Abstract
RNA nanotechnology harnesses RNA structural motifs to build nano-sized architectures which assemble through selective base pair interactions. Here, we report the crystal structure-guided design of highly stable RNA nanotriangles that self-assemble cooperatively from short oligonucleotides. The crystal structure of an 81 nucleotide nanotriangle determined at 2.6 Å reveals the yet smallest circularly closed nano-object made entirely of double-stranded RNA. Assembly of the nanotriangle architecture involved RNA corner motifs that were derived from ligand-responsive RNA switches which offer the opportunity to control self-assembly and dissociation.
Keywords: RNA structures, X-ray diffraction, self-assembly, RNA switch, nanostructure design
Graphical Abstract
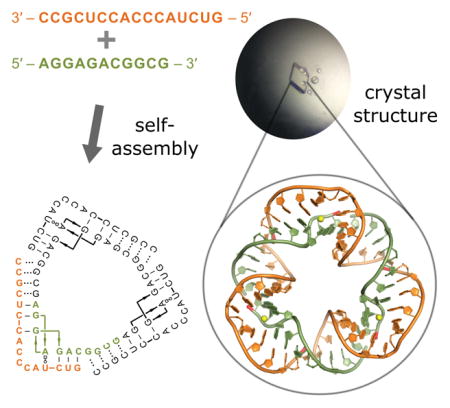
RNA nanostructure design and characterization: Two short oligonucleotides self-assemble cooperatively in solution to form the yet smallest circularly closed nanotriangle made entirely of double-stranded RNA. This nano-object formed crystals, and its structure was determined by X-ray diffraction at 2.6 Å resolution.
Nucleic acid has been used extensively to build nano-sized objects by controlling assembly through designed base pair interactions. Complex nano-objects have been obtained by recursive folding of long nucleic acid sequences through inclusion of alternating double- and single-stranded regions, junctions and helper oligonucleotides. The design of nano-objects has exploited structural motifs observed in crystal structures.[1–14] We have previously used short oligonucleotides to construct a self-assembling RNA nanosquare of 100 nucleotides.[12] Here we describe the crystal structure-guided design of RNA nanotriangles that self-assemble in a cooperative process from multiple copies of short oligonucleotides. Crystal structure analysis of an RNA triangle containing 81 nucleotides reveals the yet smallest circularly closed nano-object made entirely of double-stranded RNA. Nanotriangle self-assembly and dissociation is sequence-dependent and may be modulated by ligands which bind recognition motifs incorporated into the RNA architectures.
Recently, the simplest known ligand-responsive RNA switches have been discovered in the internal ribosome entry sites (IRES) of positive strand RNA viruses of the Flavi- and Picornaviridae families. These RNA switch motifs are located in subdomain IIa of IRES elements and regulate viral protein synthesis through a ligand-dependent conformational transition.[15,16] Unlike traditional riboswitches, viral RNA switches do not undergo secondary structure changes but rather a purely mechanical switching between two distinct and stable conformations. The viral RNA motifs adopt a bent conformation in the absence of a ligand while an elongated conformation is captured by a bound ligand. Structures of ligand-free IIa switches from hepatitis C virus (HCV) and Seneca Valley virus (SVV) IRES elements as well as the ligand-bound switch from the HCV IRES were previously determined by X-ray crystallography.[16–18] The ligand-free IIa switch motif from HCV has previously been used as a corner building block to design and construct a self-assembling nanosquare and a nanoprism.[12,19] We concluded that other RNA nano-objects may be obtained from viral IIa switch motifs by rational design including variation in length of the helices flanking each corner unit and adjusting the orientation of corners relative to a common plane.
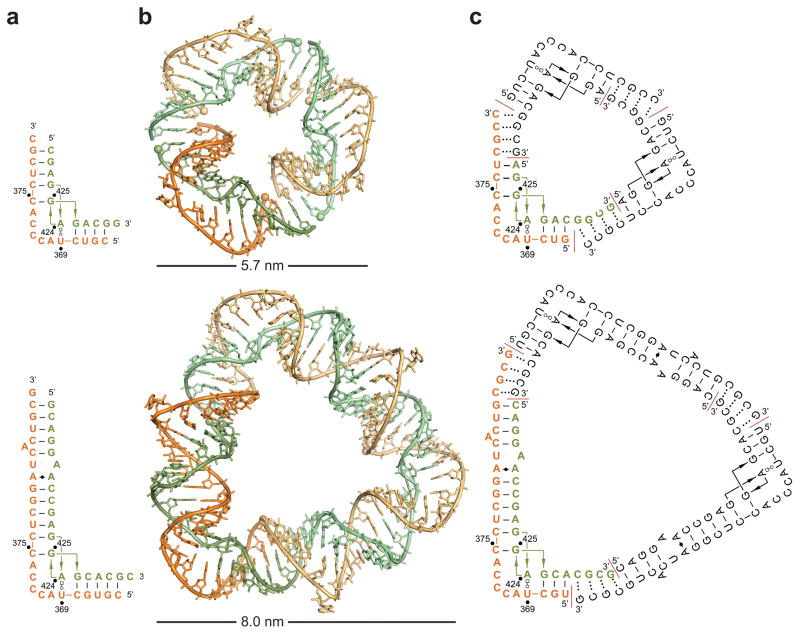
Crystal structure analysis of the IIa corner motif from the SVV IRES provided design directions to construct triangular nano-objects (Figure 1). The three-dimensional structure of the SVV motif was obtained from crystals of a short and long RNA construct, both of which showed identical corner structures.[16] Crystal packing of both constructs revealed circularly closed triangles involving pseudo-continuous stacking and intermolecular base pair formation between a 3′ terminal overhanging nucleotide on both ends of each of three identical corner units (Figure 1b; Figures S1–S3 in the Supporting Information). Self-assembling nanotriangle constructs were designed guided by the triangular arrangement seen in the SVV IIa RNA crystal packing. To promote self-assembly of corner motifs into a triangle, constructs were designed with the same total number of nucleotides and base identity but varying 3′ terminal overhang length. Secondary structure models of exemplary self-assembling constructs containing four nucleotide overhangs are shown in Figure 1c. A small triangle is constructed from two oligonucleotides, including an inner and outer strand of 11 and 16 residues, respectively (Figure 1c, top). Three copies of each the inner and outer strands are designed to self-assemble as a single small triangle of 81 nucleotides. Similarly, a large triangle is constructed from strands of 20 and 26 residues to form a nano-object of 138 nucleotides (Figure 1c, bottom).
Figure 1.
Design of self-assembling RNA nanotriangles. a) Secondary structures of short and long oligonucleotide constructs representing the subdomain IIa motif from the SVV IRES. Single nucleotide overhangs aided crystallization by facilitating RNA packing. Residue numbering refers to the SVV genome. b) Circularly closed triangles seen in the packing of both short and long IIa RNA crystal structures (PDB ID: 4P97 and 4PHY). c) Secondary structure models of self-assembling RNA triangles containing four nucleotide overhangs and designed using the crystal packing of short and long IIa constructs. Red lines indicate oligonucleotide termini.
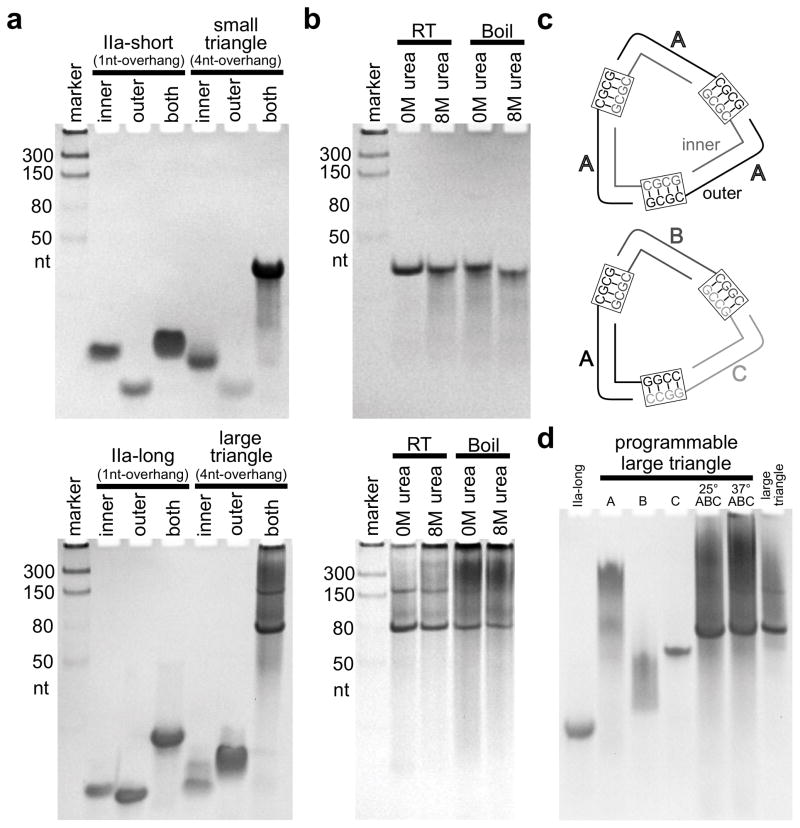
SVV short and long IIa corner motif constructs (Figure 1a) which contain single 3′ nucleotide overhangs were analyzed by native polyacrylamide gel electrophoresis (PAGE) and found to migrate as single bands consistent with their respective size (Figure 2a, 1nt-overhang both; see the Supporting Information for all experimental details). In contrast, nanotriangle RNA constructs carrying four overhanging nucleotides (Figure 1c) to promote self-assembly were found to migrate slower and consistent with the size of symmetrical triangles comprised of three identical corners (Figure 2a; 4nt-overhang both). The small triangle construct migrated as a single band, while the large triangle construct gave rise to a faster moving major band and a slower moving minor band, the latter consistent with the size of a dimer of triangles (see Figure S4 for discussion). The faster moving major band was confirmed as a single triangle of three corners by comparative analysis of an identically sized programmable triangle that contains three distinct corners (A, B, C), each with a unique single-stranded overhang sequence which allows formation exclusively of the designed triangle with an A-B-C configuration but not other assemblies (Figure 2c,d). Both RNA nanotriangles were evaluated for chemical and thermostability, and shown to resist boiling temperature as well as incubation with 8M urea (Figure 2b, see the Supporting Information for discussion).
Figure 2.
Nanotriangle assembly and stability. Structures were analyzed by native PAGE in the presence of 2.5mM MgCl2. a) Top: Assembly of subdomain IIa short crystal construct containing single nucleotide (nt) overhangs (secondary structure shown in Figure 1a, top) and self-assembling small triangle construct containing four nucleotide overhangs (secondary structure shown in Figure 1c, top). Bottom: Assembly of subdomain IIa long crystal construct containing single nucleotide overhangs (secondary structure shown in Figure 1a, bottom) and self-assembling large triangle construct containing four nucleotide overhangs (panel c, top; secondary structure shown in Figure 1c, bottom). b) Stability of small triangle (top) and large triangle (bottom) when treated with or without 8M urea at room temperature (RT) or boiling. c) Diagram of large triangle (AAA, top) and programmable large triangle (ABC, bottom). d) Assembly of programmable large triangle (ABC) from corners A, B, and C.
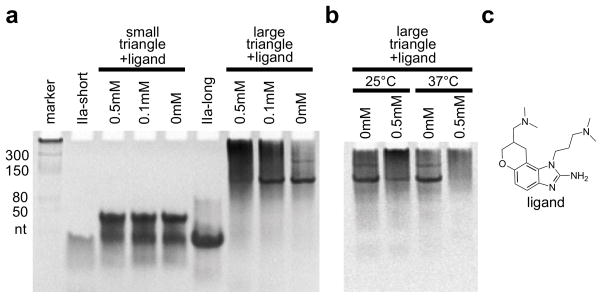
Preliminary studies were performed to determine effects of switch-binding ligands on RNA nanotriangle assembly and dissociation. Native PAGE analysis revealed a decrease in large triangle assembly efficiency in the presence of 500uM ligand[20] (Figure 3a,c). As a consequence of the ligand capture of elongated IIa RNA switches in the self-assembling constructs, end-to-end association of straightened corner units of the large triangle led to multimer formation. This was not observed when ligand was added to the small triangle, likely due to the lability of the construct which has only 7 base pairs flanking an internal loop of 5 unpaired bases and forms by all-or-nothing assembly from single strands. Dissociation of already assembled large triangles, which are less compact and stabilized by 15 base pairs flanking the internal loop, was observed upon incubation with ligand for the symmetrical (Figure 3b) as well as the programmable construct (Figure S6). These findings suggest that the RNA corner units retain their function as ligand-responsive switches when incorporated into nanotriangles. The ligand-triggered dissociation of nanotriangles and assembly of alternate structures opens potential avenues for the construction of RNA nano-objects that respond to environmental signals.
Figure 3.
Nanotriangle self-assembly efficiency and dissociation in the presence of ligand. Ligand binding of the IIa switch captures an elongated conformation of the RNA.[16] a) Assembly of the small triangle is not affected in the presence of a binding ligand. Assembly of the large triangle is partially prevented in the presence of a binding ligand. Single corner units of the large triangle construct, captured by the ligand in an elongated conformation, prevent triangle formation and promotes end-to-end multimerization into longer species. b) Dissociation of large triangles was observed when incubated post-assembly with a binding ligand. c) Structure of the benzimidazole ligand which binds to the IIa RNA switch element of the nanotriangles.
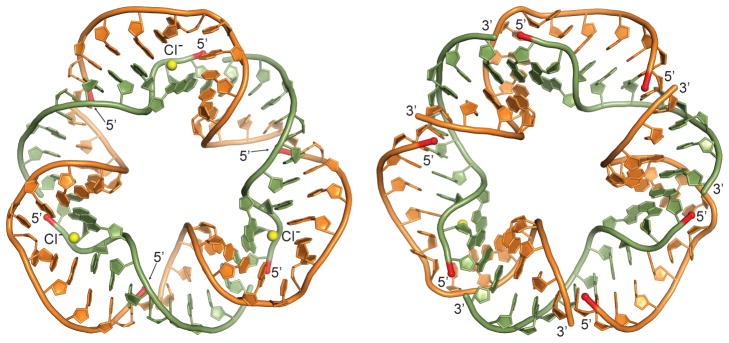
To investigate the three-dimensional structure of the small and large RNA nanotriangles, we crystallized both constructs. Well-diffracting crystals were obtained for the small triangle. Structure determination by X-ray diffraction at 2.6 Å resolution (Figures 4, S7–S10 and Table S1) revealed a circularly closed and continuously double-stranded RNA which exhibited an architecture similar to the pseudo-continuously closed small triangle seen in the crystal packing of the short IIa construct (Figures 4 and 1b). As designed, the nanotriangle comprises three identical and symmetrical corner units, each forming from an inner and outer RNA strand with four overhanging nucleotides which hybridize with neighboring corner strands. Triangle sides are composed of 11 base pairs and measure ~5 nm in length, while corners contain the IIa internal loop of 5 bases. The overall structure appears hexagonal at first glance as it is not planar but has distorted sides that twist to accommodate three 90° corner motifs in a closed triangular architecture. The resulting structure is more compact than a planar triangle of comparable side length. The 12 termini of the 6 single strands constituting the RNA triangle are located on the same face of the nanostructure (Figures 4 and S7a). This feature offers an opportunity to build more complex structures and to functionalize the nanotriangle for sensor and materials applications.
Figure 4.
Crystal structure of the self-assembling RNA nanotriangle. Views from both sides of the triangle plane are shown. The back view (left) reveals three Cl− ions (yellow spheres) bound at the Watson-Crick edge of A374 and C375. The terminal residues of all constituting oligonucleotides reside on one face of the triangle (front view, right). 5′ termini are highlighted in red. Atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB ID: 5CNR).
The design of nano-scale objects that self-assemble from short oligonucleotides remains a key challenge in the emerging field of RNA nanotechnology. While larger nano-architectures have previously been constructed from structurally complex RNA motifs or long nucleic acid sequences in conjunction with helper oligonucleotides, we aimed at creating minimalist RNA nano-objects by efficient assembly of short sequences which by themselves do not adopt stable structures. We exploited detailed insight from X-ray crystallography to design and construct two different RNA nanotriangles that self-assembled from six oligonucleotides in solution and were crystallized for structural studies. The nanotriangles display remarkable stability towards denaturation and their composition offers unique structural features that promise applications in medicine, nanomaterials engineering, and as tools to test nano-scale phenomena. As a consequence of the hierarchical self-assembly from six short oligonucleotides, the RNA nanotriangles will be readily modified by conjugation at any of the 12 component strand termini to introduce additional functionality. Since the corner units used in the construction of the nanotriangles are also ligand-dependent conformational switches, association and dissociation of the resulting RNA architectures is tunable which will enable the design of nanodevices sensitive to environmental or cellular milieus.
Supplementary Material
Acknowledgments
We thank Andrew Bergdorf for help setting up crystal screens. M. A. B. was supported by a US Department of Education GAANN fellowship. Research was supported by the UCSD Academic Senate, grant No. RM069B. Instrumentation at the Biomolecule Crystallography Facility was acquired with funding from the National Institutes of Health, grant OD011957.
Footnotes
Supporting information for this article is available on the WWW
References
- 1.Jaeger L, Leontis NB. Angew Chem Int Ed Engl. 2000;39:2521. doi: 10.1002/1521-3773(20000717)39:14<2521::aid-anie2521>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2000;112:2576. [Google Scholar]
- 2.Shu D, Huang LP, Hoeprich S, Guo P. J Nanosci Nanotechnol. 2003;3:295. doi: 10.1166/jnn.2003.160. [DOI] [PubMed] [Google Scholar]
- 3.Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, Hansma HG, Jaeger L. Science. 2004;306:2068. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 4.Nasalean L, Baudrey S, Leontis NB, Jaeger L. Nucleic Acids Res. 2006;34:1381. doi: 10.1093/nar/gkl008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bindewald E, Grunewald C, Boyle B, O’Connor M, Shapiro BA. J Mol Graph Model. 2008;27:299. doi: 10.1016/j.jmgm.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Severcan I, Geary C, Verzemnieks E, Chworos A, Jaeger L. Nano Lett. 2009;9:1270. doi: 10.1021/nl900261h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Severcan I, Geary C, Chworos A, Voss N, Jacovetty E, Jaeger L. Nat Chem. 2010;2:772. doi: 10.1038/nchem.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Afonin KA, Bindewald E, Yaghoubian AJ, Voss N, Jacovetty E, Shapiro BA, Jaeger L. Nat Nanotechnol. 2010;5:676. doi: 10.1038/nnano.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geary C, Chworos A, Jaeger L. Nucleic Acids Res. 2011;39:1066. doi: 10.1093/nar/gkq748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grabow WW, Zakrevsky P, Afonin KA, Chworos A, Shapiro BA, Jaeger L. Nano Lett. 2011;11:878. doi: 10.1021/nl104271s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohno H, Kobayashi T, Kabata R, Endo K, Iwasa T, Yoshimura SH, Takeyasu K, Inoue T, Saito H. Nat Nanotechnol. 2011;6:116. doi: 10.1038/nnano.2010.268. [DOI] [PubMed] [Google Scholar]
- 12.Dibrov SM, McLean J, Parsons J, Hermann T. Proc Natl Acad Sci U S A. 2011;108:6405. doi: 10.1073/pnas.1017999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bindewald E, Afonin K, Jaeger L, Shapiro BA. ACS Nano. 2011;5:9542. doi: 10.1021/nn202666w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geary C, Rothemund PWK, Andersen ES. Science. 2014;345:799. doi: 10.1126/science.1253920. [DOI] [PubMed] [Google Scholar]
- 15.Boerneke MA, Hermann T. RNA Biol. 2015;12:780. doi: 10.1080/15476286.2015.1054592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boerneke MA, Dibrov SM, Gu J, Wyles DL, Hermann T. Proc Natl Acad Sci U S A. 2014;111:15952. doi: 10.1073/pnas.1414678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dibrov SM, Johnston-Cox H, Weng YH, Hermann T. Angew Chem Int Ed Engl. 2007;46:226. doi: 10.1002/anie.200603807. [DOI] [PubMed] [Google Scholar]; Angew Chem. 2007;119:230. [Google Scholar]
- 18.Dibrov SM, Ding K, Brunn ND, Parker MA, Bergdahl BM, Wyles DL, Hermann T. Proc Natl Acad Sci U S A. 2012;109:5223. doi: 10.1073/pnas.1118699109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu J, Liu Z, Jiang W, Wang G, Mao C. Nat Commun. 2015;6:5724. doi: 10.1038/ncomms6724. [DOI] [PubMed] [Google Scholar]
- 20.Parsons J, Castaldi MP, Dutta S, Dibrov SM, Wyles DL, Hermann T. Nat Chem Biol. 2009;5:823. doi: 10.1038/nchembio.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.