Abstract
Mitochondria selective autophagy, known as mitophagy, plays a pivotal role in several biological processes, such as elimination of the damaged mitochondria, removal of the mitochondria from immature red blood cells and sperm. The defects in mitophagy are associated with a wide spectrum of human diseases, including neurodegenerative disease, aging, cardiac disease and autoimmune disease. However, the mechanism underlying mitophagy remains largely unclear. Here, we report the characterization of a novel splice variant of BECN1/Beclin 1, BECN1s, which is produced by an alternative splicing mechanism. BECN1s is primarily associated with the outer-membrane of mitochondria. Unlike unspliced BECN1, which is essential for nonselective macroautophagy induction, BECN1s is indispensible for mitochondria-selective autophagy. Furthermore, BECN1s plays an important role in starvation- and membrane depolarization-induced mitophagy. Taken together, our findings broaden the view of BECN1 as an important regulator in autophagy, and implicate BECN1s as a specific mitophagy mediator.
Keywords: BECN1, BECN1s, mitochondrial depolarization, mitophagy, starvation
Abbreviations
- Atg
autophagy-related
- bp
base pairs
- BECN1s
BECN1 short isoform
- CCCP
carbonyl cyanide m-chlorophenylhydrazone
- EBSS
Earle's balanced salt solution
- ECD
evolutionarily conserved domain
- GFP
green fluorescent protein
- LAMP1
lysosomal-associated membrane protein 1
- MAP1LC3/LC3
microtubule-associated protein 1 light chain 3
- PtdIns3K
class III phosphatidylinositol 3-kinase
Introduction
Mitochondrial dysfunction has severe cellular consequences and has been linked to many human diseases including neurodegenerative disease, aging, cardiac and autoimmune diseases.1-4 Thus, maintaining a healthy population of mitochondria is essential for cellular homeostasis. Selective autophagy of mitochondria, known as mitophagy, which mediates the selective removal of damaged mitochondria, plays an important role in mitochondrial quality control. Mitophagy also mediates clearance of mitochondria in developing erythrocytes5-7 and sperm-derived mitochondria as well.8
Mitophagy can be induced under many conditions including starvation,9 photodamage,9 hypoxia,10,11 ROS production,12,13 and mitochondrial uncoupler-induced mitochondrial depolarization.14,15 To date, many specific regulators of mitophagy have been identified. In yeast, the autophagy-related (Atg) protein Atg32 is required for mitochondrial autophagy. While in mammalian species, BNIP3L, a BH3-only member of the BCL2 family, is indispensable for autophagic removal of mitochondria during reticulocyte maturation.16-20 In addition, the PINK1-PARK2 pathway is responsible for elimination of damaged mitochondria in mammals.14,15,21-23 Once mitochondrial damages occur, PINK1 accumulates and recruits PARK2/PARKIN specifically to the damaged mitochondria, thereafter initiating mitophagy.
BECN1/Beclin 1, the first mammalian autophagy protein to be described, plays a central role in autophagy.24 BECN1 performs this function as part of a core complex that contains PIK3C3/VPS34 (the catalytic subunit of the class III phosphatidylinositol 3-kinase [PtdIns3K]) and PIK3R4/VPS15. BECN1 has a BCL2 binding domain (BBD), a central coiled-coil domain (CCD), and an evolutionarily conserved domain (ECD).25 Recent structural dada indicate that BECN1 has a β-α repeated, autophagy-specific (BARA) domain at the C terminus, and the ECD is a part of this domain.26 Although noncanonical BECN1-independent autophagy and mitophagy contribute to MPP+ toxicity,27 BECN1 is thought to be an essential protein for autophagy.28 Many, if not all of gene products comprise either their transcriptional variants or structurally related isoforms, for BECN1, its isoforms have not yet been reported. In this study, we identify a novel splice variant of BECN1 that we designated as BECN1s (for short isoform), which is a specific regulator of mitophagy. BECN1s, which lacks exon 10 and exon 11 of unspliced BECN1, is mainly associated with the outer-membrane of mitochondria. Although it retains the ability to activate PIK3C3, BECN1s is not essential for induction of nonselective macroautophagy. Intriguingly, BECN1s is indispensible for starvation- and mitochondrial depolarization-induced mitophagy.
Results
BECN1s is a splice variant of BECN1
When we amplified the BECN1 cDNA from HeLa cells using primers P1 and P2 that were complementary to exon 8 and exon 12 of the BECN1 gene, respectively, an additional 324 base pairs (bp) PCR product was obtained in addition to the predicted 546 bp band encoding the C terminus of BECN1 (Fig. 1A). This 324 bp PCR product was also observed in other cell lines such as U2OS, HCT116, H1299, MCF7 and SH-SY5Y (Fig. S1A). The subsequent DNA sequencing analysis showed that the lower molecular weight band was identical to BECN1 cDNA except that the entire exons 10 and 11 were missing (Fig. 1B). We named this short isoform as BECN1s. The ORF of BECN1s encoded a protein of 396 amino acids, which lacks the partial evolutionarily conserved domain (ECD) and C-terminal domains as compared with BECN1 (Fig. 1B). Careful inspection of the BECN1s coding sequence revealed that GT and AG were indeed utilized as the donor and acceptor spice sites, respectively (Fig. 1B). These findings indicate that BECN1s is a novel splice variant of BECN1.
Figure 1.
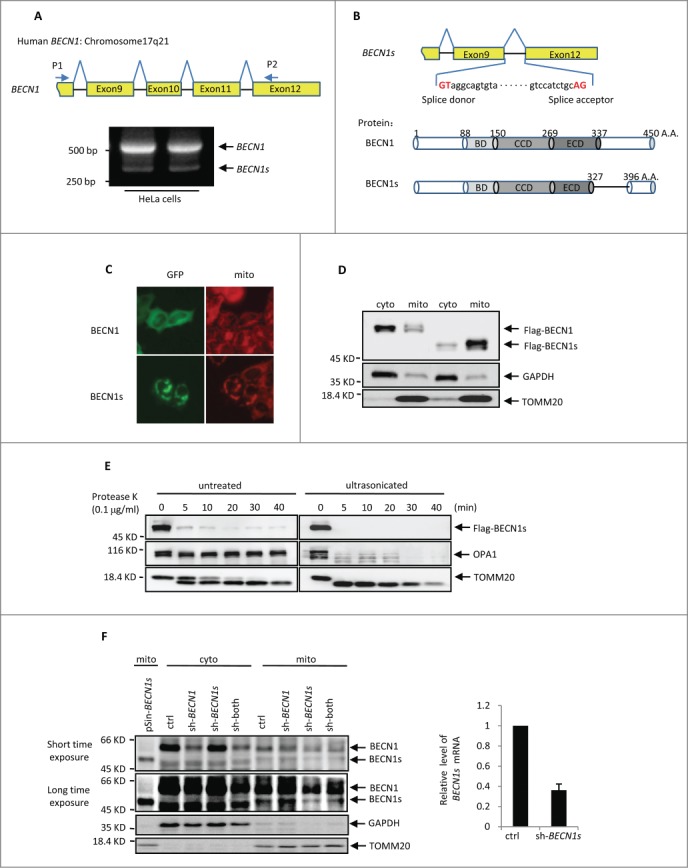
BECN1s is a splice variant of BECN1 (A) RT-PCR was performed with total RNAs extracted from HeLa cells using primer P1 and P2. The locations of P1 and P2 on BECN1 cDNA are indicated. (B) A schematic illustration of cDNA and protein sequences of the BECN1 and BECN1s. BD, BCL2 binding domain; CCD, coiled-coil domain; ECD, evolutionarily conserved domain. (C) HeLa cells expressing either GFP-BECN1 or GFP-BECN1s were stained with MitoTracker Red. The images were taken under a fluorescence microscope. (D) HEK 293T cells were transfected with either Flag-BECN1 or Flag-BECN1s. Twenty-four h after transfection, cells were subjected to cytoplasm/mitochondria subcellular fractionation. Proteins from cytoplasmic and mitochondrial fractions were analyzed by western blot. GAPDH and TOMM20 were used as markers for cytoplasmic and mitochondrial fractions, respectively. (E) HEK 293T cells were transfected with Flag-BECN1s. Twenty-four h after transfection, cells were homogenized for mitochondria isolation. The isolated mitochondria were then treated with proteinase K for the indicated periods of time before or after supersonic treatment, followed by western blot analysis to detect Flag-BECN1s levels. (F) HeLa cells expressing the indicated shRNAs and BECN1s proteins were subjected to cytosolic and mitochondrial subcellular fractionation. Sh-both indicates the shRNA targeting both BECN1 and BECN1s. Proteins from cytoplasmic and mitochondrial fractions were analyzed by western blot using anti-BECN1 antibody. GAPDH and TOMM20 were used as markers for cytoplasmic and mitochondrial fractions, respectively. The BECN1s knockdown efficiency was also shown.
We next examined the cellular localization of BECN1s. The immunofluorescence and the cytosolic and mitochondrial subcellular fractionation analyses revealed that unlike BECN1, which was evenly distributed in the cytoplasm, BECN1s predominantly colocalized with mitochondria (Fig. 1C-D). Similar results were found in U2OS and MEF cells (Fig. S1B and S1C). To further determine whether BECN1s is a membrane protein in mitochondria, isolated mitochondria were incubated with proteinase K before or after ultrasonication. As was expected, proteinase K did not affect the inner-membrane protein OPA1 when mitochondria were intact, but it destroyed OPA1 after mitochondria disruption by ultrasonication (Fig. 1E). Similar to the outer-membrane protein TOMM20, BECN1s was quickly degraded by proteinase K regardless of the mitochondrial integrity (Fig. 1E), suggesting that BECN1s is associated with the outer membrane of mitochondria.
To confirm further the authenticity of BECN1s, we designed 2 shRNAs. BECN1s shRNA, targeting the BECN1 exon9-exon12 junction, was able to specifically knock down BECN1s but not BECN1. In contrast, BECN1 shRNA, targeting BECN1 exon 10, can only knock down BECN1. The cytosolic and mitochondrial fractions of HeLa cells expressing either BECN1 shRNA or BECN1s shRNA were then analyzed by western blot with anti-BECN1 antibody recognizing the N terminus of BECN1. A specific 50-kDa band was detected in the mitochondrial fraction of the control cells, and the size of this band was comparable to that of untagged BECN1s (Fig. 1F). The intensity of this 50-kDa band was decreased by BECN1s shRNA but not by BECN1 shRNA. Also, introduction of the shRNA targeting both BECN1 and BECN1s was able to decrease the intensities of both this 50-kDa and upper BECN1 bands (Fig. 1F). Taken together, our data strongly suggest the cellular existence of BECN1s.
BECN1s is not essential for the initiation of macroauto- phagy
It has been well recognized that BECN1 plays a central role in the initiation of autophagy through interacting with and regulating class III PtdIns3K activity. The evolutionarily conserved domain of BECN1 is required for PIK3C3 binding.25 Since BECN1s lacks a partial ECD, we asked whether BECN1s could still bind to PIK3C3 and regulate its activity. By performing an immunoprecipitation assay, we showed that both BECN1 and BECN1s were able to interact with PIK3C3 (Fig. 2A), indicating that loss of the partial ECD in BECN1s does not affect its binding ability to PIK3C3. We next sought to investigate whether BECN1s could induce PIK3C3 activity like BECN1 does. We employed GFP-tagged double FYVE finger of HGS/Hrs (hepatocyte growth factor-regulated tyrosine kinase substrate) to detect the lipid phosphorylation activity of PIK3C3. Because the FYVE probe specifically binds to phosphatidylinositol 3-phosphate,29 the product of activated PIK3C3, and forms the GFP-FYVE puncta, we could measure PIK3C3 activity by detecting FYVE fluorescence. The results showed that both BECN1 and BECN1s were able to stimulate GFP-FYVE puncta formation in BECN1 knockdown HeLa cells (Fig. 2B). These combined data indicate that BECN1s still retains the ability to interact with PIK3C3 and regulate its activity.
Figure 2.
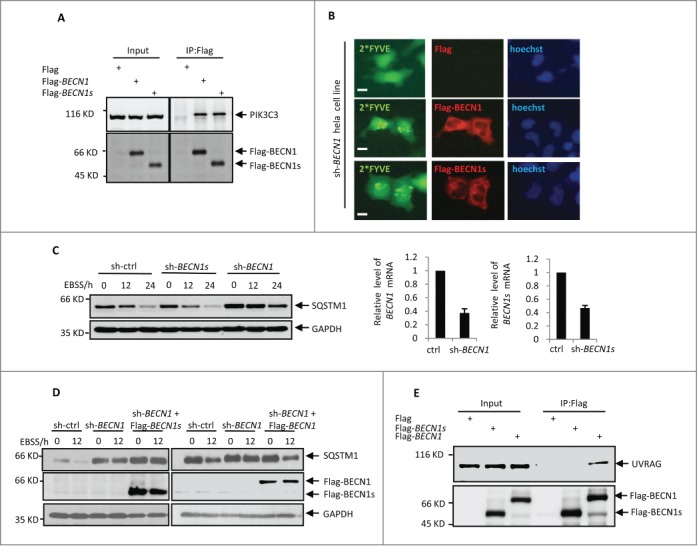
BECN1s is not involved in macroautophagy initiation. (A) HEK 293T cells were transfected with control vector, Flag-BECN1, or Flag-BECN1s as indicated. Twenty-four h later, cell lysates were immunoprecipitated with anti-Flag antibody, followed by western blot analysis with anti-PIK3C3 antibody. (B) HeLa cells with BECN1 stable knockdown were transfected with GFP-2*FYVE plus either control vector, Flag-BECN1, or Flag-BECN1s as indicated. Twenty-four h after transfection, cells were immunostained with anti-Flag antibody. The images were taken under a fluorescence microscope. Scale bar: 20 μm. (C) HeLa cells expressing control shRNA, BECN1 shRNA, or BECN1s shRNA were treated with EBSS for the indicated periods of time. Cell lysates were then analyzed by western blot with anti-SQSTM1 and anti-GAPDH antibodies. The shRNA-mediated knockdown efficiency for BECN1 and BECN1s was also shown. (D) HeLa cells expressing the indicated shRNAs and proteins were treated with EBSS for the indicated periods of time. Cell lysates were then analyzed by western blot with anti-SQSTM1, anti-Flag and anti-GAPDH antibodies. (E) HEK 293T cells were transfected with either control vector, Flag-BECN1 or Flag-BECN1s as indicated. Twenty-four h after transfection, cell lysates were immunoprecipitated with anti-Flag antibody, followed by western blot analysis with anti-UVRAG and anti-Flag antibodies.
To assess whether BECN1s is involved in starvation-induced autophagy, HeLa cells expressing either BECN1 shRNA or BECN1s shRNA were treated with or without EBSS. Consistent with the previous reports,30 BECN1 knockdown markedly impaired the EBSS-induced reduction of the autophagy marker protein SQSTM1/p62 levels (Fig. 2C), reinforcing the critical role of BECN1 in the regulation of autophagy. However, the EBSS-initiated autophagy was not inhibited by BECN1s knockdown (Fig. 2C). In addition, unlike BECN1, BECN1s failed to restore autophagy under EBSS treatment condition when endogenous BECN1 was knocked down (Fig. 2D). These data suggest that BECN1s is not essential for starvation-initiated macroautophagy.
Many proteins participate in BECN1-mediated autophagy through interacting with BECN1-PtdIns3K complex. Among these proteins, UVRAG is an essential component of the BECN1-PtdIns3K lipid kinase complex and is an important signaling checkpoint for autophagy.31,32 We therefore evaluated the binding ability of BECN1s to UVRAG. Correlated with its function, which cannot induce nonselective autophagy, BECN1s did not interact with UVRAG (Fig. 2E). We also checked whether BECN1s could interact with other BECN1 regulator proteins such as ATG14 and BCL2. The results showed that similar to BECN1, BECN1s was also able to associate with ATG14 and BCL2 (Fig. S2). These results suggest that loss function of BECN1s in mediating starvation-induced autophagy may be due to the compromised ability of BECN1s to interact with UVRAG.
BECN1s induces mitochondrial autophagy
Since BECN1s was mainly associated with mitochondria, we asked whether BECN1s regulates selective mitochondrial autophagy (mitophagy). The constructs encoding either BECN1 or BECN1s were transfected into HeLa cells. With the increasing expression of BECN1s but not BECN1, levels of the mitochondrial protein TOMM20 were concurrently decreased (Fig. 3A). In addition, ectopic expression of BECN1s also led to the decreased expression of other mitochondrial proteins, such as CYCS (cytochrome c, somatic) and VDAC1 (Fig. 3B). To verify whether mitophagy plays a key role in BECN1s-mediated reduced expression of the mitochondria proteins, an inhibitor of autophagic vacuole maturation bafilomycin A1 (BAF) was utilized. When autophagy was blocked by BAF, BECN1s-mediated decreased expression of the mitochondrial protein TIMM23 was greatly recovered (Fig. 3C), indicating that BECN1s induces mitophagy. In supporting of this, the immunofluorescence analysis showed that BECN1s was well colocalized with mitochondrial protein TOMM20, lysosomal-associated protein LAMP1 and autophagy-associated protein SQSTM1 (Fig. 3D). Like other well-known mitophagy-induced proteins such as BNIP3L and FUNDC1, BECN1s was able to reduce levels of the TOMM20 protein but not levels of endoplasmic reticulum protein GOLGA2. Again, BECN1s-induced reduction of TOMM20 levels can be reversed by BAF (Fig. 3E). To further prove that BECN1s induces mitophagy, we performed an electron microscopy assay. In this assay, carbonyl cyanide m-chlorophenylhydrazone (CCCP) was used as a positive control. CCCP is a mitochondrial uncoupler, which can trigger mitophagy by severely reducing the mitochondrial membrane potential.33 As was expected, treatment with CCCP dramatically increased the formation of the mitochondria-containing autophagic vesicles in HeLa cells (Fig. 3F). Intriguingly, ectopic expression of BECN1s also enhanced the mitochondria-containing autophagic vesicle formation (Fig. 3F). Taken together, these findings demonstrate that BECN1s specifically induces mitophagy.
Figure 3 (See previous page).
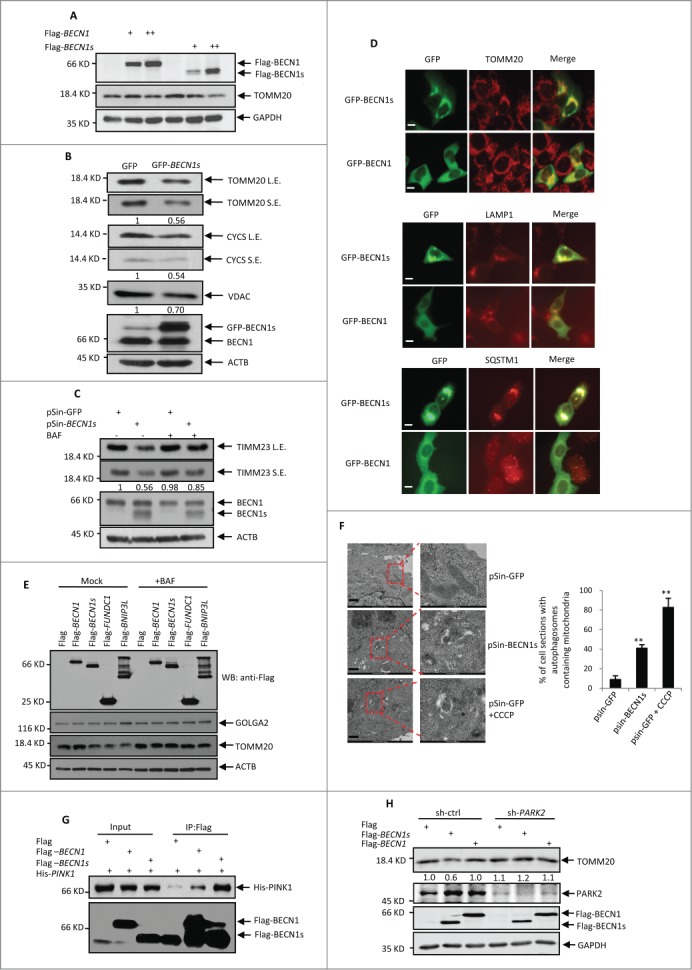
BECN1s induces mitophagy. (A) HeLa cells were transfected with the increasing amounts of constructs encoding either Flag-BECN1 or Flag-BECN1s. Twenty-four h after transfection, cell lysates were analyzed by western blot with anti-Flag, anti-TOMM20 and anti-GAPDH antibodies. (B) HeLa cells were transfected with either GFP control vector or GFP-BECN1. Twenty-four h after transfection, cell lysates were analyzed by western blot with the indicated antibodies. The blots were also qualified by using Gel-Pro analyzer software (Rockville, MD, USA). The value of each band indicates the relative expression levels of the indicated protein after normalizing to the loading control ACTIN. L.E. and SE indicate long time exposure and short time exposure, respectively. (C) HeLa cells were infected with lentiviruses expressing GFP or BECN1s. Forty-eight h after infection, cells were treated with or without BAF for another 12 h. Cell lysates were then analyzed by western blot with anti-TIMM23, anti-BECN1 and anti-ACTIN antibodies. The blot was also qualified by using Gel-Pro analyzer software (Rockville, MD, USA). The value of each band indicates the relative expression levels of TIMM23 after normalizing to the loading control ACTIN. L.E. and SE indicate long time exposure and short time exposure, respectively. (D) HeLa cells were transfected with either GFP control vector or GFP-BECN1s. Twenty-four h after transfection, cells were immunostained with anti-TOMM20, anti-LAMP1 and anti-SQSTM1 antibodies, respectively. Scale bar: 20 μm. (E) HeLa cells were transfected with the constructs encoding Flag-BECN1, Flag-BECN1s, Flag-BNIP3L and Flag-FUNDC1 as indicated. Twenty-four h after transfection, cells were treated with or without BAF for another 12 h. Cell lysates were analyzed by western blot with the indicated antibodies. (F) Cells expressing GFP or BECN1s were treated with or without Carbonyl cyanide m-chlorophenylhydrazone (CCCP) for 12 h. Cells were then subjected to electron microscopy analysis. The shown images are representatives of 3 independent experiments. Scale bar: 1 μm. The percentage of cells with autophagosomes containing mitochondria is also shown as mean±SD from 3 independent experiments. n>28 cells per experiments. **, P<0.01. (G) HEK 293T Cells were transfected with Flag-BECN1, Flag-BECN1s and His-PINK1 in the indicated combinations. Twenty-four h after transfection, cell lysates were immunoprecipitated with anti-Flag antibody, followed by western blot analysis with anti-His antibody. (H) SH-SY5Y cells expressing control shRNA or PARK2 shRNA were transfected with Flag-BECN1 or Flag-BECN1s. Forty-eight h after transfection, cell lysates were analyzed by western blot with anti-TOMM20 antibody. The blot was also qualified by using Gel-Pro analyzer software (Rockville, MD, USA). The value of each band indicates the relative expression levels of TOMM20 after normalizing to the loading control GAPDH.
It has been well recognized that the PINK1-PARK2 pathway is important for mitophagy initiation. We therefore sough to explore whether BECN1s induces mitophagy through the PINK1-PARK2 pathway. The immunoprecipitation analysis revealed that BECN1s is able to strongly interact with PINK1 (Fig. 3G), but not other mitophagy related proteins HSP90, FUNDC1 and BNIP3L (Fig. S3), indicating that BECN1s may work in concert with PINK1 to regulate mitophagy. We also showed PARK2 knockdown was able to reverse the inhibitory effect of BECN1s on TOMM20 protein levels (Fig. 3H), indicating that BECN1s-induced mitophagy is PARK2-dependent.
BECN1s plays an important role in EBSS- and CCCP-induced mitophagy
To determine the physiological significance of BECN1s-induced mitophagy, we first investigated whether BECN1s is essential for starvation-initiated mitophagy. HeLa cells expressing control shRNA, BECN1 shRNA or BECN1s shRNA were treated with EBSS, followed by western blot analysis to detect the mitochondria protein levels. In control cells, EBSS treatment dramatically decreased levels of the mitochondrial proteins TOMM20, cytochrome c and MFN1 (Fig. 4A). Knockdown of BECN1 failed to repress this decrease of mitochondrial proteins expression (Fig. 4A and S4A), indicating that BECN1 is dispensable for starvation-induced mitophagy. Nevertheless, knockdown of BECN1s greatly reversed EBSS-decreased mitochondrial proteins expression (Fig. 4A and S4A). Similar results were also obtained in U2OS cells (Fig. S4B). In line with these findings, treatment with BECN1s shRNA but not BECN1 shRNA diminished the autophagic vesicles containing mitochondria in HCT116 cells (Fig. 4B). In addition, treatment with BAF reversed EBSS-reduced TOMM20 expression in control cells but not in BECN1s knockdown cells (Fig. 4C and S4C). Also, knockdown of BECN1s did not regulate the effect of EBSS on SQSTM1 and LC3 protein levels (Fig. 4C, and S4C and 4D). These data suggest that BECN1s is essential for EBSS-induced mitophagy.
Figure 4 (See previous page).
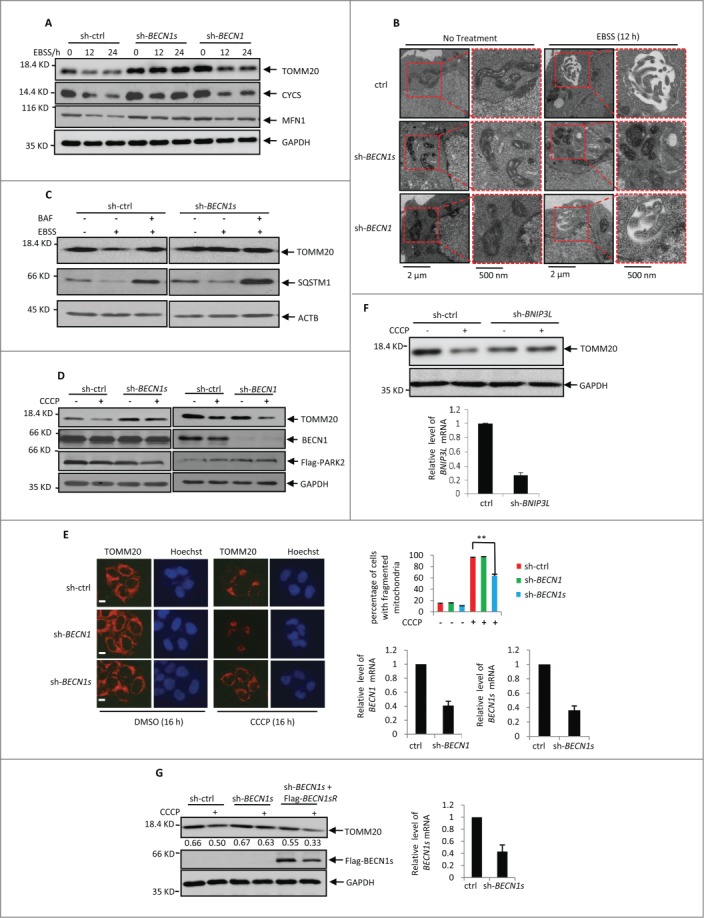
EBSS- and CCCP-induced mitophagy is BECN1s-dependent. (A) HeLa cells expressing control shRNA, BECN1 shRNA or BECN1s shRNA were treated with EBSS for the indicated periods of time. Cell lysates were analyzed by western blot with the indicated antibodies. The shRNA-mediated knockdown efficiency for BECN1 and BECN1s was shown in Fig. S4A. (B) HCT116 cells expressing the indicated shRNAs were incubated under normal or starvation conditions for 12 h, followed by electron microscopy analysis. (C) HCT116 cells transfected with indicated shRNA were cultured in normal growth medium or treated with EBSS in the absence or presence of BAF. Cell lysates were analyzed by western blot with indicated antibodies. The shRNA-mediated knockdown efficiency for BECN1s was shown in Fig. S4C. (D) HCT116 cells were transfected with Flag-PARK2 plus the indicated shRNAs. Twenty-four h after transfection, cells were treated with 10 μM CCCP for another 16 h. Cell lysates were analyzed by western blot with anti-TOMM20, anti-BECN1 and anti-GAPDH antibodies. The shRNA-mediated knockdown efficiency for BECN1 and BECN1s was shown in Fig. S4E. (E) HeLa cells expressing the indicated shRNAs were treated with 10 μM CCCP for 16 h. Cells were then immunostained with anti-TOMM20 antibody. Scale bar: 20 μm. The percentage of cells with fragmented mitochondria was calculated and shown accordingly. **, P<0.01. The shRNA-mediated knockdown efficiency for BECN1 and BECN1s was also shown. (F) HCT116 cells expressing either control shRNA or BNIP3L shRNA were treated with or without 10 μM CCCP for 16 h. Cell lysates were then analyzed by western blot with anti-TOMM20 antibody. The efficient knockdown of BNIP3L was also confirmed by real-time RT-PCR analysis. (G) Lysates from HCT116 cells expressing the indicated shRNAs and proteins were treated with 10 μM CCCP for 16 h. Cell lysates were analyzed by western blot with antibodies against TOMM20, Flag and GAPDH. The blot was quantified using Gel-Pro analyzer software (Rockville, MD, USA). The value of each band indicates the relative expression levels of TOMM20 after normalizing to the loading control GAPDH. The shRNA-mediated knockdown efficiency for BECN1s was also shown.
We next evaluated whether BECN1s plays a pivotal role in mitochondrial depolarization-induced mitopahgy. HCT116 cells expressing control shRNA, BECN1 shRNA or BECN1s shRNA were treated with CCCP, followed by western blot and immunofluorescence analyses to examine TOMM20 expression. The results showed that BECN1s knockdown but not BECN1 knockdown recovered CCCP-decreased expression of TOMM20 (Fig. 4D to E and S4E). As a positive control, BNIP3L knockdown also dramatically inhibited CCCP-induced mitophagy (Fig. 4F). More importantly, ectopic expression of BECN1s was able to restore the ability of CCCP to reduce TOMM20 levels in BECN1s knockdown HCT116 cells (Fig. 4G). Collectively, these results suggest that BECN1s is also indispensable for depolarization-induced mitophagy.
Discussion
Previous studies have discovered that many regulators participate in processes of mitophagy induced by different stress conditions, such as BNIP3L,18,34,35 PINK1,14,15,21,23 and FUNDC1.36 In this study, we provide evidence demonstrating that BECN1s, a novel BECN1 splice variant, plays an important role in mitophagy induced by starvation and mitochondrial depolarization.
Compared to BECN1, BECN1s loses the function that initiates macroautophagy. This loss of function may be due to the inability of BECN1s to interact with autophagy regulating protein, such as UVRAG. Although BECN1s is not essential for macroautophagy initiation, BECN1s is able to specifically induce mitochondria-selective autophagy. Also, BECN1s retains the ability to activate PtdIns3K, which is important for initiation of autophagy,37 despite the lack of a partial ECD and C-terminal domain of BECN1s, compared to BECN1. It has been reported that BECN1 can bind to PINK1 and induce autophagy, but this is not specific for mitophagy initiation due to the fact that BECN1 mainly locates in the cytoplasm.38 We show that BECN1s is mainly associated with the outer-membrane of mitochondria and has a strong binding affinity for PINK1, suggesting that BECN1s initiates mitophagy via the interaction with PINK1. In support of this, BECN1s-induced mitophagy is PARK2 dependent. Although BECN1s is clearly involved in the regulation of mitophagy, it would be interesting to determine whether BECN1s possesses other cellular functions in addition to mitophagy regulation.
Alternative splicing occurs as a natural phenomenon in eukaryotes, through which the biodiversity of proteins are significantly increased. In humans, around 95% of multiexonic genes are alternatively spliced.39 Our finding of BECN1s as a BECN1 splice variant indicates the complexity of the regulation of BECN1. The different functions of BECN1 and BECN1s raise an intriguing possibility that BECN1 may interplay with BECN1s under certain physiological and stress conditions, although this has to be further investigated. Since both macroautophagy and mitophagy have been linked to various human diseases, it would be important to address how cellular BECN1s expression is controlled, whether BECN1 and BECN1s expression needs to be balanced under physiological conditions, and whether BECN1s dysregulation is associated with any human diseases. In summary, our findings suggest that BECN1s is a novel mitophagy regulator, and open a new avenue for the study of BECN1-mediated autophagy where the splicing event of BECN1 should definitely be considered.
Materials and Methods
Cell culture and reagents
HeLa, U2OS, HCT116 H1299, MCF7, SH-SY5Y and HEK 293T cell lines were cultured in DMEM (Dulbecco's modified Eagle's medium) medium containing 10% fetal bovine serum. Antibodies against GAPDH (sc-365062), OPA1 (sc-393296), MFN1 (sc-166644), TOMM20 (sc-17764), TIMM23 (sc-514463), VDAC1 (sc-390996), LC3 (sc-134226) and SQSTM1/p62 (sc-28359) were purchased from Santa Cruz Biotechnology. Antibodies against BECN1 (ab92389) and PIK3C3 (ab124905) were obtained from Abcam. Anti-ACTB/β-ACTIN (A5441) and anti-Flag (F1804) antibodies were ordered from Sigma Aldrich. Anti-GFP (632381) antibody was ordered from Clontech. Proteinase K (P2308), bafilomycin A1/BAF (B1793), and CCCP (C2759) were purchased from Sigma.
Knockdown of BECN1 and BECN1s
To knock down BECN1 and BECN1s, shRNAs against BECN1 and BECN1s were cloned into a pLKO.1 vector (Addgene, 8453). The target sequences are 5′-GAG GTT GAG AAA GGC GAG A-3′ (BECN1) and 5′-TTT CAG AGG ATG GAT GTG G-3′ (BECN1s). shRNA-mediated knockdown of BECN1 or BECN1s was performed as previously described.40
Reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was isolated using Trizol (Invitrogen, 15596-02), followed by RT-PCR analysis by using a PrimeScript™ RT-PCR Kit (TakaRa, Dalian, China) according to the manufacturer's instruction. The PCR primer sequences for RT-PCR are as follows: 5′- AAT GCA ACC TTC CAC ATC TGG and 5′- TTT CTC CAC ATC CAT CCT CTG (BECN1s); 5′-TTT AAT GCA ACC TTC C (P1) and 5′-TCA TTT GTT ATA AAA TTG TGA GG (P2).
Cytosolic and mitochondrial subcellular fractionation
Cytosolic and mitochondrial subcellular fractionation was performed as previously described.4 Briefly, cells were homogenized in homogenization buffer (20 mM HEPES-KOH, pH 7.5, 10 mM KCl, 1.5 mM MgCl2, 1 mM sodium EDTA, 1 mM sodium EGTA, 1 mM dithiothreitol, 250 mM sucrose [BBI, SB0498] and protease inhibitor cocktail [Roche, 04693132001]). Membrane rupture of cells was verified by microscopy. The homogenates were subjected to centrifugation at 600×g for 5 min at 4°C. The resulting low-speed pellet fractions, which contained nuclei and unbroken cells, were discarded. The postnuclear supernatant fraction was further centrifuged at 7000×g for 10 min at 4°C to obtain cytoplasmic and mitochondrial fractions. The mitochondrial pellet was washed and solubilized in TNC buffer (10 mM Tris acetate, pH 8.0, 0.5% Nonidet P-40 [Sangon, NDB0385], 5 mM CaCl2 and protease inhibitor cocktail).
Overexpression of BECN1s in HeLa cells
To generate lentiviruses expressing BECN1s, HEK293T cells grown on a 6-cm dish were transfected with 2 μg of pSin-BECN1s construct, 1 μg of pmd2.g and 2 μg of pspax2. Twelve h after transfection, cells were cultured with DMEM medium containing 20% fetal bovine serum for an additional 24 h. The culture medium containing lentivirus particles was filtered through a 0.45-mm polyvinylidene difluoride filter and incubated with HeLa cells for 12 h, followed by selection with 5 μg/ml puromycin for another 24 h.
Measurement of PIK3C3 lipid phosphorylation activity
To detect the lipid phosphorylation activity of PIK3C3, HeLa cells were transfected with GFP-2*FYVE construct (a kind gift from Dr. Harald Stenmark, The Norwegian Radium Hospital),41 which encodes GFP-tagged double FYVE fingers of HGS/Hrs (hepatocyte growth factor-regulated tyrosine kinase substrate). The GFP-2*FYVE fingers bind specifically to the PIK3C3 product, phosphatidylinositol-3-phosphate, and form the puncta. Twenty-four h after transfection, the GFP-2*FYVE puncta were observed under a fluorescence microscope.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This work was supported by grants from National Natural Science Foundation of China (81430065, 31371388, 31371428 and 31422035); the Ministry of Science and Technology of China (2014CB910601 and 2015CB553800); the Natural Science Foundation of Anhui province (1408085J07); and a Special Research Fund for the Doctoral Program of Higher Education from Ministry of Education of China (20123402130006).
References
- 1.Johri A, Beal MF. Mitochondrial dysfunction in neurodegenerative diseases. J Pharmacol Exp Therapeutics 2012; 342:619-30; http://dx.doi.org/ 10.1124/jpet.112.192138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zungu M, Schisler J, Willis MS. All the Little Pieces. Circulation J 2011; 75:2513-21; http://dx.doi.org/ 10.1253/circj.CJ-11-0967 [DOI] [PubMed] [Google Scholar]
- 3.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature 2011; 469:221-5; PMID:21124315; http://dx.doi.org/ 10.1038/nature09663 [DOI] [PubMed] [Google Scholar]
- 4.Frezza C, Cipolat S, Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured filroblasts. Nature Protocols 2007; 2:287-95; PMID:17406588; http://dx.doi.org/ 10.1038/nprot.2006.478 [DOI] [PubMed] [Google Scholar]
- 5.Kundu M. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood 2008; 112:1493-502; PMID:18539900; http://dx.doi.org/ 10.1182/blood-2008-02-137398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J. Mitochondrial clearance is regulated by Atg7-dependent and -independent mechanisms during reticulocyte maturation. Blood 2009; 114:157-64; PMID:19417210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mortensen M. Loss of autophagy in erythroid cells leads to defective removal of mitochondria and severe anemia in vivo. Proc Natl Acad Sci USA 2010; 107:832-7; PMID:20080761; http://dx.doi.org/ 10.1073/pnas.0913170107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato M, Sato K. Degradation of Paternal Mitochondria by Fertilization-Triggered Autophagy in C. elegans Embryos. Science 2011; 334:1141-4; PMID:21998252; http://dx.doi.org/ 10.1126/science.1210333 [DOI] [PubMed] [Google Scholar]
- 9.Kim I, Rodriguez-Enriquez S, Lemasters JJ. Selective degradation of mitochondria by mitophagy. Archives Biochem Biophysics 2007; 462:245-53; http://dx.doi.org/ 10.1016/j.abb.2007.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, Bosch-Marce M, Shimoda LA, Tan YS, Baek JH, Wesley JB, Gonzalez FJ, Semenza GL. Mitochondrial autophagy is an HIF-1-dependent adaptive metabolic response to hypoxia. J Biol Chem 2008; 283:10892-903; PMID:18281291; http://dx.doi.org/ 10.1074/jbc.M800102200 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J, Mazure NM. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol 2009; 29:2570-81; PMID:19273585; http://dx.doi.org/ 10.1128/MCB.00166-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurihara Y, Kanki T, Aoki Y, Hirota Y, Saigusa T, Uchiumi T, Kang D. Mitophagy plays an essential role in reducing mitochondrial production of reactive oxygen species and mutation of mitochondrial DNA by maintaining mitochondrial quantity and quality in yeast. J Biol Chem 2012; 287:3265-72; PMID:22157017; http://dx.doi.org/ 10.1074/jbc.M111.280156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y, Nartiss Y, Steipe B, McQuibban GA, Kim PK. ROS-induced mitochondrial depolarization initiates PARK2/PARKIN-dependent mitochondrial degradation by autophagy. Autophagy 2012; 8:1462-76; PMID:22889933; http://dx.doi.org/ 10.4161/auto.21211 [DOI] [PubMed] [Google Scholar]
- 14.Matsuda N. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol 2010; 189:211-21; PMID:20404107; http://dx.doi.org/ 10.1083/jcb.200910140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geisler S. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and SQSTM1/p62. Nat Cell Biol 2010; 12:119-31; PMID:20098416; http://dx.doi.org/ 10.1038/ncb2012 [DOI] [PubMed] [Google Scholar]
- 16.Schweers RL. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc Natl Acad Sci USA 2007; 104:19500-5; PMID:18048346; http://dx.doi.org/ 10.1073/pnas.0708818104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwarten M. NIX directly binds to GABARAP: a possible crosstalk between apoptosis and autophagy. Autophagy 2009; 5:690-8; PMID:19363302; http://dx.doi.org/ 10.4161/auto.5.5.8494 [DOI] [PubMed] [Google Scholar]
- 18.Sandoval H. Essential role for Nix in autophagic maturation of erythroid cells. Nature 2008; 454:232-5; PMID:18454133; http://dx.doi.org/ 10.1038/nature07006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Novak I. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep 2010; 11:45-51; PMID:20010802; http://dx.doi.org/ 10.1038/embor.2009.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding WX. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62- mediated mitochondrial priming. J Biol Chem 2010; 285:27879-90; PMID:20573959; http://dx.doi.org/ 10.1074/jbc.M110.119537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawajiri S. PINK1 is recruited to mitochondria with parkin and associates with LC3 in mitophagy. FEBS Lett 2010; 584:1073-9; PMID:20153330; http://dx.doi.org/ 10.1016/j.febslet.2010.02.016 [DOI] [PubMed] [Google Scholar]
- 22.Narendra DP. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol 2010; 8:e1000298; PMID:20126261; http://dx.doi.org/ 10.1371/journal.pbio.1000298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vives-Bauza C. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci USA 2010; 107:378-83; PMID:19966284; http://dx.doi.org/ 10.1073/pnas.0911187107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by BECN1. Nature 1999; 402,672-6; PMID:10604474; http://dx.doi.org/ 10.1038/45257 [DOI] [PubMed] [Google Scholar]
- 25.Furuya N, Yu J, Byfield M, Pattingre S, Levin B. The evolutionarily conserved domain of Beclin 1 is required for Vps34 binding, autophagy and tumor suppressor function. Autophagy 2005; 1(1):46-52; PMID:16874027; http://dx.doi.org/ 10.4161/auto.1.1.1542 [DOI] [PubMed] [Google Scholar]
- 26.Noda NN, Kobayashi T, Adachi W, Fujioka Y, Ohsumi Y, Inagaki F. Structure of the novel C-terminal domain of vacuolar protein sorting 30/autophagy-related protein 6 and its specific role in autophagy. J Biol Chem 2012; 287: 16256-66; PMID:22437838; http://dx.doi.org/ 10.1074/jbc.M112.348250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chu CT, Zhu J, Dagda R. BECN1-Independent Pathway of Damage-Induced Mitophagy and Autophagic Stress: Implications for Neurodegeneration and Cell Death. Autophagy 2007; 3(6):663-6; PMID:17622797; http://dx.doi.org/ 10.4161/auto.4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinha S, Levine B. The autophagy effector Beclin 1: a novel BH3-only protein. Oncogene 2008; 27 Suppl 1:S137-48; PMID:19641499; http://dx.doi.org/ 10.1038/onc.2009.51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stenmark H, Aasland R, Driscoll PC. The phosphatidylinositol 3-phosphate-binding FYVE finger. FEBS Lett 2002; 513:77-84; PMID:11911884; http://dx.doi.org/ 10.1016/S0014-5793(01)03308-7 [DOI] [PubMed] [Google Scholar]
- 30.Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol 2009; 11:468-76; PMID:19270693; http://dx.doi.org/ 10.1038/ncb1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi Y, Coppola D, Matsushita N, Cualing HD, Sun M, Sato Y, Liang C, Jung JU, Cheng JQ, Mulé JJ, et al.. Bif-1 interacts with Beclin 1 through UVRAG and regulates autophagy and tumorigenesis. Nat Cell Biol 2007; 9(10):1142-51; PMID:17891140; http://dx.doi.org/ 10.1038/ncb1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin 1-binding protein UVRAG. Nat Cell Biol 2006; 8:688-99; PMID:16799551; http://dx.doi.org/ 10.1038/ncb1426 [DOI] [PubMed] [Google Scholar]
- 33.Narendra D, Tanaka A, Suen DF, Youle RJ. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol 2008; 183:795-803; PMID:19029340; http://dx.doi.org/ 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, Ney PA. NIX induces mitochondrial autophagy in reticulocytes. Autophagy 2008; 4:354-6; PMID:18623629; http://dx.doi.org/ 10.4161/auto.5552 [DOI] [PubMed] [Google Scholar]
- 35.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Löhr F, Popovic D, Occhipinti A, et al.. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep 2010; 11:45-51; PMID:20010802; http://dx.doi.org/ 10.1038/embor.2009.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu L, Feng D, Chen G, Chen M, Zheng Q, Song P, Ma Q, Zhu C, Wang R, Qi W, et al.. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol 2012; 14(2):177-85; PMID:22267086; http://dx.doi.org/ 10.1038/ncb2422 [DOI] [PubMed] [Google Scholar]
- 37.Backer JM. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem J 2008; 410:1-17; PMID:18215151; http://dx.doi.org/ 10.1042/BJ20071427 [DOI] [PubMed] [Google Scholar]
- 38.Michiorri S, Gelmetti V, Giarda E, Lombardi F, Romano F, Marongiu R, Nerini-Molteni S, Sale P, Vago R, Arena G, et al.. The Parkinson-associated protein PINK1 interacts with Beclin 1 and promotes autophagy. Cell Death Differ 2010; 17:962-74; PMID:20057503; http://dx.doi.org/ 10.1038/cdd.2009.200 [DOI] [PubMed] [Google Scholar]
- 39.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet 2008; 40:1413-5; PMID:18978789; http://dx.doi.org/ 10.1038/ng.259 [DOI] [PubMed] [Google Scholar]
- 40.Wang W, Cheng B, Miao L, Mei Y, Wu M. Mutant p53-R273H gains new function in sustained activation of EGFR signaling via suppressing miR-27a expression. Cell Death Dis 2013; 4:e574; PMID:23559009; http://dx.doi.org/ 10.1038/cddis.2013.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J 2000; 19: 4577-88; PMID:10970851; http://dx.doi.org/ 10.1093/emboj/19.17.4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


