Abstract
RAB24 belongs to a family of small GTPases and has been implicated to function in autophagy. Here we confirm the intracellular localization of RAB24 to autophagic vacuoles with immuno electron microscopy and cell fractionation, and show that prenylation and guanine nucleotide binding are necessary for the targeting of RAB24 to autophagic compartments. Further, we show that RAB24 plays a role in the maturation and/or clearance of autophagic compartments under nutrient-rich conditions, but not during short amino acid starvation. Quantitative electron microscopy shows an increase in the numbers of late autophagic compartments in cells silenced for RAB24, and mRFP-GFP-LC3 probe and autophagy flux experiments indicate that this is due to a hindrance in their clearance. Formation of autophagosomes is shown to be unaffected by RAB24-silencing with siRNA. A defect in aggregate clearance in the absence of RAB24 is also shown in cells forming polyglutamine aggregates. This study places RAB24 function in the termination of the autophagic process under nutrient-rich conditions.
Keywords: aggregate, autophagosome, autophagy, basal, membrane, RAB24, starvation
Abbreviations
- Baf
bafilomycin A1
- CFP
cyan fluorescence protein
- CHO
chinese hamster ovary
- CTBP1
C-terminal-binding protein 1
- DMEM
Dulbecco's modified Eagle's medium
- EBSS
Earle's balanced salt solution
- ER
endoplasmic reticulum
- GABARAP
GABA(A) receptor-associated protein
- GDI1
GDP dissociation inhibitor 1
- GFP
green fluorescent protein
- GOSR1
golgi SNAP receptor complex member 1
- HTT
huntingtin
- LAMP1
lysosomal-associated membrane protein 1
- MAP1LC3/LC3
microtubule-associated protein 1 light chain 3
- NRK
normal rat kidney
- NSF
N-ethylmaleimide-sensitive factor
- PFA
paraformaldehyde
- PNN/DRS
pinin, desmosome associated protein
- PQ
polyglutamine
- RFP
red fluorescent protein
- siRNA
small interfering RNA
- SNAP29
synaptosomal-associated protein, 29kDa
- SNARE
SNAP receptor
- SQSTM1
sequestosome 1
- TPCK-trypsin
N-tosyl-L-phenylalanyl chloromethyl ketone-treated trypsin
- WT
wild type.
Introduction
Membrane dynamics in eukaryotic cells work via vesicle transport, which allows a strict control over the preservation of the structure and function of different organelles. These processes require a multitude of protein machineries and regulatory factors such as coat proteins and cargo receptors, sorting signals, regulatory proteins and tethers as well as docking and fusion mediators.
Membrane tethering steps generally require RAB proteins to recruit effectors to function in fission and fusion events. RABs are peripheral membrane proteins that are specifically targeted to the site of their function within cells and can thus be used as markers for organelles.1 RAB proteins are attached to their intracellular membrane site of function via a prenyl group.2 Autophagic degradation requires several membrane fusion events, and not surprisingly, many RAB proteins have been described to function in autophagy. Some RABs and other GTP binding proteins function in the early phase of autophagosome formation or regulation of autophagy (RAB4, RAB5, RAB32, RAB1B and SAR1) and some later in the membrane elongation or maturation process (RAB7, RAB8 and RAB9). Increasing research is revealing that some RABs have roles in several stages of the autophagic process; both in the formation and in the maturation, directly by interaction with the autophagic machinery (RAB33) or indirectly through endosome maturation (RAB11) as well as some other ways.3-8 RAB24 localizes in vesicles positive for the autophagosome marker MAP1LC3/LC3 (microtubule-associated protein 1 light chain 3) but so far RAB24 has not been reported to function in autophagy. The roles of small RAB GTPases and their regulators in autophagy are comprehensively summarized in recent reviews by Szatmári and Sass as well as Chua et al.9,10
RAB24 has been first characterized by Olkkonen et al. as a perinuclear protein colocalizing with Golgi markers as well as with some late endosome markers, the best match being an ER-Golgi intermediate compartment/ERGIC marker, RAB2.11 This has been suggested to be an indicator of RAB24 functioning in some sort of autophagy-related transport route between the ER-Golgi intermediate compartment and late endocytic compartments. Later, the features of RAB24 were characterized as different from a typical RAB protein in several aspects,12 raising questions about the mechanism of its function in the regulation of membrane dynamics. The second conjecture that RAB24 might be involved in autophagy came from a study where overexpressed RAB24 is shown to change localization upon amino acid starvation in Chinese hamster ovary (CHO) cells. Munafo and Colombo show that RAB24 colocalizes with LC3 and with a marker for acidic compartments, monodansylcadaverine, in vesicular structures.13 Despite the effort to characterize RAB24 and resolve its function, the results so far have not been conclusive. Although differences between RAB24 and other RAB proteins have been described, such as insufficient prenylation, lower GTPase activity and predominant occurrence in the GTP-bound state, and tyrosine phosphorylation, the biological significance of these differences is still unknown.12,14 Furthermore, although colocalization of RAB24 with autophagosome markers imply a function in autophagy,13 an effect on the process is yet to be shown.
We set out to clarify which phase of autophagy requires RAB24. Here we show that RAB24 is not needed for the formation of autophagosomes but is necessary for the clearance of autophagic compartments in full culture medium conditions. This suggests that RAB24 has a role in termination of the basal autophagic process. We also show that RAB24 needs to be prenylated and bound to a nucleotide in order to become recruited to LC3-positive autophagosomes, while tyrosine phosphorylation is less important for the recruitment.
Results
RAB24 colocalizes with LC3 in full culture medium and during amino acid starvation
To test the localization of RAB24 in LC3-positive vesicles, we used both a HeLa cell line stably expressing MYC-RAB24 and cells transiently overexpressing MYC-RAB24, concentrating on cells with moderate to low expression levels as judged by the intensity of the fluorescence signals. RAB24 was labeled with a monoclonal RAB24 antibody. LC3-positive vesicles were found at low frequency in these cells, reflecting a low level of basal autophagy.
To investigate the localization of RAB24 in nutrient-rich conditions as well as during serum and amino acid starvation, HeLa cells stably expressing MYC-RAB24 were treated in amino acid-free Earle's balanced salt solution (EBSS) for different time periods. This approach was used since the expression levels of endogenous RAB24, in all cell lines under all conditions we tested, were too low for detection by immunofluorescence staining. MYC-RAB24 and untagged RAB24 showed identical localization in immunofluorescence analysis (not shown). In full culture medium, more than 60% of LC3-positive puncta were also positive for RAB24 (Fig. 1, Fig. S1A). During serum and amino acid starvation the proportion of LC3-positive puncta positive for RAB24 slightly increased until 2 h, and then decreased back to the starting level (Fig. S1A). We also estimated the average total intensity of RAB24 signals (Fig. S1B), and the average density of RAB24 signals (Fig. S1C), in the LC3-positive puncta. RAB24 total intensity increased during starvation and reached the highest value at 2 h, while RAB24 labeling density increased slightly during starvation, but the differences were not statistically significant. We also measured the colocalization of RAB24 and LC3 using Pearson correlation coefficient (Fig. S1D). The correlation increased during starvation and reached the highest value at 4 h. This is due to the increase in the number of LC3-puncta. These findings are in agreement with the report of Munafo and Colombo13 showing that induction of autophagy changes the subcellular distribution of RAB24.
Figure 1.
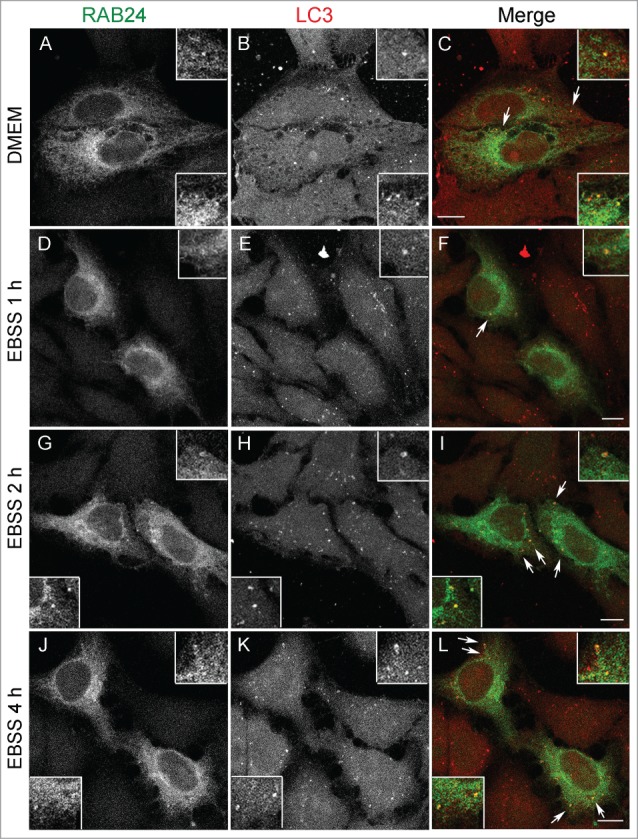
RAB24 localizes in LC3-positive vesicles. HeLa cells stably expressing myc-RAB24 were treated with a serum and amino acid-free medium (EBSS) for (A to C) 0, (D to F) 1, (G to I) 2 or (J to L) 4 h. Cells were labeled with RAB24 and LC3 antibodies and imaged with a confocal microscope. Inserts and arrows indicate vesicles positive for both proteins. See Figure S1 for quantitative analysis of the colocalization. Bar: 10 μm.
RAB24 localizes on inner and outer autophagosomal membranes
Immunofluorescence staining of RAB24 showed that in many cases RAB24 formed a ring around an LC3-positive punctum (Fig. 2A i and ii), or both RAB24 and LC3 structures were ring shaped. This suggested that RAB24 localized to the limiting membranes of autophagic vacuoles. The localization of RAB24 to autophagic compartments was confirmed by immuno-electron microscopy. Gold immunolabeling with anti-RAB24 in RAB24-transfected HeLa and NRK cells revealed that RAB24 was found both on the inner (arrowheads) and the outer (arrows) limiting membrane of the autophagosomes (Fig. 2B i and ii). To study the localization of the endogenous RAB24 protein, we used subcellular fractionation with OptiPrep gradient centrifugation and found that RAB24 distributed in the same fractions where LC3-II and SQSTM1 were predominant (Fig. 2C). SQSTM1 functions as a receptor protein in selective autophagy and it is degraded via autophagy.15 When cells were starved and treated with bafilomycin A1 (Baf), a selective inhibitor of lysosomal acidification that prevents degradation of autophagic cargo, RAB24 accumulated in the same OptiPrep fractions as LC3-II and SQSTM1 (Fig. 2C). To confirm the localization of RAB24 to both the inner and outer limiting membranes of autophagic compartments, the OptiPrep fractionation was combined with a proteinase protection assay. HeLa cells were either left untreated or incubated in EBSS in the presence of Baf for 2 h to maximally accumulate autophagosomes and to accumulate the putative RAB24 in the inner limiting membranes. The samples were fractionated using OptiPrep gradients, and the collected fractions were incubated with N-tosyl-L-phenylalanyl chloromethyl ketone (TPCK-) treated trypsin, in the absence or presence of 1% NP-40 (Fig. S2). Part of RAB24 was protected from degradation by TPCK-trypsin, confirming its localization in both the outer and inner limiting membranes of autophagic structures. LC3-II, which is known to localize to both the outer and inner limiting membranes of autophagosomes, was also partially proteinase protected. As expected, the cargo protein SQSTM1 was almost completely proteinase protected, while the early endosome antigen 1 (EEA1), a protein that associates on the cytosolic side of endosomal vesicles, was not protected. As expected, all proteins were degraded when TPCK-trypsin was added together with the detergent NP-40. We could also detect RAB24 in both the autolysosomal and dense lysosomal membrane fractions isolated from rat liver. Both of these fractions are positive for LC3-II,16,17 indicating they contain autophagic membranes, but the dense lysosome fraction is likely to represent a later maturation step in the autophagic pathway (Fig. 2D). Interestingly, RAB24 was more abundant in the dense lysosomal membranes, while more RAB7 was detected in the autolysosomal membranes.
Figure 2.
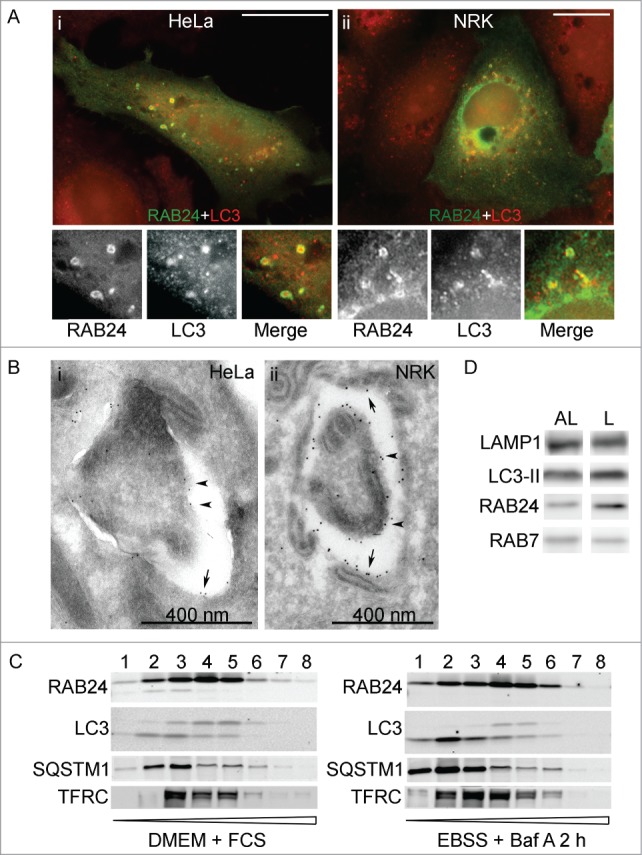
RAB24 localizes in the limiting membranes of autophagic compartments. HeLa (A i, B i, C) or NRK (A ii, B ii) cells were transfected with MYC-RAB24 and (A, C) left untreated in full-culture medium (DMEM), or (B) incubated in serum and amino acid-free EBSS for 90 min, or (C) incubated in EBSS with 100 nM Baf for 2 h. Cells were labeled with anti-RAB24 and anti-LC3 (A i and ii), or anti-RAB24 (B i and ii). (A i and ii) RAB24 label was in several cases observed to be ring-shaped, indicating it localized to the limiting membranes of the LC3-positive structures. (B i and ii) Immunoelectron microscopy confirmed the localization of RAB24 in the limiting membranes of autophagic compartments. Arrows indicate the outer limiting membrane and arrowheads the inner limiting membrane. (C) Subcellular fractionation of HeLa cells in a continuous OptiPrep gradient. The numbers above the western blot images indicate the 8 fractions collected from the top of the gradient. Highest OptiPrep concentration is in fraction 8 on the right. The fractions were detected for RAB24, LC3, SQSTM1 and transferrin receptor (TFRC). (D) Autolysosomal (AL) and dense lysosomal (L) membranes were isolated from rat liver and western blotting was used to detect RAB24, LC3, LAMP1 and RAB7 in these fractions. Bar in A i and ii: 10 μm. Gold particles in B i are 5 nm and ii 10 nm in diameter.
Targeting of RAB24 to autophagosomes requires guanine nucleotide binding and prenylation, but not tyrosine phosphorylation
To determine the regions in the RAB24 amino acid sequence critical for its targeting to autophagosomes, mutations were made to sites with known or inferred importance to RAB proteins, particularly in sites where RAB24 is known to differ from other RABs. Mutant plasmids were expressed in NRK cells that were double labeled with anti-RAB24 and anti-LC3 for confocal microscope examination. NRK cells were used in this analysis since they are more active in autophagy than HeLa cells.
RAB24 has an unusual serine (S67) in the place of glutamine in the second GTP-binding motif, which is thought to be responsible for the low GTPase activity of RAB24.12 Replacement of S67 with leucine (RAB24S67L) was found to destabilize RAB24 GTP binding. This mutant has been previously reported to be unable to retain bound GTP.13 Further, the overexpression of this mutant in CHO cells caused a delay in the development of the Coxiella-replicative compartments for up to 24 h after infection.13,18 We used a GTP-agarose binding assay to study the ability of RAB24S67L to bind nucleotides. In agreement with the previous reports, we found that the S67L mutant had a reduced ability to bind GTP-agarose as compared to the wild-type protein (Fig. 3A). When expressed in NRK cells, RAB24S67L showed a diffuse localization both in fed and starved cells (Figs. 4G to L) similar to earlier studies.13 RAB24S67L showed no colocalization with LC3 vesicles (Fig. S3).
Figure 3.
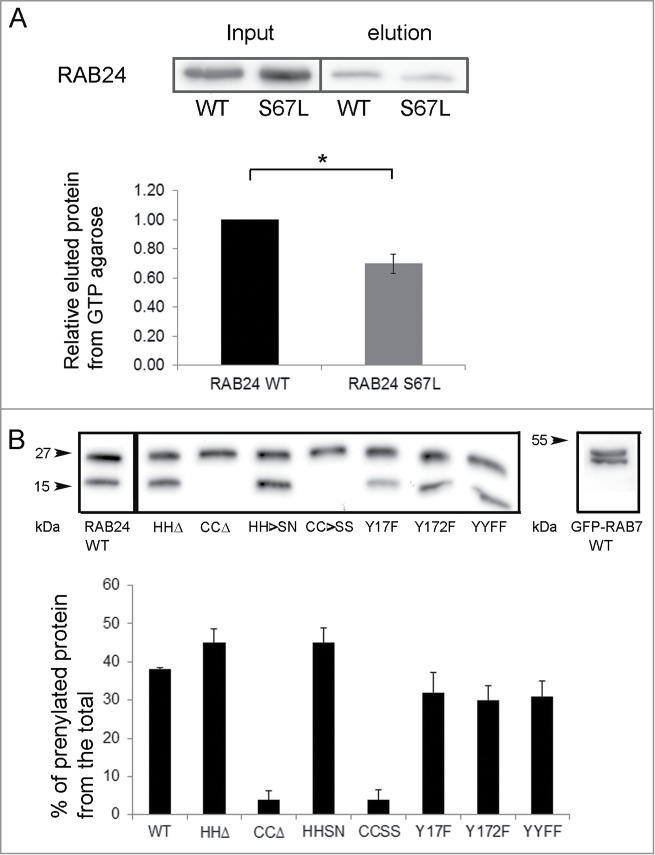
Analysis of RAB24 mutants. (A) GTP binding of RAB24S67L mutant was tested using GTP-agarose beads. Recombinant proteins were produced in E. Coli production strain BL21 and lysates were incubated with GTP-agarose gel. After washing, the bound protein was eluted from the gel with excess GTP, and western blotting was used to compare the eluted amounts of RAB24-WT and S67L. Eluted protein bands were normalized with their input bands. The columns and error bars show the mean and SEM from 4 independent experiments. (B) HeLa cells were transiently transfected with MYC-tagged wild-type or mutant MYC-RAB24 plasmids. Cell extracts were electrophoresed on urea (4 to 8 M) acrylamide (10–20%) SDS gradient gels and blotted on PVDF membranes. MYC antibody was used for detection. Prenylation-deficient mutants, CCΔ and CC→SS, lack the lower band, which represents the prenylated form of the protein. Nearly 40% of wild-type RAB24, and 45% of prenylation-competent mutants, are in the prenylated form. Tyrosine phosphorylation deficiency had a slight but nonsignificant effect on prenylation. The columns and error bars show the mean and SEM from 4 or 5 independent experiments. GFP-RAB7 is shown as a reference.
Figure 4.
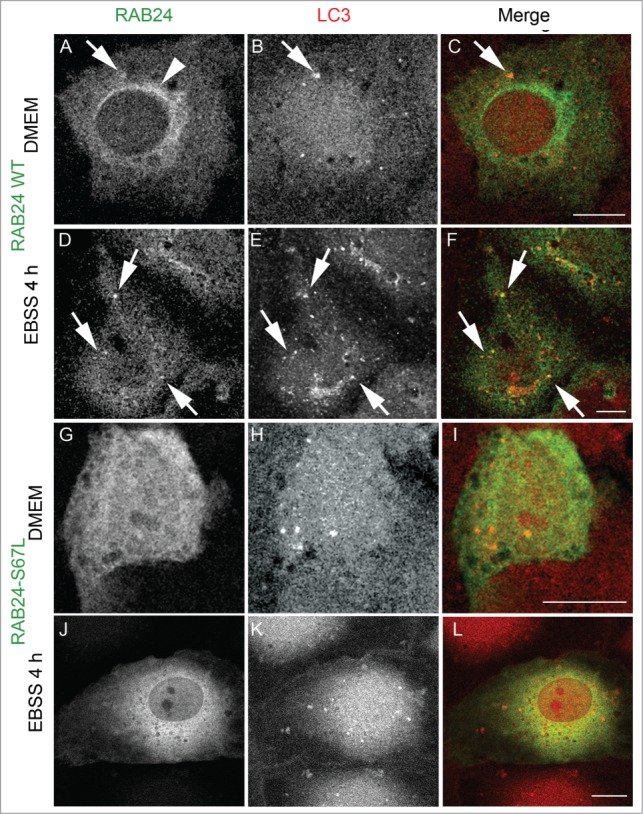
Guanine nucleotide binding is needed for the targeting of RAB24 to LC3 vesicles. WT or guanine nucleotide binding-deficient mutant RAB24S67L was expressed in NRK cells. (A to C and G to I) Cells were kept in full medium, DMEM, or (D to F and J to L) treated with a serum and amino acid-free medium, EBSS, for 4 h and after fixation labeled with antibodies against RAB24 and LC3. RAB24-WT localized to a perinuclear structure (arrowhead) and partially in LC3-positive puncta (arrows). RAB24S67L had a diffuse appearance and did not form the perinuclear staining pattern. See Figure S3 for quantification of the colocalization. Bar: 10 μm.
RAB proteins are anchored to the membrane via a prenyl group attached to their C-terminal cysteines. Recombinant RAB24 is poorly prenylated in comparison to RAB1B in a cell-free assay.12 In order to study whether prenylation of RAB24 is required for the recruitment to LC3-positive vesicles, prenylation-deficient or prenylation-competent RAB24 mutants were created. Prenylation-deficient mutants either lacked the C-terminal cysteines (CCΔ) or had the cysteines replaced with serines (CC→SS). Prenylation-competent mutants lacked the unusual histidines in the C terminus (HHΔ) or had the histidines replaced with serine and asparagine (SN). This sequence (-CCSN) in the C-tail of the HH→SN mutant mimics the sequence of RAB5, the closest relative of RAB24 by sequence comparison. MYC-tagged wild-type (WT) and mutant proteins were expressed in HeLa cells and the prenylation of RAB24 was analyzed by western blotting using a MYC (Fig. 3B) or RAB24 (not shown) antibody. The prenylated form of RAB24 was separated from the nonprenylated form in a urea gradient SDS PAGE gel where proteins migrate at a different pace according to their prenylation state19 (Fig. 3B). Unlike in previous studies, we observed that a considerable portion of WT RAB24 was prenylated. Deletion of the 2 histidines (HHΔ) and replacement of the tail with that of RAB5 (HH→SN) slightly increased the proportion of the prenylated protein, while deletion of the 2 cysteines (CCΔ) and their replacement with serines (CC→SS) abolished the prenylation completely. Examination of mutant RAB24-expressing NRK cells with confocal microscopy revealed that staining of the prenylation-deficient mutants was diffuse with no apparent localization to any membrane structures, neither in starved nor nonstarved cells (Fig. 5, Fig. S3). Prenylation-deficient mutants CC→SS and CCΔ were not targeted to LC3-positive autophagosomes even upon amino acid deprivation (Fig. 5D to F and J to L). Quantification of colocalization with LC3 showed Pearson coefficients significantly lower than with WT RAB24 in nonstarved cells as well as during starvation (Fig. S3). Interestingly, the localization of the prenylation-competent mutants HH→SN and HHΔ (Fig. 6) resembled that of WT RAB24 (Figs. 4A to F) with a clear perinuclear localization pattern under basal conditions (Fig. 6 arrow heads). Similar to WT RAB24, the prenylation-competent mutants HH→SN and HHΔ partially colocalized with LC3, both in nonstarved cells and after starvation in amino acid-free medium for 4 h (Fig. 6 arrows, Fig. S3).
Figure 5.
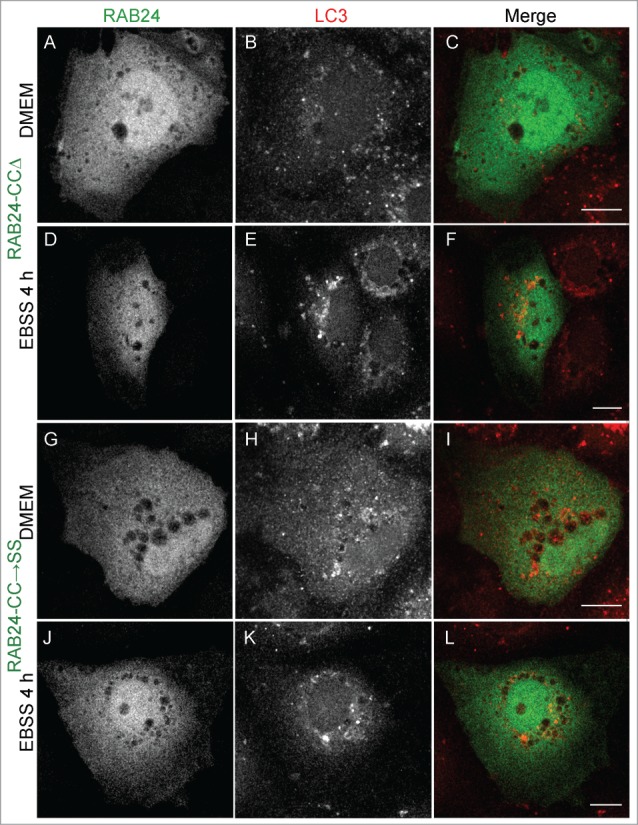
Prenylation-deficient mutants of RAB24 are not targeted to LC3 vesicles. Mutant RAB24 plasmids were expressed in NRK cells. Cells were kept in full-culture medium (DMEM) or treated with serum and amino acid-free medium (EBSS) for 4 h and after fixation labeled with antibodies against RAB24 and LC3. Prenylation-deficient mutants, CCΔ (A to F) and CCSS (G to L), had a diffuse appearance and did not form the perinuclear staining pattern observed for wild-type RAB24 (see Fig. 4A). Further, the RAB24 mutants did not localize in LC3 vesicles neither in normal medium nor in amino acid-free medium. For quantitative analysis of colocalization, see Figure S3. Bar: 10 μm.
Figure 6.
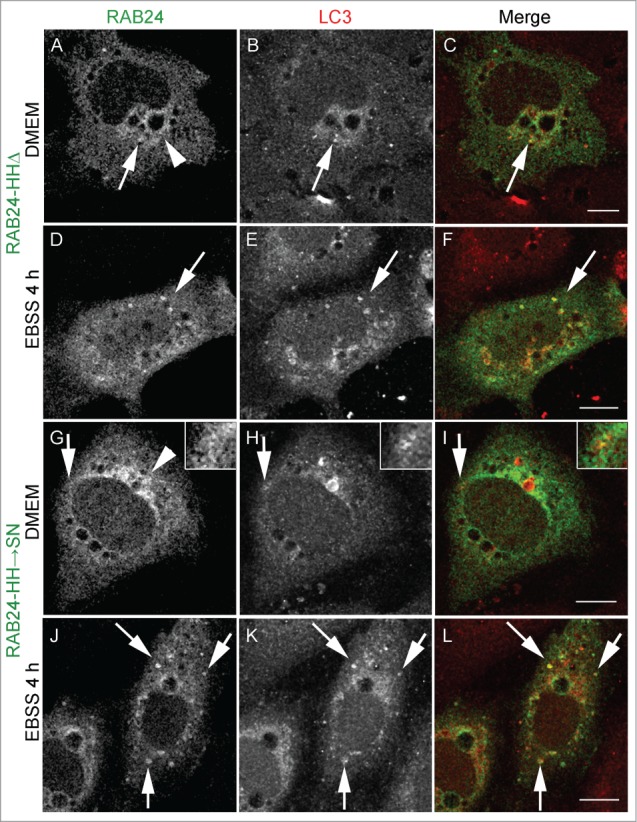
Prenylation-competent mutants of RAB24 are targeted to LC3 vesicles. Mutant RAB24 plasmids were expressed in NRK cells. Cells were kept in full-culture medium (DMEM) or treated with serum and amino acid-free medium (EBSS) for 4 h and after fixation labeled with antibodies against RAB24 and LC3. Prenylated mutants, HHΔ (A to F) and HHSN (G to L), resembled wild-type RAB24 with a perinuclear localization (arrowheads, compare with Fig. 4A). Further, both mutants partially colocalized with LC3 both in full culture-medium and after a 4 h EBSS treatment (arrows). Inserts highlight the colocalization in panels G to I. For quantitative analysis of colocalization, see Figure S3. Bar: 10 μm.
As previously shown, RAB24 has 2 tyrosine residues (Y17 and Y172), that are phosphorylated in cultured HEK293 cells.14 To clarify if this phosphorylation is needed for autophagosome targeting we replaced either one or both of these tyrosines with phenylalanine creating 3 different mutants: Y17F, Y172F and YY17,172FF. These mutants showed slightly reduced prenylation compared to WT RAB24 (Fig. 3B). As WT RAB24, all tyrosine phosphorylation-deficient mutants localized to a perinuclear compartment (Fig. 7A, arrowhead, Fig. 4A) and colocalized with LC3, although to a lesser degree in starved cells (Fig. 7 arrows, Fig. S3, Fig. S4 arrows). Collectively, these results show that guanine nucleotide binding and prenylation are needed for the targeting of RAB24 to autophagosomes whereas tyrosine phosphorylation of Y17 and Y172 is less important for autophagosome targeting.
Figure 7.
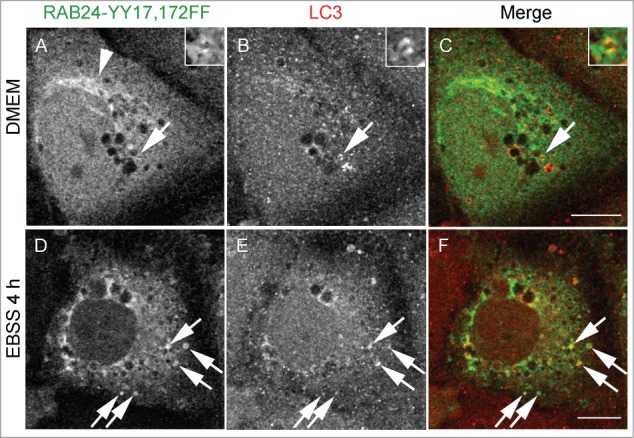
Tyrosine phosphorylation is less important for the targeting of RAB24 to LC3 vesicles. Mutant RAB24 plasmids were expressed in NRK cells. Cells were kept in full culture medium (DMEM) or treated with a serum and amino acid-free medium (EBSS) and after fixation labeled with antibodies against RAB24 and LC3. Tyrosine phosphorylation-deficient mutant YY17,172FF resembled wild-type RAB24 with perinuclear localization (arrowhead) (compare to Fig. 4A). RAB24 mutant partially colocalized with LC3 in full-culture medium (A to C, arrows) and after EBSS treatment (D to F, arrows). For quantitative analysis of colocalization, see Figure S3. Bar: 10 μm.
RAB24 is not needed for the formation or clearance of autophagosomes upon short-term amino acid deprivation
An siRNA smart pool was used to determine the effect of RAB24-silencing on autophagy in HeLa cells. The silencing was verified in each experiment by western blotting (Fig. 8A), which showed that typically 70% to 90% of RAB24 protein had disappeared. Quantitative transmission electron microscopy revealed that silencing of RAB24 did not prevent the formation of autophagosomes during a 2 h serum and amino acid starvation in EBSS (Fig. 8B, Fig. S5). Both immature and degradative autophagic compartments were observed in RAB24-depleted cells (Figs. 8B and C). However, RAB24-silenced cells had over 4 times more autophagic compartments than cells treated with control siRNA in full-culture medium (Fig. 8B, Fig. S5). Nevertheless, there was no significant difference in the amount of autophagic compartments in RAB24-silenced and control cells when the cells were incubated for 2 h in EBSS for induction of autophagy (Fig. 8B, Fig. S5). Further, the amount of autophagosomes was the same in RAB24-silenced and control cells after treatment with EBSS for 2 h followed by a chase in full medium for 2 h to allow clearance of the starvation-induced autophagic compartments (Fig. 8B, Fig. S5). These results indicate that RAB24 is not needed for the formation or clearance of autophagosomes that form during a 2-h serum and amino acid starvation, but suggest that RAB24 has a function in basal autophagy. The notable 4-fold increase in the amount of autophagic compartments in cells kept in full culture medium when RAB24 was absent (Fig. 8B, Fig. S5) could be due to induction of autophagosome formation or a blockage in the clearance of formed autophagosomes.
Figure 8.
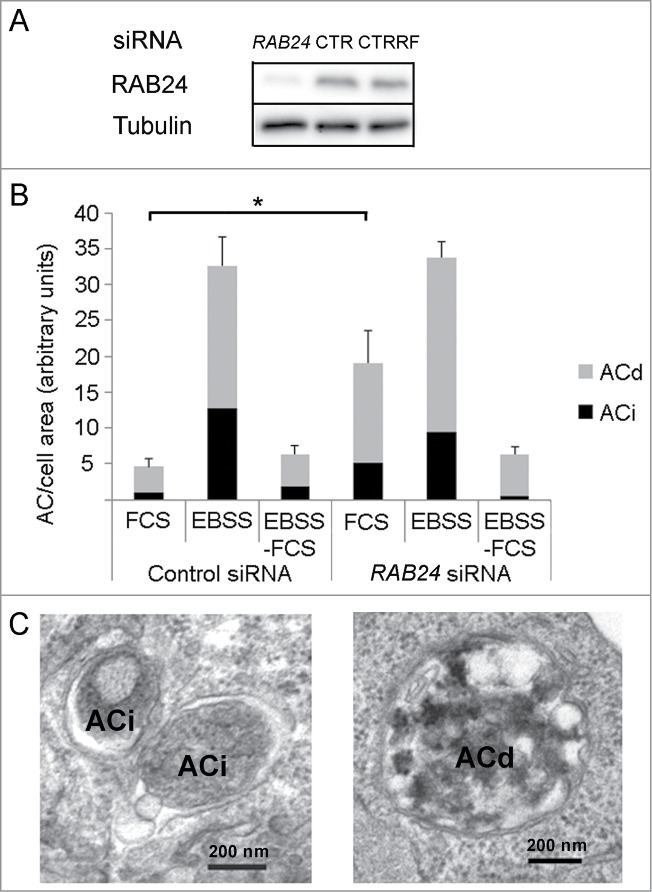
RAB24 is not needed for the formation or clearance of autophagosomes formed during 2-h serum and amino acid deprivation. (A) siRNA was used to silence RAB24 in HeLa cells and western blotting was used to monitor the protein levels. Tubulin is shown as a loading control. RAB24 silencing was approximately 95% as compared to 2 different siRNA controls, CTR, nontargeted siRNA and CTRRF, RISC-free control. (B) Quantitative electron microscopy was used to monitor the amount of autophagic compartments. The cells were either fixed without treatment (FCS), or incubated in serum and amino acid-free medium for 2 h (EBSS), or first incubated in EBSS for 2 h and then chased in full-culture medium for 2 h (EBSS-FCS), before fixation. (B and C) Immature (ACi) and degradative (ACd) autophagic compartments were quantified. Note that there was an increase in the amount of autophagic compartments in RAB24-silenced cells compared to control cells in full medium (FCS). For representative microscopy images, see Figure S5. The columns and error bars show the mean and SEM, respectively, from 3 to 4 counted grid squares, each containing a minimum of 50 to 60 cell profiles. Statistical significance was tested with Mann-Whitney U-test (P = 0.05). AC, autophagic compartment; ACd, degradative autophagic compartment; ACi, immature autophagic compartment.
RAB24 is needed for the final maturation and/or clearance of late autophagic compartments in full-culture medium
In order to study the formation rate of autophagic compartments and carry out a quantitative analysis of autophagic sequestration, it is essential to block the autophagic flux.20 This can be achieved by chemical agents that inhibit lysosomal degradation such as Baf, an inhibitor of vacuolar-type H+-ATPases. This inhibition blocks autophagic clearance by hindering the maturation of autophagosomes, thus giving information about the autophagosome formation rate.21 To clarify if the increase in the number of autophagic compartments in the absence of RAB24 was due to increased autophagosome formation or decreased clearance, we performed an siRNA experiment equivalent to Figure 8 with Baf to block lysosomal degradation and hence the clearance of autophagosomes. The cells were placed in fresh full-culture medium in the presence or absence of Baf for 2 h. To exclude the possibility that we were observing off-target effects when using the smart pool of siRNA duplexes (Fig. 8), RAB24 was depleted with 3 single siRNA oligos in addition to the pooled siRNA (Fig. 9A). The silencing was confirmed by western blotting, which showed that RAB24 levels were reduced to 17% to 45% of the endogenous level in all samples (Fig. S6). There was no significant difference in the total number of autophagosomes and autolysosomes between control and RAB24-depleted cells upon Baf treatment (Fig. 9A). The difference, however, persisted between RAB24-silenced cells and control cells that were either untreated, or changed to fresh full-medium without Baf for 2 h (Fig. 8B and 9A, Fig. S5 and S6). Approximately 4-fold more late autophagic compartments were found in RAB24-silenced cells than in control cells. While Baf treatment causes an accumulation of immature autophagic compartments containing undigested material, most structures accumulating in RAB24 depleted cells in full culture medium without Baf were degradative structures that contained electron dense internal material in an advanced phase of degradation (Fig. 8B and C, 9A and B). Further, the single siRNA oligos gave the same results as the smart pool, indicating that the observed accumulation of autophagic compartments is not likely to be an off-target effect (Fig. 9A, Fig. S6).
Figure 9.
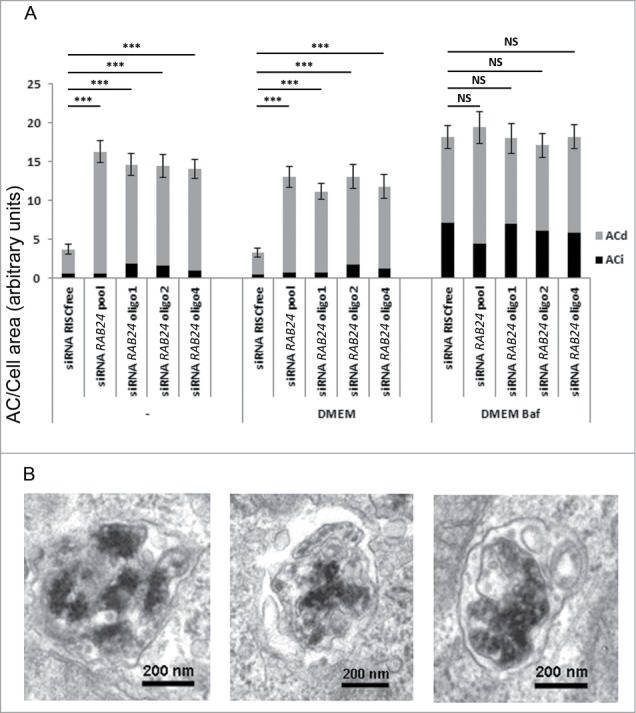
RAB24 is needed for the clearance of late autophagic compartments in full-culture medium conditions. Smart pool siRNA or single siRNA oligos (1, 2, or 4) from the pool were used to silence RAB24 in HeLa cells. The cells were either fixed without treatment, or incubated in fresh full culture medium for 2 h, or in fresh full-culture medium containing Baf (100 nM) for 2 h, before fixation. (A) Autophagic compartments were quantified by electron microscopy. Equal amounts of autophagic compartments were observed in the Baf-treated, RAB24-silenced and control cells. However, the RAB24 siRNA-transfected cells contained 4 times more autophagic compartments than the control cells in full-culture medium conditions. For representative image, see Figure S6. (A and B) The accumulating autophagic compartments in the RAB24-silenced cells were mostly degradative ACds. RAB24 silencing was 83% for the RAB24 smart pool and 55 to 69% for the single oligos. The columns and error bars show the mean and SEM, respectively, from a minimum of 40 images taken at 1500X primary magnification from 2 to 4 grid squares. The data are from one representative experiment out of 3 with similar results (P ≤ 3.341*10−6 for *** and P ≥ 0.618 for NS). AC, autophagic compartment; ACd, degradative autophagic compartment; ACi, immature autophagic compartment; NS, nonsignificant.
The tandem-tagged LC3 construct, mRFP-GFP-LC3, is used to monitor autophagosome maturation to acidic autolysosomes, as the GFP fluorescence is lost while the mRFP fluorescence is more acid resistant.21 To clarify whether the accumulating autophagic compartments seen in RAB24 depleted cells were acidic, we used a stably mRFP-GFP-LC3-expressing cell line with RAB24 siRNA transfection. We quantified the ratio of areas of yellow (neutral) and red (acidic) LC3-positive vesicles and found no difference between RAB24-silenced and control cells (Fig. S7A). This result suggests that the autophagic compartments that accumulate in RAB24-silenced cells are acidic. This is in agreement with their electron microscopic morphology that was similar to degradative autophagic compartments. Taken together, these results indicate that RAB24 is required for the final maturation or clearance of late autophagic compartments during basal conditions.
In order to further analyze the effect of RAB24-silencing on autophagic flux the degradation of long-lived proteins was analyzed in control and RAB24-silenced HeLa cells. Long-lived protein degradation mainly occurs via autophagy, and inhibition of autophagy by the genetically mediated deletion of the autophagy protein ATG5 that is required for autophagosome formation, decreases total long-lived protein degradation by approximately 40%.22 The effect of RAB24-silencing on long-lived protein degradation was studied in HeLa cells using the release of acid-soluble radioactivity from cells metabolically labeled with radioactive valine, as previously described in Bauvy et al 2009.23 In agreement with the electron microscopy analysis, the silencing of RAB24 slightly hindered the degradation of long-lived proteins in nutrient-rich conditions with no significant differences upon amino acid starvation (Fig. S7B).
Autophagic flux is also monitored using the protein levels of autophagy substrates such as polyubiquitinated proteins, SQSTM1, and the autophagosome-associated form of LC3 (LC3-II) or other orthologs and paralogs of yeast Atg8, such as GABARAP-II.21 We used these assays in control and RAB24-silenced HeLa cells. While the level of ubiquitinated proteins was slightly higher in the RAB24-silenced cells, there were no significant differences in the SQSTM1 protein levels or in the ratio of LC3-I/LC3-II and GAPARAP-I/GAPARAP-II in RAB24-silenced and control cells (Fig. S8A). Taken together, the autophagy flux assays showed that RAB24-silencing had a minor effect on autophagic flux, although the electron microscopy analysis revealed that 4 times more late-autophagic compartments accumulated in these cells. These results suggest that the block in the autophagic pathway is at a very late stage, after the autophagosomes have become degradative. These late autophagic compartments are likely to be positive for lysosomal markers such as the lysosomal membrane protein LAMP1. Thus, we monitored the expression levels of LAMP1 by western blotting and observed that the levels were higher in the RAB24-silenced than in the control cells, but the difference was not statistically significant (Fig. S8B). Using immunofluorescence labeling with anti-LAMP1 to estimate the mean LAMP1 intensity per cell we found that in RAB24-silenced cells the intensity of LAMP1 labeling was significantly higher than in control cells (Fig. S8B). This is in agreement with the electron microscopy results, indicating that late, acidic and LAMP1-positive, degradative autophagic vacuoles accumulate in RAB24-silenced cells under basal conditions. Since autophagic flux is minimally affected in these cells, it is likely that RAB24 is not required for the initiation of autophagic degradation, but more likely needed for the clearance of the late autophagic compartments after degradation has been initiated.
RAB24 facilitates the clearance of mutant HTT/huntingtin aggregates
Since nutrient-deprivation independent, i.e., quality control, autophagy strives for maintaining homeostasis and damage control rather than producing energy, we wanted to examine if RAB24 had an effect on the clearance of a specific autophagy substrate, mutant Huntingtin.24 For this purpose we used HeLa cells with stable inducible expression of cyan fluorescent protein (CFP)-tagged Huntingtin (HTT) peptides with a polyglutamine (PQ) repeat of 65 residues as the HTT aggregates formed in these cells have been found to be degraded by autophagy.22 Expression of the mutant protein is prevented by tetracycline addition. We used siRNA to study the effect of RAB24-silencing on the clearance of formed protein aggregates after cessation of PQ HTT expression by tetracycline addition. A dot blot filter trap assay was used to biochemically monitor the aggregate clearance in 65 PQ cells. Large, SDS insoluble protein aggregates are retained in cellulose acetate membrane while SDS soluble protein in cell extracts passes through the system.25 After prevention of mutant HTT synthesis there were more aggregates left in the RAB24 siRNA-transfected cells than in control cells, particularly after one-d tetracycline treatment when the silencing of RAB24 was most effective (Figs. 10A and B). Similar effects were observed with quantitative analysis of CFP-positive aggregates per nucleus in fluorescence images (Fig. 10C). Representative images are presented in Figure S9. Surprisingly the number of HTT aggregates increased in the RAB24 siRNA samples one d after tetracycline addition compared to d 0 (Fig. 10C). This is likely due to the dispersal of large aggregates into smaller ones. Further, in agreement with the filter trap assay, the clearance of HTT aggregates was decreased in RAB24 siRNA-transfected cells compared to the control siRNA-transfected cells one and 3 d after tetracycline addition (Fig. 10C and Fig. S9). After 3 d of tetracycline treatment, aggregates were still visible in fluorescence samples (Fig. 10C) but HTT adhered poorly to the cellulose acetate filter (Fig. 10A). This could be due to sequestration of HTT to autophagic compartments where it is likely to transform into a more SDS-soluble form, which nevertheless is still visible under the microscope. Taken together, these results show that RAB24 was required for efficient clearance of HTT aggregates.
Figure 10.
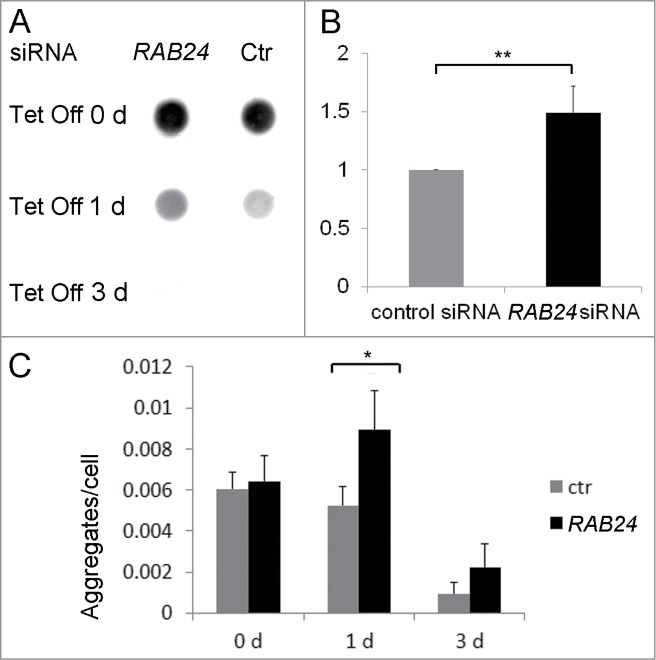
RAB24 facilitates the clearance of mutant HTT. siRNA was used to silence RAB24 in HeLa cells expressing HTT 65PQ. Two or 3 d after transfection with RAB24 or control siRNA, HTT mutant protein expression was inhibited by the addition of tetracycline (10 µg/ml) to the culture medium. The cells were given either 0, 1 or 3 d for aggregate clearance after which they were either treated for dot blot filter trap assay (A and B) or fixed and imaged by fluorescence microscopy (C). (B) Quantification of the intensity of one-d Tet Off dots on cellulose acetate membrane showed a slower aggregate clearance in RAB24-silenced cells compared to the control. The signals were negligible after 3 d in tetracycline and therefore were not quantitated. The columns and error bars show the mean and SEM from 4 independent experiments where the RAB24 silencing was 86% on average. (C) Quantitation of the HTT aggregates in fluorescence images revealed that there were more aggregates per cell in RAB24-silenced cells than in the control cells. The columns show the mean and SEM from 21 or 22 analyzed pictures, each containing several hundreds of cells (a total of 3637 to 18003 cells per sample). Representative images are presented in Figure S9. The experiment was repeated 2 times with similar results. The mean RAB24-silencing in the 2 experiments was 66% in 0-d samples and 67% in 3-d samples. Statistical significance was estimated using the Wilcoxon test for paired populations (P = 0.005).
Discussion
Our experiments confirmed the localization of RAB24 to LC3-positive vesicles, both in full-culture medium where autophagy occurs at a basal level and after amino acid starvation, over 60% of LC3-positive puncta were positive for RAB24. Further, RAB24 localization to autophagosomes was also confirmed by immuno-electron microscopy and subcellular fractionation. RAB24 decorated the autophagosome outer and inner limiting membranes but was less prominent in the lumen of the organelle. This observation suggests that RAB24 has a function on the autophagosomal membrane as opposed to being a cargo protein destined for autophagic degradation. Like LC3, part of RAB24 will be trapped inside the autophagosomes and thus degraded in autolysosomes. However, unlike LC3 levels, the levels of RAB24 protein did not decrease during starvation (our unpublished observation). This is likely due to increased protein synthesis, as we observed increased amounts of RAB24 mRNA in starved cells (our unpublished observation).
Consistent with the concept that prenylation and GTP binding of RAB proteins are important for the implementation of their function, this study also revealed that the localization of RAB24 to LC3-positive vesicles was dependent on prenylation as well as the ability to bind nucleotides. Earlier reports suggest that RAB24 is not as strongly prenylated as the RAB1 protein.12 However, this does not necessarily infer that prenylation has less of a function as a protein lipid anchor. Indeed, our results showed that a considerable proportion of the overexpressed WT RAB24 was prenylated, and that this prenylation was needed for targeting of RAB24 to intracellular organelles including autophagosomes. In addition, our results suggest that nucleotide binding is important for RAB24 targeting to autophagosomes as well as to membranes in general, since the RAB24S67L mutant showed no localization to any cytoplasmic organelles.
Ding et al. have shown that RAB24 has 2 tyrosine residues at positions 17 and 172, that can be phosphorylated.14 Phosphorylation of tyrosines 17 and 172, however, was less important for autophagosome targeting than prenylation and nucleotide binding. The double mutant YY17,172FF showed colocalization with LC3, which was similar to that seen with WT RAB24 in full-culture medium, but less colocalization than the WT in serum and amino acid-free medium. This indicates that phosphorylation or dephosphorylation of these tyrosine residues is not likely to be a signal for the translocation of RAB24 to autophagosomes under basal conditions when RAB24 is needed for autophagosome clearance. The possible role of tyrosine phosphorylation in autophagosome targeting under starvation conditions is currently unclear.
Silencing RAB24 with siRNA revealed that RAB24 was not needed for autophagosome formation or for the clearance of autophagosomes formed in amino acid-free conditions, at least during the first 2 h of starvation. However there was a significant effect of RAB24-silencing on the nonstarved cells where only basal autophagy is active: approximately 4 times more late autophagic compartments were found in the RAB24-silenced cells than in the control cells (Figs. 8B and 9A). Monitoring autophagic flux by adding Baf revealed that the accumulation was due to decreased clearance of the autophagic compartments. Further, the morphology of the accumulating autophagic compartments, the acid-sensitive mRFP-GFP-LC3 probe, and increased intensity of LAMP1 labeling in RAB24-silenced cells, suggested that the accumulating compartments were degradative, indicating that the autophagic pathway was blocked at a later stage, after autophagosomes acquired degradative capacity. This conclusion is in agreement with the results obtained with other assays of autophagic flux, including long-lived protein degradation, and levels of ubiquitinated proteins, SQSTM1, LC3-II and GABARAP-II. All these assays showed that autophagic flux is only minimally affected in RAB24-silenced cells. Taken together, our findings suggest that RAB24 functions at a late stage of the autophagic pathway, such as the final maturation, or most likely in the clearance, of late autophagic vacuoles.
RAB24 is an unusual RAB protein since it has been reported to occur mainly in the GTP-bound form in cells,12 and thus no constitutively active mutant RAB24 has been described. No RAB24 effectors are known, but some interacting proteins have been reported. Tambe et al. 26 have used FLAG or EGFP-tagged PNN/DRS (a tumor suppressor protein) as bait in coimmunoprecipitation and report RAB24 as one of the coprecipitating proteins. They also show colocalization of PNN/DRS-EGFP and RAB24, and of PNN/DRS-EGFP and the autophagosome marker LC3, in punctate structures that accumulate in low-serum culture conditions. Tambe et al. propose that both PNN/DRS and RAB24 may be involved in the progression of autophagy. Schlager et al.27 have performed a GST affinity isolation assay with GST-immobilized RAB proteins, and observe that, similar to several other RABs, RAB24 weakly binds CCDC64B/BICDR2 (a putative RAB6 effector). The strongest binding partner of CCDC64B/BICDR2 in this assay was RAB13. Since this paper concentrates on CCDC64/BICDR1 and RAB6, the putative RAB24 interaction is not elaborated any further. Fukuda et al. report that RAB24 interacts with transcriptional corepressor CTBP1 (C-terminal binding protein 1).28 In this study, a yeast 2-hybrid assay and immunoprecipitation are used to show that 2 mutant versions of RAB24 (RAB24S67L and RAB24T21N) interact with CTBP1. Schardt et al.29 performs coimmunoprecipitation of HEK293 cells overexpressing SNAP29-EYFP (a SNAP25 family SNARE protein) and ECFP-RAB24 using anti-SNAP29, and find that RAB24 coprecipitates with SNAP29. This interaction does not require the presence of GTPγS, unlike the interaction of SNAP29 with RAB3A. Since RAB24 is not expressed in the tissue of interest, it is not studied further in this paper. Behrends et al. 30 use HA-tagged RAB24 as one of the bait proteins in their proteomics study on interactions of autophagy proteins. Their mass spectrometry primary data show coprecipitation of GDI1 (GDP dissociation inhibitor 1) and GDI2, NSF (N-ethylmaleimide-sensitive factor), and PKP1 (plakophilin 1; an armadillo repeat protein implicated to function in desmosomes) with RAB24. Behrends et al. also find RAB24 among the proteins coimmunoprecipitating with the HA-tagged SNARE protein GOSR1 (golgi SNAP receptor complex member 1), but no coprecipitation is detected between RAB24 and SNAP29, CCDC64B/BICDR2, CTBP1 or PNN/DRS. In order to identify high-confidence candidate interacting proteins, Behrends et al. have performed a comparative analysis of the proteomic results, and a subsequent reciprocal proteomic analysis to validate and delineate the interaction network. This analysis places RAB24 in the NSF network together with GOSR1, SNAP29 and several other SNARE proteins, but the analysis proposes that RAB24 would have direct interaction only with GDI1, GDI2, NSF and PKP1, while the interactions with the other proteins in this subnetwork would occur via NSF. Many of these putative indirect or direct RAB24 interacting proteins have been implicated in membrane fusion, including SNAP29, GOSR1, and NSF. Interestingly, SNAP29 has a role in autophagosome fusion with endosomes or lysosomes, acting in a SNARE complex with STX17 (syntaxin 17).14,31,32 However, unlike RAB24, SNAP29 seems to be required for both basal and starvation-induced autophagy. Another difference is that double-membrane autophagosomes accumulate in cells deficient in SNAP2930 or STX17,31 whereas we report here that single-membrane late, degradative autophagic vacuoles and autolysosomes accumulate in RAB24-deficient cells. Hence, the putative RAB24 interactors are in agreement with the idea that RAB24 functions in membrane fusion events during late steps of macroautophagy. However, further studies are required to elucidate the detailed molecular mechanisms of RAB24 functions in autophagy.
Our results suggest that there might be independent regulatory pathways for the maturation and/or clearance of autophagosomes in basal and starvation induced autophagy. This suggestion is supported by reports showing that the histone deacetylase HDAC6 and VCP/p97 (valosin-containing protein) are needed for the clearance of basal, but not starvation-induced, autophagosomes.33,34 Interestingly, our results showed that in the absence of RAB24, autophagosome clearance was not efficient in full-culture medium, but on the contrary, autophagosomes were cleared in full-culture medium if they were first induced by amino acid deprivation. This result suggests that the autophagosomes formed under amino acid deficiency and full culture medium conditions are somehow different, or that partly different molecular machineries are at work when these 2 autophagosome populations are cleared.
We show here that silencing of RAB24 also slows down the clearance of HTT protein aggregates that have been shown by earlier reports to be substrates of autophagy.24 Recent results from Agler et al. reveal that a mutation in RAB24 is associated with a canine ataxia, a hereditary neurodegenerative disease.35 This mutation results in one amino acid change in the putative switch I region in RAB24 suggestive of an effect on nucleotide binding. Affected dogs exhibit Purkinje neuron loss and axonal spheroids throughout the granular layer cells, containing late autophagic vacuoles.35 This study is in agreement with our findings that nucleotide binding is important for the recruitment of RAB24 to autophagic compartments and that RAB24 is needed for autolysosome clearance.
Taken together our results indicate that RAB24 functions in nutrient-independent basal autophagy. Late, acidic and LAMP1-positive autophagic structures accumulate in RAB24-silenced cells under basal conditions. The ultrastructural morphology of the autophagic compartments accumulating in RAB24-silenced cells resembled the morphology of autolysosomes. Further, Baf experiments showed that the accumulation was due to decreased clearance of autolysosomes, while silencing of RAB24 had minor or no effects on autophagic flux. Thus we suggest that RAB24 facilitates autophagy at a late stage, after the autophagosomes have become degradative. This later stage could be the final maturation point or more likely the clearance of degradative autophagic compartments or autolysosomes. We propose that RAB24 is involved in the disassembly of autolysosomes, either in the release of digested material or in lysosomal reformation.36
Materials and Methods
Construction of mutant plasmids
The original MYC-RAB24 pCMV5 plasmid containing the cDNA of mouse Rab24 was a gift from William A. Maltese (University of Toledo, USA). Mouse and human RAB24 have 99% amino acid sequence homology, 94% nucleotide sequence homology and 87% coding region sequence homology. Point mutations were created with specific oligonucleotide primers (from Sigma Aldrich or TAG Copenhagen), PCR amplification with Pfu DNA polymerase and cloning back to the pCMV5 plasmid. Primers were based on the published sequence of mouse Rab24. The final construct sequences were verified by sequencing (GATC Biotech). The used oligonucleotide primer sequences for each mutant were as follows:
S67L forward 5′gggcatttgggacacagcaggtctcgagcgctacgaagccatgagc3′
reverse 5′gctcatggcttcgtagcgctcgagacctgctgtgtcccaaatgccc3′
HHΔ reverse 5′cccgggatcctcaacaacagctgtagaagtaagggtttgcc3′
HH→SN reverse 5′cccgggatcctcagttactacaacagctgtagaagtaagggtttgc3′
CCΔ reverse 5′acccgggatcctcagtgatggctgtagaagtaagggtttgccttctggct3′
CC→SS reverse 5′acccgggatcctcagtgatggctgctgctgtagaagtaagggtttgccttc3′
Y17F forward 5′ggtggttatgctgggcaaggaatttgtgggcaagacgagcctggtg3′
reverse 5′caccaggctcgtcttgcccacaaattccttgcccagcataaccacc3′
Y172F forward 5′ctcttccagaaagtggctgaggatttcgtcagtgtggctgctttccagg3′
reverse 5′cctggaaagcagccacactgacgaaatcctcagccactttctggaagag3′
YY17,172FF(created with Y17F and Y172F oligonucleotide primers)
Cell culture and stable cell lines
Normal rat kidney (NRK) and HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM Sigma D6546), containing 10% fetal calf serum (Sigma F7524), penicillin streptomycin solution (Sigma P0781) and L-glutamine (Sigma 67513). HeLa cells with stable expression of MYC-RAB24 were created using geneticin selection and cultured in the medium described above, with additional 200 µg/ml geneticin (Sigma, G8168). HeLa cells expressing mutant HTT fragments (exon1htt- 65QmCFP) were a gift from Ai Yamamoto (Columbia University, USA). These cells were cultured in the medium described above, with 50 µg/ml geneticin and 5.4 µg/ml hygromycin (Calbiochem, 400053) to maintain stable protein expression. The exon1htt- 65QmCFP protein expression was inhibited with an addition of 10 µg/ml tetracycline (Amresco, E709). HeLa cells expressing mRFP-GFP-LC3 were a gift from Tamotsu Yoshimori via David Rubinsztein (Osaka University, Japan and Cambridge Institute for Medical Research, UK). These cells were cultured in the medium described above, with 600 µg/ml geneticin.
Transient transfection
MYC-RAB24 plasmids were transfected with Lipofectamine 2000 (Invitrogen, 11668-019) or TransIT-LTI (Mirus, MIR 2300) reagent according to the manufacturer's instructions one d after seeding the cells. For urea polyacrylamide gel electrophoresis, RAB24 constructs were transfected with Jet Prime reagent (Polyplus Transfection, 114-15) according to the manufacturer's instructions.
Immunofluorescence
Cells were grown on glass coverslips or 24-well plates to semiconfluency. One d after seeding or transfection the cells were treated with EBSS (Gibco, 24010-043) or full-culture medium, after which the cells were fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS; 140 mM NaCl, 2.7 mM KCl, 4.6 mM Na2HPO4 2H2O, 1.5 mM KH2PO4) for 20 to 30 min at room temperature. The cells were labeled with antibodies against RAB24 (BD Biosciences, mouse anti-RAB24 612174) and LC3 (a gift from Takashi Ueno, Juntendo University, Japan), or LAMP1 (Developmental Studies Hybridoma Bank, H4A3), followed by secondary antibodies conjugated to Alexa Fluor 488, 594 or 647 (Invitrogen, A11029, A11037, A21236). After antibody labeling the cover slips were mounted on object glasses with Mowiol (Calbiochem, 475904) containing 1,4-diazabicyclo[2.2.2]octane (Dabco; Sigma, D-2522) as antifading agent and 4′,6-diamidine-2-phenyl indole (DAPI; Pierce, 62247) to stain the nuclei. On 24-well plates the cells were stained with DAPI and stored in PBS. Images of coverslips were obtained with a wide field fluorescence microscope (Olympus AX70, PlanAPO 60×/1,40 and UPlanFI 20×/0,50 Ph1 objectives, Department of Biosciences, Division of Biochemistry and Biotechnology, University of Helsinki) or a confocal microscope (Leica DM5000, HCX APO 63×/1.30 Corr CS 21 objective, Light microscopy unit, Institute of Biotechnology, University of Helsinki). 24-well plates were imaged with Cellomics CellInsight imaging unit (Thermo Scientific, Olympus LUCPlanFL N 20×/0.45 objective, Light microscopy unit, Institute of Biotechnology, University of Helsinki).
Analysis of immunofluorescence images
ImageJ software was used to assess colocalization (Colocalization finder Plugin)37 and CellProfiler software to assess RAB24 labeling of individual LC3 puncta and the intensity of LAMP1 immunolabeling per cell.38
RAB24 labeling in LC3-positive structures was analyzed with CellProfiler software. Confocal images of 0.25 μm thick optical sections from the middle of the cell were used for the analyses. Data was collected from a minimum of 20 cells per sample. In the CellProfiler38 pipeline, DAPI (nuclei) and Alexa Fluor 488 (RAB24) images were first smoothed with a Gaussian filter. Nuclei were identified with IdentifyPrimaryObjects module using default automatic threshold settings, and cell borders were identified with IdentifySecondaryObjects module with manual thresholding. To detect spots in the Alexa Fluor 594 (LC3) images, a Laplacian of Gaussian filter was first applied39 with the RunImageJ37 module, followed by Otsu thresholding with a manually set correction factor. The spots were then filtered by size filtering and mean intensity (using manually set values), and possible false spots outside cell regions were discarded. LC3 puncta in RAB24-expressing cells were analyzed. RAB24 intensity (integrated intensity) in LC3-positive puncta was normalized with the RAB24 intensity (integrated intensity) in the parent cell and RAB24 average pixel intensity (mean intensity) in LC3-positive puncta was normalized with the RAB24 average pixel intensity (mean intensity) in the parent cell. For analysis of the proportion of LC3 puncta positive for RAB24, an individual LC3-positive punctum was considered RAB24-positive if its average RAB24 pixel intensity was higher than the average pixel intensity in the parent cell, including the LC3-positive puncta. Colocalization between RAB24 and LC3 fluorescence labels was measured with ImageJ37 software colocalization finder Plugin with selection restrained to pixel ratio of 25 to 100%. Confocal images of 0.25-μm thick optical sections from the middle of the cell were used for the analyses. 9 to 21 images, containing a minimum of 20 cells, were analyzed from each sample.
Subcellular fractionation and TPCK-trypsin protection assay
Autolysosomal and dense lysosomal membrane fractions were isolated from rat liver as described previously.16,17 Both fractions are positive for LC3-II.16,17 For subcellular fractionation and TPCK-trypsin protection assay HeLa cells were grown to near confluency on 15-cm plates. Part of the cell population was starved with EBSS containing 100 nM Baf (AH Diagnostics, BML-CM110-0100) and part was changed to fresh complete medium. After 2 h, cells were washed in PBS, trypsinized and counted to normalize the amount of cells in each condition. Cells from each condition were suspended in 4 ml HES buffer (15 mM HEPES [Sigma, H3375]-KOH, pH 7.5, 1 mM EDTA [Sigma, E6758], 166 mM sucrose [Sigma, S0389] with protease inhibitors [cOmplete, EDTA-free; Roche, 04 693 132 001]). Cells were lysed by repeated passage through the tight fitting pestle of a Dounce homogenizer (D9063, pestle B) or by passages through a ball-bearing device (Isobiotec) with a 10-μm opening. Nuclei, intact cells and cell debris were then pelleted by centrifugation at 1000 g, 4°C, for 10 min. The supernatant fraction was further centrifuged at 70,000 g, 4°C, for 30 min to pellet all major remaining subcellular compartments. The pellet fraction was resuspended in 200 μl HES buffer and layered on top of a continuous 4 ml 10-40% OptiPrep (Sigma, D1556) gradient and centrifuged at 70,000 g, 4°C, for 16 h. Alternatively, 5 concentrations of OptiPrep were prepared in HES buffer: 10%, 14.375%, 18.75%, 23.125% and 27.5%. The membrane pellet was suspended in 300 μl HES buffer containing 18.75% OptiPrep and the OptiPrep solutions were layered to form a discontinuous gradient that was centrifuged at 70,000 g, 4°C, for 16 h. Eight fractions were collected from the top of the gradients. For the proteinase protection assay, the fractions were divided into 3 portions. One set was added to 3x SDS-PAGE sample buffer directly, one was incubated with 1 mg/ml TPCK-treated trypsin (Sigma, T1426) at 37°C for 20 min, and the final set of samples was incubated with 1% NP-40 (Sigma, I8896) together with 1 mg/ml trypsin at 37°C for 20 min. To inactivate proteases, 1 mM PMSF (Sigma, P7626) was added to all samples before the addition of 3x SDS-PAGE sample buffer.
GTP-agarose binding
RAB24-WT and S67L mutant protein was expressed from the pET16b vector (Novagen, 69662-3) in E. coli protein expression strain BL21 at +37°C under antibiotic selection. Protein production was induced with 40 μg/ml IPTG (Promega, V3951) at OD600 0.6-0.8 and carried on for 3 or 4 h. Bacterial cells were harvested by centrifugation at 3000 g, at +4°C, for 30 min. Protein production was confirmed with SDS-PAGE and Coomassie Blue staining. Bacterial pellets were suspended in binding buffer (20 mM HEPES, 150 mM NaCl, 10 mM MgCl2, pH 8.0, and cOmplete protease inhibitor cocktail) and sonicated on ice with Sonifier Cell Disruptor B-30 (Branson Sonic Power Co.). Lysates were centrifuged 21100 g, +4°C, for 15 min. Aliquots of the lysates were stored for immunoblot analysis as input samples. Then, 100 to 1000 μg of each lysate was mixed with 200 μl (approximately 50 μl bed volume) of pre-equilibrated GTP-agarose gel (Sigma, G9768) and incubated in a rotator at +4°C for 1 h. The agarose beads were pelleted by centrifugation, washed in binding buffer, resuspended in 50 μl of the same buffer containing 10 mM GTP (Sigma, G5884) and incubated on ice for about 1 h with occasional vortexing. The agarose beads were pelleted by centrifugation, and the supernatant fraction containing the eluted protein was transferred into a new tube and used for SDS-PAGE and western blotting. Bands of eluted protein were quantified and normalized to bands of input samples.
siRNA
Cells were grown on 12-well plates or 3.5-cm plates with coverslips to semiconfluency. One d after seeding, the cells were transfected with RAB24 siRNA smart pool, single oligos of the smart pool, RISC-free control siRNA, or nontargeted siRNA (Dharmacon, Thermo scientific, M-008828-01, D-008828-01, D-008828-02, D-008828-04 and controls D-001220-01-05 or D-001206-13-05, respectively) according to the manufacturer's instructions with Dharmafect reagent (Dharmacon, T-2001-02) and grown for 48 or 72 h. Before fixation or preparation of cell extracts, the cells were treated depending on the experiment. Silencing was verified by immunoblotting, and a minimum of 55% silencing was considered acceptable for further analysis of the samples. Silencing was on average 80% for the smart pool and 55% to 69% for single oligos.
Western blotting
After experimental treatments, cells were collected by scraping and pelleted by centrifugation. Pelleted cells were lysed with lysis buffer (PBS containing 2% NP-40, 0.2% SDS [GIBCO, 15525-017], 1 mM EDTA and cOmplete protease inhibitor cocktail). Cells were lysed on ice for 30 min and centrifuged at 16,060-21,100 g, 4°C, for 4 to 5 min. Protein concentration was measured with a BCA Protein Assay kit (Thermo Scientific, 23228) according to the manufacturer's instructions. Gel samples were prepared with addition of either Laemmli sample buffer or urea sample buffer (0.2 M Tris-HCl, pH 6.8, 8% SDS, 2% mercaptoethanol, 0.004% bromidephenolblue, 8 M urea). Cells were either boiled for 5 min at +95°C, or incubated for 10 min at +37°C degrees (for urea gels). Samples were run on SDS-PAGE gels or 4-8 M urea 10-20% acrylamide gradient gels (urea gels). Protein gels were blotted on Roti-PVDF membranes (Roth, T830.1), which were blocked in 5% milk powder in Tris-buffered saline (10 mM Tris-HCl, pH 7.6, 150 mM NaCl) containing 0.05% Tween 20 (Sigma, P1379) (TBST) and labeled with anti-RAB24 (BD Biosciences, 612174), anti-MYC (Abcam, ab9106), anti-TUBB/tubulin (anti-β-tubulin E7 from the Developmental Studies Hybridoma Bank (DSHB), University of Iowa, IO, USA), anti-RAB7 (Suzanne Pfeffer, Stanford University School of Medicine, Stanford, CA, USA; Cell Signaling Technology, D95F2), anti-ubiquitin (Dako, Z0458), anti-SQSTM1 (BD Biosciences, 610832), anti-LC3B (Sigma, L7543, Cell Signaling Technology, 2775S), anti-LAMP1 (Yoshitaka Tanaka, Kyushu Universit, Japan; hLAMP-1 clone H4A3 from Developmental Studies Hybridoma Bank Iowa, IO, USA) or anti-GABARAP (Abgent, AP1821a) antibodies and goat anti-mouse/rabbit-HRP secondary antibodies (Jackson Immuno Research Laboratories 115-035-003/111-035-003). Detection was done using Immobilon Western HRP substrate kit (Millipore, WBKLS0500).
Sample preparation for electron microscopy
HeLa cells were treated with normal full-culture medium without or with Baf (Enzo Life Sciences BML-CM110) or EBSS for starvation. Cells were fixed with 1% glutaraldehyde in 0.2 M HEPES, pH 7.4, for 2 h at room temperature. After 30 min of fixation the cells were scraped from the culture wells in fixative and pelleted at 16,060 g for 5 min. Cell pellets remained in the fixative for another 90 min. The supernatant was changed to 0.2 M Hepes without disturbing the fixed pellet. Fixed pellets were washed with PBS and post fixed with 1% osmium tetroxide for 1 h at room temperature. Pellets were washed with water, incubated in 2% uranyl acetate for one h at room temperature in the dark and dehydrated with increasing concentrations of ethanol followed by propyleneoxide (Sigma, 471968). The pellets were then incubated in propyleneoxide-Durcupan (Fluka, 44612) mixture for 2 h at room temperature and infiltrated with 100% Durcupan overnight. The pellets were placed in beam capsules, infiltrated with Durcupan for a further 5 h, and polymerized at +60°C for 2 d. Thin sections (80 nm) were cut using an ultramicrotome, collected onto electron microscopy grids and stained with uranyl acetate and lead citrate. Quantitative counting of autophagic compartments was performed using a transmission electron microscope (JEOL 1200 EX II or JEOL 1400 EX, Electron microscopy unit, Institute of Biotechnology, University of Helsinki) as described previously.40 For immuno-electron microscopy NRK or HeLa cells were grown on 3.5-cm plates to semiconfluency. MYC-RAB24 plasmid was transfected with Lipofectamine 2000 or TransIT-LTI (Mirus, MIR 2300) reagent according to the manufacturer's instructions one day after seeding the cells. One day after transfection the cells were treated with EBSS or normal culture medium, after which the cells were fixed in 4% PFA in 0.2 M HEPES, pH 7.4, for 1 h at room temperature. Cells were washed with 2% PFA in 0.2 M HEPES and scraped from the plates, collected by centrifugation and left at +4°C overnight. The pellets were embedded in 10% gelatin (BD Biosciences, 214340), infiltrated with 20% polyvinyl-pyrrolidone (PVP; Sigma, PVP10), 2.3 M sucrose (MP Biomedicals, 821713) at +4°C overnight, placed on ultramicrotome sample holders and frozen in liquid nitrogen. Samples were cut into thin (80 nm) sections at −100°C using a cryo ultramicrotome (Leica) and picked up on electron microscopy grids (Electron Microscopy Sciences, G200-Cu). Immunolabeling was performed with a monoclonal antibody against RAB24 (BD Biosciences, 612174) and a 5-nm or 10-nm gold-conjugated secondary antibody (British BioCell, 15735-1 or 15846).
Dot blot
HeLa cells expressing CFP-tagged HTT were grown on 3.5-cm plates to semiconfluency. Cells were transfected with siRNA as described above and 48 h after transfection tetracycline was added to stop mutant HTT expression. Alternatively, 72 h after transfection cells were seeded onto new plates and tetracycline was added simultaneously. Cells were collected 0, 1 or 3 d after tetracycline addition (48, 72 and 120 h or 72, 96 and 144 h incubation after siRNA transfection) and the pelleted cells were snap-frozen in liquid nitrogen. Pellets were then treated for filter trap assay to analyze the amount of insoluble CFP-tagged HTT, as described previously.25 Briefly, 25 or 50 µg of each sample was filtered through cellulose acetate membrane (Whatman, 10404180) using a dot blot filtering unit (Bio-Rad Bio-Dot apparatus). After filtering, the membrane was blocked in 5% milk powder in TBST for one h at room temperature and labeled with GFP antibody (Fitzgerald Industries International, RDI-GRNFP4abr) that cross reacts with CFP, and goat anti-rabbit-HRP (Jackson Immuno Research Laboratories, 111-035-003). Detection was carried out as previously described for western blotting. The signals were quantified using Image-Pro Plus 7.0 software.
Protein measurement for dot blot
HeLa cells expressing CFP-tagged HTT aggregates were grown and harvested as described above. Protein concentrations in the samples for filter trap assay were assayed using a filter paper dye-binding method. Samples and BSA standards (bovine serum albumin; Bovogen Biologicals BSAS 0.1) were pipetted on Whatman 3MM CHR chromatography paper (3030-931) in 1×1 cm squares and air dried. Samples were fixed on the paper with 10% trichloroacetic acid for 15 min at room temperature. Whatman paper was washed twice with water and colored with Page Blue (Fermentas, R0571) solution for 1 h at room temperature. Excess color was removed by 3 15-min washes in water and the paper was air-dried. Squares containing samples and standards were cut from the paper and placed in microcentrifuge tubes. Color from the squares was extracted in 1 M CH3 COOK (Sigma, 32309) in 70% ethanol. The absorbances of samples and standards were measured in a 96-well plate at 595 nm.
Long-lived protein degradation
Long-lived protein degradation was studied in HeLa cells as previously described.23 HeLa cells were simultaneously transfected with RAB24 or control siRNA and metabolically labeled with 0.125 μCi/ml 14C-valine (Perkin Elmer NEC291EU050UC) in full-culture medium (DMEM). One d later short-lived proteins were chased out in full-culture medium with excess cold valine overnight. Cells were treated with nutrient rich (DMEM) or amino acid-free (EBSS) medium for approximately 4 h and long-lived protein degradation was measured by assessing the acid-soluble radioactivity released into the culture medium. The degradation was expressed as a ratio of acid-soluble radioactivity in culture medium divided by total radioactivity in the cells and medium.
HTT protein aggregate quantitation
HeLa cells expressing CFP-tagged HTT were grown on cover slips on 3.5-cm plates to semiconfluency. Cells were transfected with siRNA as described above, and 48 h after transfection tetracycline was added to cells to stop mutant HTT expression. Alternatively, 72 h after transfection cells were seeded onto new plates and tetracycline was added simultaneously. Cells were fixed 0, 1 and 3 d after the addition of tetracycline and embedded in Mowiol containing DAPI to stain the nuclei. Cells were imaged using a 20X objective (Zeiss Imager M2, Zeiss PlanAPO CHROMAT 20×/0,8 Ph2 objective, Institute of biotechnology, University of Helsinki) and bright aggregates and nuclei were counted using Image-Pro Plus 7.0 software. The number of aggregates was divided by the number of nuclei. Silencing of RAB24 was verified by immunoblotting at time points 0 and 3 d after addition of tetracycline (48 or 72 h and 120 or 144 h incubation after siRNA transfection).
Statistical testing
Unless otherwise stated, the Student t test was used to test statistical significance. Further details are given in the figure legends.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Ruusu Merivirta, Susanna Salmi, Mykola Domanskyy and Jenni Tamminen for generous technical help and input in the preliminary phases of this project. We also thank the Electron Microscopy Unit at the Institute of Biotechnology, University of Helsinki, for providing laboratory facilities and Harri Jäälinoja in the Light Microscopy Unit at the Institute of Biotechnology, University of Helsinki, for technical support in image analysis. We thank William Maltese, Ai Yamamoto and Tamotsu Yoshimori for providing the RAB24 plasmid, HTT cell lines, and mRFP-GFP-LC3 cell line, respectively.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This study was financially supported by Magnus Ehrnrooth Foundation, Biocentrum Helsinki, Helsinki University Foundations, Integrative Life Science Doctoral Program (ILS), the Nordic Autophagy Network and The Academy of Finland.
References
- 1.Pfeffer SR. Rab GTPases: Specifying and deciphering organelle identity and function. Trends Cell Biol 2001; 11:487-91; PMID:11719054 [DOI] [PubMed] [Google Scholar]
- 2.Khosravi-Far R, Lutz RJ, Cox AD, Conroy L, Bourne JR, Sinensky M, Balch WE, Buss JE, Der CJ. Isoprenoid modification of rab proteins terminating in CC or CXC motifs. Proc Natl Acad Sci U S A 1991; 88:6264-8; PMID:1648736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirota Y, Tanaka Y. A small GTPase, human Rab32, is required for the formation of autophagic vacuoles under basal conditions. Cell Mol Life Sci 2009; 66:2913-32; PMID:19593531; http://dx.doi.org/ 10.1007/s00018-009-0080-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Itoh T, Fujita N, Kanno E, Yamamoto A, Yoshimori T, Fukuda M. Golgi-resident small GTPase Rab33B interacts with Atg16L and modulates autophagosome formation. Mol Biol Cell 2008; 19:2916-25; PMID:18448665; http://dx.doi.org/ 10.1091/mbc.E07-12-1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zoppino FC, Militello RD, Slavin I, Alvarez C, Colombo MI. Autophagosome formation depends on the small GTPase Rab1 and functional ER exit sites. Traffic 2010; 11:1246-61; PMID:20545908; http://dx.doi.org/ 10.1111/j.1600-0854.2010.01086.x [DOI] [PubMed] [Google Scholar]
- 6.Jager S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen EL. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci 2004; 117:4837-48; PMID:15340014; http://dx.doi.org/ 10.1242/jcs.01370 [DOI] [PubMed] [Google Scholar]
- 7.Gutierrez MG, Munafo DB, Beron W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci 2004; 117:2687-97; PMID:15138286; http://dx.doi.org/ 10.1242/jcs.01114 [DOI] [PubMed] [Google Scholar]
- 8.Longatti A, Lamb CA, Razi M, Yoshimura S, Barr FA, Tooze SA. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. J Cell Biol 2012; 197:659-75; PMID:22613832; http://dx.doi.org/ 10.1083/jcb.201111079 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szatmari Z, Sass M. The autophagic roles of rab small GTPases and their upstream regulators: A review. Autophagy 2014; 10:1154-66; PMID:24915298; http://dx.doi.org/29395 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chua CE, Gan BQ, Tang BL. Involvement of members of the rab family and related small GTPases in autophagosome formation and maturation. Cell Mol Life Sci 2011; 68(20):3349-58; PMID:21687989; http://dx.doi.org/ 10.1007/s00018-011-0748-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olkkonen VM, Dupree P, Killisch I, Lutcke A, Zerial M, Simons K. Molecular cloning and subcellular localization of three GTP-binding proteins of the rab subfamily. J Cell Sci 1993; 106 (Pt 4):1249-61; PMID:8126105 [DOI] [PubMed] [Google Scholar]
- 12.Erdman RA, Shellenberger KE, Overmeyer JH, Maltese WA. Rab24 is an atypical member of the rab GTPase family. deficient GTPase activity, GDP dissociation inhibitor interaction, and prenylation of Rab24 expressed in cultured cells. J Biol Chem 2000; 275:3848-56; PMID:10660536 [DOI] [PubMed] [Google Scholar]
- 13.Munafo DB, Colombo MI. Induction of autophagy causes dramatic changes in the subcellular distribution of GFP-Rab24. Traffic 2002; 3:472-82; PMID:12047555; http://dx.doi.org/tra030704 [pii]. [DOI] [PubMed] [Google Scholar]
- 14.Ding J, Soule G, Overmeyer JH, Maltese WA. Tyrosine phosphorylation of the Rab24 GTPase in cultured mammalian cells. Biochem Biophys Res Commun 2003; 312:670-5; PMID:14680817; http://dx.doi.org/ 10.1016/j.bbrc.2003.10.171 [DOI] [PubMed] [Google Scholar]
- 15.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol 2005; 171:603-14; PMID:16286508; http://dx.doi.org/jcb.200507002 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ueno T, Ishidoh K, Mineki R, Tanida I, Murayama K, Kadowaki M, Kominami E. Autolysosomal membrane-associated betaine homocysteine methyltransferase. limited degradation fragment of a sequestered cytosolic enzyme monitoring autophagy. J Biol Chem 1999; 274:15222-9; PMID:10329731 [DOI] [PubMed] [Google Scholar]
- 17.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000; 19:5720-8; PMID:11060023; http://dx.doi.org/ 10.1093/emboj/19.21.5720 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutierrez MG, Vazquez CL, Munafo DB, Zoppino FC, Beron W, Rabinovitch M, Colombo MI. Autophagy induction favours the generation and maturation of the coxiella-replicative vacuoles. Cell Microbiol 2005; 7:981-93; PMID:15953030; http://dx.doi.org/ 10.1111/j.1462-5822.2005.00527.x [DOI] [PubMed] [Google Scholar]
- 19.Sanford JC, Pan Y, Wessling-Resnick M. Prenylation of Rab5 is dependent on guanine nucleotide binding. J Biol Chem 1993; 268:23773-6; PMID:8226909 [PubMed] [Google Scholar]
- 20.Rubinsztein DC, Cuervo AM, Ravikumar B, Sarkar S, Korolchuk V, Kaushik S, Klionsky DJ. In search of an “autophagomometer”. Autophagy 2009; 5:585-9; PMID:19411822 [DOI] [PubMed] [Google Scholar]
- 21.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al.. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8:445-544; PMID:22966490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol 2001; 152:657-68; PMID:11266458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bauvy C, Meijer AJ, Codogno P. Assaying of autophagic protein degradation. Methods Enzymol 2009; 452:47-61; PMID:19200875; http://dx.doi.org/ 10.1016/S0076-6879(08)03604-5 [doi]. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto A, Cremona ML, Rothman JE. Autophagy-mediated clearance of huntingtin aggregates triggered by the insulin-signaling pathway. J Cell Biol 2006; 172:719-31; PMID:16505167; http://dx.doi.org/ 10.1083/jcb.200510065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wanker EE, Scherzinger E, Heiser V, Sittler A, Eickhoff H, Lehrach H. Membrane filter assay for detection of amyloid-like polyglutamine-containing protein aggregates. Methods Enzymol 1999; 309:375-86; PMID:10507036 [DOI] [PubMed] [Google Scholar]
- 26.Tambe Y, Yamamoto A, Isono T, Chano T, Fukuda M, Inoue H. The drs tumor suppressor is involved in the maturation process of autophagy induced by low serum. Cancer Lett 2009; 283:74-83; PMID:19368996; http://dx.doi.org/ 10.1016/j.canlet.2009.03.028 [doi]. [DOI] [PubMed] [Google Scholar]
- 27.Schlager MA, Kapitein LC, Grigoriev I, Burzynski GM, Wulf PS, Keijzer N, de Graaff E, Fukuda M, Shepherd IT, Akhmanova A, et al.. Pericentrosomal targeting of Rab6 secretory vesicles by bicaudal-D-related protein 1 (BICDR-1) regulates neuritogenesis. EMBO J 2010; 29:1637-51; PMID:20360680; http://dx.doi.org/ 10.1038/emboj.2010.51 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukuda M, Kanno E, Ishibashi K, Itoh T. Large scale screening for novel rab effectors reveals unexpected broad rab binding specificity. Mol Cell Proteomics 2008; 7:1031-42; PMID:18256213; http://dx.doi.org/ 10.1074/mcp.M700569-MCP200 [doi]. [DOI] [PubMed] [Google Scholar]
- 29.Schardt A, Brinkmann BG, Mitkovski M, Sereda MW, Werner HB, Nave KA. The SNARE protein SNAP-29 interacts with the GTPase Rab3A: Implications for membrane trafficking in myelinating glia. J Neurosci Res 2009; 87:3465-79; PMID:19170188; http://dx.doi.org/ 10.1002/jnr.22005 [doi]. [DOI] [PubMed] [Google Scholar]
- 30.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature 2010; 466:68-76; PMID:20562859; http://dx.doi.org/ 10.1038/nature09204 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 2012; 151:1256-69; PMID:23217709; http://dx.doi.org/ 10.1016/j.cell.2012.11.001 [doi]. [DOI] [PubMed] [Google Scholar]
- 32.Takats S, Nagy P, Varga A, Pircs K, Karpati M, Varga K, Kovacs AL, Hegedus K, Juhasz G. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in drosophila. J Cell Biol 2013; 201:531-9; PMID:23671310; http://dx.doi.org/ 10.1083/jcb.201211160 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee JY, Koga H, Kawaguchi Y, Tang W, Wong E, Gao YS, Pandey UB, Kaushik S, Tresse E, Lu J, et al.. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J 2010; 29:969-80; PMID:20075865; http://dx.doi.org/ 10.1038/emboj.2009.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tresse E, Salomons FA, Vesa J, Bott LC, Kimonis V, Yao TP, Dantuma NP, Taylor JP. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy 2010; 6:217-27; PMID:20104022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agler C, Nielsen DM, Urkasemsin G, Singleton A, Tonomura N, Sigurdsson S, Tang R, Linder K, Arepalli S, Hernandez D, et al.. Canine hereditary ataxia in old english sheepdogs and gordon setters is associated with a defect in the autophagy gene encoding RAB24. PLoS Genet 2014; 10:e1003991; PMID:24516392; http://dx.doi.org/ 10.1371/journal.pgen.1003991 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, et al.. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 2010; 465:942-6; PMID:20526321; http://dx.doi.org/ 10.1038/nature09076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rasband WS, Image J, U. S. National Institutes of Health, Bethesda, Maryland, USA, http://imagej.nih.gov/ij/, 1997-2014 [Google Scholar]
- 38.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al.. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol 2006; 7:R100; PMID:17076895; http://dx.doi.org/gb-2006-7-10-r100 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sage D, Neumann FR, Hediger F, Gasser SM, Unser M. Automatic tracking of individual fluorescence particles: Application to the study of chromosome dynamics. IEEE Trans Image Process 2005; 14:1372-83; PMID:16190472 [DOI] [PubMed] [Google Scholar]
- 40.Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. Monitoring autophagy by electron microscopy in mammalian cells. Methods Enzymol 2009; 452:143-64; PMID:19200881; http://dx.doi.org/ 10.1016/S0076-6879(08)03610-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


