Abstract
Oncolytic viruses are known to stimulate the antitumor immune response by specifically replicating in tumor cells. This is believed to be an important aspect of the durable responses observed in some patients and the field is rapidly moving toward immunotherapy. As a further means to engage the immune system, we engineered a virus, vesicular stomatitis virus (VSV), to encode the proinflammatory cytokine interferon-γ. We used the 4T1 mammary adenocarcinoma as well as other murine tumor models to characterize immune responses in tumor-bearing animals generated by treatment with our viruses. The interferon-γ-encoding virus demonstrated greater activation of dendritic cells and drove a more profound secretion of proinflammatory cytokines compared to the parental virus. From a therapeutic point of view, the interferon-γ virus slowed tumor growth, minimized lung tumors, and prolonged survival in several murine tumor models. The improved efficacy was lost in immunocompromized animals; hence the mechanism appears to be T-cell-mediated. Taken together, these results demonstrate the ability of oncolytic viruses to act as immune stimulators to drive antitumor immunity as well as their potential for targeted gene therapy.
Introduction
The capability to evade the immune system is essential for tumor establishment and has been identified as a hallmark of cancer.1 Conversely, an effective antitumor immune response against tumor-associated antigens is believed to prevent relapse of the disease.2 Various proinflammatory cytokines stimulate immunity and their exogenous administration has been used for the treatment of many cancers.3 Among these, interferon-γ (IFNγ), has a myriad of roles in both arms of immunity.4 It is mainly produced by T cells, natural killer (NK) cells, and macrophages and has effects on all nucleated cells of the body.5 It is a powerful immune modulator that has antiproliferative, proapoptotic, and antiangiogenic functions.5,6 It also induces adaptive immunity through increased antigen presentation and T-cell activation.7 The pivotal role of IFNγ in tumor immunosurveillance is exemplified by the observation that mice deficient for the IFNγ receptor develop tumors more rapidly and at a higher frequency.8 Many observations show that the presence of IFNγ might be beneficial in the treatment of various cancers. For examples, in human breast cancer samples, the expression of IFNγ and its receptor were reported to be increased in benign lesions compared to malignant lesions.9 Because of these observations, along with its many protective functions against cancer and its immune-stimulating properties, IFNγ has long been considered a promising candidate for the treatment of various malignancies.5 It was demonstrated using the 4T1 murine breast cancer model that intratumoral (IT) administration of IFNγ reduces metastases to the lungs and liver.10 In the clinic, it has been shown that that local injection of IFNγ in skin lesions of breast cancer patients resulted in partial regression.11 Also, recombinant human IFNγ-1b has successfully been used to delay progression of microbial infections in chronic granulomatous disease,12 but its use in human cancer treatment has given mixed results.13–16 One of the reasons for the limited efficacy was attributed to the very short half-life of IFNγ in serum and therefore, local expression is a more promising approach.17 One successful strategy to deliver genes to the tumor site is to use viruses. Indeed, the direct IT injection of virus expressing IFNγ demonstrated efficacy in two different xenograft models.18 Tumor-specific oncolytic viruses (OV) encoding transgenes have also been shown to be an efficient tool to drive sustained local transgene expression.19,20
OVs like the vesicular stomatitis virus (VSV) were selected to specifically destroy tumor cells while leaving normal tissues unaffected.21 While normal cells are protected by type I IFNs, cancer cells frequently acquire defects in this pathway. Thus, normal, but not tumor cells, will be protected against VSV.22 To further attenuate VSV, a deletion in its M protein (VSVΔ51), which prevents the blockade of the IFN response, has been developed for clinical use.23 OVs destroy tumors by several mechanisms. Of these, the direct oncolysis of cancer cells is an important aspect.21 Nevertheless, the potential of VSV as an immunotherapy agent is under-studied.19 By destroying cancer cells, viruses liberate tumor-associated antigens in the presence of pathogen-associated molecular patterns, which stimulates antitumor immunity.24 This can result in long-lasting immune responses that target both primary tumors and metastases.25 In this study, we engineered VSVΔ51 to express murine IFNγ. In murine tumor models, we demonstrate in vivo expression of functional IFNγ within the tumor microenvironment. Compared to the parental virus, VSVΔ51-IFNγ elicited secretion of proinflammatory factors in the blood, enhanced activation of dendritic cells (DCs), and generated a greater tumor-specific immune response.
Results
IFNγ expression does not affect the ability of VSV to replicate and kill tumor cells
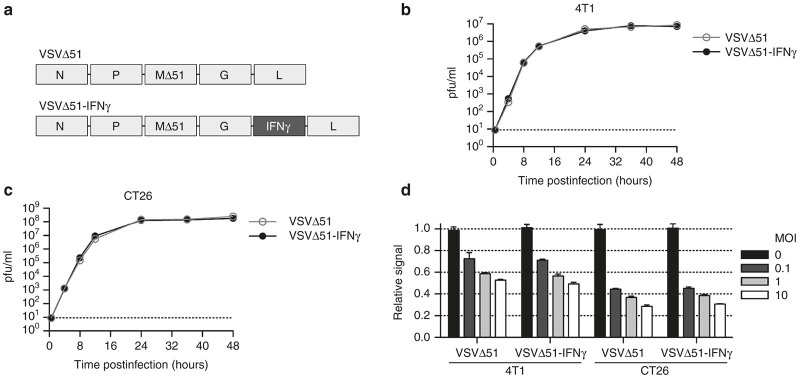
We engineered the virus to encode the murine IFNγ gene between the G and L genes (Figure 1a) based on the previous reports of successful transgene expression in this location without impairing virus replication.26 We assessed its replication kinetics compared to the parental virus (VSVΔ51) in two different tumor cell lines. Cells were infected at a low multiplicity of infection (MOI) in vitro to generate multi-step growth curves. In both the 4T1 mammary and the CT26 colon carcinoma cell lines, we obtained nearly overlapping growth curves when comparing the two viruses (Figure 1b,c), demonstrating that the expression of IFNγ by VSVΔ51 does not affect its replication in these cell lines. To investigate the cytolytic capabilities of the viruses, we infected the same cell lines at various MOIs and quantified their metabolic activity. As expected, we observed a dose-dependent killing of the cells (Figure 1d). The results obtained showed no difference for the two viruses; thus, the expression of IFNγ does not influence viral cytotoxicity. Notably, our data indicate a greater sensitivity of the CT26 cells compared with the 4T1 cells as approximately 10-fold more virus (Figure 1b,c) and 25% more killing (Figure 1d) are observed for the CT26 cells.
Figure 1.
The IFNγ-encoded virus is not impaired in its replication kinetics and cytotoxicity. (a) Schematic representation of the VSVΔ51 and VSVΔ51-IFNγ viral backbones. (b) Replication kinetics of VSVΔ51 and VSVΔ51-IFNγ in the 4T1 and (c) CT26 tumor cell lines upon infection at a multiplicity of infection (MOI) of 0.1. The dashed line indicates the limit of detection. The graphs show virus titers obtained from duplicate samples. (d) In vitro cell viability assay of 4T1 and CT26 cells infected for 24 hours at various MOIs. Results were normalized to the average of the values obtained for the corresponding noninfected cells. Results represent values obtained from triplicate samples. There is no statistically significant difference between the viruses in b, c, and d, according to the two-way analysis of variance test.
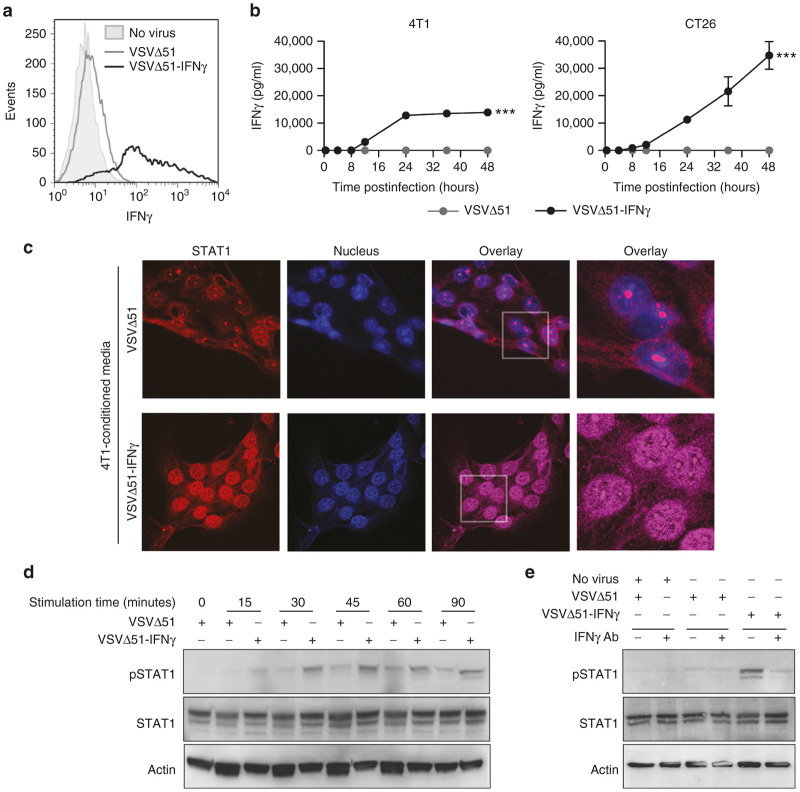
To confirm transgene expression, 4T1 and CT26 cells were infected with virus and the expression of IFNγ was assessed by flow cytometry. IFNγ was detected only in cells infected with VSVΔ51-IFNγ (Figure 2a). To quantify the amount of IFNγ produced, supernatants from infected cells were collected at various time points after infection and the concentration of IFNγ was assessed by enzyme-linked immunosorbent assay (ELISA). We observed secretion of significant amounts of IFNγ in both cell lines infected with VSVΔ51-IFNγ while none was detected from cells infected with the parental virus (Figure 2b). The IFNγ produced by 4T1 cells plateaued at 14,000 pg/ml at 24 hours, while CT26 cells reached 35,000 pg/ml at 48 hours postinfection. These findings are in agreement with the increased replication and killing by VSV in CT26 cells as compared to 4T1 cells (Figure 1b–d).
Figure 2.
Virus-expressed IFNγ is secreted and functional. (a) Flow cytometry histogram showing the intracellular IFNγ staining of 4T1 cells uninfected or infected with VSVΔ51 or VSVΔ51-IFNγ for 18 hours at a multiplicity of infection (MOI) of 0.01. (b) IFNγ concentrations determined by enzyme-linked immunosorbent assay of the supernatants of 4T1 and CT26 cells infected with VSVΔ51 or VSVΔ51-IFNγ at an MOI of 0.01 for various periods of time. Results represent values obtained from duplicate infections. ***P < 0.001 (two-way analysis of variance). (c) Confocal microscopy pictures of 4T1 cells stained with a STAT1-specific antibody and 4’,6-diamidino-2-phenylindole. Supernatant from 4T1 cells uninfected or infected for 24 hours at an MOI of 3 was collected and transferred onto fresh 4T1 cells for 60 minutes upon virus removal (virus-cleared conditioned media). The gray boxes indicate the region that is enlarged in the right panels. (d) and (e) Western blot analysis of phosphorylated-STAT1 (pSTAT1), STAT1 and actin. Virus-cleared conditioned media was transferred onto fresh 4T1 cells for various periods of time. (e) For half the conditions, the supernatant was incubated with an IFNγ-blocking antibody prior to a 20-minute incubation with the fresh cells.
IFNγ encoded by VSV is functional
Upon IFNγ binding to its receptor, the signal transducer and activator of transcription (STAT1) is phosphorylated and translocates to the nucleus to induce transcription of target genes.4 Thus, to assess the functionality of IFNγ, we evaluated the phosphorylation of STAT1 as well as its nuclear translocation mediated by virus-cleared supernatants of infected tumor cells. The concentration of IFNγ was determined by ELISA and diluted to 100 pg/ml, a concentration that is sufficient to mediate STAT1 phosphorylation (pSTAT1) in 4T1 and CT26 cells. To assess the subcellular localization of STAT1 upon stimulation, we performed confocal microscopy on cells incubated for 60 minutes with the same conditioned media. While only a small number of cells incubated with the VSVΔ51-conditioned supernatant demonstrated a nuclear localization of STAT1, all of the cells from the IFNγ virus-conditioned media displayed nuclear localization of STAT1 (Figure 2c). By western blot, we detected increased levels of pSTAT1 for both conditioned supernatants compared to control supernatant (Figure 2d). However, pSTAT1 was detected earlier for the VSVΔ51-IFNγ supernatant (15 versus 30 minutes) and the bands showed a greater signal for all time points tested. In order to validate that the signal obtained for pSTAT1 was the result of direct IFNγ stimulation, we preincubated the conditioned supernatants with an IFNγ-blocking antibody. After a 20-minute stimulation, only the cells incubated with the VSVΔ51-IFNγ-conditioned media showed a band corresponding to pSTAT1 (Figure 2e), consistent with the results obtained in Figure 2d. As expected, IFNγ blockade prevented the phosphorylation of STAT1 (Figure 2e, right lane), confirming the role of IFNγ in the upregulation of pSTAT1. Similar results were obtained with the CT26 cell line (not shown). Interestingly, a flow cytometry experiment demonstrated an increase in MHCI (an IFNγ responsive gene) surface expression for cells infected in vitro with the IFNγ virus for 24 hours (Supplementary Figure S1). Taken together, the data demonstrate the secretion of biologically active IFNγ by VSVΔ51-IFNγ upon infection of tumor cells.
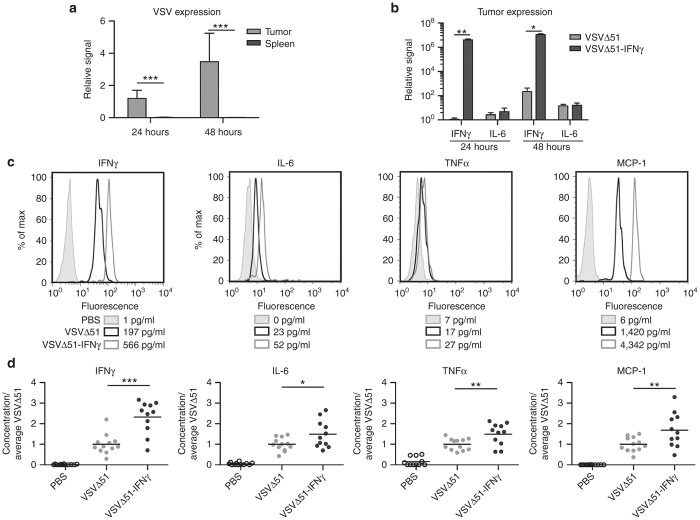
In order to evaluate the localization of the viruses following a single IT injection in our model, we performed a complete biodistribution by plaque assay and found the virus to be restricted to the tumors at both 24 and 48 hours post-treatment (Supplementary Figure S2a). We also performed quantitative polymerase chain reaction (qPCR) on spleen of the same animals since this technique is more sensitive and the spleen filters the blood and offers a good readout of potential leakage of virus into the bloodstream. Our results demonstrate that, at both time points, the virus was barely detectable in the spleens of the animals (Figure 3a). To measure the expression of IFNγ and IL-6 (an IFNγ responsive gene) in the tumors, animals were treated IT (single dose) with either virus and the expression of the cytokines was measured by qPCR 24 and 48 hours post-treatment. Our data show that, although the mRNA levels of IFNγ are slightly more than 10,000-fold higher in the tumors from the animals treated with VSVΔ51-IFNγ, the expression of IL-6 remains the same for both viruses (Figure 3b). Since we expect IL-6 and other cytokines to be mostly upregulated by immune cells, we sought to determine the systemic levels of IFNγ and various proinflammatory factors that should be upregulated in the presence of IFNγ. To do so, 4T1 tumor-bearing mice were treated IT (single dose) with phosphate-buffered saline (PBS), VSVΔ51, or VSVΔ51-IFNγ and were sacrificed 24 and 48 hours postinjection to collect blood and tumor samples. qPCR was used to quantify the amount of virus present in the tumors prior to cytokine analysis. Similar amounts of viral genomes were detected in both groups (Supplementary Figure S2b). If the transgene is functional in vivo, different proinflammatory factors should be increased compared to the control group. Secreted proinflammatory factors should be detected in the blood if produced in sufficient amounts, thus resulting in broader immune activation. To test this, we isolated serum from treated animals and IFNγ, interleukin-6 (IL-6), tumor necrosis factor-α (TNFα) and monocyte chemoattractant protein-1 (MCP-1) concentrations were determined using cytometric bead arrays. All the factors tested were higher in both virus-treated groups compared with PBS-treated animals (Figure 3c,d), consistent with an immune response to viral infection.27 Importantly, IFNγ levels were higher in the serum of VSVΔ51-IFNγ-treated 4T1 tumor-bearing mice, reaching levels around 600 pg/ml compared to 200 pg/ml for the VSVΔ51-treated group (Figure 3c), thus confirming expression of the viral transgene (Figure 3b). The concentrations of IL-6, MCP-1, and TNFα were also elevated compared with the parental virus (Figure 3c,d), confirming functionality of the virus-encoded IFNγ. Similar results were obtained with CT26 tumor-bearing mice (Supplementary Figure S2c).
Figure 3.
VSVΔ51-IFNγ induces increased serum levels of various proinflammatory cytokines. (a) VSV N mRNA levels in the tumors and spleens of intratumorally-treated animals 24 and 48 hours post-treatment (single injection) were assessed by quantitative polymerase chain reaction (qPCR) (n = 3). Results are expressed as fold increase over the average obtained for VSVΔ51 at 24 hours in the tumors. (b) IL-6 and IFNγ mRNA levels in the tumors of phosphate-buffered saline (PBS) or virus-treated animals 24 and 48 hours after a single intratumoral treatment were assessed by qPCR (n = 4). Results are expressed as fold increase over the average obtained for VSVΔ51 at 24 hours in the tumors. (c) Cytometric bead array histograms showing one representative sample per group. Levels of various proinflammatory cytokines in the serum of 4T1 tumor-bearing mice treated IT with PBS or virus 24 hours after treatment were analyzed. The numbers in the legend indicate the cytokine concentration in the serum. (d) Relative concentration of each cytokine for all animals tested. The data represents a combination of two different experiments. *P < 0.1, **P < 0.01, ***P < 0.001 (unpaired one-tailed T-test).
The expression of IFNγ by VSV differentially shapes the immune landscape
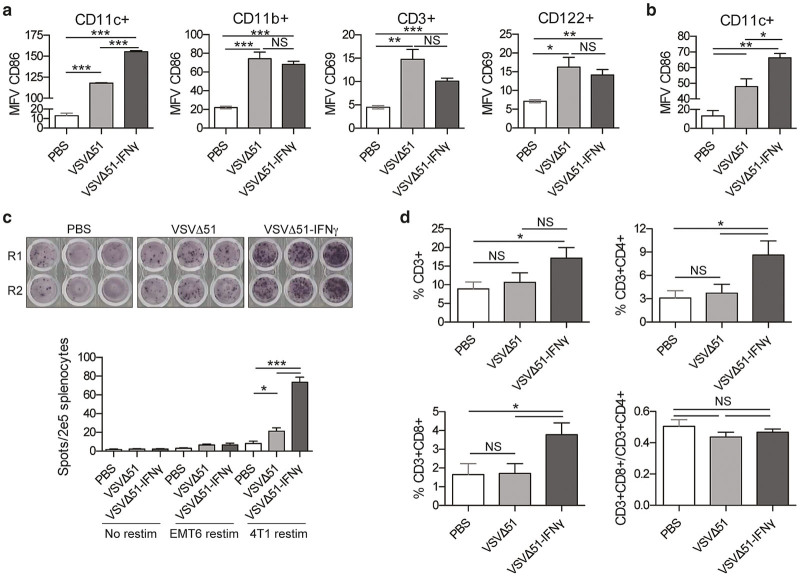
To determine whether expression of IFNγ by the virus would lead to a greater activation of immune cells, we performed a flow cytometric survey of cell populations obtained from the spleen of treated mice. Supplementary Figure S3a shows that similar amounts of VSVΔ51 and VSVΔ51-IFNγ were detected in the lungs of the 4T1 lung tumor-bearing animals 24 hours following administration of a single IV treatment. For CD3+ T cells as well as CD122+ NK cells, the expression levels of the CD69 early activation marker was assessed.28,29 Both viruses triggered the activation of T and NK cells, but no significant difference was observed in their activation levels when treated with the IFNγ-expressing virus at this time point (Figure 4a, right panels). For DCs (CD11c+) as well as monocytes, macrophages and granulocytes (CD11b+), the expression of the early activation marker CD86 was measured.30 While both viruses activated DCs and granulocytes, we consistently observed an increase in CD86 mean fluorescence value on CD11c+ splenocytes from mice treated with VSVΔ51-IFNγ compared to the parental virus (Figure 4a, left panels). Remarkably, the same phenomenon was observed 24 hours after IT injection (Figure 4b). Similar results were also observed for CT26 lung tumor-bearing mice (Supplementary Figure S3b). Interestingly, in the CT26 tumor model, NK-cell activation was still detectable after 48 hours, but went back to basal levels 96 hours post-treatment (Supplementary Figure S3c).
Figure 4.
IFNγ-expressing virus induces greater activation of dendritic cells. (a) 4T1 lung tumor-bearing mice were treated intravenously with a single dose of virus or phosphate-buffered saline (PBS) (n = 4). Twenty-four hours post-treatment, splenocytes were harvested and stained for CD11c, CD11b, CD86, CD3, CD122, and CD69 24 hours post-treatment. Samples were analyzed by flow cytometry. The graphs show the mean fluorescence values. Data are representative of two different experiments. (b) 4T1 fat-pad tumor-bearing mice were treated intratumorally with one dose of virus or PBS. Twenty-four hours post-treatment, the splenocytes were stained for CD11c and CD86. (c) IFNγ ELISPOT of splenocytes restimulated ex vivo with 4T1 tumor cells. 4T1 fat-pad tumor bearing mice were treated with PBS, VSVΔ51, or VSVΔ51-IFNγ. Ten days post-treatment, splenocytes were harvested and the ELISPOT was performed. R1 and R2 are two replicates from the same mouse. The bar chart represents the counts obtained for each group. (d) Single cell suspensions were obtained from tumors of the same mice and stained for CD3, CD4, and CD8. Samples were analyzed by flow cytometry. The graphs show the percentage of the cells within the tumor that were positive for the different markers. NS: P > 0.1, *P < 0.1, **P < 0.01, ***P < 0.001 (unpaired one-tailed T-test).
To evaluate the antitumor immune response generated by the viruses, an IFNγ ELISPOT was performed with splenocytes isolated from immunocompetent animals following virus treatment. The spots observed following ex vivo restimulation with 4T1 target cells reflect tumor-specific activation. Our results show that, in absence of restimulation, or when the splenocytes are restimulated with a tumor cell line to which the animals were never exposed (EMT6), most animals showed no activation of splenocytes (Figure 4c and Supplementary Figure S3d). However, there was a threefold increase in the number of spots detected for the VSVΔ51-IFNγ-treated animals compared with the control virus (Figure 4c, bottom panel), demonstrating a greater antitumor immune response. Because the spots appear only upon restimulation with 4T1 cells, the immune cells that are responsible for the IFNγ secretion are likely tumor-specific T cells. We therefore investigated the presence of T cells within the tumors. We found an increase in the percentage of CD3+ cells within the tumor with VSVΔ51-IFNγ treatment (Figure 4d). Percentages of both CD4+ and CD8+ T cells were augmented two- to threefold. However, the ratio of effector to helper cells was unchanged (Figure 4d, bottom right panel). Taken together, these data indicate that VSVΔ51-IFNγ induces a greater antitumor immune response.
VSVΔ51-IFNγ shows greater efficacy in vivo in various murine tumor models
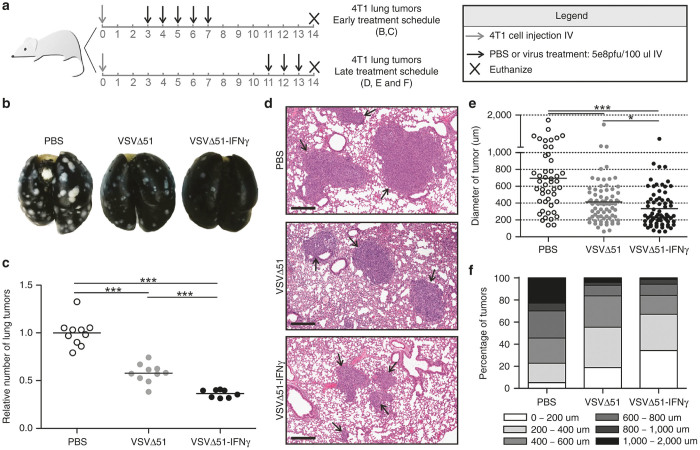
To determine if the increased secretion of proinflammatory factors, the enhanced activation of DCs and the greater antitumor immune response translated into improved efficacy in vivo, we first assessed the effect of VSVΔ51-IFNγ following systemic delivery in the 4T1 lung-tumor model. Given the extreme aggressiveness of this model and difficulty in observing survival advantages, alternate readouts of efficacy were utilized. We used both early and a late treatment schedules (illustrated in Figure 5a) and looked at lung tumors. When mice were treated early post-tumor seeding, analysis of the lungs revealed fewer tumors for both virus-treated groups compared with the control group (Figure 5b,c), with VSVΔ51-IFNγ being more efficient, reducing the average amount of lung tumor nodules (61 for VSVΔ51-IFNγ compared to 185 for the untreated mice and 112 for the control virus) (Supplementary Figure S4a). This reduction in tumor numbers was also accompanied by a modest prolongation of survival (Supplementary Figure S4b). When mice were treated 11 days post-tumor seeding, lung tumors showed a reduction in size following treatment with both viruses when assessed by H&E staining. Representative pictures are shown in Figure 5d. When all the tumors from the H&E of the lungs were measured, we observed a significant difference in the mean tumor diameter with both viruses performing better than PBS and the IFNγ-encoding virus showing the smallest tumors (average of 329 µm for VSVΔ51-IFNγ, 412 µm for VSVΔ51, and 687 µm for control mice) (Figure 5e). Furthermore, when the tumors were subdivided into groups according to their size and the percentage of tumors belonging to each group was calculated, it was clear that the tumors from mice treated with the IFNγ-expressing virus were much smaller than the ones from the other groups (Figure 5f).
Figure 5.
VSVΔ51-IFNγ reduces the number and size of lung tumors in the 4T1 tumor model. (a) Schematic representation of the treatment schedules used in this figure. (b) Representative pictures of lungs from mice treated with PBS, VSVΔ51 or VSVΔ51-IFNγ. Lungs were perfused with india ink to reveal the tumors (appear white). (c) Total surface lung tumor counts of all mice. (d) Representative pictures showing the average size of tumors observed in each treatment group (scale bar: 300 μm). Mice from the late treatment schedule (n = 3 or 4) were sacrificed, the lungs were fixed in formalin and paraffin embedded sections were stained with hematoxylin and eosin. (e) Graph showing the largest diameter of each tumor observed in d. (f) The results obtained in e were subdivided according to their size and represented as percentage of the tumors within the group belonging to each subdivision. *P < 0.1, ***P < 0.0001 (Mann-Whitney U-test).
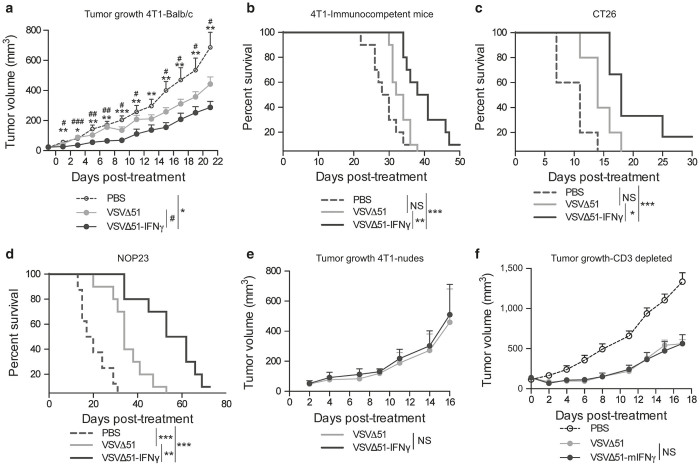
We investigated the efficacy of VSVΔ51-IFNγ in several subcutaneous and orthotopic tumor models following IT administration. The 4T1 and CT26 tumor models were investigated, as well as NOP23, another breast cancer model that can be implanted orthotopically in the mammary fat pad. The treatment schedules were optimized for each tumor model and are summarized in Supplementary Figure S4c. Tumors were measured over time and tumor volume was calculated. Figure 6a shows that VSVΔ51-IFNγ was the most efficient at controlling tumor growth in the 4T1 model. Indeed, the differences observed between the PBS and the VSVΔ51-IFNγ-treated group were highly significant within 2 days after initiation of treatment. The reduction in tumor size also translated to a prolongation of survival, with one animal being cured (Figure 6b). A significant prolongation of survival was also observed for the CT26 and NOP23 tumor models (Figure 6c,d). To assess whether the improved efficacy observed with VSVΔ51-IFNγ compared to VSVΔ51 was immune-mediated, we compared the efficacy of the two viruses in terms of tumor growth and survival of 4T1 tumor-bearing nude mice, which lack T cells. In contrast to the results obtained in immunocompetent mice (Figure 6a,b), 4T1 tumors in nude mice showed no difference in growth rate or survival following treatment with the two viruses (Figure 6e and Supplementary Figure S4d). To further support this finding, we repeated the experiment in immunocompetent mice but with depletion of the T-cell compartment (CD3 depletion). As expected, our data show the loss of the survival advantage in absence of T cells (Figure 6f and Supplementary Figure S4e). Thus, T cells are required for the tumor control mediated by VSVΔ51-IFNγ.
Figure 6.
VSVΔ51-IFNγ demonstrates better efficacy compared to the parental virus. (a) Tumor volumes of 4T1 fat-pad tumors of mice treated with phosphate-buffered saline (PBS), VSVΔ51 or VSVΔ51-IFNγ (n = 9 or 10). The animals were treated daily when the average tumor diameter reached 5 mm, for a total of five injections (first dose IV and subsequent doses IT). Mice were sacrificed when they reached end-point. NS: P > 0.1, *P < 0.1, **P < 0.01, ***P < 0.001 (unpaired one-tailed T-test). (b) Kaplan-Meier survival curves were generated from the same experiment. (c) and (d) Kaplan-Meier survival curves of CT26 and NOP23 tumor-bearing mice treated or not with VSVΔ51 or VSVΔ51-IFNγ. The CT26 tumor-bearing animals were treated daily with three injections (first dose IV and subsequent doses IT) and the NOP23 tumor-bearing animals were treated IT every second day starting at day 18 for a total of six injections. (e) Tumor volumes of 4T1 fat-pad tumors of nude mice treated with VSVΔ51 or VSVΔ51-IFNγ (n = 5). The animals were treated daily when the average tumor diameter reached 5 mm, for a total of five injections (first dose IV and subsequent doses IT). (f) Tumor volumes of 4T1 fat-pad tumors of Balb/c mice treated with PBS or either viruses following T-cell depletion (n = 7). T cells were depleted when the average tumor diameter reached 5 mm and the virus treatment started 24 hours later, for a total of five injections (first dose IV and subsequent doses IT). For survival experiments; NS: P > 0.1, *P < 0.1, **P < 0.01, ***P < 0.001 (Mantel-Cox test).
Discussion
In this study, we demonstrate that IFNγ expression by VSVΔ51 induces a more potent antitumor immune response leading to an improved efficacy compared with VSVΔ51 in various murine tumor models. VSVΔ51-IFNγ produces high amounts of functional transgene both in vitro and in vivo. While in vitro only the VSVΔ51-IFNγ-infected cells produced IFNγ (Figure 2a,b), the signal observed in the serum of VSVΔ51-treated animals (Figure 3c,d) can be explained by the presence of various immune cells that can induce and secrete the cytokine upon virus recognition. Accordingly, virus injection was sufficient to trigger secretion of IFNγ and activate immune cells without the requirement for transgene expression (Figures 3d and 4a,b). Although detectable amounts of IFNγ were produced in response to treatment with the parental virus, the improved efficacy observed in various tumor models (Figures 5 and 6) demonstrates the benefits of virus mediated overexpression of IFNγ. These findings suggest that specific production of the cytokine within the tumor microenvironment is beneficial for the antitumor immune response. In addition, the treatment might also be improved by the sustained virus-driven overexpression of IFNγ.
We suggest that VSVΔ51-IFNγ, in addition to its direct effects on tumor cells, induces a greater tumor-specific immune response by increasing MHCI antigen presentation by tumor cells (Supplementary Figure S1) as well as enhancing DC maturation (Figure 4a,b). This antitumor immunity would control tumor growth and even cure some of the animals (Figure 6). Consistent with this theory, we observed a greater antitumor immune response for the mice treated with VSVΔ51-IFNγ compared to the parental virus (Figure 4c). Although total splenocytes were used for the ELISPOT assay, the cells secreting IFNγ are likely T cells. Indeed, mainly NK cells, macrophages and T cells secrete IFNγ.4 Of these, only the T cells require antigen stimulation to produce the cytokine. Concurrent with this, NK-cell activation, as measured by CD69 expression at cell surface, was undetectable 96 hours after treatment (Supplementary Figure S3c). Furthermore, efficacy was completely abrogated when monitoring tumor growth and survival of immunocompromised animal or following T-cell depletion of immunocompetent animals (Figure 6e,f and Supplementary Figure S4d,e). Given that nude mice are T-cell-deficient and that T-cell depletion also led to this effect, the loss of the phenotype also points to a role of the adaptive arm of immunity for the improved efficacy conferred by the IFNγ-producing virus. The roles of mature DCs and IFNγ in the generation of efficient T-cell responses are well established. Of note, IFNγ was shown to increase proliferation of CD4+ T cells31 as well as numbers of CD8+ T cells in the context of acute viral infection32 and we showed increased presence of T cells in the tumors of the VSVΔ51-IFNγ-treated animals (Figure 4d).
Our results show the induction of various proinflammatory factors in vivo in response to treatment with our virus (Figure 3). Of these, MCP-1 is known to be induced by IFNγ33. While MCP-1 was found to be produced by various cells, macrophages and DCs are the main source.34 Although we did not observe any difference in the early activation of CD11b+ cells (Figure 4a), IFNγ is known for its importance in conditioning macrophages for activation35 and thus, these cells might have important roles to play in different contexts. For instance, the lungs contain a high amount of resident macrophages, which might have an important role to play in this particular setting. On the other hand, we consistently observed a greater activation of DCs following treatment with VSVΔ51-IFNγ (Figure 4a,b and Supplementary Figure S3b), these cells might contribute to the increased concentration of MCP-1 detected in the serum of the treated animals. Furthermore, MCP-1 is mostly known for its role in attracting monocytic cells; however, it was also shown to attract adoptively-transferred T cells to tumors producing the chemokine.36 Given the local IT production of IFNγ following treatment with our engineered virus (Figure 3b), MCP-1 should be locally produced in the tumor and lead to a similar situation. Consistent with this theory, our results show an increase in IT T cells following administration of VSVΔ51-IFNγ compared to the parental virus (Figure 4d). Other IFNγ-induced factors such as IP-10, which are also known to attract T cells, could be involved in the observed phenotype as well.37
Despite the fact that IFNγ is well known for its antiviral activity,38 we did not observe any difference in the amount virus recovered from the tumors 24 hours postinjection (Supplementary Figure S2a,b). Although we did not investigate this, expression of the cytokine by the virus may result in inhibition of viral replication at later time points or in normal tissues, thus acting as a safety mechanism. This strategy would be similar to VSV expressing the antiviral cytokine IFNβ, which was shown to have a better safety profile compared with the parental virus.39 One of the main causes of toxicity following treatment with wild-type VSV is neuro-pathogenesis which results from the replication of the virus in the central nervous system and can lead to paralysis or death.40 Although type I IFNs are required for the control of VSV infection in peripheral tissues, the lack of induction of these cytokines in the central nervous system seems to be the reason for the virus proliferation observed in the brain.41 Interestingly, IFNγ was shown to be a key player in the antiviral response to VSV in the central nervous system.42 Given the established protective role of IFNγ in the brain, one interesting application for VSVΔ51-IFNγ would be for the treatment of glioblastoma and other brain cancers. Further supporting this idea, administration of recombinant IFNγ has been tested in different clinical trials for glioblastoma. Although the results were disappointing, local administration in combination with other therapies was proposed to be a promising avenue.43 In this study, we demonstrate the improved antitumor efficacy of VSVΔ51-IFNγ compared with VSVΔ51 in various tumor models. These findings further support the notion that engineering OVs to express cytokines can provide additional benefits for cancer immunotherapy.
Materials and Methods
Cell lines and culture
Vero kidney epithelial, 4T1 murine mammary carcinoma, EMT6 murine mammary carcinoma, and CT26 murine colon carcinoma cell lines were purchased from the American Type Culture Collection (Manassas, VA). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Corning cellgro, Manassas, VA) supplemented with 10% fetal bovine serum (FBS) (Sigma life science, St-Louis, MO) and maintained at 37 °C with 5% CO2. NOP23 cells were maintained in high-glucose DMEM supplemented with 10% FBS, 1% insulin transferrin selenium, and 1% pen strep (Fisher Scientific, Waltham, MA).
Viral constructs
The IFNγ gene (1–155) was amplified by PCR on cDNA obtained from RNA extracted from LPS stimulated C57BL/6 murine splenocytes. The forward and reverse primers were designed to include XhoI and NheI restriction sites. The digested PCR fragment was cloned into the XhoI and NheI digested VSVΔ51 viral backbone, as previously described.23
Virus rescue
Rescues were performed as previously described.44,45 Briefly, Vero cells were infected with vaccinia virus expressing T7 polymerase at an MOI of 3 for 2 hours. The virus inoculum was then removed and cells were transfected with T7-driven plasmids encoding VSV N, P, and L genes, as well as the VSVΔ51-IFNγ plasmid. Supernatants were collected approximately 48 hours after transfection and passed through a 0.22-μm filter (MillexGP, Carrigtwohill, Ireland) to remove vaccinia virus.
Virus purification
Vero cells were infected at an MOI of 0.01 for 24 hours and culture supernatants were collected and filtered using a 0.2-μm bottle top filter (Millipore, Etobicoke, Canada). The supernatant was then centrifuged at 30,100 g for 1h30 and the pellet was resuspended in Dulbecco’s phosphate-buffered saline (Corning cellgro, Manassas, VA). The purified virus was kept at −80 °C.
Titering
Viral titers were obtained by plaque assay. Briefly, serial dilutions of virus were prepared in serum-free DMEM. 100 μl of the dilutions were transferred to monolayers of Vero cells in six-well plates and incubated at 37 °C for 1 hour. After the incubation, cells were overlayed with 0.5% agarose in DMEM supplemented with 10% FBS. Plates were incubated for 24 hours at 37 °C with 5% CO2 and plaques were counted.
ELISA and ELISPOT
The concentration of IFNγ was obtained using the Quantikine ELISA SixPak, mouse IFNγ (R&D systems, Minneapolis, MN) following the manufacturer’s protocol. The IFNγ-secreting cells were assessed using the ELISPOT mouse IFNγ kit (Mabtech, Cincinnati, OH) according to the manufacturer’s protocol.
Cell viability assay
The metabolic activity of the cells was assessed using alamarBlue (BIO-RAD, Mississauga, Ontario, Canada) according to the manufacturer’s protocol. Briefly, 24 hours postseeding in a 96-well plate (Corning, Manassas, VA), the cells were infected with various amounts of virus. At the indicated time point, 10 μl of alamarBlue was added to each well and the fluorescence was measure at 590 nm upon excitation at 530 nm using a Fluoroskan Ascent FL (Thermo Labsystems, Beverly, MA).
Microscopy
Cells were cultured on coverslips for 24 hours prior to incubation with the conditioned media. Following a 60-minute incubation, the cells were washed with cold PBS and fixed using ice-cold methanol:acetone (1:1). Blocking buffer (5% FBS, 0.3% triton, PBS) and antibody dilution buffer were used (1% bovine serum albumin, 0.3% triton, PBS). The cells were stained using a rabbit anti-STAT1 (Cell signalling technology, Danvers, MA) and a goat anti-rabbit-594 secondary antibody (Molecular Probes, Waltham, MA). Prolong gold anti-fade with 4’,6-diamidino-2-phenylindole (Molecular Probes) was used to mount the coverslips onto slides. The images were analyzed using the LSM510 confocal inverted microscope (Zeiss, Oberkochen, Germany).
Western blot
Cells were pelleted and lysed on ice for 30 minutes using 1% NP-40, complete protease inhibitor cocktail (Roche, Mississauga, Ontario, Canada) and phosphatase inhibitor sodium orthovanadate (Calbiochem, Etobicoke, Ontario, Canada) supplemented lysis buffer. Lysates were centrifuged for 5 minutes at 16,000 g and the supernatants were mixed with dithiothreitol-supplemented loading buffer. The samples were migrated on NuPAGE 4–12% Bis-Tris gels (Novex by Life Technology, Carlsbad, CA) and transferred onto PVDF membranes (GE Healthcare, Buckinghanshire, UK) prior to blocking with 50% fetal bovine serum. The membranes were probed for phospho-STAT1, STAT1 and actin (all from Cell Signalling Technology, Danvers, MA). Membranes were then probed with a goat anti-rabbit peroxidase-conjugated antibody (Cederlane, Burlington, Ontario, Canada) and SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific, Rockford, IL) was used to reveal the signal. The gels were analyzed using a FluorChem FC2 (Alpha Innotech, San Leandro, CA).
In vitro IFNγ stimulation
Cells were uninfected or infected with VSVΔ51 or VSVΔ51-IFNγ for 24 hours at an MOI of 3 in serum-free DMEM. Supernatant was collected and filtered using a 0.22-μm filter (MillexGP, Carrigtwohill, IRL) to remove dead cells. To remove the virus from the conditioned media, the cell-cleared supernatant was filtered again by centrifugation at 4,000 g for 45 minutes using Amicon Ultra 50K centrifugal filters (Millipore, Carrigtwohill, IRL). Prior to incubation with the virus-conditioned media, fresh cells were starved for 1h in serum-free DMEM. The cells were then stimulated using the virus-free conditioned media that was preneutralized or not by a 20 minutes incubation with an IFNγ-blocking antibody. Cells were stimulated at 37 °C for 20 minutes and lysed on ice immediately after incubation.
Quantitative PCR
Lungs from treated tumor-bearing mice were homogenized in 500 μl of Dulbecco’s phosphate-buffered saline. An aliquot was taken and RNA was extracted using RNeasy RNA extraction kit (QIAGEN, Toronto, Ontario, Canada) according to the manufacturer’s protocol. RNA and purity was assessed using a NanoDrop ND-1000 spectrophotometer (Thermoscientific, Waltham, MA) and 500 ng of each sample was used for reverse transcription using SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA) and Oligo(dT)20 primers (Fisher Scientific). qPCR primers used in this study: VSV N gene forward 5′-GATAGTACCGGAGGATTGACGACTA-3′ and reverse: 5′-TCAAACCATCCGAGCCATTC-3′ and IFNγ gene forward 5′-CTCTTCCTCATGGCTGTTTCT-3′ and reverse 5′-TTCTTCCACATCTATGCCACTT-3′. For the qPCR, SYBR green PCR master mix (Applied Biosystems, Carlsbad, CA) reagent was used according to the manufacturer’s protocol and the samples were analyzed using a Rotor-Gene RG3000A (Corbett Research, Mortlake, Australia).
Tumor dissociation
Single cell suspensions were obtained using the mouse tumor cocktail mix, gentle MACS C tubes and gentle MACS dissociator (Miltenyi, San Diego, CA) according to the manufacturer’s protocol.
Flow cytometry
Cells were harvested and cell surface staining was performed on ice for 30 minutes. Cells were then washed twice with cold fluorescence-activated cell sorting (FACS) buffer (3% FBS, 0.09% sodium azide, PBS). For surface staining, cells were incubated with combinations of anti-CD122-FITC, -CD69-PE, -CD3-PECY7, -CD11c-APC, and -CD86-FITC (BD Biosciences, San Jose, CA) for 30 minutes. Cells were then washed twice and resuspended in FACS buffer for analysis using a Cyan ADP 9 flow cytometer (Beckman Coulter, Mississauga, Ontario, Canada).
For splenocyte stainings, mice were sacrificed and the spleens were collected at the indicated time points. The spleens were mashed through 70 μm cell strainers (Fisher, Waltham, MA) in FACS buffer (3% serum, 0.09% sodium azide, PBS). Lysis of red blood cells was performed using ACK lysis buffer (0.15M sodium chloride, 10 mmol/l potassium bicarbonate, 0.1 mmol/l disodium ethylenediaminetetraacetic acid, pH7.2). Upon resuspension in FACS buffer, the splenocytes were counted and 1 million cells per condition were stained following the same procedure describes above.
Blood collection and cytometric bead array
Blood samples were collected using lithium–heparin-coated capillary tubes (Sarstedt, Newton, NC). Samples were kept at room temperature and centrifugated at 16,000 g for 5 minutes to collect the serum. To determine the concentration of IFNγ, TNFα, MCP-1, and IL-6, cytometric bead array (BD Biosciences) was used according to the manufacturer’s protocol.
In vivo experiments and tumor models
All experiments were performed in accordance with institutional guidelines for animal care (University of Ottawa). For the mouse tumor models, Balb/c and nude mice (Charles River Laboratories, Wilmington, MA) were used. For the 4T1 tumor models, 2 × 105 cells were injected in the second right fatpad (orthotopic model) or 1 × 105 cell were injected intravenously (lung tumor model). For the CT26 tumor model, 4 × 105 cells were injected subcutaneously in the right flank. For the NOP23 model, 1 × 106 cells were injected into the mammary fat pad of MMTV/neuOT-I/OT-II mice which are bred on the C57BL/6 background.46
For intravenous and IT injections, the virus preparations were diluted to the appropriate concentration in a total volume of 100 μl of PBS and injected IT or intravenously using insulin syringes (The Stevens Co, Montreal, Québec, Canada).
For T-cell depletion, anti-CD3e antibody (BioXcell, West Lebanon, NH) was injected IP 24 hours prior to the beginning of virus treatment.
Tumor measurements
The length and width of the tumors were measured using digital calipers (Fowler, Newton, MA) and the tumor volume was calculated using the following formula (width2*length)/2. The mice were considered end-point when they showed respiratory distress, displayed significant weight lost or when the tumor volume reached 1,500 mm3.
Histological analysis
At the indicated time points, the mice were euthanized and the lungs were collected and fixed in 10% buffered formalin phosphate (Fisher Scientific) for 48 hours. The tissues were embedded in paraffin and sections were stained using hematoxylin and eosin. The tumor diameters were measured from sections scanned with the Aperio ScanScope using ImageScope software.
India ink lung perfusion
After euthanization, the ribcage was opened and the trachea was exposed. India ink solution (50% india ink (Super black Statesville, NC), 50% PBS (Corning cellgro)) was injected directly into the trachea using a 23 gauge needle (BD Biosciences). Immediately after perfusion, the lungs were removed and fixed by immersion into Feketeye solution (70% ethanol, 10% formaldehyde, 5% glacial acetic acid) to reveal the tumors.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 5.0 software. Statistical significance was calculated using Student’s T-test with Welch’s correction, one-way or two-way analysis of variance or Mann-Whitney U-test as indicated in the figure legends. Error bars represent standard error of the mean.
Acknowledgments
J.C.B. is supported by the Terry Fox Foundation (TFF), the Canadian Cancer Society Research Institute (CCSRI), the Ontario Institute for Cancer Research (OICR), the Canadian Institute for Health Research (CIHR), the Ottawa Regional Cancer Foundation and the Ottawa Hospital Foundation. M-C.B-D., D.G.R., V.G., M.M., and M.S. were supported by fellowships and scholarships from the CIHR. K.T-B. was supported by a fellowship from Susan G. Komen. S.A. received a fellowship from the Canadian Breast Cancer Foundation (CBCF). J-S.D. was supported by the TFF, the CIHR and the CCSRI. Brad Nelson is supported by the Canadian Oncolytic Virus Consortium (COVCO), the CIHR, the TFF and the British Columbia Cancer Foundation. Conception and design: M-C.B-D. and J.C.B.; In vitro experiments: M-C.B-D., D.G.R., L.E.St-G., and M.S.; In vivo experiments: M-C.B-D., D.G.R., T.F., K.T-B., M.M., and J.P.; writing: M-C.B-D. and D.G.R.; review: V.A.J., S.A., J-S.D., and B.N.
The authors declare no conflict of interest.
References
- Hanahan, D and Weinberg, RA (2011). Hallmarks of cancer: the next generation. Cell 144: 646–674. [DOI] [PubMed] [Google Scholar]
- Payne, KK, Toor, AA, Wang, XY and Manjili, MH (2012). Immunotherapy of cancer: reprogramming tumor-immune crosstalk. Clin Dev Immunol 2012: 760965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie, C (2008). Interferon gamma, a possible therapeutic approach for late-stage prostate cancer? Anticancer Res 28(5B): 2843–2849. [PubMed] [Google Scholar]
- Schroder, K, Hertzog, PJ, Ravasi, T and Hume, DA (2004). Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol 75: 163–189. [DOI] [PubMed] [Google Scholar]
- Ikeda, H, Old, LJ and Schreiber, RD (2002). The roles of IFN gamma in protection against tumor development and cancer immunoediting. Cytokine Growth Factor Rev 13: 95–109. [DOI] [PubMed] [Google Scholar]
- Alshaker, HA and Matalka, KZ (2011). IFN-γ, IL-17 and TGF-β involvement in shaping the tumor microenvironment: The significance of modulating such cytokines in treating malignant solid tumors. Cancer Cell Int 11: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi, MR and Merlino, G (2011). The two faces of interferon-γ in cancer. Clin Cancer Res 17: 6118–6124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan, DH, Shankaran, V, Dighe, AS, Stockert, E, Aguet, M, Old, LJ et al. (1998). Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci USA 95: 7556–7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Tuñón, I, Ricote, M, Ruiz A, A, Fraile, B, Paniagua, R and Royuela, M (2007). Influence of IFN-gamma and its receptors in human breast cancer. BMC Cancer 7: 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- duPre’, SA, Redelman, D and Hunter, KW Jr (2008). Microenvironment of the murine mammary carcinoma 4T1: endogenous IFN-gamma affects tumor phenotype, growth, and metastasis. Exp Mol Pathol 85: 174–188. [DOI] [PubMed] [Google Scholar]
- Habif, DV, Ozzello, L, De Rosa, CM, Cantell, K and Lattes, R (1995). Regression of skin recurrences of breast carcinomas treated with intralesional injections of natural interferons alpha and gamma. Cancer Invest 13: 165–172. [DOI] [PubMed] [Google Scholar]
- Bemiller, LS, Roberts, DH, Starko, KM and Curnutte, JT (1995). Safety and effectiveness of long-term interferon gamma therapy in patients with chronic granulomatous disease. Blood Cells Mol Dis 21: 239–247. [DOI] [PubMed] [Google Scholar]
- Miller, CH, Maher, SG and Young, HA (2009). Clinical Use of Interferon-gamma. Ann N Y Acad Sci 1182: 69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulitzky, W, Gastl, G, Aulitzky, WE, Herold, M, Kemmler, J, Mull, B et al. (1989). Successful treatment of metastatic renal cell carcinoma with a biologically active dose of recombinant interferon-gamma. J Clin Oncol 7: 1875–1884. [DOI] [PubMed] [Google Scholar]
- Small, EJ, Weiss, GR, Malik, UK, Walther, PJ, Johnson, D, Wilding, G et al. (1998). The treatment of metastatic renal cell carcinoma patients with recombinant human gamma interferon. Cancer J Sci Am 4: 162–167. [PubMed] [Google Scholar]
- Alberts, DS, Marth, C, Alvarez, RD, Johnson, G, Bidzinski, M, Kardatzke, DR et al. GRACES Clinical Trial Consortium. (2008). Randomized phase 3 trial of interferon gamma-1b plus standard carboplatin/paclitaxel versus carboplatin/paclitaxel alone for first-line treatment of advanced ovarian and primary peritoneal carcinomas: results from a prospectively designed analysis of progression-free survival. Gynecol Oncol 109: 174–181. [DOI] [PubMed] [Google Scholar]
- Balachandran, S and Adams, GP (2013). Interferon-γ-induced necrosis: an antitumor biotherapeutic perspective. J Interferon Cytokine Res 33: 171–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, RY, Zhu, YH, Zhou, L, Zhao, P, Li, HL, Zhu, LC et al. (2012). Adenovirus-mediated delivery of interferon-γ gene inhibits the growth of nasopharyngeal carcinoma. J Transl Med 10: 256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichty, BD, Breitbach, CJ, Stojdl, DF and Bell, JC (2014). Going viral with cancer immunotherapy. Nat Rev Cancer 14: 559–567. [DOI] [PubMed] [Google Scholar]
- Vile, RG (2014). How to train your oncolytic virus: the immunological sequel. Mol Ther 22: 1881–1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeyaullah, M, Patro, M, Ahmad, I, Ibraheem, K, Sultan, P, Nehal, M et al. (2012). Oncolytic viruses in the treatment of cancer: a review of current strategies. Pathol Oncol Res 18: 771–781. [DOI] [PubMed] [Google Scholar]
- Stojdl, DF, Lichty, B, Knowles, S, Marius, R, Atkins, H, Sonenberg, N et al. (2000). Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nat Med 6: 821–825. [DOI] [PubMed] [Google Scholar]
- Stojdl, DF, Lichty, BD, tenOever, BR, Paterson, JM, Power, AT, Knowles, S et al. (2003). VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 4: 263–275. [DOI] [PubMed] [Google Scholar]
- Bridle, BW, Hanson, S and Lichty, BD (2010). Combining oncolytic virotherapy and tumour vaccination. Cytokine Growth Factor Rev 21: 143–148. [DOI] [PubMed] [Google Scholar]
- Lichty, B (2012). Oncolytic viruses: a step into cancer immunotherapy. Virus Adapt Treat 4: 1–21. [Google Scholar]
- Schnell, MJ, Buonocore, L, Whitt, MA and Rose, JK (1996). The minimal conserved transcription stop-start signal promotes stable expression of a foreign gene in vesicular stomatitis virus. J Virol 70: 2318–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phung, TT, Sugamata, R, Uno, K, Aratani, Y, Ozato, K, Kawachi, S et al. (2011). Key role of regulated upon activation normal T-cell expressed and secreted, nonstructural protein1 and myeloperoxidase in cytokine storm induced by influenza virus PR-8 (A/H1N1) infection in A549 bronchial epithelial cells. Microbiol Immunol 55: 874–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms, PE and Ellis, TM (1996). Utility of flow cytometric detection of CD69 expression as a rapid method for determining poly- and oligoclonal lymphocyte activation. Clin Diagn Lab Immunol 3: 301–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel, LA, Sun, MM, Geurs, TL, Carayannopoulos, LN and French, AR (2013). Markers of nonselective and specific NK cell activation. J Immunol 190: 6269–6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman, P and Eriksson, H (2007). Early activation markers of human peripheral dendritic cells. Hum Immunol 68: 324–333. [DOI] [PubMed] [Google Scholar]
- Reed, JM, Branigan, PJ and Bamezai, A (2008). Interferon gamma enhances clonal expansion and survival of CD4+ T cells. J Interferon Cytokine Res 28: 611–622. [DOI] [PubMed] [Google Scholar]
- Whitmire, JK, Tan, JT and Whitton, JL (2005). Interferon-gamma acts directly on CD8+ T cells to increase their abundance during virus infection. J Exp Med 201: 1053–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, ZH, Han, Y, Wei, T, Aras, S, Chaturvedi, P, Tyler, S et al. (2001). Regulation of monocyte chemoattractant protein (MCP)-1 transcription by interferon-gamma (IFN-gamma) in human astrocytoma cells: postinduction refractory state of the gene, governed by its upstream elements. FASEB J 15: 383–392. [DOI] [PubMed] [Google Scholar]
- Deshmane, SL, Kremlev, S, Amini, S and Sawaya, BE (2009). Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res 29: 313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosser, DM (2003). The many faces of macrophage activation. J Leukoc Biol 73: 209–212. [DOI] [PubMed] [Google Scholar]
- Brown, CE, Vishwanath, RP, Aguilar, B, Starr, R, Najbauer, J, Aboody, KS et al. (2007). Tumor-derived chemokine MCP-1/CCL2 is sufficient for mediating tumor tropism of adoptively transferred T cells. J Immunol 179: 3332–3341. [DOI] [PubMed] [Google Scholar]
- Dufour, JH, Dziejman, M, Liu, MT, Leung, JH, Lane, TE and Luster, AD (2002). IFN-gamma-inducible protein 10 (IP-10; CXCL10)-deficient mice reveal a role for IP-10 in effector T cell generation and trafficking. J Immunol 168: 3195–3204. [DOI] [PubMed] [Google Scholar]
- Liu, SY, Sanchez, DJ, Aliyari, R, Lu, S and Cheng, G (2012). Systematic identification of type I and type II interferon-induced antiviral factors. Proc Natl Acad Sci USA 109: 4239–4244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmon, CL, Saloura, V, Fridlender, ZG, Wongthida, P, Diaz, RM, Thompson, J et al. (2009). Expression of IFN-beta enhances both efficacy and safety of oncolytic vesicular stomatitis virus for therapy of mesothelioma. Cancer Res 69: 7713–7720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan, VS, Furr, SR, Sterka, DG Jr, Nelson, DA, Moerdyk-Schauwecker, M, Marriott, I et al. (2010). Vesicular stomatitis virus infects resident cells of the central nervous system and induces replication-dependent inflammatory responses. Virology 400: 187–196. [DOI] [PubMed] [Google Scholar]
- Trottier, MD, Lyles, DS and Reiss, CS (2007). Peripheral, but not central nervous system, type I interferon expression in mice in response to intranasal vesicular stomatitis virus infection. J Neurovirol 13: 433–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesler, DA and Reiss, CS (2002). The role of IFN-gamma in immune responses to viral infections of the central nervous system. Cytokine Growth Factor Rev 13: 441–454. [DOI] [PubMed] [Google Scholar]
- Kane, A and Yang, I (2010). Interferon-gamma in brain tumor immunotherapy. Neurosurg Clin N Am 21: 77–86. [DOI] [PubMed] [Google Scholar]
- Lawson, ND, Stillman, EA, Whitt, MA and Rose, JK (1995). Recombinant vesicular stomatitis viruses from DNA. Proc Natl Acad Sci USA 92: 4477–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun, J, McManus, D, Lefebvre, C, Hu, K, Falls, T, Atkins, H et al. (2010). Identification of genetically modified Maraba virus as an oncolytic rhabdovirus. Mol Ther 18: 1440–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall, EM, Milne, K, Martin, ML, Watson, PH, Theiss, P and Nelson, BH (2007). Spontaneous mammary tumors differ widely in their inherent sensitivity to adoptively transferred T cells. Cancer Res 67: 6442–6450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.