Abstract
Autophagy is a pathway mediating vacuolar degradation and recycling of proteins and organelles, which plays crucial roles in cellular physiology. To ensure its proper cytoprotective function, the induction and amplitude of autophagy are tightly regulated, and defects in its regulation are associated with various diseases. Transcriptional control of autophagy is a critical aspect of autophagy regulation, which remains largely unexplored. In particular, very few transcription factors involved in the activation or repression of autophagy-related gene expression have been characterized. To identify such regulators, we analyzed the expression of representative ATG genes in a large collection of DNA-binding mutant deletion strains in growing conditions as well as after nitrogen or glucose starvation. This analysis identified several proteins involved in the transcriptional control of ATG genes. Further analyses showed a correlation between variations in expression and autophagy magnitude, thus identifying new positive and negative regulators of the autophagy pathway. By providing a detailed analysis of the regulatory network of the ATG genes our study paves the way for future research on autophagy regulation and signaling.
Keywords: autophagy, gene expression, stress, transcription factor, yeast
Abbreviations
- Atg
autophagy related
- Cvt
cytoplasm-to-vacuole targeting
- GFP
green fluorescent protein.
Introduction
Macroautophagy (hereafter referred to as autophagy) is a conserved pathway during which portions of the cytoplasm, organelles, or pathogens are engulfed by a double-membrane structure called the phagophore, which, after completion, forms into an autophagosome that fuses with the vacuole. Upon vacuolar fusion, the autophagy cargo as well as the inner membrane of the autophagosome are degraded. Molecules resulting from autophagy degradation are then recycled back to the cytosol and reused by the cell to maintain homeostasis in unfavorable conditions.1
Autophagy is highly upregulated upon nutrient limitation as well as multiple stress conditions where it serves cytoprotective functions and promotes cell survival.2 As a consequence, the autophagy pathway plays critical roles in mammalian development, cellular physiology and the immune response,3-5 and defects in autophagy are associated with severe pathologies such as cancer or metabolic diseases.6-7 A basal level of autophagy is also maintained in physiological conditions. For example, in mammals basal autophagy is involved in the degradation of damaged organelles and protein aggregates, which can otherwise lead to various neurodegenerative disorders such as Huntington, Alzheimer and Parkinson diseases.9 In yeast, along with a low level of constitutive autophagy the cytoplasm-to-vacuole targeting (Cvt) pathway, a biosynthetic type of selective autophagy, delivers resident enzymes to the vacuole during growth.8
High, uncontrolled, autophagy activity can also be detrimental for cells as it can lead to cell death or possibly promote the replication/spread of microbes or cancer cells that, in some instances, can use autophagy as a source of nutrients or to survive in unfavorable conditions.10-13 Therefore, to support proper cellular functions, the magnitude of autophagy has to be finely controlled, notably by regulating the autophagy-related (Atg) proteins, the core components of the autophagy machinery. Numerous studies have provided significant advances on the understanding of the posttranslational modifications of several Atg proteins, which affects their localization and activity, as well as protein-protein interactions; key players of these regulatory networks have been characterized (for review see ref. 14,15). The mechanisms involved in transcriptional regulation of ATG genes remains, however, largely unknown. Upon stress conditions, the expression of most of the ATG genes is upregulated and this correlates with an increase in autophagy activity. Recent studies show that the transcriptional regulation of ATG genes is critical for autophagy: the level of Atg8, for example, correlates with autophagosome size,16 that of Atg9 correlates with their number,17 and the amount of Atg7 modulates autophagy amplitude.18 In higher eukaryotes the family of FOXO transcription factors, GATA1 as well as the master regulator TFEB, activate autophagy,19-21 while ZKSCAN3 and GATA4 are involved in the repression of mammalian autophagy-related genes.22-23 In yeast, recent studies characterized Ume6 and Pho23 as transcriptional repressors of ATG genes and negative regulators of autophagy.24,17 Nevertheless, very few transcription factors involved in the regulation of the expression of ATG genes in either yeast or mammals are known. To identify such regulators we conducted a screen in which DNA-binding protein deletion strains were tested for the expression of ATG genes. This revealed Rph1 as a master transcriptional repressor of autophagy.18 Here we report on the overall results of the screen, which identify new autophagy regulators, either activators or repressors of ATG gene expression. Candidates showing the highest variations of expression were further characterized to assay their impact on autophagy activity and the Cvt pathway. Together, our results constitute a valuable resource in the understanding of the transcription of autophagy-related genes as well as overall autophagy regulation and provide new directions in autophagy research.
Results
A screen for DNA-binding proteins regulating the expression of ATG genes
In order to identify new transcriptional regulators of auto-phagy the expression of a subset of ATG genes was analyzed in comparison between wild-type cells and a collection of DNA-binding mutants (Fig. 1A). To select the target ATG genes used in this study we integrated multiple criteria: (i) Most, but not all, ATG genes are upregulated after nutrient starvation,17 targets should show a strong induction of expression, indicating that they are under transcriptional regulation in these conditions. (ii) Variation in the expression of the selected genes should translate into a detectable effect with regard to autophagy; i.e., the amount of the corresponding protein modulates the level of autophagy activity. (iii) Target genes should be representative of different steps of the autophagy pathway and notably of selective autophagy.
Figure 1.
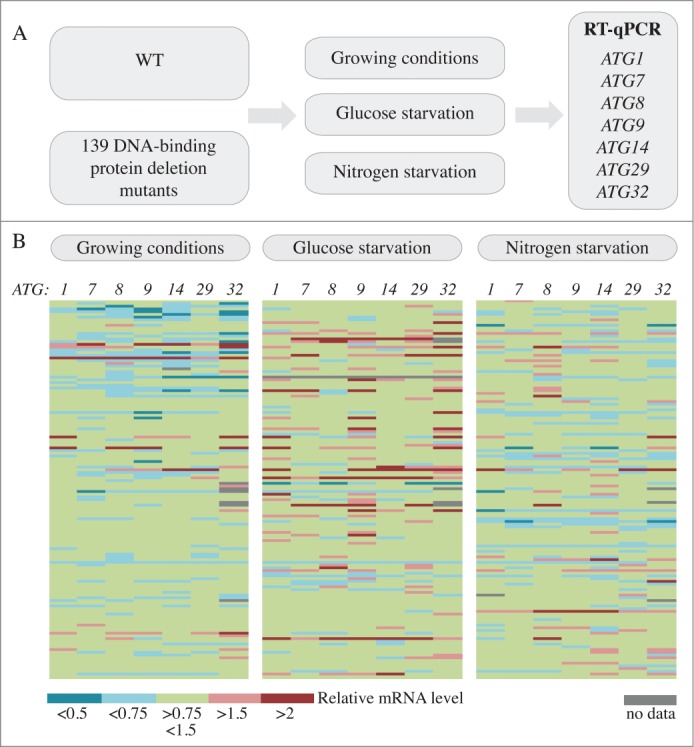
A screen for DNA-binding proteins involved in the regulation of ATG gene expression. (A) Schematic of the screen. (B) Color graph illustrating the results of the screen. Each line represents the mRNA level of an independent mutant relative to the wild type in the same condition, which was set to 1.
Based on these points, we selected ATG1, ATG7, ATG8, ATG9, ATG14, ATG29, and ATG32: ATG1 and ATG29 as markers of autophagy induction, ATG9 for the supply of lipids to the expanding phagophore, ATG14 for vesicle nucleation, ATG7, ATG8 and ATG9 for vesicle expansion and completion, and ATG32 as a marker of mitophagy, the selective autophagic degradation of mitochondria.25 Furthermore, previous work from our lab showed that the level of Atg7, Atg8 and Atg9 correlates with autophagy activity,16-18 indicating that modulation in the expression of the corresponding genes should affect the magnitude of autophagic degradation.
An increase in mRNA level upon autophagy activation could result either from the release of a repressive factor or the recruitment of an activator. Therefore gene expression was analyzed by RT-qPCR in nutrient-replete conditions (growing conditions), where the deletion of a repressor should lead to mRNA enrichment, as well as after nitrogen starvation and glucose starvation where the deletion of a positive factor would result in a reduction in the mRNA level compared to the wild type.
Among the 139 strains analyzed, 61 strains showed a reduction of 1.5X of the induction of at least one ATG gene compared to the induction in wild-type cells after nitrogen starvation, and among these, 5 strains showed a more than 2-fold reduction (Fig. 1B, Table S1). Upon glucose starvation, 37 strains showed a reduction of 1.5X of the induction of at least one ATG gene compared to the induction seen in wild-type cells, and among these one strain showed a more than 2-fold reduction (Fig. 1B, Table S2). Finally, 13 strains showed an induction of 1.5 fold or more in the expression of at least one ATG gene and 6 showed an increase of more than 2 fold in growing conditions compared to the wild type (Fig. 1B, Table S3). Note that among the hits, the ume6Δ and rph1Δ cells showed enriched mRNA levels of ATG genes consistent with previous publications (Table S3, Fig. S1).24,18 The results for all 139 strains in the different conditions are listed in Tables S1 to S3.
As a proof of concept and to assay the level of confidence of the results obtained in the screen, we repeated the analysis with the candidate strains that showed the highest variation compared to wild type, that is, an increase in expression of more than 2X in growing conditions (potential repressors) or a reduction of more than 2X after starvation (potential activators). The deletion of SPT10, FYV5, SFL1, ZAP1 and YRM1 led to a significant increase in the expression of at least one ATG gene; sko1Δ cells showed a similar pattern although the differences were not quite statistically significant (Fig. S1A). The deletion of MBP1 resulted in a large reduction in the induction of expression of most of the analyzed target genes after glucose starvation (Fig. S1B), whereas the deletion of GCN4, GLN3, GAT1 and SWI5 decreased the induction of the expression of at least one ATG gene after nitrogen starvation (Fig. S1C). Together these results confirmed the validity of the results obtained during our screen and identified Spt10, Fyv5, Sfl1, Sko1, Zap1 and Yrm1 as potential transcriptional repressors of autophagy, and Mbp1, Gcn4, Gln3, Gat1 and Swi5 as potential transcriptional activators of autophagy (Table 1).
Table 1.
DNA binding proteins identified as potential regulators of ATG gene expression
| Gene ID | Deletion phenotype | Descriptiona | |
|---|---|---|---|
| SPT10 | YJL127C | Increased expression in growing conditions | Putative histone acetylase |
| FYV5 | YCL058C | Protein involved in the regulation of the mating pathway | |
| SFL1 | YOR140W | Transcriptional repressor and activator | |
| SKO1 | YNL167C | Basic leucine zipper transcrption factor of the ATF/CREB family | |
| ZAP1 | YJL056C | Zinc-regulated transcription factor | |
| YRM1 | YOR172W | Zinc finger transcription factor | |
| MBP1 | YDL056W | Decreased induction upon glucose starvation | Transcription factor |
| GCN4 | YEL009C | Decreased induction upon nitrogen starvation | bZIP transcriptional activator |
| GLN3 | YER040W | Transcriptional activator | |
| GAT1 | YFL021W | Transcriptional activator, GATA1 type zinc finger domain | |
| SWI5 | YDR146C | Transcription factor |
Gene descriptions are from the Saccharomyces Genome Database, available at http://www.yeastgenome.org
Analysis of transcriptional activators of autophagy
To further characterize the proteins identified during the screen and to verify the screen results we decided to delete our candidate genes in another yeast strain background to eliminate potential strain-dependent phenotypes and also to confirm that the expression defects were due to the correct gene deletion. For all but the MBP1 deletion, we used the SEY6210 background expressing the modified Pho8Δ60 protein that forms the basis of an assay to monitor autophagy activity.26 The deletion of MBP1 was made in the W303 background, which shows a better response to glucose starvation-induced autophagy (our unpublished observation). Among the potential transcriptional activators, 2 mutants, swi5Δ and mbp1Δ, now showed comparable ATG expression with the wild type after nitrogen and glucose starvation, respectively, indicating that the corresponding gene was unlikely to be involved in autophagy regulation or that the phenotype was strain specific (Fig. S2A and B). In contrast, the deletion of GCN4, GLN3 and GAT1 recapitulated the results observed during the initial screen. In gcn4Δ cells, we observed a reduction in the induction of ATG1 expression as well as Atg1 protein level compared to wild-type cells after nitrogen starvation (Fig. 2A–B, compare lane 2 to 5 and lane 3 to 6). Under these conditions the expression of ATG7, ATG8, ATG9, ATG29, and ATG32 was induced to a lesser extent in gln3Δ and gat1Δ cells compared to the wild type (Fig. 2C, lower panel) suggesting that the corresponding proteins are transcriptional activators of ATG genes. In addition, the deletion of GLN3 resulted in an accumulation of ATG8 and ATG29 mRNAs in growing conditions (Fig. 2C, upper panel) indicating that Gln3 is directly or indirectly involved in the repression of these genes when autophagy is kept at a basal level. Gln3 and Gat1 are both part of the GATA family of transcription factors and bind to similar DNA motifs.27-30 The double deletion gat1Δ gln3Δ did not have an additive effect on ATG expression after nitrogen starvation compared to the single gln3Δ or gat1Δ strain (Fig. 2C) suggesting that the 2 proteins may recognize the same consensus sites on ATG promoters and work together to induce their transcription.
Figure 2.
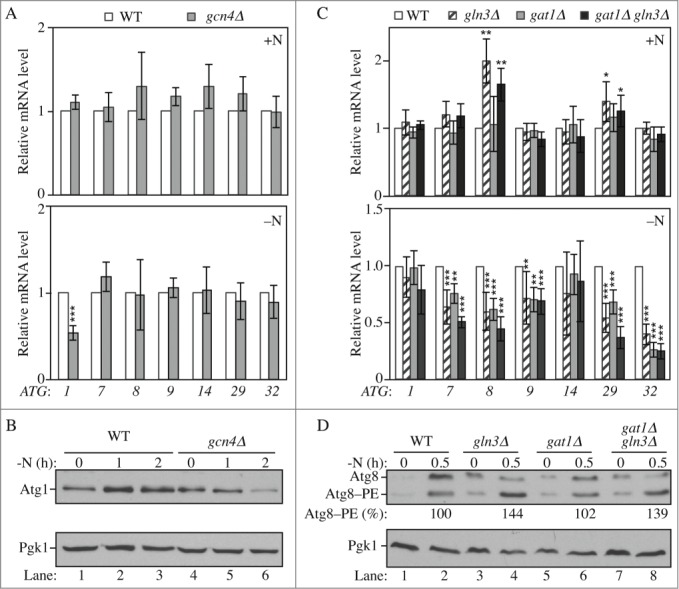
Gln3, Gat1 and Gcn4 are transcriptional modulators of ATG genes upon nitrogen starvation. (A-B) Gcn4 is required for the proper induction of ATG1 upon nitrogen starvation. (A) Wild-type (WLY176; SEY6210) and gcn4Δ (YAB387) cells were grown in YPD (+N) until mid-log phase (upper panel) and then starved for nitrogen (-N) for 1 h (lower panel). mRNA levels were quantified by RT-qPCR. The mRNA level of individual ATG genes was normalized to the mRNA level of the corresponding gene in wild-type cells, which was set to 1. The data represent the average of at least 3 independent experiments. (B) Wild-type and gcn4Δ cells were grown in YPD until mid-log phase and then starved for nitrogen (-N) for the indicated times. Protein extracts were analyzed by western blot with anti-Atg1 and anti-Pgk1 (loading control) antisera. (C-D) Gln3 and Gat1 are required for the proper induction of ATG7, ATG8, ATG9, ATG29 and ATG32 after nitrogen starvation; the deletion of GLN3 increases the expression of ATG8 and ATG29 in growing conditions. (C) Wild-type (WLY176), gln3Δ (YAB385), gat1Δ (YAB384) and gat1Δ gln3Δ (YAB386) cells were grown and mRNA analyzed as in (A). The data represent the average of at least 3 independent experiments. (D) For the analysis of Atg8 protein level, wild-type (WLY176; SEY6210), gln3Δ (YAB385), gat1Δ (YAB384) and gat1Δ gln3Δ (YAB386) cells were grown as in (B). Protein extracts were analyzed by western blot with anti-Atg8 and anti-Pgk1 (loading control) antisera. The percent Atg8–PE of total Atg8 is indicated.
To determine the effect of such transcriptional variation on protein level we focused on Atg8, as the corresponding gene was one of the most significantly affected in the mutants and because the level of this protein modulates autophagy activity.16 Consistent with the mRNA data, the level of Atg8 was higher in gln3Δ and gat1Δ gln3Δ cells compared to the wild type in growing conditions (Fig. 2D, compare lane 1 to lanes 3 and 7). Shortly after starvation we did not observe a major difference in the overall amount of Atg8 although there was a relative enrichment of the pool of Atg8 conjugated to phosphatidylethanolamine (PE), Atg8–PE, in gln3Δ and gat1Δ gln3Δ cells (Fig. 2D, compare lane 2 to lanes 4 and 8). This suggests a more rapid autophagy induction in these cells due to the increased expression of the ATG genes in growing conditions. Together these results reveal the role of Gln3 in the repression of some ATG genes in growing conditions and show that Gat1, Gcn4 and Gln3 are activators required for the induction of ATG gene expression after nitrogen starvation.
To determine how these transcriptional defects affect autophagy flux, we monitored autophagy activity using the Pho8Δ60 assay. In this assay, we use a modified phosphatase precursor, Pho8Δ60, which can only be delivered from the cytoplasm to the vacuole by the autophagy pathway. Inside the vacuole, Pho8Δ60 is activated; therefore, levels of phosphatase activity of Pho8Δ60 reflect the rate of nonselective autophagy.26 Consistent with a reduction of ATG gene expression after nitrogen starvation, the GAT1 deletion strain showed a 20% decrease, whereas the GCN4 deletion resulted in a 50% reduction in Pho8Δ60-dependent phosphatase activity compared to wild-type cells (Fig. 3A); atg1Δ cells were used as a negative control and showed no induction of autophagy. In contrast, and despite the defects in ATG expression after starvation, the deletion of GLN3 and the double GAT1 GLN3 deletion did not show any defects in autophagy activity upon nitrogen starvation (Fig. 3A). We hypothesized that this could result from the increased expression, notably of ATG8, observed in growing conditions in these strains. This may cause an increase in autophagy activity shortly after induction and would therefore counteract the potential repressive effect of the mutations over the time course of this assay. Such a phenotype is seen, for example, in the ume6Δ strain, where higher basal Atg8 levels result in a more rapid increase in autophagy activity in starvation conditions.24
Figure 3.
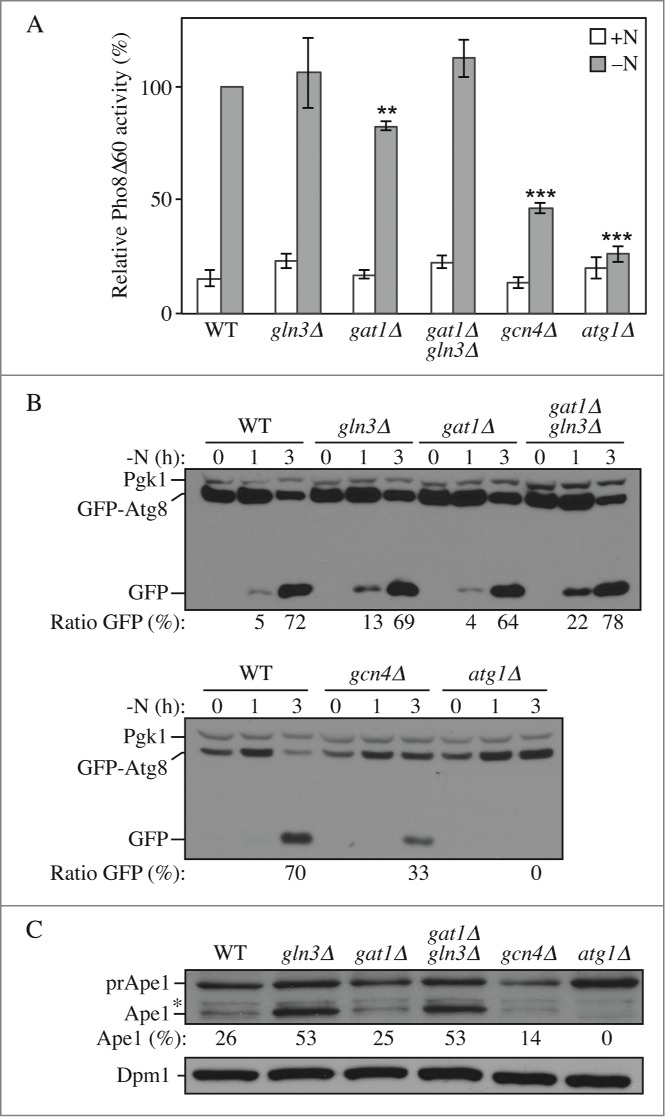
Gln3, Gat1 and Gcn4 affect autophagy activity. Wild-type (WLY176), gln3Δ (YAB385), gat1Δ (YAB384), gat1Δ gln3Δ (YAB386), gcn4Δ (YAB387) and atg1Δ (WLY192) cells were grown in YPD (+N) and then starved for nitrogen for the indicated times. (A) Autophagy activity as measured by the Pho8Δ60 assay is decreased in gat1Δ and gcn4Δ cells. Cells were starved for nitrogen for 3 h (-N). The Pho8Δ60 activity was measured and normalized to the activity of the wild-type cells after starvation, which was set to 100%. n=3 independent experiments. (B) Autophagy as measured by the GFP-Atg8 processing assay is increased shortly after starvation in gln3Δ and gat1Δ gln3Δ cells, but decreased in gat1Δ and gcn4Δ cells. Cells were transformed with an integrating plasmid carrying a GFP-Atg8 construct under the control of the CUP1 promoter. Cells were collected and protein extracts analyzed by western blot with anti-YFP antibody and anti-Pgk1 (loading control) antiserum. The percentage of free GFP:total GFP is indicated. (C) The Cvt pathway as measured by the maturation of prApe1 is increased in gln3Δ and gat1Δ gln3Δ cells but decreased in gcn4Δ cells. Cells were grown in nutrient-rich medium until mid-log phase and then collected. Protein extracts were analyzed by western blot with anti-Ape1 antiserum and anti-Dpm1 (loading control) antibody. prApe1, precursor form; Ape1, mature form. The percentage of Ape1:total Ape1 is indicated. *, Nonspecific band.
To extend our analysis we used the GFP-Atg8 processing assay. During autophagy, Atg8 is lipidated by conjugation with PE and recruited to the phagophore, the precursor to the autophagosome. After the fusion of the autophagosome with the vacuole, the GFP-Atg8 chimera is hydrolyzed: Atg8 is rapidly degraded while the more stable GFP moiety will accumulate. Therefore, the accumulation of free GFP reflects the magnitude of autophagy cargo delivery.31 After 3 h of nitrogen starvation the mutants displayed a similar autophagy phenotype compared to that obtained from the Pho8Δ60 assay (Fig. 3B). The ratio of free GFP:total GFP was decreased compared to that of the wild-type cells in the gat1Δ and gcn4Δ strains, respectively. The deletion of GLN3 and the double GAT1 GLN3 deletion did not display a major difference compared to wild-type cells after 3 h, but showed higher GFP-Atg8 processing after 1 h of nitrogen starvation, supporting the hypothesis that higher expression of at least ATG8 in growing conditions in these strains increased autophagy activity in the short term after its induction.
We also analyzed the effects of transcriptional variation of ATG genes on the Cvt pathway, a selective type of autophagy used for the delivery of the resident hydrolase aminopeptidase I (Ape1) to the vacuole,32 by monitoring the processing of precursor (pr)Ape1. Higher expression of ATG8 and ATG29 in gln3Δ and gat1Δ gln3Δ cells led to an increase in the Cvt pathway as shown by the accumulation of mature Ape1 compared to wild-type cells (Fig. 3C). The GAT1 deletion was similar to wild type, whereas the GCN4 deletion resulted in a reduction of about 50% in the Cvt pathway. This latter result was unexpected given that gcn4Δ cells did not show any mRNA phenotype in growing conditions. We thus propose that this transcription factor might affect the expression of other genes (potentially other ATG genes) in these conditions that would account for the defects in prApe1 processing.
Analysis of transcriptional repressors of autophagy
Similar to the transcriptional activators, the phenotype of our potential repressors was tested by generating strains in a new background. Among them, 4 mutants, sfl1Δ, sko1Δ, yrm1Δ and zap1Δ now showed comparable ATG expression with the wild type in growing conditions indicating that the corresponding protein was unlikely to be a transcriptional repressor of autophagy or that its effect was strain dependent (Fig. S2C). In contrast, the deletion of FYV5 led to a significant upregulation of ATG1, ATG8, ATG9 and ATG14, whereas the deletion of SPT10 significantly increased the expression of ATG1, ATG8, ATG9, ATG29 and ATG32 in growing conditions (Fig. 4A). To test whether the effect on gene expression was reflected at the protein level we analyzed the amount of Atg8 and Atg9 proteins, which showed major differences in expression in these 2 strains compared to the wild type. Consistent with the mRNA results, the level of Atg8 and Atg9 was largely increased in spt10Δ cells (Fig. 4B); in contrast the deletion of FYV5 did not significantly affect the level of these 2 proteins. To explain these discrepancies we propose that Fyv5 may affect a multitude of cellular pathways including protein translation, which, if slowed down, may not reflect mRNA enrichment.
Figure 4.
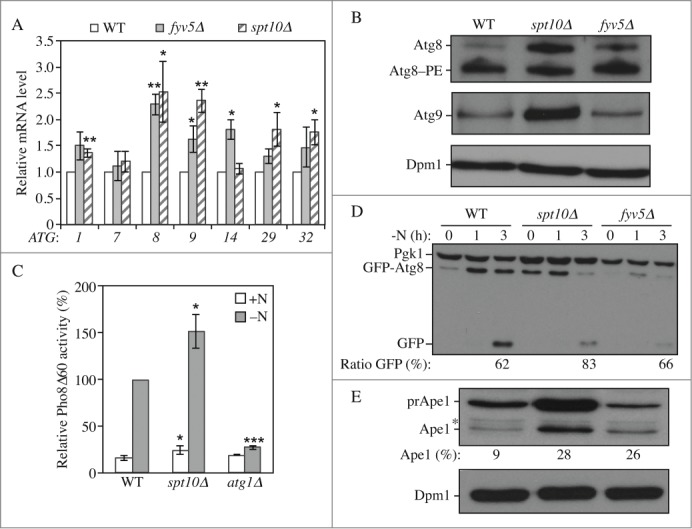
Spt10 and Fyv5 are transcriptional repressors of ATG gene expression. Wild type (WLY176), spt10Δ (YAB415), fyv5Δ (YAB414) and atg1Δ (WLY192) cells were grown in YPD (+N) until mid-log phase. (A) Spt10 and Fyv5 repress the expression of ATG genes in growing conditions. mRNA was extracted and quantified by RT-qPCR as in Figure 2. Data represent the average of at least 3 independent experiments. (B) Protein extracts were analyzed by western blot with anti-Atg8, anti-Atg9 and anti-Dpm1 (loading control) antisera and antibodies. (C-D) Autophagy is increased in spt10Δ cells. Cells were grown in YPD until mid-log phase (+N) and then starved for nitrogen (-N) for the indicated times. (C) The Pho8Δ60 activity was measured and normalized as in Figure 2 for cells that were starved for 3 h. Data represent the average of at least 3 independent experiments. (D) Cells were transformed with an integrating plasmid carrying a GFP-Atg8 construct under the control of the endogenous ATG8 promoter. Protein extracts were analyzed by western blot with anti-YFP antibody and anti-Pgk1 (loading control) antisera. The percentage of free GFP:total GFP is indicated. (E) The Cvt pathway as measured by the maturation of prApe1 is increased in spt10Δ and fyv5Δ cells. Proteins were extracted from cells grown in nutrient-rich conditions, and the extracts were analyzed by western blot with anti-Ape1 antiserum and anti-Dpm1 (loading control) antibody. prApe1, precursor form; Ape1, mature form. The percentage of Ape1:total Ape1 is indicated. *, Nonspecific band.
We next tested autophagy activity in these strains using the Pho8Δ60 assay. The deletion of SPT10 resulted in higher phosphatase activity in growing conditions as well as after starvation (Fig. 4C). Consistent with this result, autophagy measured by the GFP-Atg8 processing assay was induced to a higher extent in spt10Δ cells compared to the wild type (Fig. 4D). The deletion of FYV5 had a strong effect on the level of the Pho8Δ60 protein (Fig. S3A), which prevented the use of the corresponding assay. However, using the GFP-Atg8 processing assay we did not observe a major effect on autophagy activity in fyv5Δ cells (Fig. 4D). Finally, the analysis of prApe1 showed an increase of the Cvt pathway in spt10Δ and fyv5Δ cells compared to the wild type (Fig. 4E). It is worth noting that the spt10Δ strain displayed an increase in autophagy activity, despite the fact that there was an increase in ATG gene mRNA only in growing conditions; however, this phenotype is similar to that seen with strains such as rph1Δ18 and ume6Δ,24 which also affect transcription only in growing conditions. We think that the elevated amount of the corresponding Atg proteins provides a “jump start” in autophagy once the cells are placed in inducing conditions.
Characterization of Sfl1
We initially selected Sfl1 as a candidate because the deletion of the SFL1 gene showed higher ATG expression in the screen (Fig. S1A), although the generation of a new deletion strain showed that this was likely a false positive result (Fig. S2C). Nevertheless, analysis of a strain overexpressing the protein caught our attention: it showed higher expression of several ATG genes in growing conditions, and the expression of ATG8 was especially massively induced (Fig. 5A). In accordance with an increased level of mRNAs, the overexpression of Sfl1 resulted in an accumulation of Atg proteins as illustrated by the analysis of Atg1, Atg8 and Atg9 (Fig. 5B). As the steady state level of the Pho8Δ60 protein was severely reduced in cells overexpressing Sfl1 results using the Pho8Δ60 assay could be misleading (Fig. S3B). Instead autophagy activity was assayed using the GFP-Atg8 processing assay. The overexpression of Sfl1 led to a large increase in autophagy in growing conditions as well as shortly after starvation, but not after prolonged incubation (Fig. 5C). The observation that the autophagy activity of wild-type cells reaches that of the mutants with longer times of starvation is consistent with the fact that overexpressing Sfl1 increases the transcription of ATG genes only in nutrient-rich conditions; again, an overaccumulation of Atg proteins can support a jump-start in autophagy activity upon its induction by nitrogen starvation. After longer times of starvation, ATG levels in wild-type cells will reach that of the mutant strain, abolishing the difference in autophagy activity relative to the Sfl1 overexpressor.
Figure 5.
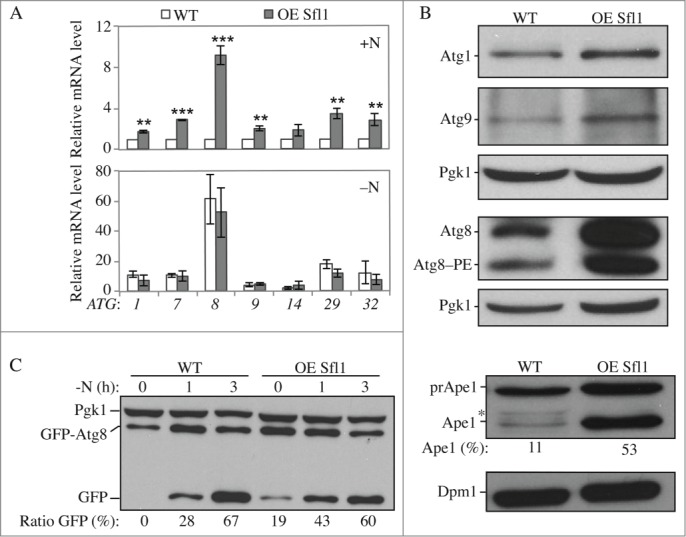
Sfl1 promotes ATG gene expression and autophagy. (A–B) The overexpression of Sfl1 induces the expression of ATG genes and proteins. Wild-type cells (WLY176; SEY6210) and cells with overexpressed (OE) Sfl1 (YAB377) were grown in YPD (+N) until mid-log phase. (A) Cells in growing conditions (+N, upper panel) and after 1 h of nitrogen starvation (-N, lower panel) were collected. mRNA levels were analyzed and quantified as in Figure 2. Data represent the average of at least 3 independent experiments. (B) Protein extracts were analyzed by western blot with anti-Atg1, anti-Atg9, anti-Atg8 and anti-Pgk1 (loading control) antisera. (C) The overexpression of Sfl1 promotes autophagy activity in growing conditions. Cells were transformed with an integrating plasmid carrying a GFP-Atg8 construct under the endogenous ATG8 promoter. Protein extracts were analyzed by western blot with anti-YFP antibody and anti-Pgk1 (loading control) antisera. The percentage of free GFP:total GFP is indicated. (D) The Cvt pathway as measured by the maturation of prApe1 is increased in cells overexpressing Sfl1. Proteins were extracted from cells grown in nutrient-rich conditions. Extracts were analyzed by western blot with anti-Ape1 antiserum and anti-Dpm1 (loading control) antibody. prApe1, precursor form; Ape1, mature form. The percentage of Ape1:total Ape1 is indicated. *, Nonspecific band.
Because overexpression of Sfl1 resulted in higher expression of ATG genes in growing conditions we hypothesized that the Cvt pathway might also be affected in these cells. Indeed, analysis of prApe1 indicated that overexpressing Sfl1 resulted in an overaccumulation of the mature form of Ape1 suggesting that the Cvt pathway was increased (Fig. 5D). Together our results suggest that Sfl1 acts as a transcriptional activator of autophagy and the Cvt pathway.
Discussion
In this study, a screen for DNA-binding proteins that modulate ATG gene expression identified several transcriptional regulators of autophagy. As a proof of concept of the screen we initially focused on the characterization of proteins for which deletion of the corresponding gene showed the highest variation of ATG gene expression. This criterion identified Gcn4, Sfl1, Gat1 and Gln3 as potential transcriptional activators of auto-phagy, and Spt10 and Fyv5 as putative repressors of ATG gene expression. It is worth noting that the transcriptional modulations we observed in the deletion strains might not result from a direct effect of the corresponding protein: the deletion of key regulators may indeed cause cellular stresses or modulate the level of an intermediate protein which would in turn affect the expression of ATG genes. Nonetheless, an analysis of the promoter regions of ATG genes identified one or multiple consensus DNA-binding sites in at least one of the ATG genes analyzed for Gat1, Gcn4, Gln3 and Sfl1 (Table S4) thus supporting a direct regulation by, at least, these transcriptional regulators. Furthermore, chromatin immunoprecipitation experiments revealed the direct binding of Gcn4 at the promoter of ATG1 (Fig. S4A and B).
Gcn4 was previously shown to modulate autophagy activity upon amino acid and nitrogen starvation conditions,33-34 although the mechanism by which this regulation is achieved was not clearly identified. Our results reveal here that Gcn4 is required for the full induction of ATG1 expression upon nitrogen starvation. In addition, in cells where the ATG1 promoter was changed for the Gcn4-insensitive PMP3 promoter, the deletion of GCN4 caused less of a reduction of autophagy activity than in the corresponding wild-type cells (35% instead of 50%; Fig. S4C and D). Together these data provide new insights into the function of Gcn4 showing that it regulates autophagy partly through its promotion of ATG1 expression after starvation, but also indicate that other GCN4-sensitive genes, not identified at this time, are involved. Gat1 and Gln3 are part of the GATA family of transcription factors, which are well-described activators of gene expression upon nutrient starvation conditions. Gln3 was previously suggested to affect the expression of ATG14;35 in our study we showed that this protein is required for the induction of several ATG genes upon nitrogen starvation, although we did not observe a significant effect on ATG14. We also report that the deletion of GLN3 increases the expression of ATG8 and ATG29 in growing conditions, suggesting that the Gln3 protein is directly or indirectly involved in the repression of some ATG genes. Previous studies showed that the mammalian GATA1 and GATA4 transcription factors are involved in the expression of several autophagy-related genes,20,23 suggesting that the function of the GATA family members in regulating autophagy is conserved from yeast to mammals.
Sfl1 is characterized as a dual transcriptional activator and repressor notably involved in the regulation of flocculation-related genes.36 In mammalian cells, HSF2, a Sfl1 homolog, was suggested to induce autophagy upon heat shock although the molecular mechanism underlying its regulation was not characterized.37 We show here that the overexpression of the Sfl1 protein greatly increases the expression of several ATG genes in yeast, especially ATG8, as well as autophagy activity in growing conditions. These results suggest that Sfl1 functions as a transcriptional activator of the autophagy pathway.
Our results also show that Spt10 and Fyv5 repress the expression of ATG genes in growing conditions and that Spt10 negatively regulates autophagy activity. Based on this finding we propose that Spt10 functions in the maintenance of autophagy at a basal level when environmental conditions are optimal, possibly through its putative histone acetylase activity.
To maintain cell homeostasis and prevent disease, rates of autophagy have to be finely tuned in response to multiple environmental conditions. Several signaling pathways regulating autophagy have been characterized and the activity of Atg proteins is controlled by multiple posttranslational modifications. In the last 10 y, an increasing number of proteins acting in the regulation of autophagy have been identified showing the high complexity of this signaling network. Previous work from our lab revealed the critical importance of transcriptional control of ATG genes for autophagy regulation, adding yet another layer of complexity to the overall regulation of the pathway.17-18,24 Results presented here provide new directions in the understanding of the transcriptional control of autophagy. It is worth noting that all of the autophagy transcriptional regulators analyzed in the present study, or that were previously characterized, affect the expression of ATG8. The level of Atg8 correlates with the size of autophagosomes and the magnitude of autophagic degradation;16 regulating the expression of ATG8 might therefore be pivotal for the modulation of autophagy activity. An in silico analysis of the consensus binding sites of ATG8 regulators indicates putative binding at multiple locations scattered throughout the ATG8 promoter, suggesting a high activity and complexity at this locus under stress conditions (Fig. S4E).
Besides the DNA-binding proteins analyzed in detail in this study, we report here on several proteins for which deletion of the corresponding gene affects ATG expression to a lesser extent. Although mild, these effects may be responsible for subtle modification of autophagy rates, which, combined with other signaling pathways, may account for large variations in autophagy activity. Furthermore, the multitude of transcriptional regulators of ATG expression certainly reflects the variety of environmental conditions upon which autophagy has to be modulated. Our study should therefore help in unraveling the multiple actors involved in autophagy regulation by identifying downstream effectors of signaling pathways. Finally, as most regulatory pathways are conserved from yeast to mammals, our study of the transcription of yeast autophagy genes may contribute valuable information that can be used for the therapeutic treatment of autophagy-related diseases.
Materials and Methods
Yeast strains, media and cell culture
Gene disruptions were performed using a standard method.38 Yeast cells were grown in YPD (1% yeast extract, 2% peptone, and 2% glucose [all wt/vol]) or synthetic minimal medium (SMD; 0.67% yeast nitrogen base [ForMedium, CYN0410], 2% glucose, supplemented with the appropriate amino acids and vitamins). Autophagy was induced in nitrogen starvation medium (SD-N; 0.17% yeast nitrogen base without amino acids [ForMedium, CYN0501], containing 2% glucose) or glucose starvation medium (0.67% yeast nitrogen base, 3% glycerol, supplemented with amino acids and vitamins). The yeast DNA-binding protein mutants analyzed during the screen came from a collection in the BY4742 background except as otherwise indicated. Other strains used in this study are listed in Table S5.
RNA and RT-qPCR
Total RNA was extracted using the NucleoSpin RNA kit (Clontech, 740955). DNase treatment was performed according to the kit instruction to eliminate genomic DNA contamination. RT-qPCR reactions were performed as previously described,18 using gene-specific primers listed in Table S6. The transcript abundance in samples was determined using a comparative threshold cycle method. The relative abundance of the reference mRNAs of TAF10, TFC1, UBC6 or SLD3 in each sample was determined and used to normalize for differences of total RNA amount according to the method described by Vandesompele et al.39
Statistical analyses
Statistical differences were assayed using one-sample t test and Student t test; *, p < 0 .05; **, p < 0 .01; ***, p < 0 .001.
Other Methods
Protein extraction, immunoblot, GFP-Atg8 processing, and Pho8Δ60-dependent phosphatase assays were performed as previously described.40-41,31 Antisera to Atg1,42 Atg9,43 Atg8,44 Pgk1 (a generous gift from Dr. Jeremy Thorner, University of California, Berkeley), Ape1,45 monoclonal Dpm1 (Life Technologies, 5C5A7) and monoclonal YFP (Clontech, JL-8) were used as previously described or according to the manufacturer's instructions.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This work was supported by NIH grant GM053396 to DJK and a Rackham predoctoral fellowship to M.J.
References
- 1.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol 2007; 9:1102-9; PMID:17909521; http://dx.doi.org/ 10.1038/ncb1007-1102 [DOI] [PubMed] [Google Scholar]
- 2.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 1993; 333:169-74; PMID:8224160; http://dx.doi.org/ 10.1016/0014-5793(93)80398-E [DOI] [PubMed] [Google Scholar]
- 3.Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell 2004; 6:463-77; PMID:15068787; http://dx.doi.org/ 10.1016/S1534-5807(04)00099-1 [DOI] [PubMed] [Google Scholar]
- 4.Deretic V, Levine B. Autophagy, immunity, and microbial adaptations. Cell Host Microbe 2009; 5:527-49; PMID:19527881; http://dx.doi.org/ 10.1016/j.chom.2009.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi CS, Shenderov K, Huang NN, Kabat J, Abu-Asab M, Fitzgerald KA, Sher A, Kehrl JH. Activation of autophagy by inflammatory signals limits IL-1beta production by targeting ubiquitinated inflammasomes for destruction. Nat Immunol 2012; 13:255-63; PMID:22286270; http://dx.doi.org/ 10.1038/ni.2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang J, Klionsky DJ. Autophagy and human disease. Cell Cycle 2007; 6:1837-49; PMID:17671424; http://dx.doi.org/ 10.4161/cc.6.15.4511 [DOI] [PubMed] [Google Scholar]
- 7.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature 2008; 451:1069-75; PMID:18305538; http://dx.doi.org/ 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lynch-Day MA, Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS Lett 2010; 584:1359-66; PMID:20146925; http://dx.doi.org/ 10.1016/j.febslet.2010.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nixon RA. The role of autophagy in neurodegenerative disease. Nat Med 2013; 19:983-97; PMID:23921753; http://dx.doi.org/ 10.1038/nm.3232 [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Azad MB, Gibson SB. Superoxide is the major reactive oxygen species regulating autophagy. Cell Death Differ 2009; 16:1040-52; PMID:19407826; http://dx.doi.org/ 10.1038/cdd.2009.49 [DOI] [PubMed] [Google Scholar]
- 11.Levine B, Yuan J. Autophagy in cell death: an innocent convict? J Clin Invest 2005; 155:2679-88; PMID:NOT_FOUND; http://dx.doi.org/ 10.1172/JCI26390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ravikumar B, Sarkar S, Davies JE, Futter M, Garcia-Arencibia M, Green-Thompson ZW, Jimenez-Sanchez M, Korolchuk VI, Lichtenberg M, Luo S, et al.. Regulation of mammalian autophagy in physiology and pathophysiology. Physiol Rev 2010; 90:1383-435; PMID:20959619; http://dx.doi.org/ 10.1152/physrev.00030.2009 [DOI] [PubMed] [Google Scholar]
- 13.Jin M, Klionsky DJ. Regulation of autophagy: Modulation of the size and number of autophagosomes. FEBS Lett 2014; 588:2457-63; PMID:24928445; http://dx.doi.org/ 10.1016/j.febslet.2014.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng Y, Yao Z, Klionsky DJ. How to control self-digestion: transcriptional, post-transcriptional, and post-translational regulation of autophagy. Trends Cell Biol 2015; 25:354-63; PMID:25759175; http://dx.doi.org/ 10.1016/j.tcb.2015.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie Y, Kang R, Sun X, Zhong M, Huang J, Klionsky DJ, Tang D. Posttranslational modification of autophagy-related proteins in macroautophagy. Autophagy 2015; 11:28-45; PMID:25484070; http://dx.doi.org/ 10.4161/15548627.2014.984267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell 2008; 19:3290-8; PMID:18508918; http://dx.doi.org/ 10.1091/mbc.E07-12-1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin M, He D, Backues SK, Freeberg MA, Liu X, Kim JK, Klionsky DJ. Transcriptional regulation by pho23 modulates the frequency of autophagosome formation. Curr Biol 2014; 24:1314-22; PMID:24881874; http://dx.doi.org/ 10.1016/j.cub.2014.04.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernard A, Jin M, González-Rodríguez P, Füllgrabe J, Delorme-Axford E, Backues SK, Joseph B, Klionsky DJ. Rph1/KDM4 mediates nutrient-limitation signaling that leads to the transcriptional induction of autophagy. Curr Biol 2015; 25:546-55; PMID:25660547; http://dx.doi.org/ 10.1016/j.cub.2014.12.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Webb AE, Brunet A. FOXO transcription factors: key regulators of cellular quality control. Trends Biochem Sci. 2015; 39:159-69; http://dx.doi.org/ 10.1016/j.tibs.2014.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang YA, Sanalkumar R, O'Geen H, Linnemann AK, Chang CJ, Bouhassira EE, Farnham PJ, Keles S, Bresnick EH. Autophagy driven by a master regulator of hematopoiesis. Mol Cell Biol 2012; 32:226-39; PMID:22025678; http://dx.doi.org/ 10.1128/MCB.06166-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al.. TFEB links autophagy to lysosomal biogenesis. Science 2011; 332:1429-33; PMID:21617040; http://dx.doi.org/ 10.1126/science.1204592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chauhan S, Goodwin JG, Manyam G, Wang J, Kamat AM, Boyd DD. ZKSCAN3 is a master transcriptional repressor of autophagy. Mol Cell 2013; 50:16-28; PMID:23434374; http://dx.doi.org/ 10.1016/j.molcel.2013.01.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kobayashi S, Volden P, Timm D, Mao K, Xu X, Liang Q. Transcription factor GATA4 inhibits doxorubicin-induced autophagy and cardiomyocyte death. J Biol Chem 2009; 285:793-804; PMID:19901028; http://dx.doi.org/ 10.1074/jbc.M109.070037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartholomew CR, Suzuki T, Du Z, Backues SK, Jin M, Lynch-Day MA, Umekawa M, Kamath A, Zhao M, Xie Z, et al.. Ume6 transcription factor is part of a signaling cascade that regulates autophagy. Proc Natl Acad Sci U S A 2012; 109:11206-10; PMID:22733735; http://dx.doi.org/ 10.1073/pnas.1200313109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell 2009; 17:98-109; PMID:19619495; http://dx.doi.org/ 10.1016/j.devcel.2009.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noda T, Klionsky DJ. The quantitative Pho8Delta60 assay of nonspecific autophagy. Methods Enzymol 2008; 451:33-42; PMID:19185711; http://dx.doi.org/ 10.1016/S0076-6879(08)03203-5 [DOI] [PubMed] [Google Scholar]
- 27.Hofman-Bang J. Nitrogen catabolite repression in Saccharomyces cerevisiae. Mol Biotechnol 1999; 12:35-73; PMID:10554772; http://dx.doi.org/ 10.1385/MB:12:1:35 [DOI] [PubMed] [Google Scholar]
- 28.Magasanik B, Kaiser CA. Nitrogen regulation in Saccharomyces cerevisiae. Gene 2002; 290:1-18; PMID:12062797; http://dx.doi.org/ 10.1016/S0378-1119(02)00558-9 [DOI] [PubMed] [Google Scholar]
- 29.Broach JR. Nutritional control of growth and development in yeast. Genetics 2012; 192:73-105; PMID:22964838; http://dx.doi.org/ 10.1534/genetics.111.135731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Conrad M, Schothorst J, Kankipati HN, Van Zeebroeck G, Rubio-Texeira M, Thevelein JM. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev 2014; 38:254-99; PMID:24483210; http://dx.doi.org/ 10.1111/1574-6976.12065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem 2004; 279:29889-94; PMID:15138258; http://dx.doi.org/ 10.1074/jbc.M404399200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science 2000; 290:1717-21; PMID:11099404; http://dx.doi.org/ 10.1126/science.290.5497.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ecker N, Mor A, Journo D, Abeliovich H. Induction of autophagy flux by amino acid deprivation is distinct from nitrogen starvation-induced macroautophagy. Autophagy 2010; 6:879-90; PMID:20647741; http://dx.doi.org/ 10.4161/auto.6.7.12753 [DOI] [PubMed] [Google Scholar]
- 34.Yang Z, Geng J, Yen WL, Wang K, Klionsky DJ. Positive or negative roles of different cyclin-dependent kinase Pho85-cyclin complexes orchestrate induction of autophagy in Saccharomyces cerevisiae. Mol Cell 2010; 23:250-64; PMID:NOT_FOUND; http://dx.doi.org/ 10.1016/j.molcel.2010.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan TF, Bertram PG, Ai W, Zheng XF. Regulation of APG14 expression by the GATA-type transcription factor Gln3p. J Biol Chem 2001; 276:6463-7; PMID:11096087; http://dx.doi.org/ 10.1074/jbc.M008162200 [DOI] [PubMed] [Google Scholar]
- 36.Robertson LS, Fink GR. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc Natl Acad Sci U S A 1998; 95:13783-7; PMID:9811878; http://dx.doi.org/ 10.1073/pnas.95.23.13783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prasad KV, Taiyab A, Jyothi D, Srinivas UK, Sreedhar AS. Heat shock transcription factors regulate heat induced cell death in a rat histiocytoma. J Biosci 2007; 32:585-93; PMID:17536178; http://dx.doi.org/ 10.1007/s12038-007-0058-4 [DOI] [PubMed] [Google Scholar]
- 38.Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998; 14:953-61; PMID:9717241; http://dx.doi.org/ [DOI] [PubMed] [Google Scholar]
- 39.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002; 3:H0034; http://dx.doi.org/ 10.1186/gb-2002-3-7-research0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yorimitsu T, Zaman S, Broach JR, Klionsky DJ. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol Biol Cell 2007; 18:4180-9; PMID:17699586; http://dx.doi.org/ 10.1091/mbc.E07-05-0485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noda T, Matsuura A, Wada Y, Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun 1995; 210:126-32; PMID:7741731; http://dx.doi.org/ 10.1006/bbrc.1995.1636 [DOI] [PubMed] [Google Scholar]
- 42.Abeliovich H, Zhang C, Dunn WA Jr, Shokat KM, Klionsky DJ. Chemical genetic analysis of Apg1 reveals a non-kinase role in the induction of autophagy. Mol Biol Cell 2003; 14:477-90; PMID:12589048; http://dx.doi.org/ 10.1091/mbc.E02-07-0413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noda T, Kim J, Huang WP, Baba M, Tokunaga C, Ohsumi Y, Klionsky DJ. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol 2000; 148:465-80; PMID:10662773; http://dx.doi.org/ 10.1083/jcb.148.3.465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang WP, Scott SV, Kim J, Klionsky DJ. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J Biol Chem 2000; 275:5845-51; PMID:10681575; http://dx.doi.org/ 10.1074/jbc.275.8.5845 [DOI] [PubMed] [Google Scholar]
- 45.Klionsky DJ, Cueva R, Yaver DS. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol 1992; 119:287-99; PMID:1400574; http://dx.doi.org/ 10.1083/jcb.119.2.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


