Over-expressing or silencing proteases or their inhibitors alter the metabolite composition of the grain and modify the germination process in barley
Abstract
Proteolysis is an essential process throughout the mobilization of storage proteins in barley (Hordeum vulgare) grains during germination. It involves numerous types of enzymes, with C1A Cys proteases the most abundant key players. Manipulation of the proteolytic machinery is a potential way to enhance grain yield and quality, and it could influence the mobilization of storage compounds along germination. Transgenic barley plants silencing or over-expressing the cathepsin F-like HvPap-1 Cys protease show differential accumulation of storage molecules such as starch, proteins, and free amino acids in the grain. It is particularly striking that the HvPap-1 artificial microRNA lines phenotype show a drastic delay in the grain germination process. Alterations to the proteolytic activities in the over-expressing and knock-down grains associated with changes in the level of expression of several C1A peptidases were also detected. Similarly, down-regulating cystatin Icy-2, one of the proteinaceous inhibitors of the cathepsin F-like protease, also has important effects on grain filling. However, the ultimate physiological influence of manipulating a peptidase or an inhibitor cannot be always predicted, since the plant tries to compensate the modified proteolytic effects by modulating the expression of some other peptidases or their inhibitors.
Barley (Hordeum vulgare) is an annual, monocotyledonous plant of the Poaceae family and one of the world’s earliest domesticated and most important crop plants. It currently represents the fourth most abundant cereal in both surface and tonnage harvested. According to FAO, the area in barley cultivation in 2013 was 49,148,479 ha, with a yield of more than 2,929 kg/ha, meaning an average world production of approximately 144 Mt (FAOSTAT, http://faostat3.fao.org/download/Q/QC/E). The high production of this crop is mainly because of its importance in the malting industry and breweries as well as to the nutritional intake of humans and livestock. Barley is considered a model species for cereal research, because its genome is fully sequenced (Mayer et al., 2012). Furthermore, it constitutes one of the best options to genetically improve cultivars to tackle climate change and guarantee cereal production (Nevo et al., 2012; Dawson et al., 2015).
During grain development and maturation, proteins involved in germination are stored in the endosperm together with starch and lipids. The cereal grain protein concentration is about 10% to 12%, which is relatively low compared to legume seeds. Nevertheless, cereals’ impact on humans and livestock nutrition is about three times higher than the effect of the most protein-rich legume seeds (Shewry and Halford, 2002). Storage, structural, metabolic, and protective proteins are present in the grains. The major part corresponds to storage proteins, which represent nearly 80% of total proteins, falling into three different fractions based on the extraction method and their solubility: albumins (water soluble), globulins (soluble in salt solutions), and prolamins (soluble in alcohol/water mixtures; Shewry et al., 1995). With the exceptions of oat and rice, the major endosperm storage proteins in all cereal grains are prolamins (Shewry and Halford, 2002), named hordeins in barley. Hordeins are highly hydrophobic molecules, rich in Pro and Gln, and with a low content of charged amino acids, particularly the essential amino acid Lys. There are three broad groups of hordeins: B (sulfur-rich), C (sulfur-poor), and D (high Mr), with several subgroups within the B-group (Shewry et al., 1995). All of them are coordinately expressed during endosperm development (Sorensen et al., 1989), and their expression is tightly regulated (Diaz et al., 2005).
Limited proteolysis mediated by peptidases is essential for the initiation of storage protein breakdown to nurture the developing embryo (Müntz, 1996). Peptidases were formerly classified as Cys- (Cys), Ser- (Ser), aspartic- (Asp), and metallo-proteases (metallic ion), according to the residue present in the active site of the enzyme. Papain-like Cys proteases (CysProt), known as C1A (family C1, clan CA) according to the MEROPS database (Rawlings et al., 2014), constitute one of the most abundant groups of proteases responsible for the degradation and mobilization of storage proteins in seeds. Their role during germination has been reported in a wide range of both monocot and dicot plants (Grudkowska and Zagdańska, 2004; Tan-Wilson and Wilson, 2012). This group of proteases has a high number of members in angiosperms, ranking from 32 in Arabidopsis (Arabidopsis thaliana) to 45 in rice, which are classified into cathepsins L-, B-, H-, and F- like (Martinez and Diaz, 2008); in barley, 41 members have been identified (Díaz-Mendoza et al., 2014). A previous analysis reported that 27 C1A CysProt are among the 42 proteases involved in the germination of barley grain (Zhang and Jones, 1995).
A complete transcriptome analysis in two tissue fractions (starchy endosperm/aleurone and embryo/scutellum) has shown the induction of a high number of CysProt genes during germination, most of them mediated by GAs (Sreenivasulu et al., 2008). Some of the barley C1A proteases expressed in grain tissues have been characterized. Among them, several cathepsin l-like proteases of the scutellar epithelium and the aleurone layer were secreted to the endosperm upon germination in response to GA (Koehler and Ho, 1990; Mikkonen et al., 1996; Martinez et al., 2009). A cathepsin H-like protease isolated from GA-induced aleurone cells was targeted to vacuoles, and a cathepsin B-like protein was expressed in the aleurone and induced by GA treatment (Holwerda and Rogers, 1992; Martínez et al., 2003). Recently, the HvPap-1, a cathepsin F-like peptidase, has been described to be involved in barley grain protein mobilization and modulated by its own propeptide and its inhibitors, the cystatins (Cambra et al., 2012). This protein is able to in vitro degrade different substrates, including barley endosperm proteins (hordeins, albumins, and globulins). It has been localized in protein bodies and vesicles of the embryo, and it is induced by GA in aleurone cells (Cambra et al., 2012).
Cystatins are the endogenous proteinaceous inhibitors of C1A CysProt. They have several roles in plants and have been extensively related to the regulation of physiological processes in seeds (Benchabane et al., 2010). In barley, the whole cystatin family has been identified and characterized (Martinez et al., 2009). The functional relationship between barley cystatins and cathepsin l- and F-like proteases has been inferred from their common implication as counterparts during hordein storage protein mobilization upon barley grain germination (Martinez et al., 2009; Cambra et al., 2012). The barley cystatin HvCPI-2 encoded by the Icy-2 gene is a good inhibitor of different barley cathepsin l- and F-like CysProt, such as HvPap-1, -4, -6, -10, and -16. It is strongly expressed in the germinating embryo and repressed by GA3 in aleurone layers. These results suggest a key role for HvCPI-2 in the regulation of the CysProt activity in barley grain (Martinez et al., 2009; Cambra et al., 2012).
Previous research indicates that a complex regulatory network including C1A CysProt and their inhibitors is involved in the regulation of the barley grain germination process. This work demonstrates how biotechnological modifications of the proteolytic machinery may affect grain composition and, consequently, germination in barley. For this purpose, in planta participation of the cathepsin F-like HvPap-1 and the cystatin HvCPI-2 proteins during grain filling and mobilization of stored proteins was analyzed in barley transgenic lines over-expressing the HvPap-1 gene or knocking-down the expression of either the HvPap-1 or Icy-2 genes.
RESULTS
Transgenic Barley Lines Over-expressing or Silencing HvPap-1 Protease or Silencing Icy-2 Cystatin
Transgenic barley plants were obtained from immature embryos after Agrobacterium coculture and selection on hygromycin-containing media. Transgenic barley plants ubiquitously over-expressing the HvPap-1 gene were generated using p6U and p6d35s binary vectors. Silencing of the HvPap-1 and Icy-2 genes was generated using the artificial microRNA (amiRNA) technology. For each construct, 30 independent primary plants were generated. Afterward, four to six T1 events were preliminarily used for molecular characterization. Homozygous material was generated via embryogenic pollen cultures (Coronado et al., 2005), and obtained homozygous plants were analyzed in depth. Two HvPap-1 over-expressing lines (OE Pap1: 919 and 937), two HvPap-1 silencing lines (KD Pap1: 1130 and 1175), and two Icy-2 silencing lines (KD Icy2: 1318 and 1399) were selected based on transgene copy number, transcript, and protein content for further studies (Supplemental Figs. S1A and S2A). Following these criteria, the over-expressing lines showed two copies of HvPap-1 gene, the endogenous and the transgene, estimated by real-time quantitative PCR (RT-qPCR) assay and the 2-∆∆Ct method (Supplemental Fig. S1B) and presented higher accumulation of mRNAs and protein than the wild type (Supplemental Fig. S1, C and D). The amiRNA lines contained a unique transgene insertion, and the expression levels of their messengers and accumulation of proteins were reduced in comparison with the wild type (Supplemental Figs. S1, B–D, and S2, B–D). However, neither mRNA accumulation nor protein content was completely knocked out in the amiRNA plants.
Grain Phenotype and Starch Accumulation Are Altered in Barley Transgenic Lines
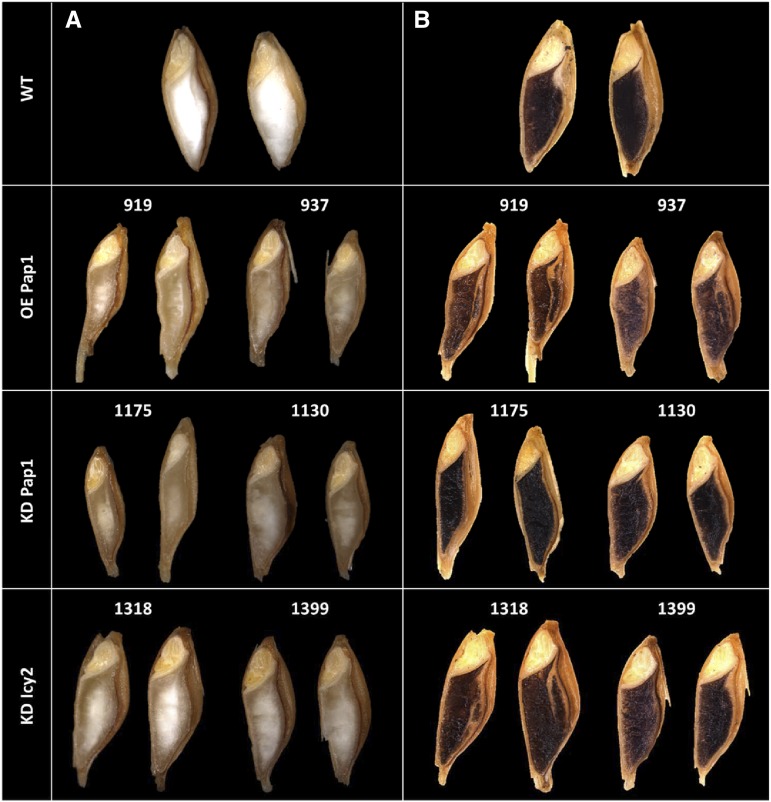
Kernels from transgenic and control plants were obtained and their grains phenotypically compared 24 h after imbibition (hai). The OE Pap1 and KD Pap1 grains were of similar size but were elongated and presented darker endosperms than the control grains. The KD Icy2 lines also showed grains with a slightly darker endosperm than the wild type (Fig. 1A). These phenotypic differences could be related to a different grain composition. The amount of starch can be inferred by the intensity of the dark-blue/black color after Lugol staining. OE Pap1 and KD Icy2 grains presented a weaker color than wild-type grains, indicating a lower amount of starch. In contrast, the staining of KD Pap1 grains was stronger than the signal observed in wild-type grains (Fig. 1B). No differences in the balance of amylose/amylopectin could be detected, since the typical amylopectin reddish coloration was not observed (Fig. 1B). Quantification of the starch content in grains corroborated that the KD Pap1 silencing lines contained significantly higher quantities of starch than the control line (Supplemental Fig. S3).
Figure 1.
Phenotype and starch staining of barley grains. A, Structure of longitudinal dissected grains 24 h after imbibition from wild-type and transgenic plants. B, Lugol's iodine staining of transgenic and wild-type barley grains.
Grain Protein Content Is Modified in Barley Transgenic Lines
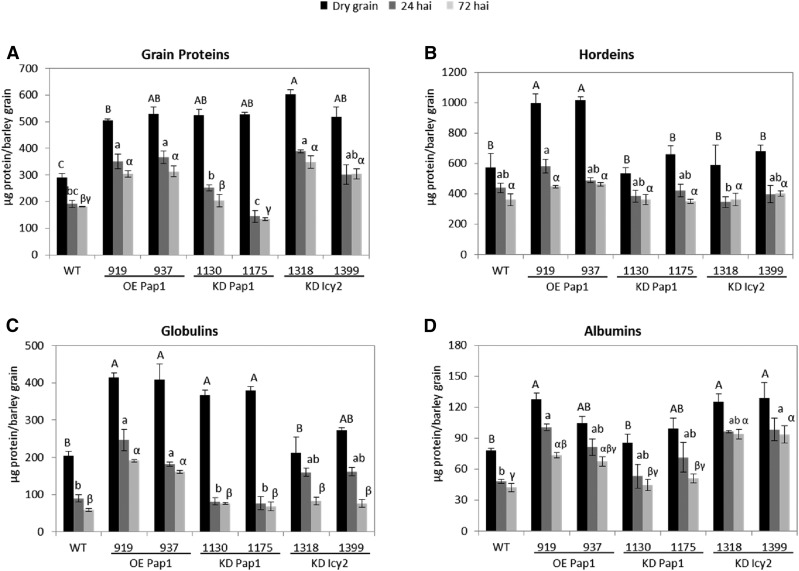
The protein quantity of deembryonated grains was also quantified (Fig. 2A). Dry grains from all transgenic lines (OE Pap1, KD Pap1, and KD Icy2) presented similar quantities of protein between them but significantly higher protein amounts per grain than the nontransformed line (Fig. 2A). These proteins are degraded as germination proceeds. At 24 and 72 h hai, the amount of protein decreased in all transgenic and nontransgenic lines. However, the rate of remaining protein was different. About 40% of the grain protein was degraded at 72 hai for all lines with the exception of KD Pap1 lines, in which about 70% of the grain protein was broken down at 72 hai (Fig. 2A). These variations in the speed of protein degradation were confirmed by electrophoretic band patterns, which were very similar for wild-type, OE Pap1, and KD Icy2 barley grains but different for KD Pap1 ones. KD Pap1 lines showed a fainter pattern for medium-high Mr protein bands (Supplemental Fig. S4A).
Figure 2.
Quantification of the protein content per grain in wild-type and transgenic lines at different hai. A, Grain proteins. B, Hordeins. C, Globulins. D, Albumins. Different letters indicate significant differences between lines for each time point (P < 0.05, honestly significant difference). Capital, small, and Greek letters are used for dry grains, 24 hai grains, and 72 hai grains, respectively.
Variations in the quantity of grain proteins could be related to alterations in the type of storage proteins in each transgenic line. Hordeins, albumins, and globulins are the main storage proteins in barley, and fractions enriched in each of these compounds were isolated. Quantification of these fractions revealed differences related to the storage and processing of these proteins (Fig. 2B-D). The total amount of hordeins was higher in OE Pap1 dry grains than in the rest of grains analyzed but was more quickly degraded in these lines, reaching similar values at 72 hai than in the other transgenic and nontransgenic grains analyzed (Fig. 2B). These specific degradative patterns are shown by the appearance of intermediated Mr bands in the electrophoretic gels for OE Pap1 (Supplemental Fig. S4B). Globulins showed a striking pattern, with a significantly higher accumulation in dry grain in both OE Pap1 and KD Pap1 transgenic lines than in the wild-type or KD Icy2 lines (Fig. 2C). However, the rate of degradation was different. In the wild type, OE Pap1 and KD Icy2 lines around 50% of stored globulins were broken down at 72 hai, whereas in the KD Pap1 lines approximately 75% of the stored albumins were degraded at the same time point. Electrophoretic gels showed this rapid degradation of most globulin bands in the KD Pap1 lines (Supplemental Fig. S4C). Finally, albumins were also accumulated differentially in the dry grains from transgenic and nontransgenic lines, with a significantly higher quantity in the KD Icy2 lines and in the 919 OE Pap1 line (Fig. 2D). However, variations on protein degradation patterns among the different lines were not clearly detected (Fig. 2D; Supplemental Fig. S4D).
Grain Metabolomic Analyses Show Changes in Amino Acid Accumulation in Barley Transgenic Lines
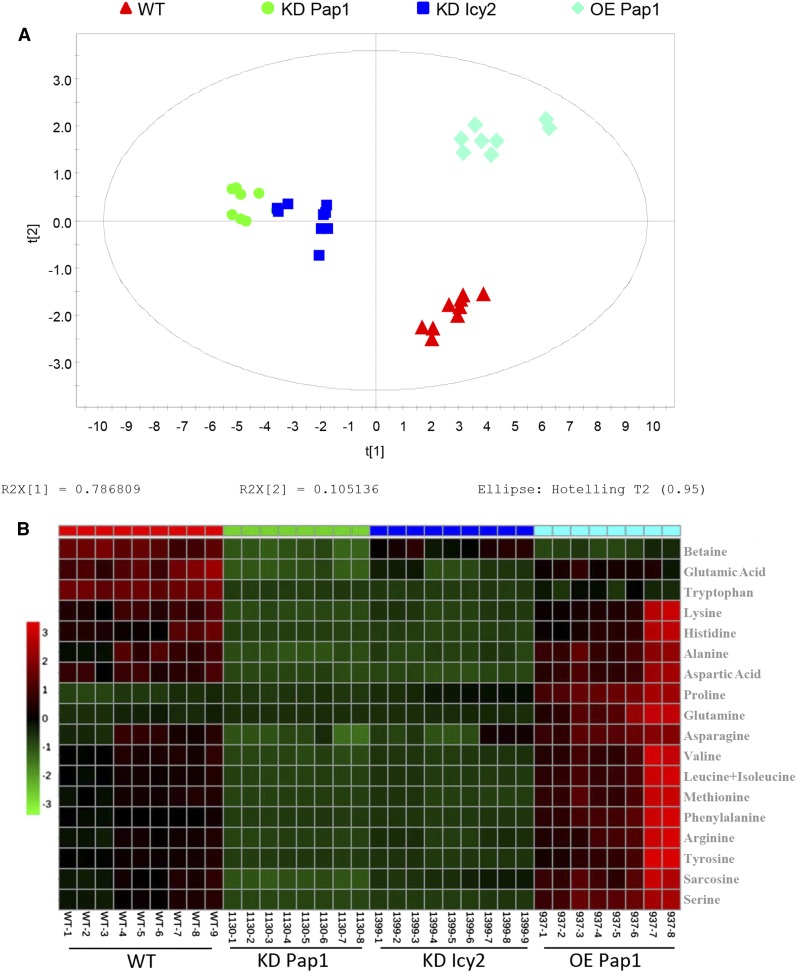
Differential protein accumulation in grains could be related to differences in the free amino acid composition in deembryonated grains. To test that, a metabolomic analysis was performed comparing the free amino acids in transgenic and nontransgenic mature grains. Principal component analysis (PCA) is an orthogonal linear transformation of possibly correlated variables into a smaller number of uncorrelated variables called principal components, where the greatest variance within the data is explained on the first coordinate. Samples that group together represent a specific phenotype. According to the PCA scores plot, it can be observed that biotechnological proteolytic modifications induced specific amino acid perturbations. Samples originating from different lines clustered into different areas in the plots (Fig. 3A). Samples from KD Pap1 1130 and KD Icy2 1399 lines clustered closely and far from wild-type grains. OE Pap1 937 samples clustered together and separately from the rest of the lines. As expected, the plot showed that variation among the different groups was more pronounced on the component 1 that accounted for the highest variation in the models. Specific differences in the amino acid composition from grain extracts are illustrated in the heatmap (Fig. 3B). The heatmap readily shows changes in the concentrations of amino acids in the grains from the different transgenic and nontransgenic lines. The quantification and significance of these changes are reported in Supplemental Table S1. In the samples coming from both KD lines, most amino acid molecules were significantly reduced, with the exception of Pro in both lines and Gln in the KD Icy2 line. On the contrary, in the OE Pap1 samples, the most remarkable difference in comparison to wild-type samples is a pronounced increase in the levels of two amino acids, Pro and Gln.
Figure 3.
PCA models and metabolite heatmap of amino acid composition of dry deembryonated grains. A, PCA scores plot of all samples included in the study (red triangles, wild-type; green dots, KD Pap1 1130; blue squares, KD Icy2 1399; turquoise diamonds, OE Pap1 937). Cumulative R2 and Q2 were 0.892 and 0.803, respectively. B, Heatmap showing the relative contributions of each amino acid in the different type of samples. The color code shows the abundance level.
Germination Is Delayed in HvPap-1 Transgenic Lines
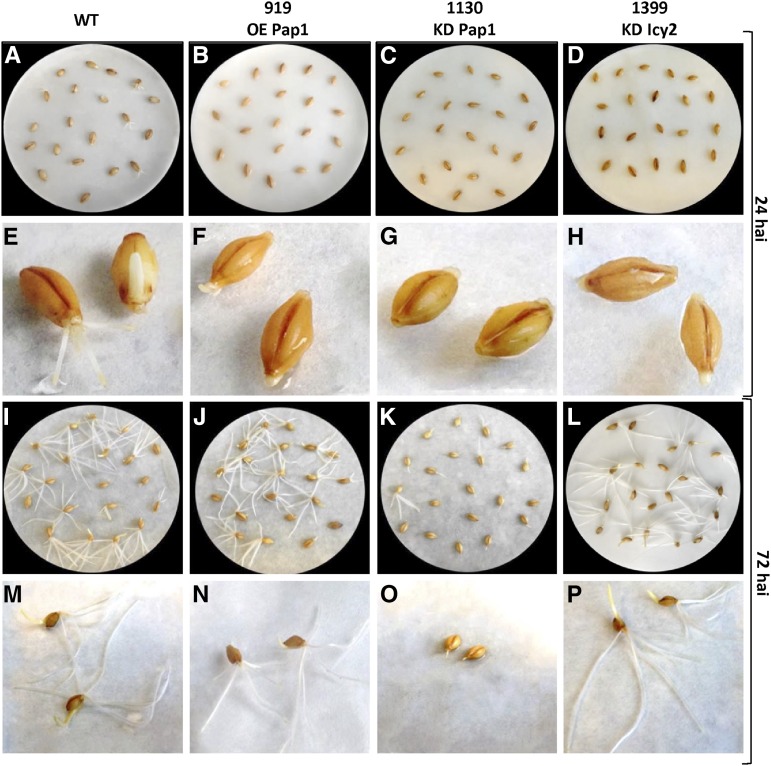
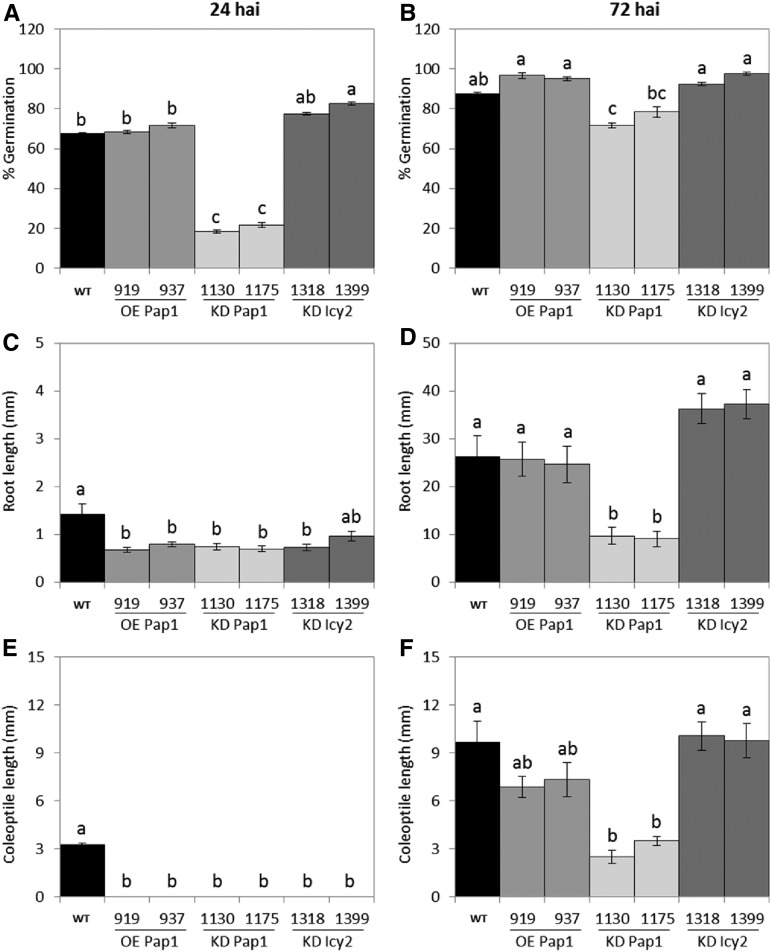
To analyze whether differences in grain composition affect germination ability, twenty barley grains per line were imbibed in watered paper on petri dishes, and germination was followed for 72 h. A striking delay was observed for KD Pap1 lines (Fig. 4). The number of germinated grains was counted at 24 and 72 hai (Fig. 5). Whereas no differences were observed in the percentage of germinated grains between wild-type, OE Pap1, and KD Icy2 lines, a lower germination rate was detected in KD Pap1 lines, especially at 24 hai (Fig. 5, A and B). Differences in development were also observed among germinated grains. All transgenic lines had shorter roots and coleoptiles than wild-type control line at 24 hai (Fig. 5, C and E). However, a higher variability was observed for the transgenic lines at 72 hai (Fig. 5, D and F). OE Pap1 grains had shorter coleoptiles, but the length of their roots was similar to the wild-type. KD Pap1 roots and coleoptiles remained shorter than in the wild type. Finally, KD Icy2 lines grew rapidly, with similar coleoptile lengths and longer roots than wild-type plants.
Figure 4.
Photographs of the germination process in wild-type and transgenic lines. A-D, I-L, General appearance of the germination process of grains 24 and 72 hai, respectively. E-H, M-P, Detailed appearance of two grains per line at 24 and 72 hai, respectively.
Figure 5.
Quantification of the germination process of wild-type and transgenic grains at 24 and 72 hai. A-B, Percentage of germinated grains. C-D, Root length. E-F, Coleoptile length. Different letters indicate significant differences between lines (P < 0.05, honestly significant difference).
Proteolytic Activities Are Affected in Dry and Germinating Grains
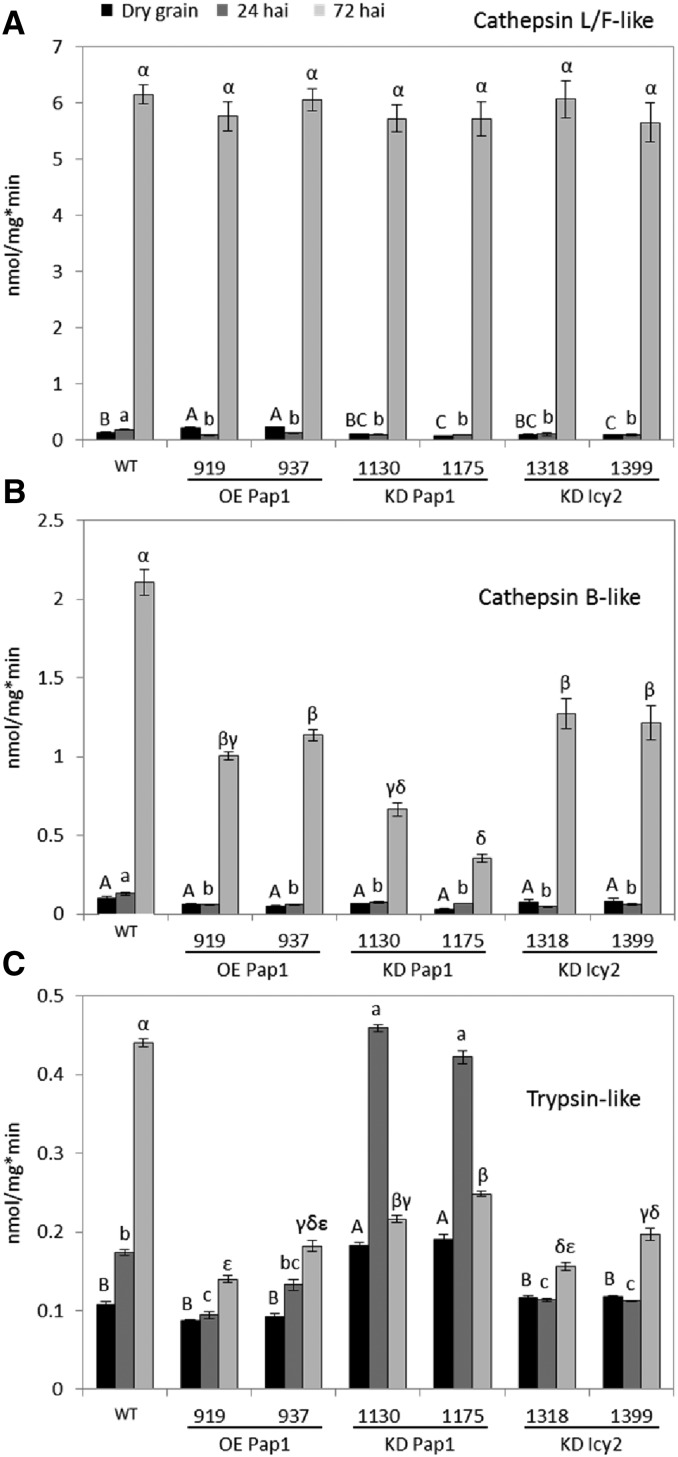
Differences in germination between wild-type and transgenic lines may be due to variations in grain composition but also to a distinct mobilization of the storage compounds. Thus, the main proteolytic activities in barley deembryonated grains (cathepsin L/F- and B-like CysProt and trypsin-like Ser protease) were analyzed at different germination times. Cathepsin L-/F-like activity was detected in all transgenic and nontransgenic lines in the dry grain, being significantly higher in the lines over-expressing the HvPap-1 gene (Fig. 6A). At 24 hai, this activity did not increase and was significantly lower in all transgenic lines than in the wild type. However, a sharp increase of cathepsin L-/F-like activity was shown in all lines at 72 hai, reaching similar levels of this peptidase activity. Dynamics of cathepsin B-like activities were similar to that of cathepsin L-/F-like (Fig. 6B), appearing in all dry grains at similar levels, without a significant increase in all transgenic lines at 24 hai, and showing a strong increase in all lines at 72 hai. However, the activity levels reached at this time point were distinct among different lines. All transgenic lines showed a significantly lower activity than the wild type, especially KD Pap1 lines. Finally, trypsin-like activity behaved differentially (Fig. 6C). It increased progressively during the germination of wild-type grains, although maximum levels were lower than CysProt activities at 72 hai. This pattern was similar for OE Pap1 and KD Icy2 grains, but the activity in these lines did not increase at 24 hai and the activity at 72 hai was significantly lower than in the wild type. In contrast, KD Pap1 lines reached more trypsin-like activity in the dry grain than wild-type; this activity peaked at 24 hai at levels similar to those observed in wild-type grains at 72 hai, and then the activity decreased.
Figure 6.
Proteolytic activities in deembryonated grains of wild-type and transgenic lines. A, Cathepsin L-/F-like CysProt activity. B, Cathepsin B-like CysProt activity. C, Trypsin-like Ser protease activity. Different letters indicate significant differences between lines for each time point (P < 0.05, honestly significant difference). Capital, small, and Greek letters are used for dry grains, 24 hai grains, and 72 hai grains, respectively.
C1A CysProt Patterns Are Altered in Transgenic Barley Grains
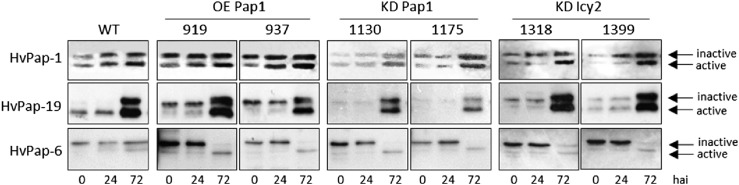
The protein profiles of CysProt HvPap-1, HvPap-6, and HvPap-19 were analyzed by immunoblot assays using total protein purified from deembryonated grains (Fig. 7). The active (mature protein) and inactive (including the inhibitory propeptide) forms of these CysProt were observed. HvPap-1 active and inactive forms increased progressively during germination in all analyzed lines, although different levels for this protein were observed. As expected, OE Pap1 plants showed a higher quantity of this protein than wild-type, whereas this protein was clearly reduced in KD Pap1 lines. The HvPap-19 active form was mainly present in wild-type dry grains. Its protein quantity increased during germination, reaching elevated levels at 72 hai in all lines. In KD Pap1 samples, the accumulation of HvPap-19 CysProt was lower. The active form of the HvPap-6 CysProt increased in all transgenic lines at 72 hai, but in wild-type grains the inactive form was predominant at the same time point.
Figure 7.
Immunoblot analyses in deembryonated grains of wild-type and transgenic lines. The accumulation of three different C1A CysProt, HvPap-1, -6, and -19, was determined at different times during grain germination (0, 24, and 72 hai).
In addition, grain embryos at 24 hai were used to analyze the expression of several proteases of the C1A family by RT-qPCR. Deembryonated grains were not used, since the quantity and quality of the RNA obtained from this tissue were insufficient for an accurate analysis. Ten proteases were analyzed: two cathepsin F-like (HvPap-1 and -2), five cathepsin l-like (HvPap-4, -6, -9, -10, and -17), one cathepsin H-like (HvPap-12), and two cathepsin B-like (HvPap-19 and -20). The results indicated a great variability in the expression levels for these genes in embryos from wild-type and transformed plants (Supplemental Fig. S5). Whereas several genes, such as HvPap-1, -4, -10, and -19, were highly expressed, others, such as HvPap-2 and -9, were poorly expressed. Comparing transgenic with wild-type embryos, several genes were repressed in all transgenic lines (HvPap-4, -9, and -10); HvPap-6 and -12 were induced in KD Icy2 lines; HvPap-17 was repressed in OE Pap1 and KD Icy2 lines; and the two cathepsin B-like genes HvPap-19 and -20 were repressed in KD Pap1 lines. As expected, HvPap-1 was induced in OE Pap1 lines and repressed in KD Pap1 lines.
C1A CysProt Are Differentially Located in Embryos of Transgenic and Wild-Type Barley Lines
Embryos were histochemically characterized at 24 hai (Supplemental Fig. S6). Deembryonated grains were not used, since the features of this tissue prevent obtaining thin sections for an accurate analysis. Higher protein content was revealed by Coomassie blue staining in OE Pap1 and KD Icy2 embryos (Supplemental Fig. S6, A–D). Starch was almost absent in OE Pap1 embryos after Lugol staining (Supplemental Fig. S6, E–H).
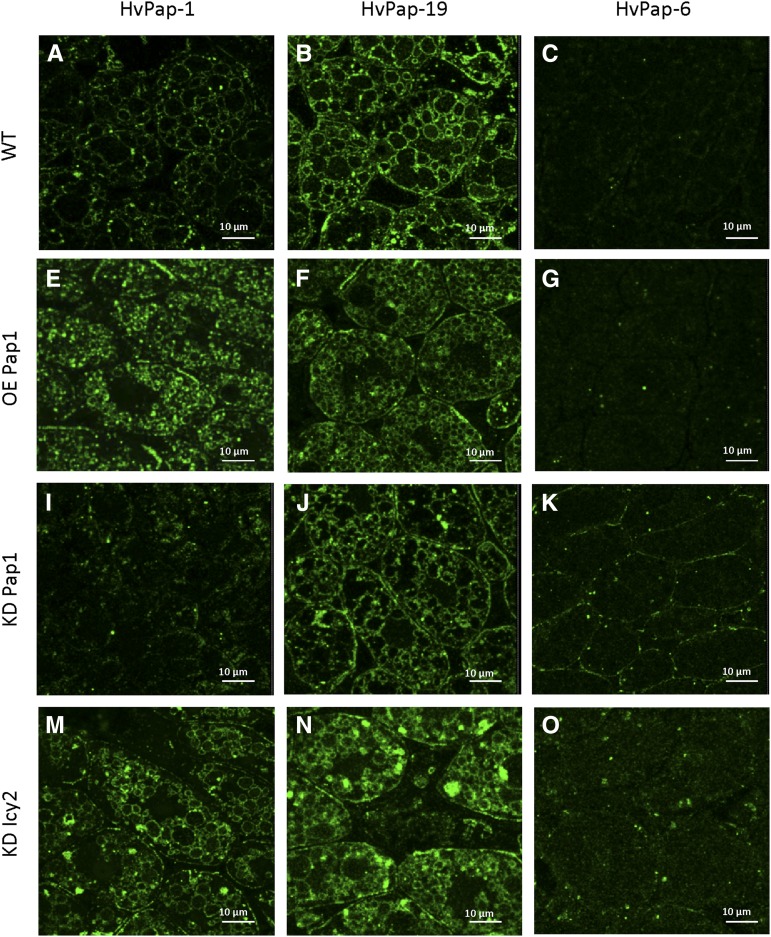
C1A CysProt HvPap-1, HvPap-19, and HvPap-6 were localized by immunofluorescence in wild-type and transgenic embryos at 24 hai (Fig. 8). A punctate or ring-shaped pattern of labeling was observed, the latter probably corresponding to the location of the proteases at the periphery of the protein bodies identified by the histochemical study. HvPap-1 was strongly detected in OE Pap1 and KD Icy2 lines. HvPap-19 was localized to all four specimens analyzed, but it was particularly strong in KD Icy2 embryos. HvPap-6 showed a weak punctate labeling in amiRNA events, KD Pap1 and KD Icy2 lines.
Figure 8.
Maximum projections of confocal series of the immuno-fluorescence localization of barley CysProt HvPap-1 (A, E, I, M), HvPap-19 (B, F, J, N), and HvPap-6 (C, G, K, O) in embryos from transgenic and nontransgenic grains at 24 hai.
DISCUSSION
Barley germination involves the activity of several proteases and amylases that hydrolyze and mobilize storage compounds. To date, the mechanisms regulating the action of these hydrolases are poorly understood. Various studies have been focused at the transcriptional level, comprising a complex network of regulatory pathways (An and Lin, 2011). The participation of several barley C1A CysProt during germination has been predicted, since members of the C1A subgroups l-, B-, H-, and F-cathepsins, such as HvPap-6, HvPap-19, HvPap-12, and particularly HvPap-1, are induced by GA treatment in barley grain (Holwerda and Rogers, 1992; Martínez et al., 2003, 2009; Cambra et al., 2012). The cathepsin F-like protease HvPap-1 was first identified in barley grains during germination by a transcriptomic analysis (Sreenivasulu et al., 2008). HvPap-1 was expressed in grain tissues during germination, and it was able to efficiently degrade stored hordeins in vitro (Cambra et al., 2012).
To demonstrate the in vivo involvement of this peptidase during germination, we generated over-expression and knock-down barley transgenic plants for the HvPap-1 gene. If HvPap-1 is one of the responsible enzymes to in vivo degrade stored proteins, a delay in the germination process should be expected for the silencing lines as well as an acceleration of the event should occur in the over-expressing ones. Several experiments were carried out to identify alterations during this process (Figs. 4 and 5). A decrease in the number of germinated grains over time was found in HvPap-1 knock-down plants, especially at 24 hai. The germination percentage was similar at 72 hai compared to the wild type, although the knock-down grains were notably in an earlier stage of development. Although we could expect an increase in the germination rate in over-expressing plants, the number of germinated grains was similar to the wild type, and, in terms of developmental stage, they even had a slight delay compared to wild type. There are two possible explanations for these observations: (1) over-expression or silencing of the HvPap-1 gene leads to significant modifications in the grain composition that subsequently affect the germination process, and (2) over-expression or silencing of the HvPap-1 gene alter the expression of some other hydrolytic activities crucial in the mobilization of stored compounds.
The capacity to store different molecules in the grain is related to these two hypotheses. An increased capacity to uptake Suc was previously related to a higher content of storage proteins in wheat (Weichert et al., 2010), and inactivation of cytosolic ADP-Glc pyrophosphorylase resulted in decreased starch and storage proteins in barley endosperms (Faix et al., 2012). Modifications of grain composition have been found for both plants with altered HvPap-1 expression, OE Pap1 and KD Pap1, in terms of a dissimilar accumulation of starch and free amino acids and a higher quantity of protein in the dry grains (Figs. 1-3). These differences and additional variations in some other stored molecules (Bowerman et al., 2015), not tested in this work, imply a differential specificity in the source of nutrients that the embryo can use to develop in a new plant.
Likewise, alteration in the genetic content of HvPap-1 implies changes in the expression of some other genes. Transcriptomic analyses of several C1A CysProt in embryos at 24 hai revealed that alterations in the expression of the HvPap-1 gene were associated with variations in the expression of some other C1A CysProt (Supplemental Fig. S5). This result is corroborated by immunolocation analyses performed in OE Pap1 and KD Pap1 embryos, in which differences in protein accumulation for HvPap-1, -6, and -19 have been found (Fig. 8). Besides, in deembryonated grains, immunoblot analyses show that the accumulation of the same HvPap-1, -6, and -19 C1A CysProt varies between the different lines (Fig. 7).
Alterations in the expression for these proteolytic enzymes should be correlated to variations in the enzymatic activity showed by the deembryonated grain during germination. Enzymatic activities measurements confirmed this circumstance (Fig. 6) and highlight the fact that enzymatic compensations would be involved in the response to alterations in the proteolytic mechanisms of the plant. However, an increase in the activity of the proteolytic machinery was not translated into a higher germination rate. KD Pap1 lines had a lower cathepsin L-/F-like and B-like enzymatic activity at 24 hai than wild-type plants, but, in contrast, they had a higher trypsin activity. Although C1A CysProt is the main activity related to degradation of stored proteins during late germination, trypsin activities are important during early germination events (Wrobel and Jones, 1992). Thus, a quick and effective degradation of stored proteins could be correlated to a rapid germination in the KD Pap1 lines, which has not been shown. In fact, several other hydrolytic enzymes are necessary for a proper degradation of the stored compounds, and genes encoding enzymes involved in the degradation of cell wall, lipids, starch, and nucleic acids are also transcribed during this process (Sreenivasulu et al., 2008). Modifications in the levels of these enzymes will also affect the onset and speed of barley grain germination.
Furthermore, the proteolytic activity in the grain should be correlated to the degradation of different fractions of stored proteins. Several peptidases with cathepsin L-/F-like proteolytic activity had the capacity to degrade stored hordeins in vitro (Martinez et al., 2009; Cambra et al., 2012), and they are probably involved in the mobilization of the amino acid content of albumins and globulins. Different C1A CysProt could have specific capacities to degrade storage proteins. HvPap-1 has the ability to mainly degrade hordeins (Cambra et al., 2012), and a higher rate of degradation for this protein fraction was observed in OE Pap1 lines (Fig. 2). However, as mentioned above, a direct relationship between gene over-expression and physiological activity cannot be concluded, since perturbations in the expression of other proteolytic enzymes may change the expected effect on protein degradation.
Thus, the changes observed in the speed of the germination process in both OE Pap1 and KD Pap1 lines should be globally considered as a consequence of both grain composition and the machinery necessary to mobilize the stored compounds.
Cystatins are key members in the regulation of C1A CysProt during barley grain germination (Martinez et al., 2009; Cambra et al., 2012). According to this, silencing of a cystatin in the grain could lead to an acceleration of the germination process, since inhibition of the C1A CysProt would be minor. The results obtained in KD Icy2 lines reinforce the importance of the complex network modulating mobilization of stored proteins. Compensating effects implying proteases such as HvPap-6 and HvPap-19 and probably some other enzymes (Figs. 7 and 8; Supplemental Fig. S5) led to altered proteolytic activities (Fig. 6) and modified grain composition in this line (Figs. 1-3). These perturbations would explain the distinct germination process observed in KD Icy2 grains (Figs. 4 and 5).
Free amino acids are major determinants related to grain processing, quality, and food safety (Halford et al., 2015). Although most of the nitrogen in the grain is incorporated into proteins, free amino acids are crucial during germination. Genetic modifications leading to variations in the accumulation of storage proteins may alter the amino acid composition in the grain. For example, antisense C-hordein barley grains had metabolic changes leading to alterations in amino acid biosynthesis (Schmidt et al., 2015). Extensive changes have been detected in the free amino acid composition in all the different transgenic barley grains (Fig. 3). The lowest accumulation of most amino acids in both the KD Pap1 and KD Icy2 lines could be also related with the slow start of germination showed in both lines, which was most remarkable in KD Pap1 line. Besides, the OE Pap1 line strongly accumulates the amino acids Pro and Gln. Hordeins are enriched in these two amino acids. Their abundance in the dry grain could be used as a source to get a higher accumulation of hordeins in OE Pap1 lines. However, a higher concentration of this storage protein does not correlate to an accelerated germination process.
In conclusion, the importance of HvPap-1 and HvCPI-2 proteins during grain germination has been demonstrated. Delayed germination phenotype observed in silencing HvPap-1 plants agrees with a role for this protease in degrading grain-stored proteins. However, caution should be taken when plants are modified over-expressing or silencing a peptidase or an inhibitor, since the plant tries to compensate for the modified proteolytic effect by modulating the expression of some other peptidases or their inhibitors. The nonexpected phenotypes during grain germination for over-expressing HvPap-1 and silencing Icy-2 plants are examples of this proteolytic reprogramming. Future work will aim to carry out similar analysis on other C1A proteases and their inhibitors in order to demonstrate the existence of a regulatory network during the germination process.
MATERIALS AND METHODS
Plant Material
The samples used in this work were obtained from barley (Hordeum vulgare) spring type cv Golden Promise plants, grown at 22°C under a 16-h-light/8-h-dark photoperiod in soil (Sanyo/Panasonic MLR-350-H). Barley transgenic lines over-expressing or silencing the barley HvPap-1 gene (OE Pap1 and KD Pap1, respectively) and silencing the barley Icy-2 gene (KD Icy2) were generated in collaboration with the IPK Gatersleben, Plant Reproductive Biology Group (Hensel et al., 2009). To generate over-expression lines, the HvPap-1 gene was transferred into the intermediate vector pUbi-AB (DNA-Cloning-Service, Hamburg, Germany) and cloned using the SfiI restriction sites into the p6U binary vector (DNA-Cloning-Service, Hamburg, Germany). This binary vector included the HPT (HYGROMYCIN PHOSPHOTRANSFERASE) selectable marker gene driven by the Zea mays UBIQUITIN-1 promoter with first intron. The HvPap-1 gene was driven by the same promoter. KD Pap1 and KD Icy2 knock-down lines were produced by amiRNA technology developed in the MicroRNA Designer Web platform (WMD3, http://wmd3.weigelworld.org). The amiRNA constructs were engineered from pNW55 vector replacing the 21 bases of the natural Osa-MIR528 miRNA by 21 bases to silence specifically the HvPap-1 or Icy-2 genes (5′ TTATGCGGCATTGATACCGGT 3′ and 5′ TAAATTATTGTGTGGGGACTC 3′, respectively). The final products of 554 bp were cloned into the p6d35S binary vector (DNA-Cloning-Service, Hamburg, Germany) using the pUbi-AB vector as intermediary. This binary vector included the HPT selectable marker gene driven by the doubled enhanced CaM35S-promoter. Immature embryos of barley cv Golden Promise were transformed with the binary vectors using the Agrobacterium tumefaciens strain AGL1 as described (Hensel et al., 2008). Homozygous transgenic plants were obtained by double haploid technology (Marthe et al., 2015). The presence of the antibiotic resistance marker was confirmed by PCR using specific primer pairs (Hensel et al., 2008). Homozygous transgenic lines for inserted gene constructions were characterized and used for grain analyses.
Analysis of the Copy Number in Transgenic Barley Lines
Total DNA was isolated from leaves of barley transgenic and nontransformed control lines. The RT-qPCR conditions were 40 cycles with 15 s at 95°C, 1 min at 60°C, and 5 s at 65°C. FastStart Universal SYBR Green Master (Rox, Roche) was used in a total volume of 20 μL. PCR amplification was performed in multiplates (Bio-Rad MLL9601) using PCR sealers (Bio-Rad MSB1001). The reactions were carried out in a C1000 thermal cycler with CFX96 optical reaction module, and results were analyzed with the CFX Manager Software 2.0 (Bio-Rad). Copy number was calculated by the 2 -∆∆Ct method to study gene expression (Livak and Schmittgen, 2001) adapted to estimate copy number (Ingham et al., 2001; Li et al., 2008). Cyclophilin and 4-hydroxyphenyl-pyruvate dioxygenase (HvCycl and Hv4Hppd genes) were used as endogenous and calibrator genes, respectively (Falk et al., 2002; Burton et al., 2004). The primers used are shown in Supplemental Table S2. Analysis of dissociation curves was performed to check gene-specific amplification and reactions were performed in triplicate.
Grain Phenotype and Starch Analyses
Grains from wild-type and transgenic barley plants were harvested and imbibed in double-distilled water. After 24 h, grains were longitudinally dissected and starch was stained with Lugol's iodine staining reagent (Sigma). Stained and nonstained grains were visualized and photographed with a Leica MZ10 F stereo microscope and a Leica DFC420C CCD camera. For total starch quantification, six dry deembryonated grains from transgenic and control lines were ground, and 10 mg was assayed with STA20 Kit (Sigma) following the manufacturer’s recommendations. Wheat starch, included in the commercial kit, was used as control. Dilutions were carried out as necessary to fit into linearity of the Glc standard curve. Measurements were performed three times for each sample. After calculations, starch content was expressed as grams of transformed starch per 100 g of initial kernel weight. Three independent biological replicates were used.
Metabolomic Analyses
Samples were obtained from dry deembryonated grains. Then 50 mg of sample was resuspended in 500 µL MeOH and disrupted by three cycles of frost/defrosting and processed with TissueLyser LT (Qiagen). After centrifugation at 19,300g, 20 min, the supernatant was transferred to a new tube and evaporated to dryness in a SpeedVac. The metabolite extracts were resuspended in 0.1 mmol L−1 formic acid containing 0.2 mmol L−1 Met sulfone (internal standard) by 1 min vortex mixing and then centrifuged (19,300g, 15 min). Clear solution was analyzed by capillary electrophoresis coupled to a mass spectrometer detector. Nineteen nitrogen metabolites, mainly amino acids and other related compounds (Ala, sarcosine, Ser, Pro, Val, betaine, Ile, Leu, Asn, Asp, Gln, Lys, Glu, Met, His, Phe, Arg, Tyr, and Trp), were analyzed as previously described (Moraes et al., 2011) with slight modifications. A CE System (7100 Agilent) coupled to a time-of-flight mass spectrometry (MS) system (Agilent 6224) was used. The separation occurred in a fused-silica capillary (Agilent; total length, 100 cm; i.d., 50 μm). Separation was under normal polarity with a background electrolyte containing 1.0 mol L−1 formic acid in 10% (v/v) methanol at 20°C. Sheath liquid (6 µL min−1) was methanol/water (1/1, v/v) containing 1.0 mmol L−1 formic acid with two reference masses to allow correction and higher accurate mass in the MS. Samples were hydrodynamically injected at 50 mbar for 35 s and stacked by injecting background electrolyte at 100 mbar for 10 s. The optimized MS parameters were: fragmentor 150 V, Skimmer 65 V, octopole 750 V, nebulizer pressure 10 psi, drying gas temperature at 200°C, and flow rate 10.0 L min−1. The capillary voltage was 3,500 V. Data were acquired in positive ESI mode with a full scan from mass-to-charge ratio 87 to 1,000 at a rate of 1.41 scan/s. The resulting CE-MS data files were cleaned of background noise and unrelated ions by the Targeted Feature Extraction tool with Profinder software (B.06.00, Agilent). Data were extracted using data mining algorithm based on the software. This software contains a list of standards used with their exact monoisotopic mass, migration time, and molecular formula. Metabolites were previously identified in the wild-type barley samples by comparison of their migration time and spectra with pure standards. After their quantitation with standard solutions, differences for individual metabolites were evaluated by a t test comparison of every case group vs wild type (where P values < 0.05 were considered significant). SIMCA-P+ 12.0.1 (Umetrics) and MetaboAnalyst v. 3.0 (Xia et al., 2012) were used for PCA plotting and Heatmap, respectively.
Grain Protein Analysis
Plant protein extracts were obtained from 20 dry deembryonated grains and from 20 deembryonated grains after 24 and 72 h of seed imbibition. Three independent biological replicates were used. Samples were ground and around 100 mg was resuspended in 500 μL of protein extraction buffer (50 mm sodium phosphate, pH 6.0, 15 mm NaCl, and 2 mm EDTA, pH 8.0). The suspension was centrifuged at 15,600g for 15 min at 4°C. The protein content of the supernatants was quantified (Bradford, 1976) with bovine serum albumin (BSA) as a standard. Protein content was analyzed using SDS-PAGE technique, and 20 µg of protein extracts was denatured using dissociation buffer (12.5 mm Tris-HCl, pH 6.8, 0.25% [w/v] SDS, 2.5% [v/v] β-mercaptoethanol, 0.01% [w/v] bromophenol blue, and 3.75% [v/v] glycerol) for 5 min at 90°C. Electrophoretic detection of proteins was performed using denaturing polyacrylamide gels at 15% (w/v). The electrophoresis was carried out using a BioRad Mini Protein Electrophoresis Cell system for 90 min at 130 V, and prestained SDS-PAGE standard broad range (BioRad) was used as a weight molecular pattern. Gels were submerged into a staining solution 0.3% (w/v) Coomassie Blue G-250 in 40% (v/v) methanol and 10% (v/v) acetic acid to stain the proteins and were distained in 25% (v/v) methanol and 10% (v/v) acetic acid.
Fractionation and Analysis of Barley Grain Storage Proteins
Albumins and globulins were sequentially extracted from 20 dry barley deembryonated grains (Shi and Xu, 2009) and 20 deembryonated grains after 24 and 72 h of water imbibition. Deembryonated grains were completely crushed in a mortar, resuspended in distilled water, and continuously stirred for 12 h at 4°C. Soluble and insoluble fractions were separated by centrifugation at 5,900g for 30 min at 4°C. The supernatant, enriched in albumins, was isolated. Globulins were extracted from the pellet by adding 5% (w/v) NaCl in distilled water at 4°C. After a second centrifugation step at 5,900g for 30 min, the recovered soluble fraction was enriched in globulins. Hordeins were extracted from a similar set of deembryonated grains after incubation in a buffer containing 55% (v/v) 2-propanol and 1% (v/v) 2-mercaptoethanol, for 1 h at 60°C and centrifugation for 10 min at 13,300g (Martinez et al., 2009). Protein concentrations and their electrophoretic bands patterns were analyzed as above described. Three independent biological replicates were used.
Germination Assays
Twenty barley grains from each transgenic line and the wild-type cultivar were placed into a 150-mm petri dish with moistened, sterile filter paper. The germination process was followed by counting the number of germinated grains at 24 and 72 hai, considering as germinated grains those that had broken the grain coat or testa. Photographs were taken with an Olympus VR-320 digital camera. The length of the emerging roots and coleoptiles was quantified using a measuring ruler. Three independent biological replicates were used.
Enzymatic Activity Assays
The enzymatic activity of the same deembryonated grain protein extracts used for grain protein quantification was determined using the fluorogenic substrates Z-FR-AMC (N-carbobenzoxy-Phe-Arg-7-amido-4-methylcoumarin [AMC]) and Z-RR-AMC (N-carbobenzoxy-Arg-Arg-AMC), susceptible to degradation by cathepsin L-/F- and B-like proteases, respectively. Mixtures of proteases and substrates were incubated in 100 mm sodium phosphate, pH 6.0, buffer containing 10 mm Cys, 1 mm EDTA, and 0.01% (v/v) Brij35 at 30°C. Trypsin-like activity was also analyzed using the fluorogenic substrate Z-R-AMC (N-carbobenzoxy-Arg-AMC), and assays were incubated in the buffer Tris-HCl 0.1 M, pH 7.5, at 30°C. Emitted fluorescence was measured with a 365-nm excitation and 465-nm emission wavelength filter. Triplicate assays were performed for determination of each value and the average was calculated. Blanks were used to account for spontaneous breakdown of substrates, and results were expressed as nmol of hydrolyzed substrate by mg of protein by min (nmol/mg*min). The system was calibrated with known amounts of AMC hydrolysis product in a standard reaction mixture.
Immunoblot Analyses
The same deembryonated grain protein extracts obtained for total protein quantification were used. After separation on an SDS-polyacrylamide gel, proteins were electrotransferred onto a nitrocellulose membrane (GE Healthcare). Immunoblotting was performed with specific crude anti-HvPap-1, -6, and -19 peptide polyclonal antibodies produced in rabbits by Pineda Antibody Services. The sequences used to produce them were: HvPap-1-IgG, NH2-CSGFAPSRFKEKPYWIIKN-CONH2; HvPap-6-IgG, NH2-CGWSPVKDVNDPHVQEL-CONH2; and HvPap-19-IgG, NH2-CQEKKHFSIDAYQVNSDPHD-CONH2. Optimal dilution of the antibody was incubated in phosphate buffered saline (PBS; 137 mm NaCl, 27 mm KCl, 10 mm Na2HPO4, 2 mm KH2PO4, pH 7.4) with 5% (w/v) milk. Peroxidase-conjugated antirabbit IgG (Sigma) diluted at 1:10,000 was used as secondary antibody for detection with ECL Plus (GE Healthcare). The specificity of these antibodies was confirmed by immunoblotting using the purified recombinant proteases HvPap-1, -6, and -19 (Supplemental Fig. S7).
RT-qPCR Analyses
For RT-qPCR studies, isolated embryos from 24-h germinated grains were used. Twenty grains and three independent biological replicates per line were used. All samples were frozen in liquid nitrogen and stored at -80°C until used for RNA extraction. Samples were ground using a mortar and a pestle. Total RNA was extracted by the phenol/chloroform method, followed by precipitation with 3 m LiCl (Oñate-Sánchez and Vicente-Carbajosa, 2008) and digestion with DNase. cDNAs were obtained from 2 μg of RNA using the RevertAid H Minus First Strand cDNA Synthesis Kit (Thermo Scientific) following the manufacturer’s instructions. RT-qPCR analyses were performed for duplicated samples by means of a CFX96 Real-time system (BioRad) using SYBR Green as a detection system. Relative expression values normalized to Cyclophilin gene (Burton et al., 2004) and expressed as 2-∆Ct. The primers used for PCR amplification are described in Supplemental Table S3.
Embryo Structural Analysis and Immuno-Fluorescence Detection
Embryos from wild-type and transgenic plants were fixed in a freshly prepared solution of 4% (w/v) formaldehyde in PBS first at room temperature under vacuum until the specimens sank, then overnight at 4°C. Next day, the samples were washed twice in PBS for 15 min. Dehydration in a series of increasing concentrations of methanol, resin infiltration, embedding, and polymerization was performed by Progressive Lowering of Temperature in a Leica AFS. From methanol 30% to 70% (v/v), the temperature dropped from 0 to -20°C at a speed of -70°C/h, with changes every 30 min. Subsequent steps were performed at -20°C. Further dehydration was achieved in 100% methanol for 90 min, with three changes. Resin infiltration was done in a series of mixtures of methanol:LRwhite (Agar Scientific) with increasing concentrations of the resin (2:1; 1:1; 1:2, v:v), for 1 h each, and finally pure resin with 0.5% (w/v) bezoin-methyl-ether as a catalyst overnight. Polymerization in plastic capsules was accomplished under UV light at -20°C for 2 d and at 22°C for 1 d. One- to 2-μm-thin sections were cut from the polymerized blocks in a Leica EM UC6 ultramicrotome, carefully collected on water drops on 10-well Teflon-printed slides (Fisher Scientific Inc.), and allowed to dry down and stored at room temperature until further use.
To assess any possible structural rearrangements at the subcellular level in the transgenic samples versus the wild type, the sections were stained with 0.3% (w/v) Coomassie Brilliant Blue (Coomassie G-250, BioRad) or 20% (v/v) lugol (Lugol's iodine staining reagent, Sigma), rinsed in distilled water, mounted, and observed on a Zeiss Axiophot microscope under bright field. Photographs were taken with a Leica DFC300 FX CCD camera using the Leica Application Suite 2.8.1 build 1554 acquisition software.
For immuno-fluorescence, the sections on the 10-well slides were hydrated with PBS for 5 min and unspecific binding sites were blocked by 10-min incubation with 5% (w/v) BSA in PBS. Then, they were incubated with 20-µL drops/well of either a rabbit-raised antibody to the CysProt HvPap-1, HvPap-6, or HvPap-19, applied 1/50 in PBS, for 1 h at room temperature in a humid chamber. After two washes of 15 min in PBS, an Alexa Fluor 488 antirabbit antibody (Molecular Probes) was applied in a 1/25 solution in 2.5% (v/v) BSA in PBS for 45 min at room temperature in a humid chamber and darkness. Subsequent to another two washes of PBS for 15 min each, the slides were mounted in a 50:50 solution of glycerol:PBS. Serial sections across the specimens were collected on a Leica SP8 confocal microscope using the laser excitation line of 488 nm to detect the proteases. All series were captured under the same conditions (pinhole size, gain, offset, magnification). The management of the series was performed with either the LAS-AF-Lite 3.1.0_8587 or Fiji software. To composite the corresponding figure in Adobe Photoshop CS3, the maximum projections of the green channel were overlaid for each treatment shown. Only the automatic levels were adjusted.
Data Analysis
Statistical differences among treatments or lines were analyzed by one-way ANOVA followed by Tukey’s (honestly significant difference) multiple comparison test performed using the software R Project (v.3.1.2) package.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers HvPap-1 (BN000093), HvPap-2 (AM941116), HvPap-4 (AM941118), HvPap-6 (AM941120), HvPap-9 (U19359), HvPap-10 (U19384), HvPap-12 (X05167), HvPap-16 (AM941126), HvPap-17 (Z97022), HvPap-19 (AJ310426), HvPap-20 (AM941127), HvIcy-2 (AJ748337), HvCycl (AK253120), Hv4Hppd (AJ000693).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Selection of HvPap-1 transgenic homozygous lines of barley generated by double haploid technology.
Supplemental Figure S2. Selection of Icy-2 transgenic homozygous lines of barley generated by double haploid technology.
Supplemental Figure S3. Starch content of de-embryonated dry grains.
Supplemental Figure S4. Protein patterns of de-embryonated grains at different germination times.
Supplemental Figure S5. RT-qPCR analyses of the mRNA accumulation in embryos.
Supplemental Figure S6. Structural characterization of embryos at 24 hours after imbibition.
Supplemental Figure S7. Immuno-blot of purified recombinant barley CysProt.
Supplemental Table S1. Amino acid composition of dry de-embryonated grains.
Supplemental Table S2. Primer sequences used for RT-qPCR amplification to analyse copy number.
Supplemental Table S3. Primer sequences used for the amplification of barley genes in RT-qPCR expression assays.
Supplementary Material
Acknowledgments
We thank Jose Rodrigo and Javier de Felipe (CTB-UPM, Pozuelo de Alarcon, Madrid, Spain) for the use of the ultramicrotome. We appreciate the excellent technical assistance of Carola Bollmann, Sabine Sommerfeld, Sibylle Freist, Andrea Mueller, and Ingrid Otto.
Glossary
- AMC
7-amido-4-methylcoumarin
- amiRNA
artificial microRNA
- BSA
bovine serum albumin
- CysProt
papain-like Cys protease
- hai
hours after imbibition
- MS
- PBS
phosphate buffered saline
- PCA
principal component analysis
- RT-qPCR
real-time quantitative PCR
Footnotes
This work was financially supported by the Ministerio de Economia y Competitividad of Spain (projects AGL2011-23650 and BIO2014-53508-R) and European Commission FP7 Marie Curie action CoFund program.
References
- An YQ, Lin L (2011) Transcriptional regulatory programs underlying barley germination and regulatory functions of gibberellin and abscisic acid. BMC Plant Biol 11: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benchabane M, Schlüter U, Vorster J, Goulet MC, Michaud D (2010) Plant cystatins. Biochimie 92: 1657–1666 [DOI] [PubMed] [Google Scholar]
- Bowerman AF, Newberry M, Dielen AS, Whan A, Larroque O, Pritchard J, Gubler F, Howitt CA, Pogson BJ, Morell MK, et al. (2015) Suppression of glucan, water dikinase in the endosperm alters wheat grain properties, germination and coleoptile growth. Plant Biotechnol J 10.1111/pbi.12394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Burton RA, Shirley NJ, King BJ, Harvey AJ, Fincher GB (2004) The CesA gene family of barley. Quantitative analysis of transcripts reveals two groups of co-expressed genes. Plant Physiol 134: 224–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cambra I, Martinez M, Dáder B, González-Melendi P, Gandullo J, Santamaría ME, Diaz I (2012) A cathepsin F-like peptidase involved in barley grain protein mobilization, HvPap-1, is modulated by its own propeptide and by cystatins. J Exp Bot 63: 4615–4629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronado M-J, Hensel G, Broeders S, Otto I, Kumlehn J (2005) Immature pollen-derived doubled haploid formation in barley cv. Golden Promise as a tool for transgene recombination. Acta Physiol Plant 27: 9 [Google Scholar]
- Dawson IK, Russell J, Powell W, Steffenson B, Thomas WT, Waugh R (2015) Barley: a translational model for adaptation to climate change. New Phytol 206: 913–931 [DOI] [PubMed] [Google Scholar]
- Diaz I, Martinez M, Isabel-LaMoneda I, Rubio-Somoza I, Carbonero P (2005) The DOF protein, SAD, interacts with GAMYB in plant nuclei and activates transcription of endosperm-specific genes during barley seed development. Plant J 42: 652–662 [DOI] [PubMed] [Google Scholar]
- Díaz-Mendoza M, Velasco-Arroyo B, González-Melendi P, Martínez M, Díaz I (2014) C1A cysteine protease-cystatin interactions in leaf senescence. J Exp Bot 65: 3825–3833 [DOI] [PubMed] [Google Scholar]
- Faix B, Radchuk V, Nerlich A, Hümmer C, Radchuk R, Emery RJ, Keller H, Götz KP, Weschke W, Geigenberger P, et al. (2012) Barley grains, deficient in cytosolic small subunit of ADP-glucose pyrophosphorylase, reveal coordinate adjustment of C:N metabolism mediated by an overlapping metabolic-hormonal control. Plant J 69: 1077–1093 [DOI] [PubMed] [Google Scholar]
- Falk J, Krauß N, Dähnhardt D, Krupinska K (2002) The senescence associated gene of barley encoding 4-hydroxyphenylpyruvate dioxygenase is expressed during oxidative stress. J Plant Physiol 159: 1245–1253 [Google Scholar]
- Grudkowska M, Zagdańska B (2004) Multifunctional role of plant cysteine proteinases. Acta Biochim Pol 51: 609–624 [PubMed] [Google Scholar]
- Halford NG, Curtis TY, Chen Z, Huang J (2015) Effects of abiotic stress and crop management on cereal grain composition: implications for food quality and safety. J Exp Bot 66: 1145–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel G, Kastner C, Oleszczuk S, Riechen J, Kumlehn J (2009) Agrobacterium-mediated gene transfer to cereal crop plants: current protocols for barley, wheat, triticale, and maize. Int J Plant Genomics 2009: 835608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel G, Valkov V, Middlefell-Williams J, Kumlehn J (2008) Efficient generation of transgenic barley: the way forward to modulate plant-microbe interactions. J Plant Physiol 165: 71–82 [DOI] [PubMed] [Google Scholar]
- Holwerda BC, Rogers JC (1992) Purification and characterization of aleurain : a plant thiol protease functionally homologous to Mammalian cathepsin h. Plant Physiol 99: 848–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham DJ, Beer S, Money S, Hansen G (2001) Quantitative real-time PCR assay for determining transgene copy number in transformed plants. Biotechniques 31: 132–134, 136–140 [DOI] [PubMed] [Google Scholar]
- Koehler SM, Ho TH (1990) Hormonal regulation, processing, and secretion of cysteine proteinases in barley aleurone layers. Plant Cell 2: 769–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Brunner AM, Shevchenko O, Meilan R, Ma C, Skinner JS, Strauss SH (2008) Efficient and stable transgene suppression via RNAi in field-grown poplars. Transgenic Res 17: 679–694 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Marthe C, Kumlehn J, Hensel G (2015) Barley (Hordeum vulgare L.) transformation using immature embryos. Methods Mol Biol 1223: 71–83 [DOI] [PubMed] [Google Scholar]
- Martinez M, Cambra I, Carrillo L, Diaz-Mendoza M, Diaz I (2009) Characterization of the entire cystatin gene family in barley and their target cathepsin L-like cysteine-proteases, partners in the hordein mobilization during seed germination. Plant Physiol 151: 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez M, Diaz I (2008) The origin and evolution of plant cystatins and their target cysteine proteinases indicate a complex functional relationship. BMC Evol Biol 8: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez M, Rubio-Somoza I, Carbonero P, Díaz I (2003) A cathepsin B-like cysteine protease gene from Hordeum vulgare (gene CatB) induced by GA in aleurone cells is under circadian control in leaves. J Exp Bot 54: 951–959 [DOI] [PubMed] [Google Scholar]
- Mayer KF, Waugh R, Brown JW, Schulman A, Langridge P, Platzer M, Fincher GB, Muehlbauer GJ, Sato K, Close TJ, et al. ; International Barley Genome Sequencing Consortium (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491: 711–716 [DOI] [PubMed] [Google Scholar]
- Mikkonen A, Porali I, Cercos M, Ho TH (1996) A major cysteine proteinase, EPB, in germinating barley seeds: structure of two intronless genes and regulation of expression. Plant Mol Biol 31: 239–254 [DOI] [PubMed] [Google Scholar]
- Moraes EP, Rupérez FJ, Plaza M, Herrero M, Barbas C (2011) Metabolomic assessment with CE-MS of the nutraceutical effect of Cystoseira spp extracts in an animal model. Electrophoresis 32: 2055–2062 [DOI] [PubMed] [Google Scholar]
- Müntz K. (1996) Proteases and proteolytic cleavage of storage proteins in developing and germinating dicotyledoneous seeds. J Exp Bot 47: 605–622 [Google Scholar]
- Nevo E, Fu YB, Pavlicek T, Khalifa S, Tavasi M, Beiles A (2012) Evolution of wild cereals during 28 years of global warming in Israel. Proc Natl Acad Sci USA 109: 3412–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oñate-Sánchez L, Vicente-Carbajosa J (2008) DNA-free RNA isolation protocols for Arabidopsis thaliana, including seeds and siliques. BMC Res Notes 1: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Waller M, Barrett AJ, Bateman A (2014) MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res 42: D503–D509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Rizzi V, Gaziola SA, Medici LO, Vincze E, Kozak M, Lea PJ, Azevedo RA (2015) Lysine metabolism in antisense C-hordein barley grains. Plant Physiol Biochem 87: 73–83 [DOI] [PubMed] [Google Scholar]
- Shewry PR, Halford NG (2002) Cereal seed storage proteins: structures, properties and role in grain utilization. J Exp Bot 53: 947–958 [DOI] [PubMed] [Google Scholar]
- Shewry PR, Napier JA, Tatham AS (1995) Seed storage proteins: structures and biosynthesis. Plant Cell 7: 945–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C, Xu LL (2009) Characters of cysteine endopeptidases in wheat endosperm during seed germination and subsequent seedling growth. J Integr Plant Biol 51: 52–57 [DOI] [PubMed] [Google Scholar]
- Sorensen MB, Cameron-Mills V, Brandt A (1989) Transcriptional and post-transcriptional regulation of gene expression in developing barley endosperm. Mol Gen Genet 217: 195–201 [Google Scholar]
- Sreenivasulu N, Usadel B, Winter A, Radchuk V, Scholz U, Stein N, Weschke W, Strickert M, Close TJ, Stitt M, et al. (2008) Barley grain maturation and germination: metabolic pathway and regulatory network commonalities and differences highlighted by new MapMan/PageMan profiling tools. Plant Physiol 146: 1738–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan-Wilson AL, Wilson KA (2012) Mobilization of seed protein reserves. Physiol Plant 145: 140–153 [DOI] [PubMed] [Google Scholar]
- Weichert N, Saalbach I, Weichert H, Kohl S, Erban A, Kopka J, Hause B, Varshney A, Sreenivasulu N, Strickert M, et al. (2010) Increasing sucrose uptake capacity of wheat grains stimulates storage protein synthesis. Plant Physiol 152: 698–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel R, Jones BL (1992) Appearance of endoproteolytic enzymes during the germination of barley. Plant Physiol 100: 1508–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS (2012) MetaboAnalyst 2.0--a comprehensive server for metabolomic data analysis. Nucleic Acids Res 40: W127-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Jones BL (1995) Characterization of germinated barley endoproteolytic enzymes by two dimensional gel electrophoresis. J Cereal Sci 21: 145–153 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.