Abstract
Adenosine deaminases acting on RNA (ADARs) convert adenosine to inosine in double-stranded RNA. This A-to-I editing occurs not only in protein-coding regions of mRNAs, but also frequently in non-coding regions that contain inverted Alu repeats. Editing of coding sequences can result in the expression of functionally altered proteins that are not encoded in the genome, whereas the significance of Alu editing remains largely unknown. Certain microRNA (miRNA) precursors are also edited, leading to reduced expression or altered function of mature miRNAs. Conversely, recent studies indicate that ADAR1 forms a complex with Dicer to promote miRNA processing, revealing a new function of ADAR1 in the regulation of RNA interference.
Adenosine to inosine (A-to-I) RNA editing was originally discovered as a mysterious enzymatic activity causing unwinding of double-stranded RNA (dsRNA) in Xenopus laevis oocytes and embryos1. Soon after, it became clear that this activity is carried out by an adenosine deaminase acting on RNA (ADAR)2,3. These discoveries established the field of A-to-I RNA editing4,5. Initially, a limited number of editing sites were discovered serendipitously in protein-coding regions of mRNAs, when comparing human genomic DNA versus cDNA sequences. However, the development of deep sequencing and recent advancements in bioinformatics made it possible to screen A-to-I RNA-editing sites globally. Surprisingly, the most frequent and widespread targets of A-to-I RNA editing are dsRNAs made from inverted Alu repetitive elements (Alu dsRNAs), which are located within introns and untranslated regions6–19.
Precursors of certain microRNAs (miRNAs) also undergo A-to-I RNA editing, which negatively regu lates the expression and function of the mature miRNAs20–24. Conversely, recent studies indicate that ADAR1 forms a complex with Dicer to promote miRNA processing and RNA interference (RNAi) efficacy25,26. This Review summarizes our current knowledge on A-to-I RNA editing and ADARs in mammals. Its focus, however, is on the significance of non-coding, repetitive RNA editing and on the interactions between the RNA-editing and RNAi mechanisms. For reviews focusing on the relevance of A-to-I RNA editing to brain functions, viral infection and human diseases, see REFS 4,5,27–35. For reviews on other types of RNA editing, see REFS 36–39.
Mechanism and regulation of RNA editing
A-to-I RNA editing is mediated by ADAR family members, which are conserved in the animal kingdom40 (FIG. 1a).
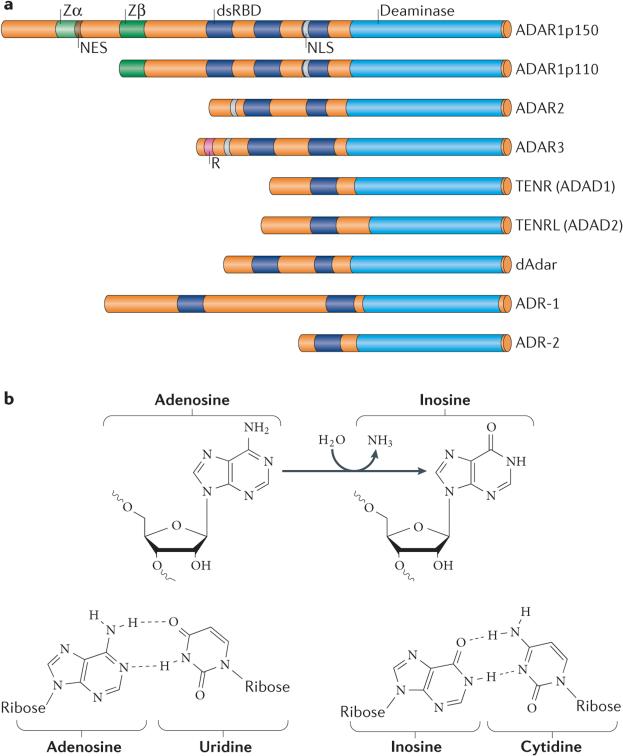
Figure 1. Deamination of adenosine to inosine by adenosine deaminases acting on RNA (ADAR) proteins.
a | Three human ADAR family members (ADAR1, ADAR2 and ADAR3), two human ADAD (adenosine deaminase domain-containing) family members (TENR and TENRL), Drosophila melanogaster dAdar, and two Caenorhabditis elegans ADAR proteins (ADR-1 and ADR-2), share common functional domains. These include two or three repeats of the double-stranded RNA (dsRNA)-binding domain (dsRBD) and a catalytic deaminase domain. Certain structural features, such as Z-DNA-binding domains (Zα and Zβ) and the Arg-rich, single-stranded RNA (ssRNA)-binding R domain, are unique to particular ADAR members. b | ADARs catalyse a hydrolytic deamination reaction that converts adenosine to inosine (top). Whereas adenosine base-pairs with uridine, inosine behaves like a guanosine, as it base-pairs with cytidine in a Watson–Crick-bonding configuration (bottom). NES, nuclear export signal; NLS, nuclear localization signal. Part b reprinted with permission from REF. 5, Annual Reviews.
ADARs and their domain structures
Vertebrates have three ADAR genes, ADAR1 (REF. 41), ADAR2 (REF. 42) and ADAR3 (REFS 43,44). ADARs have common functional domains (FIG. 1a). The dsRNA-binding domain (dsRBD) (~65 amino acids), which has an α-β-β-β-α configuration, makes direct contact with dsRNA45. The carboxy-terminal region contains the deaminase domain that forms the catalytic centre of an ADAR. Certain structural features are unique to particular ADAR members: ADAR1 contains two Z-DNA-binding domains (Zα and Zβ)46, whereas ADAR3 contains an Arg-rich single-stranded RNA (ssRNA)-binding domain (R domain) at its amino-terminal region43. The functional significance of these unique domains is not well understood. The enzymatic activities of ADAR1 (REF. 41) and ADAR2 (REF. 42) have been demonstrated, but the A-to-I RNA-editing activity of ADAR3 is yet to be shown43,44,47. ADAR1 is ubiquitously expressed41, whereas ADAR2 is most highly expressed in the brain, but is also expressed in other tissues42. ADAR3 expression is restricted to the brain43,44. In addition to these three ADARs, testis nuclear RNA-binding protein (TENR; also known as ADAD1), which is specifically expressed in testes and is required for spermatogenesis48, and TENR-like (TENRL; also known as ADAD2), which is expressed in the brain49, have sequence and domain-structure similarity to ADAR but have no deaminase activity, owing to the lack of amino acid residues that are crucial for the catalytic reaction. In Drosophila melanogaster, only a single ADAR2-like gene, dAdar, is present, and in Caenorhabditis elegans, two ADAR genes, adr1 and adr2, are known (FIG. 1a). ADARs are absent in all protozoa, yeast and plants40.
Mechanism of deamination and editing-site selectivity
During the A-to-I RNA editing process, adenosine is converted to inosine by hydrolytic deamination at the C6 position2,3 (FIG. 1b). The translation machinery reads the inosine as if it were guanosine, base-pairing it with cytosine (FIG. 1b). In this manner, A-to-I RNA editing can result in the incorporation of amino acids that are not directly encoded in the genome. X-ray crystallographic analysis of the catalytic domain of human ADAR2 revealed that His394, Glu396, Cys451 and C516 are involved in the coordination of a zinc ion and formation of the catalytic centre50. A base-flipping mechanism probably places the targeted adenosine in the catalytic pocket for the deamination reaction50. Structural studies also revealed the presence of inositol hexakisphosphate (InsP6) buried within the enzyme core, surrounded by many Arg and Lys residues and located very close to the catalytic centre. The InsP6 molecule is predicted to have a crucial role during the deamination reaction, although its exact function is currently not known50.
ADAR acts on both inter- and intramolecular dsRNAs of >20 bp in length51. More than half of all adenosines of long (>100 bp), fully base-paired dsRNAs can be edited by ADARs. By contrast, only a few adenosines of short and/or partially base-paired dsRNAs are selectively edited, perhaps indicating that the secondary structure of substrates dictates editing-site selectivity52. For example, site selectivity in the glutamate receptor GRIA2 (formerly known as GluR2 and GluRB) precursor mRNA (pre-mRNA) at the Q/R site requires an intramolecular dsRNA structure that is formed between the exonic sequence around the editing site and a downstream intronic complementary sequence termed the ECS (editing site complementary sequence)53. Owing to this requirement for the intron, A-to-I editing at this site is believed to occur in the nucleus, either before or simultaneously with splicing. Although no strict sequence specificity is required for A-to-I RNA editing, a preference for editing adenines neighbouring 5′ uridine and 3′ guanosine has been reported54. Certain sites are edited by ADAR1 only or ADAR2 only, whereas other sites are edited equally well by both54–57 (TABLES 1, 2).
Table 1.
A-to-I editing in selected mammalian protein-coding sequences and its functional consequences
| Gene | Protein | Recoding | ADAR responsible | Function | Refs |
|---|---|---|---|---|---|
| GRIA2 | GluR2 subunit of AMPA glutamate receptor | Q→R | ADAR2 | Change in Ca2+ permeability | 132 |
| R→G | ADAR1, ADAR2 | Change in receptor desensitization | 133 | ||
| GRIA3 | GluR3 subunit of AMPA glutamate receptor | R→G | ADAR1, ADAR2 | Change in receptor desensitization | 133 |
| GRIA4 | GluR4 subunit of AMPA glutamate receptor | R→G | ADAR1, ADAR2 | Change in receptor desensitization | 133 |
| GRIK1 | GluR5 subunit of kainate glutamate receptor | Q→R | ADAR1, ADAR2 | Change in Ca2+ permeability | 134 |
| GRIK2 | GluR6 subunit of kainate glutamate receptor | Q→R | ADAR1, ADAR2 | Change in Ca2+ permeability | 135, 136 |
| I→V | ADAR1, ADAR2 | ||||
| Y→C | ADAR2 | ||||
| HTR2C | Serotonin receptor 2C | I→V, M | ADAR1 | Change in G protein-coupling functions | 74 |
| N→S, G, D | ADAR1, ADAR2 | ||||
| I→V | ADAR2 | ||||
| KCNA1 | Voltage-gated K+ channel (Kv1.1) | I→V | ADAR2 | Change in channel inactivation | 75 |
| GABRA3 | GABAA receptor, subunit α3 | I→M | ADAR1, ADAR2 | Kinetics of activation and inactivation, receptor trafficking | 76 |
| BLCAP | Bladder cancer-associated protein | Y→C | ADAR1, ADAR2 | Not determined | 137, 138 |
| Q→R | ADAR1, ADAR2 | ||||
| K→R | ADAR1, ADAR2 | ||||
| CYFIP2 | Cytoplasmic FMR1-interacting protein 2 | K→E | ADAR2 | Not determined | 137 |
| FLNA | Filamin-α | Q→R | ADAR1, ADAR2 | Not determined | 137 |
| FLNB | Filamin-β | M→V | ADAR1, ADAR2 | Liver cancer progression | 139 |
| COPA | Coatomer protein complex subunit-α | I→V | ADAR2 | Suppression of liver cancer | 139 |
| IGFBP7 | Insulin-like growth factor-binding protein 7 | K→R | – | Proteolytic cleavage sensitivity | 137 |
| R→G | |||||
| AR | Androgen receptor | T→A | ADAR1, ADAR2 | Prostate cancer progression | 140 |
| Inhibition of interaction with androgen ligands | |||||
| AZIN1 | Antizyme inhibitor 1 | S→G | ADAR1 | Liver cancer progression | 141 |
| Change in affinity for antizyme | |||||
| NEIL1 | DNA repair enzyme NEI-like protein 1 | K→R | ADAR1 | Change in efficiency or specificity of damaged base removal | 142 |
| GLI1 | Glioma-associated oncogene 1 | R→G | ADAR1, ADAR2 | Increased transcription enhancement | 143 |
| RHOQ | RAS homology family member Q | N→S | – | Colorectal cancer metastasis | 144 |
| Disruption of interaction with Rap–RapGAP |
ADAR, adenosine deaminases acting on RNA; GABAA, γ-aminobutyric acid type A.
Table 2.
A-to-I editing of mammalian microRNAs (miRNAs)
| miRNA* | Position‡ | Editing levels (%)§ | ADAR responsible | Refs | |
|---|---|---|---|---|---|
| Human | Mouse | ||||
| let-7g | +7 | 10 | 0 | – | 21 |
| + 10 | 30 | 20 | ADAR2 | ||
| let-7-2-5P | +10 | 10 | 0 | – | 21 |
| miR-27a-5P | −6 | 30 | 0 | – | 21 |
| +1 | 50 | 20 | ADAR2 | ||
| +7 | 10 | 0 | – | ||
| miR-33a-5P | +10 | 30 | 0 | – | 21 |
| miR-34b-5P | +11 | – | 40 | – | 124 |
| miR-99a-5P | +1 | 20 | 20 | ADAR2 | 21 |
| miR-99b-3P | −1 | 10 | 10 | ADAR1 | 21 |
| +3 | 50 | 10 | ADAR1 | ||
| miR-122-5p | −7 | 30 | – | ADAR2 | 145 |
| miR-142-5P | +4 | – | 5 | ADAR1 | 24 |
| +5 | – | 5 | ADAR1 | ||
| miR-142-3P | +4 (+40)∥ | – | 10 | ADAR1, ADAR2 | 24 |
| miR-151-3P | −1 | – | 10 | ADAR1 | 22 |
| +3 | 40 | 30 | ADAR1 | ||
| miR-153-1-3P | +7 | 10 | 0 | – | 21 |
| miR-153-2-3P | +7 | 30 | 0 | – | 21 |
| miR-197-3P | −34 | 30 | – | – | 21 |
| miR-203-3P | +21 | 60 | – | ADAR2 | 21 |
| miR-214-3p | +6 | 10 | – | ADAR2 | 145 |
| miR-376a-1-5P | +3 (+4)∥ | 50 | – | ADAR2 | 23 |
| miR-376a-1-3P | +6 (+44)∥ | 40 | 0 | ADAR1 | 23 |
| miR-376a-2-5P | +4 | 90 | 50 | ADAR2 | 23 |
| miR-376a-2-3P | +6 (+44)∥ | 100 | 0 | ADAR1 | 23 |
| miR-376b-3p | +6 (+44)∥ | 95 | 50 | ADAR1 | 23 |
| miR-379-5P | +5 | 60 | 20 | ADAR2 | 21 |
| miR-381-3P | +4 | 6 | 13 | – | 115, 116 |
| miR-411-5P | +5 | 80 | 60 | ADAR1 | 21 |
| miR-423 + 5P | −4 | 40 | 20 | ADAR1 | 21 |
| miR-497-5P | +2 | 6 | 10 | – | 115, 116 |
| miR-532-5P | +15 | 10 | 0 | – | 21 |
| miR-589-3P | +6 | 70 | – | – | 115 |
| miR-607-3P | +6 | 70 | – | – | 21 |
| +17 | 80 | – | – | ||
| +20 | 80 | – | – | ||
| miR-652-5P | −10 | 40 | 0 | – | 21 |
| miR-3099-3P | +7 | – | 80 | – | 124 |
ADAR, adenosine deaminase acting on RNA.
Only editing sites verified by sequencing of specific primary mRNAs (pri-miRNAs) or detected independently more than once by deep sequencing of mature miRNAs are listed.
The 5′ end of the human mature miRNA sequence registered at the miRBase database is counted as +1.
Editing levels indicate fractions of edited miRNAs over edited and unedited miRNAs. The highest editing level reported either in total brain tissue, in sub-regions of the brain or in cultured cells is presented.
Alternative numbering used in certain references is indicated in parentheses.
Regulation of ADAR expression and localization
Transcription from separate promoters generates two isoforms of ADAR1, a full-length, interferon-inducible ADAR1p150 and a shorter and constitutively expressed ADAR1p110, which lacks the N-terminal portion of the protein, including the Zα domain58,59 (FIG. 1a). ADAR2 expression is positively regulated by the transcription activator CREB (cyclic adenosine monophosphate response element-binding protein) in the brain60, and by c-Jun N-terminal kinase 1 (JNK1; also known as MAPK8) in pancreatic β-cells61. Interestingly, CREB also suppresses transcription from the ADAR1p110 promoter in metastatic melanomas62. Homodimerization is required for the A-to-I RNA-editing activities of ADAR1 and ADAR2 (REFS 63–65). The third dsRBD of ADAR1 and the first dsRBD of ADAR2 are required for their homodimerization26,64. ADAR3 is unable to homodimerize, which may underlie its lack of A-to-I RNA editing activity63.
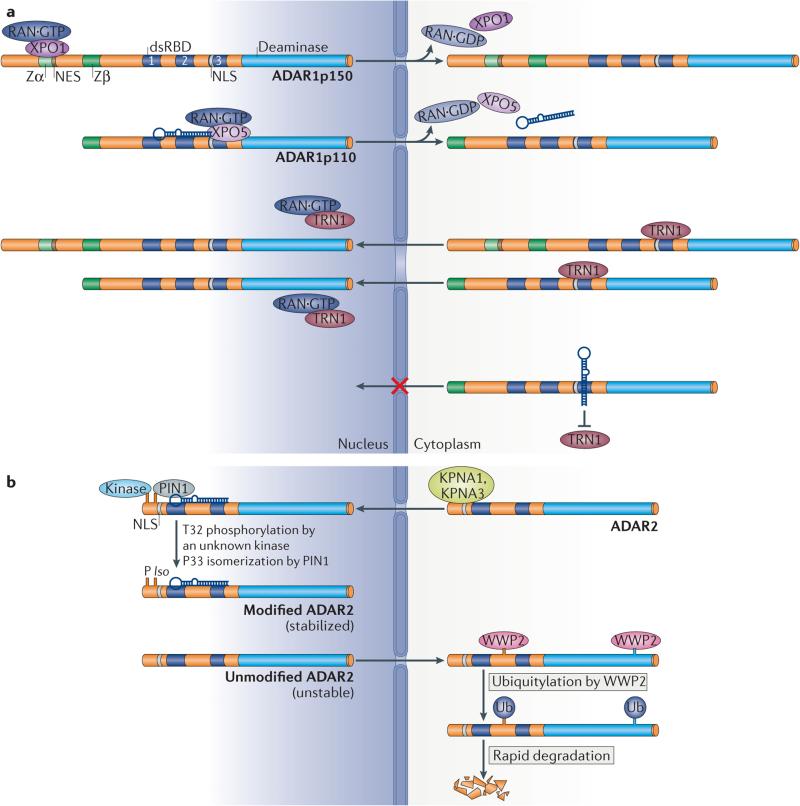
Both ADAR1p150 and ADAR1p110 shuttle between the nucleus and the cytoplasm66–68 (FIG. 2a). Binding of the nuclear export factor exportin 1 (XPO1; also known as CRM1) to the nuclear export signal (NES) located within the Zα domain, together with RAN·GTP, regulates nuclear export of ADAR1p150 (REF. 69). The nuclear localization signal (NLS) located in the third dsRBD is responsible for localization of ADAR1 in the nucleus and nucleolus66,68. Nuclear export of ADAR1p110 is mediated by XPO5–RAN·GTP and is regulated by dsRNA binding to the dsRBDs, whereas nuclear import of ADAR1p110 is mediated by binding of transportin 1 (TRN1) to the third dsRBD, which is inhibited by binding of dsRNA68,70. The predominantly nucleolar localization of ADAR2 is regulated by the binding of karyopherin subunit α1 (KPNA1) and KPNA3 (REF. 71) to an Arg-rich NLS in the N-terminal region66,71,72 (FIG. 2b). Post-translational modification regulates the nuclear localization and stability of ADAR2. Phosphorylation of Thr32 activates ADAR2 interaction with the prolyl-isomerase PIN1 in a dsRNA-binding dependent manner, which isomerizes Pro33 and posi tively controls the nuclear localization and stability of ADAR2. By contrast, the E3 ubiquitin ligase WWP2 promotes the degradation of ADAR2 in the cytoplasm73 (FIG. 2b).
Figure 2. Cellular localization of adenosine deaminases acting on RNA 1 (ADAR1) and ADAR2.
a | Exportin 1 (XPO1) binds to the nuclear export signal (NES) located within the Zα domain of ADAR1p150 and regulates its nuclear export together with RAN·GTP. Nuclear export of ADAR1p110 is mediated by XPO5–RAN·GTP and regulated by double-stranded RNA (dsRNA) binding to its dsRNA-binding domains (dsRBDs). The nuclear localization signal (NLS) located in dsRBD3 is responsible for localization of both ADAR1p150 and p110 in the nucleus and nucleolus. Nuclear import of ADAR1p110 is mediated by binding of transportin 1 (TRN1) to dsRBD3, which is inhibited by binding of dsRNA. b | The nuclear and nucleolar localization of ADAR2 is regulated by binding of karyopherin subunit α1 (KPNA1) and KPNA3 to an NLS located in the amino-terminal region. Phosphorylation of Thr32 by a currently unknown kinase enables interaction of ADAR2 with the prolyl-isomerase PIN1 in a dsRNA-binding-dependent manner, which isomerizes Pro33 and positively controls the nuclear localization and stability of ADAR2. The E3 ubiquitin ligase WWP2 promotes rapid degradation of ADAR2 in the cytoplasm, which is why ADAR2 is usually not detected in the cytoplasm. Ub, ubiquitin.
Editing of protein-coding sequences
Transcripts of a relatively small number of genes are edited within their coding regions; this is termed recoding-type editing28,29. These include physiologically important mammalian genes, such as those encoding the glutamate receptor subunit GluR2 (REF. 53), the G protein-coupled serotonin receptor 5-HT2CR74, the potassium channel Kv1.1 (REF. 75) and the α3 subunit of GABAA (γ-aminobutyric acid type A) receptor (GABRA3)76. Recoding-type editing often dramatically alters protein functions28,29 (TABLE 1). For instance, editing of the Q/R site (recoding of Gln to Arg) located in the channel-pore-loop domain of GluR2 results in a channel that is impermeable to Ca2+ (REF. 53), whereas editing of five adenosines located in the second intracellular loop domain of 5-HT2CR changes the G protein-coupling functions of the receptor74. Editing of the I/V site of Kv1.1 substantially reduces the inactivation rate of this voltage-gated channel75, and the trafficking and proper localization of GABRA3 are reduced by editing of its I/M site76. Recent transcriptome deep sequencing and global screening for editing sites have revealed that the recoding type of A-to-I editing also occurs in genes other than those encoding neurotransmitter receptors and ion channels. However, recoding-type editing to any significant degree (>20% editing) is rare; the functions of ~80 mammalian genes in total might be regulated by A-to-I editing7,13,15,17–19.
Deficiency in A-to-I RNA editing
Adar2-null mutant mice die several weeks after birth following frequent epileptic seizures, which are caused by neuronal death owing to excess influx of Ca2+. This is a result of severe deficiency in the editing of an almost exclusive ADAR2 target, GRIA2 pre-mRNA, at its Q/R site56. Deficient editing of the Q/R site seems to underlie the loss of motor neurons in patients with sporadic amyotrophic lateral sclerosis (ALS)77, and death of motor neurons and other symptoms of ALS are indeed detected in motor neuron-specific knockouts of Adar2 in mice78. Furthermore, deficiency in GluR2 Q/R-site editing and the consequent excess Ca2+ influx have been proposed to activate the kinase AKT and lead to glioblastoma proliferation79, as well as neuronal death in forebrain ischemia60. The inactivation of ADAR1 in mice results in an embryonic-lethal phenotype that is characterized by defective erythropoiesis, aberrant activation of interferon signalling and widespread apoptosis55,57,80–82. ADAR1 seems to protect organisms from the deleterious effects of interferon activation, which is relevant to many human pathological processes, such as chronic inflammation and autoimmune disorders80. The embryonic lethal phenotype of Adar1-null mice can be rescued by the simultaneous inactivation of mitochondrial antiviral signalling adaptor protein (MAVS)83 or melanoma differentiation-associated protein 5 (MDA5; also known as IFIH1)81, the upstream genes involved in the interferon activation pathway. This indicates the relevance of ADAR1 function in the regulation of the interferon pathway to the embryonic-lethal phenotype of Adar1-null mice81,83. Endogenous long dsRNAs produced from inverted retrotransposon repeats of the LINE (long interspersed nuclear elements) and SINE (short interspersed nuclear elements) families and located in the 3′ untranslated regions (UTRs) of several genes, such as Krüppel-like factor 1 (Klf1), optineurin (Optn), and OPA-interacting protein 5 homologue (Oip5), have been proposed as crucial ADAR1-substrate RNAs. Failure to edit these dsRNAs may lead to the activation of cytosolic dsRNA sensing by MDA5 and of MAVS-mediated interferon signalling in Adar1-null mouse embryos81.
Dysfunction of A-to-I editing causes human diseases
Nine ADAR1 mutations were found in a subset of patients with Aicardi–Goutières syndrome (AGS)84. AGS is an autosomal-recessive inflammatory disorder that affects the brain and skin and is characterized by an aberrant immune response and increased interferon-α expression84. Failed editing of certain endogenous dsRNAs such as Alu by the mutant ADAR1, and the consequent activation of interferon signalling, seem to underlie the pathogenesis of AGS83,85. Indeed, the overproduction of interferons is detected in the brains and spinal cords of conditional Adar1-null mice85. A large number of ADAR1 mutations (>130) are also associated with dyschromatosis symmetrica hereditaria (DSH), which is an autosomal-dominant disorder that is mainly found in Asian individuals and is characterized by hypo- and hyper-pigmentation of the skin86. ADAR1 haploinsufficiency, as well as dominant-negative effects of the mutant ADAR1, are likely to underlie the pathogenesis of DSH65,86.
The 5-HT2CR pre-mRNA is edited at five sites by ADAR1 and ADAR2 (REFS 55–57), and the combinatorial editing of these sites results in the expression of 24 protein isoforms. Mutant mouse lines expressing either the unedited or the fully edited forms of 5-HT2CR have been established87,88. Mutant mice expressing fully edited 5-HT2CR had significantly decreased fat mass and increased energy expenditure, resulting from hyperactivation of the sympathetic nervous system, suggesting that 5-HT2CR mRNA editing has a regulatory role in lipolysis and metabolism87. The same 5-HT2CR mRNA-editing mutant mice exhibit a phenotype similar to Prader–Willi syndrome (PWS), which is characterized by obesity and a range of developmental abnormalities88. However, the mutant mice lacked certain PWS-associated symptoms such as obesity, and thus the relevance of over-editing of 5-HT2CR mRNA to PWS needs further investigation. The 5-HT2CR mRNA-editing mutant mice (both over- and under-edited) exhibit anti-depressive and exaggerated anxiety- like behaviours, indicating a relevance of 5-HT2CR mRNA editing to psychiatric disorders89. An association of altered editing patterns of 5-HT2CR with several psychiatric disorders, including anxiety, depression, bipolar disorder, schizophrenia and suicide, as well as with autism, has been reported, although the findings have often been inconsistent owing to the difficulty of analysing post-mortem human brain samples28,29,90.
Alu dsRNA editing and its implications
Recent advances in sequencing technology, as well as in bioinformatics analyses of sequence databases, have made it possible to globally screen for previously unknown A-to-I editing sites in normal tissues and in various cancers. This has resulted in the identification of many millions of new editing sites in human transcripts6–19. Surprisingly, almost all of these new sites reside in introns and 3′ UTRs that harbour Alu dsRNAs.What is the fate of RNAs carrying highly edited Alu repeats? Are there any proteins that specifically recognize and interact with these highly edited RNAs? Are there any functions for highly edited Alu dsRNAs?
Exonization of intronic Alu sequences
An inosine is recognized by the splicing machinery as a guanosine. Thus, A-to-I editing of Alu sequences can generate splice donor and acceptor sites. For instance, AU-to-IU editing will generate a sequence that could be recognized as the canonical 5′ splice donor site GU (IU as GU), and AA-to-AI editing will generate a sequence that could be recognized as the 3′ splice acceptor site AG (AI as AG). Self-editing by ADAR2 of an intronic sequence of its own pre-mRNA indeed results in the creation of an alternative 3′ splice acceptor site and the suppression of ADAR2 expression, thereby functioning as a negative autoregulatory mechanism91,92. A-to-I editing-mediated exonization of Alu sequences (FIG. 3a) has been reported for several genes, including G protein-coupled receptor 107 (REF. 6), nuclear prelamin A93 and seryl-tRNA synthetase19.
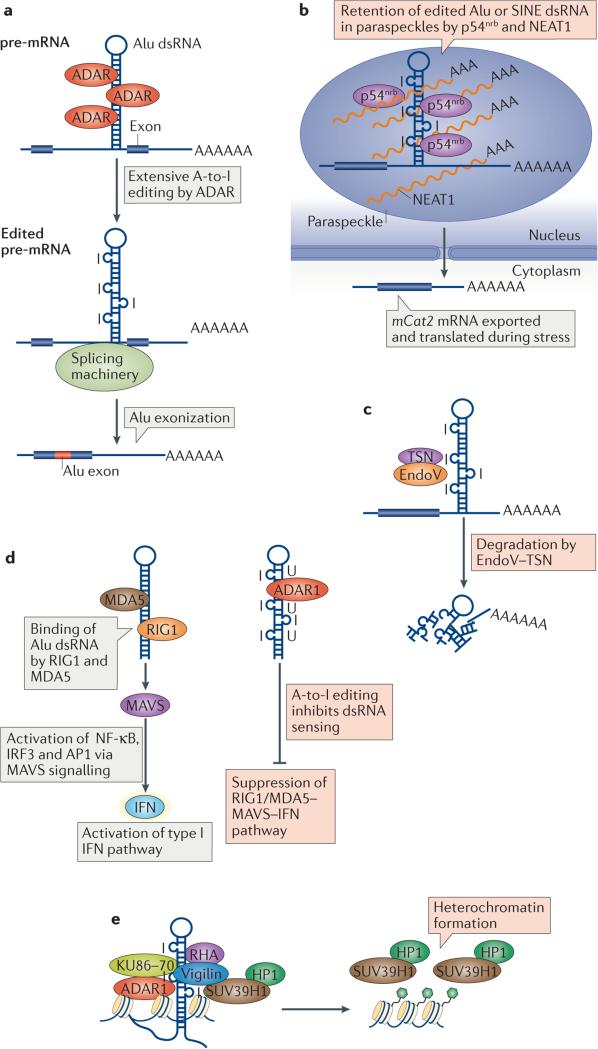
Figure 3. Editing of Alu double-stranded RNAs (dsRNAs) and its consequences.
a | Genome-wide inverted Alu repeats in introns and in 3′ untranslated regions (UTRs) form intramolecular RNA duplexes (Alu dsRNAs), which are edited by adenosine deaminases acting on RNA (ADARs). Inosines at intronic Alu dsRNAs are recognized by the splicing machinery as guanosines, thus effectively creating new splice sites, which results in the inclusion of intronic Alu sequences in the mature mRNAs (Alu exonization). b | Extensively edited Alu dsRNAs are retained in nuclear paraspeckles by a mechanism involving p54nrb and the long non-coding RNA nuclear paraspeckle assembly transcript 1(NEAT1).The formation of paraspeckles is absolutely dependent on NEAT1.p54nrb, and perhaps also NEAT1, bind specifically to inosine-containing RNAs such as extensively edited Alu dsRNA. Under certain conditions, such as stress, the paraspeckle-trapped RNA may be released into the cytoplasm for translation, as seen with the mouse Ctn RNA. Editing of short interspersed nuclear element (SINE) dsRNA within the 3′ UTR of the Ctn RNA leads to its nuclear retention. When cells are placed under stress, Ctn RNA is cleaved and polyadenylated at an alternative site, resulting in the loss of the edited SINE sequences and release of the mRNA from the nucleus as the protein-coding mCat2 (cationic amino acid transporter 2) mRNA. c | Extensively edited Alu dsRNAs may be degraded by endonuclease V (EndoV) together with Tudor-SN (TSN), thereby controlling the expression levels of genes harbouring Alu repeats. d | Extensively edited Alu dsRNA containing multiple and consecutive I·U mismatched wobble pairs (I·U-dsRNA) may suppress the interferon (IFN) signalling pathway, which would otherwise be activated by unedited Alu dsRNAs. Unedited long dsRNAs (viral and cellular) are potent inducers of IFN signalling. I·U-dsRNA may competitively inhibit the binding of dsRNA to retinoic acid-inducible gene 1(RIG1)or melanoma differentiation-associated protein 5 (MDA5), both of which are cytosolic sensors for dsRNA and upstream regulators of the mitochondrial antiviral signalling adaptor protein (MAVS)-mediated IFN activation pathway. e | Extensively edited Alu dsRNAs containing multiple inosines may contribute to heterochromatin formation and gene silencing. Vigilin binds to RNA containing multiple inosines, such as extensively edited Alu dsRNAs. Vigilin has been shown to form a complex with ADAR1, RNA helicase A (RHA), and 86 kDa subunit of Ku antigen (KU86)–KU70, which recruits the histone methyltransferase SUV39H1. SUV39H1 catalyses the methylation of H3 Lys9 (H3K9me), an epigenetic mark that is recognized by heterochromatin protein 1 (HP1), leading to the formation of heterochromatin and to gene silencing. AP1, activator protein-1; IRF3, interferon regulatory factor 3; NF-κB, nuclear factor-κB; pre-mRNA, precursor mRNA. Part a adapted from REF. 146, Nature Publishing Group.
Retention of edited Alu dsRNAs in paraspeckles
The inosine-specific RNA-binding protein p54nrb was proposed to mediate the specific retention in nuclear paraspeckles of mRNAs containing extensively edited Alu dsRNA94 (FIG. 3b). At least 333 human genes contain Alu dsRNA sequences in their 3′ UTRs95. A-to-I editing and p54nrb-dependent nuclear retention of transcripts of one such human gene, nicolin 1, has been demonstrated95. Another example is the mouse Ctn gene96. Ctn transcripts contain a long dsRNA formed of inverted repeats of mouse SINEs in the 3′ UTR96. Under conditions of stress, Ctn RNA is post-transcriptionally cleaved and polyadenylated at an alternative site, resulting in the loss of the edited SINE sequences and the release of the protein-coding mCat2 (also known as Slc7a2) mRNA to the cytoplasm, where it is translated into cationic amino acid transporter 2 proteins96 (FIG. 3b). More recent studies have revealed that the nuclear paraspeckle assembly transcript 1 (NEAT1), a long non-coding RNA, is also required for the formation of nuclear paraspeckles and for the retention mechanism. Human embryonic stem cells (ES cells) lack NEAT1 and do not form paraspeckles. Accordingly, LIN28 mRNAs containing extensively edited Alu dsRNA in their 3′ UTRs can be detected in the cytoplasm despite the presence of p54nrb, indicating that both p54nrb and NEAT1 are required for the retention mechanism97.
Degradation of edited Alu dsRNAs by endonuclease V
A ribonuclease activity that specifically cleaves both RNA strands of a dsRNA that contains multiple I·U base pairs (that is, extensively edited Alu dsRNA) has been reported98. Endonuclease V (EndoV) was recently identified as this ribonuclease99. Tudor staphylococcal nuclease (Tudor-SN) seems to promote the activity of EndoV as a cofactor100. Thus, A-to-I editing of Alu dsRNAs may lead to degradation by EndoV together with Tudor-SN, which in turn might control the expression levels of genes harbouring Alu dsRNA (FIG. 3c). A-to-I hyperedited RNAs are easily detected in steady-state mRNA pools, and thus their degradation is not constitutive and must be regulated, perhaps by compartmentalization of EndoV and Tudor-SN: cytoplasmic as well as nucleolar localization of EndoV has been reported99, whereas Tudor-SN localizes to stress granules in stress conditions101,102. Interestingly, ADAR1p150 seems to bind to extensively edited dsRNA via its Zα domain and localizes together with Tudor-SN to stress granules in stress conditions, although the roles of ADAR1p150 and Tudor-SN, as well as the fate of the extensively edited dsRNA in stress granules, remain to be established102,103. Thus, EndoV–Tudor-SN could regulate the expression of genes containing extensively edited Alu dsRNA in special circumstances, for instance, during stress or viral infection.
Suppression of the interferon response
The interferon signalling pathway is activated in response to infection by pathogens such as viruses and bacteria, as well as by long dsRNAs such as synthetic poly(I:C). A biological function for extensively edited dsRNAs in the suppression of the interferon signalling pathway has been proposed, using a synthetic dsRNA that contains multiple and consecutive IU-mismatched wobble base pairs (IU-dsRNA)104. IU-dsRNA, which resembles extensively edited Alu dsRNA, forms a unique configuration104 and inhibits the activation of interferon signalling and of interferon-stimulated genes (ISGs) induced by poly(I:C) dsRNAs in cultured cell lines. The IU-dsRNA was proposed to competitively inhibit binding to poly(I:C) dsRNA by retinoic acid-inducible gene 1 (RIG1; also known as DDX58) or MDA5, both of which are cytosolic sensors for dsRNA and upstream regulators of the MAVS-mediated interferon activation pathway104 (FIG. 3d). The same IU-dsRNA was shown to suppress the interferon pathway that is aberrantly activated in Adar1-null mouse embryonic fibroblasts83. Recent studies identified LINE and SINE dsRNAs present in 3′ UTRs of Klf1, Optn, and Oip5 as candidate sources of endogenous IU-dsRNAs81. Failure in hyper-editing these dsRNAs by ADAR1 may lead to their sensing by RIG1 and MDA5, and consequent activation of MAVS-mediated interferon signalling, which is perhaps relevant to Adar1-null mouse phenotypes, as well as to the pathology of AGS81.
Heterochromatin formation and gene silencing
The involvement of RNAi and its components (such as endogenous short interfering RNAs (endo-siRNAs) and PIWI-interacting RNAs) in the establishment of heterochromatin and in silencing the expression of repetitive sequences and transposons is well known in plants, fission yeast and various other eukaryotes. However, it has been debated whether similar mechanisms operate in mammalian cells105. Interestingly, vigilin, which is a multi-KH-domain protein, binds to inosine-containing RNAs such as extensively edited Alu dsRNA and forms a complex with ADAR1, the KU86 (86 kDa subunit of Ku antigen; also known as XRCC5)–KU70 heterodimer (which is involved in the repair of DNA double-strand breaks), ATP-dependent RNA helicase A (RHA) and heterochromatin protein 1 (HP1)106. The D. melanogaster homologue of vigilin, Ddp1, localizes to heterochromatin and is essential for gene silencing in flies. Vigilin also interacts with SUV39H1, which methylates histone H3 on Lys9 (H3K9me). The H3K9me epigenetic mark provides a binding site for HP1, which mediates the formation of heterochromatin and gene silencing107 (FIG. 3e). Association of Alu elements with H3K9me and their involvement in heterochromatin formation have been reported108. These findings suggest a possible contribution of extensively edited Alu dsRNAs to heterochromatin formation and gene silencing of a region enriched in Alu sequences.
By contrast, antagonistic effects of dAdar (which is the only known D. melanogaster ADAR gene) on hetero chromatic gene silencing of Hoppel transposable elements were reported. It was proposed that dAdar edits a long dsRNA generated from Hoppel loci, thereby preventing Dicer from processing it into endo-siRNAs, which are required for RNAi-mediated hetero chromatin formation and gene silencing109. However, the involvement of Dicer in this transposon-silencing mechanism and the generation of endo-siRNAs from Hoppel elements remain to be shown.
Editing of miRNAs and its consequences
Primary miRNA (pri-miRNA) transcripts fold to form dsRNA (hairpin) structures, which are processed in the nucleus into precursor-miRNAs (pre-miRNAs) of ~70 nt in length by the RNase III protein Drosha, in complex with the pri-miRNA recognition factor DGCR8 (FIG. 4a). Pre-mRNAs are then exported to the cytoplasm, where they are processed further by another RNase III protein, Dicer, in complex with TAR RNA-binding protein (TRBP; also known as TARBP2) to generate double-stranded, mature miRNAs of ~22 nt in length. Mature miRNAs are loaded onto Argonaute (AGO) proteins and together form the core of the RNA-induced silencing complex (RISC). The miRNA guide strand (the functional strand retained by AGO) directs the RISC to the target mRNAs, causing translation repression or mRNA decay. Nucleotides 2–8 of the guide strand, known as the seed sequence, are particu larly important in directing the selection of mRNA targets110. miRNA-mediated gene silencing has crucial roles in many biological processes, such as tissue differentiation, cell proliferation, embryonic development and apoptosis, and its misregulation can result in human diseases111,112. Certain pri-miRNAs undergo A-to-I editing (TABLE 2), which affects their biogenesis and function.
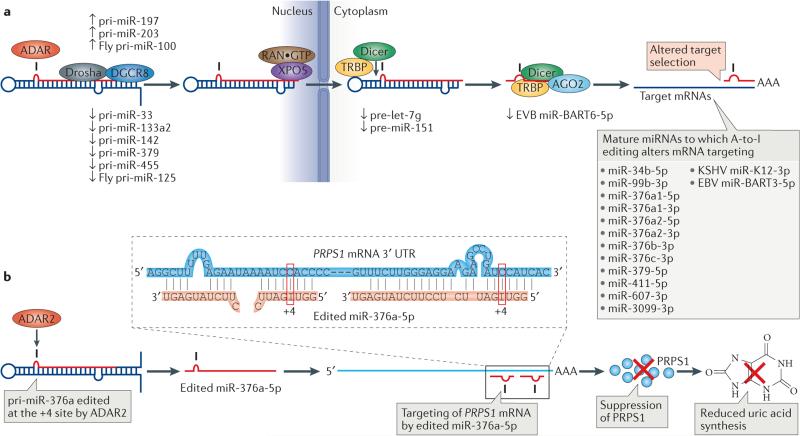
Figure 4. Regulation of microRNA (miRNA) processing, expression and selectivity by RNA editing.
a | Primary miRNAs (pre-miRNAs) are processed by the Drosha–DGCR8 complex into precursor miRNAs (pri-miRNAs) in the nucleus, exported to the cytoplasm by exportin 5 (XPO5)–RAN·GTP and processed by the Dicer–TAR RNA-binding protein (TRBP) complex into mature miRNA duplexes. One strand of this duplex is then loaded onto the RNA-induced silencing complex (RISC), which results in the degradation or the inhibition of translation of target mRNAs. Editing can affect any of the miRNA biogenesis steps, including Drosha cleavage, Dicer cleavage and RISC loading, as well as miRNA target selection. Known examples of miRNA editing and their consequences are shown. b | Silencing of phosphoribosyl pyrophosphate synthetase 1 (PRPS1) by miR-376a-5p edited at the +4 site by ADAR2 (adenosine deaminases acting on RNA2) and the consequent suppression of uric acid synthesis. A single A-to-I nucleotide change in the seed sequence of miR-376a-5p results in redirection of target gene selection. One of those genes, specifically targeted by the edited miR-376a-5p, is PRPS1, which encodes an essential enzyme involved in purine metabolism and the uric acid synthesis pathway. Repression of PRPS1 by the edited miR-376a-5p results in reduced expression of uric acid in certain tissues, such as brain, in which uric acid levels need to be tightly regulated. The 3′ untranslated region (UTR) of PRPS1 mRNA has two target sites for the edited miR-376a-5p (inset). AGO2, Argonaute 2; EBV, Epstein–Barr virus; KSHV, Kaposi sarcoma-associated herpes virus.
Approximately 20% of pri-miRNAs are edited in the adult human brain21. In addition, editing of several pri-miRNAs encoded by DNA viruses (Epstein–Barr virus (EBV) and Kaposi sarcoma-associated herpes virus HHV-8) has been reported20,113,114. It was anticipated that next-generation sequencing of small RNAs would reveal many new A-to-I editing sites in miRNAs. However, only a small number of new sites that are edited at significant frequency (>5% editing) were identified in mature miRNAs, indicating that the expression of edited mature miRNAs is relatively rare115,116. This may be because editing of pri-miRNAs results mostly in inhibition of miRNA biogenesis.
Suppression of miRNA biogenesis
ADARs can suppress miRNA maturation at different processing stages by editing-dependent and editing-independent mechanisms. The recognition of pri-miRNA hairpin structures by the Drosha–DGCR8 complex can be affected by A-to-I editing, as was first demonstrated for pri-miR-142. Editing of pri-miR-142 at the +4 and +5 positions (counting from the 5′ end of the mature miRNA sequence) by ADAR1 and ADAR2 inhibits its cleavage by Drosha–DGCR8 (REF. 24) (FIG. 4a). As expected, the expression of miR-142-5p is substantially higher in the spleen of Adar1- and of Adar2-null mice compared with wild-type mice24. Although editing of pri-miR-142 prevents its processing to pre-miR-142, no accumulation of edited pri-miR-142 was detected in HEK293 cells ectopically overexpressing ADAR1 and ADAR2, owing to their degradation by EndoV–Tudor-SN99. Degradation of edited fly pri-miR-125, presumably by EndoV–Tudor-SN, was also reported117. Thus, A-to-I editing-dependent degradation of pri-miRNAs could be considered to be a control mechanism of miRNA biogenesis and activity. Inhibition of cleavage by Drosha was also reported for pri-miR-33, pri-miR-133a2 and pri-miR-379 (REF. 21). By contrast, editing increases Drosha cleavage — although very slightly — for pri-miR-197, and substantially for pri-miR-203 (REF. 21), as well as for D. melanogaster pri-miR-100 (REF. 117). Editing of pri-miR-455 at the +2 and +17 positions by ADAR1 and suppression of the Drosha cleavage step were also reported in human melanocytes62. Suppression of ADAR1 expression, and thus reduced editing of these sites, results in increased expression of miR-455-5p and suppression of its target, the tumour suppressor cytoplasmic polyadenylation element-binding protein 1 (CPEB1), which could be relevant to metastasis of melanomas62.
ADAR1 edits the −1 and +3 positions of pri-miR-151 in certain tissues, such as amygdala, cerebral cortex and lung, which results in a complete block of pre-miR-151 cleavage by Dicer–TRBP and inhibition of miR-151-3p expression22 (FIG. 4a). Binding of the Dicer–TRBP complex to unedited and to edited pre-miR-151 are comparable, indicating that Dicer cleavage, not binding, is inhibited by editing. Partial prevention of pre-let-7g cleavage by Dicer due to editing of the +4 position by ADAR2 was also reported21.
Loading of miRNA onto AGO2-containing RISC can also be inhibited by editing. ADAR1 edits the +20 position of pri-miR-BART6, a miRNA encoded by the EBV, which results in the inhibition of miR-BART6-5p loading onto RISC and thus of its function; this, in turn, affects the latency state of EBV20 (FIG. 4a). When not edited, miR-BART6-5p specifically targets the human Dicer mRNA, which is evidence that EBV has developed a unique strategy to suppress host RNAi20. Thus, editing of pri-miR-BART6 by ADAR1 could have evolved as a human counteractive strategy against the suppression of RNAi by EBV.
Owing to its dsRNA-binding capacity, ADAR2 seems to sequester and thus inhibit the processing of pri-miR-376a1 and pri-miR-376a2, most probably at the Drosha cleavage step118. Editing-independent suppression by ADAR2 of miRNA processing and its tumour-promoting role in glioblastoma were also proposed119. Expression of miRNAs is indeed altered in Adar2-null mouse embryos, most probably through an editing-independent mechanism120. ADAR1 also suppresses the expression of many miRNAs, including the stem cell self-renewal-promoting miR-302 family of miRNAs, in an RNA editing-independent manner, which is essential for neural differentiation of human ES cells121. Editing-independent suppression of miR-222 expression by ADAR1 and the consequent upregulation of ICAM1, and the relevance of this to melanoma immune resistance, have also been reported122. Finally, global screening for ADAR1-binding sites suggested that ADAR1 might compete with DGCR8 for binding to many pri-miRNAs123. Thus, a larger subset of miRNAs than those edited might be affected by both ADAR1 and ADAR2, independently of their catalytic functions.
Alteration of miRNA target specificity
In some cases pri-miRNA editing does not inhibit miRNA maturation, leading to the expression of edited mature miRNAs that can be loaded onto AGO2–RISC. However, as editing — even at a single site — can alter the base pairing properties of the miRNA, it can also affect recognition of its target mRNA, especially if editing takes place within the seed sequence of the miRNA.
Members of the miR-376 cluster are transcribed as one transcript and processed to individual pre-miRNAs23. In miR-376a, at least two main sites are edited, corresponding to +4 (located in the 5p seed sequence) and +44 (or alternatively numbered as the +6 site; located in the 3p seed sequence). ADAR2 edits the +4 site, whereas ADAR1 edits the +44 site. Interestingly, edited miR-376a targets an almost completely different set of genes than unedited miR-376a. One of the targets specific to the miR-376a-5p edited at the +4 site is phosphoribosyl pyrophosphate synthetase 1 (PRPS1), which is an essential enzyme involved in purine metabolism and in the uric acid synthesis pathway (FIG. 4b). A human disorder characterized by gout and neuro-developmental impairment with hyperuricaemia is caused by substantially increased PRPS1 expression, indicating the importance of tightly regulated expression of this enzyme. Adar2-null mice have both PRPS1 and uric acid levels upregulated approximately twofold in the cortex. No increase in PRPS1 or uric acid levels was detected in the livers of Adar2-null mice, consistent with the fact that the +4 site is barely edited in wild-type mouse liver23. This proves that editing of the +4 site of pri-miR-376a by ADAR2 tightly regulates uric acid levels in a tissue-specific manner by redirecting miRNA target specificity23 (FIG. 4b). Editing of miR-376 cluster miRNAs increases from embryonic day 19 (E19) in mouse embryos, suggesting that it may be important for embryo development124. Furthermore, the differential silencing of RAS-related protein RAP2A and of the E3 ubiquitin ligase AMFR by unedited and edited miR-376a-5p, respectively, were reported to affect glioblastoma metastasis125. Seed sequence editing of several other mature miRNAs has also been reported21,114,124 and is likely to alter target gene specificity (FIG. 4a). Silencing of the tumour suppressor Dice1 (deleted in cancer 1; also known as Ints6) by the EBV-encoded miR-BART3-5p is antagonized owing to editing of pri-miR-BART3 at the +5 site (seed sequence) by ADAR1 (REF. 113).
Regulation of RNAi by ADARs
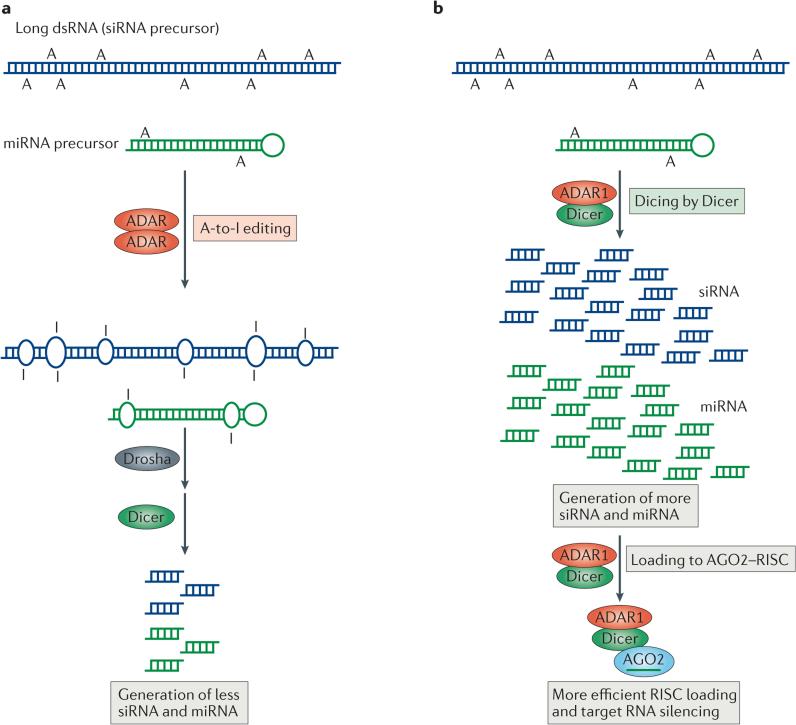
Both A-to-I RNA-editing and RNAi pathways act on dsRNA. It has been speculated that the A-to-I editing and RNAi may interact with each other by competing for shared dsRNA substrates126. Indeed, the two processes antagonistically interact (FIG. 5a) (see the discussion on miRNA editing in the previous section and the discussion below). Conversely, a stimulative interaction between RNA editing and RNAi machinery proteins exists, as ADAR1 forms a complex with Dicer and promotes its activity (FIG. 5b).
Figure 5. Regulation of RNA interference (RNAi) by adenosine deaminases acting on RNA (ADARs).
Two different types of interaction between RNA-editing and RNAi pathways are known, one antagonistic and the other stimulative. a | In antagonistic interactions, ADAR–ADAR homodimers edit long double-stranded RNA (dsRNA) and certain microRNA (miRNA) precursors. Editing changes the dsRNA structure and makes it less accessible to Drosha and/or Dicer, which consequently decreases the efficacy of RNAi by reducing the production of short interfering RNAs (siRNAs) and miRNAs. b | In the case of stimulative interactions, ADAR1, as part of a Dicer–ADAR1 heterodimer, promotes RNAi by increasing the Dicer cleavage reaction rate, thereby generating more siRNAs and miRNAs and enhancing RISC (RNA-induced silencing complex) loading and target mRNA silencing. AGO2, Argonaute 2. Figure adapted with permission from REF. 26, Elsevier.
Suppression of RNAi
Analyses of ADAR-null C. elegans strains have revealed the presence of an antagonistic interaction between A-to-I RNA-editing and RNAi pathways127,128. C. elegans strains that contain homozygous deletions of both adr1 and adr2 genes have a chemotaxis-defective phenotype128, which is rescued by crossing these worms with RNAi-defective worms, indicating that enhanced RNAi leading to the suppression of a chemotaxis gene underlies the chemotaxis-defective phenotype of ADAR-null worms127,128. A-to-I RNA editing of a dsRNA made of a chemotaxis gene transcript could perhaps inhibit its silencing by RNAi127,128 (FIG. 5a). However, details of the putative interaction between RNA editing and RNAi, as well as the identity of the chemotaxis gene, remain unknown.
It has recently been reported that endo-siRNAs derived from loci enriched with inverted repeats and transposons are dramatically upregulated in ADAR-null mutant worms. A-to-I RNA editing of dsRNA regions of transcripts derived from these loci seems to inhibit their entry into the RNAi silencing pathway and to consequently suppress synthesis of endo-siRNAs from these transcripts129. A separate study suggested that biogenesis of not only endo-siRNAs, but also of miRNAs, is significantly affected in ADAR-null mutant worms130 (FIG. 4a). In the fruit fly white+ eye reporter system, antagonistic effects on RNAi were observed by the introduction of human ADAR1p150, but not of ADAR1p110 or ADAR2. Interestingly, it is not the A-to-I editing but the dsRNA-binding activity of ADAR1p150 that seems to be responsible for its RNAi-antagonistic function118.
ADAR1 interacts with Dicer and promotes its activity
Analysis of proteins that interact with epitope-tagged Dicer or ADAR1 has revealed a robust and direct interaction between Dicer and ADAR1 (REF. 26). Although both ADAR1p150 and ADAR1p110 can form the complex in vitro, ADAR1p110 seems to be the true partner of Dicer in vivo25. ADAR1 distinguishes between its functions in RNA editing and in RNAi by the formation of two different complexes: ADAR1–ADAR1 homodimers in the nucleus for RNA editing, and Dicer–ADAR1 heterodimers in the cytoplasm. ADAR1 in complex with Dicer has no A-to-I RNA-editing activity, perhaps reflecting the fact that homodimerization is required for its A-to-I RNA-editing activity63. In addition to Dicer, ADAR1 interacts indirectly with AGO2 through its interaction with Dicer, resulting in the formation of Dicer–ADAR1–AGO2 complexes of ~450 kDa in size26. Dicer contains a DEAD-box RNA helicase domain in its N-terminal region, followed by DUF283 and PAZ domains, two catalytic RNase III domains, and a dsRNA-binding domain at the C-terminus. The second dsRBD (dsRBD2) of ADAR1, and the DEAD-box RNA helicase and DUF283 domains of Dicer, are required for the formation of the Dicer–ADAR1 complex26.
In principle, ADAR1 as part of the Dicer–ADAR1 complex could be inhibitory with respect to Dicer function. In fact, however, it increases the Vmax of Dicer-mediated cleavage of pre-miRNAs fourfold in comparison with the reaction with Dicer alone26. Similar analyses on Dicer cleavage of a long dsRNA indicated that ADAR1 also promotes processing of endo-siRNAs, showing that ADAR1 upregulates the turnover rate of Dicer and substantially increases the overall rate of miRNA and endo-siRNA production26 (FIG. 5b). miRNAs generated by the Dicer–ADAR1 complex were found to be fully functional when tested in various miRNA silencing assays26. In addition, ADAR1 substantially pro- motes RISC assembly and loading of miRNAs26 (FIG. 5b). Interestingly, neither the dsRNA-binding nor the deamin ase activities of ADAR1 is required for promoting the miRNA-processing and RISC-loading activities of Dicer. The catalytic activity of Dicer is auto-inhibited by its DEAD-box RNA helicase domain131. Thus, the enhancement of Dicer activity by ADAR1 may be due to ADAR1 binding to the Dicer DEAD-box RNA helicase domain, thereby blocking its auto-inhibitory effect26.
ADAR1 upregulates miRNA expression in mouse embryos
Analysis of miRNA expression levels in mouse embryos indicates that a rapid and dramatic increase of miRNA production occurs globally at around E11–E12 (REFS 25,26), which is likely to be essential for embryo development. This developmental stage-specific increase in miRNA production seems to be caused by concomitant upregulation of Dicer and ADAR1p110 (REF. 26). Global suppression of miRNA production is detected in Adar1-null mouse embryos, which die at around E12 (REFS 25,26). In contrast to the rapid increase in ADAR1p110 expression, TRBP expression remains very low around this period, perhaps indicating that the contribution of ADAR1 is more important than that of TRBP in the miRNA-mediated RNAi mechanism, at least during embryonic development of the E11–E12 stage26.
The considerable upregulation of miRNA production at the E11–E12 stage cannot occur in Adar1-null mouse embryos, owing to the lack of the Dicer–ADAR1p110 complex25,26. This seems to result in dysregulated expression of many genes, which would otherwise be repressed by these miRNAs during normal development, as dramatic changes of global gene expression patterns are detected in the Adar1-null embryos. The target genes have different functions, but cell death control and activation of interferon signalling are two of the most significant functions represented26. Thus, deficiency in the RNAi function of ADAR1 may underlie, at least partly, the embryonic lethality of Adar1-null mice around E12 (REFS 25,26).
Concluding remarks and outlook
It is now almost 30 years since the discovery of A-to-I RNA editing mediated by ADAR4,5. Since then, considerable progress has been made in understanding the editing mechanism, characterizing invertebrate and vertebrate ADAR genes, identifying numerous A-to-I editing sites in a wide range of coding and non-coding RNAs, and unravelling the relevance of A-to-I RNA editing to human diseases and revealing its interactions with RNAi pathways.
Nonetheless, we realize that many important questions in this field remain to be answered. For example, what is the selective advantage that initially drove the evolution of A-to-I RNA editing in the animal kingdom? ADAR genes are absent in plant, fungi and yeast genomes. In these organisms, very powerful RNAi pathways utilize dsRNA and play a major part in many important processes, such as silencing of transposons and heterochromatin formation. Did A-to-I RNA editing evolve as a mechanism to assist or replace RNAi? ADAR expression levels are not necessarily correlated well with A-to-I RNA-editing levels of target RNAs within a given tissue or developmental stage, indicating the presence of a currently unidentified mechanism (or mechanisms) that determines editing levels. Certain ADAR gene family members seem to have functions in addition to A-to-I RNA editing, for example, the function of ADAR1 in RNAi. However, it is not known how the balance between the A-to-I RNA-editing and the RNAi functions of ADAR1 is regulated. Exciting findings are likely to be made in the field of A-to-I RNA editing, through future investigations addressing these questions.
Alu
A type of retrotransposon of the short interspersed nuclear elements (SINE) family found in primate genomes. There are about 1.4 million copies of Alu in the human genome.
Z-DNA
A left-handed form of DNA that is different from the common A and B structural isoforms of DNA. Its biological functions are largely unknown.
Deamination
The chemical process that replaces a primary amino group by a hydroxyl group, resulting in conversion of one nucleoside to another.
Inositol hexakisphosphate
(InsP6). An intracellular organic compound that is found throughout the animal kingdom and is affiliated with a wide range of important physiological activities such as modulation of haemoglobin structure and function.
Retrotransposon
A class of genetic elements that includes endogenous retroviruses and transposable elements, which propagate in the genome through an intermediate RNA stage.
Nuclear paraspeckles
Discrete, irregularly shaped nuclear compartments. Usually, approximately 10–30 paraspeckles are present in the interphase mammalian nucleus. Their function is not known, but they may trap certain proteins in the nucleus.
Wobble base pairs
Pairs of nucleotides other than G:C and A:U, such as thermodynamically less stable I:U and G:U pairs. Wobble base pairs, like Watson–Crick base pairs, participate in RNA folding and the formation of secondary structures.
Endogenous short interfering RNAs
(endo-siRNAs). siRNAs derived from endogenous double-stranded transcripts and repetitive elements such as Alu or other retrotransposons.
RNase III protein
A double-stranded RNA (dsRNA)-specific endonuclease that cleaves dsRNA into short fragments with a 3′ overhang and a recessed 5′ phosphate. The RNA interference (RNAi) factors Drosha and Dicer are such proteins.
RNA-induced silencing complex
(RISC). A complex containing short interfering RNAs (siRNAs) or microRNAs (miRNAs) and an Argonaute protein, which mediates the degradation or translation inhibition of target mRNAs that have high sequence complementarity to the small RNAs.
Acknowledgements
The author thanks John M. Murray for critical reading of the manuscript. This work was supported in part by grants from the U.S. National Institutes of Health, Ellison Medical Foundation, Macula Vision Research Foundation and the Commonwealth Universal Research Enhancement Program of the Pennsylvania Department of Health.
Footnotes
Competing interests statement
The author declares no competing financial interests.
DATABASES
National Cancer Institute Drug Dictionary: http://www.cancer.gov/drugdictionary
Online Mendelian Inheritance in Man (OMIM): http://www.omim.org/ ADAR1 | ADAR2 | ADAR3
RADAR editing sites database: http://rnaedit.com
DARNED editing sites database: http://beamish.ucc.ie
miRNA editing sites databases: http://www.cs.tau.ac.il/~mirnaed/
miRBase database: http://www.mirbase.org/
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Bass BL, Weintraub H. A developmentally regulated activity that unwinds RNA duplexes. Cell. 1987;48:607–613. doi: 10.1016/0092-8674(87)90239-x. [DOI] [PubMed] [Google Scholar]
- 2.Bass BL, Weintraub H. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell. 1988;55:1089–1098. doi: 10.1016/0092-8674(88)90253-x. [DOI] [PubMed] [Google Scholar]
- 3.Wagner RW, Smith JE, Cooperman BS, Nishikura K. A double-stranded RNA unwinding activity introduces structural alterations by means of adenosine to inosine conversions in mammalian cells and Xenopus eggs. Proc. Natl Acad. Sci. USA. 1989;86:2647–2651. doi: 10.1073/pnas.86.8.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hogg M, Paro S, Keegan LP, O'Connell MA. RNA editing by mammalian ADARs. Adv. Genet. 2011;73:87–120. doi: 10.1016/B978-0-12-380860-8.00003-3. [DOI] [PubMed] [Google Scholar]
- 5.Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010;79:321–349. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Athanasiadis A, Rich A, Maas S. Widespread A-to-I RNA editing of Alu-containing mRNAs in the human transcriptome. PLoS Biol. 2004;2:e391. doi: 10.1371/journal.pbio.0020391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bazak L, et al. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 2014;24:365–376. doi: 10.1101/gr.164749.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blow M, Futreal PA, Wooster R, Stratton MR. A survey of RNA editing in human brain. Genome Res. 2004;14:2379–2387. doi: 10.1101/gr.2951204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fumagalli D, et al. Principles governing A-to-I RNA editing in the breast cancer transcriptome. Cell Rep. 2015;13:277–289. doi: 10.1016/j.celrep.2015.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han L, et al. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell. 2015;28:515–528. doi: 10.1016/j.ccell.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim DD, et al. Widespread RNA editing of embedded Alu elements in the human transcriptome. Genome Res. 2004;14:1719–1725. doi: 10.1101/gr.2855504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levanon EY, et al. Systematic identification of abundant A-to-I editing sites in the human transcriptome. Nat. Biotechnol. 2004;22:1001–1005. doi: 10.1038/nbt996. [DOI] [PubMed] [Google Scholar]
- 13.Li JB, et al. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science. 2009;324:1210–1213. doi: 10.1126/science.1170995. [DOI] [PubMed] [Google Scholar]
- 14.Paz-Yaacov N, et al. Elevated RNA editing activity is a major contributor to transcriptomic diversity in tumors. Cell Rep. 2015;13:267–276. doi: 10.1016/j.celrep.2015.08.080. [DOI] [PubMed] [Google Scholar]
- 15.Peng Z, et al. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat. Biotechnol. 2012;30:253–260. doi: 10.1038/nbt.2122. [DOI] [PubMed] [Google Scholar]
- 16.Porath HT, Carmi S, Levanon EY. A genome-wide map of hyper-edited RNA reveals numerous new sites. Nat. Commun. 2014;5:4726. doi: 10.1038/ncomms5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramaswami G, et al. Accurate identification of human Alu and non-Alu RNA editing sites. Nat. Methods. 2012;9:579–581. doi: 10.1038/nmeth.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramaswami G, et al. Identifying RNA editing sites using RNA sequencing data alone. Nat. Methods. 2013;10:128–132. doi: 10.1038/nmeth.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sakurai M, Yano T, Kawabata H, Ueda H, Suzuki T. Inosine cyanoethylation identifies A-to-I RNA editing sites in the human transcriptome. Nat. Chem. Biol. 2010;6:733–740. doi: 10.1038/nchembio.434. [References 6–19 describe the global identification of numerous A-to-I editing sites in Alu repeats.] [DOI] [PubMed] [Google Scholar]
- 20.Iizasa H, et al. Editing of Epstein–Barr virus-encoded BART6 microRNAs controls their Dicer targeting and consequently affects viral latency. J. Biol. Chem. 2010;285:33358–33370. doi: 10.1074/jbc.M110.138362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kawahara Y, et al. Frequency and fate of microRNA editing in human brain. Nucleic Acids Res. 2008;36:5270–5280. doi: 10.1093/nar/gkn479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer–TRBP complex. EMBO Rep. 2007;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawahara Y, et al. Redirection of silencing targets by adenosine-to-inosine editing of mi RNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [This article shows that A-to-I editing of a miRNA-376a precursor results in alteration of the target genes of the miRNA.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang W, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishikura K, Sakurai M, Ariyoshi K, Ota H. Antagonistic and stimulative roles of ADAR1 in RNA silencing. RNA Biol. 2013;10:1240–1247. doi: 10.4161/rna.25947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ota H, et al. ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell. 2013;153:575–589. doi: 10.1016/j.cell.2013.03.024. [The authors demonstrate that ADAR1 forms a complex with Dicer to promote miRNA processing and RNAi efficacy.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galeano F, Tomaselli S, Locatelli F, Gallo A. A-to-I RNA editing: the “ADAR” side of human cancer. Semin. Cell Dev. Biol. 2012;23:244–250. doi: 10.1016/j.semcdb.2011.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Hood JL, Emeson RB. Editing of neurotransmitter receptor and ion channel RNAs in the nervous system. Curr. Top. Microbiol. Immunol. 2012;353:61–90. doi: 10.1007/82_2011_157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jepson JE, Reenan RA. RNA editing in regulating gene expression in the brain. Biochim. Biophys. Acta. 2008;1779:459–470. doi: 10.1016/j.bbagrm.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 30.Rosenthal JJ, Seeburg PH. A-to-I RNA editing: effects on proteins key to neural excitability. Neuron. 2012;74:432–439. doi: 10.1016/j.neuron.2012.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samuel CE. ADARs: viruses and innate immunity. Curr. Top. Microbiol. Immunol. 2012;353:163–195. doi: 10.1007/82_2011_148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slotkin W, Nishikura K. Adenosine-to-inosine RNA editing and human disease. Genome Med. 2013;5:105. doi: 10.1186/gm508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tariq A, Jantsch MF. Transcript diversification in the nervous system: A to I RNA editing in CNS function and disease development. Front. Neurosci. 2012;6:99. doi: 10.3389/fnins.2012.00099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomaselli S, Galeano F, Locatelli F, Gallo A. ADARs and the balance game between virus infection and innate immune cell response. Curr. Issues Mol. Biol. 2014;17:37–52. [PubMed] [Google Scholar]
- 35.Rosenthal JJ. The emerging role of RNA editing in plasticity. J. Exp. Biol. 2015;218:1812–1821. doi: 10.1242/jeb.119065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takenaka M, Zehrmann A, Verbitskiy D, Hartel B, Brennicke A. RNA editing in plants and its evolution. Annu. Rev. Genet. 2013;47:335–352. doi: 10.1146/annurev-genet-111212-133519. [DOI] [PubMed] [Google Scholar]
- 37.Smith HC, Bennett RP, Kizilyer A, McDougall WM, Prohaska KM. Functions and regulation of the APOBEC family of proteins. Semin. Cell Dev. Biol. 2012;23:258–268. doi: 10.1016/j.semcdb.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aphasizhev R, Aphasizheva I. Mitochondrial RNA processing in trypanosomes. Res. Microbiol. 2011;162:655–663. doi: 10.1016/j.resmic.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gott JM, Emeson RB. Functions and mechanisms of RNA editing. Annu. Rev. Genet. 2000;34:499–531. doi: 10.1146/annurev.genet.34.1.499. [DOI] [PubMed] [Google Scholar]
- 40.Jin Y, Zhang W, Li Q. Origins and evolution of ADAR-mediated RNA editing. IUBMB Life. 2009;61:572–578. doi: 10.1002/iub.207. [DOI] [PubMed] [Google Scholar]
- 41.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Molecular cloning of cDNA for double-stranded RNA adenosine deaminase, a candidate enzyme for nuclear RNA editing. Proc. Natl Acad. Sci. USA. 1994;91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melcher T, et al. A mammalian RNA editing enzyme. Nature. 1996;379:460–464. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- 43.Chen CX, et al. A third member of the RNA-specific adenosine deaminase gene family, ADAR3, contains both single-and double-stranded RNA binding domains. RNA. 2000;6:755–767. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Melcher T, et al. RED2, a brain-specific member of the RNA-specific adenosine deaminase family. J. Biol. Chem. 1996;271:31795–31798. doi: 10.1074/jbc.271.50.31795. [DOI] [PubMed] [Google Scholar]
- 45.Stefl R, Xu M, Skrisovska L, Emeson RB, Allain FH. Structure and specific RNA binding of ADAR2 double-stranded RNA binding motifs. Structure. 2006;14:345–355. doi: 10.1016/j.str.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 46.Herbert A, et al. A Z-DNA binding domain present in the human editing enzyme, double-stranded RNA adenosine deaminase. Proc. Natl Acad. Sci. USA. 1997;94:8421–8426. doi: 10.1073/pnas.94.16.8421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider MF, Wettengel J, Hoffmann PC, Stafforst T. Optimal guideRNAs for re-directing deaminase activity of hADAR1 and hADAR2 in trans. Nucleic Acids Res. 2014;42:e87. doi: 10.1093/nar/gku272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schumacher JM, Lee K, Edelhoff S, Braun RE. Distribution of Tenr, an RNA-binding protein, in a lattice-like network within the spermatid nucleus in the mouse. Biol. Reprod. 1995;52:1274–1283. doi: 10.1095/biolreprod52.6.1274. [DOI] [PubMed] [Google Scholar]
- 49.McKee AE, et al. A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Dev. Biol. 2005;5:14. doi: 10.1186/1471-213X-5-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Macbeth MR, et al. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science. 2005;309:1534–1539. doi: 10.1126/science.1113150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nishikura K, et al. Substrate specificity of the dsRNA unwinding/modifying activity. EMBO J. 1991;10:3523–3532. doi: 10.1002/j.1460-2075.1991.tb04916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lehmann KA, Bass BL. The importance of internal loops within RNA substrates of ADAR1. J. Mol. Biol. 1999;291:1–13. doi: 10.1006/jmbi.1999.2914. [DOI] [PubMed] [Google Scholar]
- 53.Higuchi M, et al. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron–exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 54.Lehmann KA, Bass BL. Double-stranded RNA adenosine deaminases ADAR1 and ADAR2 have overlapping specificities. Biochemistry. 2000;39:12875–12884. doi: 10.1021/bi001383g. [DOI] [PubMed] [Google Scholar]
- 55.Hartner JC, et al. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- 56.Higuchi M, et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- 57.Wang Q, et al. Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J. Biol. Chem. 2004;279:4952–4961. doi: 10.1074/jbc.M310162200. [DOI] [PubMed] [Google Scholar]
- 58.George CX, Samuel CE. Human RNA-specific adenosine deaminase ADAR1 transcripts possess alternative exon 1 structures that initiate from different promoters, one constitutively active and the other interferon inducible. Proc. Natl Acad. Sci. USA. 1999;96:4621–4626. doi: 10.1073/pnas.96.8.4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patterson JB, Samuel CE. Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol. Cell. Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Peng PL, et al. ADAR2-dependent RNA editing of AMPA receptor subunit GluR2 determines vulnerability of neurons in forebrain ischemia. Neuron. 2006;49:719–733. doi: 10.1016/j.neuron.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 61.Yang L, et al. c-Jun amino-terminal kinase-1 mediates glucose-responsive upregulation of the RNA editing enzyme ADAR2 in pancreatic beta-cells. PLoS ONE. 2012;7:e48611. doi: 10.1371/journal.pone.0048611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shoshan E, et al. Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma growth and metastasis. Nat. Cell Biol. 2015;17:311–321. doi: 10.1038/ncb3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cho DS, et al. Requirement of dimerization for RNA editing activity of adenosine deaminases acting on RNA. J. Biol. Chem. 2003;278:17093–17102. doi: 10.1074/jbc.M213127200. [DOI] [PubMed] [Google Scholar]
- 64.Poulsen H, et al. Dimerization of ADAR2 is mediated by the double-stranded RNA binding domain. RNA. 2006;12:1350–1360. doi: 10.1261/rna.2314406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Valente L, Nishikura K. RNA binding-independent dimerization of adenosine deaminases acting on RNA and dominant negative effects of nonfunctional subunits on dimer functions. J. Biol. Chem. 2007;282:16054–16061. doi: 10.1074/jbc.M611392200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Desterro JM, et al. Dynamic association of RNA-editing enzymes with the nucleolus. J. Cell Sci. 2003;116:1805–1818. doi: 10.1242/jcs.00371. [DOI] [PubMed] [Google Scholar]
- 67.Fritz J, et al. RNA-regulated interaction of transportin-1 and exportin-5 with the double-stranded RNA-binding domain regulates nucleocytoplasmic shuttling of ADAR1. Mol. Cell. Biol. 2009;29:1487–1497. doi: 10.1128/MCB.01519-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Strehblow A, Hallegger M, Jantsch MF. Nucleocytoplasmic distribution of human RNA-editing enzyme ADAR1 is modulated by double-stranded RNA-binding domains, a leucine-rich export signal, and a putative dimerization domain. Mol. Biol. Cell. 2002;13:3822–3835. doi: 10.1091/mbc.E02-03-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Poulsen H, Nilsson J, Damgaard CK, Egebjerg J, Kjems J. CRM1 mediates the export of ADAR1 through a nuclear export signal within the Z-DNA binding domain. Mol. Cell. Biol. 2001;21:7862–7871. doi: 10.1128/MCB.21.22.7862-7871.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barraud P, Banerjee S, Mohamed WI, Jantsch MF, Allain FH. A bimodular nuclear localization signal assembled via an extended double-stranded RNA-binding domain acts as an RNA-sensing signal for transportin 1. Proc. Natl Acad. Sci. USA. 2014;111:E1852–E1861. doi: 10.1073/pnas.1323698111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Maas S, Gommans WM. Identification of a selective nuclear import signal in adenosine deaminases acting on RNA. Nucleic Acids Res. 2009;37:5822–5829. doi: 10.1093/nar/gkp599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sansam CL, Wells KS, Emeson RB. Modulation of RNA editing by functional nucleolar sequestration of ADAR2. Proc. Natl Acad. Sci. USA. 2003;100:14018–14023. doi: 10.1073/pnas.2336131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Marcucci R, et al. Pin1 and WWP2 regulate GluR2 Q/R site RNA editing by ADAR2 with opposing effects. EMBO J. 2011;30:4211–4222. doi: 10.1038/emboj.2011.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Burns CM, et al. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 75.Bhalla T, Rosenthal JJ, Holmgren M, Reenan R. Control of human potassium channel inactivation by editing of a small mRNA hairpin. Nat. Struct. Mol. Biol. 2004;11:950–956. doi: 10.1038/nsmb825. [DOI] [PubMed] [Google Scholar]
- 76.Daniel C, Wahlstedt H, Ohlson J, Bjork P, Ohman M. Adenosine-to-inosine RNA editing affects trafficking of the γ-aminobutyric acid type A (GABAA) receptor. J. Biol. Chem. 2011;286:2031–2040. doi: 10.1074/jbc.M110.130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kawahara Y, et al. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004;427:801. doi: 10.1038/427801a. [DOI] [PubMed] [Google Scholar]
- 78.Hideyama T, et al. Induced loss of ADAR2 engenders slow death of motor neurons from Q/R site-unedited GluR2. J. Neurosci. 2010;30:11917–11925. doi: 10.1523/JNEUROSCI.2021-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ishiuchi S, et al. Ca2+-permeable AMPA receptors regulate growth of human glioblastoma via Akt activation. J. Neurosci. 2007;27:7987–8001. doi: 10.1523/JNEUROSCI.2180-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hartner JC, Walkley CR, Lu J, Orkin SH. ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat. Immunol. 2009;10:109–115. doi: 10.1038/ni.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liddicoat BJ, et al. RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science. 2015;349:1115–1120. doi: 10.1126/science.aac7049. [The authors show that A-to-I editing by ADAR1 is required for preventing sensing by MDA5 of long dsRNAs made from repetitive elements, with consequences for the embryonic lethality of Adar1-null mice.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.XuFeng R, et al. ADAR1 is required for hematopoietic progenitor cell survival via RNA editing. Proc. Natl Acad. Sci. USA. 2009;106:17763–17768. doi: 10.1073/pnas.0903324106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mannion NM, et al. The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep. 2014;9:1482–1494. doi: 10.1016/j.celrep.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rice GI, et al. Mutations in ADAR1 cause Aicardi–Goutieres syndrome associated with a type I interferon signature. Nat. Genet. 2012;44:1243–1248. doi: 10.1038/ng.2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang S, et al. Adenosine deaminase acting on RNA 1 limits RIG-I RNA detection and suppresses IFN production responding to viral and endogenous RNAs. J. Immunol. 2014;193:3436–3445. doi: 10.4049/jimmunol.1401136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Suzuki N, et al. Ten novel mutations of the ADAR1 gene in Japanese patients with dyschromatosis symmetrica hereditaria. J. Invest. Dermatol. 2007;127:309–311. doi: 10.1038/sj.jid.5700528. [DOI] [PubMed] [Google Scholar]
- 87.Kawahara Y, et al. Dysregulated editing of serotonin 2C receptor mRNAs results in energy dissipation and loss of fat mass. J. Neurosci. 2008;28:12834–12844. doi: 10.1523/JNEUROSCI.3896-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Morabito MV, et al. Mice with altered serotonin 2C receptor RNA editing display characteristics of Prader–Willi syndrome. Neurobiol. Dis. 2010;39:169–180. doi: 10.1016/j.nbd.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mombereau C, Kawahara Y, Gundersen BB, Nishikura K, Blendy JA. Functional relevance of serotonin 2C receptor mRNA editing in antidepressant-and anxiety-like behaviors. Neuropharmacology. 2010;59:468–473. doi: 10.1016/j.neuropharm.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Eran A, et al. Comparative RNA editing in autistic and neurotypical cerebella. Mol. Psychiatry. 2013;18:1041–1048. doi: 10.1038/mp.2012.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rueter SM, Dawson TR, Emeson RB. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- 92.Feng Y, Sansam CL, Singh M, Emeson RB. Altered RNA editing in mice lacking ADAR2 autoregulation. Mol. Cell. Biol. 2006;26:480–488. doi: 10.1128/MCB.26.2.480-488.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lev-Maor G, et al. RNA-editing-mediated exon evolution. Genome Biol. 2007;8:R29. doi: 10.1186/gb-2007-8-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang Z, Carmichael GG. The fate of dsRNA in the nucleus: a p54nrb-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell. 2001;106:465–475. doi: 10.1016/s0092-8674(01)00466-4. [DOI] [PubMed] [Google Scholar]
- 95.Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–1705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Prasanth KV, et al. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 97.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Scadden AD, Smith CW. Specific cleavage of hyper-edited dsRNAs. EMBO J. 2001;20:4243–4252. doi: 10.1093/emboj/20.15.4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Morita Y, et al. Human endonuclease V is a ribonuclease specific for inosine-containing RNA. Nat. Commun. 2013;4:2273. doi: 10.1038/ncomms3273. [The authors show that EndoV is the ribonuclease specific to inosine-containing RNAs.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Scadden AD. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat. Struct. Mol. Biol. 2005;12:489–496. doi: 10.1038/nsmb936. [DOI] [PubMed] [Google Scholar]
- 101.Scadden AD. Inosine-containing dsRNA binds a stress-granule-like complex and downregulates gene expression in trans. Mol. Cell. 2007;28:491–500. doi: 10.1016/j.molcel.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Weissbach R, Scadden AD. Tudor-SN and ADAR1 are components of cytoplasmic stress granules. RNA. 2012;18:462–471. doi: 10.1261/rna.027656.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ng SK, Weissbach R, Ronson GE, Scadden AD. Proteins that contain a functional Z-DNA-binding domain localize to cytoplasmic stress granules. Nucleic Acids Res. 2013;41:9786–9799. doi: 10.1093/nar/gkt750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vitali P, Scadden AD. Double-stranded RNAs containing multiple IU pairs are sufficient to suppress interferon induction and apoptosis. Nat. Struct. Mol. Biol. 2010;17:1043–1050. doi: 10.1038/nsmb.1864. [The authors show that I·U dsRNA resembling extensively edited RNA can suppress interferon signalling, which is activated by unedited long dsRNA.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 2013;14:100–112. doi: 10.1038/nrg3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wang Q, Zhang Z, Blackwell K, Carmichael GG. Vigilins bind to promiscuously A-to-I-edited RNAs and are involved in the formation of heterochromatin. Curr. Biol. 2005;15:384–391. doi: 10.1016/j.cub.2005.01.046. [The authors report that vigilin in complex with ADAR1 binds to inosine-containing RNAs, revealing a possible role for A-to-I editing in heterochromatic gene silencing in mammalian cells.] [DOI] [PubMed] [Google Scholar]
- 107.Zhou J, Wang Q, Chen LL, Carmichael GG. On the mechanism of induction of heterochromatin by the RNA-binding protein vigilin. RNA. 2008;14:1773–1781. doi: 10.1261/rna.1036308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kondo Y, Issa JP. Enrichment for histone H3 lysine 9 methylation at Alu repeats in human cells. J. Biol. Chem. 2003;278:27658–27662. doi: 10.1074/jbc.M304072200. [DOI] [PubMed] [Google Scholar]
- 109.Savva YA, et al. RNA editing regulates transposon-mediated heterochromatic gene silencing. Nat. Commun. 2013;4:2745. doi: 10.1038/ncomms3745. [The authors report that A-to-I editing of dsRNAs derived from retrotransposons antagonizes silencing by RNAi of retrotransposons in flies, revealing a possible role for A-to-I editing in the suppression of heterochromatic gene silencing.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 111.Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Croce CM, Calin GA. mi RNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 113.Lei T, et al. Perturbation of biogenesis and targeting of Epstein–Barr virus-encoded miR-BART3 microRNA by adenosine-to-inosine editing. J. Gen. Virol. 2013;94:2739–2744. doi: 10.1099/vir.0.056226-0. [DOI] [PubMed] [Google Scholar]
- 114.Pfeffer S, et al. Identification of microRNAs of the herpesvirus family. Nat. Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- 115.Alon S, et al. Systematic identification of edited microRNAs in the human brain. Genome Res. 2012;22:1533–1540. doi: 10.1101/gr.131573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chiang HR, et al. Mammalian microRNAs: experimental evaluation of novel and previously annotated genes. Genes Dev. 2010;24:992–1009. doi: 10.1101/gad.1884710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chawla G, Sokol NS. ADAR mediates differential expression of polycistronic microRNAs. Nucleic Acids Res. 2014;42:5245–5255. doi: 10.1093/nar/gku145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Heale BS, et al. Editing independent effects of ADARs on the miRNA/siRNA pathways. EMBO J. 2009;28:3145–3156. doi: 10.1038/emboj.2009.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tomaselli S, et al. Modulation of microRNA editing, expression and processing by ADAR2 deaminase in glioblastoma. Genome Biol. 2015;16:5. doi: 10.1186/s13059-014-0575-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vesely C, Tauber S, Sedlazeck FJ, von Haeseler A, Jantsch MF. Adenosine deaminases that act on RNA induce reproducible changes in abundance and sequence of embryonic mi RNAs. Genome Res. 2012;22:1468–1476. doi: 10.1101/gr.133025.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen T, et al. ADAR1 is required for differentiation and neural induction by regulating microRNA processing in a catalytically independent manner. Cell Res. 2015;25:459–476. doi: 10.1038/cr.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Galore-Haskel G, et al. A novel immune resistance mechanism of melanoma cells controlled by the ADAR1 enzyme. Oncotarget. 2015;6:28999–29015. doi: 10.18632/oncotarget.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bahn JH, et al. Genomic analysis of ADAR1 binding and its involvement in multiple RNA processing pathways. Nat. Commun. 2015;6:6355. doi: 10.1038/ncomms7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ekdahl Y, Farahani HS, Behm M, Lagergren J, Ohman M. A-to-I editing of microRNAs in the mammalian brain increases during development. Genome Res. 2012;22:1477–1487. doi: 10.1101/gr.131912.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Choudhury Y, et al. Attenuated adenosine-to-inosine editing of microRNA-376a* promotes invasiveness of glioblastoma cells. J. Clin. Invest. 2012;122:4059–4076. doi: 10.1172/JCI62925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bass BL. Double-stranded RNA as a template for gene silencing. Cell. 2000;101:235–238. doi: 10.1016/s0092-8674(02)71133-1. [DOI] [PubMed] [Google Scholar]
- 127.Tonkin LA, Bass BL. Mutations in RNAi rescue aberrant chemotaxis of ADAR mutants. Science. 2003;302:1725. doi: 10.1126/science.1091340. [This article shows the interaction between RNAi and RNA-editing pathways in vivo in mutant worm strains.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tonkin LA, et al. RNA editing by ADARs is important for normal behavior in Caenorhabditis elegans. EMBO J. 2002;21:6025–6035. doi: 10.1093/emboj/cdf607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wu D, Lamm AT, Fire AZ. Competition between ADAR and RNAi pathways for an extensive class of RNA targets. Nat. Struct. Mol. Biol. 2011;18:1094–1101. doi: 10.1038/nsmb.2129. [The authors report that A-to-I editing of dsRNAs derived from transposons and pseudogenes suppresses the generation of endo-siRNAs from these loci and their entry into the RNAi pathway.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Warf MB, Shepherd BA, Johnson WE, Bass BL. Effects of ADARs on small RNA processing pathways in C. elegans. Genome Res. 2012;22:1488–1498. doi: 10.1101/gr.134841.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Ma E, MacRae IJ, Kirsch JF, Doudna JA. Autoinhibition of human dicer by its internal helicase domain. J. Mol. Biol. 2008;380:237–243. doi: 10.1016/j.jmb.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Sommer B, Kohler M, Sprengel R, Seeburg PH. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 133.Lomeli H, et al. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 134.Sailer A, et al. Generation and analysis of GluR5(Q636R) kainate receptor mutant mice. J. Neurosci. 1999;19:8757–8764. doi: 10.1523/JNEUROSCI.19-20-08757.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Egebjerg J, Heinemann SF. Ca2+ permeability of unedited and edited versions of the kainate selective glutamate receptor GluR6. Proc. Natl Acad. Sci. USA. 1993;90:755–759. doi: 10.1073/pnas.90.2.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kohler M, Burnashev N, Sakmann B, Seeburg PH. Determinants of Ca2+ permeability in both TM1 and TM2 of high affinity kainate receptor channels: diversity by RNA editing. Neuron. 1993;10:491–500. doi: 10.1016/0896-6273(93)90336-p. [DOI] [PubMed] [Google Scholar]
- 137.Levanon EY, et al. Evolutionarily conserved human targets of adenosine to inosine RNA editing. Nucleic Acids Res. 2005;33:1162–1168. doi: 10.1093/nar/gki239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Galeano F, et al. Human BLCAP transcript: new editing events in normal and cancerous tissues. Int. J. Cancer. 2010;127:127–137. doi: 10.1002/ijc.25022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chan TH, et al. A disrupted RNA editing balance mediated by ADARs (Adenosine DeAminases that act on RNA) in human hepatocellular carcinoma. Gut. 2014;63:832–843. doi: 10.1136/gutjnl-2012-304037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Martinez HD, et al. RNA editing of androgen receptor gene transcripts in prostate cancer cells. J. Biol. Chem. 2008;283:29938–29949. doi: 10.1074/jbc.M800534200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chen L, et al. Recoding RNA editing of AZIN1 predisposes to hepatocellular carcinoma. Nat. Med. 2013;19:209–216. doi: 10.1038/nm.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yeo J, Goodman RA, Schirle NT, David SS, Beal PA. RNA editing changes the lesion specificity for the DNA repair enzyme NEIL1. Proc. Natl Acad. Sci. USA. 2010;107:20715–20719. doi: 10.1073/pnas.1009231107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shimokawa T, et al. RNA editing of the GLI1 transcription factor modulates the output of Hedgehog signaling. RNA Biol. 2013;10:321–333. doi: 10.4161/rna.23343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Han SW, et al. RNA editing in RHOQ promotes invasion potential in colorectal cancer. J. Exp. Med. 2014;211:613–621. doi: 10.1084/jem.20132209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liu WH, et al. ADAR2-mediated editing of miR-214 and miR-122 precursor and antisense RNA transcripts in liver cancers. PLoS ONE. 2013;8:e81922. doi: 10.1371/journal.pone.0081922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Nishikura K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat. Rev. Mol. Cell Biol. 2006;7:919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]