Abstract
Autophagy is an evolutionarily conserved and exquisitely regulated self-eating cellular process with important biological functions. Phosphatidylinositol 3-kinases (PtdIns3Ks) and phosphoinositide 3-kinases (PI3Ks) are involved in the autophagic process. Here we aim to recapitulate how 3 classes of these lipid kinases differentially regulate autophagy. Generally, activation of the class I PI3K suppresses autophagy, via the well-established PI3K-AKT-MTOR (mechanistic target of rapamycin) complex 1 (MTORC1) pathway. In contrast, the class III PtdIns3K catalytic subunit PIK3C3/Vps34 forms a protein complex with BECN1 and PIK3R4 and produces phosphatidylinositol 3-phosphate (PtdIns3P), which is required for the initiation and progression of autophagy. The class II enzyme emerged only recently as an alternative source of PtdIns3P and autophagic initiator. However, the orthodox paradigm is challenged by findings that the PIK3CB catalytic subunit of class I PI3K acts as a positive regulator of autophagy, and PIK3C3 was thought to be an amino acid sensor for MTOR, which curbs autophagy. At present, a number of PtdIns3K and PI3K inhibitors, including specific PIK3C3 inhibitors, have been developed for suppression of autophagy and for clinical applications in autophagy-related human diseases.
Keywords: autophagy, MTOR, PIK3C3, PI3K, PI3K inhibitor
Abbreviations
- 3-MA
3-methyladenine
- AMBRA1
autophagy/Beclin 1 regulator 1
- ATG
autophagy related
- ATM
ATM serine/threonine kinase
- ATR
ATR serine/threonine kinase
- BH3
Bcl-2 homology 3
- CCD
coiled-coil domain
- CDK
cyclin-dependent kinase
- CISD2
CDGSH iron sulfur domain 2
- class I/II PI3K
class I/II phosphoinositide 3-kinase
- class III PtdIns3K
class III phosphatidylinositol 3-kinase
- DAPK1
death-associated protein kinase 1
- DDR
DNA damage response
- EIF4EBP1
eukaryotic translation initiation factor 4E binding protein 1
- ER
endoplasmic reticulum
- FOXO
forkhead box O
- FYCO1
FYVE and coiled-coil domain containing 1
- GAP
GTPase-activating protein
- HMGB1
high mobility group box 1
- HSPA1A
heat shock 70kDa protein 1A
- IR
ionizing radiation
- MAPK8
mitogen-activated protein kinase 8
- MEFs
mouse embryonic fibroblasts
- MTMR
myotubularin-related protein
- MTOR
mechanistic target of rapamycin (serine/threonine kinase)
- MTORC1
mechanistic target of rapamycin complex 1
- MVBs
spherical multivesicular bodies
- NRBF2
nuclear receptor binding factor 2
- PAS
phagophore assembly site
- PDPK1
3-phosphoinositide dependent protein kinase 1
- PE
phosphatidylethanolamine
- PIKFYVE
phosphoinositide kinase, FYVE finger containing
- PINK1
PTEN-induced putative kinase 1
- PtdIns3K-C1
class III phosphatidylinositol 3-kinase complex I
- PtdIns3K-C2
class III phosphatidylinositol 3-kinase complex II
- PKB
protein kinase B
- PRKA
protein kinase, AMP-activated
- PRKD1
protein kinase D1
- PRKDC
protein kinase, DNA-activated, catalytic polypeptide
- PtdIns5P
phosphatidylinositol 5-phosphate
- PtdIns4P
phosphatidylinositol 4-phosphate
- PtdIns(4,5)P2
phosphatidylinositol 4,5-bisphosphate
- PtdIns3P
phosphatidylinositol 3-phosphate
- PtdIns(3,5)P2
phosphatidylinositol 3,5-bisphosphate
- PtdIns(3,4,5)P3
phosphatidylinositol 3,4,5-trisphosphate
- RHEB
Ras homolog enriched in brain
- RPS6KB1
ribosomal protein S6 kinase, 70kDa, polypeptide 1
- SQSTM1
sequestosome 1
- TECPR1
tectonin β-propeller repeat containing 1
- TFEB
transcription factor EB
- TRIM28
tripartite motif containing 28
- TSC
tuberous sclerosis
- UV
ultraviolet
- UVRAG
UV radiation resistance associated
- WDFY3
WD repeat and FYVE domain containing 3
- WIPI
WD repeat domain, phosphoinositide interacting
- ZFYVE1
zinc finger, FYVE domain containing 1.
Introduction
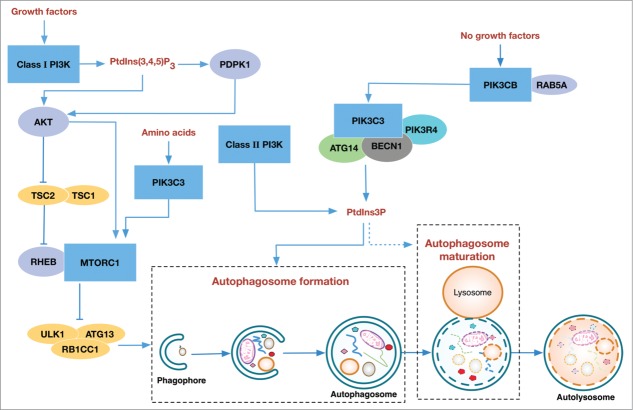
Macroautophagy (hereafter referred as autophagy) is a cellular degradation process conserved in eukaryotic cells in which intracellular organelles and proteins are engulfed by double-membrane compartments termed phagophores that mature into autophagosomes, which subsequently fuse with lysosomes for degradation. Basal autophagy constitutively occurs as a housekeeping mechanism to clear “cytotoxic” components such as misfolded proteins and damaged organelles.1 This process is enhanced under nutrient-limiting conditions to provide the needed nutrients for cell survival.2-4 Given that intracellular components are broken down during this process, essential elements must be spared and the initiation and progression of autophagy must be tightly controlled, which is ensured by intricate regulatory mechanisms via numerous kinases and signaling pathways. One of the most pivotal groups of kinases controlling autophagy are the phosphatidylinositol 3-kinases (PtdIns3Ks) and the phosphoinositide 3-kinases (PI3Ks). In this review, we will discuss the multilateral and multipronged regulation of autophagy by 3 classes of PtdIns3K and PI3K enzymes (Fig. 1).
Figure 1.
Regulation of autophagy by 3 classes of PtdIns3K and PI3Ks. The main regulatory pathways are illustrated: class I PI3K responds to growth factor signaling and produces PtdIns(3,4,5)P3 which negatively regulates autophagy via the AKT-MTORC1-ULK1 complex, whereas the class III PtdIns3K, mainly the ATG14-containing PtdIns3K-C1, as well as class II PI3K, contributes to the initiation and progression of autophagy through the production of PtdIns3P. PtdIns3P positively regulates the formation of autophagosome. However, the function of PtdIns3P in the maturation step is controversial (depicted in dotted line): its presence is required for the recruitment of effector proteins for the maturation of autophagosome, but its turnover is also essential for the fusion of the autophagosome with the lysosome. Moreover, the PIK3CB subunit of class I PI3K activates PtdIns3K-C1 through RAB5A when growth factor is unavailable, thus promoting autophagy. In addition, PIK3C3 may also act as a sensor of amino acid and an activator of MTORC1.
The Process of Autophagy and its Machinery
The entire process of autophagy can be viewed as being comprised of the following stages: initiation, nucleation, expansion, maturation, and degradation. Numerous autophagy-related (ATG) genes have been identified and the corresponding ATG proteins are sequentially activated to regulate different stages of autophagy. These proteins can be grouped into various complexes based on their functions: the ULK1 kinase complex, the ATG9-ATG2-WIPI1/Atg18 complex, PIK3C3/Vps34-BECN1-PIK3R4/p150/Vps15 complex, and 2 ubiquitin-like conjugation systems, which include the ATG12–ATG5 and LC3/GABARAP proteins.5–7 In mammalian cells, under autophagy-stimulating conditions, the ULK1 complex (mainly composed of ULK1, ATG13, ATG101 and RB1CC1/FIP200) undergoes a series of phosphorylation/dephosphorylation events, which lead to the activation of this complex and the initiation of autophagy.8 After the initiation step, a cup-shaped, primary double-membrane structure, the phagophore, is gradually formed.8,9 The nucleation of the phagophore membrane takes place at a site termed the phagophore assembly site (PAS) in yeast,10 whereas an additional site, the omegasome, may play a similar role in mammalian cells.9 The recruitment of the ULK1 and PIK3C3 complexes to the nucleation site is the initial step for autophagy,11-15 and ULK1 relocalization is a prerequisite for ATG14-PIK3C3 complex targeting.11,14,15 The PIK3C3 complex generates phosphatidylinositol 3-phosphate (PtdIns3P) at the nucleation site,16 which creates a PtdIns3P-enriched milieu, reinforcing the localization of the ULK1 complex and other ATG proteins at the growing membrane9,12 and triggering the assembly of PtdIns3P-binding effector proteins, which is critical for the subsequent elongation of the phagophore and maturation of the autophagosome.7,17 The ATG12 ubiquitin-like conjugation system mediates the conjugation of ATG12 to ATG5, which then binds to ATG16L1.9,14 The latter, which resides at the elongating membrane, facilitates the subsequent modification of LC3 into a phosphatidylethanolamine (PE)-linked form.5,9,14,15 The lipidated LC3 then accumulates on phagophore and autophagosomal membranes, which presumably plays a role in membrane bending, curvature, and closure,9,18 tethering of unidentified precursor structures of autophagosomes,8 and selective targeting of autophagy substrates.9,19 During phagophore expansion, the ATG9-ATG2-WIPI1 complex is likely involved in recruiting lipids to the growing phagophore.20-22 In the final step, the trafficking and fusion of autophagosomes with lysosomes is facilitated by PtdIns3P-binding proteins, PIK3C3 complexes, and interacting partners, as well as RAB proteins.7,16,23 The dynamics of the autophagic process, from the growth and closure of membrane structures to the acquisition of membrane-bound proteins, can be viewed as a membrane trafficking process, reminiscent of the endosomal trafficking pathway. It is not surprising that cells utilize an overlapping set of phospholipids, lipid kinases, and membrane-bound proteins in both the autophagy and endosomal pathways.
Three Classes of PtdIns3K and PI3K enzymes
In mammalian cells, PtdIns3Ks and PI3Ks are grouped into 3 classes based on their structural features and substrate specificities, as summarized in Table 1. Class I PI3Ks are heterodimeric kinases composed of a catalytic subunit (PIK3CA, PIK3CB, PIK3CD, or PIK3CG) and a regulatory subunit, and they are further subdivided to class IA (with PIK3CA, PIK3CB, or PIK3CD as the catalytic subunit) and class IB (with PIK3CG as the catalytic subunit).24,25 Class I PI3Ks can phosphorylate PtdIns, PtdIns4P and PtdIns(4,5)P2 in vitro, but they may preferentially use PtdIns(4,5)P2 to produce PtdIns(3,4,5)P3 in vivo.24 Class II PI3Ks are monomeric kinases which have 3 isoforms (PIK3C2A, PIK3C2B and PIK3C2G), and they prefer PtdIns and PtdIns4P as substrates to produce PtdIns3P and PtdIns(4,5)P2.26 Both class I PI3K and class II PI3K are downstream effectors of receptor tyrosine kinases and G-protein-coupled receptors.24 Note that both class I and II enzymes can function as either phosphatidylinositol 3-kinases (phosphorylating the 3′ hydroxyl of phosphatidylinositol) or phosphoinositide 3-kinases (i.e., adding an additional phosphate to a phosphoinositide) in vitro, but are referred to as PI3Ks for simplicity. Class III PtdIns3K forms large multi-subunit complexes with PIK3C3 as the catalytic subunit and PIK3R4 as the regulatory subunit, and their composition diversifies in different biological context. Class III PtdIns3K generates PtdIns3P, and functions as the most pivotal positive regulator of autophagy.24,27
Table 1.
Composition and substrate preference of 3 classes of PI3Ks in mammalian cells
| Subunits |
||||||
|---|---|---|---|---|---|---|
| Class | Catalytic subunit | Regulatory subunit | Lipid substrates | Products | Function in autophagy | |
| Class I PI3K | Class IA |
PIK3CAPIK3CBPIK3CD |
PIK3R1PIK3R2PIK3R3 |
PtdInsPtdIns4PPtdIns(4,5)P2 | PtdIns3PPtdIns(4,5)P2PtdIns(3,4,5)P3 | Anti-autophagy |
| |
Class IB |
PIK3CG |
PIK3R5PIK3R6 |
|
|
|
| Class II PI3K |
PIK3C2APIK3C2BPIK3C2G |
— |
PtdInsPtdIns4P |
PtdIns3PPtdIns(3,4)P2 |
Pro-autophagy |
|
| Class III PtdIns3K | PIK3C3 | PIK3R4 | PtdIns | PtdIns3P | Pro-autophagy | |
Dash (−) indicates that it does not exist in mammalian cells.
MTOR (mechanistic target of rapamycin), ATM serine/threonine kinase (ATM), ATR serine/threonine kinase (ATR), and PRKDC (protein kinase, DNA-activated, catalytic polypeptide) constitute the PI3K-related kinase family, sometimes referred to as class IV PI3K, based on their sequence homology with other PI3Ks.28,29 The mechanistic role of MTOR in autophagy regulation will be reviewed in details below. ATM, ATR, and PRKDC are key signaling transducers in the DNA damage response (DDR) network.30,31 They are activated by ultraviolet (UV) light, ionizing radiation (IR), and DNA damaging chemicals, relay the DNA damage signal to DNA repair proteins, TP53/p53 and other transcription effectors, checkpoint kinases, and other cell cycle regulators to facilitate DNA damage repair and to make cell fate decisions.30-32 Accumulating evidence has unveiled the complex crosstalk between DDR pathways and autophagy. Activation of ATM33-36 by various insults induces autophagy, and TP5337-39 and MTOR35,36,39-41 are 2 representative downstream signaling nodes connecting the DDR pathways to autophagy. The descriptions of autophagy regulation by ATR are rather scarce, but there is strong evidence showing that ATR is a potential upstream regulator of autophagy.42 With regard to PRKDC, its connection to the autophagy pathway is less clear, albeit there is evidence suggesting the presence of such a link.38,43,44 The mobilization of autophagy via these DDR transducers can be prosurvival38,39,45-48 or antisurvival;38,39,48,49 and the dual roles that autophagy plays in cell fate determination under DNA damage stress conditions add to the complexity of its involvement in DDR, and may influence the outcome of anticancer chemotherapy. The interplay between DDR and autophagy is a burgeoning area of research in the autophagy field, linking autophagy with genomic integrity. In light of the fact that class IV PI3Ks are an unconventional class of PI3Ks, we do not cover their regulatory functions in autophagy in this review.
Differential Regulation of Autophagy by PtdIns3K and PI3Ks
In mammalian cells, 3 classes of PI3K enzymes localize at distinct subcellular compartments and specialize in controlling disparate stages of autophagy in response to diverse stimuli. The first observation of differential regulation of autophagy by class I PI3K and class III PtdIns3K was reported by Petiot et al.50 in human colon cancer cells. This distinct regulatory mechanism is now recognized as the canonical paradigm: whereas class I PI3Ks act as the negative regulator of autophagy at the plasma membrane, class III PtdIns3K is the indispensable kinase producing PtdIns3P at the precursor membrane which drives autophagy progression (Fig. 1). However, the real regulatory network of autophagy by PtdIns3K and PI3Ks is far more complicated than what this simplified picture represents, and the role of class II PI3K in autophagy is just beginning to emerge.
Regulation of autophagy by class I PI3Ks
The signaling cascade through which class I PI3K-AKT-MTORC1 regulates autophagy has been well characterized (Fig. 1). Upon ligand-receptor binding (e.g., growth factors binding to respective receptors), class I PI3Ks translocate onto the inner leaflet of the plasma membrane, where they phosphorylate PtdIns(4,5)P2 to generate PtdIns(3,4,5)P3.51 PtdIns(3,4,5)P3 recruits proteins bearing PH domains. Among these proteins, the serine/threonine protein kinase AKT/PKB has a pivotal role in class I PI3K signaling networks. AKT binds to the PtdIns(3,4,5)P3-rich region on the plasma membrane along with another PH domain-containing kinase, PDPK1/PDK1 (3-phosphoinositide dependent protein kinase 1). AKT then undergoes phosphorylation mediated by PDPK1 and MTORC2, rendering AKT fully active.51 Activated AKT in turn regulates various downstream targets to promote nutrient uptake, metabolism, cell growth, and proliferation.51,52
One of the most important signaling pathways downstream of class I PI3K and AKT in regulation of autophagy is MTOR complex 1 (MTORC1).52,53 MTORC1 is composed of MTOR, RPTOR, MLST8, AKT1S1/PRAS40, and DEPTOR, and it integrates a variety of signals, including growth factors, amino acids, and glucose via distinct signaling pathways.54 Amino acids regulate MTORC1 via their control of the nucleotide status of RRAG proteins and lysosomal localization of MTORC1.55-57 Glucose availability modulates MTORC1 through PRKA/AMPK (protein kinase, AMP-activated), which is activated by an increase in the cytosolic AMP/ATP ratio.56,58,59 The input of growth factor signals from the plasma membrane is transmitted by class I PI3K-AKT to the tuberous sclerosis (TSC) complex, resulting in the phosphorylation of TSC2 by AKT and thereby the inactivation of TSC1-TSC2. TSC1-TSC2 inactivation abolishes its GTPase-activating protein (GAP) activity toward RHEB (Ras homolog enriched in brain), and preserves RHEB in its GTP-bound state, which is required for MTORC1 activation.56,59,60 In addition to the TSC1-TSC2-RHEB-MTORC1 arm, AKT can directly regulate MTORC1 by phosphorylating and displacing AKT1S1, a negative regulator of MTORC1.54,59 Active MTORC1 docks at lysosomes, promoting protein synthesis, and cell growth through EIF4EBP1/4E-BP1 (eukaryotic translation initiation factor 4E binding protein 1) and RPS6KB1/S6K1 (ribosomal protein S6 kinase, 70kDa, polypeptide 1), and inhibits autophagy via phosphorylation of ULK1 and ATG13.53,54 Phosphorylation of the latter proteins by MTORC1 destabilizes the ULK1-ATG13-ATG101-RB1CC1 complex and inhibits ULK1 kinase activity, leading to inhibition of autophagy initiation.53 When MTORC1 is inhibited, ULK1 and ATG13 become dephosphorylated and the ULK1 complex is engaged as an autophagy initiator and recruiter for other effectors such as ATG14.27 Moreover, ULK1 is subject to ubiquitination mediated by AMBRA1 (autophagy/beclin-1 regulator 1) under proautophagic conditions, stabilizing this kinase and increasing its activity, whereas AMBRA1 is under the regulation of MTORC1.61 In addition, MTORC1 negatively regulates lysosomal biogenesis and function via impeding the nuclear translocation of TFEB (transcription factor EB);23,62-64 therefore, MTORC1 influences the degradative capacity of lysosomes, which provides another link between class I PI3K and autophagy.
The second pathway underlying the regulatory function of class I PI3K and AKT in autophagy is related to the fact that AKT promotes glucose uptake and glycolysis, which can contribute to ATP production.27,52 As mentioned above, when the cytosolic AMP/ATP ratio is elevated, PRKA is activated and suppresses MTORC1, either by TSC2 activation or RPTOR inhibition through phosphorylation.58 Conversely, the preservation of ATP level by enhanced glucose utilization prevents the induction of autophagy. In addition to the effect through MTORC1, PRKA is also able to activate autophagy via phosphorylation of ULK1, when cellular energy becomes scarce.65 Therefore, promotion of glucose metabolism by AKT prevents the activation of PRKA and autophagy induction.
The third possible mechanism associated with the regulatory effect of class I PI3K and AKT is via the FOXO (forkhead box O) nuclear transcription factors. It has been well established that AKT phosphorylates FOXOs, resulting in their cytoplasmic retention and inactivation.66 FOXOs are a family of transcription factors in charge of the transcription of a series of autophagy-related genes, such as ATG12, BCEN1 and GABARAPL1.67 Thus, class I PI3K controls not only the initiation of autophagy, but also the replenishment of autophagic machinery during autophagy.
Fourth, it is intriguing that the activity of MTORC1 is negatively associated with that of class III PtdIns3K, and this relationship is specific to the ATG14-containing class III PtdIns3K complex,68 indicating that class I PI3K signaling is coordinated with class III PtdIns3K activity.
However, there is an exception to this paradigm: the PIK3CB/p110β catalytic subunit of class I PI3K. In contrast to the role of class I PI3K, which generally suppresses autophagy, PIK3CB acts as a positive regulator of autophagy. Pik3cb-deficient mouse embryonic fibroblasts (MEFs), liver and heart display impaired autophagic function, whereas overexpression of PIK3CB activates autophagy.69 Subsequently, it was found that upon growth factor withdrawal, PIK3CB dissociates from the growth receptor complex and interacts with RAB5A/RAB5, keeping this small GTPase in the GTP-bound state.70 GTP-loaded RAB5A plays a role in the activation of PIK3C3.71 Through association with RAB5A, PIK3CB enhances PIK3C3 activity and promotes autophagy.70 In contrast to the class I PI3K holokinases, which carry the signals from growth factors and indirectly curb autophagy, the PIK3CB subunit acts as a sensor of growth factor availability and stimulates autophagy under circumstances of growth factor limitation.
Regulation of autophagy by class III PtdIns3K
The primary clue for the dependence of autophagy on PtdIns3K activity was yielded by the study showing that wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes, although it was unclear which class of PI3K accounted for autophagy regulation.72 The first revelation of the relevance of PIK3C3/Vps34 to autophagy was provided by a study in the yeast Hansenula polymorpha, showing that its PIK3C3 homolog participates in pexophagy, a selective form of autophagy.73 After the dissection of the specific functions of 2 Vps34 complexes in autophagy and vacuolar protein sorting,74 the involvement of mammalian class III PtdIns3K in autophagy was subsequently uncovered in a study which described the opposing functions of class I and III PI3K and PtdIns3K enzymes.50 Class III PtdIns3K forms different protein complexes, which are organized in a divergent and multilayered fashion (Fig. 2). The stratification can be viewed based on their interaction and stability: PIK3C3-PIK3R4-BECN1, the most stable and prevalent complex, constitutes the core; while ATG14 and UVRAG (UV radiation resistance associated) are combined with the core in PtdIns3K complex I (PtdIns3K-C1; ATG14-containing PtdIns3K complex) and PtdIns3K complex II (PtdIns3K-C2; UVRAG-containing PtdIns3K complex), respectively;75-78 KIAA0226/Rubicon is a regulatory subunit that is thought to stably bind to UVRAG-containing complexes;77,78 AMBRA1, NRBF2 (nuclear receptor binding factor 2) and other accessory factors transiently or unstably interact with either BECN1 (e.g., AMBRA1)79,80 or ATG14 (e.g., NRBF2),81-83 thus comprising the peripheral components.84,85 UVRAG also exists in a separate complex excluding ATG14, which holds an important role in the regulation of autophagy at the maturation step.84,86 Atg14/ATG14 and Vps38/UVRAG are present exclusively in 2 Vps34/PIK3C3 complexes (PtdIns3K complex I and II in yeast, and class III PtdIns3K-C1 and PtdIns3K-C2 in mammalian cells, respectively), which are organized in similar V-shape conformations with Vps15/PIK3R4 at the base.87 In yeast, PtdIns3K complex I and II play a role in autophagy and the vacuolar protein sorting pathway, respectively.74 In mammalian cells, PtdIns3K-C1 and PtdIns3K-C2 mainly function in autophagy and endosomal trafficking, respectively;75,88,89 however, both are implicated in autophagy.75,76,86 In the PIK3C3 complexes, PIK3R4 is required for the kinase activity of PIK3C3,89–92 and it serves as a bridge linking PIK3C3 with ATG14-BECN1, or with UVRAG-BECN1.87 For clarity, the compositions of Vps34/PIK3C3 complexes in yeast and mammals and their functional specificity are summarized in Table 2. In the following sections, we will discuss in more detail how each of the components in class III PtdIns3K complexes functions as a point in the autophagy-regulatory network under different circumstances and the composition of the corresponding complexes (Fig. 2).
Figure 2.
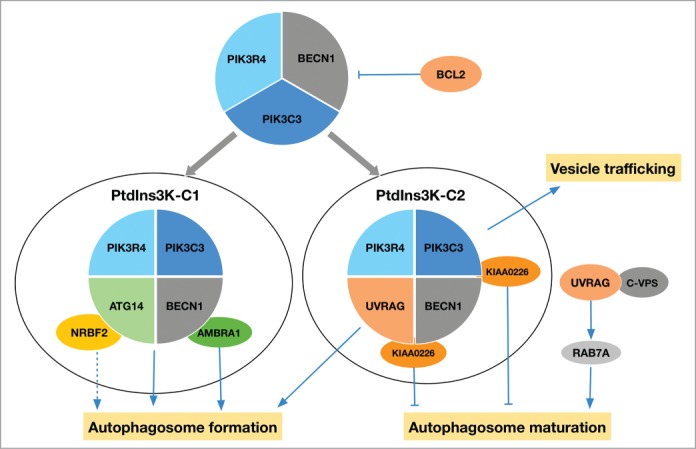
Diversified composition of class III PtdIns3K complexes and their roles in autophagy. PIK3C3-PIK3R4-BECN1 is the core complex; a proportion of the core complex is combined with ATG14 or UVRAG in a mutually exclusive manner, constituting PtdIns3K-C1 and PtdIns3K-C2, respectively. Both PtdIns3K-C1 and PtdIns3K-C2 contribute to autophagosome formation, and UVRAG can regulate autophagosome maturation independent of BECN1 through interacting with the class C VPS complex (C-VPS) and activating RAB7A. BCL2, by binding to BECN1, inhibits the formation of class III PtdIns3K complexes. KIAA0226 is considered as a negative regulator of autophagosome maturation, which interacts with PtdIns3K-C2 through directly binding to PIK3C3, BECN1, and UVRAG. As peripheral proteins, AMBRA1 and NRBF2 both interact with PtdIns3K-C1. AMBRA1 activates autophagy at an early step, while NRBF2 plays regulatory roles in autophagosome formation (dotted line denotes that there is controversy about the effect of NRBF2 on autophagy). Additional peripheral components of class III PtdIns3K complexes have also been reported, which are not included in the diagram.
Table 2.
Composition and functional specificity of Vps34/PIK3C3 complexes in yeast and mammals
| Yeast |
Mammals |
|||
|---|---|---|---|---|
| Complex | PtdIns3K complex I | PtdIns3K complex II | PtdIns3K-C1 | PtdIns3K-C2 |
| Core kinase | Vps34 | Vps34 | PIK3C3 | PIK3C3 |
| Regulatory subunits | Vps30 | Vps30 | BECN1 | BECN1 |
| Vps15 | Vps15 | PIK3R4 | PIK3R4 | |
| Atg38 | NRBF2 | |||
| Defining component | Atg14 | Vps38 | ATG14 | UVRAG |
| Biological function | Autophagy | Vacuole protein sorting | Autophagy | Endosomal trafficking and autophagy* |
*The role of PtdIns3K-C2 in autophagy is still controversial.
PtdIns3P is a key phospholipid required for the progression of autophagy
PtdIns3P acts as a signaling molecule and platform for the assembly and coordination of various effector proteins during the autophagic process. Under basal conditions, PtdIns3P is detected using a 2 × FYVE probe on early endosomes, spherical multivesicular bodies (MVBs), and the nucleus in mammalian cells, reflecting its role in vesicle trafficking.93 Upon autophagy induction, this phospholipid migrates and becomes punctate, periodically forming an omega-shaped structure, which presumably corresponds to the omegasome.16 The dynamic evolution of the PtdIns3P-positive compartment was monitored by ZFYVE1/DFCP1 (zinc finger, FYVE domain containing 1) using live cell imaging.16 This structure expands following amino acid withdrawal, and at a certain time point LC3 accumulates and co-localizes with ZFYVE1. The membrane structure positive for both ZFYVE1 and LC3 dilates with time until LC3 exits from the PtdIns3P-enriched membrane, after which the ring-shape becomes increasingly invisible. Although the origin of the phagophore remains a question of extensive debate (with the endoplasmic reticulum [ER], mitochondria, Golgi apparatus, and plasma membrane all being candidates as a membrane source for autophagy17), this study demonstrated that the PtdIns3P-containing punctate structures are in close association with the ER.16 The spatial and temporal dynamic involvement of PtdIns3P-positive structure suggests a critical role of PtdIns3P for autophagosome biogenesis.16 Indeed, production of PtdIns3P serves as the most primary signal for autophagosome formation through recruiting a number of effectors. As the very early event of autophagy initiation, most of the ULK1 complexes form punctate structures spatially overlapping with omegasomes; however, only when PtdIns3Ps are generated and enriched at the loci is the punctation of ULK1 stabilized and enhanced.12 The other PtdIns3P effectors at the early stages include ZFYVE1, WIPI (WD repeat domain, phosphoinositide interacting) proteins, and WDFY3/ALFY (WD repeat and FYVE domain containing 3).7,27 The role of ZFYVE1 in autophagy remains elusive, but the punctation of ZFYVE1 is one of the early observable events during nucleation.16 WIPI proteins (WIPI1 to WIPI4), mammalian homologs of yeast Atg18, may have the ability to sense and regulate PtdIns3P levels and participate in the initiation and maturation of the omegasome.7,27,94,95 WDFY3 is implicated in cargo selection and delivery in selective autophagy.17,27,96 It is further conjectured that the PtdIns3P synthesized in situ, together with its effector proteins, signals the de novo synthesis of other phospholipids (e.g., phosphatidylcholine, phosphatidylethanolamine, and phosphatidylserine), which constitute the autophagosomal membrane.97 At the late stage of autophagy, additional PtdIns3P effectors interact with PtdIns3P. TECPR1 (tectonin β-propeller repeat containing 1), associating with both PtdIns3P and the ATG12–ATG5 complex, mediates the fusion of autophagosomes with lysosomes,98 whereas FYCO1 (FYVE and coiled-coil domain containing 1) is also involved in the maturation and trafficking of autophagosomes.17,99 The stringent requirement of phospholipids in the progression of autophagy points to the indispensable role of their producers, PtdIns3Ks and PI3Ks, in the autophagic pathway.
However, it should be mentioned that PtdIns3P is not persistently and equally concentrated in both leaflets of autophagosomal membrane during the course of autophagy.16,100,101 The completion of autophagy requires the clearance of PtdIns3P from the autophagosome, enabling the disassembly of its effectors and the fusion of the autophagosome with the lysosome. The dephosphorylation of PtdIns3P is carried out by phosphoinositide 3-phosphatases in the myotubularin protein family.102 Among these phosphatases, the involvement of MTMR14/Jumpy (myotubularin-related protein 14),103,104 MTMR3,105 MTMR6 and MTMR7103 in autophagy has been documented. The yeast PtdIns3P phosphatase Ymr1 is essential for the late stage of autophagy, and the absence of this protein results in the accumulation of autophagosomes in the cytoplasm; the failure in the turnover of PtdIns3P and dissociation of Atg proteins from the autophagosomal membrane presumably accounts for the defect in autophagosome-vacuole fusion in the ymr1C397S phosphatase-inactive strain.101 Therefore, it is clear that the balance of PtdIns3P, which is tightly controlled by both kinases and phosphatases, may determine the timing and extent of autophagic flux. In addition to the dose, the asymmetric distribution of PtdIns3P on the 2 faces of the double autophagosomal membrane is also critical, which may be related to the recruitment of effectors to specialized sites as well as the remodeling of the associated membrane. For the latter notion, the study by Cheng et al.100 may provide supporting evidence. An intensive delineation of PtdIns3P distribution on autophagosomal membranes indicated that in yeast, PtdIns3P is located in both leaflets of the autophagosome, with more abundance in the lumenal leaflet than the cytoplasmic leaflet. Conversely, mammalian PtdIns3P is exclusively distributed in the cytoplasmic leaflet during autophagy. It was further shown that Ymr1 partially accounts for the PtdIns3P asymmetry, and, interestingly, abrogation of this asymmetry results in the aberrant structure of autophagosomal vesicles.100 The regulatory role of phosphatases in autophagy is equally important to that of PtdIns3Ks and PI3Ks; however, the scope of this review precludes in-depth discussion on the functions of the former in autophagy.
The list of autophagy-relevant phosphoinositides does not end with PtdIns3P. PtdIns4P, PtdIns(4,5)P2, PtdIns5P, and PtdIns(3,5)P2 have also been described to participate in autophagy (e.g., selective autophagy, and/or a certain stage/stages of macroautophagy; for a detailed review, see ref. 17).17,106 Intriguingly, PtdIns5P, produced by the type III PtdInsP 5-kinase PIKFYVE (phosphoinositide kinase, FYVE finger containing) or the phosphatase MTMR3 plays a predominant role in glucose starvation-induced autophagy, in which the role of PtdIns3P is dispensable.106 In the case of autophagy induced in HBSS starvation medium, PtdIns5P also has a compensatory effect to sustain autophagy if PtdIns3P is depleted.106 In particular, the noncanonical autophagy pathways stimulated by nonstarvation conditions are likely to be mediated by PtdIns3P-independent mechanisms.107-109 This emerging body of evidence points to the need to revisit the requirement of PtdIns3P for autophagy; in spite of the discrepancy, PtdIns3P is still widely accepted as a key phospholipid for the autophagic process, which remains one of the main focuses in this review.
Regulation of autophagy by PIK3C3 via formation of protein complexes and direct post-translational modifications
PIK3C3 is so far recognized as the predominant source of PtdIns3P, and it holds a fundamental role in both endosomal trafficking and autophagy.90 Pik3c3 knockout mice are embryonic lethal,110 whereas tissue specific ablation of Pik3c3 blocks autophagic degradation and protein turnover, and severely debilitates liver and heart function.111
As the core catalytic subunit in the class III PtdIns3K complex, PIK3C3 per se is a point of regulatory convergence by diverse autophagy-modulating mechanisms. A study by Kim et al.112 demonstrated the presence of 2 different regulatory mechanisms controlling the role of PIK3C3 in autophagy via: (i) formation of different protein complexes and (ii) direct phosphorylation of PIK3C3. In their study, PIK3C3 is present in 2 types of class III PtdIns3K complexes, nonautophagic and proautophagic; and glucose starvation selectively elevates the kinase activity of ATG14- or UVRAG-containing class III PtdIns3K complexes (proautophagic), but suppresses that of PIK3C3 in complexes lacking these 2 proteins (nonautophagic). This dichotomous regulation is enabled by the preferential phosphorylation of free PIK3C3 (T163/S165) or BECN1 (S91/S94) in proautophagic class III PtdIns3K complexes by PRKA.112 In addition to this selected modification under starvation, it is interesting that PIK3C3 activity is probably modulated in sync with cell cycle progression, supported by the evidence that CDK1 (cyclin-dependent kinase 1) and CDK5 phosphorylate PIK3C3 at distinct sites, which represses its activity.113 Additionally, PRKD1/PKD (protein kinase D1) phosphorylates PIK3C3 under conditions of oxidative stress,114 and is dependent on acetylated HSPA1A (heat shock 70kDa protein 1A), whereas TRIM28/KAP1 (tripartite motif containing 28) is able to mediate PIK3C3 SUMOylation in response to autophagy-inducing stress,115 both of which activate autophagy.
PIK3C3 is widely accepted as a positive regulator of autophagy; however, the function of PIK3C3 in amino acid sensing adds another layer of complexity to its regulatory role in autophagy. The amino acid sensing model through Ca2+-CALM/calmodulin-PIK3C3 has been reported previously.116 In this model, PIK3C3 is part of the MTOR complex; amino acids stimulate an elevation in the intracellular Ca2+ level and Ca2+-CALM/calmodulin binding, which increases PIK3C3 activity; consequently, PtdIns3Ps produced by PIK3C3 activate MTOR by replacing FKBP8/FKBP38 with RHEB.116 This model is consistent with earlier studies showing that PIK3C3 is required for MTOR-RPS6KB1 signaling activity,117,118 and is supported by the evidence that amino acid restimulation of MTOR activity is impaired in Pik3c3 KO MEFs111 and MTOR activity is remarkably ablated in Pik3c3 KO embryos.110 However, several issues are still under dispute concerning the amino acid-Ca2+-CALM-PIK3C3-MTOR signaling model. For instance, the increase in intracellular Ca2+ by amino acid stimulation as shown in the study116 was contradicted by the result of another investigation demonstrating that amino acid withdrawal, rather than addition, resulted in cytosolic Ca2+ increase.119 Second, the dependence of PIK3C3 activity on Ca2+-CALM116 is refuted by the evidence that neither Ca2+ chelator nor CALM inhibitor blocks PIK3C3 activity.91 More importantly, the effect of amino acid withdrawal on different PIK3C3 complexes remains unsettled: contradictory observations were reported showing the activation of general PIK3C3116,118,120 but suppression of ATG14-interacting PIK3C368,120 after amino acid deprivation. It should be noted that the study by Russel et al.120 may offer a potential explanation: the opposing effects of amino acid withdrawal on class III PtdIns3K activity in PIK3C3-BECN1 and PIK3C3-ATG14 immunoprecipitates were demonstrated, and the presence of ATG14 is likely to determine the specific activation of the ATG14-containing PIK3C3 complex mediated by ULK1 phosphorylation of BECN1. Together with the evidence of differential regulation of PIK3C3 complexes by energy deprivation,112 a hypothesis that may reconcile the contradictory roles of PIK3C3 in MTOR signaling and autophagy could be proposed: certain PIK3C3-interacting components, for instance ATG14, may specify the differential regulation of distinct PIK3C3 complexes by nutrients, or determine the spatial distribution of PIK3C3 that confers divergent biological functions. Alternatively, the requirement of PIK3C3 for MTORC1 activity may be ascribed to the release of amino acids by autophagic degradation. Our group121 and others122 have demonstrated that the generation of amino acids by autophagy is essential for MTORC1 activity; thus, the impairment of autophagy by Pik3c3 knockdown or inhibition may influence this feedback circuit. Additionally, since a proportion of PIK3C3 complexes are involved in the endosomal system and are plausibly important for proper lysosomal function,75,76,111,123 Pik3c3 ablation may affect the activation of MTORC1 by amino acids at the lysosome, which is dependent on the function of several lysosomal proteins55-57 and normal lysosomal degradation.122,124 In contrast, yeast Vps34 is the sole PtdIns3K and it functions only in vacuolar trafficking/autophagy but not in the TORC1 regulatory network;125 whether and why mammalian PIK3C3 has evolved to harbor bifurcate functions in MTOR modulation and autophagy is a mystifying question. This perplexing issue notwithstanding, the paradoxical roles of PIK3C3 in MTOR stimulation in response to amino acid and autophagy induction highlight PIK3C3 as a key node in the cell signaling network, worthy of intensive study.
Regulation of autophagy by PIK3C3 via BECN1
BECN1 (the mammalian homolog of yeast Vps30/Atg6) is among the first mammalian homologs of yeast Atg proteins discovered. It binds to BCL2 family-antiapoptotic proteins through its BCL2 homology 3 (BH3) domain, interacts with PIK3C3 via its coiled-coil domain (CCD) and evolutionarily conserved domain, and associates with UVRAG or ATG14 through the CCD.126,127 Similar to Pik3c3 deletion, knockout of Becn1 results in embryonic mortality.128 BECN1 is required for PIK3C3 activity and autophagy, and deficiency in BECN1 results in impaired autophagic function.128,129 Binding of BCL2 to BECN1 blocks autophagy, as BCL2 shields BECN1 from class III PtdIns3K complexes and prevents autophagy.126,127 Nutrient-rich conditions promote BCL2-BECN1 binding and inhibit autophagy,130 whereas various stresses disrupt this interaction and unfetter BECN1 as an autophagy initiator.131,132 The phosphorylation of BCL2 by MAPK8/JNK1 (mitogen-activated protein kinase 8) under starvation conditions,131 and CDKN2A/p19Arf/ARF at the mitochondria,132 reduce BCL2 and BECN1 interaction, resulting in the activation of autophagy. Modification of BECN1 itself is another way to modulate autophagy. BECN1 phosphorylation by ULK1 at Ser14 is one of the key initiating events required for autophagy induction upon starvation or MTOR inhibition, leading to activation of the ATG14-containing class III PtdIns3K complexes.120 The aforementioned PRKA phosphorylation of BECN1 under glucose starvation also enhances the activity of ATG14- or UVRAG-containing class III PtdIns3K complexes.112 Of note, BECN1 is an important downstream target of AKT, which inhibits autophagy, and aberrant BECN1 phosphorylation by AKT is implicated in tumorigenesis.133 In this scenario, AKT-mediated phosphorylation of BECN1 increases the binding of BECN1 with YWHA/14–3–3 proteins, which tethers BECN1 to the intermediate filament protein VIM/vimentin, leading to the suppression of autophagy.133 Thus, AKT modulates the indirect interaction of BECN1 with intermediate filaments, mirroring the AMBRA1-regulated subcellular localization of BECN1-PIK3C3 to another kind of cytoskeleton, microtubules (to be discussed below).80 In addition, DAPK1 (death-associated protein kinase 1) phosphorylates BECN1 at its BH3 domain and releases BECN1 from BCL2, which promotes autophagy.134 EGFR can also phosphorylate BECN1, but is inhibitory for PIK3C3 activity and autophagy.135 There are also different circumstances that result in the linkage of BECN1 to diverse types of ubiquitin chains, which exert distinct effects on BECN1 and autophagic activity.136,137 For instance, ubiquitination of BECN1 through Lys-11-linked138 or Lys-48-linked139 ubiquitin chains promotes the degradation of BECN1, which possibly decreases autophagy, whereas Lys-63-linked ubiquitination enhances autophagy through dissociating BECN1 from BCL2.140 More importantly, BECN1 serves as a platform to coordinate various regulators of autophagy, which has been termed the “BECN1 interactome.”85,141 These secondary interacting proteins of class III PtdIns3K complexes, some of which are viewed as the peripheral components of class III PtdIns3K complexes, include AMBRA1, HMGB1 (high mobility group box 1), BIRC5/survivin, PINK1 (PTEN induced putative kinase 1), CISD2/NAF-1 (CDGSH iron sulfur domain 2), and the abovementioned regulators including BCL2; these proteins can either promote (e.g., AMBRA1, HMGB1, BIRC5, PINK1) or inhibit (e.g., CISD2, BCL2) autophagy.85
Regulation of autophagy by PIK3C3 via ATG14
As shown in Figure 2, ATG14 is an integral component of the class III PtdIns3K complex, and its activity is a prerequisite for the proper function of autophagy. The coiled-coil region of ATG14 binds to the CCD region of BECN1 as well as the N-terminal C2 domain of PIK3C3.75,77 Interaction between ATG14 and PIK3C3 can elevate the activity of the latter when BECN1 is present.77 Deletion of ATG14/Atg14 impairs autophagosome formation,75,77,78,142 characterized by accumulation of LC3-II and ubiquitinated protein inclusions.75,77 During autophagosome formation, ATG14 translocates to phagophore and/or omegasome membranes,143 and it is likely to target a proportion of PIK3C3 to the ER, which is crucial for PtdIns3P production and autophagy induction.142,144 Indeed, the Barkor/Atg14(L) autophagosome targeting sequence/BATS domain of ATG14 has been confirmed to be required for targeting the PIK3C3 complex to the phagophore and/or omegasome membrane and for the initiation of autophagosome formation.145 Exogenously expressed ATG14 colocalizes with LC3, ATG12 and ATG5, indicating that ATG14 may engage in the recruitment of LC3, ATG12, and ATG5 at an early step of autophagy.75,77,78 More interestingly, ATG14 likely mediates the differential regulation of PIK3C3 in different class III PtdIns3K complexes. ATG14 may be the determining factor dictating the selective inhibition of the ATG14-containing class III PtdIns3K complex by MTORC1 under nutrient-rich conditions,68 and the activation of PtdIns3K-C1 by ULK1120 and proautophagic class III PtdIns3K complexes by PRKA upon nutrient starvation.112
Regulation of autophagy by PIK3C3 via UVRAG
Human UVRAG cDNA was first isolated and discovered to be able to confer partial UV-resistance on a xeroderma pigmentosum cell line.146 Mutation of UVRAG is frequently observed in various human cancers and may be responsible for the etiology of these cancers.147-150 UVRAG is critical for DNA repair and chromosomal stability, especially in response to radiation.151-153 Using GST affinity isolation, UVRAG was identified as a binding partner of BECN1-PIK3C3, which directly interacts with BECN1 via its CCD.76 UVRAG also binds to the N-terminal C2 domain of PIK3C3.75 UVRAG overexpression enhances the activity of autophagy, the presence of UVRAG can enhance class III PtdIns3K kinase activity, and UVRAG is required for autophagosome formation.76 The essential role of UVRAG in autophagy was further confirmed in Uvrag-deficient mice, in which autophagic flux is abated.154 Additionally, UVRAG was reported to be a target of AKT1, which inhibits autophagy through reduction of UVRAG.155 Interestingly, UVRAG, as a PtdIns3P-binding effector per se, shuttles between ER and autophagic membranes, coordinating different membrane trafficking events, and maintaining organelle homeostasis under various conditions.156 In a PtdIns3P-dependent manner, UVRAG is bound to the ER tethering protein complex RINT1-ZW10-NBAS, involved in Golgi-to-ER transport; however, when autophagy is induced, UVRAG is dissociated from the ER and is bound to the SH3GLB1/Bif-1-BECN1-PIK3C3 complex.156
UVRAG is necessary for vesicle tethering and fusion in the endocytic system.86 The role of UVRAG in vesicle trafficking extends to autophagic membrane trafficking; however, the function of UVRAG in autophagosome-lysosome fusion seems not to require BECN1.86 In a BECN1-independent manner, UVRAG reinforces the association of the class C VPS complex with the autophagosome and activates RAB7A/RAB7, thereby facilitating autophagosome maturation and degradation of autophagic cargo by lysosomes.86 It should be noted that the UVRAG-containing class III PtdIns3K complex is more abundant than the ATG14-containing complex,77,78 probably due to its constitutive function under normal conditions. Finally, there are studies arguing against the requirement of UVRAG for autophagy, which show that UVRAG does not colocalize with LC3 under starvation conditions, and its knockdown does not lead to a defect in autophagy.75,157,158 Although controversy remains regarding the mechanistic role of UVRAG in the regulation of autophagy, its implication in the autophagic process cannot be overlooked, and how it functions in different stages of autophagy awaits further elucidation (Fig. 2)
Regulation of autophagy by PIK3C3 via other interacting partners
In addition to BECN1, ATG14, and UVRAG as discussed above, there are several other important proteins in the class III PtdIns3K complexes that are implicated in the autophagic process; due to limited space, herein we only cover a few of them briefly.
KIAA0226/Rubicon is an important negative regulator in the maturation step of autophagy as well as endocytic trafficking. KIAA0226 was identified as a stable partner interacting with a proportion of BECN1-PIK3C3-UVRAG complexes,77,78 but whether it co-exists with ATG14 remains controversial.77 Overexpression of KIAA0226 retards LC3-II and SQSTM1/p62 (sequestosome 1) degradation and autophagosome/lysosome maturation, whereas knockdown of this protein results in a decline of LC3-II and SQSTM1 as well as an increase in the number of autolysosomes.77,78
AMBRA1 plays a positive role in autophagy presumably by regulating the BECN1-PIK3C3 interaction.79,80 AMBRA1 also acts as a tether controlling BECN1-PIK3C3 subcellular localization. Under normal conditions, AMBRA1 anchors the BECN1-PIK3C3 core complex to microtubules by interacting with dynein light chains 1/2; upon induction of autophagy, ULK1 phosphorylates AMBRA1 and releases the core complex, enabling the latter to translocate to the ER and initiate autophagosome formation.80 In addition, as mentioned above, AMBRA1 is negatively regulated by MTOR; together with the E3-ligase TRAF6, AMBRA1 engages in ULK1 ubiquitination, which promotes ULK1 activity under autophagy-inducing conditions.61
NRBF2 was identified as a protein involved in autophagy, which is present only in the ATG14-containing class III PtdIns3K complex in mammalian cells.81-83,159 The yeast ortholog of NRBF2 is presumed to be Atg38, although there are sequence diversities.82,160 Likewise, Atg38 is recognized as a component of the Atg14-containing Vps34 complex I in yeast, which is required for the autophagy pathway.160 Intriguingly, in yeast Atg38 is responsible for the stability of Vps34 complex I, which connects Vps34-Vps15 with Atg14-Vps30,160 reminiscent of the function of NRBF2 in mediating ATG14, PIK3C3-PIK3R4, and BECN1 interaction in mammalian cells.82 However, it is a question under debate whether NRBF2 positively81,83 or negatively82 regulates autophagy in higher eukaryotes.
The emerging role of class II PI3Ks in autophagy
Since PtdIns3P is the major phosphorylated lipid that drives the progression of autophagy, it is plausible that multiple sources of PtdIns3P exist in eukaryotic cells. Notably, in vitro studies demonstrated that PtdIns is the preferred target of class II PI3K, although this kinase also displays activity toward PtdIns4P.161 It is therefore plausible that class II PI3K is a contributor to the PtdIns3P pool in addition to class III PtdIns3K. In support of this hypothesis, there is emerging evidence that class II PI3K is a significant producer of PtdIns3P, and class II PI3K holds a wide spectrum of functions ranging from cell signaling, to cell migration and vesicle trafficking.161-166 One hint implying a role of class II PI3K in autophagy was provided by the studies on cell corpse clearance by phagocytosis in C. elegans.167,168 In this process, autophagy is required for phagosome maturation and clearance of apoptotic cells; most importantly, class II PI3K was identified as a complementary kinase producing PtdIns3P on phagosomes, indispensable for the degradation of cell corpses in C. elegans.167,168 However, studies from several Pik3c3 knockout animal models suggest that PtdIns3P generation and autophagic function rely predominantly on PIK3C3 activity.111,169,170 The question of whether class III PtdIns3K is the exclusive kinase responsible for PtdIns3P generation during autophagy remained unsolved until a recent study by Devereaux et al. revealed an alternative source of PtdIns3P in Pik3c3 knockout MEFs.171 In Pik3c3 KO MEFs, it is surprising that PtdIns3P is still detectable using a higher affinity PtdIns3P probe and the PtdIns3P-binding protein WIPI1, and autophagosomes are still observed under starvation conditions; class II PI3K was confirmed as another contributor to the PtdIns3P pool during autophagy, and depletion of class II PI3K further decreases autophagic flux, which is inhibited by Pik3c3 deletion.171 Thus, cells may ensure the adequate and timely production of autophagic PtdIns3P by involving more than one producer (Fig. 1).
PtdIns3K and PI3K Inhibitors and their Application in Autophagy and Diseases
Development of potent PtdIns3K and PI3K inhibitors has been a focus of research for years, and great progress has been made in the development of tools to constrain the activity of PI3Ks. The fact that aberrant signaling activity of PtdIns3Ks and PI3Ks is correlated with various pathological conditions and mutations in these enzymes is frequently observed in cancer makes these lipid kinases one of the most sought-after drug targets.24,172 Based on their target selectivity, the available PtdIns3K and PI3K inhibitors can be classified as pan-PI3K inhibitors, dual PI3K-MTOR inhibitors and class or isoform-selective inhibitors, as summarized in Table 3.24,26,173-175
Table 3.
Examples of PtdIns3K and PI3K inhibitors, their status in clinical development and effects on autophagy
| Selectivity | Name of the inhibitor | Molecular targets | Effects on on autophagy | Status in clinical trials | References |
|---|---|---|---|---|---|
| Pan-class | Wortmannin | PtdIns3K and PI3K enzymes, as well as PI3K-related kinases | Inhibition | Failed to enter clinical trial | 26,72,175,177,178,181,190-192 |
| LY294002 | PtdIns3K and PI3K enzymes, as well as PI3K-related kinases | Dual effect: activation and inhibition | Failed to enter clinical trial | 26,72,175,177 | |
| SF1126 | PtdIns3K and PI3K enzymes, as well as PI3K-related kinases | Unknown | Phase I | 177,194-201 | |
| Class I-specific | PX-866 | PIK3CA, PIK3CB, PIK3CD, PIK3CG | Activation | Phase I/II | National Cancer Institute (USA) http://www.cancer.gov/clinicaltrials/24,202-211 |
| XL147 | PIK3CA, PIK3CB, PIK3CD, PIK3CG | Activation | Phase I/II | 24,212-219 | |
| BKM120 | PIK3CA, PIK3CB, PIK3CD, PIK3CG | Activation | Phase I/ II/III | National Cancer Institute (USA) http://www.cancer.gov/clinicaltrials/24,26,220-224 | |
| BAY80–6946 | PIK3CA, PIK3CB | Unknown | Phase I/ II | National Cancer Institute (USA) http://www.cancer.gov/clinicaltrials/24,26,225 | |
| Dual PI3K and MTOR | NVP-BGT226 | PIK3CA, PIK3CB, PIK3CD, PIK3CG, MTOR | Activation | Phase I/ II | 24,173,182,226,227 |
| BEZ235 | PIK3CA, PIK3CB, PIK3CD, PIK3CG, MTOR | Activation | Phase I/ II | National Cancer Institute (USA) http://www.cancer.gov/clinicaltrials/24,228-233 | |
| XL765 | PIK3CA, PIK3CB, PIK3CD, PIK3CG, MTOR | Activation | Phase I/ II | National Cancer Institute (USA) http://www.cancer.gov/clinicaltrials/24,234-238 | |
| Class I-Isoform specific | BYL719 | PIK3CA | Unknown | Phase I/ II | National Cancer Institute (USA) http://www.cancer.gov/clinicaltrials/ |
| INK1117 | PIK3CA | Unknown | Phase I | National Cancer Institute (USA) http://www.cancer.gov/clinicaltrials/ | |
| GSK2636771 | PIK3CB | Unknown | Phase I/II | National Cancer Institute (USA) http://www.cancer.gov/clinicaltrials/ | |
| SAR260301 | PIK3CB | Unknown | Phase I | 173 | |
| IPI-145 | PIK3CG, PIK3CD | Unknown | Phase I/ II/ III | National Cancer Institute (USA) http://www.cancer.gov/clinicaltrials/239 | |
| GDC-0941 | PIK3CA, PIK3CD | Activation | Phase I/ II | National Cancer Institute (USA) http://www.cancer.gov/clinicaltrials/175,219,240-244 | |
| GS-1101 | PIK3CD | Unknown | Phase I/ II/ III | National Cancer Institute (USA) http://www.cancer.gov/clinicaltrials/245–251 | |
| Class III-specific | Compound 31((2S)8-[(3R)3-Methylmorpholin-4-yl]-1-(3-methyl-2-oxobutyl)-2-(trifluoromethyl)-3,4-dihydro2H-pyrimido[1,2α]pyrimidin-6-one) | PIK3C3 | Unknown | NA | 186 |
| PIK-III | PIK3C3 | Inhibition | NA | 185 | |
| SAR405 | PIK3C3 | Inhibition | NA | 183 | |
| VPS34-IN1 | PIK3C3 | Unknown | NA | 184 |
NA: Not available.
Pan-PI3K inhibitors have potent inhibitory effect on all class I PI3Ks, but their off-target effects on other PI3K-related kinases such as MTOR confound their application and therefore display unfavorable side effects in clinical trials.175,176 Wortmannin and LY294002 are among the early-discovered pan-PI3K inhibitors; however, they are not ideal for clinical applications because of nonselectivity and poor pharmacological properties.24,26,175,177 In light of these limitations, novel pan-PI3K inhibitors (e.g.,, BKM120, BAY80–6946) have been developed and demonstrate improved pharmaceutical potential and decreased toxicity in clinical trials.24,26,177 Additionally, since wortmannin and LY294002 impose inhibitory effects on class III PtdIns3K, they are also used to block autophagy.72,178 3-methyladenine (3-MA), first discovered as an autophagy inhibitor,179 shows functional similarity with LY294002, which may exert dual effects on class I and class III PtdIns3Ks.178,180 In comparison to 3-MA, which has been shown to have a dual effect on autophagy due to its transient inhibitory effect on class III PtdIns3K, wortmannin is a preferred autophagy inhibitor based on its persistent inhibition of class III PtdIns3K.181 Nevertheless, caution must be taken when these autophagy inhibitors are used, with nutrient conditions and biological contexts taken into account. Dual PI3K inhibitors are designed to inhibit both catalytic subunit of all class I PI3Ks and MTOR (e.g., NVP-BGT226, XL765).173,175,182 They are more effective in dampening PI3K-MTOR signaling, because they act on 2 nodes of this pathway and prevent the feedback activation of IRS1-PI3K by MTOR in the case of inhibiting MTOR alone. Isoform-specific PI3K inhibitors only target a specific isoform of PI3Ks, for instance, the PIK3CA inhibitor INK1117, the PIK3CB inhibitor GSK2636771, and the PIK3CD inhibitor GS-1101, which display fewer side effects and increased selectivity compared to the other classes of inhibitors in clinical trials.26,175
Inspiring progress has been made recently in the development of effective and specific inhibitors targeting PIK3C3.183-186 An early study resolved the molecular structure of PIK3C3, and revealed that PIK3C3 has a smaller and more rigid ATP binding pocket than that of PIK3CG, which restricts the binding of classical PI3K inhibitors.187 This molecular dissection of PIK3C3 paved the way for the subsequent development of novel inhibitors that fit well within the PIK3C3 ATP cavity. A number of recent studies reported new PIK3C3 inhibitors, including VPS34-IN1,184 Compound 31 (a tetrahydropyrimidopyrimidinone derivative),186 PIK-III,185 and SAR405;183 among them, Compound 31 and SAR405 exhibit the highest potency (IC50 PIK3C3 equals 2 nM and 1.2 nM, respectively) and favorable selectivity toward PIK3C3.183,186 Notably, PIK-III and SAR405, both of which bind the ATP binding pocket of PIK3C3, exert efficient inhibition on autophagy as well as LC3 lipidation.183,185 The application of PIK-III led to the identification of a novel substrate of selective autophagy, NCOA4, which mediates the degradation of ferritin and turnover of iron.185 Moreover, SAR405 displays a potential anticancer therapeutic property, as it shows synergistic antiproliferative activity in combination with the MTOR inhibitor everolimus.183 These PIK3C3 inhibitors will prove to be valuable tools in delineating the function of class III PtdIns3K in each stage of autophagy as well as other cellular processes; also, they are promising pharmaceutical candidates for augmenting the activity of current anticancer drugs as well as having a potential application to other diseases.
Development of selective inhibitors targeting class II PI3Ks are significantly lagging, which impedes the elucidation of this cryptic class of PI3Ks. In response to pan-PI3K inhibitors, PIK3C2B and PIK3C2G exhibit similar sensitivity to wortmannin as class I PI3Ks and class III PtdIns3Ks, but PIK3C2A only responds to higher doses of wortmannin and LY294002.161,166 Several inhibitors that demonstrate increased effectivity and selectivity toward specific isoforms of class II PI3Ks have been described recently,188,189 offering hope for developing more selective inhibitors of class II PI3Ks. Since this class of PI3Ks is equally important in malignant transformation and carcinogenesis,161,175 finding specific and effective class II PI3K inhibitors are among the important research goals for cell signaling studies and cancer treatment.
Concluding Remarks and Perspective
Autophagy has been an intensive research area in the last decade and the mechanisms of how autophagy is regulated have been intensively studied. Meanwhile, possible strategies have been proposed for manipulating this crucial pathway for the purpose of curing diseases and promoting health. PtdIns3Ks and PI3Ks are the cardinal type of kinases imperative for the proper functioning of autophagy. Generally, class I PI3Ks keep autophagy at bay in response to upstream stimulation; class III PtdIns3Ks, as well as class II PI3Ks, are required for the initiation and progression of autophagy. However, there are still gaps and discrepancies in our knowledge on the regulation of autophagy by these enzymes. For instance, how PIK3C3 reconciles its amino acid sensing function with its kinase activity under different conditions, and whether class II PI3Ks play a more pronounced role in autophagy and how they are regulated by certain autophagy-inducing stresses remain to be settled. Additionally, the contribution of phospholipids other than PtdIns3P to the biogenesis of the autophagosome as well as the later stages of autophagy also requires more attention from researchers. The progress in studying the involvement of lipid kinases in autophagy not only greatly facilitates our understanding of the molecular mechanisms controlling the autophagic process, but also promotes the discovery of pharmaceutical and therapeutic strategies targeting autophagy to cure related human diseases. Among them, development of class-specific PtdIns3K and PI3K inhibitors remains an important research objective in the realm of autophagy research.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgment
The authors thank Mr. YB Ong for the technical support in preparation of the figures.
Funding
X Yu is supported by a research scholarship from the National University of Singapore. The work in SHM’s lab is supported by research grants from the National Medical Research Council (NMRC) Singapore (NMRC-CIRG/1346/2012 and NMRC/CIRG/1373/2013). The work in LYC’s lab is supported by grants from the National Medical Research Council (NMRC) Singapore (NMRC/BNIG/2013/2013), and Singapore Ministry of Education Academic Research Fund (T1–2011 Sep-05 and T1–2014 Apr −05).
References
- 1.Mizushima N. Autophagy: process and function. Genes Dev 2007; 21:2861-73; PMID:18006683; http://dx.doi.org/ 10.1101/gad.1599207 [DOI] [PubMed] [Google Scholar]
- 2.Yamada E, Singh R. Mapping autophagy on to your metabolic radar. Diabetes 2012; 61:272-80; PMID:22275084; http://dx.doi.org/ 10.2337/db11-1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh R, Cuervo AM. Autophagy in the cellular energetic balance. Cell Metab 2011; 13:495-504; PMID:21531332; http://dx.doi.org/ 10.1016/j.cmet.2011.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shen HM, Codogno P. Autophagic cell death: Loch Ness monster or endangered species? Autophagy 2011; 7:457-65; PMID:21150268; http://dx.doi.org/ 10.4161/auto.7.5.14226 [DOI] [PubMed] [Google Scholar]
- 5.Tanida I. Autophagosome formation and molecular mechanism of autophagy. Antioxid Redox Signal 2011; 14:2201-14; PMID:20712405; http://dx.doi.org/ 10.1089/ars.2010.3482 [DOI] [PubMed] [Google Scholar]
- 6.Shibutani ST, Yoshimori T. A current perspective of autophagosome biogenesis. Cell Res 2014; 24:58-68; PMID:24296784; http://dx.doi.org/ 10.1038/cr.2013.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simonsen A, Tooze SA. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J Cell Biol 2009; 186:773-82; PMID:19797076; http://dx.doi.org/ 10.1083/jcb.200907014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y. Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat Rev Mol Cell Biol 2009; 10:458-67; PMID:19491929; http://dx.doi.org/ 10.1038/nrm2708 [DOI] [PubMed] [Google Scholar]
- 9.Carlsson SR, Simonsen A. Membrane dynamics in autophagosome biogenesis. J Cell Sci 2015; 128:193-205; PMID:25568151; http://dx.doi.org/ 10.1242/jcs.141036 [DOI] [PubMed] [Google Scholar]
- 10.Suzuki K, Ohsumi Y. Current knowledge of the pre-autophagosomal structure (PAS). FEBS Lett 2010; 584:1280-6; PMID:20138172; http://dx.doi.org/ 10.1016/j.febslet.2010.02.001 [DOI] [PubMed] [Google Scholar]
- 11.Itakura E, Mizushima N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 2010; 6:764-76; PMID:20639694; http://dx.doi.org/ 10.4161/auto.6.6.12709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karanasios E, Stapleton E, Manifava M, Kaizuka T, Mizushima N, Walker SA, Ktistakis NT. Dynamic association of the ULK1 complex with omegasomes during autophagy induction. J Cell Sci 2013; 126:5224-38; PMID:24013547; http://dx.doi.org/ 10.1242/jcs.132415 [DOI] [PubMed] [Google Scholar]
- 13.Wirth M, Joachim J, Tooze SA. Autophagosome formation–the role of ULK1 and Beclin1-PI3KC3 complexes in setting the stage. Semin Cancer Biol 2013; 23:301-9; PMID:23727157; http://dx.doi.org/ 10.1016/j.semcancer.2013.05.007 [DOI] [PubMed] [Google Scholar]
- 14.Russell RC, Yuan HX, Guan KL. Autophagy regulation by nutrient signaling. Cell Res 2014; 24:42-57; PMID:24343578; http://dx.doi.org/ 10.1038/cr.2013.166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koyama-Honda I, Itakura E, Fujiwara TK, Mizushima N. Temporal analysis of recruitment of mammalian ATG proteins to the autophagosome formation site. Autophagy 2013; 9:1491-9; PMID:23884233; http://dx.doi.org/ 10.4161/auto.25529 [DOI] [PubMed] [Google Scholar]
- 16.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 2008; 182:685-701; PMID:18725538; http://dx.doi.org/ 10.1083/jcb.200803137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dall'Armi C, Devereaux KA, Di Paolo G. The role of lipids in the control of autophagy. Curr Biol 2013; 23:R33-45; PMID:23305670; http://dx.doi.org/ 10.1016/j.cub.2012.10.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell 2008; 19:3290-8; PMID:18508918; http://dx.doi.org/ 10.1091/mbc.E07-12-1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy 2011; 7:279-96; PMID:21189453; http://dx.doi.org/ 10.4161/auto.7.3.14487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J Cell Sci 2013; 126:2534-44; PMID:23549786; http://dx.doi.org/ 10.1242/jcs.122960 [DOI] [PubMed] [Google Scholar]
- 21.Orsi A, Razi M, Dooley HC, Robinson D, Weston AE, Collinson LM, Tooze SA. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell 2012; 23:1860-73; PMID:22456507; http://dx.doi.org/ 10.1091/mbc.E11-09-0746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young AR, Chan EY, Hu XW, Kochl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J Cell Sci 2006; 119:3888-900; PMID:16940348; http://dx.doi.org/ 10.1242/jcs.03172 [DOI] [PubMed] [Google Scholar]
- 23.Shen HM, Mizushima N. At the end of the autophagic road: an emerging understanding of lysosomal functions in autophagy. Trends Biochem Sci 2014; 39:61-71; PMID:24369758; http://dx.doi.org/ 10.1016/j.tibs.2013.12.001 [DOI] [PubMed] [Google Scholar]
- 24.Akinleye A, Avvaru P, Furqan M, Song Y, Liu D. Phosphatidylinositol 3-kinase (PI3K) inhibitors as cancer therapeutics. J Hematol Oncol 2013; 6:88; PMID:24261963; http://dx.doi.org/ 10.1186/1756-8722-6-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, Bilanges B. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol 2010; 11:329-41; PMID:20379207; http://dx.doi.org/ 10.1038/nrm2882 [DOI] [PubMed] [Google Scholar]
- 26.McNamara CR, Degterev A. Small-molecule inhibitors of the PI3K signaling network. Fut Med Chem 2011; 3:549-65; PMID:21526896; http://dx.doi.org/ 10.4155/fmc.11.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farrell F O, Rusten TE, Stenmark H. Phosphoinositide 3-kinases as accelerators and brakes of autophagy. FEBS J 2013; 280:6322-37; PMID:23953235; http://dx.doi.org/ 10.1111/febs.12486 [DOI] [PubMed] [Google Scholar]
- 28.Foster FM, Traer CJ, Abraham SM, Fry MJ. The phosphoinositide (PI) 3-kinase family. J Cell Sci 2003; 116:3037-40; PMID:12829733; http://dx.doi.org/ 10.1242/jcs.00609 [DOI] [PubMed] [Google Scholar]
- 29.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov 2009; 8:627-44; PMID:19644473; http://dx.doi.org/ 10.1038/nrd2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Durocher D, Jackson SP. DNA-PK, ATM and ATR as sensors of DNA damage: variations on a theme? Curr Opin Cell Biol 2001; 13:225-31; PMID:11248557; http://dx.doi.org/ 10.1016/S0955-0674(00)00201-5 [DOI] [PubMed] [Google Scholar]
- 31.Falck J, Coates J, Jackson SP. Conserved modes of recruitment of ATM, ATR and DNA-PKcs to sites of DNA damage. Nature 2005; 434:605-11; PMID:15758953; http://dx.doi.org/ 10.1038/nature03442 [DOI] [PubMed] [Google Scholar]
- 32.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature 2009; 461:1071-8; PMID:19847258; http://dx.doi.org/ 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen JH, Zhang P, Chen WD, Li DD, Wu XQ, Deng R, Jiao L, Li X, Ji J, Feng GK, et al.. ATM-mediated PTEN phosphorylation promotes PTEN nuclear translocation and autophagy in response to DNA-damaging agents in cancer cells. Autophagy 2015; 11:239-52; PMID: 25701194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zou Y, Wang Q, Li B, Xie B, Wang W. Temozolomide induces autophagy via ATMAMPKULK1 pathways in glioma. Mol Med Rep 2014; 10:411-6; PMID: 24737504 [DOI] [PubMed] [Google Scholar]
- 35.Liang N, Jia L, Liu Y, Liang B, Kong D, Yan M, Ma S, Liu X. ATM pathway is essential for ionizing radiation-induced autophagy. Cell Signal 2013; 25:2530-9; PMID:23993957; http://dx.doi.org/ 10.1016/j.cellsig.2013.08.010 [DOI] [PubMed] [Google Scholar]
- 36.Tripathi DN, Chowdhury R, Trudel LJ, Tee AR, Slack RS, Walker CL, Wogan GN. Reactive nitrogen species regulate autophagy through ATM-AMPK-TSC2-mediated suppression of mTORC1. Proc Natl Acad Sci U S A 2013; 110:E2950-7; PMID:23878245; http://dx.doi.org/ 10.1073/pnas.1307736110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duan X, Ponomareva L, Veeranki S, Choubey D. IFI16 induction by glucose restriction in human fibroblasts contributes to autophagy through activation of the ATM/AMPK/p53 pathway. PloS One 2011; 6:e19532; PMID:21573174; http://dx.doi.org/ 10.1371/journal.pone.0019532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhuang W, Qin Z, Liang Z. The role of autophagy in sensitizing malignant glioma cells to radiation therapy. Acta Biochim Biophys Sin 2009; 41:341-51; PMID:19430698; http://dx.doi.org/ 10.1093/abbs/gmp028 [DOI] [PubMed] [Google Scholar]
- 39.Vessoni AT, Filippi-Chiela EC, Menck CF, Lenz G. Autophagy and genomic integrity. Cell Death Differ 2013; 20:1444-54; PMID:23933813; http://dx.doi.org/ 10.1038/cdd.2013.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alexander A, Cai SL, Kim J, Nanez A, Sahin M, MacLean KH, Inoki K, Guan KL, Shen J, Person MD, et al.. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc Natl Acad Sci U S A 2010; 107:4153-8; PMID:20160076; http://dx.doi.org/ 10.1073/pnas.0913860107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alexander A, Kim J, Walker CL. ATM engages the TSC2/mTORC1 signaling node to regulate autophagy. Autophagy 2010; 6:672-3; PMID:20581436; http://dx.doi.org/ 10.4161/auto.6.5.12509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mori C, Yamaguchi Y, Teranishi M, Takanami T, Nagase T, Kanno S, Yasui A, Higashitani A. Over-expression of ATR causes autophagic cell death. Genes Cell 2013; 18:278-87; http://dx.doi.org/ 10.1111/gtc.12034 [DOI] [Google Scholar]
- 43.Daido S, Yamamoto A, Fujiwara K, Sawaya R, Kondo S, Kondo Y. Inhibition of the DNA-dependent protein kinase catalytic subunit radiosensitizes malignant glioma cells by inducing autophagy. Cancer Res 2005; 65:4368-75; PMID:15899829; http://dx.doi.org/ 10.1158/0008-5472.CAN-04-4202 [DOI] [PubMed] [Google Scholar]
- 44.Zhuang W, Li B, Long L, Chen L, Huang Q, Liang ZQ. Knockdown of the DNA-dependent protein kinase catalytic subunit radiosensitizes glioma-initiating cells by inducing autophagy. Brain Res 2011; 1371:7-15; PMID:21108935; http://dx.doi.org/ 10.1016/j.brainres.2010.11.044 [DOI] [PubMed] [Google Scholar]
- 45.Yoon JH, Ahn SG, Lee BH, Jung SH, Oh SH. Role of autophagy in chemoresistance: regulation of the ATM-mediated DNA-damage signaling pathway through activation of DNA-PKcs and PARP-1. Biochem Pharmacol 2012; 83:747-57; PMID:22226932; http://dx.doi.org/ 10.1016/j.bcp.2011.12.029 [DOI] [PubMed] [Google Scholar]
- 46.Chen LH, Loong CC, Su TL, Lee YJ, Chu PM, Tsai ML, Tsai PH, Tu PH, Chi CW, Lee HC, et al.. Autophagy inhibition enhances apoptosis triggered by BO-1051, an N-mustard derivative, and involves the ATM signaling pathway. Biochem Pharmacol 2011; 81:594-605; PMID:21184746; http://dx.doi.org/ 10.1016/j.bcp.2010.12.011 [DOI] [PubMed] [Google Scholar]
- 47.Zanotto-Filho A, Braganhol E, Klafke K, Figueiro F, Terra SR, Paludo FJ, Morrone M, Bristot IJ, Battastini AM, Forcelini CM, et al.. Autophagy inhibition improves the efficacy of curcumin/temozolomide combination therapy in glioblastomas. Cancer Lett 2015; 358:220-31; PMID:25542083; http://dx.doi.org/ 10.1016/j.canlet.2014.12.044 [DOI] [PubMed] [Google Scholar]
- 48.Liu H, He Z, Simon HU. Targeting autophagy as a potential therapeutic approach for melanoma therapy. Semin Cancer Biol 2013; 23:352-60; PMID:23831275; http://dx.doi.org/ 10.1016/j.semcancer.2013.06.008 [DOI] [PubMed] [Google Scholar]
- 49.Fiorito F, Ciarcia R, Granato GE, Marfe G, Iovane V, Florio S, De Martino L, Pagnini U. 2,3,7,8-tetrachlorodibenzo-p-dioxin induced autophagy in a bovine kidney cell line. Toxicology 2011; 290:258-70; PMID:22015590; http://dx.doi.org/ 10.1016/j.tox.2011.10.004 [DOI] [PubMed] [Google Scholar]
- 50.Petiot A, Ogier-Denis E, Blommaart EF, Meijer AJ, Codogno P. Distinct classes of phosphatidylinositol 3'-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J Biol Chem 2000; 275:992-8; PMID:10625637; http://dx.doi.org/ 10.1074/jbc.275.2.992 [DOI] [PubMed] [Google Scholar]
- 51.Hemmings BA, Restuccia DF. PI3K-PKB/Akt pathway. Cold Spring Harb Perspect Biol 2012; 4:a011189; PMID:22952397; http://dx.doi.org/ 10.1101/cshperspect.a011189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shanware NP, Bray K, Abraham RT. The PI3K, metabolic, and autophagy networks: interactive partners in cellular health and disease. Annu Rev of Pharmacol Toxicol 2013; 53:89-106; PMID:23294306; http://dx.doi.org/ 10.1146/annurev-pharmtox-010611-134717 [DOI] [PubMed] [Google Scholar]
- 53.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett 2010; 584:1287-95; PMID:20083114; http://dx.doi.org/ 10.1016/j.febslet.2010.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci 2009; 122:3589-94; PMID:19812304; http://dx.doi.org/ 10.1242/jcs.051011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bar-Peled L, Sabatini DM. Regulation of mTORC1 by amino acids. Trends Cell Biol 2014; 24:400-6; PMID:24698685; http://dx.doi.org/ 10.1016/j.tcb.2014.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jewell JL, Guan KL. Nutrient signaling to mTOR and cell growth. Trends Biochem Sci 2013; 38:233-42; PMID:23465396; http://dx.doi.org/ 10.1016/j.tibs.2013.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science 2011; 334:678-83; PMID:22053050; http://dx.doi.org/ 10.1126/science.1207056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 2009; 10:307-18; PMID:19339977; http://dx.doi.org/ 10.1038/nrm2672 [DOI] [PubMed] [Google Scholar]
- 59.Yang Q, Guan KL. Expanding mTOR signaling. Cell Res 2007; 17:666-81; PMID:17680028; http://dx.doi.org/ 10.1038/cr.2007.64 [DOI] [PubMed] [Google Scholar]
- 60.Liao Y, Hung MC. Physiological regulation of Akt activity and stability. Am J Transl Res 2010; 2:19-42; PMID:20182580 [PMC free article] [PubMed] [Google Scholar]
- 61.Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, et al.. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol 2013; 15:406-16; PMID:23524951; http://dx.doi.org/ 10.1038/ncb2708 [DOI] [PubMed] [Google Scholar]
- 62.Zhou J, Tan SH, Nicolas V, Bauvy C, Yang ND, Zhang J, Xue Y, Codogno P, Shen HM. Activation of lysosomal function in the course of autophagy via mTORC1 suppression and autophagosome-lysosome fusion. Cell Res 2013; 23:508-23; PMID:23337583; http://dx.doi.org/ 10.1038/cr.2013.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, et al.. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 2012; 31:1095-108; PMID:22343943; http://dx.doi.org/ 10.1038/emboj.2012.32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal 2012; 5:ra42; PMID:22692423; http://dx.doi.org/ 10.1126/scisignal.2002790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 2011; 13:132-41; PMID:21258367; http://dx.doi.org/ 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Calnan DR, Brunet A. The FoxO code. Oncogene 2008; 27:2276-88; PMID:18391970; http://dx.doi.org/ 10.1038/onc.2008.21 [DOI] [PubMed] [Google Scholar]
- 67.Eijkelenboom A, Burgering BM. FOXOs: signalling integrators for homeostasis maintenance. Nat Rev Mol Cell Biol 2013; 14:83-97; PMID:23325358; http://dx.doi.org/ 10.1038/nrm3507 [DOI] [PubMed] [Google Scholar]
- 68.Yuan HX, Russell RC, Guan KL. Regulation of PIK3C3/VPS34 complexes by MTOR in nutrient stress-induced autophagy. Autophagy 2013; 9:1983-95; PMID:24013218; http://dx.doi.org/ 10.4161/auto.26058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dou Z, Chattopadhyay M, Pan JA, Guerriero JL, Jiang YP, Ballou LM, Yue Z, Lin RZ, Zong WX. The class IA phosphatidylinositol 3-kinase p110-beta subunit is a positive regulator of autophagy. J Cell Biol 2010; 191:827-43; PMID:21059846; http://dx.doi.org/ 10.1083/jcb.201006056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dou Z, Pan JA, Dbouk HA, Ballou LM, DeLeon JL, Fan Y, Chen JS, Liang Z, Li G, Backer JM, et al.. Class IA PI3K p110beta subunit promotes autophagy through Rab5 small GTPase in response to growth factor limitation. Mol Cell 2013; 50:29-42; PMID:23434372; http://dx.doi.org/ 10.1016/j.molcel.2013.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Murray JT, Panaretou C, Stenmark H, Miaczynska M, Backer JM. Role of Rab5 in the recruitment of hVps34/p150 to the early endosome. Traffic 2002; 3:416-27; PMID:12010460; http://dx.doi.org/ 10.1034/j.1600-0854.2002.30605.x [DOI] [PubMed] [Google Scholar]
- 72.Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelarova H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem 1997; 243:240-6; PMID:9030745; http://dx.doi.org/ 10.1111/j.1432-1033.1997.0240a.x [DOI] [PubMed] [Google Scholar]
- 73.Kiel JA, Rechinger KB, van der Klei IJ, Salomons FA, Titorenko VI, Veenhuis M. The Hansenula polymorpha PDD1 gene product, essential for the selective degradation of peroxisomes, is a homologue of Saccharomyces cerevisiae Vps34p. Yeast 1999; 15:741-54; PMID:10398343; http://dx.doi.org/ 10.1002/(SICI)1097-0061(19990630)15:9%3c741::AID-YEA416%3e3.0.CO;2-O [DOI] [PubMed] [Google Scholar]
- 74.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol 2001; 152:519-30; PMID:11157979; http://dx.doi.org/ 10.1083/jcb.152.3.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell 2008; 19:5360-72; PMID:18843052; http://dx.doi.org/ 10.1091/mbc.E08-01-0080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol 2006; 8:688-99; PMID:16799551; http://dx.doi.org/ 10.1038/ncb1426 [DOI] [PubMed] [Google Scholar]
- 77.Zhong Y, Wang QJ, Li X, Yan Y, Backer JM, Chait BT, Heintz N, Yue Z. Distinct regulation of autophagic activity by Atg14L and Rubicon associated with Beclin 1-phosphatidylinositol-3-kinase complex. Nat Cell Biol 2009; 11:468-76; PMID:19270693; http://dx.doi.org/ 10.1038/ncb1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al.. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol 2009; 11:385-96; PMID:19270696; http://dx.doi.org/ 10.1038/ncb1846 [DOI] [PubMed] [Google Scholar]
- 79.Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, et al.. Ambra1 regulates autophagy and development of the nervous system. Nature 2007; 447:1121-5; PMID:17589504 [DOI] [PubMed] [Google Scholar]
- 80.Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M, Pagliarini V, Matteoni S, Fuoco C, Giunta L, et al.. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. J Cell Biol 2010; 191:155-68; PMID:20921139; http://dx.doi.org/ 10.1083/jcb.201002100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lu J, He L, Behrends C, Araki M, Araki K, Jun Wang Q, Catanzaro JM, Friedman SL, Zong WX, Fiel MI, et al.. NRBF2 regulates autophagy and prevents liver injury by modulating Atg14L-linked phosphatidylinositol-3 kinase III activity. Nat Commun 2014; 5:3920; PMID:24849286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhong Y, Morris DH, Jin L, Patel MS, Karunakaran SK, Fu YJ, Matuszak EA, Weiss HL, Chait BT, Wang QJ. Nrbf2 protein suppresses autophagy by modulating Atg14L protein-containing Beclin 1-Vps34 complex architecture and reducing intracellular phosphatidylinositol-3 phosphate levels. J Biol Chem 2014; 289:26021-37; PMID:25086043; http://dx.doi.org/ 10.1074/jbc.M114.561134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cao Y, Wang Y, Abi Saab WF, Yang F, Pessin JE, Backer JM. NRBF2 regulates macroautophagy as a component of Vps34 Complex I. Biochem J 2014; 461:315-22; PMID:24785657; http://dx.doi.org/ 10.1042/BJ20140515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex–at the crossroads of autophagy and beyond. Trends Cell Biol 2010; 20:355-62; PMID:20356743; http://dx.doi.org/ 10.1016/j.tcb.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fu LL, Cheng Y, Liu B. Beclin-1: autophagic regulator and therapeutic target in cancer. Int J Biochem Cell Biol 2013; 45:921-4; PMID:23420005; http://dx.doi.org/ 10.1016/j.biocel.2013.02.007 [DOI] [PubMed] [Google Scholar]
- 86.Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, et al.. Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol 2008; 10:776-87; PMID:18552835; http://dx.doi.org/ 10.1038/ncb1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baskaran S, Carlson LA, Stjepanovic G, Young LN, Kim do J, Grob P, Stanley RE, Nogales E, Hurley JH. Architecture and dynamics of the autophagic phosphatidylinositol 3-kinase complex. ELife 2014; 3; PMID:25490155; http://dx.doi.org/ 10.7554/eLife.05115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Itakura E, Mizushima N. Atg14 and UVRAG: mutually exclusive subunits of mammalian Beclin 1-PI3K complexes. Autophagy 2009; 5:534-6; PMID:19223761; http://dx.doi.org/ 10.4161/auto.5.4.8062 [DOI] [PubMed] [Google Scholar]
- 89.Thoresen SB, Pedersen NM, Liestol K, Stenmark H. A phosphatidylinositol 3-kinase class III sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates cytokinesis and degradative endocytic traffic. Exp Cell Res 2010; 316:3368-78; PMID:20643123; http://dx.doi.org/ 10.1016/j.yexcr.2010.07.008 [DOI] [PubMed] [Google Scholar]
- 90.Yan Y, Backer JM. Regulation of class III (Vps34) PI3Ks. Biochem Soc Trans 2007; 35:239-41; PMID:17371248; http://dx.doi.org/ 10.1042/BST0350242 [DOI] [PubMed] [Google Scholar]
- 91.Yan Y, Flinn RJ, Wu H, Schnur RS, Backer JM. hVps15, but not Ca2+/CaM, is required for the activity and regulation of hVps34 in mammalian cells. Biochem J 2009; 417:747-55; PMID:18957027; http://dx.doi.org/ 10.1042/BJ20081865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stack JH, Herman PK, Schu PV, Emr SD. A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J 1993; 12:2195-204; PMID:8387919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J 2000; 19:4577-88; PMID:10970851; http://dx.doi.org/ 10.1093/emboj/19.17.4577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Efe JA, Botelho RJ, Emr SD. Atg18 regulates organelle morphology and Fab1 kinase activity independent of its membrane recruitment by phosphatidylinositol 3,5-bisphosphate. Mol Biol Cell 2007; 18:4232-44; PMID:17699591; http://dx.doi.org/ 10.1091/mbc.E07-04-0301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Polson HE, de Lartigue J, Rigden DJ, Reedijk M, Urbe S, Clague MJ, Tooze SA. Mammalian Atg18 (WIPI2) localizes to omegasome-anchored phagophores and positively regulates LC3 lipidation. Autophagy 2010; 6:506-22; PMID:20505359; http://dx.doi.org/ 10.4161/auto.6.4.11863 [DOI] [PubMed] [Google Scholar]
- 96.Filimonenko M, Isakson P, Finley KD, Anderson M, Jeong H, Melia TJ, Bartlett BJ, Myers KM, Birkeland HC, Lamark T, et al.. The selective macroautophagic degradation of aggregated proteins requires the PI3P-binding protein Alfy. Mol Cell 2010; 38:265-79; PMID:20417604; http://dx.doi.org/ 10.1016/j.molcel.2010.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Girardi JP, Pereira L, Bakovic M. De novo synthesis of phospholipids is coupled with autophagosome formation. Med Hypotheses 2011; 77:1083-7; PMID:21963355; http://dx.doi.org/ 10.1016/j.mehy.2011.09.008 [DOI] [PubMed] [Google Scholar]
- 98.Chen D, Fan W, Lu Y, Ding X, Chen S, Zhong Q. A mammalian autophagosome maturation mechanism mediated by TECPR1 and the Atg12-Atg5 conjugate. Mol Cell 2012; 45:629-41; PMID:22342342; http://dx.doi.org/ 10.1016/j.molcel.2011.12.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pankiv S, Alemu EA, Brech A, Bruun JA, Lamark T, Overvatn A, Bjorkoy G, Johansen T. FYCO1 is a Rab7 effector that binds to LC3 and PI3P to mediate microtubule plus end-directed vesicle transport. J Cell Biol 2010; 188:253-69; PMID:20100911; http://dx.doi.org/ 10.1083/jcb.200907015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cheng J, Fujita A, Yamamoto H, Tatematsu T, Kakuta S, Obara K, Ohsumi Y, Fujimoto T. Yeast and mammalian autophagosomes exhibit distinct phosphatidylinositol 3-phosphate asymmetries. Nat Commun 2014; 5:3207; PMID:24492518 [DOI] [PubMed] [Google Scholar]
- 101.Cebollero E, van der Vaart A, Zhao M, Rieter E, Klionsky DJ, Helms JB, Reggiori F. Phosphatidylinositol-3-phosphate clearance plays a key role in autophagosome completion. Curr Biol 2012; 22:1545-53; PMID:22771041; http://dx.doi.org/ 10.1016/j.cub.2012.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Vergne I, Deretic V. The role of PI3P phosphatases in the regulation of autophagy. FEBS Lett 2010; 584:1313-8; PMID:20188094; http://dx.doi.org/ 10.1016/j.febslet.2010.02.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vergne I, Roberts E, Elmaoued RA, Tosch V, Delgado MA, Proikas-Cezanne T, Laporte J, Deretic V. Control of autophagy initiation by phosphoinositide 3-phosphatase Jumpy. EMBO J 2009; 28:2244-58; PMID:19590496; http://dx.doi.org/ 10.1038/emboj.2009.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dowling JJ, Low SE, Busta AS, Feldman EL. Zebrafish MTMR14 is required for excitation-contraction coupling, developmental motor function and the regulation of autophagy. Hum Mol Genet 2010; 19:2668-81; PMID:20400459; http://dx.doi.org/ 10.1093/hmg/ddq153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Taguchi-Atarashi N, Hamasaki M, Matsunaga K, Omori H, Ktistakis NT, Yoshimori T, Noda T. Modulation of local PtdIns3P levels by the PI phosphatase MTMR3 regulates constitutive autophagy. Traffic 2010; 11:468-78; PMID:20059746; http://dx.doi.org/ 10.1111/j.1600-0854.2010.01034.x [DOI] [PubMed] [Google Scholar]
- 106.Vicinanza M, Korolchuk VI, Ashkenazi A, Puri C, Menzies FM, Clarke JH, Rubinsztein DC. PI(5)P regulates autophagosome biogenesis. Mol Cell 2015; 57:219-34; PMID:25578879; http://dx.doi.org/ 10.1016/j.molcel.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mauthe M, Jacob A, Freiberger S, Hentschel K, Stierhof YD, Codogno P, Proikas-Cezanne T. Resveratrol-mediated autophagy requires WIPI-1-regulated LC3 lipidation in the absence of induced phagophore formation. Autophagy 2011; 7:1448-61; PMID:22082875; http://dx.doi.org/ 10.4161/auto.7.12.17802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Scarlatti F, Maffei R, Beau I, Codogno P, Ghidoni R. Role of non-canonical Beclin 1-independent autophagy in cell death induced by resveratrol in human breast cancer cells. Cell Death Differ 2008; 15:1318-29; PMID:18421301; http://dx.doi.org/ 10.1038/cdd.2008.51 [DOI] [PubMed] [Google Scholar]
- 109.Zhu JH, Horbinski C, Guo F, Watkins S, Uchiyama Y, Chu CT. Regulation of autophagy by extracellular signal-regulated protein kinases during 1-methyl-4-phenylpyridinium-induced cell death. Am J Pathol 2007; 170:75-86; PMID:17200184; http://dx.doi.org/ 10.2353/ajpath.2007.060524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhou X, Takatoh J, Wang F. The mammalian class 3 PI3K (PIK3C3) is required for early embryogenesis and cell proliferation. PloS One 2011; 6:e16358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jaber N, Dou Z, Chen JS, Catanzaro J, Jiang YP, Ballou LM, Selinger E, Ouyang X, Lin RZ, Zhang J, et al.. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc Natl Acad Sci U S A 2012; 109:2003-8; PMID:22308354; http://dx.doi.org/ 10.1073/pnas.1112848109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, Guan KL. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 2013; 152:290-303; PMID:23332761; http://dx.doi.org/ 10.1016/j.cell.2012.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Furuya T, Kim M, Lipinski M, Li J, Kim D, Lu T, Shen Y, Rameh L, Yankner B, Tsai LH, et al.. Negative regulation of Vps34 by Cdk mediated phosphorylation. Mol cell 2010; 38:500-11; PMID:20513426; http://dx.doi.org/ 10.1016/j.molcel.2010.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Eisenberg-Lerner A, Kimchi A. PKD is a kinase of Vps34 that mediates ROS-induced autophagy downstream of DAPk. Cell Death Differ 2012; 19:788-97; PMID:22095288; http://dx.doi.org/ 10.1038/cdd.2011.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yang Y, Fiskus W, Yong B, Atadja P, Takahashi Y, Pandita TK, Wang HG, Bhalla KN. Acetylated hsp70 and KAP1-mediated Vps34 SUMOylation is required for autophagosome creation in autophagy. Proc Natl Acad Sci U S A 2013; 110:6841-6; PMID:23569248; http://dx.doi.org/ 10.1073/pnas.1217692110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gulati P, Gaspers LD, Dann SG, Joaquin M, Nobukuni T, Natt F, Kozma SC, Thomas AP, Thomas G. Amino acids activate mTOR complex 1 via Ca2+/CaM signaling to hVps34. Cell Metab 2008; 7:456-65; PMID:18460336; http://dx.doi.org/ 10.1016/j.cmet.2008.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Byfield MP, Murray JT, Backer JM. hVps34 is a nutrient-regulated lipid kinase required for activation of p70 S6 kinase. J Biol Chem 2005; 280:33076-82; PMID:16049009; http://dx.doi.org/ 10.1074/jbc.M507201200 [DOI] [PubMed] [Google Scholar]
- 118.Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P, Byfield MP, Backer JM, Natt F, Bos JL, et al.. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci U S A 2005; 102:14238-43; PMID:16176982; http://dx.doi.org/ 10.1073/pnas.0506925102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ghislat G, Patron M, Rizzuto R, Knecht E. Withdrawal of essential amino acids increases autophagy by a pathway involving Ca2+/calmodulin-dependent kinase kinase-beta (CaMKK-beta). J Biol Chem 2012; 287:38625-36; PMID:23027865; http://dx.doi.org/ 10.1074/jbc.M112.365767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol 2013; 15:741-50; PMID:23685627; http://dx.doi.org/ 10.1038/ncb2757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yu X, Long YC. Autophagy modulates amino acid signaling network in myotubes: differential effects on mTORC1 pathway and the integrated stress response. FASEB J 2015; 29:394-407; PMID:25376834; http://dx.doi.org/ 10.1096/fj.14-252841 [DOI] [PubMed] [Google Scholar]
- 122.Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, et al.. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 2010; 465:942-6; PMID:20526321; http://dx.doi.org/ 10.1038/nature09076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Backer JM. The regulation and function of Class III PI3Ks: novel roles for Vps34. Biochem J 2008; 410:1-17; PMID:18215151; http://dx.doi.org/ 10.1042/BJ20071427 [DOI] [PubMed] [Google Scholar]
- 124.Li M, Khambu B, Zhang H, Kang JH, Chen X, Chen D, Vollmer L, Liu PQ, Vogt A, Yin XM. Suppression of lysosome function induces autophagy via a feedback down-regulation of MTOR complex 1 (MTORC1) activity. J Biol Chem 2013; 288:35769-80; PMID:24174532; http://dx.doi.org/ 10.1074/jbc.M113.511212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Juhasz G, Hill JH, Yan Y, Sass M, Baehrecke EH, Backer JM, Neufeld TP. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol 2008; 181:655-66; PMID:18474623; http://dx.doi.org/ 10.1083/jcb.200712051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sahni S, Merlot AM, Krishan S, Jansson PJ, Richardson DR. Gene of the month: BECN1. J Clin Pathol 2014; 67:656-60; PMID:24811486; http://dx.doi.org/ 10.1136/jclinpath-2014-202356 [DOI] [PubMed] [Google Scholar]
- 127.Toton E, Lisiak N, Sawicka P, Rybczynska M. Beclin-1 and its role as a target for anticancer therapy. J Physiol Pharmacol 2014; 65:459-67; PMID:25179078 [PubMed] [Google Scholar]
- 128.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A 2003; 100:15077-82; PMID:14657337; http://dx.doi.org/ 10.1073/pnas.2436255100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al.. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 2003; 112:1809-20; PMID:14638851; http://dx.doi.org/ 10.1172/JCI20039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pattingre S, Tassa A, Qu X, Garuti R, Liang XH, Mizushima N, Packer M, Schneider MD, Levine B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005; 122:927-39; PMID:16179260; http://dx.doi.org/ 10.1016/j.cell.2005.07.002 [DOI] [PubMed] [Google Scholar]
- 131.Wei Y, Pattingre S, Sinha S, Bassik M, Levine B. JNK1-mediated phosphorylation of Bcl-2 regulates starvation-induced autophagy. Mol Cell 2008; 30:678-88; PMID:18570871; http://dx.doi.org/ 10.1016/j.molcel.2008.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pimkina J, Humbey O, Zilfou JT, Jarnik M, Murphy ME. ARF induces autophagy by virtue of interaction with Bcl-xl. J Biol Chem 2009; 284:2803-10; PMID:19049976; http://dx.doi.org/ 10.1074/jbc.M804705200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang RC, Wei Y, An Z, Zou Z, Xiao G, Bhagat G, White M, Reichelt J, Levine B. Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 2012; 338:956-9; PMID:23112296; http://dx.doi.org/ 10.1126/science.1225967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zalckvar E, Berissi H, Mizrachy L, Idelchuk Y, Koren I, Eisenstein M, Sabanay H, Pinkas-Kramarski R, Kimchi A. DAP-kinase-mediated phosphorylation on the BH3 domain of beclin 1 promotes dissociation of beclin 1 from Bcl-XL and induction of autophagy. EMBO Rep 2009; 10:285-92; PMID:19180116; http://dx.doi.org/ 10.1038/embor.2008.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wei Y, Zou Z, Becker N, Anderson M, Sumpter R, Xiao G, Kinch L, Koduru P, Christudass CS, Veltri RW, et al.. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell 2013; 154:1269-84; PMID:24034250; http://dx.doi.org/ 10.1016/j.cell.2013.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Abrahamsen H, Stenmark H, Platta HW. Ubiquitination and phosphorylation of Beclin 1 and its binding partners: tuning class III phosphatidylinositol 3-kinase activity and tumor suppression. FEBS Lett 2012; 586:1584-91; PMID:22673570; http://dx.doi.org/ 10.1016/j.febslet.2012.04.046 [DOI] [PubMed] [Google Scholar]
- 137.Reidick C, El Magraoui F, Meyer HE, Stenmark H, Platta HW. Regulation of the Tumor-Suppressor Function of the Class III Phosphatidylinositol 3-Kinase Complex by Ubiquitin and SUMO. Cancers 2014; 7:1-29; PMID:25545884; http://dx.doi.org/ 10.3390/cancers7010001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Platta HW, Abrahamsen H, Thoresen SB, Stenmark H. Nedd4-dependent lysine-11-linked polyubiquitination of the tumour suppressor Beclin 1. Biochem J 2012; 441:399-406; PMID:21936852; http://dx.doi.org/ 10.1042/BJ20111424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Xu C, Liu J, Hsu LC, Luo Y, Xiang R, Chuang TH. Functional interaction of heat shock protein 90 and Beclin 1 modulates Toll-like receptor-mediated autophagy. FASEB J 2011; 25:2700-10; PMID:21543763; http://dx.doi.org/ 10.1096/fj.10-167676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shi CS, Kehrl JH. TRAF6 and A20 regulate lysine 63-linked ubiquitination of Beclin-1 to control TLR4-induced autophagy. Sci Signal 2010; 3:ra42; PMID:20501938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol 2010; 22:140-9; PMID:20097051; http://dx.doi.org/ 10.1016/j.ceb.2010.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, Noda T, Yoshimori T. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol 2010; 190:511-21; PMID:20713597; http://dx.doi.org/ 10.1083/jcb.200911141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pfisterer SG, Bakula D, Frickey T, Cezanne A, Brigger D, Tschan MP, Robenek H, Proikas-Cezanne T. Lipid droplet and early autophagosomal membrane targeting of Atg2A and Atg14L in human tumor cells. J Lipid Res 2014; 55:1267-78; PMID:24776541; http://dx.doi.org/ 10.1194/jlr.M046359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Obara K, Ohsumi Y. Atg14: a key player in orchestrating autophagy. Int J Cell Biol 2011; 2011:713435; PMID:22013444; http://dx.doi.org/ 10.1155/2011/713435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Fan W, Nassiri A, Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L). Proc Nat Acad Sci U S A 2011; 108:7769-74; PMID:21518905; http://dx.doi.org/ 10.1073/pnas.1016472108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Teitz T, Penner M, Eli D, Stark M, Bakhanashvili M, Naiman T, Canaani D. Isolation by polymerase chain reaction of a cDNA whose product partially complements the ultraviolet sensitivity of xeroderma pigmentosum group C cells. Gene 1990; 87:295-8; PMID:2332174; http://dx.doi.org/ 10.1016/0378-1119(90)90316-J [DOI] [PubMed] [Google Scholar]
- 147.Bekri S, Adelaide J, Merscher S, Grosgeorge J, Caroli-Bosc F, Perucca-Lostanlen D, Kelley PM, Pebusque MJ, Theillet C, Birnbaum D, et al.. Detailed map of a region commonly amplified at 11q13–>q14 in human breast carcinoma. Cytogenet Cell Genet 1997; 79:125-31; PMID:9533029; http://dx.doi.org/ 10.1159/000134699 [DOI] [PubMed] [Google Scholar]
- 148.Goi T, Kawasaki M, Yamazaki T, Koneri K, Katayama K, Hirose K, Yamaguchi A. Ascending colon cancer with hepatic metastasis and cholecystolithiasis in a patient with situs inversus totalis without any expression of UVRAG mRNA: report of a case. Surg Today 2003; 33:702-6; PMID:12928850; http://dx.doi.org/ 10.1007/s00595-002-2567-y [DOI] [PubMed] [Google Scholar]
- 149.Liang C, Feng P, Ku B, Oh BH, Jung JU. UVRAG: a new player in autophagy and tumor cell growth. Autophagy 2007; 3:69-71; PMID:17106237; http://dx.doi.org/ 10.4161/auto.3437 [DOI] [PubMed] [Google Scholar]
- 150.Ionov Y, Nowak N, Perucho M, Markowitz S, Cowell JK. Manipulation of nonsense mediated decay identifies gene mutations in colon cancer Cells with microsatellite instability. Oncogene 2004; 23:639-45; PMID:14737099; http://dx.doi.org/ 10.1038/sj.onc.1207178 [DOI] [PubMed] [Google Scholar]
- 151.Zhao Z, Oh S, Li D, Ni D, Pirooz SD, Lee JH, Yang S, Lee JY, Ghozalli I, Costanzo V, et al.. A dual role for UVRAG in maintaining chromosomal stability independent of autophagy. Dev Cell 2012; 22:1001-16; PMID:22542840; http://dx.doi.org/ 10.1016/j.devcel.2011.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Park JM, Tougeron D, Huang S, Okamoto K, Sinicrope FA. Beclin 1 and UVRAG confer protection from radiation-induced DNA damage and maintain centrosome stability in colorectal cancer cells. PloS One 2014; 9:e100819; PMID:24956373; http://dx.doi.org/ 10.1371/journal.pone.0100819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhao Z, Ni D, Ghozalli I, Pirooz SD, Ma B, Liang C. UVRAG: at the crossroad of autophagy and genomic stability. Autophagy 2012; 8:1392-3; PMID:22885520; http://dx.doi.org/ 10.4161/auto.21035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Song Z, An L, Ye Y, Wu J, Zou Y, He L, Zhu H. Essential role for UVRAG in autophagy and maintenance of cardiac function. Cardiovasc Res 2014; 101:48-56; PMID:24081163; http://dx.doi.org/ 10.1093/cvr/cvt223 [DOI] [PubMed] [Google Scholar]
- 155.Yang W, Ju JH, Lee KM, Nam K, Oh S, Shin I. Protein kinase B/Akt1 inhibits autophagy by down-regulating UVRAG expression. Exp Cell Res 2013; 319:122-33; PMID:23200933; http://dx.doi.org/ 10.1016/j.yexcr.2012.11.014 [DOI] [PubMed] [Google Scholar]
- 156.He S, Ni D, Ma B, Lee JH, Zhang T, Ghozalli I, Pirooz SD, Zhao Z, Bharatham N, Li B, et al.. PtdIns(3)P-bound UVRAG coordinates Golgi-ER retrograde and Atg9 transport by differential interactions with the ER tether and the beclin 1 complex. Nat Cell Biol 2013; 15:1206-19; PMID:24056303; http://dx.doi.org/ 10.1038/ncb2848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lorincz P, Lakatos Z, Maruzs T, Szatmari Z, Kis V, Sass M. Atg6/UVRAG/Vps34-containing lipid kinase complex is required for receptor downregulation through endolysosomal degradation and epithelial polarity during Drosophila wing development. BioMed Res Int 2014; 2014:851349; PMID:25006588; http://dx.doi.org/ 10.1155/2014/851349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Knaevelsrud H, Ahlquist T, Merok MA, Nesbakken A, Stenmark H, Lothe RA, Simonsen A. UVRAG mutations associated with microsatellite unstable colon cancer do not affect autophagy. Autophagy 2010; 6:863-70; PMID:20724836; http://dx.doi.org/ 10.4161/auto.6.7.13033 [DOI] [PubMed] [Google Scholar]
- 159.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature 2010; 466:68-76; PMID:20562859; http://dx.doi.org/ 10.1038/nature09204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Araki Y, Ku WC, Akioka M, May AI, Hayashi Y, Arisaka F, Ishihama Y, Ohsumi Y. Atg38 is required for autophagy-specific phosphatidylinositol 3-kinase complex integrity. J Cell Biol 2013; 203:299-313; PMID:24165940; http://dx.doi.org/ 10.1083/jcb.201304123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Falasca M, Maffucci T. Role of class II phosphoinositide 3-kinase in cell signalling. Biochem Soc Trans 2007; 35:211-4; PMID:17371240; http://dx.doi.org/ 10.1042/BST0350229 [DOI] [PubMed] [Google Scholar]
- 162.Arcaro A, Zvelebil MJ, Wallasch C, Ullrich A, Waterfield MD, Domin J. Class II phosphoinositide 3-kinases are downstream targets of activated polypeptide growth factor receptors. Mole Cell Biol 2000; 20:3817-30; PMID:10805725; http://dx.doi.org/ 10.1128/MCB.20.11.3817-3830.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Maffucci T, Cooke FT, Foster FM, Traer CJ, Fry MJ, Falasca M. Class II phosphoinositide 3-kinase defines a novel signaling pathway in cell migration. J Cell Biol 2005; 169:789-99; PMID:15928202; http://dx.doi.org/ 10.1083/jcb.200408005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Biswas K, Yoshioka K, Asanuma K, Okamoto Y, Takuwa N, Sasaki T, Takuwa Y. Essential role of class II phosphatidylinositol-3-kinase-C2alpha in sphingosine 1-phosphate receptor-1-mediated signaling and migration in endothelial cells. J Biol Chem 2013; 288:2325-39; PMID:23192342; http://dx.doi.org/ 10.1074/jbc.M112.409656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wen PJ, Osborne SL, Morrow IC, Parton RG, Domin J, Meunier FA. Ca2+-regulated pool of phosphatidylinositol-3-phosphate produced by phosphatidylinositol 3-kinase C2alpha on neurosecretory vesicles. Mol Biol Cell 2008; 19:5593-603; PMID:18843041; http://dx.doi.org/ 10.1091/mbc.E08-06-0595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Traer CJ, Foster FM, Abraham SM, Fry MJ. Are class II phosphoinositide 3-kinases potential targets for anticancer therapies? Bull Cancer 2006; 93:E53-8; PMID:16777618 [PubMed] [Google Scholar]
- 167.Lu N, Shen Q, Mahoney TR, Neukomm LJ, Wang Y, Zhou Z. Two PI 3-kinases and one PI 3-phosphatase together establish the cyclic waves of phagosomal PtdIns(3)P critical for the degradation of apoptotic cells. PLoS Biol 2012; 10:e1001245; PMID:22272187; http://dx.doi.org/ 10.1371/journal.pbio.1001245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Cheng S, Wu Y, Lu Q, Yan J, Zhang H, Wang X. Autophagy genes coordinate with the class II PI/PtdIns 3-kinase PIKI-1 to regulate apoptotic cell clearance in C. elegans. Autophagy 2013; 9:2022-32; PMID:24165672; http://dx.doi.org/ 10.4161/auto.26323 [DOI] [PubMed] [Google Scholar]
- 169.Bechtel W, Helmstadter M, Balica J, Hartleben B, Kiefer B, Hrnjic F, Schell C, Kretz O, Liu S, Geist F, et al.. Vps34 deficiency reveals the importance of endocytosis for podocyte homeostasis. J Am Soc Nephrol 2013; 24:727-43; PMID:23492732; http://dx.doi.org/ 10.1681/ASN.2012070700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Parekh VV, Wu L, Boyd KL, Williams JA, Gaddy JA, Olivares-Villagomez D, Cover TL, Zong WX, Zhang J, Van Kaer L. Impaired autophagy, defective T cell homeostasis, and a wasting syndrome in mice with a T cell-specific deletion of Vps34. J Immunol 2013; 190:5086-101; http://dx.doi.org/ 10.4049/jimmunol.1202071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Devereaux K, Dall'Armi C, Alcazar-Roman A, Ogasawara Y, Zhou X, Wang F, Yamamoto A, De Camilli P, Di Paolo G. Regulation of mammalian autophagy by class II and III PI 3-kinases through PI3P synthesis. PloS One 2013; 8:e76405; PMID:24098492; http://dx.doi.org/ 10.1371/journal.pone.0076405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Rodon J, Dienstmann R, Serra V, Tabernero J. Development of PI3K inhibitors: lessons learned from early clinical trials. Nat Rev Clin Oncol 2013; 10:143-53; PMID:23400000; http://dx.doi.org/ 10.1038/nrclinonc.2013.10 [DOI] [PubMed] [Google Scholar]
- 173.Brana I, Siu LL. Clinical development of phosphatidylinositol 3-kinase inhibitors for cancer treatment. BMC Med 2012; 10:161; PMID:23232172; http://dx.doi.org/ 10.1186/1741-7015-10-161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer 2009; 9:550-62; PMID:19629070; http://dx.doi.org/ 10.1038/nrc2664 [DOI] [PubMed] [Google Scholar]
- 175.Martini M, Ciraolo E, Gulluni F, Hirsch E. Targeting PI3K in cancer: any good news? Front Oncol 2013; 3:108; PMID:23658859; http://dx.doi.org/ 10.3389/fonc.2013.00108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Eisenreich A, Rauch U. PI3K inhibitors in cardiovascular disease. Cardiovasc Ther 2011; 29:29-36; PMID:20626398; http://dx.doi.org/ 10.1111/j.1755-5922.2010.00206.x [DOI] [PubMed] [Google Scholar]
- 177.Cleary JM, Shapiro GI. Development of phosphoinositide-3 kinase pathway inhibitors for advanced cancer. Curr Oncol Rep 2010; 12:87-94; PMID:20425592; http://dx.doi.org/ 10.1007/s11912-010-0091-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yang YP, Hu LF, Zheng HF, Mao CJ, Hu WD, Xiong KP, Wang F, Liu CF. Application and interpretation of current autophagy inhibitors and activators. Acta Pharmacol Sin 2013; 34:625-35; PMID:23524572; http://dx.doi.org/ 10.1038/aps.2013.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Nat Acad Sci U S A 1982; 79:1889-92; PMID:6952238; http://dx.doi.org/ 10.1073/pnas.79.6.1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Xing C, Zhu B, Liu H, Yao H, Zhang L. Class I phosphatidylinositol 3-kinase inhibitor LY294002 activates autophagy and induces apoptosis through p53 pathway in gastric cancer cell line SGC7901. Acta Biochim Biophys Sin 2008; 40:194-201; PMID:18330473; http://dx.doi.org/ 10.1111/j.1745-7270.2008.00393.x [DOI] [PubMed] [Google Scholar]
- 181.Wu YT, Tan HL, Shui G, Bauvy C, Huang Q, Wenk MR, Ong CN, Codogno P, Shen HM. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem 2010; 285:10850-61; PMID:20123989; http://dx.doi.org/ 10.1074/jbc.M109.080796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Markman B, Tabernero J, Krop I, Shapiro GI, Siu L, Chen LC, Mita M, Melendez Cuero M, Stutvoet S, Birle D, et al.. Phase I safety, pharmacokinetic, and pharmacodynamic study of the oral phosphatidylinositol-3-kinase and mTOR inhibitor BGT226 in patients with advanced solid tumors. Ann Oncol 2012; 23:2399-408; PMID:22357447; http://dx.doi.org/ 10.1093/annonc/mds011 [DOI] [PubMed] [Google Scholar]
- 183.Ronan B, Flamand O, Vescovi L, Dureuil C, Durand L, Fassy F, Bachelot MF, Lamberton A, Mathieu M, Bertrand T, et al.. A highly potent and selective Vps34 inhibitor alters vesicle trafficking and autophagy. Nat Chem Biol 2014; 10:1013-9; PMID:25326666; http://dx.doi.org/ 10.1038/nchembio.1681 [DOI] [PubMed] [Google Scholar]
- 184.Bago R, Malik N, Munson MJ, Prescott AR, Davies P, Sommer E, Shpiro N, Ward R, Cross D, Ganley IG, et al.. Characterization of VPS34-IN1, a selective inhibitor of Vps34, reveals that the phosphatidylinositol 3-phosphate-binding SGK3 protein kinase is a downstream target of class III phosphoinositide 3-kinase. Biochem J 2014; 463:413-27; PMID:25177796; http://dx.doi.org/ 10.1042/BJ20140889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Dowdle WE, Nyfeler B, Nagel J, Elling RA, Liu S, Triantafellow E, Menon S, Wang Z, Honda A, Pardee G, et al.. Selective VPS34 inhibitor blocks autophagy and uncovers a role for NCOA4 in ferritin degradation and iron homeostasis in vivo. Nat Cell Biol 2014; 16:1069-79; PMID:25327288; http://dx.doi.org/ 10.1038/ncb3053 [DOI] [PubMed] [Google Scholar]
- 186.Pasquier B, El-Ahmad Y, Filoche-Romme B, Dureuil C, Fassy F, Abecassis PY, Mathieu M, Bertrand T, Benard T, Barriere C, et al.. Discovery of (2S)-8-[(3R)-3-methylmorpholin-4-yl]-1-(3-methyl-2-oxobutyl)-2-(trifluoromethyl)- 3,4-dihydro-2H-pyrimido[1,2-a]pyrimidin-6-one: a novel potent and selective inhibitor of Vps34 for the treatment of solid tumors. J Med Chem 2015; 58:376-400; PMID:25402320; http://dx.doi.org/ 10.1021/jm5013352 [DOI] [PubMed] [Google Scholar]
- 187.Miller S, Tavshanjian B, Oleksy A, Perisic O, Houseman BT, Shokat KM, Williams RL. Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34. Science 2010; 327:1638-42; PMID:20339072; http://dx.doi.org/ 10.1126/science.1184429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.El-Said OM, Hamed MM, Laufer S, Abadi AH. Design and synthesis of novel quinazoline derivatives and their evaluation as PI3Ks inhibitors. Chem Pharm Bull 2014; 62:1166-72; PMID:25450624; http://dx.doi.org/ 10.1248/cpb.c14-00560 [DOI] [PubMed] [Google Scholar]
- 189.Freitag A, Prajwal P, Shymanets A, Harteneck C, Nurnberg B, Schachtele C, Kubbutat M, Totzke F, Laufer SA. Development of first lead structures for phosphoinositide 3-kinase-C2gamma inhibitors. J Med Chem 2015; 58:212-21; PMID:24983663; http://dx.doi.org/ 10.1021/jm5006034 [DOI] [PubMed] [Google Scholar]
- 190.Marone R, Cmiljanovic V, Giese B, Wymann MP. Targeting phosphoinositide 3-kinase: moving towards therapy. Bioch Biophys Acta 2008; 1784:159-85; PMID:17997386; http://dx.doi.org/ 10.1016/j.bbapap.2007.10.003 [DOI] [PubMed] [Google Scholar]
- 191.Powis G, Bonjouklian R, Berggren MM, Gallegos A, Abraham R, Ashendel C, Zalkow L, Matter WF, Dodge J, Grindey G, et al.. Wortmannin, a potent and selective inhibitor of phosphatidylinositol-3-kinase. Cancer Res 1994; 54:2419-23; PMID:8162590 [PubMed] [Google Scholar]
- 192.Wymann MP, Bulgarelli-Leva G, Zvelebil MJ, Pirola L, Vanhaesebroeck B, Waterfield MD, Panayotou G. Wortmannin inactivates phosphoinositide 3-kinase by covalent modification of Lys-802, a residue involved in the phosphate transfer reaction. Mol Cell Biol 1996; 16:1722-33; PMID:8657148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002). J Biol Chem 1994; 269:5241-8; PMID:8106507 [PubMed] [Google Scholar]
- 194.Garlich JR, De P, Dey N, Su JD, Peng X, Miller A, Murali R, Lu Y, Mills GB, Kundra V, et al.. A vascular targeted pan phosphoinositide 3-kinase inhibitor prodrug, SF1126, with antitumor and antiangiogenic activity. Cancer Res 2008; 68:206-15; PMID:18172313; http://dx.doi.org/ 10.1158/0008-5472.CAN-07-0669 [DOI] [PubMed] [Google Scholar]
- 195.Chiorean EG, Mahadevan D, Harris WB, Von Hoff DD, Younger AE, Rensvold DM, Shelton CF, Hennessy BT, Garlich JR, Ramanathan RK. Phase I evaluation of SF1126, a vascular targeted PI3K inhibitor, administered twice weekly IV in patients with refractory solid tumors. J Clin Oncol 2009; 48(18):3319-27; PMID:2292118422921184 [Google Scholar]
- 196.Mahadevan D, Chiorean EG, Harris WB, Von Hoff DD, Stejskal-Barnett A, Qi W, Anthony SP, Younger AE, Rensvold DM, Cordova F, et al.. Phase I pharmacokinetic and pharmacodynamic study of the pan-PI3K/mTORC vascular targeted pro-drug SF1126 in patients with advanced solid tumours and B-cell malignancies. Eur J Cancer 2012; 48:3319-27; PMID:22921184; http://dx.doi.org/ 10.1016/j.ejca.2012.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Schwertschlag US, Chiorean EG, Anthony SP, Sweeney CJ, Borad MJ, Von Hoff DD, Garlich JR, Shelton CF, Ramanathan RK. Phase 1 pharmacokinetic (PK) and pharmacodynamic(PD) evaluation of SF1126 a vascular targeted pan phosphoinositide 3-kinase (PI3K) inhibitor in patients with solid tumors. J Clin Oncol 2008; 26 [Google Scholar]
- 198.Mahadevan D, Chiorean EG, Harris W, Von Hoff DD, Younger A, Rensvold DM, Cordova F, Qi W, Shelton CF, Becker MD, et al.. Phase I study of the multikinase prodrug SF1126 in solid tumors and B-cell malignancies. J Clin Oncol 2011; 29(14):1876-84; PMID:21483007; http://dx.doi.org/ 10.1200/JCO.2010.32.7171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Garlich JR, Becker MD, Shelton CF, Qi WQ, Liu XB, Cooke L, Mahadevan D. Phase I Study of Novel Prodrug Dual PI3K/mTOR Inhibitor SF1126 In B-Cell Malignancies. Blood 2010; 116:744 [Google Scholar]
- 200.Lonial S, Harvey RD, Francis D, Gul E, Jagannath S, Farag S, Hoth D, Frumento R, Garlich JR, Trudel S. Preliminary Results of a Phase I Study of the Pan-PI3 Kinase Inhibitor SF1126 in Patients with Relapsed and Refractory Myeloma. Blood 2009; 114:1492-3; http://dx.doi.org/ 10.1182/blood-2009-04-217281 [DOI] [Google Scholar]
- 201.Majchrzak A, Witkowska M, Smolewski P. Inhibition of the PI3K/Akt/mTOR signaling pathway in diffuse large B-cell lymphoma: current knowledge and clinical significance. Molecules 2014; 19:14304-15; PMID:25215588; http://dx.doi.org/ 10.3390/molecules190914304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Jimeno A, Shirai K, Choi M, Laskin J, Kochenderfer M, Spira A, Cline-Burkhardt V, Winquist E, Hausman D, Walker L, et al. A randomized, phase 2 trial of cetuximab with or without PX-866, an irreversible oral phosphatidylinositol 3-kinase Inhibitor, in patients with relapsed or metastatic head and neck squamous cell cancer. Ann Oncol 2014; 26:556-561; PMID:25524478; http://dx.doi.org/24926548 10.1093/annonc/mdu574. [DOI] [PubMed] [Google Scholar]
- 203.Levy B, Spira A, Becker D, Evans T, Schnadig I, Camidge DR, Bauman JE, Hausman D, Walker L, Nemunaitis J, et al.. A randomized, phase 2 trial of Docetaxel with or without PX-866, an irreversible oral phosphatidylinositol 3-kinase inhibitor, in patients with relapsed or metastatic non-small-cell lung cancer. J Thorac Oncol 2014; 9:1031-5; PMID:24926548; http://dx.doi.org/ 10.1097/JTO.0000000000000183 [DOI] [PubMed] [Google Scholar]
- 204.Bowles DW, Senzer N, Hausman D, Peterson S, Vo A, Walker L, Cohen RB, Jimeno A. A multicenter phase 1 study of PX-866 and cetuximab in patients with metastatic colorectal carcinoma or recurrent/metastatic squamous cell carcinoma of the head and neck. Invest New Drugs 2014; 32:1197-203; PMID:24916771; http://dx.doi.org/ 10.1007/s10637-014-0124-3 [DOI] [PubMed] [Google Scholar]
- 205.Bowles DW, Ma WW, Senzer N, Brahmer JR, Adjei AA, Davies M, Lazar AJ, Vo A, Peterson S, Walker L, et al.. A multicenter phase 1 study of PX-866 in combination with docetaxel in patients with advanced solid tumours. Br J Cancer 2013; 109:1085-92; PMID:23942080; http://dx.doi.org/ 10.1038/bjc.2013.474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Hong DS, Bowles DW, Falchook GS, Messersmith WA, George GC, O'Bryant CL, Vo AC, Klucher K, Herbst RS, Eckhardt SG, et al.. A multicenter phase I trial of PX-866, an oral irreversible phosphatidylinositol 3-kinase inhibitor, in patients with advanced solid tumors. Clin Cancer Res 2012; 18:4173-82; PMID:22693357; http://dx.doi.org/ 10.1158/1078-0432.CCR-12-0714 [DOI] [PubMed] [Google Scholar]
- 207.Ihle NT, Williams R, Chow S, Chew W, Berggren MI, Paine-Murrieta G, Minion DJ, Halter RJ, Wipf P, Abraham R, et al.. Molecular pharmacology and antitumor activity of PX-866, a novel inhibitor of phosphoinositide-3-kinase signaling. Mol Cancer Ther 2004; 3:763-72; PMID:15252137 [PubMed] [Google Scholar]
- 208.Koul D, Shen R, Kim YW, Kondo Y, Lu Y, Bankson J, Ronen SM, Kirkpatrick DL, Powis G, Yung WK. Cellular and in vivo activity of a novel PI3K inhibitor, PX-866, against human glioblastoma. Neuro Oncol 2010; 12:559-69; PMID:20156803; http://dx.doi.org/ 10.1093/neuonc/nop058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Pitz MW, Eisenhauer EA, MacNeil MV, Thiessen B, Easaw JC, Macdonald DR, Eisenstat DD, Kakumanu AS, Salim M, Chalchal H, et al. Phase II study of PX-866 in recurrent glioblastoma. Neuro-oncology 2015; PMID:25605819; http://dx.doi.org/24166903 10.1093/neuonc/nou365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Hotte SJ, Eisenhauer EA, Joshua AM, Kumar V, Ellard S, Gregg RW, Macfarlane RJ, Winquist E, Torri V, Ruether JD, et al.. NCIC CTG, IND-205: a phase II study of PX-866 in patients with recurrent or metastatic castration-resistant prostate cancer (CRPC). J Clin Oncol 2013; 31 [Google Scholar]
- 211.Jimeno A, Herbst RS, Falchook GS, Messersmith WA, Hecker S, Peterson S, Hausman DF, Kurzrock R, Eckhardt SG, Hong DS. Final results from aphase I, dose-escalation study of PX-866, an irreversible, pan-isoform inhibitor of PI3 kinase. J Clin Oncol 2010; 28 [Google Scholar]
- 212.Shapiro GI, Rodon J, Bedell C, Kwak EL, Baselga J, Brana I, Pandya SS, Scheffold C, Laird AD, Nguyen LT, et al.. Phase I safety, pharmacokinetic, and pharmacodynamic study of SAR245408 (XL147), an oral pan-class I PI3K inhibitor, in patients with advanced solid tumors. Clin Cancer Res 2014; 20:233-45; PMID:24166903; http://dx.doi.org/ 10.1158/1078-0432.CCR-13-1777 [DOI] [PubMed] [Google Scholar]
- 213.Traynor AM, Kurzrock R, Bailey HH, Attia S, Scheffold C, van Leeuwen B, Wu B, Falchook GS, Moulder SL, Wheler J. A phase I safety and pharmacokinetic (PK) study of the PI3K inhibitor XL147 (SAR245408) in combination with paclitaxel (P) and carboplatin (C) in patients (pts) with advanced solid tumors. J Clin Oncol 2010; 28 [Google Scholar]
- 214.Moldovan C, Soria J, LoRusso P, Guthrie T, Song C, Nguyen LT, Martini J, Infante JR, Burris HA. A phase I safety and pharmacokinetic (PK) study of the PI3K inhibitor XL147 (SAR245408) in combination with erlotinib in patients (pts) with advanced solid tumors. J Clin Oncol 2010; 28 [Google Scholar]
- 215.Shapiro G, Kwak E, Baselga J, Rodon J, Scheffold C, Laird AD, Bedell C, Edelman G. Phase I dose-escalation study of XL147, a PI3K inhibitor administered orally to patients with solid tumors. J Clin Oncol 2009; 27 [Google Scholar]
- 216.Edelman G, Bedell C, Shapiro G, Pandya SS, Kwak EL, Scheffold C, Nguyen LT, Laird A, Baselga J, Rodon J. A phase I dose-escalation study of XL147 (SAR245408), a PI3K inhibitor administered orally to patients (pts) with advanced malignancies. J Clin Oncol 2010; 28:2191-7; PMID:2035132920351329 [Google Scholar]
- 217.Calvo E, Edelman G, Baselga J, Kwak E, Scheffold C, Nguyen L, Shapiro GI. A phase I dose-escalation study of the safety, pharmacokinetics and pharmacodynamics of XL147, a novel PI3K inhibitor administered orally to patients with advanced solid tumors. Ejc Suppl 2008; 6:69; http://dx.doi.org/ 10.1016/S1359-6349(08)72150-5 [DOI] [Google Scholar]
- 218.Matulonis U, Vergote I, Backes F, Martin LP, McMeekin S, Birrer M, Campana F, Xu Y, Egile C, Ghamande S. Phase II study of the PI3K inhibitor pilaralisib (SAR245408; XL147) in patients with advanced or recurrent endometrial carcinoma. Gynecol Oncology 2015; 136:246-53; PMID:25528496; http://dx.doi.org/ 10.1016/j.ygyno.2014.12.019 [DOI] [PubMed] [Google Scholar]
- 219.Corcelle EA, Puustinen P, Jaattela M. Apoptosis and autophagy: targeting autophagy signalling in cancer cells -'trick or treats'? FEBS J 2009; 276:6084-96; PMID:19788415; http://dx.doi.org/ 10.1111/j.1742-4658.2009.07332.x [DOI] [PubMed] [Google Scholar]
- 220.Burger MT, Pecchi S, Wagman A, Ni ZJ, Knapp M, Hendrickson T, Atallah G, Pfister K, Zhang Y, Bartulis S, et al.. Identification of NVP-BKM120 as a Potent, Selective, Orally Bioavailable Class I PI3 Kinase Inhibitor for Treating Cancer. ACS Med Chem Lett 2011; 2:774-9; PMID:24900266; http://dx.doi.org/ 10.1021/ml200156t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Bendell JC, Rodon J, Burris HA, de Jonge M, Verweij J, Birle D, Demanse D, De Buck SS, Ru QC, Peters M, et al.. Phase I, dose-escalation study of BKM120, an oral pan-Class I PI3K inhibitor, in patients with advanced solid tumors. J Clin Oncol 2012; 30:282-90; PMID:22162589; http://dx.doi.org/ 10.1200/JCO.2011.36.1360 [DOI] [PubMed] [Google Scholar]
- 222.Rodon J, Brana I, Siu LL, De Jonge MJ, Homji N, Mills D, Di Tomaso E, Sarr C, Trandafir L, Massacesi C, et al.. Phase I dose-escalation and -expansion study of buparlisib (BKM120), an oral pan-Class I PI3K inhibitor, in patients with advanced solid tumors. Invest New Drugs 2014; 32:670-81; PMID:24652201 [DOI] [PubMed] [Google Scholar]
- 223.Bedard PL, Tabernero J, Janku F, Wainberg ZA, Paz-Ares L, Vansteenkiste J, Van Cutsem E, Perez-Garcia J, Stathis A, Britten CD, et al. A phase Ib dose-escalation study of the oral pan-PI3K inhibitor buparlisib (BKM120) in combination with the oral MEK1/2 inhibitor trametinib (GSK1120212) in patients with selected advanced solid tumors. Clin Cancer Res 2015; 21:730-8; PMID:25500057; http://dx.doi.org/23721513 10.1158/1078-0432.CCR-14-1814. [DOI] [PubMed] [Google Scholar]
- 224.Zang C, Eucker J, Liu H, Coordes A, Lenarz M, Possinger K, Scholz CW. Inhibition of pan-class I phosphatidyl-inositol-3-kinase by NVP-BKM120 effectively blocks proliferation and induces cell death in diffuse large B-cell lymphoma. Leuk Lymphoma 2014; 55:425-34; PMID:23721513; http://dx.doi.org/ 10.3109/10428194.2013.806800 [DOI] [PubMed] [Google Scholar]
- 225.Patnaik A, Ramanathan RK, Dubowy R, Tolcher AW, Papadopoulos KP, Beeram M, Mountz JM, Rasco D, Lotze MT, Laymon C, et al.. The evolution of Pi3 kinase inhibitors: results of a first-in-human phase I study of Bay80-6946 in advanced solid tumors. Ann Oncol 2012; 23:16; http://dx.doi.org/ 10.1093/annonc/mds439 [DOI] [Google Scholar]
- 226.Glienke W, Maute L, Wicht J, Bergmann L. The dual PI3K/mTOR inhibitor NVP-BGT226 induces cell cycle arrest and regulates Survivin gene expression in human pancreatic cancer cell lines. Tum Biol 2012; 33:757-65; PMID:22170433; http://dx.doi.org/ 10.1007/s13277-011-0290-2 [DOI] [PubMed] [Google Scholar]
- 227.Chang KY, Tsai SY, Wu CM, Yen CJ, Chuang BF, Chang JY. Novel phosphoinositide 3-kinase/mTOR dual inhibitor, NVP-BGT226, displays potent growth-inhibitory activity against human head and neck cancer cells in vitro and in vivo. Clin Cancer Res 2011; 17:7116-26; PMID:21976531; http://dx.doi.org/ 10.1158/1078-0432.CCR-11-0796 [DOI] [PubMed] [Google Scholar]
- 228.Ahnert JR, Burris HA, Schellens JHM, Schuler M, Goodman O, Britten C, Richards D, Demanse D, Silva A, Baselga J. Phase I dose-escalation study of the oral dual MTOR/PI3K inhibitor BEZ235, solid dispersion system (SDS) sachet formulation, in patients with advanced solid tumors. Eur J Cancer 2012; 48:112; http://dx.doi.org/ 10.1016/S0959-8049(12)72164-0 [DOI] [Google Scholar]
- 229.Burris H, Rodon J, Sharma S, Herbst RS, Tabernero J, Infante JR, Silva A, Demanse D, Hackl W, Baselga J. First-in-human phase I study of the oral PI3K inhibitor BEZ235 in patients (pts) with advanced solid tumors. J Clin Oncol 2010; 28 [Google Scholar]
- 230.Dirix L, Schuler M, Machiels J, Hess D, Awada A, Steeghs N, Paz-Ares L, von Moos R, Rabault B, Rodon J. Phase Ib dose-escalation study of Bez235 or Bkm120 in combination with paclitaxel (Ptx) in patients with advanced solid tumors. Ann Oncol 2012; 23:157-8 [Google Scholar]
- 231.Krop IE, Saura C, Ahnert JR, Becerra C, Britten CD, Isakoff SJ, Demanse D, Hackl W, Quadt C, Silva AP, et al.. A phase I/IB dose-escalation study of BEZ235 in combination with trastuzumab in patients with PI3-kinase or PTEN altered HER2+metastatic breast cancer. J Clin Oncol 2012; 30 [Google Scholar]
- 232.Salkeni MA, Beg MS, Olowokure OO, Fathallah H, Thomas H, Mercer CA, Charif M, Chaudhary RT, Karim NA, Warnick R, et al.. A dose escalation, single arm, phase Ib/II combination study of BEZ235 with everolimus to determine the safety, pharmacodynamics, and pharmacokinetics in subjects with advanced solid malignancies including glioblastoma multiforme. J Clin Oncol 2012; 30 [Google Scholar]
- 233.Seitz C, Hugle M, Cristofanon S, Tchoghandjian A, Fulda S. The dual PI3K/mTOR inhibitor NVP-BEZ235 and chloroquine synergize to trigger apoptosis via mitochondrial-lysosomal cross-talk. Int J Cancer 2013; 132:2682-93; PMID:23151917; http://dx.doi.org/ 10.1002/ijc.27935 [DOI] [PubMed] [Google Scholar]
- 234.Janne PA, Cohen RB, Laird AD, Mace S, Engelman JA, Ruiz-Soto R, Rockich K, Xu J, Shapiro GI, Martinez P, et al.. Phase I safety and pharmacokinetic study of the PI3K/mTOR inhibitor SAR245409 (XL765) in combination with erlotinib in patients with advanced solid tumors. J Thoracic Oncol 2014; 9:316-23; PMID:24496004; http://dx.doi.org/ 10.1097/JTO.0000000000000088 [DOI] [PubMed] [Google Scholar]
- 235.Papadopoulos KP, Tabernero J, Markman B, Patnaik A, Tolcher AW, Baselga J, Shi W, Egile C, Ruiz-Soto R, Laird AD, et al.. Phase I safety, pharmacokinetic, and pharmacodynamic study of SAR245409 (XL765), a novel, orally administered PI3K/mTOR inhibitor in patients with advanced solid tumors. Clin Cancer Res 2014; 20:2445-56; PMID:24583798; http://dx.doi.org/ 10.1158/1078-0432.CCR-13-2403 [DOI] [PubMed] [Google Scholar]
- 236.LoRusso P, Markman B, Tabernero J, Shazer R, Nguyen L, Heath E, Patnaik A, Papadopoulos K. A phase I dose-escalation study of the safety, pharmacokinetics (PK), and pharmacodynamics of XL765, a PI3K/TORC1/TORC2 inhibitor administered orally to patients (pts) with advanced solid tumors. J Clin Oncol 2009; 27 [Google Scholar]
- 237.Markman B, LoRusso PM, Patnaik A, Heath E, Laird AD, van Leeuwen B, Papadopoulos KP, Baselga J. A phase I dose-escalation study of the safety, pharmacokinetics and pharmacodynamics of XL765, a novel inhibitor of PI3K and mTOR, administered orally to patients with solid tumors. Ejc Suppl 2008; 6:68-9; http://dx.doi.org/ 10.1016/S1359-6349(08)72148-7 [DOI] [Google Scholar]
- 238.Ghadimi MP, Lopez G, Torres KE, Belousov R, Young ED, Liu J, Brewer KJ, Hoffman A, Lusby K, Lazar AJ, et al.. Targeting the PI3K/mTOR axis, alone and in combination with autophagy blockade, for the treatment of malignant peripheral nerve sheath tumors. Mol Cancer Ther 2012; 11:1758-69; PMID:22848094; http://dx.doi.org/ 10.1158/1535-7163.MCT-12-0015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Flinn IW, Horwitz SM, Patel M, Younes A, Porter JR, Sweeney J, Allen K, Kelly P, Kahl BS. Clinical safety and activity in a phase 1 trial of IPI-145, a potent inhibitor of phosphoinositide-3-kinase-delta,gamma, in patients with advanced hematologic malignancies. Blood 2012; 120 [Google Scholar]
- 240.Sarker D, Kristeleit R, Mazina KE, Ware JA, Yan Y, Dresser M, Derynck MK, De-Bono J. A phase I study evaluating the pharmacokinetics (PK) and pharmacodynamics (PD) of the oral pan-phosphoinositide-3 kinase (PI3K) inhibitor GDC-0941. J Clin Oncol 2009; 27 [Google Scholar]
- 241.Baird RD, Kristeleit RS, Sarker D, Olmos D, Sandhu SK, Yan Y, Koeppen H, Levy GG, Jin J, De Bono JS. A phase I study evaluating the pharmacokinetics (PK) and pharmacodynamics (PD) of the oral pan-phosphoinositide-3 kinase (PI3K) inhibitor GDC-0941. J Clin Oncol 2010; 28 [Google Scholar]
- 242.Baird R, Kristeleit R, Sarker D, Olmos D, Sandhu S, Levy GG, Mazina KE, Yan Y, Ware J, De Bono J. A phase I study evaluating the pharmacokinetics (Pk) and pharmacodynamics (Pd) of the oral pan-phosphoinositide-3 kinase (Pi3k) inhibitor Gdc-0941. Ann Oncol 2010; 21:170-1 [Google Scholar]
- 243.LoRusso P, Sarker D, Von Hoff D, Tibes R, Derynck MK, Ware JA, Yan Y, Demetri GD, de Bono JS, Wagner AJ. Pharmacokinetics and pharmacodynamic biomarkers for the pan-PI3K inhibitor GDC-0941: initial phase I evaluation. Ejc Suppl 2008; 6:70-1; http://dx.doi.org/ 10.1016/S1359-6349(08)72155-4 [DOI] [Google Scholar]
- 244.Sarker D, Ang JE, Baird R, Kristeleit R, Shah K, Moreno V, Clarke PA, Raynaud FI, Levy G, Ware JA, et al.. First-in-human phase I study of pictilisib (GDC-0941), a potent pan-class I phosphatidylinositol-3-kinase (PI3K) inhibitor, in patients with advanced solid tumors. Clin Cancer Res 2015; 21:77-86; PMID:25370471; http://dx.doi.org/ 10.1158/1078-0432.CCR-14-0947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.de Vos S, Schreeder MT, Flinn IW, Coutre SE, Leonard JP, Wagner-Johnston ND, Fowler NH, Boccia RV, Barrientos JC, Boyd T, et al.. A Phase 1 study of the selective phosphatidylinositol 3-kinase-delta (PI3K delta) inhibitor, Cal-101 (GS-1101), in combination with rituximab and/or bendamustine in patients with previously treated, indolent non-hodgkin lymphoma (iNHL). Blood 2011; 118:1160 [Google Scholar]
- 246.De Vos S, Sehn LH, Mulligan SP, Dreyling MH, Rummel MJ, Zinzani PL, Johnson DM, Evans S, Dansey RD, Godfrey WR, et al.. A phase III, randomized, double-blind, placebo-controlled study evaluating the efficacy and safety of idelalisib (GS-1101) in combination with bendamustine and rituximab for previously treated indolent non-Hodgkin lymphomas (iNHL). J Clin Oncol 2013; 31 [Google Scholar]
- 247.Eradat HA, Coutre SE, Barrientos JC, Rai KR, Farber CM, Hillmen P, Sharman JP, Ghia P, Coiffier B, Walewski JA, et al.. A phase III, randomized, double-blind, placebo-controlled study evaluating the efficacy and safety of idelalisib (GS-1101) in combination with bendamustine and rituximab for previously treated chronic lymphocytic leukemia (CLL). J Clin Oncol 2013; 31 [Google Scholar]
- 248.Flinn I, Kimby E, Cotter FE, Giles FJ, Janssens A, Pulczynski EJ, Ysebeart L, Pluta A, Marco JAG, Taylor K, et al.. A phase III, randomized, controlled study evaluating the efficacy and safety of idelalisib (GS-1101) in combination with ofatumumab for previously treated chronic lymphocytic leukemia (CLL). J Clin Oncol 2013; 31 [Google Scholar]
- 249.Furman RR, Barrientos JC, Sharman JP, De Vos S, Leonard J, Coutre SE, Schreeder MT, Wagner-Johnston ND, Boyd TE, Fowler NH, et al.. A phase I/II study of the selective phosphatidylinositol 3-kinase -delta (PI3K delta) inhibitor, GS-1101 (CAL-101), with ofatumumab in patients with previously treated chronic lymphocytic leukemia (CLL). J Clin Oncol 2012; 30; PMID:2254760222547602 [Google Scholar]
- 250.O'Brien SM, Lamanna N, Kipps TJ, Flinn I, Zelenetz AD, Burger JA, Holes L, Johnson DM, Gu J, Dansey RD, et al.. A phase II study of the selective phosphatidylinositol 3-kinase delta (PI3K delta) inhibitor idelalisib (GS-1101) in combination with rituximab (R) in treatment-naive patients (pts) >= 65 years with chronic lymphocytic leukemia (CLL) or small lymphocytic lymphoma (SLL). J Clin Oncol 2013; 31 [Google Scholar]
- 251.Sharman J, de Vos S, Leonard JP, Furman RR, Coutre SE, Flinn IW, Schreeder MT, Barrientos JC, Wagner-Johnston ND, Boyd T, et al.. A Phase 1 study of the selective phosphatidylinositol 3-kinase-delta (PI3K delta) inhibitor, CAL-101 (GS-1101), in combination with rituximab and/or bendamustine in patients with relapsed or refractory chronic lymphocytic leukemia (CLL). Blood 2011; 118:779-80 [Google Scholar]