Abstract
In spite of adjuvant chemotherapy, a significant fraction of patients with localized breast cancer (BC) relapse after optimal treatment. We determined the occurrence of cytoplasmic MAP1LC3B/LC3B (microtubule-associated protein 1 light chain 3B)-positive puncta, as well as the presence of nuclear HMGB1 (high mobility group box 1) in cancer cells within surgical BC specimens by immunohistochemistry, first in a test cohort (152 patients) and then in a validation cohort of localized BC patients who all received adjuvant anthracycline-based chemotherapy (1646 patients). Cytoplasmic LC3B+ puncta inversely correlated with the intensity of SQSTM1 staining, suggesting that a high percentage cells of LC3B+ puncta reflects increased autophagic flux. After setting optimal thresholds in the test cohort, cytoplasmic LC3B+ puncta and nuclear HMGB1 were scored as positive in 27.2% and 28.6% of the tumors, respectively, in the validation cohort, while 8.7% were considered as double positive. LC3B+ puncta or HMGB1 expression alone did not constitute independent prognostic factors for metastasis-free survival (MFS) in multivariate analyses. However, the combined positivity for LC3B+ puncta and nuclear HMGB1 constituted an independent prognostic factor significantly associated with prolonged MFS (hazard ratio: 0.49 95% confidence interval [0.26–0.89]; P = 0.02), and improved breast cancer specific survival (hazard ratio: 0.21 95% confidence interval [0.05–0.85]; P = 0.029). Subgroup analyses revealed that within patients with poor-prognosis BC, HMGB1+ LC3B+ double-positive tumors had a better prognosis than BC that lacked one or both of these markers. Altogether, these results suggest that the combined positivity for LC3B+ puncta and nuclear HMGB1 is a positive predictor for longer BC survival.
Keywords: breast cancer, histology, HMGB1, LC3, pathology, prognostic, tumor microarray
Abbreviations
- BC
breast cancer
- BCSS
breast cancer-specific survival
- CI
confidence interval
- HMGB1
high mobility group box 1
- HR
hazard ratio
- LC3B (MAP1LC3B/LC3B)
microtubule-associated protein 1 light chain 3B
- MFS
metastasis free survival
- OS
overall survival
- PBS
phosphate-buffered saline
- SQSTM1/p62
sequestosome 1
- TLR4
toll-like receptor 4
- TMAs
tissue microarrays.
Introduction
Major advances have been achieved in the management of early breast cancer (BC) during recent decades. Chemotherapy and endocrine therapy have improved BC survival to the point that >75% to 80% of BC patients are expected to be cured nowadays. However, there is still a significant fraction of patients who eventually succumb to their disease due to distant relapse. Therefore, it is important to identify patients with a high likelihood of metastatic relapse with the scope of enrolling them in clinical trials that combine molecular diagnosis with targeted therapies. Conversely, the identification of patients with low risk of relapse could allow for the avoidance of toxic adjuvant chemotherapies. Hence, the prediction of the residual risk of relapse is considered as an urgent unmet medical need, as emphasized at the 2013 Saint Gallen Consensus conference.1 BC is a heterogeneous malignancy that has been subjected to subclassification over the past decade, based on gene expression analyses.2-5 To date, 4 clinically distinct BC subtypes have been consistently described according to their hormonal status and HER2 oncogene amplification.5 However, within each disease entity, significant differences in clinical outcome exist, prompting the search for additional biometrics.6
Autophagy has been linked to BC pathogenesis, since the first publication of a report that the gene coding for the essential autophagy-related protein BECN1/BECLIN 1 is often subjected to loss of heterozygosity in BC.7 Indeed, mice that are haploinsufficient for Becn1 (genotype: Becn1+/−) exhibit an autophagy defect and are more prone to develop a variety of cancers including BC than autophagy-competent control animals.8-10 Low expression of the mRNA coding for BECN1 is also a negative prognostic marker in BC patients.11 These data indicate that autophagy may act as a tumor suppressor mechanism in BC, as it also has been reported for Kras-induced pancreas and lung cancers.12,13 A recent study has demonstrated that HMGB1 (high mobility group box 1), one of the most abundant nonhistone chromatin-binding proteins, can regulate autophagy if it is released from the nucleus by disrupting the autophagy-inhibitory interaction between BECN1 and BCL2.14,15 HMGB1 is also a ligand of TLR4 (toll-like receptor 4) and stimulates anticancer immune responses.16 Importantly, a loss-of-function allele of TLR4 has a negative prognostic impact on BC.16 Similarly, autophagy has been linked to the release of ATP from stressed and dying tumor cells,17,18 allowing this factor to interact with purinergic receptors (in particular with P2RX7 [purinergic receptor P2X, ligand-gated ion channel 7]), which is also involved in anticancer immunosurveillance.19 Loss-of-function alleles of both TLR4 and P2RX7 both negatively affect the prognosis of BC patients treated with adjuvant chemotherapy and interact epistatically,16,19 providing yet another possible functional link between autophagy-related ATP release and extracellular HMGB1. Both pathways, may affect BC prognosis by affecting immunosurveillance.
Here, we explored the clinical implications of the relationship between HMGB1 and LC3B by using previously validated immunohistochemical methods.20,21 We determined the presence of nuclear HMGB1 and cytoplasmic LC3B puncta in 2 cohorts of BC patients, establishing that both markers do not correlate among each other, but can still predict the fate of women affected by BC.
Results
Prognostic significance of HMGB1 and LC3B in the training cohort
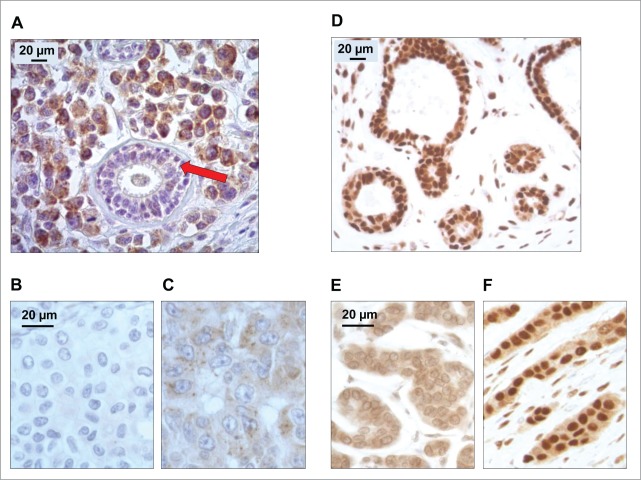
Validated immunohistochemical methods for the detection of LC3B20 and HMGB121 revealed major heterogeneity among distinct BC samples (Fig. 1A–F), as first determined on the training cohort of 152 BC treated with adjuvant anthracycline-based chemotherapy (for patient characteristics see Table S1). The antibody directed against the N-terminal domain of the human LC3B isoform yielded a specific signal predominantly localized in the cytoplasm.22,23 The number of cytoplasmic LC3B+ puncta per cell was increased in tumors, relative to surrounding healthy tissues (Fig. 1A). Within the tumor beds, tumor infiltrating lymphocytes and stromal cells could also exhibit cytoplasmic LC3B+ dots.20 We concentrated our analysis on malignant cells only, quantifying the percentage of cancer cells with clearly visible cytoplasmic LC3B+ puncta (see Fig. 1A–C for representative images, Fig. 2A for frequency distributions). The intensity of the diffuse cytoplasmic LC3B staining was also heterogeneous in distinct tumors (Fig. S1 and S2).
Figure 1.
Patterns of LC3B and HMGB1 immunohistochemical staining of breast adenocarcinomas in the training cohort. (A) LC3B puncta in a breast adenocarcinoma: nonmalignant breast gland (red arrow) staining negatively for LC3B, in the proximity of tumor cells with intense LC3B positivity. (B) Representative aspect of a breast adenocarcinoma without any detectable cytoplasmic LC3B puncta. (C) Cytoplasmic LC3B puncta in a breast adenocarcinoma that was considered as positive for LC3B staining. (D) Representative strong nuclear HMGB1 staining in normal mammary glands. (E) Representative aspect of a breast adenocarcinoma without any detectable nuclear staining (tumor considered as negative for nuclear HMGB1 expression). (F) Homogeneous nuclear HMGB1 staining in a breast adenocarcinoma considered as positive for HMGB1 nuclear expression.
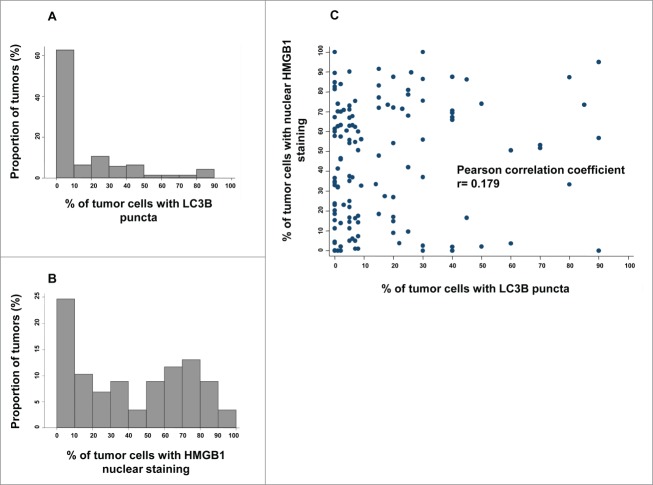
Figure 2.
LC3B and HMGB1 staining quantitative analysis in the training cohort. (A) Distribution frequencies of percent of breast cancer cells harboring LC3B cytoplasmic puncta in the training cohort. (B) Distribution frequencies of percent of breast cancer cells with HMGB1-positive nuclei in the training cohort. (C) Correlation between percent of breast cancer cells with LC3B puncta, and percent of breast cancer cells with HMGB1 nuclear staining in each sample of the training cohort.
Theoretically, LC3B puncta can increase in number because more autophagic material is sequestered in autophagosomes (increased autophagic flux) or because autophagosomes fail to be degraded after their fusion with lysosomes (decreased autophagic flux). To discriminate between these 2 possibilities, we stained the training cohort of BC samples with an antibody specific for SQSTM1 (sequestosome 1), followed by quantification of the frequency of cancer cells with absent, weak, moderate, or strong cytoplasmic staining for SQSTM1 (Fig. S3A). SQSTM1 is a well-characterized autophagic substrate the abundance of which decreases when autophagic flux increases.24 We observed that the amount of LC3B puncta inversely correlated with the intensity of the SQSTM1 staining in BC tissues (Fig. S3B and C). Therefore, we conclude that a high number of LC3B puncta indeed reflects an elevated autophagic flux in this pathology. In contrast, no correlation was found between HMGB1 status and SQSTM1 staining (Fig. S3B–D).
The anti-HMGB1 antibody strongly labeled nuclei from healthy epithelial, immune, and stromal cells, as well as from most in situ adenocarcinomas in a rather homogeneous manner across tissue sections.25 However, the mean percentage of HMGB1 nuclear expression was much lower in larger tumors (see Fig. 1D–F for representative images). We determined the percentage of cancer cells exhibiting clear positivity for nuclear HMGB1 for each sample (see Fig. 2B for frequency distributions). There was no correlation between the frequency of cells with HMGB1+ nuclei and that of cells with LC3B+ puncta (Pearson correlation coefficient = 0.179) (Fig. 2C), indicating that these 2 variables can be considered independently from each other. Next, we calculated the optimal cut-off point for LC3B and HGMB1 positivity that would allow for patient survival stratification, as 10% and 50%, respectively (Fig. S4).
Based on the log-rank outcome approach, any cutpoint in the range of 37% to 67% for HMGB1 and 8% to 15% for LC3B reached significance levels of 0.001 and 0.05, respectively. To limit the risks associated to the selection of a cutpoint close to the borders of the above ranges and for the sake of simplicity, we set the cut-off points at 50% and 10% for HMGB1 and LC3, respectively. Our choice is encouraged by the examination of the martingale plots where the chosen cutpoints fell in the middle of the steepest slope indicative of an optimal cutpoint model. As a result, we considered tumors as LC3B+ if they contained >10% malignant cells with LC3B+ puncta. HMGB1+ tumors were defined as those containing >50% cancer cells with HMGB1+ nuclei.
Importantly, the correlation between the intensity of the diffuse cytoplasmic LC3B staining and the presence of LC3B puncta was poor (Fig. S5). Moreover, based on the same statistical methods for adequate cutoff determination, we did not find a meaningful cutpoint value for LC3B intensity that would allow for BC risk stratification (Fig. S6). For this reason, we only evaluated the presence of LC3B puncta (rather than LC3B intensity) as a prognostic factor in the validation cohort.
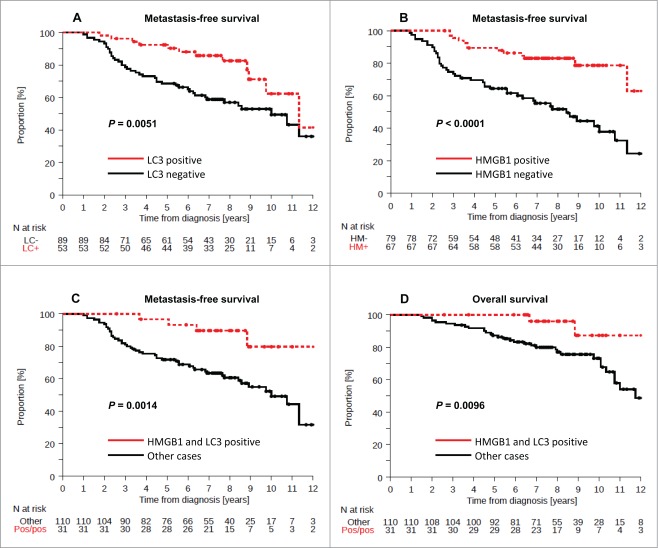
The presence of LC3B puncta (which was found in 37% of the cases) was associated with prolonged metastasis free survival (MFS) in univariate (P = 0.007) but not in multivariate (P = 0.157) analyses (Fig. 3A, Table 1). Forty-six percent of BC in the training cohort were scored as HMGB1+ and exhibited a favorable outcome, as determined by univariate analysis (Fig. 3B, P < 0.0001) (Table 1). In contrast, 22% of BC were considered double-positive (LC3B+ HMGB1+) and exhibited a rather good prognosis with regard to MFS and OS (Fig. 3C, P = 0.0014, Fig. 3D, P = 0.0096). Based on these results, we postulated that the combined analysis of LC3B and HMGB1 might allow for risk stratification of BC patients, and we tested this hypothesis on an independent, larger cohort.
Figure 3.
Kaplan-Meier survival plots of the training cohort. Metastasis-free survival (MFS) in the training cohort according to (A) cytoplasmic LC3B puncta in breast cancer cells (>10% = positive, or <10% = negative), (B) nuclear HMGB1 expression in breast cancer cells (>50% = positive, or <50% = negative). Breast cancer-specific survival (BCSS) in the PACS04 trial according to (B) cytoplasmic LC3B puncta and (D) nuclear HMGB1 expression. P values were calculated using the log-rank test.
Table 1.
Univariate and multivariate (incorporating LC3 or HMGB1) analysis (Cox regression) for factors associated with MFS in the training cohort
| Univariate analysis |
Multivariate analysis (LC3) |
Multivariate analysis (HMGB1) |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Event/n | Event/n | Event/n | |||||||||||||||
| 60/151 | HR | IC 95% | p-value | 60/151 | HR | IC 95% | p-value | 60/151 | HR | IC 95% | p-value | ||||||
| HMGB1 | HMGB1 | ||||||||||||||||
| Negative | 43/79 | 1 | <0.0001 | Negative | 43/79 | 1 | 0.001 | ||||||||||
| Positive | 14/67 | 0.28 | [0.15;0.53] | Positive | 14/67 | 0.33 | [0.17;0.64] | ||||||||||
| LC3 | LC3 | ||||||||||||||||
| Negative | 43/89 | 1 | 0.007 | Negative | 43/89 | 1 | 0.157 | ||||||||||
| Positive | 12/53 | 0.41 | [0.22;0.78] | Positive | 12/53 | 0.61 | [0.31;1.21] | ||||||||||
| pT stage | pT stage | pT stage | |||||||||||||||
| T1 | 32/88 | 1 | 0.3569 | T1 | 27/80 | 1 | 0.8775 | T1 | 30/84 | 1 | 0.4852 | ||||||
| T2 | 23/56 | 1.32 | [0.77;2.28] | T2 | 23/55 | 1.07 | [0.58;1.96] | T2 | 23/56 | 0.69 | [0.37;1.29] | ||||||
| T3 | 2/3 | 1.95 | [0.47;8.2] | T3 | 2/3 | 1.39 | [0.3;6.55] | T3 | 2/3 | 1.67 | [0.35;7.99] | ||||||
| T4 | 3/4 | 2.46 | [0.75;8.11] | T4 | 3/4 | 1.67 | [0.45;6.16] | T4 | 2/3 | 1.47 | [0.3;7.19] | ||||||
| pN stage | pN stage | pN stage | |||||||||||||||
| N0 | 14/42 | 1 | 0.0094 | N0 | 12/40 | 1 | 0.0764 | N0 | 13/41 | 1 | 0.0037 | ||||||
| N1 | 30/85 | 0.95 | [0.5;1.81] | N1 | 29/81 | 1.56 | [0.75;3.26] | N1 | 28/81 | 1.39 | [0.66;2.92] | ||||||
| N2 | 12/19 | 1.79 | [0.82;3.88] | N2 | 11/17 | 2.27 | [0.91;5.7] | N2 | 12/19 | 2.7 | [1.08;6.73] | ||||||
| N3 | 4/5 | 4.84 | [1.57;14.94] | N3 | 3/4 | 6.56 | [1.46;29.51] | N3 | 4/5 | 9.78 | [2.51;38.1] | ||||||
| Tumor grade | Tumor grade | Tumor grade | |||||||||||||||
| 1–2 | 24/80 | 1 | <0.0001 | 1–2 | 22/75 | 1 | 0.003 | 1–2 | 22/77 | 1 | 0.001 | ||||||
| 3 | 36/70 | 2.69 | [1.58;4.57] | 3 | 33/67 | 2.81 | [1.43;5.53] | 3 | 35/69 | 3.14 | [1.59;6.19] | ||||||
| Luminal | Luminal | Luminal | |||||||||||||||
| No | 17/38 | 1 | 0.101 | No | 16/37 | 1 | 0.688 | No | 17/38 | 1 | 0.668 | ||||||
| Yes | 43/113 | 0.62 | [0.35;1.1] | Yes | 39/105 | 0.87 | [0.44;1.73] | Yes | 40/108 | 1.16 | [0.58;2.33] | ||||||
Prognostic significance of HMGB1 and LC3B in the validation cohort
The validation cohort involved tissue microarrays that were stained for the detection of LC3B and HMGB1 (for representative images see Fig. S7), making it possible to obtain quantitative data from 1581 patients that had been included in a prospective clinical trial evaluating different regimens of anthracyline-based adjuvant chemotherapy (see Patients and Methods). The median follow-up was 4.95 y (95% confidence interval [CI] = [4.93; 4.97] y). MFS events were observed in 253 patients (16%), and 140 (8.8%) had died, among whom 119 deaths were from BC. Within this cohort, we defined a subgroup of patients with poor prognosis (with tumor diameters of >2 cm, >3 cancer cell-involved axillary lymph nodes, an SBR score of III, and triple-negative tumors), following established criteria.26
Using the same criterion for LC3B positivity as for the training cohort, 430 tumors (27.2%) were considered as positive for LCB puncta (LC3B+). These LC3B+ breast tumors were more frequent in HER2-overexpressing BC (P = 0.005, Table 2). LC3B positivity was associated with a lower risk of death from BC in univariate analysis: (BC-specific survival [BCSS]: HR = 0.61 [0.39-0.97], logrank test P = 0.035), but not with significantly improved MFS (HR = 0.76 [0.56–1.02], logrank test P = 0.07) (Fig. 4A, B). Moreover, this marker could be considered as an independent prognostic factor of improved MFS (HR: 0.74 [0.55–1] P = 0.049) and BCSS (HR = 0.60 [0.38–0.96] P = 0.035), as determined by multivariate analysis (Table 3). Considering MFS, we did not find any significant interaction between trastuzumab treatment and LC3B status (interaction test P = 0.36). Moreover, in patients bearing HER2+ tumors, trastuzumab administration was well balanced between tumors that were evaluated as positive or negative for LC3B puncta (interaction test: P = 0.368). As a result, we believe that trastuzumab treatment did not influence the results concerning the status of LC3B puncta.
Table 4.
Univariate and multivariate* analysis (Cox regression) for factors associated with MFS
| Univariate analysis |
Multivariate analysis |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| |
|
Event/n |
HR |
IC 95% |
p-value |
Event/n |
HR |
IC 95% |
p-value |
| 241/1524 | 232/1476 | ||||||||
| HMGB1 & LC3 | |||||||||
| Other cases | 230/1392 | 1 | 0.018 | 221/1348 | 1 | 0.02 | |||
| Positive / Positive | 11/132 | 0.48 | [0.26;0.88] | 11/128 | 0.49 | [0.26;0.89] | |||
| Treatment arm | |||||||||
| 6 FEC | 120/758 | 1 | 0.907 | 117/736 | 1 | 1 | 0.897 | ||
| 6 ED | 121/766 | 0.99 | [0.77;1.27] | 115/740 | 0.9 | [0.69;1.16] | |||
| Age (years) | |||||||||
| <50 | 105/687 | 1 | 0.739 | ||||||
| >= 50 | 136/837 | 1.04 | [0.81;1.35] | ||||||
| Menopausal status | |||||||||
| Premenopausal | 77/594 | 1 | 0.696 | ||||||
| Post menopausal | 81/586 | 1.06 | [0.78;1.45] | ||||||
| Tumor size (mm) | |||||||||
| <=20 mm | 86/753 | 1 | <0.0001 | 83/737 | 1 | 0.0001 | |||
| 20–50 mm | 119/667 | 1.64 | [1.24;2.16] | 117/653 | 1.23 | [0.92;1.63] | |||
| >50 mm | 35/93 | 3.64 | [2.46;5.4] | 32/86 | 2.54 | [1.67;3.85] | |||
| Involved lymph nodes | |||||||||
| N1 : 1 – 3 | 95/1037 | 1 | <0.0001 | 94/1009 | 1 | <0.0001 | |||
| N2 : 4 – 9 | 95/377 | 2.98 | [2.25;3.97] | 88/360 | 2.43 | [1.81;3.27] | |||
| N3 : >=10 | 51/110 | 6.3 | [4.49;8.86] | 50/107 | 4.96 | [3.48;7.07] | |||
| Tumor grade | |||||||||
| 1/2 | 94/884 | 1 | <0.0001 | 93/875 | 1 | 0.001 | |||
| 3 | 140/605 | 2.45 | [1.89;3.18] | 139/601 | 1.69 | [1.25;2.28] | |||
| ER | |||||||||
| Negative | 77/331 | 1 | <0.0001 | ||||||
| Positive | 164/1193 | 0.54 | [0.41;0.7] | ||||||
| PR | |||||||||
| Negative | 104/466 | 1 | <0.0001 | ||||||
| Positive | 118/932 | 0.52 | [0.4;0.68] | ||||||
| HER2+++ | |||||||||
| No | 166/1222 | 1 | <0.0001 | ||||||
| Yes | 75/302 | 2 | [1.52;2.63] | ||||||
| BC molecular subtype | |||||||||
| TN | 36/148 | 1 | <0.0001 | 35/146 | 1 | 0.0051 | |||
| HER2+++ | 75/302 | 1 | [0.67;1.49] | 74/298 | 0.91 | [0.6;1.36] | |||
| Luminal | 129/1072 | 0.44 | [0.3;0.63] | 123/1032 | 0.58 | [0.38;0.87] | |||
including HMGB1/LC3 combined variable
Table 2.
Patients and tumor characteristics in PACS04 trial (N = 1581) according to HMGB1 and LC3 tumor status
| HMGB1 |
LC3 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Negative | Positive | p valueChi2 | Negative | Positive | p-valueChi2 | |||||
| Treatment arm | ||||||||||
| 6 FEC | 564 (50%) | 220 (48.6%) | 0.606 | 568 (49.3%) | 225 (52.3%) | 0.285 | ||||
| 6 ED | 564 (50%) | 233 (51.4%) | 584 (50.7%) | 205 (47.7%) | ||||||
| Age (years) | ||||||||||
| <50 | 522 (46.3%) | 187 (41.3%) | 0.071 | 520 (45.1%) | 193 (44.9%) | 0.928 | ||||
| >= 50 | 606 (53.7%) | 266 (58.7%) | 632 (54.9%) | 237 (55.1%) | ||||||
| Menopausal status | ||||||||||
| Premenopausal | 454 (51.1%) | 163 (48.1%) | 0.349 | 441 (49.7%) | 174 (52.7%) | 0.342 | ||||
| Post menopausal | 435 (48.9%) | 176 (51.9%) | 447 (50.3%) | 156 (47.3%) | ||||||
| Tumor size (mm) | ||||||||||
| <=20 mm | 527 (47.1%) | 255 (56.7%) | 0.003 | 573 (50%) | 211 (49.7%) | 0.904 | ||||
| 20–50 mm | 518 (46.3%) | 170 (37.8%) | 501 (43.8%) | 185 (43.5%) | ||||||
| >50 mm | 74 (6.6%) | 25 (5.6%) | 71 (6.2%) | 29 (6.8%) | ||||||
| Involved lymph nodes | ||||||||||
| N1 : 1 – 3 | 769 (68.2%) | 311 (68.7%) | 0.549 | 778 (67.5%) | 299 (69.5%) | 0.739 | ||||
| N2 : 4 – 9 | 272 (24.1%) | 114 (25.2%) | 291 (25.3%) | 101 (23.5%) | ||||||
| N3 : >=10 | 87 (7.7%) | 28 (6.2%) | 83 (7.2%) | 30 (7%) | ||||||
| Tumor grade | ||||||||||
| 1/2 | 618 (56.3%) | 308 (69.1%) | <0.0001 | 674 (59.8%) | 235 (56.1%) | 0.187 | ||||
| 3 | 479 (43.7%) | 138 (30.9%) | 453 (40.2%) | 184 (43.9%) | ||||||
| ER | ||||||||||
| Negative | 280 (24.8%) | 65 (14.4%) | <0.0001 | 262 (22.7%) | 86 (20%) | 0.241 | ||||
| Positive | 848 (75.2%) | 388 (85.7%) | 890 (77.3%) | 344 (80%) | ||||||
| PR | ||||||||||
| Negative | 376 (36.7%) | 107 (24.9%) | <0.0001 | 351 (33.3%) | 139 (34.8%) | 0.573 | ||||
| Positive | 648 (63.3%) | 322 (75.1%) | 704 (66.7%) | 260 (65.2%) | ||||||
| HER2+++ | ||||||||||
| No | 884 (78.4%) | 382 (84.3%) | 0.007 | 939 (81.5%) | 323 (75.1%) | 0.005 | ||||
| Yes | 244 (21.6%) | 71 (15.7%) | 213 (18.5%) | 107 (24.9%) | ||||||
| BC molecular subtype | ||||||||||
| TN | 244 (21.7%) | 71 (15.7%) | <0.0001 | 213 (18.5%) | 107 (24.9%) | 0.016 | ||||
| HER2+++ | 130 (11.6%) | 25 (5.5%) | 118 (10.3%) | 36 (8.4%) | ||||||
| Luminal | 752 (66.8%) | 357 (78.8%) | 819 (71.2%) | 287 (66.7%) | ||||||
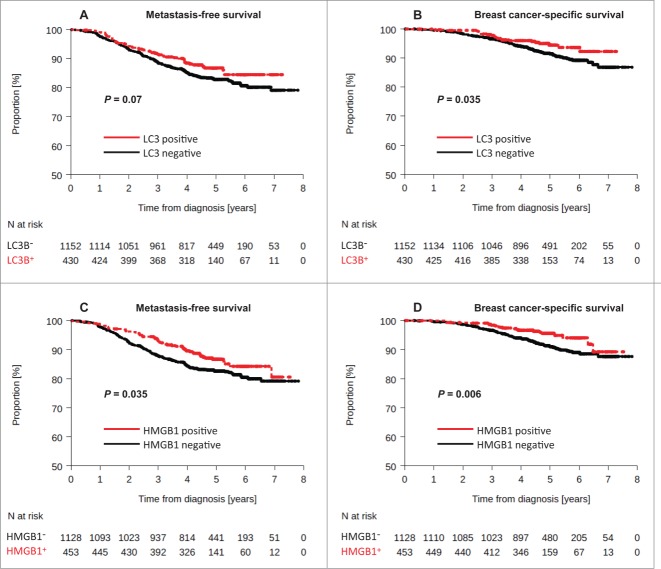
Figure 4.
Kaplan-Meier survival plots for the validation cohort after analysis of LC3B and HMGB1. Metastasis-free survival (MFS) of BC patients included in the PACS04 trial after stratification of patients according to (A) cytoplasmic LC3B puncta in BC cells (>10% = positive, or<10% = negative), or (C) nuclear HMGB1 expression in breast cancer cells (>50% = positive, or <50% = negative). Breast cancer-specific survival (BCSS) in the PACS04 trial according to (B) cytoplasmic LC3B puncta and (D) nuclear HMGB1 expression. P values were calculated using the log-rank test.
Table 3.
Multivariate analyses (Cox regression, including LC3 variable) for factors associated with MFS and BCSS in the validation cohort
| MFS |
BCSS |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Event/n | Death/n | |||||||||||
| 242/1532 | HR | IC 95% | p-value | 113/1532 | HR | IC 95% | p-value | |||||
| LC3 | ||||||||||||
| Negative | 186/1118 | 1 | 0.049 | 91/1118 | 1 | 0.035 | ||||||
| Positive | 56/414 | 0.74 | [0.55;1] | 22/414 | 0.6 | [0.38;0.96] | ||||||
| Treatment arm | ||||||||||||
| 6 FEC | 124/769 | 1 | 0.294 | 55/769 | 1 | 0.969 | ||||||
| 6 ED | 118/763 | 0.87 | [0.68;1.13] | 58/763 | 1.01 | [0.69;1.46] | ||||||
| Tumor size (mm) | ||||||||||||
| <=20 mm | 85/768 | 1 | <0.0001 | 41/768 | 1 | 0.1383 | ||||||
| 20–50 mm | 123/671 | 1.28 | [0.97;1.7] | 59/671 | 1.14 | [0.76;1.72] | ||||||
| >50 mm | 34/93 | 2.61 | [1.74;3.92] | 13/93 | 1.91 | [1.01;3.62] | ||||||
| Involved lymph nodes | ||||||||||||
| N1 : 1 – 3 | 99/1047 | 1 | <0.0001 | 45/1047 | 1 | <0.0001 | ||||||
| N2 : 4 – 9 | 91/375 | 2.38 | [1.78;3.18] | 43/375 | 2.39 | [1.56;3.65] | ||||||
| N3 : >=10 | 52/110 | 5.19 | [3.67;7.35] | 25/110 | 5.01 | [3.01;8.33] | ||||||
| Tumor grade | ||||||||||||
| 1/2 | 97/899 | 1 | 0.001 | 35/899 | 1 | 0.001 | ||||||
| 3 | 145/633 | 1.62 | [1.2;2.17] | 78/633 | 2.17 | [1.37;3.43] | ||||||
| BC molecular subtype | ||||||||||||
| TN | 38/152 | 1 | 0.0014 | 28/152 | 1 | <0.0001 | ||||||
| HER2+++ | 77/316 | 0.81 | [0.54;1.2] | 36/316 | 0.47 | [0.29;0.79] | ||||||
| Luminal | 127/1064 | 0.52 | [0.35;0.77] | 49/1064 | 0.3 | [0.18;0.5] | ||||||
Using the same criterion for HMGB1 positivity as for the training set, 453 patients (28.6%) harbored HMGB1+ tumors. Positivity for HMGB1 was associated with a lower risk of metastatic relapse or death from BC in univariate analysis (MFS: HR = 0.73 [0.55–0.98], logrank test P = 0.035), and BCSS: HR = 0.52 [0.32–0.84], logrank test P = 0.006) (Fig. 4C, D). HMGB1 negativity was significantly associated with characteristics of poor prognosis at diagnosis, such as larger tumor size (P = 0.003), higher tumor grade (P < 0.0001), and aggressive molecular subtype (such as HER2-overexpressing or triple-negative tumors) (P < 0.0001) (Table 2). Thus, HMGB1 positivity did not constitute an independent prognostic factor either, as confirmed by multivariate Cox regression analyses (MFS: HR = 0.85 [0.63–1.15], P = 0.29, and BCSS: HR = 0.67 [0.41–1.08] P = 0.102).
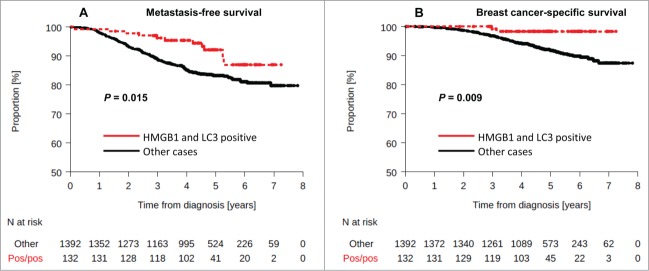
Double positivity for LC3B and HMGB1 was observed in 8.7% of samples. No interaction between HMGB1 and LC3B staining, and molecular subtype was observed (interaction test, P = 1.00). The LC3B+ HMGB1+ status was associated with a lower rate of metastatic relapse or death (Fig. 5A, B) both in univariate (MFS: HR = 0.48 [0.26–0.88], logrank test P = 0.018, BCSS: HR = 0.19 [0.05–0.77], logrank test P = 0.02) or multivariate analysis (MFS: HR = 0.49 [0.26–0.89] P = 0.02), BCSS: HR = 0.21 [0.05–0.85] P = 0.029). The 4-y MFS were 95.3% and 85.1% in patients with HMGB1+ and LC3B+ tumors and in the other patients group, respectively.
Figure 5.
Kaplan-Meier curves for the validation cohort stratified after combined analysis of LC3B and HMGB1. Metastasis-free survival (A) and breast-cancer specific survival (B) in the PACS04 trial according to positivity for both cytoplasmic LC3B puncta and nuclear HMGB1expression in breast cancer cells. P values were calculated using the log-rank test.
To further assess the potential utility of LC3B and HMGB1 as biomarkers, we explored whether dual positivity could complement currently used prognostication tools. The subgroup of patients with good prognosis (tumor size <2 cm, N+ <3, positive hormone receptors with tumor grade <III) was associated with a low risk of metastatic relapse, irrespective of the expression of LC3B and HMGB1 (Fig. 6A). However, although double positivity for LC3B and HMGB1 appeared to be associated with better MFS, independently from clinical prognostic factors (Fig. 6B), within the subgroup of patients with poor prognosis, double positivity for LC3B and HMGB1 constituted a useful tool of stratification (Fig. 6A). Within this subgroup, patients with LC3B+HMGB1+ tumors had an excellent outcome (HR = 0.50 [0.27–0.95] P = 0.034) with 4-year MFS of 94.24% [86.71%; 97.56%] vs. 81.41% [78.83%; 83.7%] for all other patients with poor prognosis.
Figure 6.
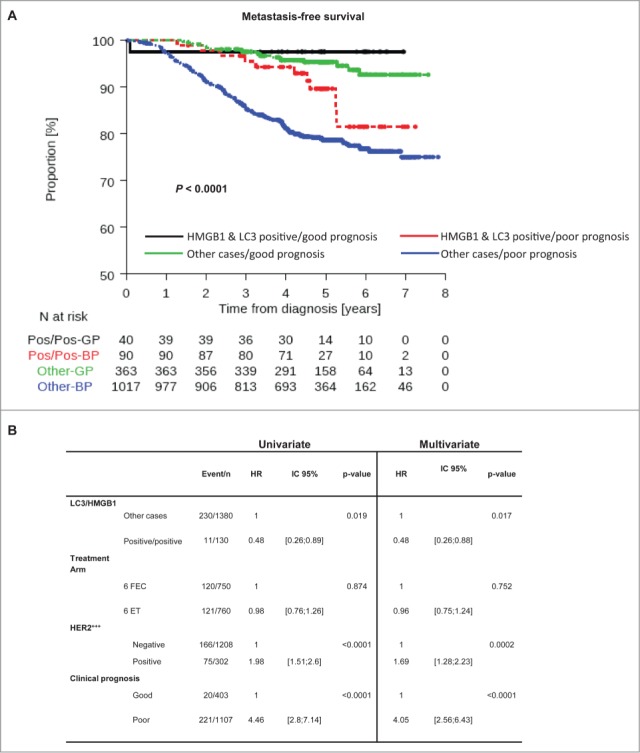
LC3B and HMGB1 double positivity as a complement to current prognostication tools. (A) Kaplan-Meier curves for metastasis-free survival (MFS) in the PACS04 trial stratified according to the presence of both cytoplasmic LC3B puncta and nuclear HMGB1 in breast cancer cells, as well as clinical prognostic status (good or poor). P values were calculated using the log-rank test. (B) Univariate and multivariate analyses (by Cox regression) of factors associated with MFS in the PACS04 trial. GP, good prognosis; BP, bad prognosis.
Within the limitations of the size of the cohort and the length of the follow-up, these results validate the working hypothesis that positivity for LC3B and HMGB1 constitutes a positive prognostic biomarker.
Discussion
Here, we demonstrate the potential clinical utility of quantifying cytoplasmic LC3B puncta and nuclear HMGB1 expression. Patients that lack both markers below a critical threshold (<10% cells with LC3B+ puncta; <50% cells with nuclear HMGB1) have a particularly poor prognosis, especially if they fall into the group of patients that are considered to have a bad prognosis, based on classical histopathological criteria. These findings have been obtained on a training cohort allowing to determine optimal cut-off levels that distinguish positive and negative groups and then confirmed on a much larger cohort.
A number of studies have addressed LC3B staining patterns in BC. LC3B staining is reported to be generally higher in tumor tissues than in normal tissues.27,28 In estrogen receptor-negative BC, no association has been found between LC3B expression and androgen receptor expression and HER2 positivity.29 LC3B staining is particularly intense in BC that already have metastasized into lymph nodes as compared to tumors that have not yet disseminated.30 Moreover, LC3B staining is correlated with the expression of the proliferation marker MKI67/Ki-67.27,30 In a cohort of patients with locally advanced BC that received neoadjuvant chemotherapy, a high intensity of LC3B staining was described as a (negative) independent prognostic factor for relapse-free and overall survival,27 in particular in triple-negative cancers.31 A recent meta-analysis confirmed that high expression of LC3B predicts adverse OS in BC.32 However, none of the aforementioned studies was based on the quantification of LC3B+ puncta, and rather scored the overall expression level of LC3B, meaning that the evaluation criteria was rather different from those reported here. We observed that the intensity of LC3B did not correlate with the presence of LC3B+ puncta. Importantly, the presence of LC3B+ puncta did correlate with a reduction in the abundance of cytoplasmic SQSTM1 expression, suggesting that negativity for LC3B+ puncta does reflect reduced autophagic flux.
HMGB1 has also been evaluated in BC in previous studies showing that more advanced tumors tend to reduce HMGB1 expression.21 Indeed, cancers that are transplanted into mice are HMGB1+ when they are small and their HMGB1 expression level is reduced as they grow.21 The molecular mechanisms of this phenomenon are still elusive. After neoadjuvant chemotherapy with epirubicin and docetaxel, HMGB1 levels in the plasma increase, especially in those patients that reached a pathological complete response.33,34 These results are in apparent accordance with our present observation that patients with low HMGB1 expression in their cancer exhibit a worse outcome than patients with high HMGB1 expression.21 It will be interesting and necessary to combine the analysis of HMGB1 expression with that of TLR4 (which must be expressed on dendritic cells to mediate HMGB1-elicited anticancer immune responses).16 If extracellular HMGB1 truly acts via TLR4 to stimulate immunosurveillance, both factors (as well as loss-of-function alleles of TLR4) would be expected to be epistatic with respect to patient prognosis.
It is noteworthy that nuclear HMGB1 expression and LC3B+ puncta did not correlate among each other, in spite of prior observations that the release of HMGB1 from the nucleus into the cytoplasm may favor autophagy.14,15 This discrepancy might be explained by the pronounced biological heterogeneity of distinct tumors, each of which is genetically distinct. Alternatively, the functional interaction between HMGB1 and autophagy might be restricted to certain cell types, as this has been suggested by a recent work showing that HMGB1 is dispensable for autophagy to occur in hepatocytes and cardiomyocytes in vivo.35 We must emphasize that the absent correlation between nuclear HMGB1 staining and cytoplasmic LC3B+ puncta that we report here does not invalidate the possibility that HMGB1 regulates autophagy in specific tissues.
Irrespective of these considerations, it appears clear that HMGB1 and LC3B can be considered as independent biomarkers that, if analyzed together, allow for an improved risk stratification of BC patients, especially in poor prognosis patients. One of the limitations of this approach, though, is the rather low proportion of BC patients that are double positive for nuclear HMGB1 and cytoplasmic L3C3B+ puncta. It will be important to further evaluate the likely immunological impact of these parameters by correlating double positivity with the composition of the immune infiltrate, and to evaluate these parameters (as well as their evolution) with regard to the sensitivity of BC xenografts to chemotherapy. This kind of approach will finally allow to weigh the relative contribution of cell-autonomous and immunological factors determining patient prognosis. In addition, therapeutic measures designed to compensate for defective HMGB1 expression by providing synthetic ligands of TLR4 are in preclinical development.21 Along the same line, it will be interesting to investigate whether pharmacological agents that induce autophagy in vivo36,37 may improve therapeutic responses in BC.
Patients and Methods
Training cohort of patients
The training cohort included 152 patients treated at Georges François Leclerc Cancer Center (Dijon, France) for localized HER2-negative BC between 1998 and 2007. All patients received 6 cycles of anthracyclines-based adjuvant chemotherapy (FEC100: epirubicin 100 mg/m2, cyclophosphamide 500 mg/m2, and 5-fluorouracil 500 mg/m2). After adjuvant chemotherapy, all patients with hormone receptor-positive disease received endocrine therapy for 5 y. Radiation therapy was given to all patients treated with conservative surgery and was performed after mastectomy if recommended according to the local risk of relapse. For each patient formalin-fixed paraffin-embedded (FFPE) tissue blocks of BC surgical specimen were retrieved from the department of pathology, and 4-µm-thick slides were stained for hematoxylin, as well as for LC3B and HMGB1. The study was approved by the local ethics committee, and all patients gave written informed consent at the time of the diagnosis for the use of tumor samples for research purposes (national French authorization number: AC-2008-69).
Design of the working hypothesis
We filed the working hypothesis that HMGB1 and LC3B may be valuable factors predicting the residual risk of relapse at the Win tumor biomarker registery (http://win.biomarkerregistry.org/study.jsp?id=505 on October 24, 2012). To prospectively validate the prognostic significance of these immunogenic cell death (ICD) markers in determining the residual risk of relapse in localized BC, we analyzed 1581 tissue microarrays from patients with node-positive LBC enrolled in the PACS04 Phase III trial (clinicaltrials.gov NCT 00054587).
Validation cohort of patients
The PACS04 trial included 3,010 patients with nonmetastatic node-positive BC between 2001 and 2004. This randomized phase III trial compared 6 cycles of FEC100 with 6 cycles of ED75 (epirubicin + docetaxel, each at 75 mg/m2). Radiotherapy was prescribed after conservative surgery, and according to local guidelines. Adjuvant hormone therapy was prescribed to patients with tumors that were positive for hormone receptors. Patients overexpressing HER2 (19% of patients) were randomized to either one year of trastuzumab, or placebo. From the 3,010 participants enrolled in the PACS04 trial, 1836 patients gave written consent for parallel biomarker studies on tumor blocks. These surgical specimens were centrally available at the UNICANCER tumor bank (Department of Pathology of Jean Perrin Cancer Center, Clermont-Ferrand, France). Tumor tissue was identified within the surgical specimen, and processed for their inclusion in tissue microarrays for 1836 patients included in the trial. For 255 patients, material processing did not result in sufficient cancer tissue, so that HMGB1 and LC3B expression was finally evaluated in 1581 tumors only.
Construction of tissue microarrays
FFPE tissue blocks from BC surgical specimen were retrieved from the UNICANCER tumor bank. Hematoxylin/eosin-stained slides from each block were reviewed by a pathologist to identify tumor areas. Tissue microarrays (TMAs) were constructed with 0.6 mm diameter tissue cores from representative tumor areas from FFPE blocks. Cores were taken from viable representative tumor areas (only on breast surgical specimen) and then transferred to a paraffin block using a semiautomated tissue array instrument. Triplicate tissue cores were taken from each specimen, resulting in a composite TMA blocks. Control tissues from normal breast were also included. Multiple 4-μm-thick sections were cut for staining.
Immunohistochemical detection of HMGB1
Formalin-fixed, paraffin-embedded cancer tissue sections were deparaffinized with 3 successive passages through xylene, and rehydrated through decreasing concentrations (100%, 95%, 80%, 70% and 50%) of ethanol. Antigen retrieval was carried out by heating slides for 20 min in pH 6.0 citrate buffer (10 mM Sodium Citrate, 0.05% Tween 20) at 98°C. Endogenous peroxidase activity was inhibited with 3% hydrogen peroxidase (DAKO, S2001) for 10 min. Sections were then saturated 20 min with Protein Block Serum Free (DAKO, X0909). Without washing, the primary antibody, a polyclonal rabbit anti HMGB1 antibody (Thermo Fisher Scientific, Pierce, PA1-16926), was incubated overnight, followed by the secondary antibody (EnVision-Rabbit, DAKO K4011) for 30 min, and streptavidin-horseradish peroxidase (DAKO, P0397) was added for an additional 30 min. Peroxidase activity was revealed by means of daminobenzidine substrate (DAB, DAKO, K3468), and the sections were counterstained with Mayer hematoxylin (DAKO, S3309).21
Immunohistochemical detection of LC3B
We previously described a validated immunohistochemical protocol for the detection of LC3B puncta in human FFPE cancer specimens.20 Briefly, immunohistochemical staining of cancer tissue sections was performed using the Novolink kit (Menarini Diagnostics, RE7140-K). Deparaffinized and heated tissue sections (as above) were allowed to cool down (45 min, room temperature) and mounted on Shandon Sequenza coverplates (Thermo Fisher Scientific, 72-199-50) in distilled water, and then washed twice for 5 min with 0.1% Tween 20 (Sigma-Aldrich, P1379) v/v in phosphate-buffered saline (PBS, Life Technologies 10010-023). Thereafter, sections were incubated for 5 min with the Peroxidase Block reagent (Novolink kit, Menarini Diagnostics, RE7140-K), and subsequently washed twice for 5 min with 0.1% Tween 20 (v/v in PBS). Following a 5-min-long incubation at room temperature with the Protein Block reagent (Novolink kit, Menarini Diagnostics, RE7140-K), tissue sections were washed twice for 5 min with 0.1% Tween 20 (v/v in PBS), and then incubated overnight at 4°C with a primary antibody specific for LC3B (clone 5F10, Nanotools, 0231-100), dissolved in 1% bovine serum albumin (Sigma Aldrich, A2058) w/v in tris-buffered saline (TBS, Sigma-Aldrich, T5912) at the final concentration of 25 µg/mL. This antibody recognizes both the soluble (LC3-I) and the membrane-bound (LC3-II) form of LC3B. After 2 washes in 0.1% Tween 20 (v/v in PBS), sections were incubated for 30 min with the Post Primary Block reagent (Novolink kit, Menarini Diagnostics, RE7140-K), washed again as before and incubated for 30 min with secondary antibodies coupled to horseradish peroxidase. Upon 2 additional washes, secondary antibodies were revealed with the liquid DAB Substrate Chromogen system (Novolink kit, Menarini Diagnostics, RE7140-K) with 10 min treatment. Finally, slides were washed in distilled water, and counterstained with hematoxylin.
Immunohistochemical detection of SQSTM1
Immunoperoxidase staining for SQSTM1 (guinea pig polyclonal antibody, Progen Biotechnik, GP62-C) was performed using the Ventana Benchmark XT automated slide preparation system.38 Briefly, tissue sections (4 to 5-mm thickness) were deparaffinized (EZ-Prep, Ventana Medical Systems, 950–102) at 75°C followed by antigen retrieval (Cell Conditioning 1, Ventana Medical Systems, 950–124) at 95 to 100°C. Antibodies were incubated at room temperature for 2 h, at a 1:400 dilution. Antibody staining was developed using the UltraView Universal DAB detection system (Ventana Medical Systems, 760–500), and counterstained with hematoxylin.
Pathological assessment
For HMGB1 staining, the pattern of expression (nuclear or not) was evaluated in tumor cells: strong nuclear staining of at least 50% of the tumor cells was considered positive for HMGB1 tumor expression (see Results and Fig. S1). The intensity of diffuse cytoplasmic LC3B staining was evaluated in the training cohort on the basis of the proportion of stained cells.39 The proportion of stained cells was evaluated (from 0 to 100%), and immunostaining intensity was graded as 0 (negative), 1 (weak), 2 (moderate), or 3 (strong). Moreover, the presence of LC3B puncta in tumor cells and the percentage of tumor cells with detectable LC3B puncta were assessed, independently of the intensity of diffuse cytoplasmic staining (Fig. S1). The correlation between LC3B diffuse cytoplasmic staining intensity and percent of tumor cells with puncta was evaluated. Adequate cutoff values, were determined both for diffuse cytoplasmic, and puncta staining in order to determine the best prognostic factor. For the training cohort, considering the possibility of tumor heterogeneity, all these LC3B pathological evaluations were done on the whole tumor area and at least 15 high-power fields (x20). For the validation cohort (TMA), pathological evaluations were performed on the whole tumor area on each TMA spot. HMGB1 and LC3B expressions were independently assessed by a trained histologist (SL) and a trained BC-pathologist (FPL), both blinded for clinicopathological data. Discrepancies between the 2 observers were reviewed jointly with a second trained BC pathologist (LA) to reach consensus. HMGB1 expression and LC3B expression were evaluated separately and correlated with clinicopathological parameters assessed on surgical specimen, and long-term outcome. Positive hormone receptor (HR) status was defined as 10% of tumor cells expressing either estrogen or progesterone receptor. Positive HER2 status was defined by immunohistochemistry or by fluorescent in situ hybridization.
Statistical analysis
The association between HMGB1 expression, LC3B puncta detection, and disease characteristics was examined using the Chi2 test, the Fisher exact probability test, or Mann-Whitney test, as appropriate. MFS was defined as the time elapsed from the date of randomization to the date of the first distant recurrence, or death from any cause, whichever occurred first. BCSS was defined as the time elapsed from the date of randomization until death from BC. Patients who were alive and relapse-free at the last contact were censored at the last follow-up date. Overall survival (OS) was defined as the time from date of randomization until death from any cause. Survivors or patients that were lost to follow-up were censored at the last follow-up date. Median follow-up and 95% CI was calculated using the reverse Kaplan-Meier method. The Kaplan-Meier method was used to calculate survival probabilities according to HMGB1 and/or LC3B tumor status. The log-rank test was used for comparison of survival curves. The Cox proportional hazards regression was used for univariate and multivariate analyses of disease-free survival, MFS, OS, and BCSS. The assumption of proportional hazards was assessed by testing the relationship between scaled Schoenfeld residuals and the rank of the survival time at the level of each covariate in the model. No major violation of the proportional hazard assumption that would invalidate our findings was detected.
To determine adequate cutoff values allowing to patient survival stratification, methods of the martingale residuals for the MFS Cox regression model, and of the log-rank statistics all possible cutpoints, were used.40
All analyses were performed using Stata V11 software (StataCorp LP, College Station, TX). P values were 2-tailed and considered significant when <0.05. Results were reported following the recommendations for tumor marker prognostic studies (REMARK criteria). 41
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
GK is supported by the Ligue contre le Cancer (équipe labelisée); Agence National de la Recherche (ANR); Association pour la recherche sur le cancer (ARC); Cancéropôle Ile-de-France; Institut National du Cancer (INCa); Fondation Bettencourt-Schueller; Fondation de France; Fondation pour la Recherche Médicale (FRM); the European Commission (ArtForce); the European Research Council (ERC); the LabEx Immuno-Oncology; the SIRIC Stratified Oncology Cell DNA Repair and Tumor Immune Elimination (SOCRATE); the SIRIC Cancer Research and Personalized Medicine (CARPEM); and the Paris Alliance of Cancer Research Institutes (PACRI).
Supplementary Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1.Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thurlimann B, et al.. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol 2013; 24:2206-23; PMID:23917950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al.. Molecular portraits of human breast tumours. Nature 2000; 406:747-52; PMID:10963602; http://dx.doi.org/ 10.1038/35021093 [DOI] [PubMed] [Google Scholar]
- 3.Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al.. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 2001; 98:10869-74; PMID:11553815; http://dx.doi.org/ 10.1073/pnas.191367098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al.. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 2002; 347:1999-2009; PMID:12490681; http://dx.doi.org/ 10.1056/NEJMoa021967 [DOI] [PubMed] [Google Scholar]
- 5.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med 2009; 360:790-800; PMID:19228622; http://dx.doi.org/ 10.1056/NEJMra0801289 [DOI] [PubMed] [Google Scholar]
- 6.Andre F, Nowak F, Arnedos M, Lacroix L, Viens P, Calvo F. Biomarker discovery, development, and implementation in France: a report from the French National Cancer Institute and cooperative groups. Clin Cancer Res 2012; 18:1555-60; PMID:22422408; http://dx.doi.org/ 10.1158/1078-0432.CCR-11-2201 [DOI] [PubMed] [Google Scholar]
- 7.Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, Kalachikov S, Gilliam TC, Levine B. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics 1999; 59:59-65; PMID:10395800; http://dx.doi.org/ 10.1006/geno.1999.5851 [DOI] [PubMed] [Google Scholar]
- 8.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A 2003; 100:15077-82; PMID:14657337; http://dx.doi.org/ 10.1073/pnas.2436255100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al.. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 2003; 112:1809-20; PMID:14638851; http://dx.doi.org/ 10.1172/JCI20039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cicchini M, Chakrabarti R, Kongara S, Price S, Nahar R, Lozy F, Zhong H, Vazquez A, Kang Y, Karantza V. Autophagy regulator BECN1 suppresses mammary tumorigenesis driven by WNT1 activation and following parity. Autophagy 2014; 10:2036-52; PMID:25483966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang H, Sebti S, Titone R, Zhou Y, Isidoro C, Ross TS, Hibshoosh H, Xiao G, Packer M, Xie Y, et al.. Decreased mRNA Expression in Human Breast Cancer is Associated with Estrogen Receptor-Negative Subtypes and Poor Prognosis. EBioMedicine 2015; 2:255-63; PMID:25825707; http://dx.doi.org/ 10.1016/j.ebiom.2015.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenfeldt MT, O'Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, Au A, Rai TS, Zheng L, Ridgway R, et al.. p53 status determines the role of autophagy in pancreatic tumour development. Nature 2013; 504:296-300; PMID:24305049; http://dx.doi.org/ 10.1038/nature12865 [DOI] [PubMed] [Google Scholar]
- 13.Rao S, Tortola L, Perlot T, Wirnsberger G, Novatchkova M, Nitsch R, Sykacek P, Frank L, Schramek D, Komnenovic V, et al.. A dual role for autophagy in a murine model of lung cancer. Nat Commun 2014; 5:3056; PMID:24445999; http://dx.doi.org/ 10.1038/ncomms4056 [DOI] [PubMed] [Google Scholar]
- 14.Tang D, Kang R, Livesey KM, Cheh CW, Farkas A, Loughran P, Hoppe G, Bianchi ME, Tracey KJ, Zeh HJ 3rd, et al.. Endogenous HMGB1 regulates autophagy. J Cell Biol 2010; 190:881-92; PMID:20819940; http://dx.doi.org/ 10.1083/jcb.200911078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang D, Kang R, Livesey KM, Kroemer G, Billiar TR, Van Houten B, Zeh HJ 3rd, Lotze MT. High-mobility group box 1 is essential for mitochondrial quality control. Cell Metab 2011; 13:701-11; PMID:21641551; http://dx.doi.org/ 10.1016/j.cmet.2011.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A, Mignot G, Maiuri MC, Ullrich E, Saulnier P, et al.. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat Med 2007; 13:1050-9; PMID:17704786; http://dx.doi.org/ 10.1038/nm1622 [DOI] [PubMed] [Google Scholar]
- 17.Michaud M, Martins I, Sukkurwala AQ, Adjemian S, Ma Y, Pellegatti P, Shen S, Kepp O, Scoazec M, Mignot G, et al.. Autophagy-dependent anticancer immune responses induced by chemotherapeutic agents in mice. Science 2011; 334:1573-7; PMID:22174255; http://dx.doi.org/ 10.1126/science.1208347 [DOI] [PubMed] [Google Scholar]
- 18.Martins I, Wang Y, Michaud M, Ma Y, Sukkurwala AQ, Shen S, Kepp O, Métivier D, Galluzzi L, Perfettini JL, et al.. Molecular mechanisms of ATP secretion during immunogenic cell death. Cell Death Differ 2014; 21:79-91; PMID:23852373; http://dx.doi.org/ 10.1038/cdd.2013.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghiringhelli F, Apetoh L, Tesniere A, Aymeric L, Ma Y, Ortiz C, Vermaelen K, Panaretakis T, Mignot G, Ullrich E, et al.. Activation of the NLRP3 inflammasome in dendritic cells induces IL-1beta-dependent adaptive immunity against tumors. Nat Med 2009; 15:1170-8; PMID:19767732; http://dx.doi.org/ 10.1038/nm.2028 [DOI] [PubMed] [Google Scholar]
- 20.Ladoire S, Chaba K, Martins I, Sukkurwala AQ, Adjemian S, Michaud M, Poirier-Colame V, Andreiuolo F, Galluzzi L, White E, et al.. Immunohistochemical detection of cytoplasmic LC3 puncta in human cancer specimens. Autophagy 2012; 8:1175-84; PMID:22647537; http://dx.doi.org/ 10.4161/auto.20353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamazaki T, Hannani D, Poirier-Colame V, Ladoire S, Locher C, Sistigu A, Prada N, Adjemian S, Catani JP, Freudenberg M, et al.. Defective immunogenic cell death of HMGB1-deficient tumors: compensatory therapy with TLR4 agonists. Cell Death Differ 2014; 21:69-78; PMID:23811849; http://dx.doi.org/ 10.1038/cdd.2013.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, et al.. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 2008; 4:151-75; PMID:18188003; http://dx.doi.org/ 10.4161/auto.5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanida I, Waguri S. Measurement of autophagy in cells and tissues. Methods Mol Biol 2010; 648:193-214; PMID:20700714; http://dx.doi.org/ 10.1007/978-1-60761-756-3_13 [DOI] [PubMed] [Google Scholar]
- 24.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agnello M, Agostinis P, Aguirre-Ghiso JA, Ahn HJ, et al.. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8:445-544; PMID:22966490; http://dx.doi.org/ 10.4161/auto.19496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ladoire S, Hannani D, Vetizou M, Locher C, Aymeric L, Apetoh L, Kepp O, Kroemer G, Ghiringhelli F, Zitvogel L, et al.. Cell-death-associated molecular patterns as determinants of cancer immunogenicity. Antioxid Redox Signal 2013; 20:1098-116; PMID:23394620; http://dx.doi.org/ 10.1089/ars.2012.5133 [DOI] [PubMed] [Google Scholar]
- 26.Donegan WL. Tumor-related prognostic factors for breast cancer. CA Cancer J Clin 1997; 47:28-51; PMID:8996077 [DOI] [PubMed] [Google Scholar]
- 27.Chen S, Jiang YZ, Huang L, Zhou RJ, Yu KD, Liu Y, Shao ZM. The residual tumor autophagy marker LC3B serves as a prognostic marker in local advanced breast cancer after neoadjuvant chemotherapy. Clin Cancer Res 2013; 19:6853-62; PMID:24141623; http://dx.doi.org/ 10.1158/1078-0432.CCR-13-1617 [DOI] [PubMed] [Google Scholar]
- 28.Suman S, Das TP, Reddy R, Nyakeriga AM, Luevano JE, Konwar D, Pahari P, Damodaran C. The pro-apoptotic role of autophagy in breast cancer. Br J Cancer 2014; 111:309-17; PMID:24945999; http://dx.doi.org/ 10.1038/bjc.2014.203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim JY, Jung WH, Koo JS. Expression of Autophagy-Related Proteins According to Androgen Receptor and HER-2 Status in Estrogen Receptor-Negative Breast Cancer. PLoS One 2014; 9:e105666; PMID:25140630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lazova R, Camp RL, Klump V, Siddiqui SF, Amaravadi RK, Pawelek JM. Punctate LC3B expression is a common feature of solid tumors and associated with proliferation, metastasis, and poor outcome. Clin Cancer Res 2012; 18:370-9; PMID:22080440; http://dx.doi.org/ 10.1158/1078-0432.CCR-11-1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao H, Yang M, Zhao J, Wang J, Zhang Y, Zhang Q. High expression of LC3B is associated with progression and poor outcome in triple-negative breast cancer. Med Oncol 2013; 30:475; PMID:23371253; http://dx.doi.org/ 10.1007/s12032-013-0475-1 [DOI] [PubMed] [Google Scholar]
- 32.He Y, Zhao X, Subahan NR, Fan L, Gao J, Chen H. The prognostic value of autophagy-related markers beclin-1 and microtubule-associated protein light chain 3B in cancers: a systematic review and meta-analysis. Tumour Biol 2014; 35:7317-26; PMID:24838948; http://dx.doi.org/ 10.1007/s13277-014-2060-4 [DOI] [PubMed] [Google Scholar]
- 33.Arnold T, Michlmayr A, Baumann S, Burghuber C, Pluschnig U, Bartsch R, Steger G, Gnant M, Bergmann M, Bachleitner-Hofmann T, et al.. Plasma HMGB-1 after the initial dose of epirubicin/docetaxel in cancer. Eur J Clin Invest 2013; 43:286-91; PMID:23410002; http://dx.doi.org/ 10.1111/eci.12043 [DOI] [PubMed] [Google Scholar]
- 34.Stoetzer OJ, Fersching DM, Salat C, Steinkohl O, Gabka CJ, Hamann U, Braun M, Feller AM, Heinemann V, Siegele B, et al.. Circulating immunogenic cell death biomarkers HMGB1 and RAGE in breast cancer patients during neoadjuvant chemotherapy. Tumour Biol 2013; 34:81-90; PMID:22983919; http://dx.doi.org/ 10.1007/s13277-012-0513-1 [DOI] [PubMed] [Google Scholar]
- 35.Huebener P, Gwak GY, Pradere JP, Quinzii CM, Friedman R, Lin CS, Trent CM, Mederacke I, Zhao E, Dapito DH, et al.. High-mobility group box 1 is dispensable for autophagy, mitochondrial quality control, and organ function in vivo. Cell Metab 2014; 19:539-47; PMID:24606906; http://dx.doi.org/ 10.1016/j.cmet.2014.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov 2012; 11:709-30; PMID:22935804; http://dx.doi.org/ 10.1038/nrd3802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madeo F, Pietrocola F, Eisenberg T, Kroemer G. Caloric restriction mimetics: towards a molecular definition. Nat Rev Drug Discov 2014; PMID:25212602 [DOI] [PubMed] [Google Scholar]
- 38.Lee HS, Daniels BH, Salas E, Bollen AW, Debnath J, Margeta M. Clinical utility of LC3 and p62 immunohistochemistry in diagnosis of drug-induced autophagic vacuolar myopathies: a case-control study. PLoS One 2012; 7:e36221; PMID:22558391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi J, Jung W, Koo JS. Expression of autophagy-related markers beclin-1, light chain 3A, light chain 3B and p62 according to the molecular subtype of breast cancer. Histopathology 2013; 62:275-86; PMID:23134379; http://dx.doi.org/ 10.1111/his.12002 [DOI] [PubMed] [Google Scholar]
- 40.Hothorn T, Lausen B.. On the exact distribution of maximally selected rank statistics. Computational Statistics & Data Analysis 2003; 43:121-37; http://dx.doi.org/ 10.1016/S0167-9473(02)00225-6 [DOI] [Google Scholar]
- 41.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. Reporting recommendations for tumor marker prognostic studies (REMARK). J Natl Cancer Inst 2005; 97:1180-4; PMID:16106022; http://dx.doi.org/ 10.1093/jnci/dji237 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.