Abstract
Phosphatidic acid (PA) and diacylglycerol (DAG) are key signaling molecules and important precursors for the biosynthesis of all glycerolipids found in eukaryotes. Research conducted in the model organism Saccharomyces cerevisiae has been at the forefront of the identification of the enzymes involved in the metabolism and transport of PA and DAG. Both these lipids can alter the local physical properties of membranes by introducing negative curvature, but the anionic nature of the phosphomonoester headgroup in PA sets it apart from DAG. As a result, the mechanisms underlying PA and DAG interaction with other lipids and proteins are notoriously different. This is apparent from the analysis of the protein domains responsible for recognition and binding to each of these lipids. We review the current evidence obtained using the PA-binding proteins and domains fused to fluorescent proteins for in vivo tracking of PA pools in yeast. In addition, we present original results for visualization of DAG pools in yeast using the C1 domain from mammalian PKCδ. An emerging first cellular map of the distribution of PA and DAG pools in actively growing yeast is discussed.
Keywords: phosphatidic acid, diacylglycerol, fluorescent binding probes, glycerolipid metabolism, signaling
Introduction
Lipids are active components of signal transduction pathways that regulate cell survival and cell death. Key to the operative role of lipids as signaling molecules is their low abundance in membranes and the informational outcomes of their interactions with proteins and other lipids. The efficacy of signals mediated by lipid messengers is intimately interconnected with their metabolism. The localization of biosynthetic and catabolic enzymatic systems that produce lipid messengers is a first determinant of their cellular distribution. In addition, lipid messengers can be traded between membranes of different cellular compartments, and some can undergo transbilayer and lateral movements in a given membrane. Of particular interest for this review is the spatiotemporal regulation of two glycerolipid messengers, phosphatidic acid (PA) and diacylglycerol (DAG) (Tables 1 and 2). Pioneering studies conducted 35 years ago on the role of DAG as activator of the serine/threonine protein kinase C (PKC) have revolutionized our view of lipids as second messengers and bioactive molecules.1–4 This recognition was comparatively delayed for PA,5–7 which has more recently emerged as an intracellular lipid mediator of a large number of effector proteins.8 Besides their role in signal transduction pathways, PA and DAG are the precursors of the entire array of cellular glycerolipids, including membrane phospholipids (PLs) and triglycerides.9 Although production of PA and DAG as lipid messengers has been initially associated with PL turnover through regulation of phospholipases,10 it is clear now that biosynthetic systems are also critical components of these signaling circuits.9,11–14 Furthermore, the regulated interconversion of PA and DAG in space and time provides the versatility needed for the signal to be dynamic and transient. The aim of this study is to review the current evidence supporting the existence of different PA and DAG pools in eukaryotes, focusing on studies performed in budding yeast. In addition, we analyze the emerging cellular map of their localization obtained using fluorescent probes combined with live microscopy and the impact that alteration of metabolic pathways has on their distribution.
Table 1.
Lipid shapes and curvature.
| LIPID | SHAPE | CURVATURE | |
|---|---|---|---|
| Lysolipids | Inverted cone |

|
Positive |
| PC, PI, PS | Cylindrical |

|
Neutral |
| PA, DAG, PE | Cone |

|
Negative |
Notes: Lipids introducing positive, neutral, and negative curvatures into bilayers have inverted cone, cylindrical, and cone shapes, respectively.16 Cylindrical shape depends on the unsaturation and the length of acyl chains. In the case of PA, changes in pH and Ca2+ concentrations can modulate its shape.
Abbreviations: DAG, diacylglycerol; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PS, phosphatidylserine.
Table 2.
Similarities and differences between PA and DAG in budding yeast.
| PHOSPHATIDIC ACID | DIACYLGLYCEROL | |
|---|---|---|
| Structure |
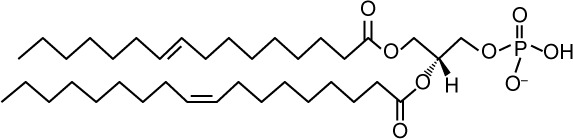
|
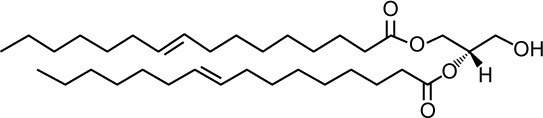
|
| Lipid shape | Cone induces negative curvature | |
| Speciesa,24,25,96 | 26:0; 26:1 28:0; 28:1 30:0; 30:1; 30:2 32:1; 32:2 34:1; 34:2 36:1; 36:2 |
|
| Headgroup charge | Negative Deprotonated: −2 Protonated: −1 |
Neutral |
| Abundanceb | 8–10 mol% (cell extract)24,96 2 mol% (TGN/E)45,97 10 mol% (PM)45 4% total PL (PM)98 0.6–2% total PL (mito)98 4% total PL (ER)98 2% total PL (vacuole)98 1.8% total PL (LD)35 |
4–6 mol% (cell extract)24,96 10 mol% (TGN/E)45,97 n.d. (PM)45 |
| Transbilayer movement26 | No | Rapid “flip flop” without protein assistance |
| Interaction with effector proteins | Electrostatic/hydrogen bond switch model65 No consensus sequence8 |
Specific, mediated by C1 consensus domain55,59 |
| Proposed roles | pH sensor68 Fusion (prospore formation)74,99 Mediates membrane recruitment of Pah1 (ER),50 Opi1 (ER),11 Spo20 (PM)73 Mitochondrial protein biogenesis100 |
Fusion (vacuole)20 Regulates protein secretion44 Membrane recruitment of Pkc1 is controversial60,61 |
Notes:
Common PA and DAG species, most abundant species are in bold.
Unless otherwise indicated, abundance is expressed as mol% of all yeast lipids, including glycerolipids, sphingolipids, and ergosterol.
Abbreviations: PL, phospholipids; TGN/E, trans-Golgi network/endosomes; PM, plasma membrane; LD, lipid droplet; mito, mitochondria; n.d., non detected.
PA and DAG: Structural Similarities and Critical Differences
PA and DAG are simple glycerolipids with acyl chains attached to the sn-1 and sn-2 positions of the glycerol backbone through ester linkages. The free sn-3 hydroxyl group represents the headgroup of DAG, while a phosphate is attached to this sn-3 position in PA as a phosphomonoester. This anionic headgroup is a defining feature of PA that distinguishes it from DAG and other PLs.15 In both PA and DAG, the polar headgroup has a smaller transversal section than that of the hydrophobic moiety, resulting in a cone molecular shape under physiological conditions (Tables 1 and 2).16,17 Therefore, small amounts of PA or DAG in bilayers introduce negative curvature, and this change in membrane architecture plays a critical role in the recruitment of proteins and in the fusion and fission processes of cellular membranes.15,17 Interestingly, DAG is able to promote the fusion of protein-free liposomes18 and has been identified as an essential lipid responsible for vacuolar fusion in yeast.19,20
It is important to recognize that eukaryotes possess a remarkable diversity of PA and DAG species, varying in acyl chain length and number of double bonds.10,21 Moreover, unique and sometimes opposite biological effects have been associated with PA or DAG, depending on their degree of saturation/unsaturation.2,12,22 This holds true even in the case of budding yeast Saccharomyces cerevisiae,23 which is a relatively simple eukaryote with a discrete complement of glycerolipid acyl chains mostly composed of C16:0, C16:1, C18:0, and C18:1.24 Lipidomic analysis of wild-type yeast grown in standard defined medium conditions revealed that PA and DAG represented a minimum of 8 and 4 mol% of total lipids, respectively, and the most abundant species contained at least one monounsaturated fatty acyl (MUFA) chain (16:1–18:1, 16:1–16:1, 16:0–16:1, and 16:0–18:1; Table 2).24 Interestingly, a recent study showed that while MUFA-containing PA species dominated all phases of yeast growth in rich medium, DAG displayed a different profile during exponential growth phase, where species containing MUFA were almost as abundant as those containing saturated fatty acyl chains.25 This unique profile of DAG in the exponential phase probably reflects a pool of DAG derived from phosphatidylinositol (PI), which is the PL that serves as precursor for the synthesis of complex sphingolipids (SLs) in the Golgi during active growth.25 Throughout postdiauxic and stationary growth phases, MUFA-containing DAG species become dominant, coinciding with the profile observed for PA species and divergent from those of PI, which display a MUFA/saturated fatty acyl ratio close to one, regardless of the phase of growth.25
Another unique feature of DAG is that in great contrast to PA, DAG moves freely across transbilayer membranes in a process called flip flop without protein assistance.26,27 Biophysical studies using model membranes have shown that insertion of DAG induces a change from lamellar to hexagonal structures and this is dependent on the amount of DAG present in the bilayer.26,28 Recent findings report that a pool of DAG is constitutively present at basal level in the outer leaflet of the plasma membrane of mammalian cells, and the movement of this DAG pool to the inner leaflet can be reduced by the presence of sphingomyelin in the outer leaflet.29,30 These studies demonstrate that the distribution of DAG in cells can be asymmetrical and regulated by its surrounding environment.
Sources of PA and DAG in Budding Yeast
De novo synthesis of PA is initiated by two sequential acylations of glycerol 3-phosphate (G3-P) using acyl-CoA as acyl donor. G3-P acyltransferases catalyze the acylation at position sn-1 producing lysophosphatidic acid (LysoPA). LysoPA is further acylated at the sn-2 position by a separate acyltransferase to produce PA. Alternatively, the pathway can be initiated by acylation of dihydroxyacetone phosphate (DHAP), which requires an extra step for the reduction of 1-acyl DHAP to LysoPA. PA can be either (i) dephosphorylated to produce DAG for the synthesis of phosphatidylcholine (PC) and phosphatidylethanolamine (PE) through the Kennedy pathway or for the synthesis of triacylglycerol for energy storage or (ii) converted to cytidine diphosphate (CDP)-DAG for the synthesis of PI as occurring in all eukaryotic cell types and for the synthesis of phosphatidylserine (PS) in yeast.9 PS can in turn be decarboxylated to PE, and PE can be sequentially methylated to produce PC. The endoplasmic reticulum (ER) is the main organelle where de novo synthesis of PA and DAG takes place. It is known that PA made in the ER is transported into mitochondria for the synthesis of mitochondrial lipids phosphatidylglycerol and cardiolipin. The recently identified lipid transfer complex Ups1–Mdm35 mediates this movement and is conserved from yeast to humans.31,32 In addition to ER contact sites, mitochondria also interact with the vacuole (yeast lysosome) through the newly discovered vacuole and mitochondria patch complex.33 Mitochondria depend on having one of these contact sites, as the deficiency of one causes expansion of the other, and deletion of both is lethal.33 Lipid droplets (LDs) originate from the ER and are also known to closely interact with mitochondria as well as peroxisomes.34 Interestingly, the complete biosynthetic pathway to make PA from G3-P or DHAP associates with LDs,35–37 opening the possibility that the enzymes of this route could piggyback on LDs to reach mitochondria. Vacuoles on the other hand could provide a source of PA and DAG from glycerolipid turnover as this organelle is known to host phospholipases (PI-phospholipase C [PLC] and PC-PLD)19,33 and to breakdown LDs (Fig. 1).38
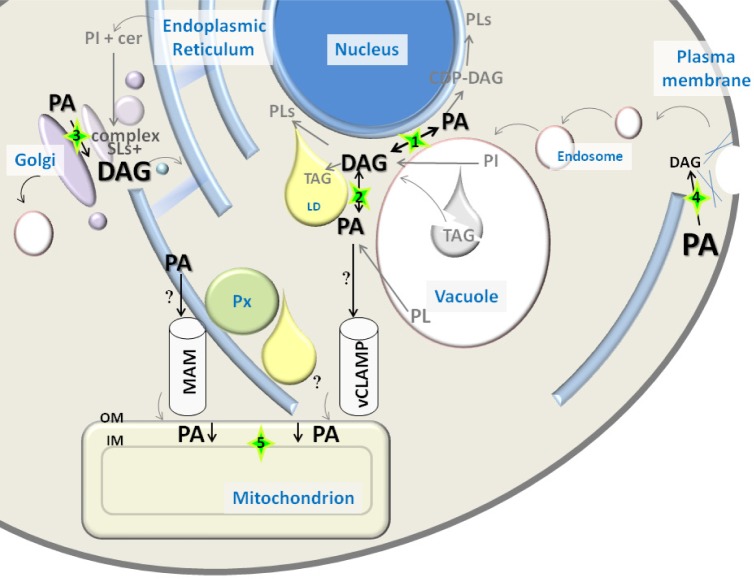
Figure 1.
PA and DAG distribution in yeast. PA and DAG font size reflects abundance of pools of these lipids in different cellular compartments, based on data obtained by combined subcellular fractionation and lipidomics approaches (Table 2). Relevant contact sites between organelles for PA/DAG conversion or transport are highlighted with a green star. Arrows involved in PA/DAG conversions reflect the presence of associated enzymatic activities in that compartment: (1) Nuclear vacuolar junction, presence of PA phosphatase Pah1 and endoplasmic reticulum (ER) localized Dgk1; (2) Vacuole, presence of PA phosphatase Dpp1 and phospholipases. It is not clear if Dgk1 localizes to the vacuole; (3) Golgi, presence of PA phosphatase Lpp1. Large pool of DAG is produced by complex sphingolipid biosynthesis; (4) Plasma membrane/actin patches (blue lines), presence of PA phosphatase App1; (5) PA transport into mitochondria is mediated by Ups1-Mdm35. Putative sources of mitochondrial PA are denoted by a question mark.
Abbreviations: CDP-DAG, CDP-diacylglycerol; Cer, ceramide; DAG, diacylglycerol; ER, endoplasmic reticulum; IM, inner membrane; LD, lipid droplet; MAM, mitochondria-associated membrane; OM, outer membrane; PA, phosphatidic acid; PI, phosphatidylinositol; PL, phospholipid; Px, peroxisome; SLs, sphingolipids; TAG, triacylglycerol; vCLAMP, vacuole and mitochondria patch.
An additional DAG pool that results from synthesis of complex SLs is located in the Golgi apparatus. The yeast pathway consumes PI and ceramide made in the ER, while producing inositol-based complex SLs and DAG in the Golgi. It has been proposed that this pool of DAG regulates the G1-to-S transition and proliferation of yeast cells.39 Interestingly, this biosynthetic pathway has the potential of controlling the balance between DAG and ceramide pools, which are two bioactive lipids with antagonistic effects on cell proliferation.40 Ceramide is made in the ER and transported to the Golgi by mechanisms involving vesicular and nonvesicular transport,41 but is not clear if DAG produced in the Golgi is traded in this process. Involvement of DAG in Golgi-to-ER transport through the formation of COPI vesicles has been described in mammalian cells.42 The importance of this pool of DAG is underscored by the essential role of the PI/PC transfer protein Sec14, which regulates protein secretion and DAG homeostasis in the Golgi.43,44 While DAG levels are high in the Golgi (10 mol%) and decrease along the secretory pathway, PA is gradually enriched, displaying moderate abundance in the Golgi (2 mol%) and reaching its maximum concentration at the plasma membrane (10 mol%) (Fig. 1).45 The PA pool detected along the secretory pathway is probably the result of PL turnover catalyzed mainly by Spo14, the yeast phospholipase D (PLD) that generates PA from PC,46 which has been shown to localize to endosomes and plasma membrane in this organism.47,48 It is worth noting that a pathway dependent on PL-remodeling enzymes generates PA in the Golgi of mammalian cells.49 Although these enzymes are conserved in S. cerevisiae, their potential contribution to a Golgi-localized PA pool in yeast has not been studied.
PA and DAG Interconversion
The dephosphorylation of PA to form DAG and the conversion of DAG to PA are catalyzed by the enzymatic activities of phosphatidate phosphatases (PAPs) and DAG kinases, respectively. Investigations conducted in yeast by Carman et al have been at the vanguard of the identification of genes coding for these critical enzymes that control PA and DAG balance in eukaryotes.13,50,51 Four PAP enzymes have been identified to date in yeast. Two integral membrane PAPs are localized to the vacuole (Dpp1) and Golgi (Lpp1), while the other two (Pah1 and App1) are soluble enzymes recruited to membranes to access PA.50,51 Association of Pah1 with membranes is highly regulated through phosphorylation and plays a critical role in the de novo glycerolipid biosynthetic pathway.9,50 Pah1 has been detected at the nucleus–vacuole (NV) junction,52 vacuolar membrane,53 and in the nucleus.54 App1 is the latest PAP discovered, proposed to play a role in endocytosis due to its association with cortical actin patches.51 In contrast to the many PAP isoforms, only one DAG kinase (Dgk1) localized mainly to the ER has been identified in yeast.13 The presence of such array of enzymes in all these cellular compartments further supports the existence of distinct PA and DAG pools at these locations (Fig. 1).
PA and DAG Effector Proteins of Budding Yeast
Although PA and DAG are alike in the fact that they are nonbilayer-forming lipids that introduce negative curvature in cellular membranes, recruitment of proteins by these signaling lipids is notoriously different. An outstanding difference among PA and DAG effector proteins is the conservation of their lipid-binding domains. In the case of DAG, a conserved binding module known as C1 domain (protein kinase C homology 1)55 mediates its specific recognition, while PA-binding proteins do not have a conserved primary amino acid sequence for lipid recognition (Table 3), making it exceptionally difficult to identify them using bioinformatics approaches.
Table 3.
Sequence of binding domains used to visualize pools of PA and DAG in yeast.
| PROTIN | REF | |
|---|---|---|
| PA binding | ||
| Opi1 basic domain [109–138] |
KRQKLSRAIAKGKDNLKEYKLNMSIESKKR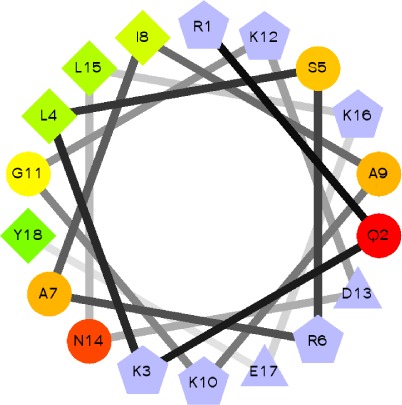
|
11 |
| Spo20[51–91] (no consensus) | MDNCSGSRRRDRLHVKLKSLRNKIHKQLHPNCRFDDATKTSDDKC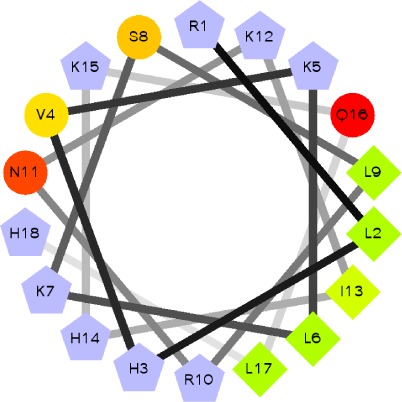
|
73 |
| DAG binding | ||
| C1-PKCδ pKCδC1A pKCδC1B Consensus |

|
102, 103 |
Notes: Minimum sequence that binds PA is in bold, while amino acids that have been shown to be directly involved in PA recognition are underlined. Helical wheel representations of the amphipathic helices were obtained using EMBOSS-Pepwheel.101 The C1δ used in this review is shown. The light and dark gray shades in the tandem C1 domain indicate the sequence of the C1A and C1B regions, respectively. Amino acids conserved in the C1 domains of PKC isoforms α, β, γ, ε, ζ, and θ are highlighted in red.
The conserved DAG-binding module consists of a cysteine-rich zinc finger-like domain, first identified in rat PKCγ.56 Mutagenesis and deletion studies have demonstrated that the C1 domain is essential for DAG binding.2,3,57 Most proteins have two C1 domains, namely C1A and C1B (Table 3), while some can have up to three.55 Each C1 domain generally consists of approximately 50 amino acid residues,55 and to date close to 60 different C1 domains have been identified in proteins ranging from lipid and protein kinases to scaffolding proteins.58 Structural studies revealed that C1 domains are composed of a helix folded around two Zn2+ ions and two antiparallel β-sheets, forming two flexible loops that generate a binding pocket into which DAG is inserted.59 It has been proposed that DAG recruits effector proteins by altering their surface nature and by stabilizing membrane-inserted states rather than by inducing conformational changes.59 The only yeast protein that contains a C1 domain is the yeast ortholog of the mammalian PKC named Pkc1, but the competence of this enzyme to bind DAG still remains controversial.60,61
In great contrast to DAG, the phosphomonoester of the PA headgroup is the first key characteristic of PA that aids in protein-binding specificity.15 Over 20 PA-binding proteins have been described in mammalian, yeast, and plant cells.62,63 In general, the identified PA-binding regions of PA effectors usually contain clustered basic, positively charged amino acid residues. One exception to this requirement of basic residues is the ABI1 plant protein phosphatase 2C-like protein.64 Effector proteins containing amino acids with primary amines, such as lysine and arginine, can act as hydrogen bond donors, while the phosphomonoester of PA acts as a hydrogen bond acceptor, as proposed by the electrostatic/hydrogen bond switch theory.65 In this model, hydrogen bonding results in the destabilization of the proton of the phosphate headgroup causing the proton to dissociate, therefore increasing the negative charge of PA from −1 to −2.15,65 Because of this unique hydrogen-bonding property, it is speculated that PA has the ability to sense changes in physiological pH of cellular compartments, thereby altering the affinity of PA effector proteins.66 The pKa of the phosphomonoester is between 6.9 and 7.9, meaning that the phosphate headgroup can be either protonated or deprotonated, depending on its localization and cellular conditions, such as pH and calcium concentration.15,67 According to the electrostatic/hydrogen bond switch model, PA-binding proteins will have an overall higher affinity for deprotonated, more negatively charged PA than the protonated form.65,66 The overall charge of PA will change as pH increases or decreases, which in turn will be detected by PA effectors to fine-tune biological outcomes. This phenomenon gives PA a unique signaling function and has been proposed to operate in the case of the yeast transcriptional repressor Opi1 (see Opi1 section).68,69
PA can be considered the only cone-shaped anionic lipid in the cell, differentiating it from other anionic lipids such as PS and PI (Table 1). Exposed hydrophobic residues on effector proteins would be able to insert into the membrane bilayer in the space created between the lipid headgroups.62,69 This penetration would contribute not only to increased binding but also to acyl chain recognition by the PA effector protein.23
The two most studied PA-binding proteins in yeast are Opi1 and Spo20 (Table 3). Interestingly, the PA-binding sequences for both of these proteins have a possible overlap with a nuclear localization signal, which suggests that nuclear traffic and PA recognition may be coupled.62
Opi1
Opi1 is a transcriptional repressor, which is normally held at the cytoplasmic leaflet of the ER membrane when cells are grown in the absence of inositol.11 Cooperative binding to PA and the ER membrane protein Scs2 is responsible for Opi1 association with the ER. Upon addition of inositol, synthesis of PI is induced via the CDP-DAG pathway, which consumes PA. This results in depletion of the initial pool of PA in the ER.11 As PA levels decrease, Opi1 is no longer retained at the ER membrane and translocates to the nucleus. Once in the nucleus, Opi1 represses the Ino2/4 transcriptional activator complex,70 which controls the expression of more than 30 genes, many of them involved in glycerolipid metabolism and inositol biosynthesis.11,70
A region of 30 residues containing 11 lysines and arginines in the N-terminal domain of Opi1 was found to be critical in binding PA (Table 3).11 Using point mutations to alanine, six of these basic residues were identified to have the strongest effect on PA binding, most likely because of electrostatic interactions.11 In addition, it has recently been shown that Opi1 preferentially binds shorter chain and more saturated PA species.23 This finding indicates that not only the headgroup but also the acyl chains of PA are critical in the binding of Opi1 to the ER membrane. It is hypothesized that the headgroup recognition is only the first level of interaction between PA and effector proteins. The second level is due to the cone shape of PA to allow for the insertion and subsequent interaction with the acyl chains.15,23 A member of the Opi1 transcription factor family in the yeast Yarrowia lipolytica, Yas3, has also been recently identified to bind to PA.71 Yas3 and Opi1 share only partial sequence similarity in their PA-binding region; however, basic amino acid residues were also identified in Yas3 to be critical for PA binding.71
The binding of Opi1 to PA provides further support of the PA electrostatic/hydrogen bond switch-binding model.15 Under standard growth conditions, yeast cells maintain a close to neutral (~7.0) cytosolic pH, however cytoplasmic acidification can be induced in response to starvation.72 Under conditions of cytosolic acidification, it was found that many lipid metabolic genes were repressed.68 This was shown to be caused by decreased Opi1 binding to PA, leading to the proposal that decreased pH alters the protonation state of PA, resulting in a reduced lipid–protein binding and nuclear translocation.68 Further support to the hypothesis of PA acting as a pH biosensor and a regulator of lipid synthesis in response to nutrient availability69 came from recent investigations showing that membrane recruitment of Pah1 is also regulated by changes in cytosolic pH in response to growth signals.52
Spo20
Spo20 is a meiosis-specific subunit of the t-target soluble N-ethylmaleimide sensitive factor attachment protein receptor (t-SNARE) complex, which is involved in the fusion of vesicles for the formation of the prospore membrane during yeast sporulation.73 Under standard growth conditions, Spo20 is localized to the nucleus; however, once the sporulation cascade begins, it translocates to the plasma membrane, likely by binding to PA.73 A stretch of 40 amino acids, Spo20[51–91] (Table 3) containing both hydrophobic and basic residues were predicted to form an amphipathic α-helix, which positively regulates Spo20 by binding to PA.73 Within this helix, the following five residues were identified to be critical for PA binding: a leucine residue that is also required for Spo20 activity and four basic residues (Table 2).73 For proper function, Spo20 requires an active Spo14.73 This specific pool of PA generated by this PLD appears to be required to properly localize Spo20 during sporulation.74 This involvement of PA in the regulation of membrane fusion interactions has been identified in multiple independent studies in both mammalian and yeast cells.63
Visualizing PA and DAG Pools in Yeast
Opi1, Spo20, and their PA-binding domains fused to green fluorescent protein (GFP) have been extensively used to track PA pools in yeast.13,68,73,75 GFP–Spo20[51–91] localizes to the plasma membrane and nucleus in actively growing cells.73,75 In addition, this PA-binding reporter localizes to aberrant enlarged perinuclear ER when yeast or bacterial DAG kinase is overproduced.13,73 The localization of this fluorescent probe was shown to be altered in conditions of cytosolic acidification.68 On the other hand, full-length Opi1 associates with the perinuclear ER when cells are grown in the absence of inositol, but this localization is dependent not only on its PA-binding domain but also on binding to Scs2.69 In contrast to full-length Opi1, and similar to GFP–Spo20[51–91], the basic domain of Opi1 fused to GFP localizes to the plasma membrane in addition to the ER. Therefore, the PA pool located in the plasma membrane could be considered the predominant PA pool in yeast, and cooperative binding to both PA and Scs2 is needed for Opi1 association with the ER membrane. Interestingly, cytosol acidification altered the distribution of both GFP–Spo20[51–91] and Opi1–GFP.11,68 Recent work by Kohlwein et al revealed additional differences in the localization of Opi1 and the Spo20-based reporter to domains of the nuclear ER that are responsible for LD assembly.75 While Opi1 was enriched in these domains upon incubation of yeast with oleate, no such enrichment was detected for the GFP–Spo20[51–91] probe.75 Differences in the recognition of PA acyl chains between these PA-binding reporters could explain their dissimilar behavior.23,75 It should also be noted that recent studies suggest that while Spo20-based membrane sensors respond to small variations of PA, this interaction is greatly influenced by the membrane environment.76 Furthermore, it has been recently postulated that differences in surface tension or membrane curvature can drive the spatial segregation of proteins containing amphipathic helices, such as Opi1 and Spo20, independent of changes in PA.77
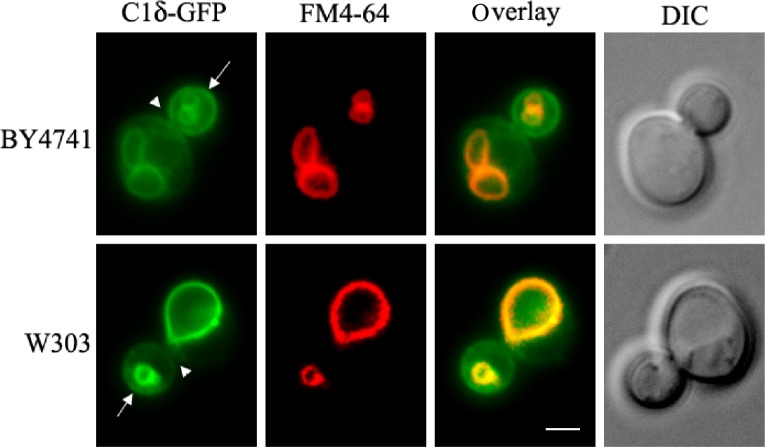
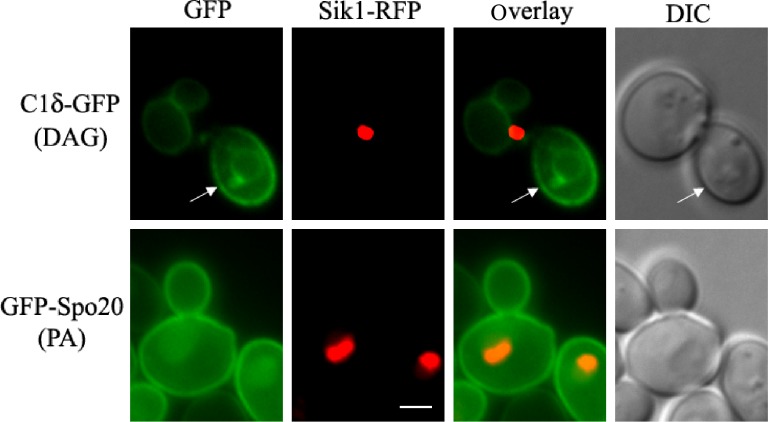
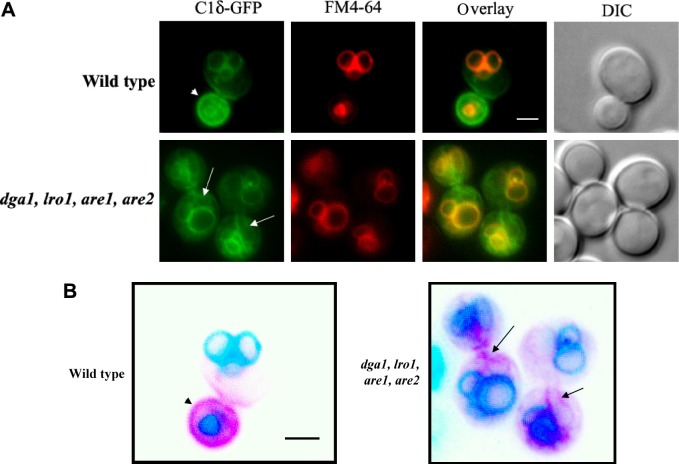
Many DAG-sensing probes have been developed to monitor DAG signaling in mammalian cells. Several C1 domains have been fused to fluorescent proteins that can sense DAG in cells.78,79 To our surprise, no record of the use of such tools for tracking DAG pools in live yeast cells could be found in the literature, aside from vacuolar fusion in vitro assays.20 Similar to mammalian PKCs, yeast Pkc1 has a C1 domain; however, its responsiveness to DAG still needs to be conclusively determined. Localization of Pkc1 fused to GFP has been monitored in live yeast cells.80,81 Pkc1–GFP showed a dynamic spatiotemporal localization at the sites of polarized growth.80 Interestingly, the C1 domain of Pkc1 was sufficient for its peripheral localization to buds and bud neck, suggesting that membrane association is probably mediated by recognition of a lipid component that could be DAG.81 In a recent study, several mammalian PKC isoforms were heterologously expressed in a yeast mutant deficient in Pkc1.82 Among all PKCs tested, PKCδ was the only one able to suppress the phenotype displayed by the lack of Pkc1 in relation to the control of the DNA integrity checkpoint.82 The C1 domain of PKCδ (C1δ) is in fact very well characterized and known to bind DAG with high affinity and has been successfully used as a probe fused to GFP in order to monitor DAG in live cells.83–86 We have recently cloned C1δ into a centromeric yeast expression vector under a constitutive promoter for in vivo visualization of DAG pools. Localization of C1δ–GFP was first monitored in different wild-type strains commonly used by the yeast community, grown under standard conditions (Fig. 2). The main localization of this DAG-binding reporter in actively growing cells was the vacuolar membrane, further confirmed by colocalization with FM4-64, which is a fluorescent dye that gets confined to this compartment after entering cells via endocytosis (Fig. 2).87 This localization is consistent with studies done using similar probes to show that DAG localizes to the membrane of isolated vacuoles from yeast cells.19,20 In fact, DAG has been identified as one of the regulatory lipids required for yeast homotypic vacuole fusion.20 Importantly, no changes in vacuolar morphology of wild-type cells due to expression of C1δ–GFP were detected (data not shown). In addition to vacuolar membrane localization, the C1δ probe also associated with the plasma membrane of daughter cells and at the bud neck (Fig. 2). This resembles the localization of yeast Pkc1,80,81 suggesting that this enzyme may indeed be recognizing a unique DAG pool important for polarized growth. In contrast to GFP–Spo20[51–91], C1δ–GFP did not show nuclear localization as determined by colocalization analysis using the nucleolar marker Sik1-RFP (Fig. 3). We then decided to challenge the validity of using this DAG-binding reporter in yeast, by testing if its localization would change in conditions where lipid homeostasis and levels of DAG are known to be altered. Cells lacking the four enzymes responsible for the final step in the synthesis of triglycerides in yeast, namely DAG acyltransferase Dga1, the transacylase Lro1, and the steryl acyltransferases Are1 and Are2 (4Δ quadruple mutant),88–93 accumulate DAG and have defects in both nuclear and peripheral ER membrane organization.52,89 Localization of the C1δ–GFP probe drastically changed when expressed in 4Δ quadruple mutant cells, clearly revealing its association with the expanded nuclear and cortical ER in addition to the vacuolar membrane (Fig. 4). Additionally, the polarized distribution of C1δ–GFP to the plasma membrane of buds and daughter cells significantly decreased in the quadruple mutant (Fig. 4). These results further support the use of C1δ–GFP as a valuable tool to track cytosolic accessible pools of DAG in yeast.
Figure 2.
C1δ–GFP localization in wild-type yeast. The tandem C1A–C1B domains of PKCδ (Table 2) fused to GFP (C1δ–GFP)102 were cloned into the p416-GPD (CEN, URA3) vector for constitutive expression in yeast. Wild-type cells (BY4741 and W303-1A) carrying this plasmid were grown to mid-log phase in synthetic defined selective medium at 30°C. For vacuolar membrane staining, cells were incubated with 16 μM FM4-64 (Molecular Probes®) for 15 minutes at 30°C and allowed to grow for an additional 30 minutes before the preparation of slides for imaging immediately after. Cells were imaged live using a Zeiss Imager Z.1 epifluorescence microscope. Arrows point to the nonvacuolar GFP signal detected in the plasma membrane of the bud, while arrowheads point to the bud neck. Scale bar = 2 μm.
Abbreviation: DIC, differential interference contrast.
Figure 3.
Comparison of PA and DAG distribution in yeast. Wild-type yeast (BY4741) cells expressing the nucleolar protein Sik1 fused to RFP from the endogenous locus104 was transformed with plasmids carrying C1δ–GFP (CEN, URA3) or GFP–Spo20[51–91] (2 μ, URA3; kindly provided by Neiman et al)73 for the detection of DAG or PA, respectively. Transformants were grown to mid-log phase in synthetic defined selective medium at 30°C. Cells were imaged live using a Zeiss Imager Z.1 epifluorescence microscope. Arrows point to the nonvacuolar GFP signal detected in the plasma membrane of the bud. Scale bar = 2 μm.
Abbreviation: DIC, differential interference contrast.
Figure 4.
Changes in the cellular distribution of DAG in cells that lack triglyceride biosynthetic enzymes. (A) A mutant yeast strain lacking the genes coding for Dga1, Lro1, Are1, and Are2 (quadruple mutant, kindly provided by Athenstaedt)93 and its isogenic wild type (BY4741) were transformed with a plasmid carrying C1δ–GFP (CEN, URA3) for the detection of DAG pools. Transformants were grown to mid-log phase in synthetic defined selective medium at 30°C. Cells were imaged live using a Zeiss Imager Z.1 epifluorescence microscope. (B) Inverted color image of the overlay displayed in (A), highlighting the details of the nonvacuolar (magenta) signal associated with the C1δ–GFP probe in wild-type and quadruple mutants. Arrowhead points to the GFP signal detected in the plasma membrane of the bud in wild-type cells, while arrows indicate nonvacuolar GFP signal associated with enlarged ER in mutant cells. Scale bar = 2 μm.
Abbreviation: DIC, differential interference contrast.
Concluding Remarks
A comparison of the cellular distribution map for PA and DAG based on the use of fluorescent reporters revealed unique compartments demarked by the presence of each of these lipids, with the vacuolar membrane emerging as a large reservoir of DAG. It is important to keep in mind that GFP–Spo20[51–91] and C1δ–GFP detect pools of PA and DAG that are facing the cytosolic leaflet of the membranes in the compartments identified but may not have access to PA or DAG metabolically trapped due to channeling within biosynthetic systems. In addition, GFP–Spo20[51–91] localizes to the nucleus, so it could potentially bind to PA of the nuclear envelope localized to the leaflet facing the nucleoplasm. In contrast to the even distribution of PA along the plasma membrane of both daughter and mother cells in actively growing (log phase) conditions, DAG displayed a polarized distribution, enriched in buds and bud neck-associated plasma membrane. The polarized DAG distribution was lost in cells with impaired TAG synthesis (4Δ quadruple mutant), which are known to have growth defects leading to the loss of cell viability when reaching the stationary phase of growth.89 It would be interesting to investigate if the DAG pool detected at the plasma membrane of buds is derived from TAG breakdown.
The region where the nuclear envelope juxtaposes with the vacuolar membrane has been named NV junction.94 The structural components of this ER contact site are well characterized and have been shown to control selective microautophagy.95 When nutrients are scarce, NV junctions can expand and proliferate, triggering the degradation of portions of the nucleus in the vacuole.95 The NV junction is a place where Pah1 and Dgk1 may alternate localization and where the PA/DAG ratio could emerge as a critical parameter that integrates ER biosynthetic and vacuolar catabolic lipid pathways. This could also be a critical region for the biogenesis of LDs as we frequently see a LD located at each corner of the NV junction in actively growing cells in the presence of glucose.36
Both PA and DAG are nonbilayer-forming lipids that contribute negative curvature to membranes. Therefore, it is not obvious how changes in the PA/DAG ratio would affect the intrinsic curvature. The most evident consequence of altering a PA/DAG balance is that it would impact the negative surface potential of the membrane and the recruitment of effector proteins. In addition, it could have implications on the asymmetric distribution of these lipids, as the conversion of PA to DAG would allow movement of this lipid precursor to the other leaflets of the membrane. Consistent with this rationale is the idea that phosphorylation of DAG would then lock PA on one leaflet of the membrane. Further investigations using these and new tools that could distinguish between PA and DAG pools in each leaflet of a bilayer in vivo, combined with the power of yeast genetics, should aid in addressing these questions and hypotheses.
Acknowledgments
The authors would like to thank Karin Athenstaedt, Aaron Neiman, and Greg Fairn for their kind gifts of plasmids and strains and M. L. Sosa Ponce for her technical contributions.
Footnotes
ACADEMIC EDITOR: Tim Levine, Editor in Chief
PEER REVIEW: Five peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1634 words, excluding any confidential comments to the academic editor.
FUNDING: This study was supported by an Natural Sciences and Engineering Research Council of Canada (NSERC) discovery grant to VZ and an NSERC-Canada Graduate Scholarship Master’s Program (NSERC-CGSM) award to BNS. The authors confirm that the funders had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert single-blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Conceived and designed the experiments: SG and VZ. Analyzed the data: SG, BNS, and VZ. Wrote the first draft of the article: SG and BNS. Contributed to the writing of the article: SG, BNS, and VZ. Agreed the manuscript results and conclusions: SG, BNS, and VZ. Jointly developed the structure and arguments for the article: SG, BNS, and VZ. Made critical revisions and approved the final version: SG, BNS, and VZ. All authors reviewed and approved the final manuscript.
REFERENCES
- 1.Inoue M, Kishimoto A, Takai Y, Nishizuka Y. Studies on a cyclic nucleotide-independent protein kinase and its proenzyme in mammalian tissues. II. Proenzyme and its activation by calcium-dependent protease from rat brain. J Biol Chem. 1977;252(21):7610–7616. [PubMed] [Google Scholar]
- 2.Takai Y, Kishimoto A, Kikkawa U, Mori T, Nishizuka Y. Unsaturated diacylglycerol as a possible messenger for the activation of calcium-activated, phospholipid-dependent protein kinase system. Biochem Biophys Res Commun. 1979;91(4):1218–1224. doi: 10.1016/0006-291x(79)91197-5. [DOI] [PubMed] [Google Scholar]
- 3.Kishimoto A, Takai Y, Mori T, Kikkawa U, Nishizuka Y. Activation of calcium and phospholipid-dependent protein kinase by diacylglycerol, its possible relation to phosphatidylinositol turnover. J Biol Chem. 1980;255(6):2273–2276. [PubMed] [Google Scholar]
- 4.Castagna M, Takai Y, Kaibuchi K, Sano K, Kikkawa U, Nishizuka Y. Direct activation of calcium-activated, phospholipid-dependent protein kinase by tumor-promoting phorbol esters. J Biol Chem. 1982;257(13):7847–7851. [PubMed] [Google Scholar]
- 5.English D. Phosphatidic acid: a lipid messenger involved in intracellular and extracellular signalling. Cell Signal. 1996;8(5):341–347. doi: 10.1016/0898-6568(95)00076-3. [DOI] [PubMed] [Google Scholar]
- 6.English D, Cui Y, Siddiqui RA. Messenger functions of phosphatidic acid. Chem Phys Lipids. 1996;80(1–2):117–132. doi: 10.1016/0009-3084(96)02549-2. [DOI] [PubMed] [Google Scholar]
- 7.Munnik T. Phosphatidic acid: an emerging plant lipid second messenger. Trends Plant Sci. 2001;6(5):227–233. doi: 10.1016/s1360-1385(01)01918-5. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Devaiah SP, Zhang W, Welti R. Signaling functions of phosphatidic acid. Prog Lipid Res. 2006;45(3):250–278. doi: 10.1016/j.plipres.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Henry SA, Kohlwein SD, Carman GM. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics. 2012;190(2):317–349. doi: 10.1534/genetics.111.130286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodgkin MN, Pettitt TR, Martin A, Michell RH, Pemberton AJ, Wakelam MJ. Diacylglycerols and phosphatidates: which molecular species are intracellular messengers? Trends Biochem Sci. 1998;23(6):200–204. doi: 10.1016/s0968-0004(98)01200-6. [DOI] [PubMed] [Google Scholar]
- 11.Loewen CJR, Gaspar ML, Jesch SA, et al. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science. 2004;304(5677):1644–1647. doi: 10.1126/science.1096083. [DOI] [PubMed] [Google Scholar]
- 12.Zhang C, Wendel AA, Keogh MR, Harris TE, Chen J, Coleman RA. Glycerolipid signals alter mTOR complex 2 (mTORC2) to diminish insulin signaling. Proc Natl Acad Sci U S A. 2012;109(5):1667–1672. doi: 10.1073/pnas.1110730109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han G-S, O’Hara L, Carman GM, Siniossoglou S. An unconventional diacylglycerol kinase that regulates phospholipid synthesis and nuclear membrane growth. J Biol Chem. 2008;283(29):20433–20442. doi: 10.1074/jbc.M802903200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster DA. Phosphatidic acid and lipid-sensing by mTOR. Trends Endocrinol Metab. 2013;24(6):272–278. doi: 10.1016/j.tem.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kooijman EE, Burger KNJ. Biophysics and function of phosphatidic acid: a molecular perspective. Biochim Biophys Acta. 2009;1791(9):881–888. doi: 10.1016/j.bbalip.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Israelachvili JN, Marcelja S, Horn RG. Physical principles of membrane organization. Q Rev Biophys. 1980;13(2):121–200. doi: 10.1017/s0033583500001645. [DOI] [PubMed] [Google Scholar]
- 17.Goñi FM, Alonso A. Structure and functional properties of diacylglycerols in membranes. Prog Lipid Res. 1999;38(1):1–48. doi: 10.1016/s0163-7827(98)00021-6. [DOI] [PubMed] [Google Scholar]
- 18.Villar AV, Alonso A, Goñi FM. Leaky vesicle fusion induced by phosphatidylinositol-specific phospholipase C: observation of mixing of vesicular inner monolayers. Biochemistry. 2000;39(46):14012–14018. doi: 10.1021/bi992515c. [DOI] [PubMed] [Google Scholar]
- 19.Jun Y, Fratti RA, Wickner W. Diacylglycerol and its formation by phospholipase C regulate Rab- and SNARE-dependent yeast vacuole fusion. J Biol Chem. 2004;279(51):53186–53195. doi: 10.1074/jbc.M411363200. [DOI] [PubMed] [Google Scholar]
- 20.Fratti RA, Jun Y, Merz AJ, Margolis N, Wickner W. Interdependent assembly of specific regulatory lipids and membrane fusion proteins into the vertex ring domain of docked vacuoles. J Cell Biol. 2004;167(6):1087–1098. doi: 10.1083/jcb.200409068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wakelam MJ. Diacylglycerol—when is it an intracellular messenger? Biochim Biophys Acta. 1998;1436(1–2):117–126. doi: 10.1016/s0005-2760(98)00123-4. [DOI] [PubMed] [Google Scholar]
- 22.Marignani PA, Epand RM, Sebaldt RJ. Acyl chain dependence of diacylglycerol activation of protein kinase C activity in vitro. Biochem Biophys Res Commun. 1996;225(2):469–473. doi: 10.1006/bbrc.1996.1196. [DOI] [PubMed] [Google Scholar]
- 23.Hofbauer HF, Schopf FH, Schleifer H, et al. Regulation of gene expression through a transcriptional repressor that senses acyl-chain length in membrane phospholipids. Dev Cell. 2014;29(6):729–739. doi: 10.1016/j.devcel.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ejsing CS, Sampaio JL, Surendranath V, et al. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci U S A. 2009;106(7):2136–2141. doi: 10.1073/pnas.0811700106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casanovas A, Sprenger RR, Tarasov K, et al. Quantitative analysis of proteome and lipidome dynamics reveals functional regulation of global lipid metabolism. Chem Biol. 2015;22(3):412–425. doi: 10.1016/j.chembiol.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 26.Contreras F-X, Sánchez-Magraner L, Alonso A, Goñi FM. Transbilayer (flip-flop) lipid motion and lipid scrambling in membranes. FEBS Lett. 2010;584(9):1779–1786. doi: 10.1016/j.febslet.2009.12.049. [DOI] [PubMed] [Google Scholar]
- 27.Hamilton JA, Bhamidipati SP, Kodali DR, Small DM. The interfacial conformation and transbilayer movement of diacylglycerols in phospholipid bilayers. J Biol Chem. 1991;266(2):1177–1186. [PubMed] [Google Scholar]
- 28.Cooke IR, Deserno M. Coupling between lipid shape and membrane curvature. Biophys J. 2006;91(2):487–495. doi: 10.1529/biophysj.105.078683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ueda Y, Ishitsuka R, Hullin-Matsuda F, Kobayashi T. Regulation of the transbilayer movement of diacylglycerol in the plasma membrane. Biochimie. 2014;107(pt A):43–50. doi: 10.1016/j.biochi.2014.09.014. [DOI] [PubMed] [Google Scholar]
- 30.Ueda Y, Makino A, Murase-Tamada K, et al. Sphingomyelin regulates the transbilayer movement of diacylglycerol in the plasma membrane of Madin-Darby canine kidney cells. FASEB J. 2013;27(8):3284–3297. doi: 10.1096/fj.12-226548. [DOI] [PubMed] [Google Scholar]
- 31.Connerth M, Tatsuta T, Haag M, Klecker T, Westermann B, Langer T. Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science. 2012;338(6108):815–818. doi: 10.1126/science.1225625. [DOI] [PubMed] [Google Scholar]
- 32.Potting C, Tatsuta T, König T, et al. TRIAP1/PRELI complexes prevent apoptosis by mediating intramitochondrial transport of phosphatidic acid. Cell Metab. 2013;18(2):287–295. doi: 10.1016/j.cmet.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 33.Elbaz-Alon Y, Rosenfeld-Gur E, Shinder V, Futerman AH, Geiger T, Schuldiner M. A dynamic interface between vacuoles and mitochondria in yeast. Dev Cell. 2014;30(1):95–102. doi: 10.1016/j.devcel.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 34.Pu J, Ha CW, Zhang S, Jung JP, Huh W-K, Liu P. Interactomic study on interaction between lipid droplets and mitochondria. Protein Cell. 2011;2(6):487–496. doi: 10.1007/s13238-011-1061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daum G, Wagner B, Arrey TN, et al. Lipid particles/droplets of the yeast Saccharomyces cerevisiae revisited: lipidome meets Proteome. Biochim Biophys Acta—Mol Cell Biol Lipids. 2011;1811(12):1165–1176. doi: 10.1016/j.bbalip.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marr N, Foglia J, Terebiznik M, Athenstaedt K, Zaremberg V. Controlling lipid fluxes at glycerol-3-phosphate acyltransferase step in yeast: unique contribution of Gat1p to oleic acid-induced lipid particle formation. J Biol Chem. 2012;287(13):10251–10264. doi: 10.1074/jbc.M111.314112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Athenstaedt K, Daum G. 1-Acyldihydroxyacetone-phosphate reductase (Ayr1p) of the yeast Saccharomyces cerevisiae encoded by the open reading frame YIL124w is a major component of lipid particles. J Biol Chem. 2000;275(1):235–240. doi: 10.1074/jbc.275.1.235. [DOI] [PubMed] [Google Scholar]
- 38.Van Zutphen T, Todde V, de Boer R, et al. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol Biol Cell. 2014;25(2):290–301. doi: 10.1091/mbc.E13-08-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cerbón J, Falcon A, Hernández-Luna C, Segura-Cobos D. Inositol phosphoceramide synthase is a regulator of intracellular levels of diacylglycerol and ceramide during the G 1 to S transition in Saccharomyces cerevisiae. Biochem J. 2005;388(1):169–176. doi: 10.1042/BJ20040475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Becker KP, Hannun YA. Protein kinase C and phospholipase D: intimate interactions in intracellular signaling. Cell Mol Life Sci. 2005;62(13):1448–1461. doi: 10.1007/s00018-005-4531-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kajiwara K, Ikeda A, Aguilera-Romero A, et al. Osh proteins regulate COPII-mediated vesicular transport of ceramide from the endoplasmic reticulum in budding yeast. J Cell Sci. 2013;127(2):376–387. doi: 10.1242/jcs.132001. [DOI] [PubMed] [Google Scholar]
- 42.Fernández-Ulibarri I, Vilella M, Lázaro-Diéguez F, et al. Diacylglycerol is required for the formation of COPI vesicles in the Golgi-to-ER transport pathway. Mol Biol Cell. 2007;18(9):3250–3263. doi: 10.1091/mbc.E07-04-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henneberry AL, Lagace TA, Ridgway ND, McMaster CR. Phosphatidylcholine synthesis influences the diacylglycerol homeostasis required for SEC14p-dependent Golgi function and cell growth. Mol Biol Cell. 2001;12(3):511–520. doi: 10.1091/mbc.12.3.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kearns BG, McGee TP, Mayinger P, et al. Essential role for diacylglycerol in protein transport from the yeast Golgi complex. Nature. 1997;387(6628):101–105. doi: 10.1038/387101a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Surma MA, Klose C, Klemm RW, Ejsing CS, Simons K. Generic sorting of raft lipids into secretory vesicles in yeast. Traffic. 2011;12(9):1139–1147. doi: 10.1111/j.1600-0854.2011.01221.x. [DOI] [PubMed] [Google Scholar]
- 46.Rose K, Rudge SA, Frohman MA, Morris AJ, Engebrecht J. Phospholipase D signaling is essential for meiosis. Proc Natl Acad Sci U S A. 1995;92(26):12151–12155. doi: 10.1073/pnas.92.26.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li X, Routt SM, Xie Z, et al. Identification of a novel family of nonclassic yeast phosphatidylinositol transfer proteins whose function modulates phospholipase D activity and Sec14p-independent cell growth. Mol Biol Cell. 2000;11(6):1989–2005. doi: 10.1091/mbc.11.6.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Routt SM, Ryan MM, Tyeryar K, et al. Nonclassical PITPs activate PLD via the Stt4p PtdIns-4-kinase and modulate function of late stages of exocytosis in vegetative yeast. Traffic. 2005;6(12):1157–1172. doi: 10.1111/j.1600-0854.2005.00350.x. [DOI] [PubMed] [Google Scholar]
- 49.Schmidt JA, Brown WJ. Lysophosphatidic acid acyltransferase 3 regulates Golgi complex structure and function. J Cell Biol. 2009;186(2):211–218. doi: 10.1083/jcb.200904147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pascual F, Carman GM. Phosphatidate phosphatase, a key regulator of lipid homeostasis. Biochim Biophys Acta—Mol Cell Biol Lipids. 2013;1831(3):514–522. doi: 10.1016/j.bbalip.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chae M, Carman GM. Characterization of the yeast actin patch protein App1p phosphatidate phosphatase. J Biol Chem. 2013;288(9):6427–6437. doi: 10.1074/jbc.M112.449629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Barbosa AD, Sembongi H, Su W-M, et al. Lipid partitioning at the nuclear envelope controls membrane biogenesis. Mol Biol Cell. 2015;26(20):3641–3657. doi: 10.1091/mbc.E15-03-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sasser T, Qiu Q-S, Karunakaran S, et al. Yeast lipin 1 orthologue pah1p regulates vacuole homeostasis and membrane fusion. J Biol Chem. 2012;287(3):2221–2236. doi: 10.1074/jbc.M111.317420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Santos-Rosa H, Leung J, Grimsey N, Peak-Chew S, Siniossoglou S. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 2005;24(11):1931–1941. doi: 10.1038/sj.emboj.7600672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hurley JH, Newton AC, Parker PJ, Blumberg PM, Nishizuka Y. Taxonomy and function of C1 protein kinase C homology domains. Protein Sci. 1997;6(2):477–480. doi: 10.1002/pro.5560060228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ono Y, Fujii T, Igarashi K, et al. Phorbol ester binding to protein kinase C requires a cysteine-rich zinc-finger-like sequence. Proc Natl Acad Sci U S A. 1989;86(13):4868–4871. doi: 10.1073/pnas.86.13.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Quest AF, Bell RM. The regulatory region of protein kinase C gamma. Studies of phorbol ester binding to individual and combined functional segments expressed as glutathione S-transferase fusion proteins indicate a complex mechanism of regulation by phospholipids, phorbol esters, and divalent cations. J Biol Chem. 1994;269(31):20000–20012. [PubMed] [Google Scholar]
- 58.Das J, Rahman GM. C1 domains: structure and ligand-binding properties. Chem Rev. 2014;114(24):12108–12131. doi: 10.1021/cr300481j. [DOI] [PubMed] [Google Scholar]
- 59.Zhang G, Kazanietz MG, Blumberg PM, Hurley JH. Crystal structure of the cys2 activator-binding domain of protein kinase C delta in complex with phorbol ester. Cell. 1995;81(6):917–924. doi: 10.1016/0092-8674(95)90011-x. [DOI] [PubMed] [Google Scholar]
- 60.Watanabe M, Chen CY, Levin DE. Saccharomyces cerevisiae PKC1 encodes a protein kinase C (PKC) homolog with a substrate specificity similar to that of mammalian PKC. J Biol Chem. 1994;269(24):16829–16836. [PubMed] [Google Scholar]
- 61.Levin DE, Fields FO, Kunisawa R, Bishop JM, Thorner J. A candidate protein kinase C gene, PKC1, is required for the S. cerevisiae cell cycle. Cell. 1990;62(2):213–224. doi: 10.1016/0092-8674(90)90360-q. [DOI] [PubMed] [Google Scholar]
- 62.Stace CL, Ktistakis NT. Phosphatidic acid- and phosphatidylserine-binding proteins. Biochim Biophys Acta. 2006;1761(8):913–926. doi: 10.1016/j.bbalip.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 63.Raghu P, Manifava M, Coadwell J, Ktistakis NT. Emerging findings from studies of phospholipase D in model organisms (and a short update on phosphatidic acid effectors) Biochim Biophys Acta. 2009;1791(9):889–897. doi: 10.1016/j.bbalip.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 64.Zhang W, Qin C, Zhao J, Wang X. Phospholipase D alpha 1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proc Natl Acad Sci U S A. 2004;101(25):9508–9513. doi: 10.1073/pnas.0402112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kooijman EE, Tieleman DP, Testerink C, et al. An electrostatic/hydrogen bond switch as the basis for the specific interaction of phosphatidic acid with proteins. J Biol Chem. 2007;282(15):11356–11364. doi: 10.1074/jbc.M609737200. [DOI] [PubMed] [Google Scholar]
- 66.Loew S, Kooijman EE, May S. Increased pH-sensitivity of protein binding to lipid membranes through the electrostatic-hydrogen bond switch. Chem Phys Lipids. 2013;169:9–18. doi: 10.1016/j.chemphyslip.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 67.Kooijman EE, Carter KM, van Laar EG, Chupin V, Burger KNJ, de Kruijff B. What makes the bioactive lipids phosphatidic acid and lysophosphatidic acid so special? Biochemistry. 2005;44(51):17007–17015. doi: 10.1021/bi0518794. [DOI] [PubMed] [Google Scholar]
- 68.Young BP, Shin JJH, Orij R, et al. Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science. 2010;329(5995):1085–1088. doi: 10.1126/science.1191026. [DOI] [PubMed] [Google Scholar]
- 69.Shin JJH, Loewen CJR. Putting the pH into phosphatidic acid signaling. BMC Biol. 2011;9:85. doi: 10.1186/1741-7007-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Carman GM, Henry SA. Phosphatidic acid plays a central role in the transcriptional regulation of glycerophospholipid synthesis in Saccharomyces cerevisiae. J Biol Chem. 2007;282(52):37293–37297. doi: 10.1074/jbc.R700038200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kobayashi S, Hirakawa K, Horiuchi H, Fukuda R, Ohta A. Phosphatidic acid and phosphoinositides facilitate liposome association of Yas3p and potentiate derepression of ARE1 (alkane-responsive element one)-mediated transcription control. Fungal Genet Biol. 2013;61:100–110. doi: 10.1016/j.fgb.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 72.Martínez-Muñoz GA, Kane P. Vacuolar and plasma membrane proton pumps collaborate to achieve cytosolic pH homeostasis in yeast. J Biol Chem. 2008;283(29):20309–20319. doi: 10.1074/jbc.M710470200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nakanishi H, de los Santos P, Neiman AM. Positive and negative regulation of a SNARE protein by control of intracellular localization. Mol Biol Cell. 2004;15(4):1802–1815. doi: 10.1091/mbc.E03-11-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu S, Wilson KA, Rice-Stitt T, Neiman AM, McNew JA. In vitro fusion catalyzed by the sporulation-specific t-SNARE light-chain Spo20p is stimulated by phosphatidic acid. Traffic. 2007;8(11):1630–1643. doi: 10.1111/j.1600-0854.2007.00628.x. [DOI] [PubMed] [Google Scholar]
- 75.Wolinski H, Hofbauer HF, Hellauer K, et al. Seipin is involved in the regulation of phosphatidic acid metabolism at a subdomain of the nuclear envelope in yeast. Biochim Biophys Acta. 2015;1851(11):1450–1464. doi: 10.1016/j.bbalip.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 76.Horchani H, de Saint-Jean M, Barelli H, Antonny B. Interaction of the Spo20 membrane-sensor motif with phosphatidic acid and other anionic lipids, and influence of the membrane environment. PLoS One. 2014;9(11):e113484. doi: 10.1371/journal.pone.0113484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grippa A, Buxó L, Mora G, et al. The seipin complex Fld1/Ldb16 stabilizes ER-lipid droplet contact sites. J Cell Biol. 2015;211(4):829–844. doi: 10.1083/jcb.201502070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Oancea E, Teruel MN, Quest AF, Meyer T. Green fluorescent protein (GFP)-tagged cysteine-rich domains from protein kinase C as fluorescent indicators for diacylglycerol signaling in living cells. J Cell Biol. 1998;140(3):485–498. doi: 10.1083/jcb.140.3.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Oancea E, Meyer T. Protein kinase C as a molecular machine for decoding calcium and diacylglycerol signals. Cell. 1998;95(3):307–318. doi: 10.1016/s0092-8674(00)81763-8. [DOI] [PubMed] [Google Scholar]
- 80.Andrews PD, Stark MJR. Dynamic, Rho1p-dependent localization of Pkc1p to sites of polarized growth. J Cell Sci. 2000;113(15):2685–2693. doi: 10.1242/jcs.113.15.2685. [DOI] [PubMed] [Google Scholar]
- 81.Denis V, Cyert MS. Molecular analysis reveals localization of Saccharomyces cerevisiae protein kinase C to sites of polarized growth and Pkc1p targeting to the nucleus and mitotic spindle. Eukaryot Cell. 2005;4(1):36–45. doi: 10.1128/EC.4.1.36-45.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Soriano-Carot M, Quilis I, Bano MC, Igual JC. Protein kinase C controls activation of the DNA integrity checkpoint. Nucleic Acids Res. 2014;42(11):7084–7095. doi: 10.1093/nar/gku373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ohashi N, Nomura W, Kato M, et al. Synthesis of protein kinase Cdelta C1b domain by native chemical ligation methodology and characterization of its folding and ligand binding. J Pept Sci. 2009;15(10):642–646. doi: 10.1002/psc.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stahelin RV, Digman MA, Medkova M, et al. Mechanism of diacylglycerol-induced membrane targeting and activation of protein kinase Cdelta. J Biol Chem. 2004;279(28):29501–29512. doi: 10.1074/jbc.M403191200. [DOI] [PubMed] [Google Scholar]
- 85.Stahelin RV, Digman MA, Medkova M, et al. Diacylglycerol-induced membrane targeting and activation of protein kinase Cε: mechanistic differences between protein kinases Cδ and C&Vegr. J Biol Chem. 2005;280(20):19784–19793. doi: 10.1074/jbc.M411285200. [DOI] [PubMed] [Google Scholar]
- 86.Sánchez-Bautista S, Corbalán-García S, Pérez-Lara A, Gómez-Fernández JC. A comparison of the membrane binding properties of C1B domains of PKC-gamma, PKCdelta, and PKCepsilon. Biophys J. 2009;96(9):3638–3647. doi: 10.1016/j.bpj.2009.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128(5):779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sorger D, Athenstaedt K, Hrastnik C, Daum G. A yeast strain lacking ipid particles bears a defect in ergosterol formation. J Biol Chem. 2004;279(30):31190–31196. doi: 10.1074/jbc.M403251200. [DOI] [PubMed] [Google Scholar]
- 89.Petschnigg J, Wolinski H, Kolb D, et al. Good fat, essential cellular requirements for triacylglycerol synthesis to maintain membrane homeostasis in yeast. J Biol Chem. 2009;284(45):30981–30993. doi: 10.1074/jbc.M109.024752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Oelkers P, Cromley D, Padamsee M, Billheimer JT, Sturley SL. The DGA1 gene determines a second triglyceride synthetic pathway in yeast. J Biol Chem. 2002;277(11):8877–8881. doi: 10.1074/jbc.M111646200. [DOI] [PubMed] [Google Scholar]
- 91.Garbarino J, Padamsee M, Wilcox L, et al. Sterol and diacylglycerol acyltransferase deficiency triggers fatty acid-mediated cell death. J Biol Chem. 2009;284(45):30994–31005. doi: 10.1074/jbc.M109.050443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schmidt C, Athenstaedt K, Koch B, Ploier B, Daum G. Regulation of the yeast triacylglycerol lipase Tgl3p by formation of nonpolar lipids. J Biol Chem. 2013;288(27):19939–19948. doi: 10.1074/jbc.M113.459610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Athenstaedt K. YALI0E32769g (DGA1) and YALI0E16797g (LRO1) encode major triacylglycerol synthases of the oleaginous yeast Yarrowia lipolytica. Biochim Biophys Acta—Mol Cell Biol Lipids. 2011;1811(10):587–596. doi: 10.1016/j.bbalip.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pan X, Roberts P, Chen Y, et al. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol Biol Cell. 2000;11(7):2445–2457. doi: 10.1091/mbc.11.7.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kvam E, Goldfarb DS. Nucleus-vacuole junctions in yeast: anatomy of a membrane contact site. Biochem Soc Trans. 2006;34(pt 3):340–342. doi: 10.1042/BST0340340. [DOI] [PubMed] [Google Scholar]
- 96.Klose C, Surma MA, Gerl MJ, Meyenhofer F, Shevchenko A, Simons K. Flexibility of a eukaryotic lipidome— insights from yeast lipidomics. PLoS One. 2012;7(4):e35063. doi: 10.1371/journal.pone.0035063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Klemm RW, Ejsing CS, Surma MA, et al. Segregation of sphingolipids and sterols during formation of secretory vesicles at the trans-Golgi network. J Cell Biol. 2009;185(4):601–612. doi: 10.1083/jcb.200901145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Horvath SE, Daum G. Lipids of mitochondria. Prog Lipid Res. 2013;52(4):590–614. doi: 10.1016/j.plipres.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 99.Mendonsa R, Engebrecht J. Phospholipase D function in Saccharomyces cerevisiae. Biochim Biophys Acta—Mol Cell Biol Lipids. 2009;1791(9):970–974. doi: 10.1016/j.bbalip.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 100.Vögtle F-N, Keller M, Taskin AA, et al. The fusogenic lipid phosphatidic acid promotes the biogenesis of mitochondrial outer membrane protein Ugo1. J Cell Biol. 2015;210(6):951–960. doi: 10.1083/jcb.201506085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16(6):276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 102.Codazzi F, Teruel MN, Meyer T. Control of astrocyte Ca(2+) oscillations and waves by oscillating translocation and activation of protein kinase C. Curr Biol. 2001;11(14):1089–1097. doi: 10.1016/s0960-9822(01)00326-8. [DOI] [PubMed] [Google Scholar]
- 103.Steinberg SF. Structural basis of protein kinase C isoform function. Physiol Rev. 2008;88(4):1341–1378. doi: 10.1152/physrev.00034.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Huh W-K, Falvo JV, Gerke LC, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425(6959):686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]