilencer-27 human siRNA set against RUNX2 were purchased from OriGene (Rockville, MD). G292 and MNNG/HOS cells were transfected with either siRUNX2 oligos or scrambled control oligos using jet-PRIME DNA transfection reagent according to the manufacturer’s protocol (BIOPARC). Subsequent JQ1 treatment was performed after 48 hr of incubation for 12 hr. Inhibition of RUNX2 expression was validated by qRT-PCR.
Statistical Analysis
All in vitro and in vivo experiments were repeated at least three times to ensure reproducibility. Two-tailed Student’s t-test was used to compare differences between two groups. One-way ANOVA test was used to compare differences among three or more groups. P values ≥0.05 were considered statistically significant.
Results
Cellular effects of JQ1 on human OS cells
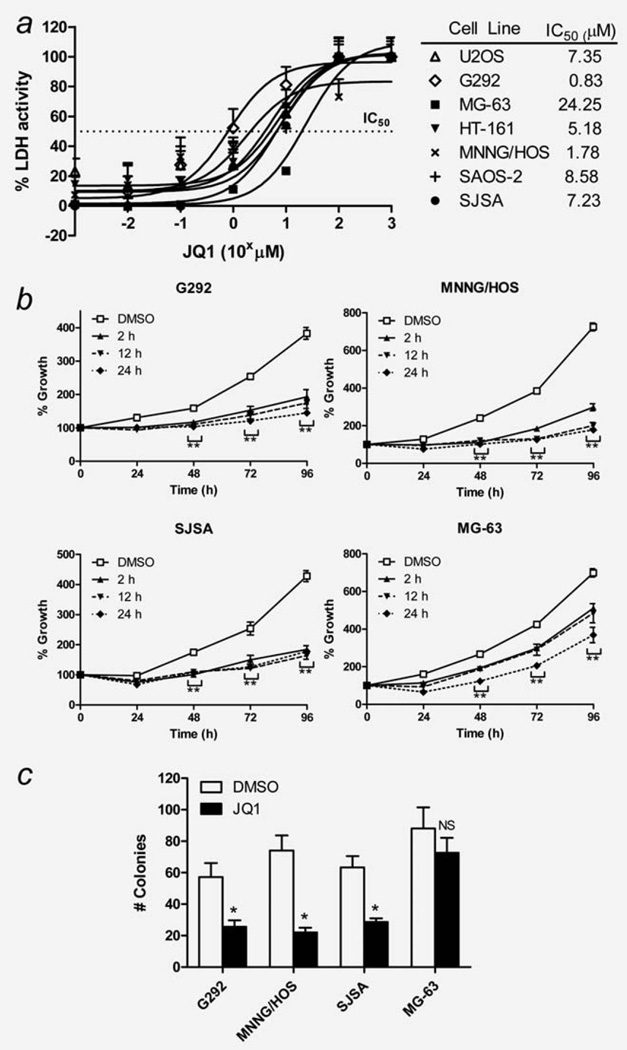
The effect of JQ1 on the proliferation and survival of seven human OS cell lines (U2OS, G292, MG-63, HT-161, MNNG/HOS, SAOS-2 and SJSA) was examined in vitro. JQ1 treatment in a range of concentrations for 72 hr displayed dose-dependent inhibition of proliferation of OS cells, with median IC50 value of 7.35 µM (range 0.83–24.25 µM, Fig. 1a). MG-63 showed the highest resistance to JQ1 (IC50 = 24.25 µM). Based on the IC50 results, we chose four OS cell lines for further in vitro studies: G292 and MNNG/HOS as the two most JQ1-sensitive cell lines, SJSA cells with moderate sensitivity and MG-63 cells as the most JQ1-resistant.
Figure 1.
Effect of JQ1 on the proliferation and survival of human osteosarcoma (OS) cells in vitro. (a) Dose-dependent antiproliferative activity of JQ1 against seven human OS cells in vitro as measured by LDH cytotoxicity assay after 72 hr exposure to JQ1. Data represent mean % LDH activity ± standard deviation (SD; error bars). (b) Time-dependent antiproliferative activity of JQ1. Cells were exposed to 7.5 µM JQ1 for 2, 12 and 24 hr, washed extensively, and cultured in JQ1-free media. Cell growth at 24–96 hr was measured by MTT assay. Data represent mean % cell growth ± SD. (c) Soft agar colony formation assay results. Cells were mixed in soft agar and cultured for 2 weeks in the presence or absence of 7.5 µM JQ1 followed by counting. Data represent mean number of colonies ± SD of three experiments done in triplicates. Asterisks (*) indicate p-value less than 0.05 (*) or 0.001 (**) by t-test. NS: not significant.
Pulse-exposure of these OS cells to 7.5 µM JQ1 for 2, 12 and 24 hr showed time-dependent JQ1 activity. Two hour pulse-exposure was sufficient to induce more than 50% growth inhibition of G292, MNNG/HOS and SJSA cells at 48, 72 and 96 hr of culture (Fig. 1b). MG-63 demonstrated the weakest, but still significant growth inhibition after 2 hr pulse exposure. Soft agar colony formation assays showed that 7.5 µM JQ1 significantly inhibited the clonogenic ability of G292, MNNG/HOS and SJSA cells, but not the MG-63 cells (Fig. 1c).
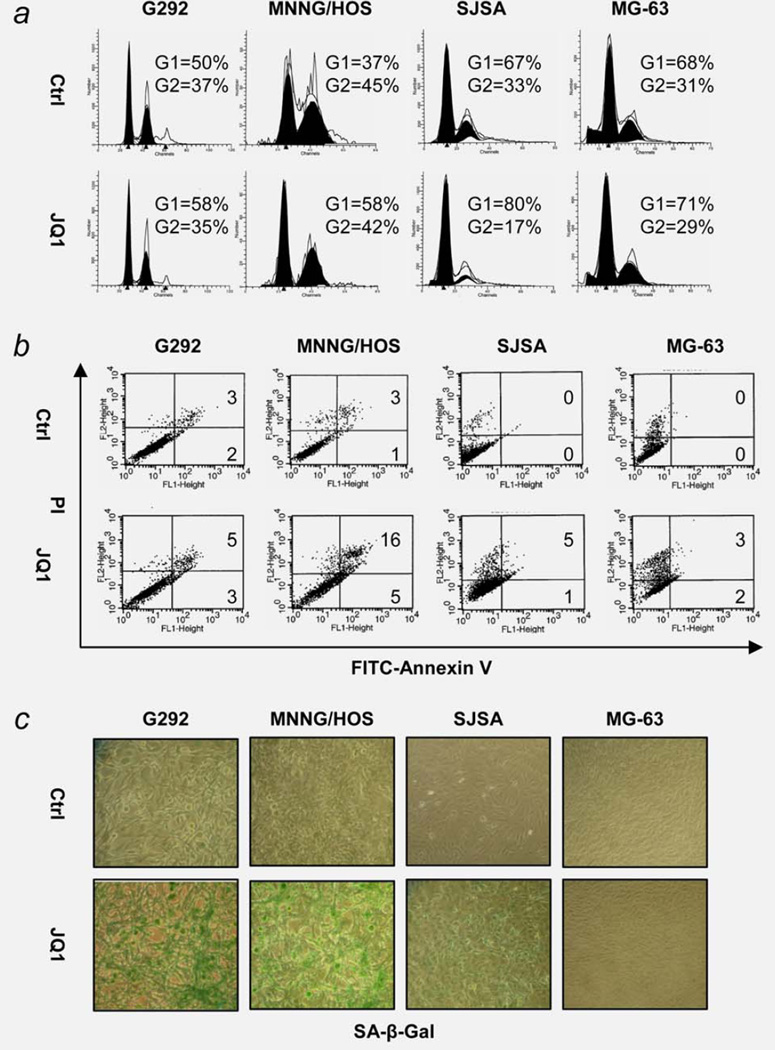
JQ1 (7.5 µM, 24 hr) increased the percentage of G1 phase cells in G292, MNNG/HOS and SJSA cells, but not MG-63 cells (Fig. 2a). Longer JQ1 exposure (7.5 µM, 72 hr) made a modest increase in apoptosis (range 5–21%; Fig. 2b). Instead, a prominent increase in premature senescence of these cells was observed when tested in the same condition by senescence-associated β-galactosidase (SA-β-gal) staining (Fig. 2c). MG-63 cells showed no increase in the number of cells undergoing senescence.
Figure 2.
Effect of JQ1 on cell cycle, apoptosis and senescence of human OS cells in vitro. (a) G1 cell cycle arrest after 24 hr exposure to 7.5 µM JQ1. Cells were stained with propidium iodide (PI) and analyzed by FACS. (b) Apoptosis of OS cells after 72 hr exposure to 7.5 µM JQ1. Cells were stained with PI and Annexin V-FITC and analyzed by FACS. Numbers indicate % gated cells in early (top) and late (bottom) apoptosis. (c) Senescence of OS cells after 72 hr exposure to 7.5 µM JQ1. Senescence-associated β-galatosidase (SA-β-gal) activity was measured by X-gal staining. Senescent cells stained dark green. Magnification ×200.
Effect of JQ1 on c-MYC and p21 expression in human OS cells
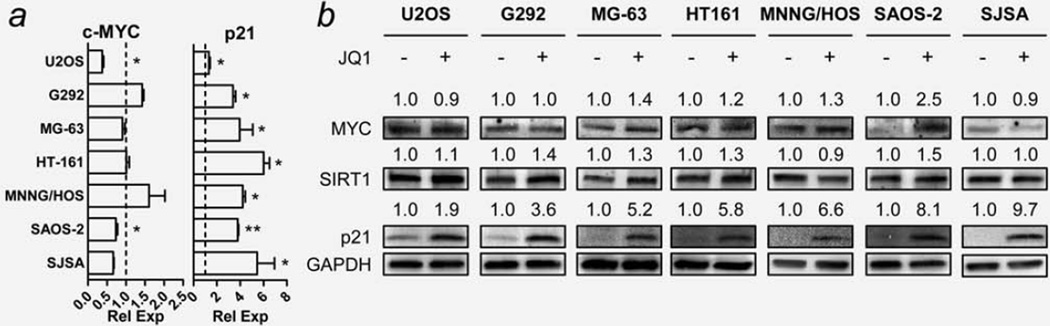
JQ1 has been reported to downregulate expression of c-MYC messenger RNA (mRNA) and upregulate levels of p21 mRNA in a variety of cancer types.5–7 We treated all seven OS cell lines with JQ1 (7.5 µM, 6 hr) and monitored changes in mRNA expression of c-MYC and p21 compared to control using qRT-PCR. Interestingly, only U2OS and SAOS-2 cells showed significant downregulation of c-MYC mRNA (Fig. 3a, left). The upregulation of p21 mRNA was observable in all seven OS cell lines (Fig. 3a, right). The protein expression of c-MYC and p21 after 12 hr exposure were also examined using Western blot analysis. No significant downregulation of c-MYC protein was observed in any of the OS cell lines (Fig. 3b). Rather, some cells showed upregulation of c-MYC protein. For example, SAOS-2 cells showed upregulation of c-MYC protein despite a downregulation of levels of MYC mRNA by JQ1. All seven cell lines showed significant upregulation of p21 protein levels in parallel with increased p21 mRNA levels (Fig. 3b). Protein levels of sirtuin 1 (SIRT1), a senescence-associated gene whose transcription is regulated by c-MYC,23 did not change in any of the OS cell lines (Fig. 3b).
Figure 3.
Effect of JQ1 on expression of c-MYC, p21 and SIRT1 in human OS cells. Cells were treated with either DMSO (control) or 7.5 µM JQ1 subjected to qRT-PCR (6 hr) and Western blot analyses (12 hr). (a) mRNA expression level of c-MYC and p21 genes in JQ1-treated cells. Dotted lines indicate the normalized expression level of each gene in DMSO-treated control cells. GAPDH was used as a loading control. Measurements were repeated in triplicates. Data represent the mean relative expression ± standard deviation (SD; error bars). Asterisks (*) indicate p-value less than 0.05 (*) or 0.001 (**) by t-test. (b) Representative images showing changes in protein level of c-MYC, SIRT1 and p21 in JQ1-treated cells. Numbers indicate relative band intensity of each gene normalized to the intensity of the genes in control (1.0). GAPDH was used as a loading control.
Synergistic effect of JQ1 and rapamycin on human OS cells in vitro
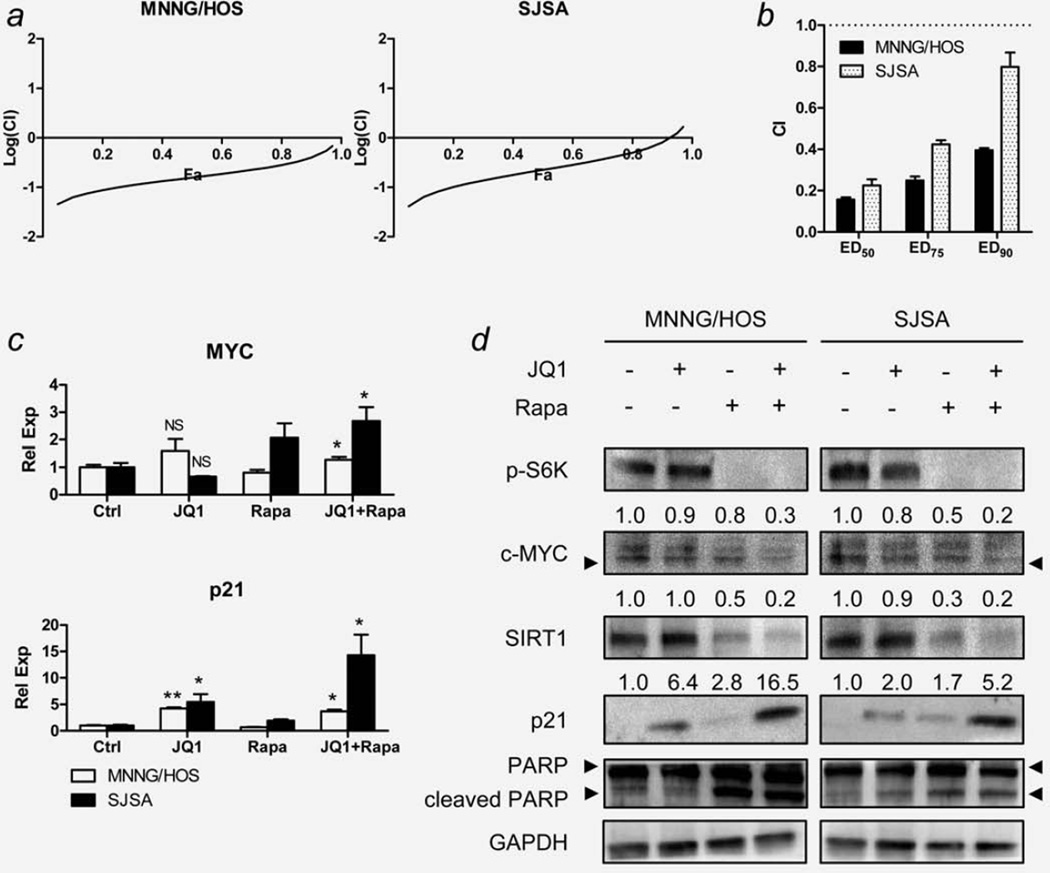
JQ1 as a single agent failed to reduce the volume and weight of the MNNG/HOS xenografts grown in immunodeficient nude mice (Supporting Information Fig. S1), which prompted us to explore combining JQ1 with rapamycin because we previously showed that the mTOR pathway is one of the major growth signaling pathways of human OS cells.14 Therefore, we hypothesized that inhibition of the mTOR pathway with rapamycin might sensitize OS cells to JQ1. To test the hypothesis, MNNG/HOS and SJSA cells were exposed to various concentrations of JQ1, rapamycin or both for 48 hr, and cell viability was examined (Supporting Information Figs. S2a–S2d and Supporting Information Table S3). In constant drug ratio analysis, average combination index (CI) values at ED50, ED75 and ED90 were all below 0.9 in both cells (Figs. 4a and 4b). In nonconstant drug ratio analysis, most ratios showed CI values less than 0.9 in both cells, as shown in isobolograms (Supporting Information Fig. S2c). Taken together, JQ1 and rapamycin displayed a strong synergism of growth inhibition of both MNNG/HOS and SJSA cells.
Figure 4.
Effect of combination of JQ1 and rapamycin on OS cells in vitro. MNNG/HOS and SJSA cells were grown in various concentrations of JQ1 and rapamycin, and the dose–response relationship was determined after 48 hr by LDH assay (Supporting Information Fig. S2a and Supporting Information Table S3). (a) Fraction affected (Fa)-Combination index (CI) plot for MNNG/HOS and SJSA cells exposed to JQ1 and rapamycin. Fa-CI plots were generated using selected data points with constant concentration ratio of JQ1 and rapamycin (1:1). (b) Changes in CI values at different drug concentrations (ED50, ED75 and ED90) of JQ1 and rapamycin in MNNG/HOS and SJSA cells. Data represent mean CI ± standard deviation (SD). (c) The effect of JQ1 (7.5 µM) and/or rapamycin (12.5 nM) on the mRNA level of c-MYC and p21 genes of MNNG/HOS and SJSA cells at 12 hr exposure. Expression levels of c-MYC and p21 were normalized to the expression level of GAPDH. Data represent mean relative expression ± SD. Asterisks (*) indicate p-value less than 0.05 (*) or 0.001 (**) by t-test. (d) Representative Western blot images showing changes in protein levels of the selected genes in the same condition as described in panel (c). Numbers indicate relative band intensity of each gene normalized to the intensity of the genes in control (1.0). GAPDH was used as a loading control. NS: not significant.
To explore the molecular effects when both drugs are combined, MNNG/HOS and SJSA cells were treated with JQ1 (7.5 µM), rapamycin (12.5 nM) or both for 24 hr. Rapamycin concentration was based on IC50 values of rapamycin in these cells (Supporting Information Fig. S2b). Decreased c-MYC protein level and enhanced upregulation of p21 protein were two of the most noticeable changes resulting from the combination of both compounds. Despite the increase in c-MYC mRNA levels (Fig. 4c), c-MYC and SIRT1 protein levels were the lowest among all groups when both compounds were combined in both MNNG/HOS and SJSA cells (Fig. 4d). The increase in p21 protein level was the greatest with the drug combination (Fig. 4d). Cleaved PARP and cleaved caspase 8, the indicators of apoptosis, increased and phospho-S6K, a downstream gene of mTOR,12 decreased in rapamycin treated cells, which did not change with addition of JQ1 (Fig. 4d, Supporting Information Fig. S2e). Overall, apoptosis induced by rapamycin was not enhanced by the addition of JQ1 (Supporting Information Figs. S2f and S2g).
Synergistic effect of JQ1 and rapamycin on MNNG/HOS human OS xenografts in vivo
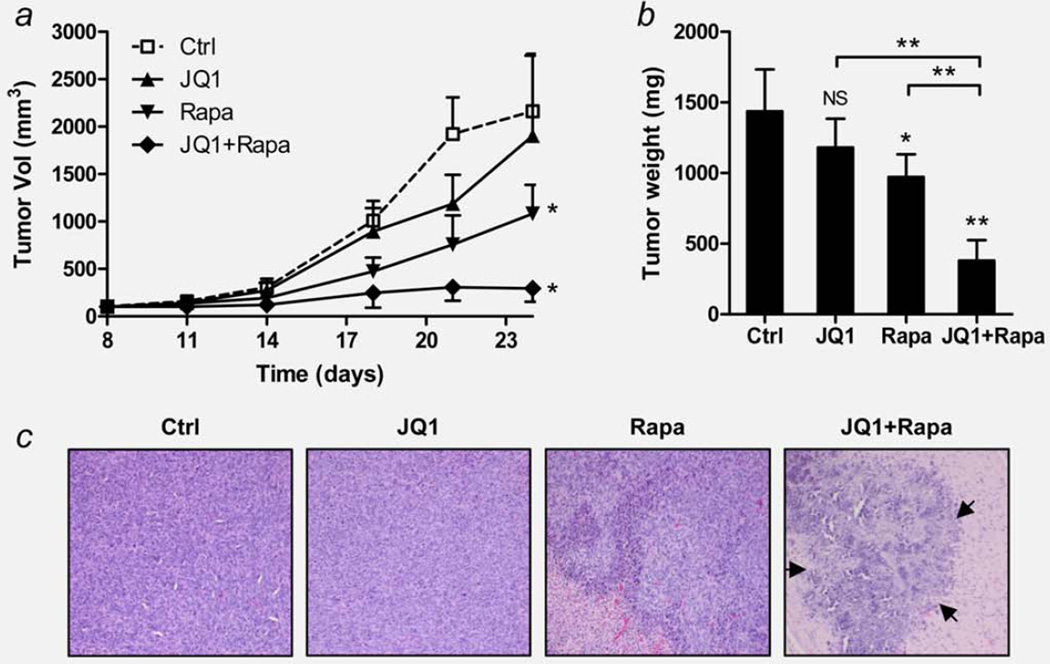
Based on the in vitro synergism of the two drugs, we tested their effects against MNNG/HOS human OS xenografts growing in immunodeficient mice (Figs. 5a and 5b). At Day 24, JQ1 as a single agent failed to reduce the tumor volume (p = 0.8996) and weight (p = 0.1526) to a significant level. Rapamycin as a single agent reduced the tumor volume by 50% versus diluent control (p<0.05), and tumor weight by 42% versus diluent control (p<0.05). The combination of the two drugs greatly reduced the tumor volume by 85% versus diluent control (p<0.05), and tumor weight by 73% (p<0.0001). At autopsy, no significant gross changes in the spleen, liver, kidney and lungs were observed. H&E staining of the tumor mass, showed large regions of necrotic tumor cells (marked by arrows) in the combination group (Fig. 5c). The average body weight of the mice in all treatment groups were significantly lower than the diluent control throughout the experiment, but no sign of toxicity was observed (Supporting Information Fig. S3).
Figure 5.
Effect of combination of JQ1 and rapamycin on human OS xenografts in vivo. MNNG/HOS xenografts were grown in athymic nude mice and treated with JQ1 and/or rapamycin 72 hr after cell injection (Day 0). Ctrl, diluent control; JQ1, JQ1 (50 mg/kg body weight, i.p. daily); Rapa, rapamycin (1.5 mg/kg, i.p. every other day). (a) Volumetric growth of MNNG/HOS xenografts. Data show mean tumor volume ± standard deviation (SD, error bars) of five mice per group. (b) Comparison of tumor weight of each group. At Day 24, mice were sacrificed and tumors were dissected and weighed. Data represent mean tumor weight ± SD of ten tumors per group. Asterisks (*) indicate p-value less than 0.05 (*) or 0.001 (**) by t-test. (c) H&E staining show representative tumor sections of each group. Arrows indicate the boundaries of necrotic tumor cells in the combination group. Magnification ×200. NS: not significant.
RUNX2 as a direct target of BRD4 inhibition by JQ1 in human OS cells in vitro
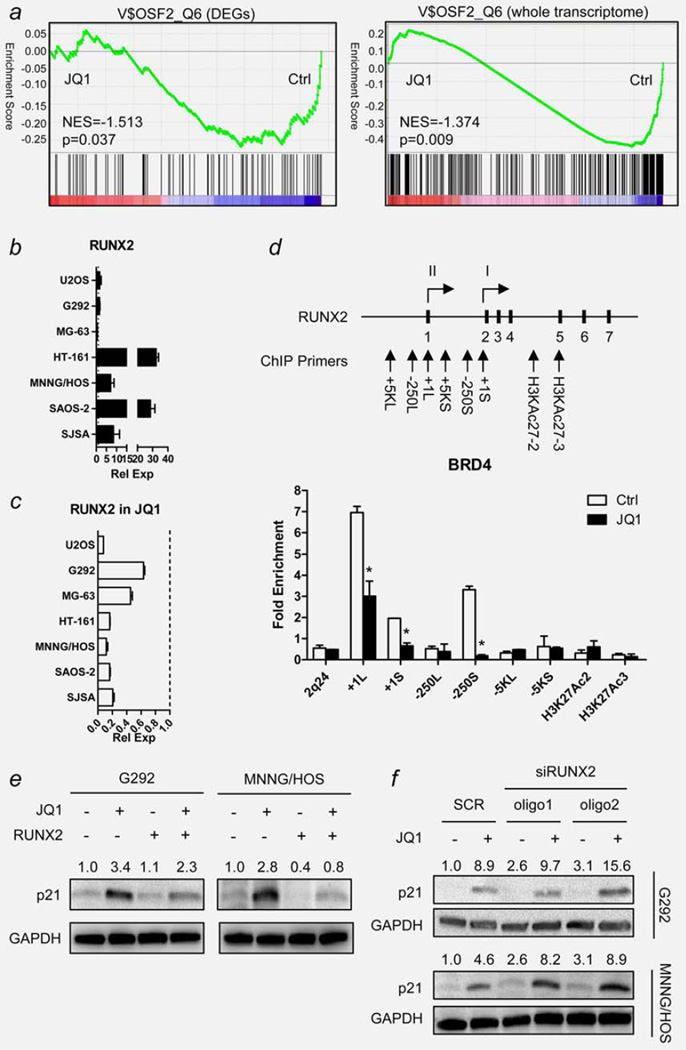
Because JQ1 failed to inhibit c-MYC expression in OS cells, cellular effects of JQ1 should be mediated by other genes. To identify the mediator of JQ1 activity in OS cells, we performed microarray analysis of MNNG/HOS cells either with or without 7.5 µM JQ1 for 6 hr. Comparison of the groups revealed many DEGs with statistical significance (Supporting Information Tables S4 and S5). Among DEGs strongly downregulated by JQ1 treatment, we chose RUNX2 as the target gene: First, GSEA demonstrated significant downregulation of genes with RUNX2 binding motif in JQ1-treated cells (p = 0.037 for DEGs only, p = 0.009 for the whole transcriptome; Fig. 6a). Second, RUNX2 gene shows several regions of frequent histone H3 acetylation sites (H3K27Ac) as identified by ENCODE project,24 showing possible involvement of BRD4 in RUNX2 transcription (Supporting Information Fig. S4). Third, transcription of Runt-family of proteins, including RUNX2, is often under control of superenhancers and P-TEFb in other cell types.25,26 Fourth, RUNX2 is known to downregulate p21 expression in OS cells, and inhibition of RUNX2 by JQ1 could partly explain the upregulation of p21 in these cells.27 Fifth, RUNX2 is overexpressed in both primary and cultured OS cells.11,28 Indeed, all OS cell lines except MG-63 showed increased expression of RUNX2 (Fig. 6b), and significant downregulation of RUNX2 mRNA level upon JQ1 treatment (7.5 µM, 6 hr; Fig. 6c).
Figure 6.
RUNX2 as a mediator of JQ1 activity in human OS cells. (a) Gene set enrichment analysis (GESA) plot showing downregulation of genes with RUNX2 binding motifs by JQ1 treatment in MNNG/HOS cells. Green lines represent the running enrichment score for the gene set as it progresses down the ranked list (x-axis) according to their differential expression score. Black vertical lines show the location of the members of the gene set. GESAs were done using either the differentially expressed genes (DEGs, left) or the whole transcriptome (right) in the gene set. (b) Endogenous mRNA expression level of RUNX2 of seven human OS cell lines measured by qRT-PCR. Data indicate relative mean expression ± standard deviation (SD, error bars). RUNX2 expression level of each cell was normalized to the RUNX2 expression level of hFOB1.19 human osteoblast cells (set to 1.0). GAPDH was used as a loading control. (c) Changes in RUNX2 mRNA expression level of seven human OS cell lines upon JQ1 treatment (7.5 µM, 6 hr). Dotted line indicates the expression level of RUNX2 mRNA of each cell without JQ1 treatment (set to 1.0). GAPDH was used as a loading control. (d) Schematic diagram showing RUNX2 gene and the location of ChIP primers used in the study. Each short, black vertical bar with a number represents one of the seven exons of RUNX2. Two arrows with Roman numerals (I and II) display the location of two transcription start sites. Bar graph below shows enrichment of PCR products from MNNG/HOS cells treated with or without JQ1 (7.5 µM, 6 hr). Data represent relative fold enrichment ± SD. Asterisk (*) indicates p-value less than 0.05 by t-test. (e) Changes in p21 protein level in RUNX2-overexpressing MNNG/HOS cells. Cells were transfected with either empty vector (EV) or pEF-BOS-RUNX2 expression vector (RUNX2). After 48 hr, cells were treated with 7.5 µM JQ1 for 12 hr and harvested for Western blot analysis. Numbers indicate relative band intensity of each gene normalized to the intensity of the genes in control (1.0). GAPDH was used as a loading control. (f) Changes in p21 protein level in MNNG/HOS cells transfected with siRNA oligos against RUNX2. Cells were transfected with either scrambled control oligos (SCR) or two kinds of siRUNX2 oligos (oligo1 and oligo2). After 48 hr, cells were treated with 7.5 µM JQ1 for 12 hr and harvested for Western blot analysis. Numbers indicate relative band intensity of each gene normalized to the intensity of the genes in control (1.0). GAPDH was used as a loading control. NES: normalized enrichment score.
To confirm that the displacement of BRD4 from the RUNX2 promoter region by JQ1 is the direct cause of the downregulation of RUNX2 mRNA level, ChIP analysis using antibody against BRD4 was performed on MNNG/HOS cells either with or without JQ1 (7.5 µM, 6 hr). Based on the H3K27Ac histone acetylation data, several regions of interest around the RUNX2 gene were chosen, which included two RUNX2 transcription start sites (+1L, +1S),28 upstream promoter regions (−250L, −250S) and two H3K27Ac histone acetylation sites (H3KAc27-2, H3KAc27-3). Additionally, three regions, 5,000 bp upstream of the two RUNX2 transcription start sites (−5KL, −5KS) and a region in chromosome 2 q-arm (2q24), were chosen as negative controls (Supporting Information Table S1). ChIP results showed that three regions, +1L, +1S and −250S, were significantly enriched in DMSO-treated control, and these enrichment diminished in JQ1-treated cells (p<0.05 for all; Fig. 6d).
To confirm further that RUNX2 is a mediator of JQ1 activity, we modulated the expression of RUNX2 in two JQ1-sensitive cell lines, G292 and MNNG/HOS (Supporting Information Fig. S5). Compared to cells with EV, cells with forced expression of RUNX2 decreased their levels of p21 protein that has been stimulated by JQ1 treatment (7.5 µM, 12 hr; Fig. 6e). In addition, RUNX2-overexpressing cells were less sensitive to JQ1 than cells with EV control as evidenced by lower relative growth inhibition by JQ1 (71% vs. 40%; Supporting Information Figs. S6a and S6b). On the contrary, cells with reduced levels of RUNX2 by siRNA-mediated knockdown were more sensitive to JQ1, showing higher levels of p21 protein induced by JQ1 treatment (7.5 µM, 12 hr) compared to cells with scrambled oligos (Fig. 6f, Supporting Information Figs. S5c, S5d, S6c and S6d). Taken together, we concluded that the antiproliferative activity of JQ1 in OS cells is partly mediated by the inhibition of RUNX2.
Discussion
In this study, we show that the BRD4 inhibition by JQ1 can moderately inhibit the growth of OS cells in vitro, but has little antiproliferative effect on OS cells growing as tumors in vivo. When combined with rapamycin, JQ1 synergistically inhibit the growth of OS cells both in vitro and in vivo. At the molecular level, JQ1 alone failed to inhibit c-MYC expression in OS cells. GSEA also confirmed that no enrichment of genes by JQ1 was identified in a gene set with c-MYC binding motifs in MNNG/HOS cells (Supporting Information Fig. S7). This is congruent with the finding in LAC cells where JQ1 also failed to inhibit c-MYC expression.2 Surprisingly, JQ1 in combination with rapamycin nearly silenced c-MYC protein expression and enhanced the upregulation of p21 protein in MNNG/HOS and SJSA cells. Paradoxically, combination of the two drugs increased c-MYC mRNA expression level in these cells. Therefore, the combinatorial effect of the two compounds to decrease c-MYC protein level may occur by either affecting translation or stabilization of c-MYC protein. This may explain the reason why we could not identify the target genes responsible for the synergism using microarray analysis, although we did identify several DEGs in JQ1 and rapamycin-treated cells (Supporting Information Tables S6 and S7). Proteome array and longer exposure to the drugs may help identify the interaction between JQ1 and rapamycin.
c-MYC is not the only example showing that JQ1 affects genes in cell-type specific manner depending on their chromatin acetylation status. One such example is c-FOS. Lockwood et al.2 reported that c-FOS was not differentially expressed in JQ1-treated LAC cells. In JQ1-treated OS cells, c-FOS was dramatically increased (Supporting Information Fig. S8a). Since c-FOS forms an activator protein 1 (AP-1) complex with c-JUN and plays an important role in OS proliferation,29 we initially assumed that upregulation of c-FOS and subsequent upregulation of AP-1 activity might explain the relative resistance of OS cells to JQ1 treatment. However, these cells also had a decrease in phospho-c-JUN protein, suggesting that overall activity of AP-1 remained unchanged (Supporting Information Fig. S8a). GSEA also confirmed that JQ1 treatment of MNNG/HOS cells did not enrich a gene set with AP-1 DNA binding motif (Supporting Information Fig. S8b). Another example of cell-specificity is IL7R. Ott et al.7 reported that JQ1 downregulated levels of IL7R in ALL cells as a direct target of BRD4 inhibition by JQ1. In JQ1-treated MNNG/HOS cells, IL7R was also dramatically decreased (Supporting Information Table S4). However, when we treated MNNG/HOS cells with human IL7 after 24 hr of serum starvation, we could not detect any changes in phosphorylation of either JAK or STAT family proteins (Supporting Information Fig. S9).
In this study, we identified RUNX2 as a direct target of BRD4 inhibition by JQ1 in human OS cells. Although less frequent than CN gain of 8q23.3-qter, recurrent CN gain of the region in chromosome 6 p-arm (6p12-p21), where RUNX2 resides, is often observed in human OS cells.11,28 Several studies also confirmed the overexpression of RUNX2 mRNA and protein in OS cells.28,30 Overexpression of RUNX2 correlated with a poor response of OS cells to chemotherapy.28 However, the contribution of RUNX2 overexpression to OS tumorigenesis has yet to be confirmed due to its complex functionality and regulatory pattern. Complicating the analysis, many OS samples have either impaired or nonfunctional retinoblastoma 1 (RB1) and p53 proteins, and these proteins are known to interact with RUNX2. Hypophosphorylated RB1 and RUNX2 together induce cell cycle exit during terminal osteoblast differentiation.28 Without functional RB1, the interaction of RUNX2 and RB1 may lead to uncontrolled cell proliferation and genetic instability.28 Regarding p53, studies have shown that loss of p53 correlated with elevated RUNX2 protein levels in OS cells.28 Considering genomic amplification of 6p12-p21, overexpresion of RUNX2 is not solely dependent on loss of p53, but the role of p53 and RUNX2 interaction remains largely unknown.
Regardless, we show that overexpression of RUNX2 could counter the effect of JQ1 in OS cells, including prevention of upregulation of p21 and lower relative growth inhibition by JQ1 (Fig. 6e, Supporting Information Fig. S6). Likewise, siRNA-mediated knockdown of RUNX2 expression in OS cells sensitized them to JQ1 treatment, marked by enhanced upregulation of p21 (Fig. 6f). Interestingly, OS cells overexpressing RUNX2 grew slower than the EV control in the presence of serum, but survived longer than controls in the absence of serum (Supporting Information Fig. S6).
Recently, similar to our investigation, a study was published that examined the effect of JQ1 in human OS cells.31 Besides the similarities of findings of the two studies, several differences were identified. They found that JQ1 downregulated c-MYC expression in OS cells at both the transcriptional and translational level, and JQ1 alone significantly inhibited the growth of OS xenografts in immunodeficient nude mice. In contrast, we noted that c-MYC was unresponsive to JQ1 treatment and the administration of rapamycin was required to enhance markedly the anticancer activity of JQ1.
In conclusion, all data show that RUNX2 is a direct target of BRD4 inhibition by JQ1 in OS cells and its inhibition partly explains the cellular effects of JQ1. When combined with rapamycin, JQ1 is a promising chemotherapeutic option to treat human OS. Possible synergism of JQ1 with other mTOR, Akt, PI3K inhibitors also deserve exploration for OS treatment.
Supplementary Material
What’s new?
Gene expression can be effectively turned on or off by addition or removal of acetyl groups. “Reader” proteins, such as the BET family of proteins, respond to these acetylation cues by recruiting transcription machinery. In this paper, the authors show that a BET protein inhibitor, JQ1, slows the growth of osteosarcoma cells in vitro, but wasn’t too effective against tumors in vivo. That is, until they tried it in combination with rapamycin: in conjunction with that drug, JQ1 killed off OS cells both in vitro and in vivo, making it potentially useful as a treatment.
Acknowledgments
Qi and Bradner in Dana-Farber Cancer Institute licensed JQ1 to Tensha Therapeutics (Cambridge, MA) for therapeutic development. Qi is a shareholder to Tensha Therapeutics. Bradner is a consultant and a shareholder to Tensha Therapeutics.
Grant sponsors: National Research Foundation Singapore and the Singapore Ministry of Education under the Research Centers of Excellence initiative, Singapore Ministry of Health’s National Medical Research Council under its Singapore Translational Research (STaR) Investigator Award, and US National Institutes of Health (to H.P.K.); Grant number: R01CA026038-35
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- 1.Andreoli F, Barbosa AJ, Parenti MD, et al. Modulation of epigenetic targets for anticancer therapy: clinicopathological relevance, structural data and drug discovery perspectives. Curr Pharm Des. 2013;19:578–613. doi: 10.2174/138161213804581918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lockwood WW, Zejnullahu K, Bradner JE, et al. Sensitivity of human lung adenocarcinoma cell lines to targeted inhibition of BET epigenetic signaling proteins. Proc Natl Acad Sci USA. 2012;109:19408–19413. doi: 10.1073/pnas.1216363109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Z, He N, Zhou Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol Cell Biol. 2008;28:967–976. doi: 10.1128/MCB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zuber J, Shi J, Wang E, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Delmore JE, Issa GC, Lemieux ME, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ott CJ, Kopp N, Bird L, et al. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood. 2012;120:2843–2852. doi: 10.1182/blood-2012-02-413021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puissant A, Frumm SM, Alexe G, et al. Targeting MYCN in neuroblastoma by BET bromodomain inhibition. Cancer Discov. 2013;3:308–323. doi: 10.1158/2159-8290.CD-12-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bandopadhayay P, Bergthold G, Nguyen B, et al. BET Bromodomain Inhibition of MYC-Amplified Medulloblastoma. Clin Cancer Res. 2014;20:912–925. doi: 10.1158/1078-0432.CCR-13-2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin JW, Squire JA, Zielenska M. The genetics of osteosarcoma. Sarcoma. 2012;2012:627254. doi: 10.1155/2012/627254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 13.Bogenmann E, Moghadam H, DeClerck YA, et al. c-myc amplification and expression in newly established human osteosarcoma cell lines. Cancer Res. 1987;47:3808–3814. [PubMed] [Google Scholar]
- 14.Lee DH, Thoennissen NH, Goff C, et al. Synergistic effect of low-dose cucurbitacin B and low-dose methotrexate for treatment of human osteosarcoma. Cancer Lett. 2011;306:161–170. doi: 10.1016/j.canlet.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee DH, Amanat S, Goff C, et al. Overexpression of miR-26a-2 in human liposarcoma is correlated with poor patient survival. Oncogenesis. 2013;2:e47. doi: 10.1038/oncsis.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chou TC, Talalay P. Quantitative analysis of dose-effect relationships:the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 17.Chou TC. Drug combination studies and their synergy quantification using the Chou–Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 18.Luong QT, O’Kelly J, Braunstein GD, et al. Antitumor activity of suberoylanilide hydroxamic acid against thyroid cancer cell lines in vitro and in vivo. Clin Cancer Res. 2006;12:5570–5577. doi: 10.1158/1078-0432.CCR-06-0367. [DOI] [PubMed] [Google Scholar]
- 19.Abramoff MDM, Magalhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics Int. 2004;11:7. [Google Scholar]
- 20.Chua SW, Vijayakumar P, Nissom PM, et al. A novel normalization method for effective removal of systematic variation in microarray data. Nucleic Acids Res. 2006;34:e38. doi: 10.1093/nar/gkl024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang YW, Yasui N, Ito K, et al. A RUNX2/PEB-P2alpha A/CBFA1 mutation displaying impaired transactivation and Smad interaction in cleidocranial dysplasia. Proc Natl Acad Sci USA. 2000;97:10549–10554. doi: 10.1073/pnas.180309597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan J, Minter-Dykhouse K, Lou Z. A c-Myc-SIRT1 feedback loop regulates cell growth and transformation. J Cell Biol. 2009;185:203–211. doi: 10.1083/jcb.200809167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Consortium EP. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 25.Lovén J, Hoke HA, Lin CY, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013;153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whyte WA, Orlando DA, Hnisz D, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Westendorf JJ, Zaidi SK, Cascino JE, et al. Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21(CIP1/WAF1) promoter. Mol Cell Biol. 2002;22:7982–7992. doi: 10.1128/MCB.22.22.7982-7992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin JW, Zielenska M, Stein GS, et al. The role of RUNX2 in osteosarcoma oncogenesis. Sarcoma. 2011;2011:282745. doi: 10.1155/2011/282745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 30.Lucero CM, Vega OA, Osorio MM, et al. The cancer-related transcription factor Runx2 modulates cell proliferation in human osteosarcoma cell lines. J Cell Physiol. 2013;228:714–723. doi: 10.1002/jcp.24218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamoureux F, Baud’huin M, Rodriguez Calleja L, et al. Selective inhibition of BET bromodomain epigenetic signalling interferes with the bone-associated tumour vicious cycle. Nat Commun. 2014;5:3511. doi: 10.1038/ncomms4511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.