Abstract
Currently, targeting the autophagic pathway is regarded as a promising new strategy for cancer drug discovery. Here, we screened the North China Pharmaceutical Group Corporation's pure compound library of microbial origin using GFP-LC3B-SKOV3 cells and identified elaiophylin as a novel autophagy inhibitor. Elaiophylin promotes autophagosome accumulation but blocks autophagic flux by attenuating lysosomal cathepsin activity, resulting in the accumulation of SQSTM1/p62 in various cell lines. Moreover, elaiophylin destabilizes lysosomes as indicated by LysoTracker Red staining and CTSB/cathepsin B and CTSD/ cathepsin D release from lysosomes into the cytoplasm. Elaiophylin eventually decreases cell viability, especially in combination with cisplatin or under hypoxic conditions. Furthermore, administration of a lower dose (2 mg/kg) of elaiophylin as a single agent achieves a significant antitumor effect without toxicity in an orthotopic ovarian cancer model with metastasis; however, high doses (8 mg/kg) of elaiophylin lead to dysfunction of Paneth cells, which resembles the intestinal phenotype of ATG16L1-deficient mice. Together, these results provide a safe therapeutic window for potential clinical applications of this compound. Our results demonstrate, for the first time, that elaiophylin is a novel autophagy inhibitor, with significant antitumor efficacy as a single agent or in combination in human ovarian cancer cells, establishing the potential treatment of ovarian cancer by this compound.
Keywords: antitumor, autophagy, cell death, elaiophylin, natural compound, ovarian cancer
Abbreviations
- 3-MA
3-methyladenine
- AO
acridine orange
- ATG5
autophagy-related 5
- ATG16L1
autophagy-related 16-like 1 (S. cerevisiae)
- BECN1
Beclin 1, autophagy-related
- CCK-8
Cell Counting Kit-8
- CQ
chloroquine
- CTSB
cathepsin B
- CTSD
cathepsin D
- DDP
cisplatin
- DMSO
dimethyl sulfoxide
- EBSS
Earle's balanced salt solution
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GFP
green fluorescent protein
- HIF1A
hypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor)
- LAMP1
lysosomal-associated membrane protein 1
- MAP1LC3B/LC3B
microtubule-associated protein 1 light chain 3 beta
- LMP
lysosomal membrane permeabilization
- MEF
mouse embryonic fibroblast
- NCPC
North China Pharmaceutical Group Corporation
- PARP1
poly (ADP-ribose) polymerase 1
- PECAM1/CD31
platelet/endothelial cell adhesion molecule 1
- RFP
red fluorescent protein
- SQSTM1
sequestosome 1
- TUNEL
terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling.
Introduction
Ovarian cancer is the fourth leading cause of cancer death in women and causes more deaths than any other female genital tract cancer.1 The high mortality rate of ovarian cancer patients is due to the difficulty in diagnosing the disease at an early stage, a tendency to relapse, and drug resistance.2 Although initial chemotherapy with cisplatin and its derivatives is effective in most patients, the relapse rate remains high and tumors become increasingly resistant to cisplatin. These facts help explain why the 5-year survival rate of this disease remains below 40%, unchanged for the past 30 y.3
Considerable effort has been devoted to identifying novel and more reliable biomarkers for early disease detection and targeted molecular therapy. This clinical phenomenon highlights the need for more efficacious therapies. Previously, dysregulation of the autophagy pathway in ovarian cancer cells was shown to participate in tumor dormancy and radio or chemoresistance.4,5 The results of a recent study indicate that autophagy mediated by MAPK1/ERK2-MAPK3/ERK1 can lead to cisplatin resistance, suggesting that inhibition of autophagy might overcome cisplatin resistance in ovarian cancer cells.6 Therefore, targeting the autophagy process in ovarian cancers may have considerable efficacy for disease treatment.
Autophagy, a lysosome-dependent pathway, is a complex catabolic process that involves the degradation of dysfunctional or useless cytoplasmic constituents.7-9 The clinical relevance of autophagy stems from its potential to protect cells against a variety of intracellular and extracellular stress signals and to favor tumor development. 10,11Paradoxically, autophagy has also been associated with cell death, because excessive or persistent autophagy can promote cell killing by depleting key organelles (e.g., endoplasmic reticulum [ER] or mitochondria), rewiring survival signals, deregulating lysosomal enzymes, and/or activating caspase-dependent apoptotic programs.12-16
Therefore, to identify novel compounds modulating autophagy in ovarian cancer, we generated SKOV3 cells that constitutively express green fluorescent protein (GFP)-tagged microtubule-associated protein 1 light chain 3 β (LC3B) (GFP-LC3B). Using GFP-LC3B as an autophagosomal marker, we established a cell-based high-throughput screening model based on iSort image cytometry that enabled us to image and quantitatively analyze GFP-LC3B in SKOV3 cells. By screening the North China Pharmaceutical Group Corporation's (NCPC) pure compound library of microbial origin, we determined that elaiophylin, a C2 symmetry 16-member macrodiolide antibiotic extracted from Streptomyces melanosporus,17 promotes autophagosome accumulation but blocks autophagic flux in the late stage of the autophagy process, ultimately inhibiting autophagy in ovarian carcinoma cells. This novel autophagy inhibitor eventually induces cell death, an effect that was amplified when combined with cisplatin or under hypoxic conditions. More excitingly, elaiophylin exerts a significant antitumor effect as a single agent in an orthotopic ovarian cancer model with metastasis, suggesting the therapeutic potential of this compound for the treatment of ovarian cancer.
Results
A functional screen to identify elaiophylin as a novel autophagy modulator from natural compounds
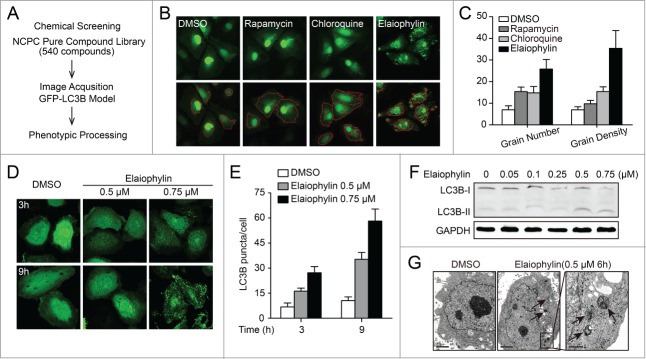
To identify novel compounds modulating autophagy in ovarian cancer, we generated SKOV3 cells constitutively expressing GFP-LC3B and screened 540 compounds from the NCPC pure compound library of microbial origin. The effect of each compound on GFP-LC3B-SKOV3 cells was monitored separately with fluorescence using iSort image cytometry. Chloroquine (CQ) and rapamycin served as positive controls and dimethyl sulfoxide (DMSO) served as the negative control. After quantitative analysis of the number and density of GFP-LC3B dots in each transfected cell, elaiophylin was identified as a potent candidate and earmarked for further analysis (Fig. 1A to C).
Figure 1.
Identification of elaiophylin as a novel autophagy modulator. (A) Schematic of functional screening for autophagy-related compounds based on automated fluorescence imaging. (B) Representative images of SKOV3 cells expressing GFP-LC3B (green) upon treatment with DMSO (negative control), chloroquine (positive control), rapamycin (positive control), or elaiophylin. iSort image cytometry was used to detect and count granules in cells and to measure the physical characteristics of granules. The bottom panels show results of image segmentation used for quantification of the GFP-LC3B puncta (red, autophagosomal GFP-LC3B). (C) The number of GFP-LC3B dots per cell was determined using iSort image cytometry after 12 h of treatment. The number of GFP-LC3B dots was counted in at least 3 independent visual fields from 3 independent wells. Results were expressed as the average grain number and grain density per cell. (D) Fluorescence imaging of the GFP-LC3B distribution in SKOV3 cells treated with elaiophylin. Cells treated with DMSO served as the negative control. (E) Quantification of data in (D), expressed as the numbers of GFP-LC3B puncta per cell at the indicated times after treatment. Error bars correspond to SEM of 3 independent experiments. (F) Elaiophylin induced autophagosome formation in SKOV3 cells, as revealed by LC3B-I to LC3B-II conversion on western blotting. (G) Effect of elaiophylin on SKOV3 cell morphology. Representative electron microscopy photomicrographs are shown (6 h). The arrow indicates autophagic vacuoles. Scale bars: 2 μm. Right panel is magnification of the boxed region (scale bars: 500 nm).
Next, we performed fluorescence assays and immunoblotting for LC3B to validate the effects of elaiophylin on autophagy. In SKOV3 cells stably expressing GFP-LC3B, elaiophylin increased numbers of fluorescent puncta per cell in a dose- and time-dependent manner (Fig. 1D to E). An enhanced LC3B conversion was further revealed by immunoblotting assays in 5 ovarian cancer cell lines (SKOV3, OVCAR3, A2780, CaOV-3, and SW626), several other human cancer cell lines (PC-3, HepG2, A549, and HeLa), and HEK-293 cells and mouse embryonic fibroblasts (MEFs) following incubation with elaiophylin. These results suggest that elaiophylin treatment universally resulted in autophagosome accumulation in mammalian cells (Fig. 1F; Fig. S1; Fig. S2). Transmission electron microscopy was used to further investigate the morphological changes induced by elaiophylin. Compared to control cells, elaiophylin-treated cells displayed representative ultrastructural morphological features of autophagosomes (double-membrane vacuolar structures containing undigested cytoplasmic contents), further highlighting the capacity of elaiophylin to promote autophagosome accumulation (Fig. 1G).
Autophagic flux was inhibited by elaiophylin treatment
Increased numbers of autophagosomes can be associated either with increased autophagosome synthesis or decreased autophagosome turnover. To distinguish between these 2 possibilities, we measured the autophagic flux. First, we analyzed autophagy in the presence of autophagy inhibitors, such as 3-methyladenine (3-MA) and CQ in GFP-LC3B-SKOV3 cells. Coincubation of elaiophylin (0.5 μM) with 3-MA (10 mM), which blocks the upstream steps of autophagy,18 reduced the accumulation of GFP-LC3B puncta. In contrast, coincubation of elaiophylin (0.5 μM) with CQ (25 μM), which blocks the downstream steps of autophagy,7 did not induce a significant increase in GFP-LC3B puncta compared with elaiophylin treatment alone (Fig. 2A and B), indicating that elaiophylin was not an autophagy inducer. Additionally, elaiophylin-treated cells did not exhibit an increase in LC3B-II levels in CQ-treated cells compared with control cells (Fig. 2C).
Figure 2.
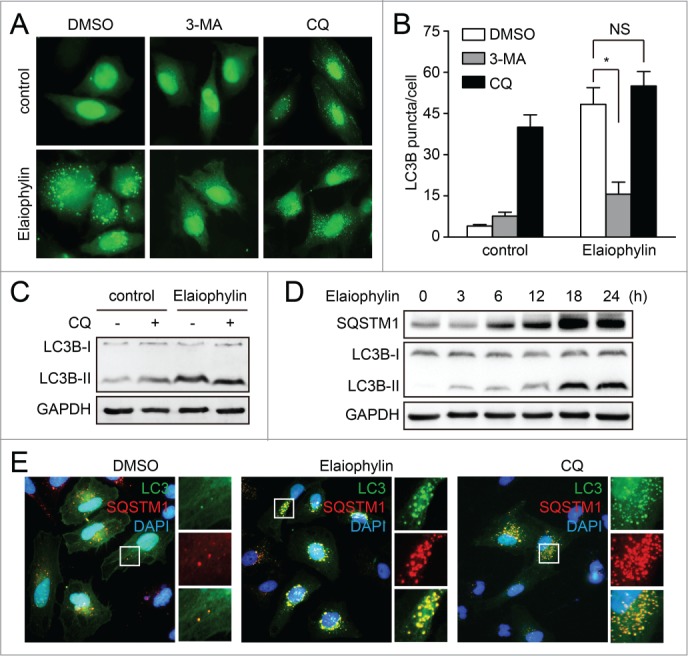
Elaiophylin inhibits autophagic flux. (A and B) GFP-LC3B-expressing SKOV3 cells were treated with DMSO, 3-MA (10 mM), or chloroquine (25 μM) alone or in the presence of elaiophylin (0.5 μM) for 12 h. GFP-LC3B puncta per cell were counted. Data are mean ± SEM for triplicate samples of at least 100 cells per sample (*P < 0.05). (C) SKOV3 cells were treated with elaiophylin (0.5 μM) alone or in the presence of chloroquine (25 μM) for 12 h. Conversion of LC3B-I to LC3B-II was evaluated by western blot. (D) SKOV3 cells were treated with elaiophylin (0.5 μM) for indicated time point. Protein expression was determined for LC3B and SQSTM1. (E) SKOV3 cells expressing GFP-LC3B were treated with DMSO, elaiophylin (0.5 μM), or chloroquine (25 μM) for 12 h, followed by staining with 4’,6-diamidino-2-phenylindole (blue) and anti-SQSTM1 (red). Panels on the right are higher-magnification images of the boxed regions. Original magnification: ×40.
To further investigate the role of elaiophylin in autophagy, we monitored the total amount of SQSTM1, which has been implicated in autophagic cargo recognition and is lost in the final stages of autophagy during autolysosome degradation.19 An increase in the amount of SQSTM1 is related to the inhibition of autophagy flux. Data presented by immunoblot revealed an accumulation of SQSTM1 following elaiophylin incubation in SKVO3 cells (Fig. 2D). Consistently, increased expression of SQSTM1 was found not only in 4 other ovarian cancer cell lines (SW626, OVCAR3, A2780, CaOV-3) but also in several other human cancer cell lines (PC-3, HepG2, A549, and HeLa), as well as in HEK-293 and mouse embryonic fibroblasts (MEFs) (Fig. S3; Fig. S4). These results suggest that elaiophylin universally interrupted autophagy flux in mammalian cells. This phenomenon was further confirmed by immunofluorescence-based analysis. As shown in Figure 2E, elaiophylin treatment led to the accumulation of SQSTM1 and GFP-LC3B puncta with extensive colocalization, similar to that seen following CQ treatment, indicating that elaiophylin inhibited the late stage of autophagy. Together, these results demonstrated that elaiophylin-induced autophagosome accumulation was due to impaired autophagic flux, indicating that elaiophylin was a potent autophagic flux inhibitor.
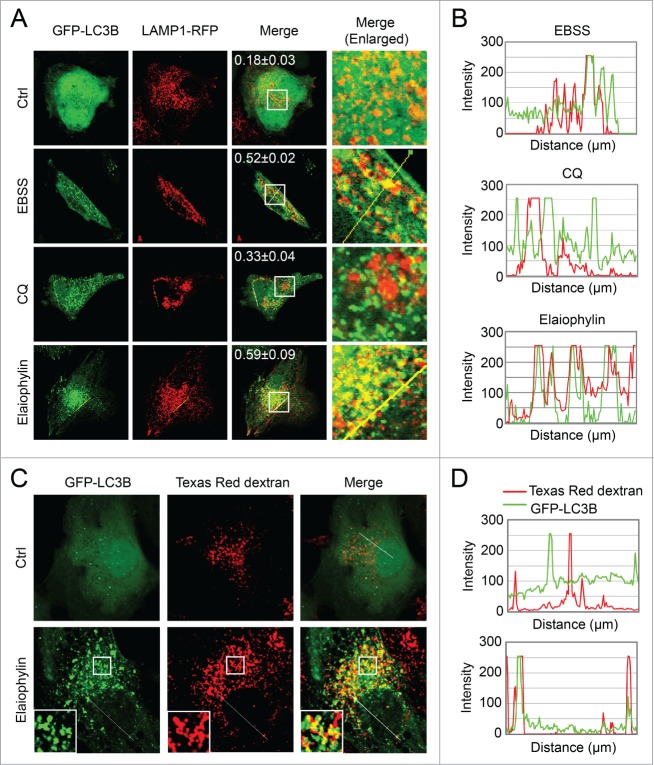
Fusion of autophagosomes with lysosomes or endosomes was not inhibited by elaiophylin
A probable explanation for the impaired autophagy flux induced by elaiophylin could be an inhibition of fusion of autophagosomes with late endosomes or lysosomes. To determine whether elaiophylin induced autophagosome fusion with lysosomes, we used confocal microscopy to assess the colocalization of GFP-LC3B with LAMP1, a marker for endosomal and lysosomal membranes (Fig. 3A and B). Notably, we found that elaiophylin induced a significant overlap between GFP-LC3B and LAMP1-RFP (Fig. 3A, Pearson correlation coefficient = 0.59) in SKOV3 cells similar to the effect of starvation (EBSS, Earle's balanced salt solution), indicating that autophagosome and lysosome fusion was not inhibited by elaiophylin treatment. In contrast, treatment with the lysosomotropic agent CQ as a negative control produced a significant separation of GFP-LC3B and LAMP1-RFP (Fig. 3A, Pearson correlation coefficient = 0.33). Furthermore, when monitoring fluid phase endocytosis by fluorescent dextran uptake, confocal analysis revealed a considerable colocalization of LC3B-positive autophagosomes with Texas Red dextran, indicating that elaiophylin did not block fluid-phase endocytosis (Fig. 3C and D).
Figure 3.
(See previous page). Elaiophylin does not block the fusion between the autophagosome with the lysosome or endosome. (A) LAMP1-RFP transfected SKOV3-GFP-LC3B cells were treated with elaiophylin (0.5 μM) or CQ (25 μM) for 12 h, or cultured in EBSS solution for 2 h. Fusion between autophagosomes (GFP-LC3B) and lysosomes (LAMP1-RFP) was evident in EBSS- and elaiophylin -treated cells (yellow in merged images). A complete separation of green and red signals was observed in CQ-treated cells. Numbers represent Pearson correlation coefficient as a statistic for quantifying colocalization calculated using ImageJ software. More than 30 cells were counted in each condition and data (mean± SD) are representative of 3 independent experiments. Original magnification: ×100. (B) The intensity profiles for both fluorescence channels of the white line positioned in (A) was measured using the Plot Profile function in the ImageJ software. (C) SKOV3-GFP-LC3B cells were incubated with Texas Red dextran (1 mg/mL) overnight followed by a 4 h chase. Cells preloaded with Texas Red dextran were then treated with or without 0.5 μM of elaiophylin for 12 h, and then subjected to confocal microscopy analysis without fixation. Insets highlight the colocalization in the main panels. Original magnification: ×100. (D) The intensity profiles for both fluorescence channels of the white line positioned in (C) was measured using the plot profile function in the ImageJ software. The regions where the peak signal for GFP-LC3B (green fluorescence) and the peak signal for Texas Red dextran (red fluorescence) overlapped were identified.
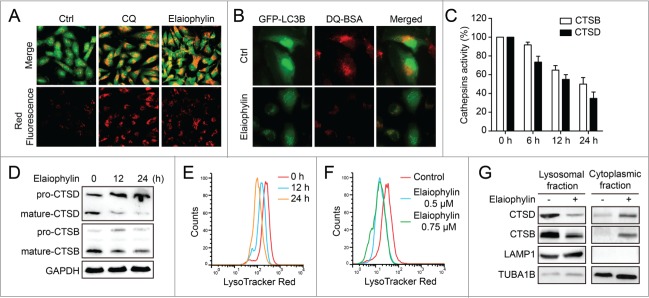
Elaiophylin blocks autophagic flux by attenuating lysosomal cathepsin activity
Because the impaired autophagic flux induced by elaiophylin was not caused by a blockade of fusion of autophagosomes with lysosomes or endosomes, it may be involved in the function of the lysosome. To this end, we examined the effects of elaiophylin on lysosomal function. First, because autophagosome and lysosome fusion is dependent on the pH in acidic compartments,20 we used acridine orange (AO; a dye that accumulates in intracellular acidic vesicles) staining to evaluate lysosomal pH. SKOV3 cells were treated for 12 h with DMSO, CQ or elaiophylin and stained with acridine orange. Both CQ and elaiophylin produced an accumulation of acidic vesicles, indicating that impaired autophagic flux induced by elaiophylin was not due to inhibition of lysosomal acidification (Fig. 4A).
Figure 4.
Elaiophylin does not interfere with acridine orange-staining of acidic compartments but downregulates lysosomal cathepsins. (A) Acridine orange (AO) staining in control, elaiophylin (0.5 μM, 12 h)- or CQ (25 μM, 12 h)-treated SKOV3 cells. The cytoplasm and nucleus essentially displayed green fluorescence, whereas the acidic compartments, including the lysosomes, displayed red fluorescence. Original magnification: ×40. (B) SKOV3-GFP-LC3B cells were incubated with DQ Red BSA (10 μg/ml) for 12 h. The cells were then washed twice with PBS before being treated with 0.5 μM elaiophylin for 12 h. The cells were fixed and analyzed for confocal microscopy. Original magnification: ×60. (C) Enzymatic activity of CTSB and CTSD in elaiophylin treated SKOV3 cells. Cells were treated with DMSO or elaiophylin (0.5 μM) at 6 h, 12 h, and 24 h as indicated. Enzymatic activity was analyzed using fluorogenic kits. Data are presented as the means ± SD from 3 independent experiments. (D) Western blotting analysis of endogenous CTSB and CTSD. (E) FACS analysis of LysoTracker Red after treatment with elaiophylin (0.5 μM) for 12 h and 24 h. (F) FACS analysis of LysoTracker Red after treatment with elaiophylin (0.5 μM or 0.75 μM) for 12 h. (G) Cell fractionation was performed to separate lysosomal and cytosolic fraction from DMSO- or elaiophylin-treated cells (24 h). CTSB and CTSD were detected by western blotting of the different fractions. A representative blot is shown, with LAMP1 as a lysosomal marker.
Next, to monitor lysosomal activity during elaiophylin treatment, cells were assayed for their ability to process DQ-BSA (a derivative of BSA), the red fluorescence of which is quenched unless it is cleaved by proteolytic enzymes.21 As shown in Figure 4B, very little dequenching of DQ-BSA occurred in elaiophylin-treated cells, indicating that intracellular proteolytic activity was inhibited in the presence of elaiophylin. Thus, we performed functional assays of lysosomal activity on elaiophylin-treated SKOV3 cells. Fluorogenic substrate assays were used to measure the enzymatic activity of CTSB and CTSD. Both CTSB and CTSD activities were reduced in a time-dependent manner following elaiophylin treatment, indicating that elaiophylin significantly inhibits the activity of cathepsins (Fig. 4C). Furthermore, we investigated the effects of elaiophylin on cathepsin processing by immunoblotting using antibodies recognizing the pro-form and the mature form of CTSB and CTSD. Consistently, elaiophylin significantly impaired the maturation of CTSB and CTSD (Fig. 4D). Taken together, our data demonstrated that elaiophylin impairs autophagic flux by inhibiting lysosomal cathepsin activity.
Elaiophylin treatment destabilized lysosomes
Because elaiophylin disrupts lysosomal degradation to inhibit the autophagy process, we next focused on examining the effect of elaiophylin on lysosomal integrity. We treated SKOV3 cells with elaiophylin for the indicated times and doses before the addition of LysoTracker Red, a dye that accumulates in acidic compartments. Elaiophylin induced a time- and dose-dependent decrease of LysoTracker Red fluorescence in SKOV3 cells, indicating that the volume of the acidic compartment decreases in a time- and dose-dependent manner following elaiophylin treatment (Fig. 4E and F). To further determine whether the reduction in the volume of the acidic compartment following elaiophylin treatment was due to lysosomal permeabilization, the localization of CTSB and CTSD was analyzed by western blotting. Treatment with elaiophylin caused a decrease in CTSB and CTSD in the lysosomal fraction and a concomitant increase in the cytoplasmic fraction (Fig. 4G). This altered distribution of CTSB and CTSD confirms the permeabilization of lysosomal membranes following elaiophylin treatment.
Elaiophylin-induced cell death was partially apoptotic
Next, we investigated the molecular modulator(s) of elaiophylin that might link autophagy inhibition to other biological processes in SKOV3 cells. Cells were treated with elaiophylin (0.5 μM) for 0, 3, 6, or 12 h prior to microarray analysis, followed by gene set enrichment analysis. Following elaiophylin treatment, a set of 409 genes belonging to the autophagy interaction network (which was defined by a proteomic approach22) were significantly upregulated (Fig. S5A). Consistent with this observation, the gene set enrichment analysis disclosed a significant enrichment of genes in the Mizushima_Autophagosome_Formation set in the elaiophylin-treated groups (Fig. S5B and S5C). These results were further validated by real-time PCR (Fig. S5D).
Interestingly, a sustained upregulation of proapoptotic genes such as those encoding DR5, BAK1, BBC3, and NOXA was identified and further validated by real-time PCR (Fig. S5E and S5F). To investigate whether elaiophylin-mediated autophagy inhibition and lysosomal dysfunction eventually affected cell survival, we used the CCK-8 assay to analyze the effects of elaiophylin on the viability of ovarian cancer cells. Elaiophylin treatment markedly induced cell death in SKOV3, A2780, and OVCAR3 cells (Fig. 5A). A synergistic effect was observed when combined with cisplatin in cisplatin-sensitive human ovarian cancer cell lines SKOV3 and A2780 (Fig. S6A and S6B). Importantly, combination elaiophylin and cisplatin treatment also significantly increased cell death in the cisplatin-resistant human ovarian cancer cell line C13* and in SKOV3-DDP, a cisplatin-resistant cell line established by increasing the concentrations of cisplatin to which parental SKOV3 cells were exposed, suggesting that elaiophylin circumvents resistance to cisplatin in ovarian cancer (Fig. S6A and S6C).
Figure 5.
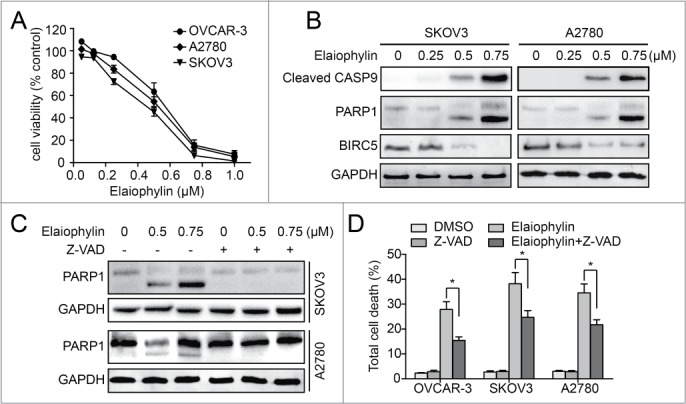
Elaiophylin-induced cell death is partially apoptotic in ovarian cancer cells. (A) Effect of elaiophylin on the viability of ovarian cancer cell lines (OVCAR3, A2780, and SKOV3). Data are mean ± SEM of 3 experiments. (B) SKOV3 and A2780 cells were treated with elaiophylin for 24 h. Protein levels of cleaved CASP9, PARP1, BIRC5, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were detected by western blot. (C) SKOV3 and A2780 cells were treated with elaiophylin in the presence or absence of Z-VAD-FMK. PARP1 and GAPDH were detected by western blot. (D) Flow cytometric analysis of cell death. SKOV3, OVCAR3, and A2780 cells were treated for 24 h with elaiophylin (0.5 μM) or DMSO in the presence or absence of Z-VAD-FMK (25 μM). Data are means ± standard deviation of 3 independent experiments (*P < 0.05).
The apoptotic cell death pathway was investigated by immunoblotting. In both the SKOV3 and A2780 cell lines, elaiophylin treatment led to significant activation of cleaved CASP9/caspase-9 and PARP1 and downregulation of BIRC5/survivin in a concentration-dependent manner (Fig. 5B). To elucidate whether caspase activation is required for elaiophylin-induced cell death, we exposed cells to the pancaspase inhibitor Z-VAD-FMK. Our immunoblot data showed that the addition of Z-VAD-FMK completely prevented PARP1 cleavage induced by elaiophylin (Fig. 5C). However, elaiophylin-induced cell death was only modestly rescued in all 3 tested ovarian cancer cell lines (Fig. 5D). With regard to cell-cycle arrest, even at doses high enough to largely inhibit cell proliferation, elaiophylin did not induce any significant change in the cell cycle (Fig. S7). Taken together, these experiments indicated that elaiophylin triggered cell death in a partially apoptotic manner.
Constitutive autophagy contributed to elaiophylin-induced cell death
To further explore the link between the inhibition of autophagy and cell viability by elaiophylin, we silenced the genes encoding key autophagy regulators ATG5 and BECN1 in SKOV3 cells. Immunoblots showed an 80% to 90% reduction in ATG5 or BECN1 levels in SKOV3 cells transfected with shRNA, revealing an effective knockdown of target proteins (Fig. 6A and B). Silencing of ATG5 or BECN1 in SKOV3 cells effectively attenuated elaiophylin-induced autophagosome accumulation, as indicated by a decrease in LC3B-II and LC3B puncta accumulation and a reduction in SQSTM1 accumulation (Fig. 6C, D and F). Importantly, downregulation of ATG5 or BECN1 obviously rescued elaiophylin-induced cleavage of PARP1 and significantly inhibited elaiophylin-induced cell death (Fig. 6C–E). These effects were specific to the reduction in ATG5 or BECN1 levels, because autophagosome accumulation and cell death were partially restored by the expression of shRNA-resistant ATG5 or BECN1 in ATG5 or BECN1 knockdown cells. These results suggest that constitutive autophagy contributed to elaiophylin-induced cell death (Fig. 6C–E).
Figure 6.
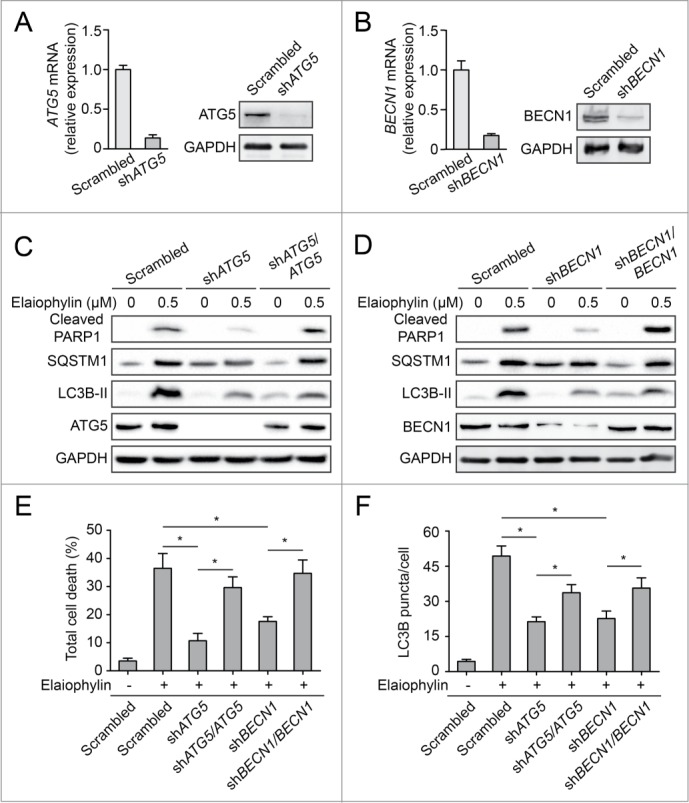
Constitutive autophagy contributes to elaiophylin-induced cell death. (A and B) Effect of shRNAs on the expression of proteins involved in autophagy. SKOV3 cells were transfected with ATG5 shRNA, BECN1 shRNA, or control (Scrambled) shRNA vectors. The expression of ATG5 and BECN1 was monitored by western blot and real-time PCR. (C and D) SKOV3 cells transfected with control shRNA, ATG5 shRNA or BECN1 shRNA, as well as with exogenous shRNA-resistant ATG5 and ATG5 shRNA, or with exogenous shRNA-resistant BECN1 and BECN1 shRNA were treated with elaiophylin (0.5 μM) as indicated for 24 h. PARP1, SQSTM1, LC3B, ATG5, and BECN1 were detected by western blot. (E) Analysis of cell death via flow cytometry of cells from Figure 6C and D. Mean ± SEM of at least 3 independent experiments is presented (*P < 0.05). (F) SKOV3-GFP-LC3B cells transfected with control shRNA, ATG5 shRNA or BECN1 shRNA, as well as with exogenous shRNA-resistant ATG5 and ATG5 shRNA, or with exogenous shRNA-resistant BECN1 and BECN1 shRNA were treated with elaiophylin (0.5 μM) for 12 h. GFP-LC3B puncta per cell were counted. Data are the mean ± SEM for triplicate samples of at least 100 cells per sample (*P < 0.05).
As elaiophylin induces lysosomal permeabilization (Fig. 4), the role of cathepsins in elaiophylin-induced cell death was explored using chemical inhibitors of cathepsins. As shown in Figure S8, both Ca074Me (CTSB inhibitor) and pepstatin A (CTSD inhibitor), partially prevented elaiophylin-induced cell death in SKOV3, indicating that cathepsins might partially involved in elaiophylin-induced cell death.
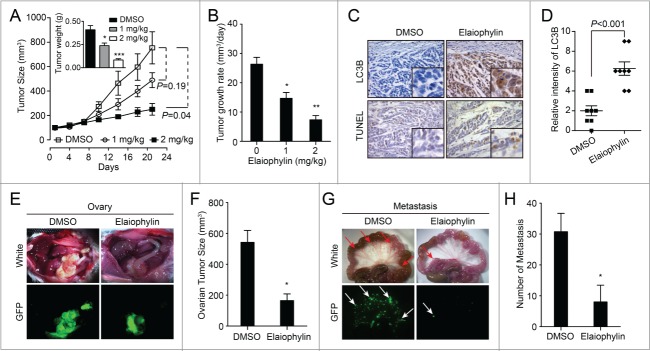
Elaiophylin exerted autophagy inhibition and antitumor efficacy in vivo
To evaluate the therapeutic benefits of elaiophylin treatment in vivo, 4-wk-old BALB/C athymic mice bearing palpable SKOV3 tumors were treated with DMSO or elaiophylin (1 mg/kg i.p. or 2 mg/kg i.p.) every 2 d. This schedule was well tolerated by the mice for 3 wk. Treatment with 2 mg/kg elaiophylin significantly suppressed tumor growth compared with DMSO treatment (Fig. 7A), resulting in a 72% decrease in the average daily tumor growth rate compared with DMSO treatment (26.3 mm3/d vs. 7.3 mm3/d; P = 0.002; Fig. 7B). The average tumor weights were calculated and indicated that significant antitumor activity occurred with 2 mg/kg elaiophylin (Fig. 7A). When an antibody against LC3B was used to visualize autophagosome formation, elaiophylin exposure significantly augmented the total number of positive cells and the signal intensity from LC3B for each cell compared with controls, indicating elaiophylin could promote autophagosome accumulation in vivo. Correspondingly, the number of apoptotic cells in tumor xenografts measured by TUNEL staining was increased in elaiophylin-treated tumors compared with DMSO-treated tumors (Fig. 7C and D). Given the enhanced cytotoxicity observed in vitro following combination therapy with cisplatin and elaiophylin, we sought to explore whether this combination could improve tumor control in vivo. Mice bearing SKOV3 xenografts were assigned to DMSO, cisplatin 4 mg/kg i.p., or cisplatin 4 mg/kg combined with elaiophylin 1 mg/kg i.p. treatment every 4 d, and tumor volume was monitored over time. The combination approach was superior to single agent treatment in the SKOV3 xenografts (P < 0.001 for comparison with cisplatin alone), indicating an even lower dose of elaiophylin could sensitize ovarian tumor cells to cisplatin in vivo (Fig. S9).
Figure 7.
Elaiophylin inhibits tumor growth in SKOV3 xenografts and orthotopic implantations. (A) Elaiophylin potently inhibited SKOV3 xenograft tumor growth in nude mice as a single-agent therapy. SKOV3 cells (5 x 106) were injected into the flanks of each mouse. When the tumors reached 100 mm3, the mice were randomized into groups of 6 to 8 mice. Mice were given i.p. injections of DMSO or 1 or 2 mg/kg elaiophylin every 2 d. Data are mean tumor size ± SEM at various time points. P values for comparative analyses between the indicated treatment groups were calculated via the generalized Mann-Whitney test. Inset bar graphs corresponds to weight of excised tumors (*P < 0.05, ***P < 0.001). (B) Average daily tumor growth rate (*P < 0.05, **P < 0.01). (C) Paraffin-embedded sections were stained with an antibody against LC3B and evaluated by the TUNEL assay. Representative photos are shown (40×). (D) Quantification of the intensity of LC3B staining as indicated in (C). Eight individual fields of each slide were randomly selected for evaluation. (E to H) SKOV3 GFP-LC3B cells were injected into the ovarian bursa of NOD/SCID mice. (E) Representative images of the development of ovarian tumors in NOD/SCID mice treated with DMSO or elaiophylin (2 mg/kg) every 2 d. Photos were taken under visible (white) or fluorescent (GFP) light. (F) Quantification of tumor volumes in (E). Tumor size was significantly decreased (*P < 0.05) in the elaiophylin-treated group compared with the DMSO-treated group. (G) Corresponding mesenteric metastasis images in (E). Photos were taken under visible (white) or fluorescent (GFP) light. The arrows indicate metastatic lesions in the mesentery. (H) Quantification of metastases in (G). Elaiophylin-treated mice displayed fewer mesenteric metastases (*P < 0.05).
To determine whether elaiophylin could impair ovarian cancer growth and metastasis as a single agent, 1.0 × 106 GFP-LC3B-SKOV3 cells were injected into the ovarian bursa of NOD/SCID mice to generate an orthotopic ovarian cancer model with metastasis. One wk after surgery, 8 mice from each cohort bearing GFP-LC3B-SKOV3 xenografts were assigned to treatment with DMSO or elaiophylin (2 mg/kg i.p.) every 2 d. Tumor growth at the primary site was significantly impaired by elaiophylin, resulting in ∼60% reduction in tumor volume (Fig. 7E and F). Furthermore, fewer mesenteric metastases were observed in elaiophylin-treated mice than in control mice (Fig. 7G and H).
To determine whether higher elaiophylin doses produce significant toxicity, mice bearing SKOV3 orthotopic ovarian cancer xenografts were given DMSO or elaiophylin at gradually increasing dosages of 4 or 8 mg/kg i.p. daily. Toxic reactions were observed only in the 8 mg/kg group and appeared as weight loss in 6/8 mice, lethargy with arched backs in 4/8 mice, and bowel obstruction in 6/8 mice. Daily dosing for the 4 mg/kg group was well tolerated, as indicated by measurements of body weight, food intake, breathing, and defecation (Fig. S11A). While histological examination of vital organs uncovered no significant differences between control- and 4 mg/kg elaiophylin-treated mice (Fig. S10), inspection of the bowel displayed extensively dilated small intestines and ilea in the 8 mg/kg elaiophylin-treated mice. Histological examination of the ilea demonstrated intact intestinal villi and crypt architecture, but reduced numbers of Paneth cells (Fig. S11B and S11C), which mimicked the phenotype of ATG16L1-deficient mice and may be a sign of autophagy deficiency.23 Thus, lower doses of elaiophylin as a single agent exerted significant antitumor activity, while higher doses led to intestinal toxicity.
Blockade of hypoxia-induced autophagy by elaiophylin sensitized cells to hypoxia
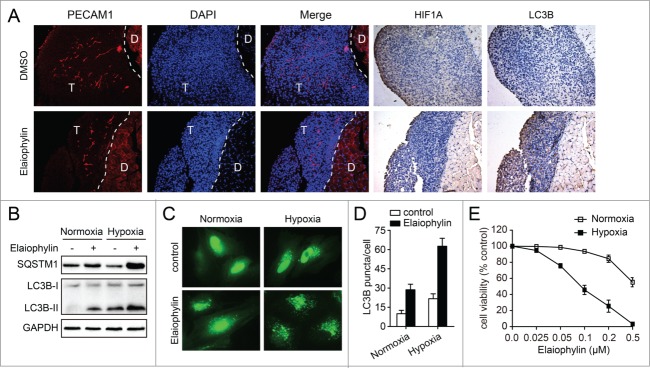
We carefully observed LC3B signal distribution in the hypoxic zones (stained for HIF1A) and vascularized areas (stained for PECAM1/CD31) of the tumors from the orthotopic ovarian cancer xenograft mouse model. Interestingly, hypoxic tumor regions, which were mainly located in the exterior regions of peritoneal metastases that are poorly vascularized, display more LC3B signal distribution compared with vascularized areas in the elaiophylin-treated group (Fig. 8A), suggesting that elaiophylin predominantly induced autophagosome accumulation to exert antitumoral activity in hypoxic cells.
Figure 8.
Inhibition of autophagy by elaiophylin addition sensitizes cells to hypoxia. (A) Representative immune-staining analysis of PECAM1, HIF1A, and LC3B in peritoneal metastases from DMSO or elaiophylin (2 mg/kg) treated mice from Figure 7E. T indicates tumor and D indicates diaphragm. (B) SKOV3 cells were treated with or without elaiophylin (0.5 μM) and cultured under normoxic or hypoxic conditions for 12 h. Cells were then harvested for detection of SQSTM1 and LC3B by western blot. (C and D) SKOV3-GFP-LC3B cells were treated with elaiophylin (0.5 μM) and cultured under normoxic or hypoxic conditions for 12 h. GFP-LC3B puncta per cell were counted. Data are the mean ± SEM for triplicate samples of at least 100 cells per sample (*P < 0.05). (E) Cell viability under normoxic and hypoxic conditions after exposure to elaiophylin for 24 h in SKOV3 cells. Data are the mean ± SEM of 3 experiments.
To further confirm that elaiophylin-induced autophagosome accumulation acted on hypoxic cells, SKOV3 cells were separately cultured under normoxia or hypoxia conditions. As shown in Figure 8B, elaiophylin increased LC3B-I to LC3B-II conversion in the hypoxia condition and blocked hypoxia-induced SQSTM1 degradation, leading to an increased level of SQSTM1 compared to the normoxia condition. These results indicate that elaiophylin disrupts hypoxia-induced autophagy. Additionally, hypoxia-induced GFP-LC3B puncta accumulation was dramatically increased by elaiophylin treatment in GFP-LC3B-SKOV3 cells (Fig. 8C and D). To further examine whether elaiophylin-mediated autophagy inhibition and lysosomal dysfunction eventually affected cell survival during hypoxia, we used the CCK-8 assay to analyze the effects of elaiophylin on the viability of SKOV3 cells. Exposure to elaiophylin caused a significant increase in cell death in hypoxia conditions even at concentrations 5-fold lower than those required to affect the survival of normoxic cells (Fig. 8E).
Discussion
Targeting the autophagic pathway is currently regarded as a promising new strategy for cancer drug discovery.24-26 In the present study, we have identified and characterized elaiophylin as a novel autophagy inhibitor. Elaiophylin, a C2 symmetry 16-member macrodiolide antibiotic, was originally isolated from Streptomyces melanosporus.17,27,28 Prior studies have explored the antibacterial and antihelminthic activity of elaiophylin; however, autophagy inhibition by elaiophylin has never been previously recognized.29 Treatment with elaiophylin leads to substantial accumulation of both autophagosome and SQSTM1 in various cells. Additionally, elaiophylin functions to impair the maturation of CTSB/D and induce subsequent lysosomal membrane permeabilization. In vitro, elaiophylin directly induces apoptosis in various cells in a dose-dependent manner through inhibition of autophagy. In vivo, administration of elaiophylin displays potent antitumor activity with a safe therapeutic window. Furthermore, elaiophylin could sensitize the antitumor effect to cisplatin both in vitro and in vivo. Interestingly, under hypoxia condition, elaiophylin induces more cell death. As such, we identify elaiophylin as a novel autophagy inhibitor with chemical structure distinct from other known inhibitors.
Currently, autophagy inhibitors can be divided into 2 categories: early-stage inhibitors and late-stage inhibitors.30,31 Early stage autophagy inhibitors, including wortmannin, LY294002, and 3-MA, mainly block the formation of autophagosomes through the inhibition of class III PtdIns 3-kinases.32,33 The late-stage autophagy inhibitors, such as CQ, hydroxylchloroquine (HCQ), bafilomycin A1, and lysosomal protease inhibitors, exert a suppressive effect downstream of autophagosome formation, including inhibition of autophagosome and lysosome fusion and/or blocking degradation of autophagic cargo inside autolysosomes.34,35 Our data demonstrate that elaiophylin belongs to the late-stage autophagy inhibitor group. Early stage autophagy inhibitors are commonly used as pharmacological tools to inhibit autophagy in vitro.30 However, late-stage autophagy inhibitors have proven to be useful adjuvants in many antitumor therapies in vivo.36,37 HCQ, a widely used late-stage autophagy inhibitor, has been evaluated in several phase I and II clinical trials in combination with chemotherapy drugs in a variety of tumor types.38-40 Furthermore, most current late-stage autophagy inhibitors are the synthetic derivatives of CQ.14,41 Notably, in this study, for the first time, we demonstrate elaiophylin, a natural compound with chemical structure distinct from CQ, as a novel late-stage autophagy inhibitor. Our results imply that screening autophagy regulators from natural compounds might be an efficient methodology to identify novel autophagy inhibitors and lead compounds for cancer therapy.
Given the ability of elaiophylin to inhibit autophagic flux and eventually induce cell death, a major concern remains whether elaiophylin-induced cell death is related to autophagy inhibition. Our present data have provided some insight into this important concern. First, as elaiophylin disrupts lysosomal degradation to inhibit the autophagy process, we have examined the possibility of a lysosomal connection in the induction of cell death. Induction of lysosomal membrane permeabilization (LMP) has been abundantly linked to cell death.42,43 In our study, elaiophylin was found to trigger LMP. Moreover, the abrogation of cathepsin activity by chemical inhibitors attenuated cell death in the presence of elaiophylin, perhaps implying that increased LMP is involved in the death-promoting effects of elaiophylin. Additionally, constitutive autophagy contributes to elaiophylin-induced cell death, because silencing of autophagy genes regulating autophagosome formation attenuated cell death following treatment with elaiophylin. Recent studies show that disruption of the late stages of autophagy leads to excessive accumulation of autophagic vacuoles containing deleterious undegraded material and has the potential to turn autophagy into a destructive process.44 It is therefore conceivable that elaiophylin is cytotoxic because the cell is sensitive to cytotoxic intermediates created by late stage inhibition of autophagy. In this case, reduced autophagosome formation could alleviate elaiophylin toxicity. This finding is in accord with a recent study that shows that silencing of autophagy genes regulating autophagosome formation protects HOSE cells from CQ-toxicity.45
Interestingly, in our present study we demonstrate that elaiophylin can efficiently sensitize cancer cells to hypoxia both in vitro and in vivo, which is in accordance with previous findings that inhibition of autophagy with autophagy inhibitors in hypoxic conditions, particularly with late autophagy inhibitors such as chloroquine, causes selective hypoxic cell killing.46 Hypoxia, a common feature of solid tumors, is well known to play important roles in the development and progression of ovarian cancer, such as inducing chemoresistance, promoting vasculogenic mimicry formation and upgrading stem cell-like properties of ovarian cancer cells.47-50 Previous studies have shown that hypoxia activates a cell survival response involving autophagy to ameliorate hypoxic stress.51 Remarkably, based on the fact that elaiophylin displays more effective antitumor activity in hypoxic regions (particularly in the exterior regions of peritoneal metastases that are poorly vascularized), our results provide a possibility for the use of elaiophylin as an adjuvant to bevacizumab treatment in further research. This is important because bevacizumab inhibits the formation of new tumor vessels, causing hypoxia in the region surrounding the tumor.52
Our findings have important clinical implications. Ovarian cancer is considered to be an intractable solid tumor. After initial treatment, a large number of cases of advanced ovarian cancer eventually become refractory or relapse due to the persistence of residual disease. These patients are considered incurable using the current available treatment options, which necessitates the identification of novel treatment modalities. Residual tumor cells may survive via autophagy and repopulate once the stressor (radio- or chemotherapy) is removed, which likely contributes to relapse.53,54 Thus, autophagy inhibition is a promising approach and is being investigated as a new target strategy for ovarian cancer treatment. Our present data highlight that elaiophylin is a novel autophagy inhibitor with unique chemical structure, providing potential for structure-based development of autophagy inhibitors for new cancer therapies.
Materials and Methods
Cell lines and cell culture
A2780 cells were obtained from the European Collection of Cell Cultures (ECACC, Salisbury, UK). The C13* cell line was a gift from Prof. Benjamin K. Tsang of the Ottawa Health Research Institute, Ottawa, Canada.55 SKOV-3 (HTB-77), OVCAR-3 (HTB-161), Caov-3 (HTB-75), SW 626 (HTB-78), PC-3 (CRL-1435), HepG2 (HB-8065), A549 (CCL-185), HeLa (CCL-2) and HEK-293 (CRL-1573) were purchased from the American Type Culture Collection. Unless otherwise noted, all cells were maintained in Dulbecco's modified Eagle's medium (Gibco/Invitrogen, 12100-046) supplemented with 10% fetal bovine serum (Gibco/Invitrogen, 10438-026) and 10 U/ml penicillin-streptomycin (Gibco/Invitrogen, 15140-122). SKOV-3 cells were maintained in McCoy 5A medium (Gibco/Invitrogen, 16600-082) and SW-626 cells in L-15 medium (Gibco/Invitrogen, 11415-064). OVCAR3 cells were maintained RPMI-1640 (Gibco/Invitrogen, 31800-089) containing 0.01 mg/ml bovine insulin and 20% fetal bovine serum. A2780 and C13* cells were maintained in RPMI-1640 containing 2 mM L-glutamine and 10% fetal bovine serum. PC-3 and A549 cells were grown in F-12K Medium (Gibco/Invitrogen, 21127-022). All cells were cultured at 37°C in a 5% (v/v) CO2 atmosphere. For hypoxia, cells were cultured under an atmosphere containing a mixture of 1% O2, 5% CO2 and 94% N2 at 37°C.
Reagents and antibodies
Elaiophylin and rapamycin were provided by North China Pharmaceutical Group Corporation New Drug R&D center. Elaiophylin was dissolved in DMSO to make a stock solution at 1mM, stored at −20°C, and diluted with proper medium before use. 3-methyladenine (M9281), pancaspase inhibitor Z-VAD-FMK (C2105), 4′, 6-diamidino-2-phenylindole (D9542), Ca074Me (C5732), pepstatin A (P5318) and chloroquine (C6628) were purchased from Sigma Aldrich. GFP-LC3B lentivirus, sh-ATG5 and sh-BECN1 were purchased from Neuron Biotech Inc. (Shanghai, China). DeadEnd™ Colorimetric TUNEL System (G7360) was purchased from Promega Biotech. LysoTracker Red (L-7528), DQ Red BSA assay (D12051), Earle balanced salt solution (EBSS; 14155063) and Texas Red dextran (D1863) were purchased from Invitrogen. LAMP1-RFP (plasmid 1817), pcDNA3-BECN1 (plasmid 21150) and pCMV-myc-ATG5 (plasmid 24922) were purchased from Addgene. Primary antibodies against MAP1LC3B (#3868), ATG5 (#12994), BECN1/beclin1 (#3495), PARP1 (#9542), and cleaved CASP9 (#7237) were from Cell Signaling Technology. Anti-CTSD (#2487-1) was from Epitomics. Anti-CTSB (ab33538) and anti-LAMP1 (ab24170) were from abcam. Anti-BIRC5/survivin (10508-1-AP), anti-GAPDH (10494-1-AP) and anti-TUBA1B (tubulin, α 1b; 11224-1-AP) were from Proteintech. SQSTM1/p62 Lck ligand (#610832) was purchased from BD Biosciences.
Cell viability assay
Cells (5,000 cells/well) were seeded in 3 replicates into 96-well plates and treated with or without elaiophylin for a desired time period at the indicated concentrations. After treatment, cell viability was evaluated using the Cell Counting Kit-8 (Dojindo Laboratories, CK04-01) assay. The cell viability was expressed as a percentage of absorbance in cells with indicated treatments to that in cells with DMSO control treatment.
Cell death assay
After each treatment, floating cells were first collected and later mixed with attached cells harvested by trypsinization. Cells were washed twice with phosphate-buffered saline (PBS; Sigma Aldrich, P3813-10PAK) and then stained with the ANXA5/annexin V-FITC and propidium iodide/PI Apoptosis Detection kit (#AP101-100; MultiSciences Biotech) according to the manufacturer's instructions. Cells were analyzed using a BD FACSCalibur flow cytometer (San Jose, CA, USA). In total, 10,000 cells were analyzed per measurement. Data were analyzed using FlowJo software.
Quantitative analysis of GFP-LC3B dots
SKOV3 cells stably expressing GFP-LC3B were seeded into PerkinElmer View 96-well plates at a density of 5,000 cells per well. Eighteen h after seeding, chemicals were added to the medium. Each chemical was tested in triplicate across a concentration gradient (0.25, 0.5, 1, 2.5 and 5 μM) to select dose-responsive hits. Cells were cultured for another 12 h at 37°C. The cells were then fixed with 3% (v/v) paraformaldehyde at room temperature for 15 min. Fixed-cells were washed once with PBS and stored at 4°C until analyzed.
Plates were scanned using iSort image cytometry, which enables the acquisition and quantitative analysis of images of GFP-LC3B dots. The “granularity” application module is used to detect and count granules in cells, permitting the direct determination of the average granule number per cell. GFP-LC3B dots were defined as bright puncta (>1.5 SD above the mean cytosolic fluorescence). Puncta with a diameter ≥0.75 μm were used as cut-off. The number of GFP-LC3B dots per cell was counted in at least 3 independent visual fields from 3 independent wells. The results were expressed as the average number of GFP-LC3B dots in each cell (mean ± standard deviation).
Wells showing a 3-fold or higher increase in the number of GFP-LC3B puncta versus control wells were re-examined to eliminate false positives. Compounds inducing a 3-fold or higher increase in the number of GFP-LC3B puncta were considered “hits.” The Z-factor of the assay was determined from the number of GFP-LC3B puncta measured in cells treated with DMSO (negative control) or chloroquine and rapamycin (positive controls).
, where σ+c, σ-c, μ+c, and μ-c are the standard deviations (σ) and the averages (μ) of the positive (+c) and negative (−c) controls. The Z factor value is 0.76 for this assay.
Microarray analysis
Identification of potential differences in elaiophylin-induced gene expression by Roche NimbleGen array analysis, total RNA was extracted from SKOV3 cells cultured in the presence of 0.5 μM of elaiophylin for 0, 3, 6, or 12 h and was subsequently subjected to microarray analysis. The Roche NimbleGen Human Genome 12 × 135K Array was used for the current study. The computer software that accompanied the microarray was used to process the data from the genechips. Gene set enrichment analysis was used to compare the elaiophylin-treated data set with gene sets in Molecular Signatures Database (MSigDB). The indicated gene symbols were derived from the NCBI GenBank database.
Transmission electron microscopy assay
Cells were seeded in 100 mm dishes and cultured overnight. The cells were treated with DMSO or elaiophylin for indicated times. After incubation, cells were harvested. Then cells were washed with cold PBS once and fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer (pH 7.2) at 4°C overnight and subsequently post-fixed in 1% osmium tetroxide for 1 h. Then samples were embedded, sectioned, doubly stained with uranyl acetate and lead citrate and finally observed under transmission electron microscopy (FEI Tecnai G2 12, Eindhoven, Netherlands).
Western blot analysis
Cells were lysed in RIPA buffer with protease inhibitors (Roche, 04693116001) and phosphatase inhibitor (Sigma, P0044). The samples were loaded onto an acrylamide gel and then transferred on a PVDF membrane. The membranes were then probed with primary antibodies at 4°C overnight, and finally incubated with a horseradish peroxidase-labeled secondary antibody (Sigma, A0545 and A6154). Detection was performed using a Pierce ECL western blotting substrate system.
Immunofluorescence staining and fluorescence microscopy
GFP-LC3B-SKOV3 cells were treated and fixed with 4% paraformaldehyde at room temperature for 10 min followed by permeabilization with 0.1% Triton X-100 (Sigma, X100) for 15 min. Fixed cells were blocked with 5% BSA (Roche, 10735086001) in PBS at 37°C for 30 min, then incubated with primary antibody against SQSTM1 (BD Biosciences, 610832) at 4°C overnight. After being washed twice with PBS, cells were incubated with Alexa Fluor-conjugated secondary antibodies (Alexa Fluor 555 goat anti-mouse; Invitrogen, A21422). Nuclei were stained for 10 min with 4′,6-diamidino-2-phenylindole (DAPI; Molecular Probes/Invitrogen, D1306). Images were captured by fluorescence microscopy (Olympus DP73, Tokyo, Japan) and by laser scanning confocal microscopy (Olympus FV1000, Tokyo, Japan).
Acridine orange staining
Cell staining with acridine orange (AO; Molecular Probes/Invitrogen, A1301) was performed according to published procedures,56 adding a final concentration of 2 μg/ml for a period of 20 min (37°C, 5% CO2). Tumor cells were incubated with elaiophylin (0.5 μM), or chloroquine (25 μM) for the indicated time before the AO was added. Photographs were obtained using an Olympus FV1000 confocal laser scanning microscope. AO produces red fluorescence (emission peak at about 650 nm) in the lysosomal compartments, and green fluorescence (emission peak between 530 and 550 nm) in the cytosolic and nuclear compartments.
Incorporation of texas red dextran
Texas Red-labeled lysine-fixable dextran (MW10, 000) was purchased from Molecular Probes/Invitrogen (D1863). GFP-LC3B-SKOV3 cells were incubated with Texas Red dextran (1 mg/ml) overnight. On the following day, the cells were transferred to fresh medium for the chase before being treated with elaiophylin (0.5 μM) for indicated time. The cells were observed with confocal microscopy without fixation.
DQ Red BSA staining
Lysosomal-dependent proteolysis was visualized with DQ Red BSA (Molecular Probes/Invitrogen, D-12051) at a concentration of 10 μg/ml for 12 h (37°C, 5% CO2). The cells were then washed 3 times with PBS before being treated with 0.5 μM elaiophylin for 12 h. Then cells were observed using an Olympus FV1000 confocal laser scanning microscope.
Cathepsin activity assay
CTSB and CTSD activity was determined using the commercial assay provided by Biovision according to the manufacturer's protocol (BioVision, K140-100/K143-100). Cathepsin activity was expressed as fold-change in relative fluorescence units (RFU) per microgram protein.
LysoTracker Red staining
Cells from different treatments were collected and then incubated in the dark for 30 min at 37°C with LysoTracker Red DND-99 (Invitrogen, 100 nM). Single-color flow cytometry was carried out on a BD FACSCalibur flow cytometer (San Jose, CA, USA), and the data were analyzed using FlowJo software.
Cell fractionation
Lysosomal fractions were extracted from cell homogenates by differential centrifugation followed by density centrifugation according to the manufacturer's protocol (Lysosome Extraction Kit; Sigma-Aldrich; LYSISO1). Briefly, cell homogenates were centrifuged for 10 min at 1000 × g at 4°C. The supernatant fraction was centrifuged for 20 min at 20,000 × g at 4°C to pellet lysosomes and other organelles, and the resulting supernatant fraction was collected as cytosolic fraction. The pellet fractions were subjected to additional centrifugation. The final pellet (lysosomal) fraction was lysed in the lysis buffer described in the procedure for western blotting. Samples were subjected to immunoblotting.
In vivo treatment
SKOV3 cells (5 × 106) resuspended in 50 μL of PBS were injected subcutaneously into the flanks of 4-wk-old BALB/C athymic mice (Beijing HFK Bioscience). Mice were housed and maintained under specific pathogen-free conditions. All animal experiments were carried out in accordance with the Guide for the Care and Use of Laboratory Animals of Tongji Hospital. When mice exhibited palpable tumors, they were randomly assigned to treatment groups (6 to 8 mice per group) to avoid treatment bias. Elaiophylin was given i.p. (1 or 2 mg/kg) every 2 d for 21 d, or DMSO was used as a control. Tumor volumes were calculated as length*(square of width)/2. After the initial treatment, the tumor size was determined every day.
GFP-LC3B-SKOV3 cells were injected orthotopically under the ovarian bursa of 6-wk-old female NOD/SCID mice (Beijing HFK Bioscience). For orthotopic injection, mice were anesthetized with a cocktail of acepromazine and torbugesic, and a dorsal incision was employed to expose the right ovary enshrouded in its oviductal fimbriae. 1.0 × 106 cells, resuspended in 10 μL of growth medium, were injected with a 30-gauge needle under the ovarian bursal membrane. One week after surgery, elaiophylin was given i.p. (2 mg/kg) every 2 d, or DMSO was used as a control. Eight wk after the injection, the mice were euthanized and necropsies were performed. Investigators were blinded to the treatment groups.
Immunohistochemistry
Tumors were resected immediately after euthanasia and fixed in 4% paraformaldehyde at 4°C for 48 h. Selected samples were embedded in paraffin, sectioned and stained with primary antibodies. The immunostaining intensity was scored as previously described.57
Statistical analysis
Data were compared using a 2-tailed Student t test, a one-way ANOVA test followed by Tukey's multiple comparison tests or a Mann-Whitney U test. A value of P < 0.05 was considered statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest needed to be disclosed.
Acknowledgments
We also thank Dr. Yang Guan of the Department of Ultrastructural Pathology (Tongji Medical College, Huazhong University of Science and Technology) for support with imaging.
Funding
This work was funded by grant from National Development Program (973) For Key Basic Research of China (Nos. 2015CB553903 and 2009CB521806); National Natural Science Foundation of China (81230038; 81101962; 81090414; 81025011; 81372801; 81072135, 81272422, 81472783) and the “863” Program of China (2012AA02A507).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- 1. Jayson GC, Kohn EC, Kitchener HC, Ledermann JA. Ovarian cancer. Lancet 2014; 384:1376-88; PMID:24767708; http://dx.doi.org/ 10.1016/S0140-6736(13)62146-7 [DOI] [PubMed] [Google Scholar]
- 2. Bast RC, Jr, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer 2009; 9:415-28; PMID:19461667; http://dx.doi.org/ 10.1038/nrc2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vaughan S, Coward JI, Bast RC, Jr, Berchuck A, Berek JS, Brenton JD, Coukos G, Crum CC, Drapkin R, Etemadmoghadam D, et al. Rethinking ovarian cancer: recommendations for improving outcomes. Nat Rev Cancer 2011; 11:719-25; PMID:21941283; http://dx.doi.org/ 10.1038/nrc3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Peracchio C, Alabiso O, Valente G, Isidoro C. Involvement of autophagy in ovarian cancer: a working hypothesis. J Ovarian Res 2012; 5:22; PMID:22974323; http://dx.doi.org/ 10.1186/1757-2215-5-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Orfanelli T, Jeong JM, Doulaveris G, Holcomb K, Witkin SS. Involvement of autophagy in cervical, endometrial and ovarian cancer. Int J Cancer J Int du Cancer 2014; 135:519-28; PMID:24122662; http://dx.doi.org/ 10.1002/ijc.28524 [DOI] [PubMed] [Google Scholar]
- 6. Wang J, Wu GS. Role of autophagy in cisplatin resistance in ovarian cancer cells. J Biol Chem 2014; 289:17163-73; PMID:24794870; http://dx.doi.org/ 10.1074/jbc.M114.558288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 8:445-544; PMID:22966490; http://dx.doi.org/ 10.4161/auto.19496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature 2008; 451:1069-75; PMID:18305538; http://dx.doi.org/ 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell 2010; 140:313-26; PMID:20144757; http://dx.doi.org/ 10.1016/j.cell.2010.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang N, Qi Y, Wadham C, Wang L, Warren A, Di W, Xia P. FTY720 induces necrotic cell death and autophagy in ovarian cancer cells: a protective role of autophagy. Autophagy 2010; 6:1157-67; PMID:20935520; http://dx.doi.org/ 10.4161/auto.6.8.13614 [DOI] [PubMed] [Google Scholar]
- 11. Zhu K, Dunner K, Jr, McConkey DJ. Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene 2010; 29:451-62; PMID:19881538; http://dx.doi.org/ 10.1038/onc.2009.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ganley IG, Wong PM, Gammoh N, Jiang X. Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Mol Cell 2011; 42:731-43; PMID:21700220; http://dx.doi.org/ 10.1016/j.molcel.2011.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gonzalez P, Mader I, Tchoghandjian A, Enzenmuller S, Cristofanon S, Basit F, Debatin KM, Fulda S. Impairment of lysosomal integrity by B10, a glycosylated derivative of betulinic acid, leads to lysosomal cell death and converts autophagy into a detrimental process. Cell Death Differ 2012; 19:1337-46; PMID:22343715; http://dx.doi.org/ 10.1038/cdd.2012.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. McAfee Q, Zhang Z, Samanta A, Levi SM, Ma XH, Piao S, Lynch JP, Uehara T, Sepulveda AR, Davis LE, et al. Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc Natl Acad Sci U S A 2012; 109:8253-8; PMID:22566612; http://dx.doi.org/ 10.1073/pnas.1118193109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Voss V, Senft C, Lang V, Ronellenfitsch MW, Steinbach JP, Seifert V, Kögel D. The pan-Bcl-2 inhibitor (-)-gossypol triggers autophagic cell death in malignant glioma. Mol Cancer Res: MCR 2010; 8:1002-16; PMID:20587533; http://dx.doi.org/ 10.1158/1541-7786.MCR-09-0562 [DOI] [PubMed] [Google Scholar]
- 16. Tormo D, Checinska A, Alonso-Curbelo D, Perez-Guijarro E, Canon E, Riveiro-Falkenbach E, Calvo TG, Larribere L, Megías D, Mulero F, et al. Targeted activation of innate immunity for therapeutic induction of autophagy and apoptosis in melanoma cells. Cancer cell 2009; 16:103-14; PMID:19647221; http://dx.doi.org/ 10.1016/j.ccr.2009.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Arai M. Azalomycins B and F, two new antibiotics. I. Production and isolation. J Antibiot 1960; 13:46-50; PMID:13848830 [PubMed] [Google Scholar]
- 18. Wu Y, Wang X, Guo H, Zhang B, Zhang XB, Shi ZJ, Yu L. Synthesis and screening of 3-MA derivatives for autophagy inhibitors. Autophagy 2013; 9:595-603; PMID:23412639; http://dx.doi.org/ 10.4161/auto.23641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Øvervatn A, Bjørkøy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem 2007; 282:24131-45; PMID:17580304; http://dx.doi.org/ 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- 20. Kawai A, Uchiyama H, Takano S, Nakamura N, Ohkuma S. Autophagosome-lysosome fusion depends on the pH in acidic compartments in CHO cells. Autophagy 2007; 3:154-7; PMID:17204842; http://dx.doi.org/ 10.4161/auto.3634 [DOI] [PubMed] [Google Scholar]
- 21. Vazquez CL, Colombo MI. Assays to assess autophagy induction and fusion of autophagic vacuoles with a degradative compartment, using monodansylcadaverine (MDC) and DQ-BSA. Methods Enzymol 2009; 452:85-95; PMID:19200877; http://dx.doi.org/ 10.1016/S0076-6879(08)03606-9 [DOI] [PubMed] [Google Scholar]
- 22. Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature 2010; 466:68-76; PMID:20562859; http://dx.doi.org/ 10.1038/nature09204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, Kishi C, Kc W, Carrero JA, Hunt S, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature 2008; 456:259-63; PMID:18849966; http://dx.doi.org/ 10.1038/nature07416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baek KH, Park J, Shin I. Autophagy-regulating small molecules and their therapeutic applications. Chem Soc Rev 2012; 41:3245-63; PMID:22293658; http://dx.doi.org/ 10.1039/c2cs15328a [DOI] [PubMed] [Google Scholar]
- 25. Hoyer-Hansen M, Jaattela M. Autophagy: an emerging target for cancer therapy. Autophagy 2008; 4:574-80; PMID:18362515; http://dx.doi.org/ 10.4161/auto.5921 [DOI] [PubMed] [Google Scholar]
- 26. Janku F, McConkey DJ, Hong DS, Kurzrock R. Autophagy as a target for anticancer therapy. Nat Rev Clin Oncol 2011; 8:528-39; PMID:21587219; http://dx.doi.org/ 10.1038/nrclinonc.2011.71 [DOI] [PubMed] [Google Scholar]
- 27. Fiedler HP, Worner W, Zahner H, Kaiser HP, Keller-Schierlein W, Muller A. Metabolic products of microorganisms. 200 Isolation and characterization of niphithricins A, B, and elaiophylin, antibiotics produced by Streptomyces violaceoniger. J Antibiot 1981; 34:1107-18; PMID:7035426; http://dx.doi.org/ 10.7164/antibiotics.34.1107 [DOI] [PubMed] [Google Scholar]
- 28. Haydock SF, Mironenko T, Ghoorahoo HI, Leadlay PF. The putative elaiophylin biosynthetic gene cluster in Streptomyces sp. DSM4137 is adjacent to genes encoding adenosylcobalamin-dependent methylmalonyl CoA mutase and to genes for synthesis of cobalamin. J Biotechnol 2004; 113:55-68; PMID:15380647; http://dx.doi.org/ 10.1016/j.jbiotec.2004.03.022 [DOI] [PubMed] [Google Scholar]
- 29. Drose S, Bindseil KU, Bowman EJ, Siebers A, Zeeck A, Altendorf K. Inhibitory effect of modified bafilomycins and concanamycins on P- and V-type adenosinetriphosphatases. Biochemistry 1993; 32:3902-6; PMID:8385991; http://dx.doi.org/ 10.1021/bi00066a008 [DOI] [PubMed] [Google Scholar]
- 30. Yang YP, Hu LF, Zheng HF, Mao CJ, Hu WD, Xiong KP, Wang F, Liu CF. Application and interpretation of current autophagy inhibitors and activators. Acta Pharmacol Sinica 2013; 34:625-35; PMID:23524572; http://dx.doi.org/ 10.1038/aps.2013.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang ZJ, Chee CE, Huang S, Sinicrope FA. The role of autophagy in cancer: therapeutic implications. Mol Cancer Therap 2011; 10:1533-41; PMID:21878654; http://dx.doi.org/ 10.1158/1535-7163.MCT-11-0047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelarova H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem/FEBS 1997; 243:240-6; PMID:9030745; http://dx.doi.org/ 10.1111/j.1432-1033.1997.0240a.x [DOI] [PubMed] [Google Scholar]
- 33. Wu YT, Tan HL, Shui G, Bauvy C, Huang Q, Wenk MR, Ong CN, Codogno P, Shen HM. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem 2010; 285:10850-61; PMID:20123989; http://dx.doi.org/ 10.1074/jbc.M109.080796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Disc 2007; 6:304-12; PMID:17396135; http://dx.doi.org/ 10.1038/nrd2272 [DOI] [PubMed] [Google Scholar]
- 35. Geng Y, Kohli L, Klocke BJ, Roth KA. Chloroquine-induced autophagic vacuole accumulation and cell death in glioma cells is p53 independent. Neuro-oncology 2010; 12:473-81; PMID:20406898; http://dx.doi.org/ 10.1093/neuonc/nop048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, Thomas-Tikhonenko A, Thompson CB. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Investigat 2007; 117:326-36; PMID:17235397; http://dx.doi.org/ 10.1172/JCI28833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rao R, Balusu R, Fiskus W, Mudunuru U, Venkannagari S, Chauhan L, Smith JE, Hembruff SL, Ha K, Atadja P, et al. Combination of pan-histone deacetylase inhibitor and autophagy inhibitor exerts superior efficacy against triple-negative human breast cancer cells. Mol Cancer Therap 2012; 11:973-83; PMID:22367781; http://dx.doi.org/ 10.1158/1535-7163.MCT-11-0979 [DOI] [PubMed] [Google Scholar]
- 38. Rosenfeld MR, Ye X, Supko JG, Desideri S, Grossman SA, Brem S, Mikkelson T, Wang D, Chang YC, Hu J, et al. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy 2014; 10:1359-68; PMID:24991840; http://dx.doi.org/ 10.4161/auto.28984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rangwala R, Chang YC, Hu J, Algazy KM, Evans TL, Fecher LA, Schuchter LM, Torigian DA, Panosian JT, Troxel AB, et al. Combined MTOR and autophagy inhibition: phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy 2014; 10:1391-402; PMID:24991838; http://dx.doi.org/ 10.4161/auto.29119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rangwala R, Leone R, Chang YC, Fecher LA, Schuchter LM, Kramer A, Tan KS, Heitjan DF, Rodgers G, Gallagher M, et al. Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma. Autophagy 2014; 10:1369-79; PMID:24991839; http://dx.doi.org/ 10.4161/auto.29118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Goodall ML, Wang T, Martin KR, Kortus MG, Kauffman AL, Trent JM, Gately S, MacKeigan JP. Development of potent autophagy inhibitors that sensitize oncogenic BRAF V600E mutant melanoma tumor cells to vemurafenib. Autophagy 2014; 10:1120-36; PMID:24879157; http://dx.doi.org/ 10.4161/auto.28594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Boya P, Kroemer G. Lysosomal membrane permeabilization in cell death. Oncogene 2008; 27:6434-51; PMID:18955971; http://dx.doi.org/ 10.1038/onc.2008.310 [DOI] [PubMed] [Google Scholar]
- 43. Repnik U, Hafner Cesen M, Turk B. Lysosomal membrane permeabilization in cell death: concepts and challenges. Mitochondrion 2014; PMID:24984038; http://dx.doi.org/ 10.1016/j.mito.2014.06.006 [DOI] [PubMed] [Google Scholar]
- 44. Degtyarev M, De Maziere A, Orr C, Lin J, Lee BB, Tien JY, Prior WW, van Dijk S, Wu H, Gray DC, et al. Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J Cell Biol 2008; 183:101-16; PMID:18838554; http://dx.doi.org/ 10.1083/jcb.200801099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Morgan MJ, Gamez G, Menke C, Hernandez A, Thorburn J, Gidan F, Staskiewicz L, Morgan S, Cummings C, Maycotte P, et al. Regulation of autophagy and chloroquine sensitivity by oncogenic RAS in vitro is context-dependent. Autophagy 2014; 10:1814-26; PMID:25136801; http://dx.doi.org/ 10.4161/auto.32135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K, Keulers T, Mujcic H, Landuyt W, Voncken JW, et al. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Investigat 2010; 120:127-41; PMID:20038797; http://dx.doi.org/ 10.1172/JCI40027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Du J, Sun B, Zhao X, Gu Q, Dong X, Mo J, Sun T, Wang J, Sun R, Liu Y. Hypoxia promotes vasculogenic mimicry formation by inducing epithelial-mesenchymal transition in ovarian carcinoma. Gynecol Oncol 2014; 133:575-83; PMID:24589413; http://dx.doi.org/ 10.1016/j.ygyno.2014.02.034 [DOI] [PubMed] [Google Scholar]
- 48. Horiuchi A, Hayashi T, Kikuchi N, Hayashi A, Fuseya C, Shiozawa T, Konishi I. Hypoxia upregulates ovarian cancer invasiveness via the binding of HIF-1alpha to a hypoxia-induced, methylation-free hypoxia response element of S100A4 gene. Int J Cancer J Int du Cancer 2012; 131:1755-67; PMID:22287060; http://dx.doi.org/ 10.1002/ijc.27448 [DOI] [PubMed] [Google Scholar]
- 49. Selvendiran K, Bratasz A, Kuppusamy ML, Tazi MF, Rivera BK, Kuppusamy P. Hypoxia induces chemoresistance in ovarian cancer cells by activation of signal transducer and activator of transcription 3. Int J Cancer J Int du Cancer 2009; 125:2198-204; PMID:19623660; http://dx.doi.org/ 10.1002/ijc.24601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liang D, Ma Y, Liu J, Trope CG, Holm R, Nesland JM, Suo Z. The hypoxic microenvironment upgrades stem-like properties of ovarian cancer cells. BMC Cancer 2012; 12:201; PMID:22642602; http://dx.doi.org/ 10.1186/1471-2407-12-201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mazure NM, Pouyssegur J. Hypoxia-induced autophagy: cell death or cell survival? Curr Opin Cell Biol 2010; 22:177-80; PMID:20022734; http://dx.doi.org/ 10.1016/j.ceb.2009.11.015 [DOI] [PubMed] [Google Scholar]
- 52. Hu YL, DeLay M, Jahangiri A, Molinaro AM, Rose SD, Carbonell WS, Aghi MK. Hypoxia-induced autophagy promotes tumor cell survival and adaptation to antiangiogenic treatment in glioblastoma. Cancer Res 2012; 72:1773-83; PMID:22447568; http://dx.doi.org/ 10.1158/0008-5472.CAN-11-3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Amaravadi RK. Autophagy-induced tumor dormancy in ovarian cancer. J Clin Investigat 2008; 118:3837-40; PMID:19033653; http://dx.doi.org/ 10.1172/JCI37667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lu Z, Luo RZ, Lu Y, Zhang X, Yu Q, Khare S, Kondo S, Kondo Y, Yu Y, Mills GB, et al. The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells. J Clin Investigat 2008; 118:3917-29; PMID:19033662; http://dx.doi.org/ 10.1172/JCI35512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Asselin E, Mills GB, Tsang BK. XIAP regulates Akt activity and caspase-3-dependent cleavage during cisplatin-induced apoptosis in human ovarian epithelial cancer cells. Cancer Res 2001; 61:1862-8; PMID:11280739 [PubMed] [Google Scholar]
- 56. Traganos F, Darzynkiewicz Z. Lysosomal proton pump activity: supravital cell staining with acridine orange differentiates leukocyte subpopulations. Methods Cell Biol 1994; 41:185-94; PMID:7532261; http://dx.doi.org/ 10.1016/S0091-679X(08)61717-3 [DOI] [PubMed] [Google Scholar]
- 57. Liu R, Li J, Zhang T, Zou L, Chen Y, Wang K, Lei Y, Yuan K, Li Y, Lan J, et al. Itraconazole suppresses the growth of glioblastoma through induction of autophagy: involvement of abnormal cholesterol trafficking. Autophagy 2014; 10:1241-55; PMID:24905460; http://dx.doi.org/ 10.4161/auto.28912 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.