Abstract
Chikungunya virus (CHIKV) causes severe, debilitating, often chronic arthralgia with high attack rates, resulting in severe morbidity and economic costs to affected communities. Since its first well-documented emergence in Asia in the 1950s, CHIKV has infected millions and, since 2007, has spread widely, probably via viremic travelers, to initiate urban transmission in Europe, the South Pacific, and the Americas. Some spread has been facilitated by adaptive envelope glycoprotein substitutions that enhance transmission by the new vector, Aedes albopictus. Although epistatic constraints may prevent the impact of these mutations in Asian strains now circulating in the Americas, as well as in African CHIKV strains imported into Brazil last year, these constraints could eventually be overcome over time to increase the transmission by A. albopictus in rural and temperate regions. Another major determinant of CHIKV endemic stability in the Americas will be its ability to spill back into an enzootic cycle involving sylvatic vectors and nonhuman primates, an opportunity exploited by yellow fever virus but apparently not by dengue viruses.
Introduction
Emergence history of chikungunya virus Chikungunya virus
(CHIKV), an alphavirus in the family Togaviridae, causes severe, debilitating and often chronic arthralgia. First discovered during a 1952–1953 outbreak in present day Tanzania, this mosquito-borne virus has since been implicated in explosive epidemics involving millions of persons in Africa, Asia, Europe, the South Pacific, and most recently the Americas [1,2]. Like all arthropod-borne viruses (arboviruses), CHIKV is zoonotic, and evolved in enzootic transmission cycles in sub-Saharan Africa involving forest-dwelling mosquito vectors and nonhuman primate (NHP) amplification hosts. Initial phylogenetic studies identified two major enzootic lineages: West African and East/Central/South African (ECSA) (Figure 1) [3]. An Asian lineage, first isolated in 1958 and implicated in outbreaks in India and South-east Asia during the 1960s, was inferred phylogenetically to have been derived from the ECSA lineage, probably following its introduction into Asia between 1879 and 1956 [4•]. However, historic records [5••,6] suggest that CHIKV had been introduced even earlier into Asia and the Americas via sailing ships, the same mechanism believed to have introduced yellow fever and dengue viruses into port cities during past centuries.
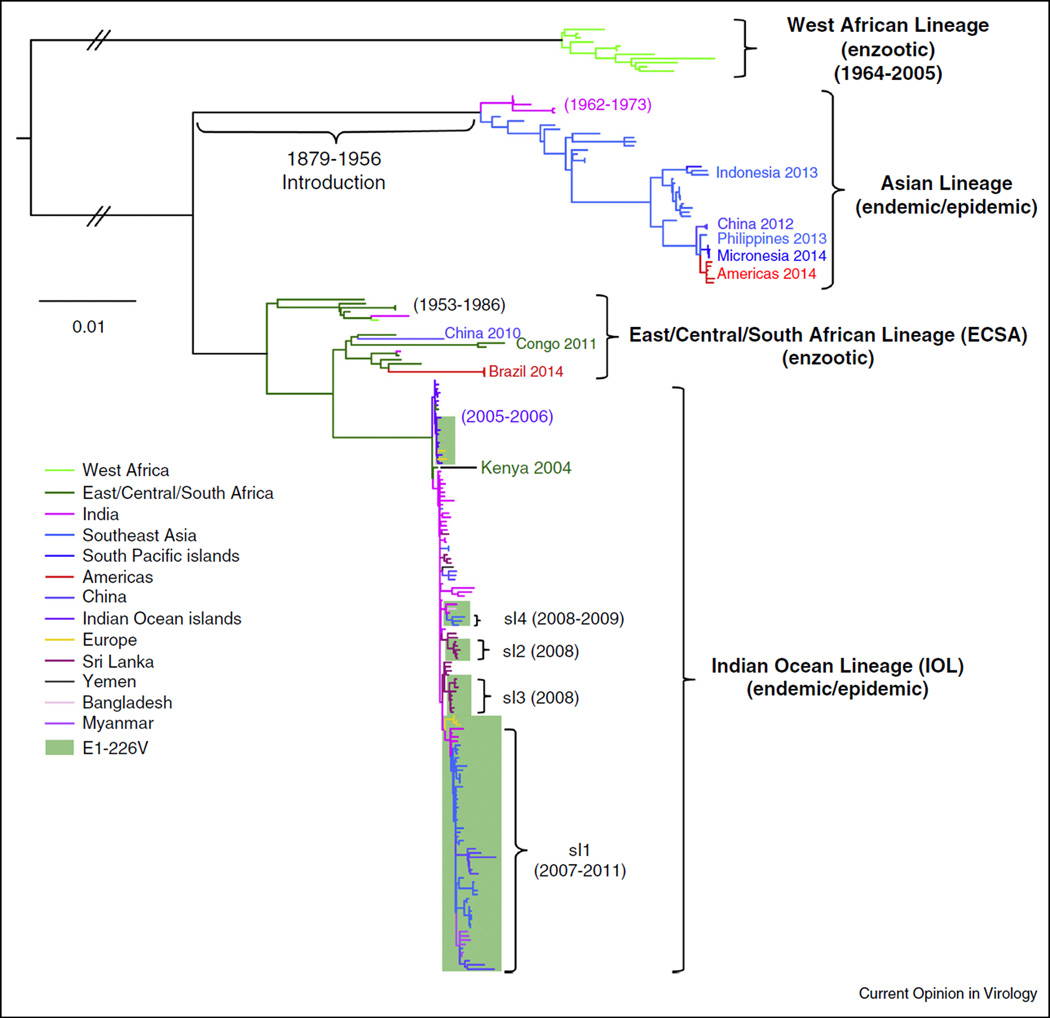
Figure 1.
Phylogenetic tree showing the relationships among enzootic and endemic/epidemic CHIKV lineages and a time estimate for introduction of the Asian lineage from Africa into Asia; sl indicates IOL sublineage with second-step A. albopictus-adaptive mutations [56]. The tree was generated using concatenated open reading frames from all complete genomic sequences found in the Genbank library using maximum likelihood implemented in PAUP 4.0 [64]. The branch colors represent countries or regions of origin for each CHIKV sample. The scale indicates nucleotide sequence divergence.
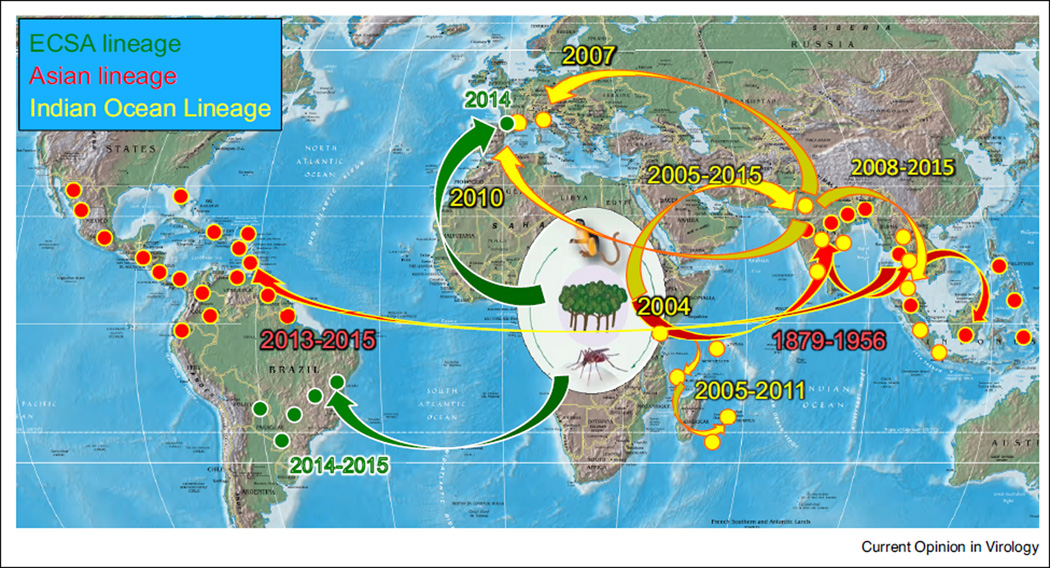
After the 1960s, CHIKV was only occasionally associated with human disease in Africa and Asia, probably due to the lack of available diagnostics and the difficulty in distinguishing human infections from dengue fever and other tropical, acute febrile diseases. However, an explosive outbreak that began in coastal Kenya in 2004 and spread first into the Indian Ocean basin, then independently into the Indian Subcontinent followed by South-east Asia (Figures 1 and 2), generated renewed attention to this reemerging virus. As millions of persons became infected, thousands of air travelers introduced this new Indian Ocean Lineage (IOL) of CHIKV, also derived from an ECSA enzootic strain (Figure 1), into nearly all regions globally, and local transmission ensued in northern Italy [7] and southern France (Figure 2) [8,9]. Al-though importations were also documented in transmission-permissive regions of the Americas, autochthonous cases were not detected in the Western Hemisphere until late 2013 on the Island of St. Martin in the Caribbean [10,11]. Subsequently, CHIKV spread to nearly all Caribbean Islands and into northern South America, Central America, and Mexico. Eleven autochthonous cases were detected in 2014 in Florida, although there was no evidence of persistence. Surprisingly, this first CHIKV strain identified in the Americas was not the IOL lineage implicated in most recent outbreaks, but belonged to the Asian lineage that had been circulating in the urban cycle at least since the 1950s and which has continued to cause outbreaks in Asia and Oceania in recent years [12,13]; importation probably occurred via a viremic traveler from Southeast Asia or Oceania, or possibly an infected mosquito transported on a flight (Figure 1) [14••].
Figure 2.
Map showing the historic spread of CHIKV from enzootic cycles in Africa to Asia, Europe and the Americas.
Following the dramatic introduction of CHIKV into the Americas in late 2013, additional transcontinental transfers continued. In June, 2014, a traveler from Angola introduced an ECSA strain into Feira de Santana, Brazil (Figure 2) [15••]. This strain could represent a major public health challenge because, like IOL strains, it could adapt for efficient transmission by Aedes (Stegomyia) albo-pictus (details below). Then, in October, 2014, a traveler from Cameroon initiated an outbreak involving 11 con-firmed cases in Montpellier, France, with A. albopictus suspected as the principal vector (Figure 2) [16].
With the increased attention on CHIKV since 2005, additional cases and outbreaks have also been detected in Africa, including in Cameroon [17,18], Gabon [19–21], Congo [http://www.ncbi.nlm.nih.gov/pubmed/25541718], Tanzania [22] and Senegal [23]. Some of these infections probably represented spillover from enzootic cycles without a switch to urban vectors, while in others urban mosquitoes such as A. albopictus were implicated in inter-human transmission.
Phylogenetic reconstruction of chikungunya virus evolution
The first phylogenetic trees of CHIKV, based on partial E1 glycoprotein gene sequences and the 3′ untranslated genome region (3′UTR), identified two main enzootic lineages: West African and ECSA [3]. These studies concluded that the Asian Lineage urban strains resulted from an introduction of an ECSA progenitor 50–430 years before 2000. More recent Bayesian coalescent estimates based on complete open reading frame sequences are 1879–1956 (Figure 1) [4•]. This study also determined that the IOL strains, which appeared in Coastal Kenya in 2004 [24], spread independently into islands in the Indian Ocean and into the Indian subcontinent in 2005. Although the IOL has not been detected in the Indian Ocean islands since 2011, it continues to circulate and evolve in South and Southeast Asia, with small outbreaks exported to Italy and France as described above. IOL divergence has included several E2 envelope glycoprotein substitutions that further enhance infectivity for A. albopictus, as discussed below (Table 1). Since the first 2013 Caribbean outbreak, Asian CHIKV strains remain highly conserved, consistent with a point source introduction, and with no evidence of adaptive evolution (Figure 1). Finally, phylogenetic placement as sisters to a 1962 Angola ECSA CHIKV strain of three isolates from Feira de Santana, Brazil (Figure 1) support the importation of this strain by a traveler from southern Africa [15••].
Table 1.
Amino acid substitutions that influence Aedes albopictus infection by CHIKV
| CHIKV Lineage | Year of first appearance |
Protein | Amino acid substitution (references) |
Approximate infectivity increase or epistatic effect |
Epistatic interactions |
|---|---|---|---|---|---|
| IOL | 2005 | E1 | A226V [55•,56,60••] | 40-fold | E1-98, E2-211 |
| IOL SL1 | 2007 | E2 | K252Q [60••] | 8-fold | ND |
| IOL SL2 (partial) | 2008 | E2 | K233E [60••] | 6-fold | ND |
| IOL SL3B | 2008 | E2/E3 | R198Q/S18F (synergistic)[60••] | 16-fold | ND |
| IOL SL4 | 2009 | E2 | L210Q [56] | 5-fold | ND |
| Asian | 1958 | E1 | A98T [57••] | Completely prevents penetrance for A. albopictus infection | E1-226V |
| ECSA | 1953 | E2 | I211T [44•] | Enables penetrance for A. albopictus infection | E1-226V |
ND — not determined; SL — sublineage.
Enzootic chikungunya virus transmission
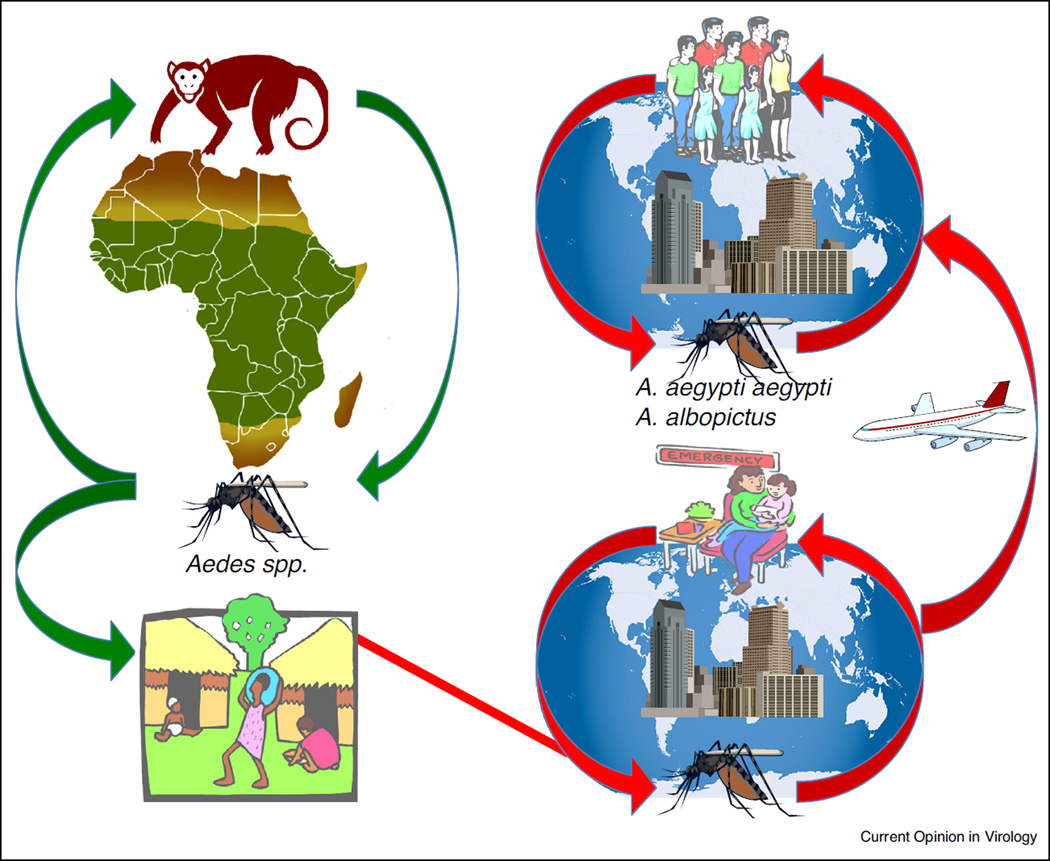
Enzootic cycles of CHIKV in sub-Saharan Africa are believed to represent its ancestral state (Figure 1). There, in forested habitats, CHIKV transmission has been documented involving several mosquito species in the genus Aedes, with nonhuman primates and possibly other vertebrates serving as amplification hosts (Figure 3). Aedes (Diceromyia) furcifer appears to be the most important enzootic vector in South Africa [25], Zimbabwe [26] and Senegal [27], while A. (Diceromyia) taylori, A. (Stegomyia) africanus [28] and A. (Stegomyia) luteocephalus have also been implicated on multiple occasions [27,29]. Of these vectors, the susceptibility to infection has only been determined experimentally for A. furcifer, which transmitted CHIKV between African green monkeys (Cercopithecus aethiops) and exhibited an oral infection threshold of less than 104.5 log infectious units.
Figure 3.
Cartoon showing enzootic and urban CHIKV transmission cycles and their connection in sub-Saharan Africa.
The role of various NHPs in enzootic CHIKV amplification has received limited attention. African green monkeys and chacma baboons (Papio ursinus) were suggested as important hosts in Zimbabwe based on high seroprevalence [26], and the former species developed viremia after experimental infection in Uganda [28]. In an enzootic Senegal focus, CHIKV was isolated from African green and Patas (Erythrocebus patas) monkeys and from the Guinea baboon (Papio papio) [27,30]. Chimpanzees were also seropositive in the Congo [31]. While a variety of other wild and domesticated vertebrates have been found seropositive in Africa and Asia, antibody titers are generally low and few experimental infections have been performed. Overall, these data suggest that NHPs are important amplification hosts but do not rule out a role for other vertebrates in enzootic maintenance.
In Asia, there is no conclusive evidence of enzootic CHIKV circulation. In Peninsular Malaysia with its history of repeated CHIKV outbreaks, 71 Bornean orangutans [32] were seronegative, and only one of 147 long-tailed macaques (Macaca fascicularis) was positive [33]. Regardless, temporary spillback of urban strains cannot be ruled out as an explanation of this or other NHP infections. Isolation of CHIKV strains genetically distinct from urban isolates and/or detection of circulation in regions remote from humans would be needed to demonstrate an independent enzootic cycle. This would be an important finding that might explain CHIKV persistence during interepidemic periods.
In summary, CHIKV appears to use several different NHPs and Aedes spp. vectors for enzootic circulation in Africa. This pattern of opportunistic vertebrate host and mosquito vectors usage has been described for other alphaviruses [34], as well as other arboviruses [35]. Flexibility in primate usage is probably critical due to the limited population sizes and turnover rates for these long-lived hosts. Furthermore, this flexibility in host and vector usage is an important factor in the ability of CHIKV to emerge via host range changes, as described below.
Vector and primate host range changes associated with urban chikungunya virus emergence
The urban CHIKV cycle presumably commences when a person entering or living near African forests becomes infected via enzootic spillover due to vectors such as A. furcifer, which enters villages and bites people (Figure 3) [23]. In Africa, the role of A. (Stegomyia) aegypti in CHIKV transmission is far from clear; the domesticated subspecies (A. aegypti aegypti) evolved convergently from the ancestral, zoophilic form (A. aegypti formosus) by acquiring a preference for artificial water containers as oviposition sites and humans as a blood and nutrition source [36]. Human migration led to the establishment of this sub-species in most tropical and subtropical regions of the world, allowing it to serve as the main global CHIKV vector [37]. Similar to A. aegypti but more recently, A. albopictus extended its range from Asia [38] into tropical and temperate zones of the Americas, Africa and Europe [39–43]. However, until recently the role of this species in CHIKV transmission was thought to be less important compared to A. aegypti, mainly due to its less prominent anthropophily. Although the current distributions in many parts of Africa of A. albopictus and the domesticated form of A. aegypti remain incompletely characterized, presumably a person infected from enzootic CHIKV spillover occasionally reaches a location where populations of these mosquitoes and their contact with people are sufficient to initiate interhuman transmission.
There is no direct evidence of any adaptive constraints on the ability of enzootic CHIKV strains to enter the urban cycle (Figure 3). However, studies of the A. albopictus-adaptive substitution E1-A226V (Table 1) suggest that acquisition of the E2-I211T substitution in ECSA strains could increase CHIKV fitness for human infection [44•]. This conclusion is based on the fact that the E2-211I residue is the most common among the enzootic ECSA strains, including those ancestral to the IOL [44•,45]; however strains involved in continuous interhuman circulation, including the IOL and Asian lineages, exhibit E2-I211T. Experimental studies failed to support a phenotype for E2-I211T (in the absence of E1-226V) in CHIKV infection of urban vectors, leaving adaptation to humans as a possible explanation for its convergent evolution before E1-A226V [44•].
In contrast to A. aegypti-borne transmission, urban IOL outbreaks involving interhuman transmission by A. albo-pictus have been consistently associated with an alanine-to-valine substitution at position 226 of the E1 glycoprotein (Table 1), including at least 6 documented occasions: Reunión island, 2006 [46••], in Indian states of Kerala, 2007 [47–49] 2009 [50], twice in Sri Lanka, 2008 [51], and once in Gabon, 2007 [52,53]. This convergent evolution strongly suggests adaptation for enhanced A. albopictus transmission. Using natural CHIKV isolates from the 2006 Reunión outbreak differing only in this residue, Vazeille et al. demonstrated that the presence of E1-226V is associated with enhanced CHIKV dissemination and higher viral RNA loads in A. albopictus [54•]. Reverse genetic experiments demonstrated that E1-A226V results in ~40–100-fold increase in CHIKV oral infectivity to A. albopictus and mediates more efficient virus transmission to mice [44•,55•,56,57••]. Subsequent studies using single-round infectious CHIKV-like particles [56] and intra-thoracically injected A. albopictus [58,59] demonstrated that E1-A226V acts primarily by increasing CHIKV oral infectivity for midgut epithelial cells, which leads to more efficient transmission.
Further research demonstrated that, in addition to E1-A226V, most A. albopictus-borne CHIKV strains have acquired additional, second-step substitutions located in the Acid Sensitive Region (ASR) of the adjacent E2 glycoprotein. This E2 region mediates a conformational change in the E2/E1 heterodimer within low pH endosomes to expose the E1 fusion peptide for entry into the cytoplasm (Table 1) [56,60••]. These substitutions appeared after acquisition of E1-A226V and have a lesser but still highly significant (typically resulting in a 5–16-fold increase in CHIKV infectivity) effect on CHIKV infection of A. albopictus, compared to the E1-A226V. The ability of CHIKV to use several different second-step adaptive mutations to more efficiently infect A. albopictus may have facilitated the rapid IOL diversification.
A critical finding related to the E1-A226V and its role in CHIKV emergence was that the valine residue had never been detected in Asian lineage CHIKV strains despite their long history of circulation in regions native to A. albopictus [4•]. An explanation for this conundrum, that not all CHIKV strains are equally susceptible to the effects of the E1-A226V substitution, was supported by reverse genetic studies. The E1-98T residue, found in all Asian strain sequences but absent from all other CHIKV lineages, completely blocks the E1-226V-enhanced oral infection phenotype in A. albopictus (Table 1) [57••], and thereby apparently prevented Asian CHIKV strains from adapting to this vector. This, in turn, may have facilitated the invasion of A. albopictus-adapted IOL strains into Southeast Asia beginning in 2006 to exploit this unoccupied vector niche [57••]. Like E1-98T, the E2-211I residue found in most ECSA strains (including the independent introduction into Feira de Santana, Brazil) also limits penetrance of E1-226V and thus CHIKV’s potential to emerge in some parts of Africa and the Americas via A. albopictus transmission [44•]. It remains to be seen if ECSA strains in Brazil will select for E2-I211T, as did early IOL strains in Kenya in 2004 [45], which will enable subsequent A. albopictus-adaptive evolution.
Prospects for expansion of enzootic chikungunya virus host range and distribution
The information summarized above may be useful in predicting the outcome of current and future CHIK outbreaks. Its historic evidence of repeated reemergence combined with the lack of evidence for adaptive barriers to initiation of the urban transmission cycle by enzootic progenitors strongly suggests that CHIKV will continue to emerge periodically and indefinitely from Africa to initiate epidemics. The ECSA origin of the past two emergences (Asian and IOL) may only reflect the geo-graphic proximity of these strains to Asia, which is highly permissive to urban circulation; but there is no evidence that West African strains are incapable of initiating inter-human transmission. Furthermore, based on our current understanding, the genomic sequences of many ECSA and West African strains may be fully capable of adapting for efficient A. albopictus transmission, as has been seen in IOL strains (Table 1). However, epistatic constraints in Asian strains [57••], which are now widespread in the Americas, and in the ECSA strain introduced last year into Brazil, suggest that these lineages may remain primarily transmitted by A. aegypti with less opportunity for movement into temperate regions where this mosquito cannot survive cold winters. The initial incrimination of A. aegypti in transmission in the region of the Americas inhabited by both urban vectors (Mexico) [61] supports this prediction. Opportunities for the use of additional urban vectors are difficult to predict in the absence of experimental data, but recent evidence that Aedes (Stegomyia) hensilli was the principal epidemic CHIKV vector on Yap Island suggests that other mosquitoes may play major roles in some regions [12].
Similarly, there is no direct evidence for a barrier to CHIKV host jumps from African NHPs into humans that accompany urban emergence. As discussed above, CHIKV may not specialize on any given NHP species and thus the use of humans may be consistent with a generalized competence for amplification in many different primates. If this is the case, CHIKV may have the potential to spill back into an enzootic South American cycle, as occurred for yellow fever virus after its importation from Africa during the slave trade [62]. However, the lack of evidence for such CHIKV spillback in Asia despite many decades if not centuries of opportunity may reflect an unrecognized barrier to the use of NHPs or sylvatic mosquito vectors in some geographic regions. The apparent failure of dengue viruses to establish enzootic trans-mission in the Americas [63] suggests such a barrier. Experimental infections of New World primates and sylvatic, primatophilic mosquitoes as well as incorporation of sylvatic regions into CHIKV surveillance will be important for determining the potential for enzootic establishment, which could stabilize CHIKV endemicity in the Americas and ensure continuous risk.
Acknowledgments
This research was supported by NIH grant AI069145 and by grant HSHQDC-13-C-B0009, from the Department of Homeland Security entitled ‘Whole Genome Approach to Microbial Forensics Technical Focus Area-3.’
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Weaver SC, Forrester NL. Chikungunya: evolutionary history and recent epidemic spread. Antiviral Res. 2015;120:32–39. doi: 10.1016/j.antiviral.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 2.Weaver SC, Lecuit M. Chikungunya virus and the global spread of a mosquito-borne disease. N Engl J Med. 2015;372:1231–1239. doi: 10.1056/NEJMra1406035. [DOI] [PubMed] [Google Scholar]
- 3.Powers AM, Brault AC, Tesh RB, Weaver SC. Re-emergence of Chikungunya and O’nyong-nyong viruses: evidence for distinct geographical lineages and distant evolutionary relationships. J Gen Virol. 2000;81:471–479. doi: 10.1099/0022-1317-81-2-471. [DOI] [PubMed] [Google Scholar]
- 4. Volk SM, Chen R, Tsetsarkin KA, Adams AP, Garcia TI, Sall AA, Nasar F, Schuh AJ, Holmes EC, Higgs S, et al. Genome-scale phylogenetic analyses of chikungunya virus reveal independent emergences of recent epidemics and various evolutionary rates. J Virol. 2010;84:6497–6504. doi: 10.1128/JVI.01603-09. A detailed phylogenetic study, which characterizes existing CHIKV lineages and rates of evolution among them.
- 5. Halstead SB. Reappearance of chikungunya, formerly called dengue, in the americas. Emerg Infect Dis. 2015;21 doi: 10.3201/eid2104.141723. Using retrospective historic analysis the author suggests that CHIKV had been introduced into Americas before 2013 but it was mistakenly identified as dengue.
- 6.Carey DE. Chikungunya and dengue: a case of mistaken identity? J Hist Med Allied Sci. 1971;26:243–262. doi: 10.1093/jhmas/xxvi.3.243. [DOI] [PubMed] [Google Scholar]
- 7.Rezza G, Nicoletti L, Angelini R, Romi R, Finarelli AC, Panning M, Cordioli P, Fortuna C, Boros S, Magurano F, et al. Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet. 2007;370:1840–1846. doi: 10.1016/S0140-6736(07)61779-6. [DOI] [PubMed] [Google Scholar]
- 8.Gould EA, Gallian P, De Lamballerie X, Charrel RN. First cases of autochthonous dengue fever and chikungunya fever in France: from bad dream to reality! Clin Microbiol Infect. 2010;16:1702–1704. doi: 10.1111/j.1469-0691.2010.03386.x. [DOI] [PubMed] [Google Scholar]
- 9.Grandadam M, Caro V, Plumet S, Thiberge JM, Souares Y, Failloux AB, Tolou HJ, Budelot M, Cosserat D, Leparc-Goffart I, et al. Chikungunya virus, southeastern France. Emerg Infect Dis. 2011;17:910–913. doi: 10.3201/eid1705.101873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leparc-Goffart I, Nougairede A, Cassadou S, Prat C, de Lamballerie X. Chikungunya in the Americas. Lancet. 2014;383:514. doi: 10.1016/S0140-6736(14)60185-9. [DOI] [PubMed] [Google Scholar]
- 11.Van Bortel W, Dorleans F, Rosine J, Blateau A, Rousset D, Matheus S, Leparc-Goffart I, Flusin O, Prat C, Cesaire R, et al. Chikungunya outbreak in the Caribbean region, December 2013 to March 2014, and the significance for Europe. Euro Surveill. 2014;19 doi: 10.2807/1560-7917.es2014.19.13.20759. [DOI] [PubMed] [Google Scholar]
- 12.Savage HM, Ledermann JP, Yug L, Burkhalter KL, Marfel M, Hancock WT. Incrimination of Aedes (Stegomyia) hensilli Farner as an epidemic vector of Chikungunya virus on Yap Island, Federated States of Micronesia, 2013. Am J Trop Med Hyg. 2015;92:429–436. doi: 10.4269/ajtmh.14-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nhan TX, Claverie A, Roche C, Teissier A, Colleuil M, Baudet JM, Cao-Lormeau VM, Musso D. Chikungunya virus imported into French polynesia, 2014. Emerg Infect Dis. 2014;20:1773–1774. doi: 10.3201/eid2010.141060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lanciotti RS, Valadere AM. Transcontinental movement of Asian genotype chikungunya virus. Emerg Infect Dis. 2014;20:1400–1402. doi: 10.3201/eid2008.140268. Phylogenetic study that analyzes the origin of the CHIKV strains that initiated the outbreak in Americas beginning in 2013. This study shows that Caribbean strains of CHIKV belong to Asian lineage and are most closely related to strains from Yap and Philippines.
- 15. Nunes MR, Faria NR, de Vasconcelos JM, Golding N, Kraemer MU, de Oliveira LF, Azevedo Rdo S, da Silva DE, da Silva EV, da Silva SP, et al. Emergence and potential for spread of Chikungunya virus in Brazil. BMC Med. 2015;13:102. doi: 10.1186/s12916-015-0348-x. Phylogenetic study which shows that, in addition to strains of the Asian lineage, the 2014 CHIKV outbreak in Brazil was caused also by strains of ECSA lineage introduced recently from Africa.
- 16.Delisle E, Rousseau C, Broche B, Leparc-Goffart I, L’Ambert G, Cochet A, Prat C, Foulongne V, Ferre JB, Catelinois O, et al. Chikungunya outbreak in Montpellier, France, September to October 2014. Euro Surveill. 2015;20 doi: 10.2807/1560-7917.es2015.20.17.21108. [DOI] [PubMed] [Google Scholar]
- 17.Peyrefitte CN, Rousset D, Pastorino BA, Pouillot R, Bessaud M, Tock F, Mansaray H, Merle OL, Pascual AM, Paupy C, et al. Chikungunya virus, Cameroon, 2006. Emerg Infect Dis. 2007;13:768–771. doi: 10.3201/eid1305.061500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Demanou M, Antonio-Nkondjio C, Ngapana E, Rousset D, Paupy C, Manuguerra JC, Zeller H. Chikungunya outbreak in a rural area of Western Cameroon in 2006: a retrospective serological and entomological survey. BMC Res Notes. 2010;3:128. doi: 10.1186/1756-0500-3-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peyrefitte CN, Bessaud M, Pastorino BA, Gravier P, Plumet S, Merle OL, Moltini I, Coppin E, Tock F, Daries W, et al. Circulation of Chikungunya virus in Gabon, 2006–2007. J Med Virol. 2008;80:430–433. doi: 10.1002/jmv.21090. [DOI] [PubMed] [Google Scholar]
- 20.Leroy EM, Nkoghe D, Ollomo B, Nze-Nkogue C, Becquart P, Grard G, Pourrut X, Charrel R, Moureau G, Ndjoyi-Mbiguino A, et al. Concurrent chikungunya and dengue virus infections during simultaneous outbreaks, Gabon, 2007. Emerg Infect Dis. 2009;15:591–593. doi: 10.3201/eid1504.080664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paupy C, Kassa Kassa F, Caron M, Nkoghe D, Leroy EM. A chikungunya outbreak associated with the vector Aedes albopictus in remote villages of Gabon. Vector Borne Zoonotic Dis. 2012;12:167–169. doi: 10.1089/vbz.2011.0736. [DOI] [PubMed] [Google Scholar]
- 22.Hertz JT, Munishi OM, Ooi EE, Howe S, Lim WY, Chow A, Morrissey AB, Bartlett JA, Onyango JJ, Maro VP, et al. Chikungunya and dengue fever among hospitalized febrile patients in northern Tanzania. Am J Trop Med Hyg. 2012;86:171–177. doi: 10.4269/ajtmh.2012.11-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Diallo D, Sall AA, Buenemann M, Chen R, Faye O, Diagne CT, Faye O, Ba Y, Dia I, Watts D, et al. Landscape ecology of sylvatic chikungunya virus and mosquito vectors in southeastern Senegal. PLoS Negl Trop Dis. 2012;6:e1649. doi: 10.1371/journal.pntd.0001649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chretien JP, Anyamba A, Bedno SA, Breiman RF, Sang R, Sergon K, Powers AM, Onyango CO, Small J, Tucker CJ, et al. Drought-associated chikungunya emergence along coastal East Africa. Am J Trop Med Hyg. 2007;76:405–407. [PubMed] [Google Scholar]
- 25.Jupp PG, McIntosh BM, Dos Santos I, DeMoor P. Laboratory vector studies on six mosquito and one tick species with chikungunya virus. Trans R Soc Trop Med Hyg. 1981;75:15–19. doi: 10.1016/0035-9203(81)90005-5. [DOI] [PubMed] [Google Scholar]
- 26.McIntosh BM, Paterson HE, McGillivray G, Desousa J. Further studies on the Chikungunya outbreak in Southern Rhodesia in 1962. I. Mosquitoes, wild primates and birds in relation to the epidemic. Ann Trop Med Parasitol. 1964;58:45–51. doi: 10.1080/00034983.1964.11686213. [DOI] [PubMed] [Google Scholar]
- 27.Diallo M, Thonnon J, Traore-Lamizana M, Fontenille D. Vectors of Chikungunya virus in Senegal: current data and transmission cycles. Am J Trop Med Hyg. 1999;60:281–286. doi: 10.4269/ajtmh.1999.60.281. [DOI] [PubMed] [Google Scholar]
- 28.Weinbren MP, Haddow AJ, Williams MC. The occurrence of Chikungunya virus in Uganda. I. Isolation from mosquitoes. Trans R Soc Trop Med Hyg. 1958;52:253–257. doi: 10.1016/0035-9203(58)90084-1. [DOI] [PubMed] [Google Scholar]
- 29.Jupp PG, McIntosh BM. Aedes furcifer and other mosquitoes as vectors of chikungunya virus at Mica, northeastern Transvaal, South Africa. J Am Mosq Control Assoc. 1990;6:415–420. [PubMed] [Google Scholar]
- 30.Monlun E, Zeller H, Le Guenno B, Traore-Lamizana M, Hervy JP, Adam F, Ferrara L, Fontenille D, Sylla R, Mondo M, et al. Surveillance of the circulation of arbovirus of medical interest in the region of eastern Senegal. Bull Soc Pathol Exot. 1993;86:21–28. [PubMed] [Google Scholar]
- 31.Osterrieth P, Deleplanque-Liegeois P, Renoirte R. Research on the Chikungunya virus in the Belgian Congo. II. Serological investigation. Ann Soc Belg Med Trop (1920) 1960;40:205–213. [PubMed] [Google Scholar]
- 32.Wolfe ND, Kilbourn AM, Karesh WB, Rahman HA, Bosi EJ, Cropp BC, Andau M, Spielman A, Gubler DJ. Sylvatic transmission of arboviruses among Bornean orangutans. Am J Trop Med Hyg. 2001;64:310–316. doi: 10.4269/ajtmh.2001.64.310. [DOI] [PubMed] [Google Scholar]
- 33.Sam IC, Chua CL, Rovie-Ryan JJ, Fu JY, Tong C, Sitam FT, Chan YF. Chikungunya virus in Macaques, Malaysia. Emerg Infect Dis. 2015;21:1683–1685. doi: 10.3201/eid2109.150439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weaver SC, Winegar R, Manger ID, Forrester NL. Alphaviruses: population genetics and determinants of emergence. Antiviral Res. 2012;94:242–257. doi: 10.1016/j.antiviral.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanley KA, Weaver SC. Arbovirus evolution. In: Domingo E, Parrish CR, Holland JJ, editors. Origin and Evolution of Viruses. Elsevier; 2008. pp. 351–392. [Google Scholar]
- 36.Powell JR, Tabachnick WJ. History of domestication and spread of Aedes aegypti — a review. Mem Inst Oswaldo Cruz. 2013;108(Suppl. 1):11–17. doi: 10.1590/0074-0276130395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsetsarkin KA, Chen R, Sherman MB, Weaver SC. Chikungunya virus: evolution and genetic determinants of emergence. Curr Opin Virol. 2011;1:310–317. doi: 10.1016/j.coviro.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith CE. The history of dengue in tropical Asia and its probable relationship to the mosquito Aedes aegypti. J Trop Med Hyg. 1956;59:243–251. [PubMed] [Google Scholar]
- 39.Gratz NG. Critical review of the vector status of Aedes albopictus. Med Vet Entomol. 2004;18:215–227. doi: 10.1111/j.0269-283X.2004.00513.x. [DOI] [PubMed] [Google Scholar]
- 40.Benedict MQ, Levine RS, Hawley WA, Lounibos LP. Spread of the tiger: global risk of invasion by the mosquito Aedes albopictus. Vector Borne Zoonotic Dis. 2007;7:76–85. doi: 10.1089/vbz.2006.0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Medlock JM, Hansford KM, Schaffner F, Versteirt V, Hendrickx G, Zeller H, Van Bortel W. A review of the invasive mosquitoes in Europe: ecology, public health risks, and control options. Vector Borne Zoonotic Dis (Larchmont, NY) 2012;12:435–447. doi: 10.1089/vbz.2011.0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gjenero-Margan I, Aleraj B, Krajcar D, Lesnikar V, Klobucar A, Pem-Novosel I, Kurecic-Filipovic S, Komparak S, Martic R, Duricic S, et al. Autochthonous dengue fever in Croatia, August–September 2010. Euro Surveill. 2011;16 [PubMed] [Google Scholar]
- 43.La Ruche G, Souares Y, Armengaud A, Peloux-Petiot F, Delaunay P, Despres P, Lenglet A, Jourdain F, Leparc-Goffart I, Charlet F, et al. First two autochthonous dengue virus infections in metropolitan France, September 2010. Euro Surveill. 2010;15:19676. [PubMed] [Google Scholar]
- 44. Tsetsarkin KA, McGee CE, Volk SM, Vanlandingham DL, Weaver SC, Higgs S. Epistatic roles of E2 glycoprotein mutations in adaption of chikungunya virus to Aedes albopictus and Ae. aegypti mosquitoes. PLoS ONE. 2009;4:e6835. doi: 10.1371/journal.pone.0006835. Using reverse genetics experiments the authors show that epistatic mutations can constrain adaptation of some strains of ECSA lineages to A. albopictus.
- 45.Kariuki Njenga M, Nderitu L, Ledermann JP, Ndirangu A, Logue CH, Kelly CH, Sang R, Sergon K, Breiman R, Powers AM. Tracking epidemic Chikungunya virus into the Indian Ocean from East Africa. J Gen Virol. 2008;89:2754–2760. doi: 10.1099/vir.0.2008/005413-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Schuffenecker I, Iteman I, Michault A, Murri S, Frangeul L, Vaney MC, Lavenir R, Pardigon N, Reynes JM, Pettinelli F, et al. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med. 2006;3:e263. doi: 10.1371/journal.pmed.0030263. Phylogenetic study of 2005–06 CHIKV strains from the Reunion Island outbreak, which first suggested that E1-A226V mutation might be associated with CHIKV adaptation to A. albopictus mosquitoes.
- 47.Dash PK, Parida MM, Santhosh SR, Verma SK, Tripathi NK, Ambuj S, Saxena P, Gupta N, Chaudhary M, Babu JP, et al. East Central South African genotype as the causative agent in reemergence of Chikungunya outbreak in India. Vector Borne Zoonotic Dis. 2007;7:519–527. doi: 10.1089/vbz.2007.7272. [DOI] [PubMed] [Google Scholar]
- 48.Arankalle VA, Shrivastava S, Cherian S, Gunjikar RS, Walimbe AM, Jadhav SM, Sudeep AB, Mishra AC. Genetic divergence of Chikungunya viruses in India (1963–2006) with special reference to the 2005–2006 explosive epidemic. J Gen Virol. 2007;88:1967–1976. doi: 10.1099/vir.0.82714-0. [DOI] [PubMed] [Google Scholar]
- 49.Cherian SS, Walimbe AM, Jadhav SM, Gandhe SS, Hundekar SL, Mishra AC, Arankalle VA. Evolutionary rates and timescale comparison of Chikungunya viruses inferred from the whole genome/E1 gene with special reference to the 2005–07 outbreak in the Indian subcontinent. Infect Genet Evol. 2009;9:16–23. doi: 10.1016/j.meegid.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 50.Niyas KP, Abraham R, Unnikrishnan RN, Mathew T, Nair S, Manakkadan A, Issac A, Sreekumar E. Molecular characterization of Chikungunya virus isolates from clinical samples and adult Aedes albopictus mosquitoes emerged from larvae from Kerala, South India. Virol J. 2010;7:189. doi: 10.1186/1743-422X-7-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hapuarachchi HC, Bandara KB, Sumanadasa SD, Hapugoda MD, Lai YL, Lee KS, Tan LK, Lin RT, Ng LF, Bucht G, et al. Re-emergence of Chikungunya virus in South-east Asia: virological evidence from Sri Lanka and Singapore. J Gen Virol. 2010;91:1067–1076. doi: 10.1099/vir.0.015743-0. [DOI] [PubMed] [Google Scholar]
- 52.Vazeille M, Moutailler S, Pages F, Jarjaval F, Failloux AB. Introduction of Aedes albopictus in Gabon: what consequences for dengue and chikungunya transmission? Trop Med Int Health. 2008;13:1176–1179. doi: 10.1111/j.1365-3156.2008.02123.x. [DOI] [PubMed] [Google Scholar]
- 53.Pages F, Peyrefitte CN, Mve MT, Jarjaval F, Brisse S, Iteman I, Gravier P, Tolou H, Nkoghe D, Grandadam M. Aedes albopictus mosquito: the main vector of the 2007 Chikungunya outbreak in Gabon. PLoS ONE. 2009;4:e4691. doi: 10.1371/journal.pone.0004691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Vazeille M, Moutailler S, Coudrier D, Rousseaux C, Khun H, Huerre M, Thiria J, Dehecq JS, Fontenille D, Schuffenecker I, et al. Two Chikungunya isolates from the outbreak of La Reunion (Indian Ocean) exhibit different patterns of infection in the mosquito, Aedes albopictus. PLoS ONE. 2007;2:e1168. doi: 10.1371/journal.pone.0001168. Experimental support for the hypothesis proposed in reference 46.
- 55. Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog. 2007;3:e201. doi: 10.1371/journal.ppat.0030201. Experimental support for the hypothesis proposed in reference 46.
- 56.Tsetsarkin KA, Weaver SC. Sequential adaptive mutations enhance efficient vector switching by chikungunya virus and its epidemic emergence. PLoS Pathog. 2011;7:e1002412. doi: 10.1371/journal.ppat.1002412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Tsetsarkin KA, Chen R, Leal G, Forrester N, Higgs S, Huang J, Weaver SC. Chikungunya virus emergence is constrained in Asia by lineage-specific adaptive landscapes. Proc Natl Acad Sci U S A. 2011;108:7872–7877. doi: 10.1073/pnas.1018344108. Using reverse genetics experiments, this study shows that an epistatic mutation exists in all strains of Asian lineage, which constrain their adaptation to A. albopictus.
- 58.Arias-Goeta C, Mousson L, Rougeon F, Failloux AB. Dissemination and transmission of the E1-226V variant of chikungunya virus in Aedes albopictus are controlled at the midgut barrier level. PLOS ONE. 2013;8:e57548. doi: 10.1371/journal.pone.0057548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsetsarkin K. Graduate School of Biomedical Sciences. Galveston: The University of Texas Medical Branch; 2011. Adaptation of chikungunya virus to Aedes albopictus mosquitoes: the role of mutations in the E1 and E2 glycoproteins; p. 279. [Google Scholar]
- 60. Tsetsarkin KA, Chen R, Yun R, Rossi SL, Plante KS, Guerbois M, Forrester N, Perng GC, Sreekumar E, Leal G, et al. Multi-peaked adaptive landscape for chikungunya virus evolution predicts continued fitness optimization in Aedes albopictus mosquitoes. Nat Commun. 2014;5:4084. doi: 10.1038/ncomms5084. This study demonstrates that multiple mutations in the E2 glycoprotein gene can further increase CHIKV fitness for transmission by A. albopictus enabling lineage diversification observed after acquisition of E1-A226V substitution.
- 61.Kautz TF, Díaz-González EE, Erasmus JH, Malo-García I, Langsjoen RM, Patterson EI, Auguste DI, Forrester NL, Maria Sanchez-Casas R, Hernandez-Avila M, et al. Chikungunya virus identified as the etiological agent of an outbreak of febrile illness in Chiapas Mexico, 2014. Emerg Infect Dis. 2015;11:2070–2073. doi: 10.3201/eid2111.150546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bryant JE, Holmes EC, Barrett AD. Out of Africa: a molecular perspective on the introduction of yellow fever virus into the Americas. PLoS Pathog. 2007;3:e75. doi: 10.1371/journal.ppat.0030075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hanley KA, Monath TP, Weaver SC, Rossi SL, Richman RL, Vasilakis N. Fever versus fever: the role of host and vector susceptibility and interspecific competition in shaping the current and future distributions of the sylvatic cycles of dengue virus and yellow fever virus. Infect Genet Evol. 2013 doi: 10.1016/j.meegid.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sunderland, MA: Sinauer Associates; 1998. [Google Scholar]