Abstract
Purpose of review
ADAMTS13 is a zinc-containing metalloprotease that cleaves von Willebrand factor (VWF). Deficiency of plasma ADAMTS13 activity is accountable for a potentially fatal blood disorder thrombotic thrombocytopenic purpura (TTP). Understanding of ADAMTS13–VWF interaction is essential for developing novel treatments to this disorder.
Recent findings
Despite the proteolytic activity of ADAMTS13 being restricted to the metalloprotease domain, the ancillary proximal C-terminal domains including the disintegrin domain, first TSP-1 repeat, cysteine-rich region, and spacer domain are all required for cleavage of VWF and its analogs. Recent studies have added to our understandings of the role of the specific regions in the disintegrin domain, the cysteine-rich domain, and the spacer domain responsible for its interaction with VWF. Additionally, regulative functions of the distal portion of ADAMTS13 including the TSP-1 2–8 repeats and the CUB domains have been proposed. Finally, fine mapping of anti-ADAMTS13 antibody epitopes have provided further insight into the essential structural elements in ADAMTS13 for VWF binding and the mechanism of autoantibody-mediated TTP.
Summary
Significant progress has been made in our understandings of the structure–function relationship of ADAMTS13 in the past decade. To further investigate ADAMTS13–VWF interactions for medical applications, these interactions must be studied under physiological conditions in vivo.
Keywords: ADAMTS13, autoantibody, protein–protein interaction, thrombosis, thrombotic thrombocytopenic purpura, von willebrand factor
INTRODUCTION
ADAMTS13, a circulating metalloprotease, was first cloned and identified as a member of A Disintegrin And Metalloprotease with ThromboSpondin type 1 repeats (ADAMTS) family in 2001 [1–3]. It is primarily synthesized in hepatic stellate cells [4–7], but is also found in other cells including endothelial cells [8,9], and megakaryocytes or platelets [10,11]. ADAMTS13 is secreted into plasma as a constitutively active enzyme at a plasma concentration of, approximately, 1 μg/ml [12–14]. It cleaves a large adhesive glycoprotein, von Willebrand factor (VWF), that plays an essential role in primary hemostasis by recruiting platelets to the site of vessel injury [15]. VWF is synthesized in vascular endothelial cells and megakaryocytes as a multimeric protein and stored in Weibel–Palade bodies of endothelial cells and the α-granules of platelets [16–18]. Plasma VWF concentration is, approximately, 10 μg/ml. Newly released VWF forms ultra large ‘string-like’ structures tethered to the cell surface that are subsequently cleaved by ADAMTS13 [19–21].
The proteolytic cleavage of ultra large VWF by ADAMTS13 is essential for maintaining the delicate balance between hemostasis and thrombosis. The inability to cleave ultra large VWF due to severe deficiency of ADAMTS13 activity results in a potentially fatal blood disorder thrombotic thrombocytopenic purpura (TTP) [22▪]. TTP can be either the result of inherited mutations in ADAMTS13, or the result of the development of autoantibodies directed against ADAMTS13 [1,23]. The absence of VWF cleavage allows for the accumulation of ultra large VWF and the aggregation of platelets that eventually cause the occlusion of arterioles and capillaries [1,23–28]. Moreover, the reduced plasma ADAMTS13 activity and increased plasma VWF are risk factors for the development of other arterial and inflammatory diseases, including myocardial infarction [29–31], ischemic stroke [32–34], preeclampsia [35], and cerebral malaria [36–40]. Therefore, a better understanding of the functional mechanisms and structures of ADAMTS13 will provide novel tools for diagnosis and treatment of these potentially fatal human diseases. In this review, we will provide comments about the recent advances in our understandings of ADAMTS13 and VWF interactions.
STRUCTURE AND FUNCTION
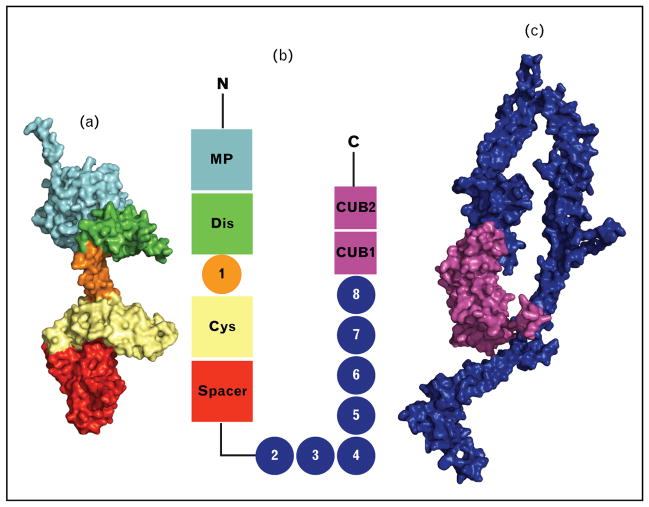
The ADAMTS family proteases are zinc-containing metalloproteases [41–43] that, unlike ADAM family proteases, are not membrane bound due to the lack of a membrane-anchor domain [44,45]. ADAMTS13, similar to other ADAMTS proteases, is comprised of a metalloprotease domain, a disintegrin-like domain, the first thrombospondin type 1 repeat, a cysteine-rich domain, and a spacer domain (Fig. 1) [1–3,7]. The C-terminal regions of ADAMTS proteases are more variable with an additional seven thrombospondin type 1 (TSP-1) repeats and two CUB (the Complement components C1r and C1s, sea urchin protein Uegf, and Bone morphogenetic protein-1) domains for ADAMTS13 (Fig. 1) [24,45,46].
FIGURE 1.
ADAMTS13 domain structure. (a) Homology model (M) and partial crystal structure (DTCS). (b) Schematic ADAMTS13 domain organization. (c) Homology model of TSP1 2–8 repeats and CUB domains. The metalloprotease domain structure is modeled from the Protein Data Bank (PDB) code 4WK7. TSP1 2–8 repeats are modeled from PDB code 1W0R, and the CUB domains are modeled from PDB code 3KQ4. TSP1 2 is attached to the spacer domain; the conformation of TSP1 2–8 and the CUB domains is unknown. The C-terminal region is included for size comparison. CUB, Complement components C1r and C1s, sea urchin protein Uegf, and Bone morphogenetic protein-1; DTCS, disintegrin-like domain, thrombospondin type-1 motif, cysteine–rich domain, and spacer domain; TSP1, thrombospondin type 1.
The propeptide on ADAMTS13 is only one-fifth of the length of a typical ADAMTS propeptide [24,47,48], which functions as a molecular chaperone and maintains the latency of newly synthesized ADAMTS proteases. This function of the propeptide in ADAMTS13 is absent; the enzymatic function and expression levels are normal with or without a pro-peptide [49].
The first ADAMTS13 domain is the metalloprotease, of which there are no crystal structures. However, closely related ADAMTS/ADAM proteases have been crystalized, and these serve as a functional and a structural template [50–52,53▪]. This domain is characterized by the perfectly conserved, adamalysin/reprolysin type, zinc-binding sequence (HEXXHXXGXXHD) [2]. When this zinc-binding sequence is absent, the protein does not have proteolytic activity [54]. This catalytic site is further characterized by a conserved downstream methionine residue. This ‘Metturn’ creates a tight turn, important to the structure of the active site, which possibly acts as a hydrophobic base beneath the zinc ion [44,55]. Between this conserved methionine and catalytic site, there is also a well conserved cysteine residue of unknown function [45,46].
Zinc-coordinating and calcium-binding residues control the cleavage activity of ADAMTS13 [56]. The zinc cation must be bound for enzymatic activity [57]. ADAMTS13 activity includes the binding of three calcium ions [2,46,57,58]. Calcium binding to the calcium three site (Asp182, Asp187, and Glu212) is essential for the structural integrity of the proteolytic site [57]. A water molecule, polarized by Glu225, is part of the conserved zinc-binding sequence, and is stabilized by the zinc ion. The hydroxide molecule created from the water molecule is hypothesized to nucleophilically attack the backbone carbonyl group between the residues Tyr1605–Met1606 allowing for the hydrolysis of that bond [56,59].
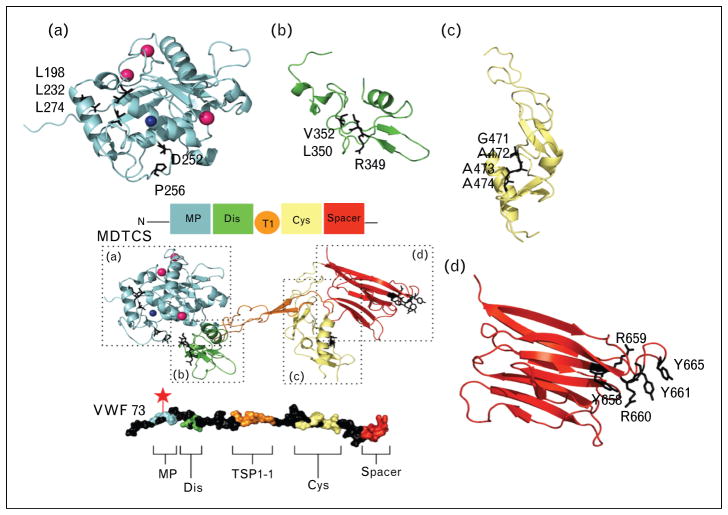
The metalloprotease domain alone does not have the ability to bind with specificity to VWF or to cleave at the VWF Tyr1605–Met1606 cleavage site [60,61]. The noncatalytic domains are necessary for substrate specificity [62]. Yet, the metalloprotease alone does exhibit cleavage activity but at a nonspecific location on VWF after 16–24 h of incubation [60]. Within the metalloprotease domain, residues Asp252 and Pro256 interact with Met1606, a residue in the cleave site on VWF (Fig. 2a) [63]. Also essential to the metalloprotease domain’s protease activity are several hydrophobic residues on ADAMTS13 together to bind to VWF [64]. In addition, residue Leu1603 on VWF, when modified has a greater negative impact, more than 100-fold activity decrease, than either of the cleavage site residues [64]. It is proposed that Leu1603 interacts with residues near the zinc ion allowing for proper cleavage of the Tyr1605–Met1606 bond (Fig. 2) [64].
FIGURE 2.
von Willebrand factor 73 exosites for ADAMTS13 and MDTCS binding residues. A colored cartoon representation of the labeled domains with a MDTCS is a homology model (M) and partial crystal structure (DTCS) shows the binding regions within MDTCS. Below, an unwound representation of von Willebrand factor (VWF) 73 is shown with exosites colored to the corresponding domain on MDTCS. The red star designates the cleavage site on VWF 73. (a) The metalloprotease domain, with the binding or active residues marked as black sticks. (b) The disintegrin domain with VWF binding residues shown as black sticks. (c) The cysteine-rich region shown with hydrophobic binding residues as black sticks. (d) The spacer domain with VWF-binding residues shown as black sticks. MDTCS, the metalloprotease domain, disintegrin-like domain, thrombospondin type-1 motif, cysteine–rich domain, and spacer domain.
The second domain on ADAMTS13 is the dis-integrin-like domain. It is named for the primary sequence similarity to the snake venom disintegrins. The disintegrin domain in ADAMTS13 lacks the hallmark cysteine signature or the RGD (arginine, glycine, asparagine) motif [45,65]. The metalloprotease domain and the disintegrin domain are functionally coupled [60,61,66,67]. With the addition of this short disintegrin domain, only 77 amino acids long, full substrate binding specificity is restored to the metalloprotease domain [60,67]. Also, despite low-relative efficiency, specific proteolytic activity is restored to the cleave site [60]. Unlike the metalloprotease domain, a crystal structure is available for the disintegrin domain as well as the following domains in the distal region [68,69]. There is little secondary structure. There are two antiparallel β-sheets, a single N-terminal α-helix, and four disulfide bridges [68]. These disulfide bridges maintain the structure of the domain [68]. The Cys322–Cys347 disulfide bridge is well conserved throughout other crystalized ADAM proteases [70,71]. Within this domain, two hydrophobic residues and one charged residue have been identified to bind to VWF. Glycine substitutions on Val352 and Leu350 produce, respectively, a four/five-fold and 10/20-fold reduction in catalytic efficiency in cleavage to VWF115 [67]. Similarly, an alanine substitution mutation introduced within the Arg349 residue decreased catalytic efficiency of cleavage of VWF115 by 20-fold [67]. The valine and leucine residues are hypothesized to interact in a hydrophobic manner and arginine in an ionically to Ala1612 and Asp1614 on VWF, respectively (Fig. 2b) [67].
The third domain in ADAMTS13 is the first TSP-1 repeat. This central TSP-1 repeat is very well conserved among ADAMTS family proteases. The domain has only three antiparallel strands, two of which are β-sheets, and the whole domain is capped by disulfide bonds on each end [68]. These strands are stabilized by a ‘CWR-layered core’ (cysteine, tryptophan, and arginine) [68,72]. The TSP-1 region is involved in substrate recognition [62]. When using a chimeric ADAMTS13/ADAMTS5 protease, replacing the cysteine rich and spacer domain in the metalloprotease domain, disintegrin-like domain, thrombospondin type-1 motif, cysteine–rich domain, and spacer domain (MDTCS), with analogous domains from ADAMTS5, reduced the enzymatic efficiency two-fold; however, replacing the cysteine-rich domain, spacer domain, along with the TSP-1 region, with ADAMTS5 analogs, the enzymatic efficiency dropped nearly nine-fold [62]. This illustrates the contribution of TSP-1 region to binding of VWF. However, these weak interactions with Gln1624–Val1630 become less imperative when the cysteine-rich and spacer domains are bound [62,66].
The cysteine-rich and disintegrin region are very structurally similar, despite the low-sequence identity (17%). The cysteine-rich domain only has a short α-helix and two pairs of double stranded antiparallel β-sheets and the structure is stabilized by six disulfide linkages [68]. Recently, residues Gly471–Val474 at the base of the variable loop within the cysteine-rich domain have been identified to form a hydrophobic pocket that appears to be involved in the binding to hydrophobic residues Ile1642, Trp1644, Ile1649, Leu1650, and Ile1651 on VWF (Fig. 2c) [73▪▪]. de Groot et al. [73▪▪] went on to postulate that the 75–200-fold reduction in proteolysis observed by Wu et al. [74] when the VWF exosite 2 is deleted, partially due to the absence of these hydrophobic interactions from the cysteine-rich domain. Additionally, they found that the regions sequentially conserved within the ADAMTS family in the cysteine-rich domain are not necessary for substrate binding [73▪▪]. Likewise, the charged region assigned the designation ‘the unique loop’, was not necessary for VWF115 cleavage [68,73▪▪].
The domain in ADAMTS13 that has the highest binding affinity for the A2 site of VWF is the spacer domain. The mechanism of VWF unwinding predicts that the exosite that binds to the spacer domain is the first exposed. This may allow the spacer domain to recognize the VWF exosite, even when VWF is only partially unfolded. The spacer domain and the cysteine-rich domain function closely with and similarly to one and other. A Leu621–Asp632 containing loop on the spacer domain has direct contact with the proximal portion of the cysteine-rich domain [68]. The spacer domain consists of 10 β-sheets that form a jellyroll topology [68]. This creates a hydrophobic cluster that is surrounded by arginine residues predicted to interact with Asp1596–Arg1659 on VWF (Fig. 2d) [68]. When ADAMTS13 is cleaved before the spacer domain (i.e., construct MDTC), there is a four-fold drop in the KD for VWF73 peptide [60]. Additionally, the proteolytic efficiency of the MDTC fragment is decreased by 20-fold [61]. Structural predictions of the arginine surrounded hydrophobic cluster have been confirmed by several functional studies. Arg660, Tyr661, and Tyr665 together are essential for VWF binding and cleavage [75,76]. These three residues are also very commonly found in the epitope site of ADAMTS13 antibodies [75,76].
The proximal domains (i.e., MDTCS) are all conserved within other ADAMTS proteases. However, within the further distal regions there are more variations between ADAMTS family proteases. These distal C-terminal regions of ADAMTS13 have not yet been crystalized, and much less is known about the structure and function.
Although the TSP-1 repeat between the disintegrin and cysteine-rich domains is well conserved within the ADAMTS proteases, the arrangement and number of the TSP-1 repeats following the spacer domain varies. Unlike the TSP1-1 repeat preceeding the spacer, the sequences of other TSP-1 repeats are not well conserved. Also, the fourth of these TSP-1 repeats has two cyseteines that are predicted to be unpaired [46]. Multiple TSP-1 repeats contain a CSVSCG (cysteine, serine, valine, serine, cysteine, glycine) motif. The second serine in this motif is glycosolated on the available side chain oxygen and the CSVSCG motif can bind the cell surface receptor CD36 [46,77].
ADAMTS13 is the only known ADAMTS protease that has two CUB domains at the distal C-terminus. The namesake protein is involved in developmental regulation [78]. Yet, the absence of the TSP-1 2–8 and the CUB domains has no negative impact upon the protease function of ADAMTS13 for VWF73 or VWF115, instead the C-terminal regions are necessary for binding globular VWF and VWF in shear conditions [79,80]. When the TSP-1 2–8 repeats and the CUB domains are truncated the remaining domains (i.e., MDTCS) still cleave VWF substrates. In fact, recent studies suggest that MDTCS may cleave VWF73 with greater efficiency (~2-fold) than full-length ADAMTS13, with respective kcat/Km values of 2.0 ± 0.6 μmol/l−1s−1 and 0.75 ± 0.16 μmol/l−1s−1 [61]. The CUB domains independently have no measurable affinity for VWF [81]. However, in the presence of shear stress, the CUB1 peptide will inhibit proteolysis of VWF [82].
The CUB domains have a negative regulatory function of ADAMTS13 activity. A full-length recombinant ADAMTS13 has been imaged through both quick-freeze, deep-etch electron microscopy and transmission electron microscopy and it appears that the distal portion of ADAMTS13 folds inwards [83▪▪,84▪▪]. This suggests that when the C-terminal portions of ADAMTS13 are not bound to VWF the TSP-1 2–8 repeats fold the CUB domains toward the spacer domain [83▪▪,84▪▪]. The removal of these CUB domains, which does not allow for the interaction of CUB domains with the spacer domain, would grant greater access to VWF by the spacer domain [83▪▪].
The C-terminal domains of ADAMTS13 may have regulatory functions entirely unrelated to the proteolytic activity. Five thiol groups within TSP-1 repeats 2–8 and CUB-1 domain potentially form disulfide bonds with VWF anchoring the enzyme to full-length VWF [85]. These disulfide interactions between ADAMTS13 and full-length VWF, as well as ADAMTS13–VWF complexes, have not been well described [79,85]. Yet, interestingly, these free thiol interactions of the distal regions appear to have antithrombotic activity independent of the proteolytic functions of ADAMTS13 [86▪▪]. Furthermore, mice lacking TSP-1 7–8 and CUB domains are more thrombogenic in high shear conditions [87], consistent with the importance of more distal C-terminal domains for function in vivo.
ANTIBODY SPECIFICITY
The presence of anti-ADAMTS13 antibodies and their epitopes has been well studied in recent years. Not all anti-ADAMTS13 antibodies disrupt the cleavage of VWF by ADAMTS13 [88]. Notably, the presence of anti-ADAMTS13 antibodies that do disrupt VWF cleavage is not always sufficient for the onset of acquired TTP, suggesting the requirement of other potential triggers for its onset [88,89▪▪].
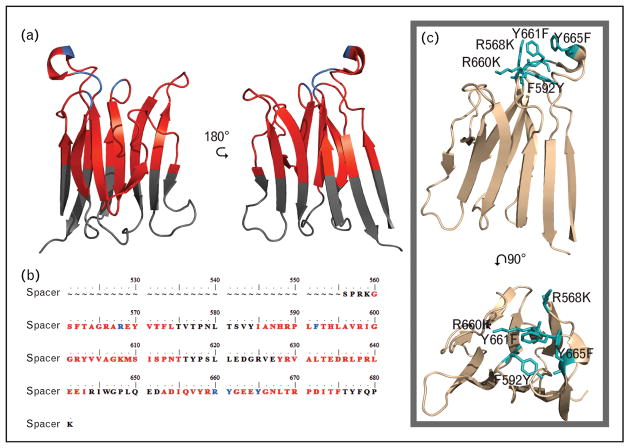
Autoantibody epitope mapping has not only further characterized the mechanism of the disease, but also uncovered the structure–function relationship of ADAMTS13. Most acquired TTP patients have multiple anti-ADAMTS13 antibodies that bind to several domains [89▪▪,90–93]. Binding epitopes, including residues Glu376, Gln159–Asp166, Tyr 305–Glu327, and Asn308 in the metalloprotease and disintegrin domains have been identified [91]. However, the most frequent antibody epitope region is found within the cysteine-rich and spacer domain, particularly the spacer domain [75,90–95,96▪▪,97]. In the mapping of the antigenic regions on ADAMTS13, the most common characteristic is the involvement of five loops located on the spacer domain (Fig. 3a). In one study using the MDTCS variant, modifications to only five residues in the spacer domain Arg660, Tyr661, Tyr 665, Arg568, and Phe592 antibody binding is strongly diminished in 46 of 48 TTP patients [90]. These residues represent three of the five-epitope loops on the spacer domain.
FIGURE 3.
Spacer domain sequence and structure (Protein Data Bank identity, 3GHM) with antibody antigenic regions marked, and gain of function (GoF) mutation. (a) Structure. (b) Sequence of the spacer domain with antigenic regions marked. Black text or regions designate no reported antibody exosite regions. Previously reported antibody binding residues are illustrated in blue. Red regions or text designates antibody exosite regions identified by conformational chemically linked peptides on scaffolds peptide technology. (c) ADAMTS13 spacer domain structure with the GoF mutations in teal.
With the understanding of the spacer domain function, and common antigenic-binding sites, gain of function (GoF) mutation (R660K/F592Y/R568K/Y661F/Y665F) was made to ADAMTS13 (Fig. 3c) [83▪▪,98]. This gain of function (GoF) mutation is both resistant to many antibody interactions and does not lose any catalytic efficiently compared with wild-type ADAMTS13 [83▪▪,98]. This and other GoF mutations offer exciting possibilities in the treatment of acquired TTP. Continuing in this direction, furthering the characterization and catalogue of anti-ADAMTS13 antibodies and their epitopes will likely lead to greater understandings of ADAMTS13 function and novel therapies for TTP.
CONCLUSION
Tremendous progress has been made in the past decade toward our understandings of the structure–functional relationship of ADAMTS13. However, animal models or humans studies may be necessary to translate the biochemical activity of ADATMS13 and its variants with their in-vivo biological functions under pathophysiological conditions.
KEY POINTS.
ADAMTS13 cleaves VWF and is essential for hemostasis.
While the metalloprotease domain contains the catalytic site, it alone is not sufficient to cleave VWF. Other ancillary domains, including the disintegrin-like domain, the first thrombospondin type 1 repeat, the cysteine-rich domain, and the spacer domain are all required for efficient catalysis of VWF to occur.
The CUB domains of ADAMTS13 may have a negative regulatory function by blocking the spacer–VWF A2 domain interaction.
Autoantibody epitope mapping may help to identify an ADAMTS13 variant that has catalytic activity in the presence of autoantibodies.
Acknowledgments
Financial support and sponsorship
The original work cited in the commentary was supported in part by grants from National Institute of Health (NIH) R01 HL115187-01A1.
Footnotes
Conflicts of interest
Authors declare no conflict of interest relevant to this work.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
- 1.Levy GG, Nichols WC, Lian EC, et al. Mutations in a member of the ADAMTS gene family cause thrombotic thrombocytopenic purpura. Nature. 2001;413:488–494. doi: 10.1038/35097008. [DOI] [PubMed] [Google Scholar]
- 2.Zheng X, Chung D, Takayama TK, et al. Structure of von Willebrand factor-cleaving protease (ADAMTS13), a metalloprotease involved in thrombotic thrombocytopenic purpura. J Biol Chem. 2001;276:41059–41063. doi: 10.1074/jbc.C100515200. [DOI] [PubMed] [Google Scholar]
- 3.Fujikawa K, Suzuki H, McMullen B, et al. Purification of human von Willebrand factor-cleaving protease and its identification as a new member of the metalloproteinase family. Blood. 2001;98:1662–1666. doi: 10.1182/blood.v98.6.1662. [DOI] [PubMed] [Google Scholar]
- 4.Uemura M, Tatsumi K, Matsumoto M, et al. Localization of ADAMTS13 to the stellate cells of human liver. Blood. 2005;106:922–924. doi: 10.1182/blood-2005-01-0152. [DOI] [PubMed] [Google Scholar]
- 5.Zhou W, Inada M, Lee TP, et al. ADAMTS13 is expressed in hepatic stellate cells. Lab Invest. 2005;85:780–788. doi: 10.1038/labinvest.3700275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niiya M, Uemura M, Zheng XW, et al. Increased ADAMTS-13 proteolytic activity in rat hepatic stellate cells upon activation in vitro and in vivo. J Thromb Haemost. 2006;4:1063–1070. doi: 10.1111/j.1538-7836.2006.01893.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soejima K, Mimura N, Hirashima M, et al. A novel human metalloprotease synthesized in the liver and secreted into the blood: possibly, the von Willebrand factor-cleaving protease? J Biochem. 2001;130:475–480. doi: 10.1093/oxfordjournals.jbchem.a003009. [DOI] [PubMed] [Google Scholar]
- 8.Turner N, Nolasco L, Tao Z, et al. Human endothelial cells synthesize and release ADAMTS-13. J Thromb Haemost. 2006;4:1396–1404. doi: 10.1111/j.1538-7836.2006.01959.x. [DOI] [PubMed] [Google Scholar]
- 9.Shang D, Zheng XW, Niiya M, et al. Apical sorting of ADAMTS13 in vascular endothelial cells and Madin-Darby canine kidney cells depends on the CUB domains and their association with lipid rafts. Blood. 2006;108:2207–2215. doi: 10.1182/blood-2006-02-002139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu L, Choi H, Bernardo A, et al. Platelet-derived VWF-cleaving metalloprotease ADAMTS-13. J Thromb Haemost. 2005;3:2536–2544. doi: 10.1111/j.1538-7836.2005.01561.x. [DOI] [PubMed] [Google Scholar]
- 11.Suzuki M, Murata M, Matsubara Y, et al. Detection of von Willebrand factor-cleaving protease (ADAMTS-13) in human platelets. Biochem Biophys Res Commun. 2004;313:212–216. doi: 10.1016/j.bbrc.2003.11.111. [DOI] [PubMed] [Google Scholar]
- 12.Feys HB, Liu F, Dong N, et al. ADAMTS-13 plasma level determination uncovers antigen absence in acquired thrombotic thrombocytopenic purpura and ethnic differences. J Thromb Haemost. 2006;4:955–962. doi: 10.1111/j.1538-7836.2006.01833.x. [DOI] [PubMed] [Google Scholar]
- 13.Rieger M, Ferrari S, Kremer Hovinga JA, et al. Relation between ADAMTS13 activity and ADAMTS13 antigen levels in healthy donors and patients with thrombotic microangiopathies (TMA) Thromb Haemost. 2006;95:212–220. doi: 10.1160/TH05-08-0550. [DOI] [PubMed] [Google Scholar]
- 14.Soejima K, Nakamura H, Hirashima M, et al. Analysis on the molecular species and concentration of circulating ADAMTS13 in Blood. J Biochem. 2006;139:147–154. doi: 10.1093/jb/mvj013. [DOI] [PubMed] [Google Scholar]
- 15.Sadler JE. Biochemistry and genetics of von Willebrand factor. Annu Rev Biochem. 1998;67:395–424. doi: 10.1146/annurev.biochem.67.1.395. [DOI] [PubMed] [Google Scholar]
- 16.Wagner DD, Marder VJ. Biosynthesis of von Willebrand protein by human endothelial cells: processing steps and their intracellular localization. J Cell Biol. 1984;99:2123–2130. doi: 10.1083/jcb.99.6.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sadler JE. von Willebrand factor. J Biol Chem. 1991;266:22777–22780. [PubMed] [Google Scholar]
- 18.Sporn LA, Chavin SI, Marder VJ, et al. Biosynthesis of von Willebrand protein by human megakaryocytes. J Clin Invest. 1985;76:1102–1106. doi: 10.1172/JCI112064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong JF, Moake JL, Nolasco L, et al. ADAMTS-13 rapidly cleaves newly secreted ultralarge von Willebrand factor multimers on the endothelial surface under flowing conditions. Blood. 2002;100:4033–4039. doi: 10.1182/blood-2002-05-1401. [DOI] [PubMed] [Google Scholar]
- 20.Dong JF, Moake JL, Bernardo A, et al. ADAMTS-13 metalloprotease interacts with the endothelial cell-derived ultra-large von Willebrand factor. J Biol Chem. 2003;278:29633–29639. doi: 10.1074/jbc.M301385200. [DOI] [PubMed] [Google Scholar]
- 21.Dong JF. Cleavage of ultra-large von Willebrand factor by ADAMTS-13 under flow conditions. J Thromb Haemost. 2005;3:1710–1716. doi: 10.1111/j.1538-7836.2005.01360.x. [DOI] [PubMed] [Google Scholar]
- 22▪.Zheng XL. ADAMTS13 and von Willebrand factor in thrombotic thrombocytopenic purpura. Annu Rev Med. 2015;66:211–225. doi: 10.1146/annurev-med-061813-013241. This review outlines the history and discovery of TTP and ADAMTS13. It also describes the biosynthesis and secretion of both VWF and ADAMTS13 and provides a brief overview of treatments and diagnosis of TTP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsai HM, Lian EC. Antibodies to von Willebrand factor-cleaving protease in acute thrombotic thrombocytopenic purpura. N Engl J Med. 1998;339:1585–1594. doi: 10.1056/NEJM199811263392203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng XL. Structure-function and regulation of ADAMTS-13 protease. J Thromb Haemost. 2013;11(Suppl 1):11–23. doi: 10.1111/jth.12221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sadler JE. A new name in thrombosis, ADAMTS13. Proc Natl Acad Sci U S A. 2002;99:11552–11554. doi: 10.1073/pnas.192448999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hosler GA, Cusumano AM, Hutchins GM. Thrombotic thrombocytopenic purpura and hemolytic uremic syndrome are distinct pathologic entities. A review of 56 autopsy cases. Arch Pathol Lab Med. 2003;127:834–839. doi: 10.5858/2003-127-834-TTPAHU. [DOI] [PubMed] [Google Scholar]
- 27.Tsai HM. Platelet activation and the formation of the platelet plug: deficiency of ADAMTS13 causes thrombotic thrombocytopenic purpura. Arterioscler Thromb Vasc Biol. 2003;23:388–396. doi: 10.1161/01.ATV.0000058401.34021.D4. [DOI] [PubMed] [Google Scholar]
- 28.Tsai HM. ADAMTS13 and microvascular thrombosis. Expert Rev Cardiovasc Ther. 2006;4:813–825. doi: 10.1586/14779072.4.6.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaikita K, Soejima K, Matsukawa M, et al. Reduced von Willebrand factor-cleaving protease (ADAMTS13) activity in acute myocardial infarction. J Thromb Haemost. 2006;4:2490–2493. doi: 10.1111/j.1538-7836.2006.02161.x. [DOI] [PubMed] [Google Scholar]
- 30.Matsukawa M, Kaikita K, Soejima K, et al. Serial changes in von Willebrand factor-cleaving protease (ADAMTS13) and prognosis after acute myocardial infarction. Am J Cardiol. 2007;100:758–763. doi: 10.1016/j.amjcard.2007.03.095. [DOI] [PubMed] [Google Scholar]
- 31.Andersson HM, Siegerink B, Luken BM, et al. High VWF, low ADAMTS13, and oral contraceptives increase the risk of ischemic stroke and myocardial infarction in young women. Blood. 2012;119:1555–1560. doi: 10.1182/blood-2011-09-380618. [DOI] [PubMed] [Google Scholar]
- 32.Bongers TN, de Maat MP, van Goor ML, et al. High von Willebrand factor levels increase the risk of first ischemic stroke: influence of ADAMTS13, inflammation, and genetic variability. Stroke. 2006;37:2672–2677. doi: 10.1161/01.STR.0000244767.39962.f7. [DOI] [PubMed] [Google Scholar]
- 33.Zhao BQ, Chauhan AK, Canault M, et al. von Willebrand factor-cleaving protease ADAMTS13 reduces ischemic brain injury in experimental stroke. Blood. 2009;114:3329–3334. doi: 10.1182/blood-2009-03-213264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujioka M, Nakano T, Hayakawa K, et al. ADAMTS13 gene deletion enhances plasma high-mobility group box1 elevation and neuroinflammation in brain ischemia-reperfusion injury. Neurol Sci. 2012;33:1107–1115. doi: 10.1007/s10072-011-0913-9. [DOI] [PubMed] [Google Scholar]
- 35.Stepanian A, Cohen-Moatti M, Sanglier T, et al. Von Willebrand factor and ADAMTS13: a candidate couple for preeclampsia pathophysiology. Arterioscler Thromb Vasc Biol. 2011;31:1703–1709. doi: 10.1161/ATVBAHA.111.223610. [DOI] [PubMed] [Google Scholar]
- 36.de Mast Q, Groot E, Asih PB, et al. ADAMTS13 deficiency with elevated levels of ultra-large and active von Willebrand factor in P. falciparum and P. vivax malaria. Am J Trop Med Hyg. 2009;80:492–498. [PubMed] [Google Scholar]
- 37.Larkin D, de Laat B, Jenkins PV, et al. Severe Plasmodium falciparum malaria is associated with circulating ultra-large von Willebrand multimers and ADAMTS13 inhibition. PLoS Pathog. 2009;5:e1000349. doi: 10.1371/journal.ppat.1000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bridges DJ, Bunn J, van Mourik JA, et al. Rapid activation of endothelial cells enables Plasmodium falciparum adhesion to platelet-decorated von Willebrand factor strings. Blood. 2010;115:1472–1474. doi: 10.1182/blood-2009-07-235150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lowenberg EC, Charunwatthana P, Cohen S, et al. Severe malaria is associated with a deficiency of von Willebrand factor cleaving protease, ADAMTS13. Thromb Haemost. 2010;103:181–187. doi: 10.1160/TH09-04-0223. [DOI] [PubMed] [Google Scholar]
- 40.Kraisin S, Naka I, Patarapotikul J, et al. Association of ADAMTS13 polymorphism with cerebral malaria. Malar J. 2011;10:366. doi: 10.1186/1475-2875-10-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kokame K, Matsumoto M, Soejima K, et al. Mutations and common polymorphisms in ADAMTS13 gene responsible for von Willebrand factor-cleaving protease activity. Proc Natl Acad Sci U S A. 2002;99:11902–11907. doi: 10.1073/pnas.172277399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Plaimauer B, Zimmermann K, Volkel D, et al. Cloning, expression, and functional characterization of the von Willebrand factor-cleaving protease (ADAMTS13) Blood. 2002;100:3626–3632. doi: 10.1182/blood-2002-05-1397. [DOI] [PubMed] [Google Scholar]
- 43.Chung DW, Fujikawa K. Processing of von Willebrand factor by ADAMTS-13. Biochemistry. 2002;41:11065–11070. doi: 10.1021/bi0204692. [DOI] [PubMed] [Google Scholar]
- 44.Lancellotti S, Basso M, De Cristofaro R. Proteolytic processing of von Willebrand factor by adamts13 and leukocyte proteases. Mediterr J Hematol Infect Dis. 2013;5:e2013058. doi: 10.4084/MJHID.2013.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barrett AJ, Rawlings ND, Woessner JF. Handbook of proteolytic enzymes. 2. Amsterdam; Boston: Elsvier Academic Press; 2004. [Google Scholar]
- 46.Crawley JT, de Groot R, Xiang Y, et al. Unraveling the scissile bond: how ADAMTS13 recognizes and cleaves von Willebrand factor. Blood. 2011;118:3212–3221. doi: 10.1182/blood-2011-02-306597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Apte SS. A disintegrin-like and metalloprotease (reprolysin-type) with thrombospondin type 1 motif (ADAMTS) superfamily: functions and mechanisms. J Biol Chem. 2009;284:31493–31497. doi: 10.1074/jbc.R109.052340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang BL. ADAMTS: a novel family of extracellular matrix proteases. Int J Biochem Cell Biol. 2001;33:33–44. doi: 10.1016/s1357-2725(00)00061-3. [DOI] [PubMed] [Google Scholar]
- 49.Majerus EM, Zheng X, Tuley EA, et al. Cleavage of the ADAMTS13 propeptide is not required for protease activity. J Biol Chem. 2003;278:46643–46648. doi: 10.1074/jbc.M309872200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shieh HS, Mathis KJ, Williams JM, et al. High resolution crystal structure of the catalytic domain of ADAMTS-5 (aggrecanase-2) J Biol Chem. 2008;283:1501–1507. doi: 10.1074/jbc.M705879200. [DOI] [PubMed] [Google Scholar]
- 51.Gerhardt S, Hassall G, Hawtin P, et al. Crystal structures of human ADAMTS-1 reveal a conserved catalytic domain and a disintegrin-like domain with a fold homologous to cysteine-rich domains. J Mol Biol. 2007;373:891–902. doi: 10.1016/j.jmb.2007.07.047. [DOI] [PubMed] [Google Scholar]
- 52.Mosyak L, Georgiadis K, Shane T, et al. Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5. Protein Sci. 2008;17:16–21. doi: 10.1110/ps.073287008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53▪.Durham TB, Klimkowski VJ, Rito CJ, et al. Identification of potent and selective hydantoin inhibitors of aggrecanase-1 and aggrecanase-2 that are efficacious in both chemical and surgical models of osteoarthritis. J Med Chem. 2014;57:10476–10485. doi: 10.1021/jm501522n. This group created an effective inhibitor for ADAMTS4/5. Hydantoin 13 was found through a screening campaign and optimized through structure-based drug design. This inhibitor is both efficacious and selective to zinc metalloproteases. The inhibitor was cocrystalized with the metalloprotease domain, which is useful as a template for homology modeling. [DOI] [PubMed] [Google Scholar]
- 54.White JM. ADAMs: modulators of cell-cell and cell-matrix interactions. Curr Opin Cell Biol. 2003;15:598–606. doi: 10.1016/j.ceb.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 55.Bode W, Gomis-Ruth FX, Stockler W. Astacins, serralysins, snake venom and matrix metalloproteinases exhibit identical zinc-binding environments (HEXXHXXGXXH and Metturn) and topologies and should be grouped into a common family, the ’metzincins’. FEBS Lett. 1993;331:134–140. doi: 10.1016/0014-5793(93)80312-i. [DOI] [PubMed] [Google Scholar]
- 56.Di Stasio E, Lancellotti S, Peyvandi F, et al. Mechanistic studies on ADAMTS13 catalysis. Biophys J. 2008;95:2450–2461. doi: 10.1529/biophysj.108.131532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gardner MD, Chion CK, de Groot R, et al. A functional calcium-binding site in the metalloprotease domain of ADAMTS13. Blood. 2009;113:1149–1157. doi: 10.1182/blood-2008-03-144683. [DOI] [PubMed] [Google Scholar]
- 58.Tsai HM. Physiologic cleavage of von Willebrand factor by a plasma protease is dependent on its conformation and requires calcium ion. Blood. 1996;87:4235–4244. [PubMed] [Google Scholar]
- 59.Bode W, Fernandez-Catalan C, Tschesche H, et al. Structural properties of matrix metalloproteinases. Cell Mol Life Sci. 1999;55:639–652. doi: 10.1007/s000180050320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ai J, Smith P, Wang S, et al. The proximal carboxyl-terminal domains of ADAMTS13 determine substrate specificity and are all required for cleavage of von Willebrand factor. J Biol Chem. 2005;280:29428–29434. doi: 10.1074/jbc.M505513200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gao W, Anderson PJ, Majerus EM, et al. Exosite interactions contribute to tension-induced cleavage of von Willebrand factor by the antithrombotic ADAMTS13 metalloprotease. Proc Natl Acad Sci U S A. 2006;103:19099–19104. doi: 10.1073/pnas.0607264104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gao W, Zhu J, Westfield LA, et al. Rearranging exosites in noncatalytic domains can redirect the substrate specificity of ADAMTS proteases. J Biol Chem. 2012;287:26944–26952. doi: 10.1074/jbc.M112.380535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.de Groot R, Lane DA, Crawley JT. The ADAMTS13 metalloprotease domain: roles of subsites in enzyme activity and specificity. Blood. 2010;116:3064–3072. doi: 10.1182/blood-2009-12-258780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xiang Y, de Groot R, Crawley JT, et al. Mechanism of von Willebrand factor scissile bond cleavage by a disintegrin and metalloproteinase with a thrombospondin type 1 motif, member 13 (ADAMTS13) Proc Natl Acad Sci U S A. 2011;108:11602–11607. doi: 10.1073/pnas.1018559108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Apte SS. A disintegrin-like and metalloprotease (reprolysin type) with thrombospondin type 1 motifs: the ADAMTS family. Int J Biochem Cell Biol. 2004;36:981–985. doi: 10.1016/j.biocel.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 66.Gao W, Anderson PJ, Sadler JE. Extensive contacts between ADAMTS13 exosites and von Willebrand factor domain A2 contribute to substrate specificity. Blood. 2008;112:1713–1719. doi: 10.1182/blood-2008-04-148759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Groot R, Bardhan A, Ramroop N, et al. Essential role of the disintegrin-like domain in ADAMTS13 function. Blood. 2009;113:5609–5616. doi: 10.1182/blood-2008-11-187914. [DOI] [PubMed] [Google Scholar]
- 68.Akiyama M, Takeda S, Kokame K, et al. Crystal structures of the noncatalytic domains of ADAMTS13 reveal multiple discontinuous exosites for von Willebrand factor. Proc Natl Acad Sci U S A. 2009;106:19274–19279. doi: 10.1073/pnas.0909755106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Akiyama M, Nakayama D, Takeda S, et al. Crystal structure and enzymatic activity of an ADAMTS-13 mutant with the East Asian-specific P475S polymorphism. J Thromb Haemost. 2013;11:1399–1406. doi: 10.1111/jth.12279. [DOI] [PubMed] [Google Scholar]
- 70.Takeda S. Three-dimensional domain architecture of the ADAM family proteinases. Semin Cell Dev Biol. 2009;20:146–152. doi: 10.1016/j.semcdb.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 71.Takeda S, Igarashi T, Mori H, et al. Crystal structures of VAP1 reveal ADAMs’ MDC domain architecture and its unique C-shaped scaffold. EMBO J. 2006;25:2388–2396. doi: 10.1038/sj.emboj.7601131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tan K, Duquette M, Liu JH, et al. Crystal structure of the TSP-1 type 1 repeats: a novel layered fold and its biological implication. J Cell Biol. 2002;159:373–382. doi: 10.1083/jcb.200206062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73▪▪.de Groot R, Lane DA, Crawley JT. The role of the ADAMTS13 cysteine-rich domain in VWF binding and proteolysis. Blood. 2015;125:1968–1975. doi: 10.1182/blood-2014-08-594556. Through the process of elimination, using glycans and mutagenesis studies, this group identified the residues within the cysteine-rich domain responsible for the majority of the binding affinity for VWF. The study isolated the binding residues on VWF, Ile1642, Trp1644, Ile1649, Leu1650, and Ile1651 and their compliment residues ADAMTS13 cysteine-rich domain, Gly471–Val474. The presence of these residues enhances the efficiency of the proteolytic cleavage of VWF by ADAMTS13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu JJ, Fujikawa K, McMullen BA, et al. Characterization of a core binding site for ADAMTS-13 in the A2 domain of von Willebrand factor. Proc Natl Acad Sci U S A. 2006;103:18470–18474. doi: 10.1073/pnas.0609190103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pos W, Crawley JT, Fijnheer R, et al. An autoantibody epitope comprising residues R660, Y661, and Y665 in the ADAMTS13 spacer domain identifies a binding site for the A2 domain of VWF. Blood. 2010;115:1640–1649. doi: 10.1182/blood-2009-06-229203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jin SY, Skipwith CG, Zheng XL. Amino acid residues Arg(659), Arg(660), and Tyr(661) in the spacer domain of ADAMTS13 are critical for cleavage of von Willebrand factor. Blood. 2010;115:2300–2310. doi: 10.1182/blood-2009-07-235101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Davis AK, Makar RS, Stowell CP, et al. ADAMTS13 binds to CD36: a potential mechanism for platelet and endothelial localization of ADAMTS13. Transfusion. 2009;49:206–213. doi: 10.1111/j.1537-2995.2008.01978.x. [DOI] [PubMed] [Google Scholar]
- 78.Bork P, Beckmann G. The CUB domain. A widespread module in developmentally regulated proteins. J Mol Biol. 1993;231:539–545. doi: 10.1006/jmbi.1993.1305. [DOI] [PubMed] [Google Scholar]
- 79.Feys HB, Anderson PJ, Vanhoorelbeke K, et al. Multistep binding of ADAMTS-13 to von Willebrand factor. J Thromb Haemost. 2009;7:2088–2095. doi: 10.1111/j.1538-7836.2009.03620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zanardelli S, Chion AC, Groot E, et al. A novel binding site for ADAMTS13 constitutively exposed on the surface of globular VWF. Blood. 2009;114:2819–2828. doi: 10.1182/blood-2009-05-224915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang P, Pan W, Rux AH, et al. The cooperative activity between the carboxyl-terminal TSP1 repeats and the CUB domains of ADAMTS13 is crucial for recognition of von Willebrand factor under flow. Blood. 2007;110:1887–1894. doi: 10.1182/blood-2007-04-083329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tao Z, Peng Y, Nolasco L, et al. Recombinant CUB-1 domain polypeptide inhibits the cleavage of ULVWF strings by ADAMTS13 under flow conditions. Blood. 2005;106:4139–4145. doi: 10.1182/blood-2005-05-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83▪▪.South K, Luken BM, Crawley JT, et al. Conformational activation of ADAMTS13. Proc Natl Acad Sci U S A. 2014;111:18578–18583. doi: 10.1073/pnas.1411979112. In this study, transmission electron microscopy analysis of wild-type ADAMTS13 suggests that the distal portions of ADAMTS13 fold toward the spacer domain. This article also showed that there is, approximately, 2.5 increase in proteolytic activity when the distal portions of ADAMTS13 are absent or blocked. This same increase in proteolytic activity was exhibited in the GoF variant (R568K/F592Y/R660K/Y661F/Y665F), suggesting this is the location of spacer domain and CUB domain interactions. Correspondingly, transmission electron microscopy analysis of the GoF variant appeared to be in a more open conformation than the wild-type ADAMTS13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84▪▪.Muia J, Zhu J, Gupta G, et al. Allosteric activation of ADAMTS13 by von Willebrand factor. Proc Natl Acad Sci U S A. 2014;111:18584–18589. doi: 10.1073/pnas.1413282112. This group used small angle radiographs scattering to show that the CUB domains interact with the proximal (MDTCS) domains of ADAMTS13. They suggest that when the distal domains bind to VWF the proximal domains are then available to interact with the A2 subunit. They also found the C-terminal portion of ADAMTS13 is responsible for the protease pH dependence. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yeh HC, Zhou Z, Choi H, et al. Disulfide bond reduction of von Willebrand factor by ADAMTS-13. J Thromb Haemost. 2010;8:2778–2788. doi: 10.1111/j.1538-7836.2010.04094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86▪▪.Bao J, Xiao J, Mao Y, et al. Carboxyl terminus of ADAMTS13 directly inhibits platelet aggregation and ultra large von Willebrand factor string formation under flow in a free-thiol-dependent manner. Arterioscler Thromb Vasc Biol. 2014;34:397–407. doi: 10.1161/ATVBAHA.113.302547. This study demonstrated through a microfluidic assay and microscopy techniques that the distal domains, TSP-1 2–8 repeats and CUB domains, prevent platelet aggregation and ultra large VWF string formation in a free-thiol-dependent manner. This demonstrates a homeostatic function of the distal domains that is independent from the proteolytic function of the proximal ADAMTS13 domains. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Banno F, Chauhan AK, Kokame K, et al. The distal carboxyl-terminal domains of ADAMTS13 are required for regulation of in vivo thrombus formation. Blood. 2009;113:5323–5329. doi: 10.1182/blood-2008-07-169359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Scheiflinger F, Knobl P, Trattner B, et al. Nonneutralizing IgM and IgG antibodies to von Willebrand factor-cleaving protease (ADAMTS-13) in a patient with thrombotic thrombocytopenic purpura. Blood. 2003;102:3241–3243. doi: 10.1182/blood-2003-05-1616. [DOI] [PubMed] [Google Scholar]
- 89▪▪.Grillberger R, Casina VC, Turecek PL, et al. Anti-ADAMTS13 IgG autoantibodies present in healthy individuals share linear epitopes with those in patients with thrombotic thrombocytopenic purpura. Haematologica. 2014;99:e58–e60. doi: 10.3324/haematol.2013.100685. In this letter, anti-ADAMTS13 antibodies from healthy plasma donors were compared with that of TTP patients. The overlap in the presence of these anti-ADAMTS13 antibodies shows that the mere presence of these antibodies is not sufficient to induce the disease state. This raises questions to the other factors involved in the onset of TTP, as well as the possibility that the presence of these antibodies precedes the disease state. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pos W, Sorvillo N, Fijnheer R, et al. Residues Arg568 and Phe592 contribute to an antigenic surface for anti-ADAMTS13 antibodies in the spacer domain. Haematologica. 2011;96:1670–1677. doi: 10.3324/haematol.2010.036327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Igari A, Nakagawa T, Moriki T, et al. Identification of epitopes on ADAMTS13 recognized by a panel of monoclonal antibodies with functional or nonfunctional effects on catalytic activity. Thromb Res. 2012;130:e79–e83. doi: 10.1016/j.thromres.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 92.Yamaguchi Y, Moriki T, Igari A, et al. Epitope analysis of autoantibodies to ADAMTS13 in patients with acquired thrombotic thrombocytopenic purpura. Thromb Res. 2011;128:169–173. doi: 10.1016/j.thromres.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 93.Klaus C, Plaimauer B, Studt JD, et al. Epitope mapping of ADAMTS13 autoantibodies in acquired thrombotic thrombocytopenic purpura. Blood. 2004;103:4514–4519. doi: 10.1182/blood-2003-12-4165. [DOI] [PubMed] [Google Scholar]
- 94.Luken BM, Turenhout EA, Kaijen PH, et al. Amino acid regions 572–579 and 657–666 of the spacer domain of ADAMTS13 provide a common antigenic core required for binding of antibodies in patients with acquired TTP. Thromb Haemost. 2006;96:295–301. doi: 10.1160/TH06-03-0135. [DOI] [PubMed] [Google Scholar]
- 95.Pos W, Luken BM, Sorvillo N, et al. Humoral immune response to ADAMTS13 in acquired thrombotic thrombocytopenic purpura. J Thromb Haemost. 2011;9:1285–1291. doi: 10.1111/j.1538-7836.2011.04307.x. [DOI] [PubMed] [Google Scholar]
- 96▪▪.Schaller M, Vogel M, Kentouche K, et al. The splenic autoimmune response to ADAMTS13 in thrombotic thrombocytopenic purpura contains recurrent antigen-binding CDR3 motifs. Blood. 2014;124:3469–3479. doi: 10.1182/blood-2014-04-561142. This group selected antibodies through phage display and EBV transformation of memory B cells were mapped using chemically linked peptides on scaffolds peptide technology. This showed with greater resolution the loops within the spacer domain common for antibody exosites. In addition, this demonstrates the similarity of antibody epitopes between TTP patents that are neither related or from the same geographic region. [DOI] [PubMed] [Google Scholar]
- 97.Luken BM, Kaijen PH, Turenhout EA, et al. Multiple B-cell clones producing antibodies directed to the spacer and disintegrin/thrombospondin type-1 repeat 1 (TSP1) of ADAMTS13 in a patient with acquired thrombotic thrombocytopenic purpura. J Thromb Haemost. 2006;4:2355–2364. doi: 10.1111/j.1538-7836.2006.02164.x. [DOI] [PubMed] [Google Scholar]
- 98.Jian C, Xiao J, Gong L, et al. Gain-of-function ADAMTS13 variants that are resistant to autoantibodies against ADAMTS13 in patients with acquired thrombotic thrombocytopenic purpura. Blood. 2012;119:3836–3843. doi: 10.1182/blood-2011-12-399501. [DOI] [PMC free article] [PubMed] [Google Scholar]