Abstract
Next-generation sequencing technology has presented an opportunity for rare variant discovery and association of these variants with disease. To address the challenges of rare variant analysis, multiple statistical methods have been developed for combining rare variants to increase statistical power for detecting associations. BioBin is an automated tool that expands on collapsing/binning methods by performing multi-level variant aggregation with a flexible, biologically informed binning strategy using an internal biorepository, the Library of Knowledge (LOKI). The databases within LOKI provide variant details, regional annotations and pathway interactions which can be used to generate bins of biologically-related variants, thereby increasing the power of any subsequent statistical test. In this study, we expand the framework of BioBin to incorporate statistical tests, including a dispersion-based test, SKAT, thereby providing the option of performing a unified collapsing and statistical rare variant analysis in one tool. Extensive simulation studies performed on gene-coding regions showed a Bin-KAT analysis to have greater power than BioBin-regression in all simulated conditions, including variants influencing the phenotype in the same direction, a scenario where burden tests often retain greater power. The use of Madsen-Browning variant weighting increased power in the burden analysis to that equitable with Bin-KAT; but overall Bin-KAT retained equivalent or higher power under all conditions. Bin-KAT was applied to a study of 82 pharmacogenes sequenced in the Marshfield Personalized Medicine Research Project (PMRP). We looked for association of these genes with 9 different phenotypes extracted from the electronic health record. This study demonstrates that Bin-KAT is a powerful tool for the identification of genes harboring low frequency variants for complex phenotypes.
1. Introduction
Examining the genetic influence of low frequency or rare variation to complex disease susceptibility may elucidate additional trait variability and disease risk which has largely remained unexplained by traditional GWAS approaches[29]. In recent years, studies on multifactorial diseases including Alzheimer's disease and prostate cancer have provided compelling evidence that rare variants are associated with complex traits and should be further examined[9, 16]. Advances in sequencing technologies and decreases in sequencing cost have provided an opportunity for rare variant discovery. However, due to the frequency of these variants, there is often low statistical power for detecting association with a phenotype, and therefore, a necessity for prohibitively large sample sizes. Collapsing or binning methods are commonly used to aggregate variants into a single genetic variable for subsequent statistical testing, reducing the degrees of freedom in the analysis and improving power[23]. BioBin[33, 34] is an automated bioinformatics tool initially developed for the multi-level collapsing of rare variants into user-designated biological features such as genes, pathways, evolutionary conserved regions (ECRs), protein families, and regulatory regions. BioBin follows a binning approach driven by prior biological knowledge by using an internal biorepository, the Library of Knowledge Integration (LOKI)[40]. LOKI combines biological information from over a dozen public databases providing variant details, regional annotations and pathway interactions. The flexible knowledge-driven binning design of BioBin allows the user to test multiple hypotheses within one unified analysis.
Rare variant association analysis of binned variants is often performed using burden or dispersion tests. Burden methods test the cumulative effect of variants within a bin and are easily applied to case-control studies as they assess the frequency of variant counts between these phenotypic groups[24]. Burden tests assume that all variants influence the trait in the same direction and magnitude of effect, and will suffer a loss of power if a mixture of protective and risk variants is present. Standard burden tests include generalized linear model regression analyses and the weighted sum statistic(WSS)[28]. Instead of testing the cumulative effect of variants within a region, dispersion or nonburden methods will test the distribution of these variants in the cases and controls thereby maintaining statistical power in the presence of a mixture of variants. The SKAT[46] package is a dispersion test that has gained widespread use as it allows for easy covariate adjustment, analyzes both dichotomous and quantitative phenotypes, and applies multiple variant weighting options. SKAT is a score-based variance component test that uses a multiple regression kernel-based approach to assess variant distribution and test for association. Both standard burden tests and the SKAT dispersion method have been well assessed in rare variant analysis.
While various tools have been specifically developed to facilitate rare variant association analysis, many methods focus either on the creation of a relevant set of variants or on the statistical analysis of already collapsed variants. This may often lead to file conversion issues for specific tools, as well as more complicated and longer analysis time. Herein we expand the framework of BioBin by integrating select statistical tests, regression and SKAT, as well as capabilities for multiple phenotype analysis (or Phenome-wide Association Studies (PheWAS)), thereby providing a comprehensive, unified bioinformatics tool for the biological binning and association analysis of rare variants. We have evaluated the commonly used regression burden analysis and SKAT in the context of BioBin with data simulations based on individuals of European descent from 1000 Genomes Project Phase I. We have also applied a BioBin-SKAT, or Bin-KAT, test to analyze nine complex human phenotypes from the Marshfield-PMRP project[31], part of the eMERGE network[14]. Our analyses highlight the utility of BioBin as a fast, comprehensive and versatile tool for the biological binning and analysis of low frequency variants in sequence data for multiple complex phenotypes and PheWAS.
2. Methods
2.1. BioBin
2.1.1. Overview of BioBin
BioBin is a unified command line bioinformatics tool written in C++ that utilizes the LOKI database for biologically inspired binning of variants, and also provides a platform for the association analysis of rare variant bins. The framework of a BioBin analysis is to determine biological features upon which data will be binned, such as genes, pathways or intergenic regions, execute bin generation using LOKI, and apply statistical association analysis to each bin. BioBin follows an allele frequency threshold binning approach using the non-major allele frequency (NMAF), defined as 1 minus the frequency of the most common allele. As NMAF and MAF are interchangeable for biallelic markers, MAF will be used in this work. BioBin allows variants below a user-specified MAF in the case or the control group to be binned thereby facilitating the aggregation of both potential risk and protective variants. BioBin was originally developed solely for the biologically informed binning of rare variants in an automated manner. To facilitate more efficient statistical analysis, we have incorporated an extensible testing infrastructure, implementing select burden and dispersion-based tests, namely regression, wilcoxon and SKAT[46] into BioBin. These are commonly used statistical tests in rare variant association analysis, and their direct implementation into BioBin streamlines the analysis, saves time, and also avoids any potential file conversion issues. Also, if an alternate statistical test is desired, BioBin may still be utilized strictly for its biologically inspired variant collapsing function. We have also integrated multiple phenotype capabilities allowing the user to efficiently perform a binned rare variant PheWAS[35, 41, 42]. BioBin analyzes each phenotype separately and uses parallel processing to increase the speed of a PheWAS analysis through a user-specified number of processors. BioBin is open source and the code is freely available at https://ritchielab.psu.edu. It is also available on demand from the authors. All supplemental files for this manuscript are available at https://ritchielab.psu.edu/publications/supplementary-data/psb-2016/biobin-on-multiple-phenotype.
2.1.2. Library of Knowledge Integration (LOKI)
BioBin collapses variants into biological features by consulting the Library of Knowledge Integration (LOKI), an internal repository containing diverse knowledge from multiple sources including NCBI dbSNP and gene Entrez[38], Kyoto Encyclopedia of Genes and Genomes (KEGG)[18], Gene Ontology (GO)[11], and Pharmacogenomics Knowledge Base (PharmGKB)[32]. LOKI integrates information from these external databases into a single local repository containing knowledge from the downloaded raw data in each database. The main data types used within LOKI are position, region, group, and source. Position refers to the chromosome and base-pair position of single variants, and region represents biological features containing a start and stop position including genes and copy number variants[33]. Sources are the external databases compiled in LOKI, while groups represent various groupings of biological features such as protein interactions, protein families and pathways. While LOKI is not distributed within the BioBin code due to size constraints, tools are provided within the source distribution allowing a user to compile and perform a local installation of LOKI by downloading data directly from the external sources. The data sources within LOKI can be individually updated as necessary in order to provide the most up-to-date information.
2.2. Simulations
Simulation testing was performed in order to evaluate regression (a standard burden test) and SKAT (a dispersion test) within the framework of a BioBin variant collapsing analysis. All tests were performed using SeqSIMLA2[4] to simulate sequence data as it allowed for the simulation of common burden and dispersion test assumptions. Randomly selected protein-coding variants with a MAF<5% in individuals of European descent from the 1000 Genomes Project Phase I[8] dataset were used as the basis for our simulations. This dataset was used to obtain a distribution of allele frequencies across the whole exome for each non-monomorphic single nucleotide variant site in the represented individuals of European descent (CEU, TSI, FIN, GBR, and IBS). This allele frequency distribution was then used to create the input for SeqSIMLA2. All simulations were performed with 100 variants as we calculated this to be an approximate average number of variants expected in a median sized 24,000bp gene[12]. For this calculation, we used known gene regions in the UCSC Human Genome Browser[19] to define the total gene region length and the 1000 Genomes Project to estimate the number of SNPs identified in these gene regions.
Simulation tests and specific parameters are shown in Table 1. Our simulations focused on two main tests: altering the odds ratio (OR) and altering the proportion of risk variants, with numerous parameters tested in each of these categories. Multiple testing parameters separated by commas in Table 1 correspond to independent simulations. The proportion of causal variants represents the percentage of disease sites of the total 100 variants being simulated. Likewise, the proportion of risk variants provides the number of risk variants of these causal sites. For instance, in our altering OR test category, when simulating 40% causal variants, we had 40 disease sites, and either 40-risk variants (when testing a 100% proportion of risk variants) or 20-risk variants and 20-protective variants (when testing a 50% proportion of risk variants). The specified OR corresponds to that of the individual causal variants. Type I error was estimated with 1,000 simulated null datasets using an OR of 1. Significance was assessed using α=0.05.
Table 1.
Simulation tests and Parameters
| Test Parameter | Altering OR | Altering Proportion of Risk Variants |
|---|---|---|
| Number of Simulations | 1000 | 1000 |
| Sample Size | 1000 cases and 1000 controls | 1000 cases and 1000 controls |
| Proportion of Causal Variants (n=100) | 40%, 10% | 40% |
| Disease Prevalence | 5% | 5%, 50% |
| Odds Ratio (OR) | 1.5, 2.0, 3.0 | 3.0 |
| Proportion of Risk Variants | 50%, 100% | 25%, 40%, 50%, 60%, 75%, 100% |
| Variant Weighting | No Weighting, Madsen and Browning | No Weighting, Madsen and Browning |
2.3. Application of Bin-KAT to natural dataset
A Bin-KAT test was used to analyze type II diabetes (TIID) and eight diagnosis indicators in 740 de-identified European American subjects from the Marshfield Clinic Personalized Medicine Research Project (PMRP) sequenced in the electronic Medical Records and Genomics (eMERGE) Network[15], as part of the eMERGE-PGX study[43]. Subjects were sequenced using PGRNseq[43], a next-generation sequencing platform designed for the targeted capture of selected pharmacogenes[43]. Case control status for TIID was determined using Mount Sinai's diabetes algorithm[20] from the Diabetes HTN CKD algorithm[37]. The eight diagnosis indicators analyzed are asthma, benign prostatic hyperplasia (BPH), cataracts, diverticulosis, gastroesophageal reflux disease (GERD), hypertension, hypothyroidism, and uterine fibrosis. For each diagnosis indicator, a subject was considered a case if diagnosed with one of the listed ICD-9 codes in Table 2 on two or more dates. Controls were defined as non-cases who did not meet the criteria of ICD-9 diagnosis on two or more dates.
Table 2.
Analyzed Phenotypes
| Phenotype | Diagnosis | Cases | Controls |
|---|---|---|---|
| TIID | Diabetes HTN CKD algorithm | 99 | 594 |
| Asthma | ICD-9 codes: Between ‘493.00’ and ‘493.92’ | 90 | 650 |
| (BPH) | ICD-9 codes: ‘600’, ‘600.0’, ‘600.00’, ‘600.01’, ‘600.09’, ‘600.2’, ‘600.20’, ‘600.21’, ‘600.9’, ‘600.90’, ‘600.91’ | 122 | 250 |
| Cataracts | ICD-9 codes: ‘366.10’, ‘366.12’, ‘366.14’, ‘366.15’, ‘366.16’, ‘366.17’, ‘366.9’ | 202 | 538 |
| Diverticulosis | ICD-9 codes: ‘562.00’, ‘562.01’, ‘562.02’, ‘562.03’, ‘562.10’, ‘562.11’, ‘562.12’, ‘562.13’ | 134 | 606 |
| GERD | ICD-9 codes: ‘530.81’, ‘530.11’ | 204 | 536 |
| Hypertension | ICD-9 codes: Between ‘401.00’ and ‘401.99’ | 374 | 366 |
| Hypothyroidism | ICD-9 codes: ‘244’, ‘244.8’, ‘244.9’, ‘245’, ‘245.2’, ‘245.8’, ‘245.9’ | 98 | 642 |
| Uterine Fibroids | ICD-9 codes: ‘218.0’, ‘218.1’, ‘218.2’, ‘218.9’, ‘654.10’, ‘654.11’, ‘654.12’, ‘654.13’, ‘654.14’ | 58 | 313 |
To highlight the multiple variant collapsing functions within BioBin, we binned variants having a MAF less than 0.05 by three features: gene, biological pathway and SNPEff[5] functional predictions with a minimum bin size of 5 variants. Gene binning analysis was performed on the 82 targeted pharmacogenes that passed QC. SNPEff functional predictions were used as a secondary collapsing strategy following gene binning. Variants annotated as having intergenic and intragenic effects by SNPEff were excluded from the analysis. Biological pathway variant binning was achieved using all pathway sources currently in the LOKI biorepository[40]. Overall Madsen and Browning[28] weighting was used to weigh binned variants inversely proportional to their MAF. SKAT was used to test for association between binned variants and each phenotype while adjusting for sex, year of birth, and median BMI.
3. Results
3.1. Simulations
We evaluated regression and SKAT within a BioBin coupled collapsing analysis using data simulations of 100 variants based on the allele frequencies of European subjects from the 1000 Genomes Project. All simulated conditions are shown in Table 1 and aim to test the assumptions of burden and dispersion methods. Table 3 displays that Type I error was well controlled in the analyses and was not being sacrificed in the regression or SKAT analysis.
Table 3.
Type I Error Results, standard error is in parentheses.
| Variant Weighting | SKAT Type I Error Rate | Regression Type I Error Rate |
|---|---|---|
| None | 0.045 (±0.011) | 0.061(±0.011) |
| Madsen-Browning | 0.037(±0.005) | 0.039(±0) |
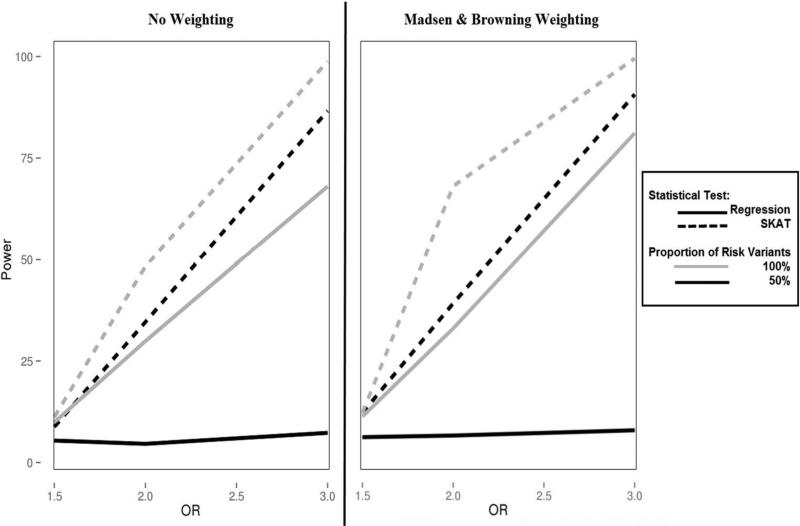
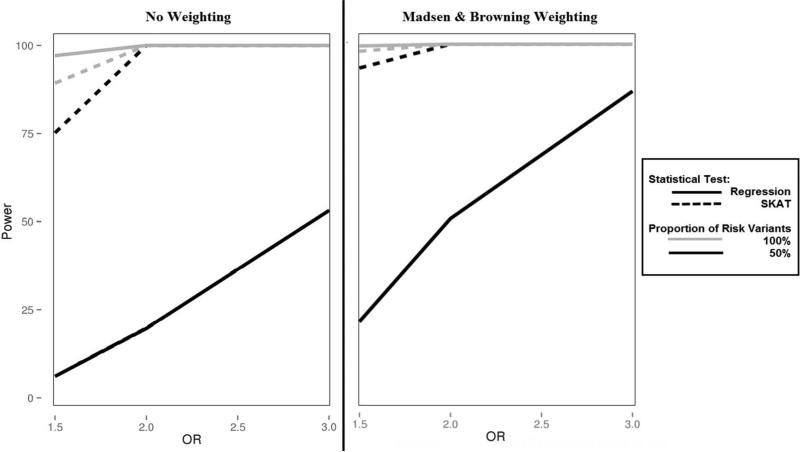
A key limitation of burden tests is loss of statistical power in the presence of a mixture of variant effects. We simulated the direction of effect by testing 100% risk variants and 50% risk, 50% protective variants. We evaluated the impact of differing directions of effect on statistical power in a Bin-KAT and BioBin-regression analysis over a varying OR range from 1.5 to 3.0. These results are shown with 10% and 40% causal variants in Figure 1 and 2, respectively. Both figures highlight the influence of variant weighting by displaying results with and without Madsen and Browning weighting.
Figure 1.
Power plot of Bin-KAT and BioBin-regression analyses with a causal variant proportion of 10%. SKAT results are represented by a dashed line; regression results have a solid line. Simulations of 100% risk variants are in grey while 50% risk variants are black.
Figure 2.
Power plot of Bin-KAT and BioBin-regression analyses with a causal variant proportion of 40%. SKAT results are represented by a dashed line; regression results have a solid line. Simulations of 100% risk variants are in grey while 50% risk variants are black.
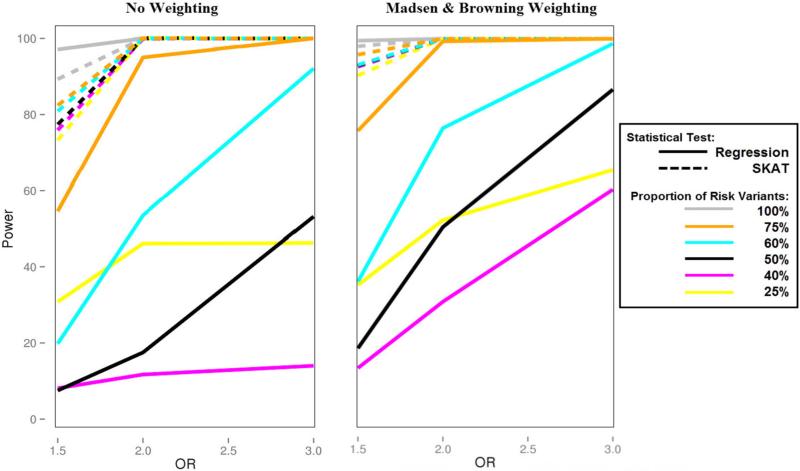
To further explore the impact of a mixture of variant effects on statistical power, we simulated data altering the proportion of risk variants over a wide range, from 25% to 100%, as seen with a disease prevalence of 5% in Figure 3. We increased this disease prevalence to 50% and present these results in Supplementary Figure 1. While a disease prevalence of 50% is high, it allowed us to create a balance in the case to control ratio and thereby symmetry in the results with comparable statistical power between 25%-75%, and 40%-60%, and a significant loss of power at 50%. This is not seen with a lower disease prevalence of 5% (Figure 3) as we are oversampling our population, so that symmetry is likely shifted.
Figure 3.
Power plot of a Bin-KAT and BioBin-regression analysis performed when altering the proportion of risk variants between 25% and 100% with a disease prevalence of 5%. SKAT results are represented by a dashed line; regression results have a solid line.
3.2 Application of Bin-KAT to natural dataset
As Bin-KAT consistently maintained greater power than a BioBin-regression, we applied this method coupled with variant weighting to simultaneously analyze 9 phenotypes in subjects of European descent from the Marshfield cohort of eMERGE-PGX project. These subjects were target sequenced for 82 pharmacogenes. We found numerous association results with p-values less than 0.05 in our gene, pathway, and SNPEff functional prediction analysis. Due to the hypothesis generating nature of this method we present all results with a p-value less than 0.05 or 0.01. As sequencing was performed on specific, targeted genes, the statistical tests are highly correlated, and therefore do not meet the independence assumptions of Bonferroni correction, which would prove too stringent in our analysis[7]. In addition, this study is exploratory in nature and all findings should be replicated in independent datasets in the future.
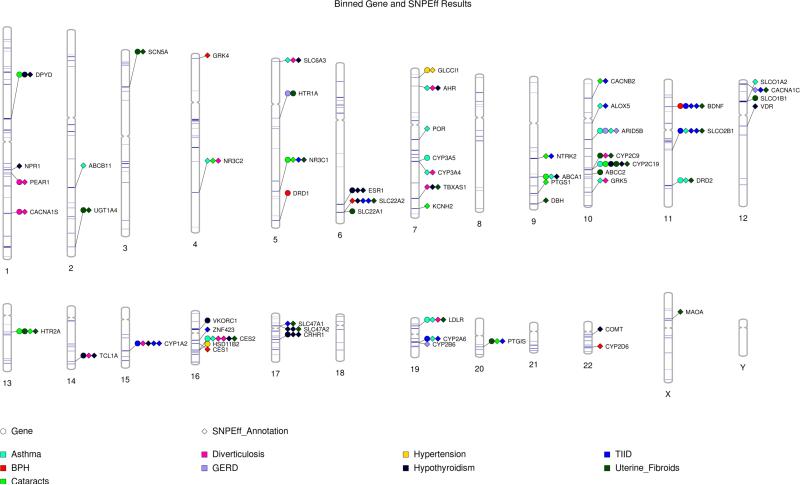
A full list of the results may be found in Supplementary Tables 1 and 2. Table 4 shows the number of results per phenotype and binned biological feature below a p-value cutoff of 0.05 for genes and SNPEff annotations, and an additional 0.01 cutoff for pathway analysis. We found significant associations with binned variants in 59 of the 82 targeted pharmacogenes. Figure 4 shows a Phenogram plot of all significant results collapsed by gene and SNPEff functional prediction displayed by chromosomal location of the gene. Details on the specific annotated SNPEff effect and impact can be found in Supplementary Table 1.
Table 4.
Number of association results per phenotype and biological feature at the specified p value cutoff. Total number of bins in each biological feature is noted in parentheses.
| Phenotype | Gene (p-value < 0.05) | Pathway (p-value<0.05) | Pathway (p-value<0.01) | SNPEff annotation (p-value <0.05) |
|---|---|---|---|---|
| Type II Diabetes | 4 (82) | 233 (8911) | 13 | 17 (458) |
| Cataracts | 5 (82) | 777 (8964) | 17 | 8 (458) |
| Hypothyroidism | 6 (82) | 324 (8991) | 6 | 19 (458) |
| Hypertension | 2 (82) | 234 (8964) | 62 | 1 (458) |
| Diverticulosis | 2 (82) | 248 (8964) | 148 | 14 (458) |
| Asthma | 6 (82) | 297 (8984) | 135 | 16 (458) |
| GERD | 2 (82) | 177 (8964) | 19 | 3 (458) |
| BPH | 2 (82) | 102 (8964) | 18 | 4 (458) |
| Uterine Fibroids | 10 (82) | 390 (8991) | 102 | 18 (458) |
Figure 4.
Phenogram plot of significant association results (p-value<0.05) in a binned gene and SNPEff functional prediction Bin-KAT analysis. The biological features are designated with different shapes, and each phenotype is represented by a different color. The target capture of the PGRNseq platform is shown by blue horizontal bands across the chromosome. The specific SNPEff effect can be found in Supplementary Table 1.
4. Discussion
In this work, we sought to expand the framework of BioBin by integrating statistical tests to provide a tool for the automated, biologically-driven binning and association analysis of rare variants. The choice of binning algorithm is often research specific, and BioBin supports this by providing variant collapsing on multiple biological levels, as well as supporting user-customized analysis. BioBin also includes multiple variant weighting schemes outside of those within a SKAT analysis, including minimum and maximum variant weighting, as well as weighting based on allele frequencies only within our phenotypic controls. Further, BioBin supports polyallelic variant sites and will incorporate all allelic information from these sites, a characteristic that is not supported by all tools. While multiple studies have performed exhaustive comparisons of burden and dispersion methods[2,6,10], we specifically chose to focus on regression and SKAT. Regression is a commonly used burden test, and several popular rare variant methods use a regression framework[1, 26, 27, 36]. SKAT was chosen due to its vast popularity as a dispersion method, its ease of covariate adjustment, and application to binary or quantitative phenotypes. Regression and SKAT have previously been compared in rare variant analysis[2, 10, 22] and here, are evaluated within the context of a biologically inspired binning method.
Simulation testing shows a Bin-KAT analysis maintains greater overall statistical power than BioBin-Regression. We found SKAT to outperform regression even in conditions where a burden analysis is assumed to have greater power than a dispersion test, such as variants influencing the phenotype in the same direction, as is presented in Figure 1 with 10% causal variants. In the 40% causal variant simulations (Figure 2), regression maintains higher power over SKAT in both weighted and unweighted tests. This suggests that the power of regression may be affected by the proportion of causal variants having the same direction of effect. However, when we encounter a mixture of both risk and protective variants, regression suffers a significant loss of power. In fact, SKAT maintains high power regardless of the proportion of risk variants simulated, and is held at 100% from an OR 2.0-3.0 (Figure 3). Our results also highlight that applying Madsen and Browning variant weighting to the binning analysis increases power.
We performed a Bin-KAT test with Madsen and Browning weighting to analyze 9 different phenotypes from Marshfield-PGX subjects who were target sequenced for specific pharmacogenes. We, and others, hypothesize that pharmacogenes related to drug response may also be associated with the diseases for which the drugs are used to treat. Using Bin-KAT, a series of significant associations were found. In the gene-binning analysis, an association between BDNF and type II diabetes (p-val 0.000437) was identified. Literature indicates that low levels of BDNF may be involved in type II diabetes pathogenesis, providing a potential explanation for the clustering of dementia, depression and type II diabetes[13, 21]. BDNF may also play a role in blood glucose metabolism and insulin resistance, a characteristic of type II diabetes[21, 30]. A number of significant results in the pathway-binning analysis performed using asthma patients included leukotriene pathways. Leukotrienes are inflammatory chemicals that can act as lipid mediators and have been well established in the pathobiology of asthma[3, 17, 44]. Leukotriene-B4 is being further investigated for its regulatory role in the development of asthma [17].
The results of this study show indications of potential pleiotropy where gene-binned variants are associated with more than one phenotype. We see this with CYP2C19, which is significantly associated with asthma, cataracts, hypothyroidism, and uterine fibroids. CYP2C19 has a highly polymorphic sequence, accounting for its variability in drug metabolism as it acts on up to 10% of clinical drugs[25]. In lung tissue, cytochrome P450 enzymes may be affected by air pollutants, and the CYP2C19*2 genotype has been implicated as a risk factor for asthma[47]. Also, linkage analysis on families with endometriosis, a disorder that may be correlated with uterine fibroids[45], indicates a potential role of CYP2C19 in endometriosis risk[39]. Association results with CYP2C19 present exciting connections that warrant further exploration. We have looked at the co-occurrence of these four phenotypes and the correlation is fairly low. Future work will aim to evaluate CYP2C19 and medication usage.
Bin-KAT serves as a powerful and versatile method for the biological binning and analysis of rare variants in sequence data. This approach was successful in the identifying novel and well-studied genes and pathways harboring low frequency variants in a multiple complex phenotype analysis. Studying the influence of low frequency variants has the potential to identify underlying risk factors, and uncover complex genotype-phenotype associations in multifactorial diseases.
Supplementary Material
Acknowledgments
We would like to thank Steven J. Schrodi, PhD, of the Bioinformatics Research Center, Marshfield Clinic in Marshfield, WI for his contributions.
Footnotes
This work is supported by NIH grant HG006389, and is also partly funded, under a grant with the Pennsylvania Department of Health using Tobacco CURE Funds. The Department specifically disclaims responsibility for any analyses, interpretations or conclusions.
References
- 1.Asimit JL, et al. ARIEL and AMELIA: Testing for an Accumulation of Rare Variants Using Next-Generation Sequencing Data. Human heredity. 2012;73(2):84–94. doi: 10.1159/000336982. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bacanu S-A, et al. Comparison of Statistical Tests for Association between Rare Variants and Binary Traits. PLoS ONE. 2012 Aug;7(8):e42530. doi: 10.1371/journal.pone.0042530. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Busse WW, et al. Leukotriene pathway inhibitors in asthma and chronic obstructive pulmonary disease. Clinical and Experimental Allergy: Journal of the British Society for Allergy and Clinical Immunology. 1999 Jun;29(Suppl 2):110–115. doi: 10.1046/j.1365-2222.1999.00019.x. 1999. [DOI] [PubMed] [Google Scholar]
- 4.Chung R-H, et al. SeqSIMLA2: simulating correlated quantitative traits accounting for shared environmental effects in user-specified pedigree structure. Genetic Epidemiology. 2015 Jan;39(1):20–24. doi: 10.1002/gepi.21850. 2015. [DOI] [PubMed] [Google Scholar]
- 5.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012 Jun;6(2):80–92. doi: 10.4161/fly.19695. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke GM, et al. A Flexible Approach for the Analysis of Rare Variants Allowing for a Mixture of Effects on Binary or Quantitative Traits. PLoS Genet. 2013 Aug;9(8):e1003694. doi: 10.1371/journal.pgen.1003694. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conneely KN, Boehnke M. So Many Correlated Tests, So Little Time! Rapid Adjustment of P Values for Multiple Correlated Tests. American Journal of Human Genetics. 2007 Dec;81(6):1158–1168. doi: 10.1086/522036. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Consortium T. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012 Nov;491(7422):56–65. doi: 10.1038/nature11632. 1000 G.P. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruchaga C, et al. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer/'s disease. Nature. 2014 Jan;505(7484):550–554. doi: 10.1038/nature12825. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dering C, et al. A comprehensive evaluation of collapsing methods using simulated and real data: excellent annotation of functionality and large sample sizes required. Frontiers in Genetics. 2014 Sep;5 doi: 10.3389/fgene.2014.00323. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dimmer EC, et al. The UniProt-GO Annotation database in 2011. Nucleic Acids Research. 2012 Jan;40(D1):D565–D570. doi: 10.1093/nar/gkr1048. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchs G, et al. 4sUDRB-seq: measuring genomewide transcriptional elongation rates and initiation frequencies within cells. Genome Biology. 2014;15(5):R69. doi: 10.1186/gb-2014-15-5-r69. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujinami A, et al. Serum brain-derived neurotrophic factor in patients with type 2 diabetes mellitus: Relationship to glucose metabolism and biomarkers of insulin resistance. Clinical Biochemistry. 2008 Jul;41(10–11):812–817. doi: 10.1016/j.clinbiochem.2008.03.003. 2008. [DOI] [PubMed] [Google Scholar]
- 14.Gottesman O, et al. The Electronic Medical Records and Genomics (eMERGE) Network: Past, Present and Future. Genetics in medicine : official journal of the American College of Medical Genetics. 2013 Oct;15(10):761–771. doi: 10.1038/gim.2013.72. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottesman O, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genetics in Medicine. 2013 Oct;15(10):761–771. doi: 10.1038/gim.2013.72. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gudmundsson J, et al. A study based on whole-genome sequencing yields a rare variant at 8q24 associated with prostate cancer. Nature Genetics. 2012 Dec;44(12):1326–1329. doi: 10.1038/ng.2437. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hallstrand TS, Henderson WR. An update on the role of leukotrienes in asthma. Current opinion in allergy and clinical immunology. 2010 Feb;10(1):60–66. doi: 10.1097/ACI.0b013e32833489c3. 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehisa M, et al. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Research. 2012 Jan;40(Database issue):D109–114. doi: 10.1093/nar/gkr988. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kent WJ, et al. The Human Genome Browser at UCSC. Genome Research. 2002 Jun;12(6):996–1006. doi: 10.1101/gr.229102. 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kho AN, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. Journal of the American Medical Informatics Association : JAMIA. 2012;19(2):212–218. doi: 10.1136/amiajnl-2011-000439. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krabbe KS, et al. Brain-derived neurotrophic factor (BDNF) and type 2 diabetes. Diabetologia. 2007 Feb;50(2):431–438. doi: 10.1007/s00125-006-0537-4. 2007. [DOI] [PubMed] [Google Scholar]
- 22.Ladouceur M, et al. The Empirical Power of Rare Variant Association Methods: Results from Sanger Sequencing in 1,998 Individuals. PLoS Genet. 2012 Feb;8(2):e1002496. doi: 10.1371/journal.pgen.1002496. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee S, et al. Optimal tests for rare variant effects in sequencing association studies. Biostatistics (Oxford, England) 2012 Sep;13(4):762–775. doi: 10.1093/biostatistics/kxs014. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee S, et al. Rare-Variant Association Analysis: Study Designs and Statistical Tests. American Journal of Human Genetics. 2014 Jul;95(1):5–23. doi: 10.1016/j.ajhg.2014.06.009. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee S-J. Clinical Application of CYP2C19 Pharmacogenetics Toward More Personalized Medicine. Frontiers in Genetics. 3. 2013 Feb; doi: 10.3389/fgene.2012.00318. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. American Journal of Human Genetics. 2008 Sep;83(3):311–321. doi: 10.1016/j.ajhg.2008.06.024. 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin D-Y, Tang Z-Z. A General Framework for Detecting Disease Associations with Rare Variants in Sequencing Studies. American Journal of Human Genetics. 2011 Sep;89(3):354–367. doi: 10.1016/j.ajhg.2011.07.015. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madsen BE, Browning SR. A Groupwise Association Test for Rare Mutations Using a Weighted Sum Statistic. PLoS Genet. 2009 Feb;5(2):e1000384. doi: 10.1371/journal.pgen.1000384. 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maher B. Personal genomes: The case of the missing heritability. Nature News. 2008 Nov;456(7218):18–21. doi: 10.1038/456018a. 2008. [DOI] [PubMed] [Google Scholar]
- 30.Marchelek-Myśliwiec M, et al. Insulin resistance and brain-derived neurotrophic factor levels in chronic kidney disease. Annals of Clinical Biochemistry. 2015 Mar;52(Pt 2):213–219. doi: 10.1177/0004563214533514. 2015. [DOI] [PubMed] [Google Scholar]
- 31.McCarty CA, et al. Informed consent and subject motivation to participate in a large, population-based genomics study: the Marshfield Clinic Personalized Medicine Research Project. Community Genetics. 2007;10(1):2–9. doi: 10.1159/000096274. 2007. [DOI] [PubMed] [Google Scholar]
- 32.McDonagh EM, et al. From pharmacogenomic knowledge acquisition to clinical applications: the PharmGKB as a clinical pharmacogenomic biomarker resource. Biomarkers in Medicine. 2011 Dec;5(6):795–806. doi: 10.2217/bmm.11.94. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore CB, et al. BioBin: a bioinformatics tool for automating the binning of rare variants using publicly available biological knowledge. BMC Medical Genomics. 2013 May;6(Suppl 2):S6. doi: 10.1186/1755-8794-6-S2-S6. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moore CB, et al. Low Frequency Variants, Collapsed Based on Biological Knowledge, Uncover Complexity of Population Stratification in 1000 Genomes Project Data. PLoS Genet. 2013 Dec;9(12):e1003959. doi: 10.1371/journal.pgen.1003959. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore CB, et al. Phenome-wide Association Study Relating Pretreatment Laboratory Parameters With Human Genetic Variants in AIDS Clinical Trials Group Protocols. Open Forum Infectious Diseases. 2014 Dec;2:1. doi: 10.1093/ofid/ofu113. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morgenthaler S, Thilly WG. A strategy to discover genes that carry multi-allelic or mono-allelic risk for common diseases: A cohort allelic sums test (CAST). Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2007 Feb;615(1–2):28–56. doi: 10.1016/j.mrfmmm.2006.09.003. 2007. [DOI] [PubMed] [Google Scholar]
- 37.Nadkarni GN, et al. Development and validation of an electronic phenotyping algorithm for chronic kidney disease. AMIA Annual Symposium Proceedings. 2014 Nov;:907–916. 2014. [PMC free article] [PubMed] [Google Scholar]
- 38.NCBI Resource Coordinators 2013 Database resources of the National Center for Biotechnology Information. Nucleic Acids Research. 2013 Jan;41(Database issue):D8–D20. doi: 10.1093/nar/gks1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Painter JN, et al. High-density fine-mapping of a chromosome 10q26 linkage peak suggests association between endometriosis and variants close to CYP2C19. Fertility and Sterility. 2011 Jun;95(7):2236–2240. doi: 10.1016/j.fertnstert.2011.03.062. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pendergrass SA, et al. Genomic analyses with biofilter 2.0: knowledge driven filtering, annotation, and model development. BioData Mining. 2013 Dec;6(1):25. doi: 10.1186/1756-0381-6-25. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pendergrass SA, et al. Phenome-Wide Association Study (PheWAS) for Detection of Pleiotropy within the Population Architecture using Genomics and Epidemiology (PAGE) Network. PLoS Genetics. 2013 Jan;9:1. doi: 10.1371/journal.pgen.1003087. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pendergrass SA, et al. The Use of Phenome-Wide Association Studies (PheWAS) for Exploration of Novel Genotype-Phenotype Relationships and Pleiotropy Discovery. Genetic epidemiology. 2011 Jul;35(5):410–422. doi: 10.1002/gepi.20589. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rasmussen-Torvik LJ, et al. Design and anticipated outcomes of the eMERGE-PGx project: a multicenter pilot for preemptive pharmacogenomics in electronic health record systems. Clinical Pharmacology and Therapeutics. 2014 Oct;96(4):482–489. doi: 10.1038/clpt.2014.137. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sampson A, Holgate S. Leukotriene modifiers in the treatment of asthma. BMJ : British Medical Journal. 1998 Apr;316(7140):1257–1258. doi: 10.1136/bmj.316.7140.1257. 1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uimari O, et al. Do symptomatic endometriosis and uterine fibroids appear together? Journal of Human Reproductive Sciences. 2011;4(1):34–38. doi: 10.4103/0974-1208.82358. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu MC, et al. Rare-Variant Association Testing for Sequencing Data with the Sequence Kernel Association Test. The American Journal of Human Genetics. 2011 Jul;89(1):82–93. doi: 10.1016/j.ajhg.2011.05.029. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yildirim Yaroğlu H, et al. CYP2C19 gene polymorphism may be a risk factor for bronchial asthma. Medical Principles and Practice: International Journal of the Kuwait University, Health Science Centre. 2011;20(1):39–42. doi: 10.1159/000322077. 2011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.