Abstract
Introduction
Current tools for automated skull stripping, normalization, and segmentation of non-human primate (NHP) brain MRI studies typically demonstrate high failure rates. Many of these failures are due to a poor initial estimate for the affine component of the transformation. The purpose of this study is to introduce a multi-atlas approach to overcome these limitations and drive the failure rate to near zero.
Materials and Methods
A library of study-specific templates (SST) spanning three Old World primate species (Macaca fascicularis, M. mulatta, Chlorocebus aethiops) was created using a previously described unbiased automated approach. Several modifications were introduced to the methodology to improve initial affine estimation at the study-specific template level, and at the individual subject level. These involve performing multiple separate normalizations to a multi-atlas library of templates and selecting the best performing template on the basis of a covariance similarity metric. This template was then used as an initialization for the affine component of subsequent skull stripping and normalization procedures. Normalization failure rate for SST generation and individual-subject segmentation on a set of 150 NHP was evaluated on the basis of visual inspection.
Results
The previous automated template creation procedure results in excellent skull stripping, segmentation, and atlas labeling across species. Failure rate at the individual-subject level was approximately 1%, however at the SST generation level it was 17%. Using the new multi-atlas approach, failure rate was further reduced to zero for both SST generation and individual subject processing.
Conclusions
We describe a multi-atlas library registration approach for driving normalization failures in NHP to zero. It is straightforward to implement, and can have application to a wide variety of existing tools, as well as in difficult populations including neonates and the elderly. This approach is also an important step towards developing fully automated high-throughput processing pipelines that are critical for future high volume multi-center NHP imaging studies for studies of drug abuse and brain health.
Keywords: Non-human primate, segmentation, cynomolgus, vervet, rhesus, INIA19, voxel based morphometry, MRI
Introduction
Accurate normalization, segmentation, and skull-stripping in non-human primate (NHP) MRI scans are notoriously difficult challenges for automated algorithms. Human tools typically perform very poorly with NHP scans and require fine-tuning before they can be applied on primate data (McLaren et al. 2010; McLaren et al. 2009; Maldjian et al. 2014). The large differences in the extracranial musculature and skull morphology between humans and NHP, and between NHP species are a large part of the problem for these algorithms, which ultimately rely on intensity based similarities between an object and the target. The brain represents a relatively small portion of the information in these NHP images. This results in frequent normalization failures for these algorithms which rely on minimization of error and invariably target the bulky and quite variable extracranial NHP tissues. These failures are quite obvious on visual inspection. Existing NHP-specific tools also can demonstrate high failure rates, even for experienced users. Our own experience with several NHP-specific tools (Wang et al. 2014; Fedorov et al. 2011) has demonstrated failure rates as high as 50%.
We recently introduced a fully automated methodology for creation of NHP study-specific templates, label-atlases, and 6-class tissue probability maps to improve performance of automated skull stripping, segmentation, and normalization(Maldjian et al. 2014). This method utilizes an unbiased diffeomorphic population averaging technique to generate a study specific template (SST), and a diffeomorphic normalization approach for skull stripping on the basis of a target template mask (B. Avants and Gee 2004; B. B. Avants et al. 2008). A study-specific 6-class segmentation tissue probability map (TPM) and label atlas is then created using the inverse transforms. For individual subjects, the initial skull stripping and affine transformation is estimated on the basis of a diffeomorphic registration to the SST. Final skull stripping and segmentation is performed within VBM8 (http://dbm.neuro.uni-jena.de/vbm.html) using the 6-class study-specific TPM. The VBM8 technique uses an adaptive Maximum A Posterior technique without needing constant a priori information on tissue probabilities, resulting in excellent tissue class separation.
Our experience using this method in several hundred NHP normalizations and segmentations has demonstrated a marked improvement over existing NHP tools, with very low failure rates (Maldjian et al. 2014). While these may occur at the individual-subject level, a failure at the SST creation level can be especially problematic, as further processing of a cohort is halted until the SST can be created. When normalization errors occur for human studies, the common advice is to manually re-orient the object image, and attempt the normalization/segmentation again. Invariably, this approach is successful at overcoming the problem as it provides a better initialization for the affine component of the transformation. Some of this can be alleviated by the use of standard scan orientation protocols. The failure of these normalization approaches is often due to a poor initial estimate of the affine transformation component of the registration procedure. While manual re-orientation is a viable solution, it is not practical in the context of developing fully automated robust registration procedures. For multimodal studies, header information is critical when coregistering or carrying transformations forward to functional MRI studies or diffusion tensor imaging (DTI) studies. Multiple series are often acquired with different voxel sizes, slice coverage, and plane of acquisition, in which image header information can be used to coregister between series and with the computed structural transformation. Altering this information in the structural image by manual re-orientation introduces additional software intervention, potential errors in data provenance, sources of potential bias, and serious liabilities for functional image analyses when not appropriately tracked. The purpose of this study is to describe a method for driving the NHP fully automated image normalization error rate to zero.
Materials and Methods
Overview of Procedures
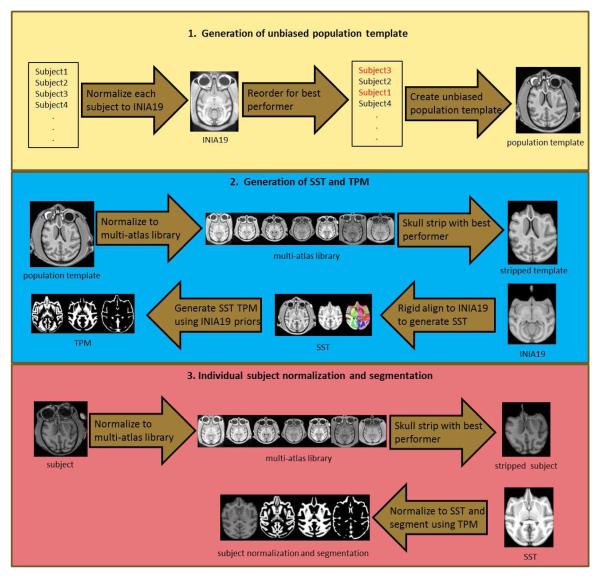
A flow chart of the procedures is provided in Figure 1.The methodology uses a variety of strategies to target the affine normalization component of the registration procedure. The main strategy is to select the best initial affine transformation based on registrations to a library of SSTs and corresponding brain masks. The best performer is then used to generate a native space skull-stripped brain image. Note that the best performer may be an interspecies registration (e.g., rhesus target SST for a vervet object). The normalization of the skull-stripped brain to the appropriate skull-stripped template brain (e.g., vervet skull-stripped object to vervet skull-stripped SST) can then proceed without difficulty. The use of interspecies templates is not a confounding variable, since the purpose is to create a native space skull stripping, without altering the morphology of the object scan. This identical strategy can be employed during the generation of a new SST, or when processing individual subjects against an SST. A variation of this strategy can be used during the initial creation of a new SST. In this step, an unbiased population template averaging procedure is used based on a list of native space subjects. The output is in the space of the initial subject in the list. Since the order of the subjects provided to the algorithm does not matter for generating an unbiased template, the list can be re-ordered on the basis of which native space image provides the best performance in matching to the template. The template remains unbiased in that all subjects are used in generating the template, and no single subject is serving as the template or target. The output space for final orientation of the template is not considered a bias, although this can have an effect on subsequent normalization performance. The resulting affine transformation can additionally be used as an initialization for the final diffeomorphic registration procedure. Similarly, the affine transformation for each individual subject from the population template generation procedure can be used as an initialization for the final normalization/segmentation of the individual subject with the SST.
Fig. 1.
Flow chart of methodology. Top row: Generation of population template uses best performer (cross-correlation metric) from individual normalizations to INIA19 template to reorder the input list of images. Unbiased population template is then created from all images in the list. Middle row: Generation of SST uses multi-atlas library to create skull-stripped template. This template can then be rigidly aligned to skull-stripped INIA19 template to generate SST and TPM. Note that SST atlas labels are generated using stacked diffeomorphic transform from INIA19 space to native population template space then to rigidly aligned SST space. Similar procedure is used to generate INIA19 priors in rigid-aligned SST space for segmentation and TPM generation. Bottom row: Individual subject normalization uses multi-atlas library to generate native space skull-stripped image. This image can then be normalized and segmented using the SST and corresponding TPM.
MRI scans
Previously acquired MRI scans of 150 monkeys including vervets (Chlorocebus aethiops), cynomolgus (Macaca fascicularis), and rhesus (Macaca mulatta) from the Wake Forest University Primate Center (Winston-Salem, NC) were used in this study. These scans were part of multiple ongoing studies funded by the National Institute on Alcohol Abuse and Alcoholism (NIAAA), National Institute on Drug Abuse (NIDA), and the National Heart, Lung and Blood Institute (NHLBI). Separate MRI cohorts for scanning included 30 vervets, 16 cynomolgus, 6 rhesus, 41 cynomolgus, 42 cynomolgus, and 15 vervet monkeys. All procedures were conducted in compliance with State and Federal laws, standards of the US Department of Health and Human Services, and guidelines established by the Wake Forest University Health Sciences Institutional Animal Care and Use Committee as well as the National Institute of Health Guide for the Care and Use of Laboratory Animals (NIH Publications No. 80-23).
Imaging data was obtained under general anesthesia. Briefly, the animals were sedated with 10-15 mg/kg ketamine and transported to the MRI Center where anesthesia was maintained via inhaled 1.5-3% isoflurane (Czoty and Nader 2015; Czoty et al. 2013; Czoty and Nader 2012). The animals were artificially ventilated to control physiological parameters across animals. Expired C02, oxygen saturation, heart rate, respiratory rate, and isoflurane concentration were monitored using an anesthesia monitor and pulse oximeter. Body temperature was maintained using warm blankets. At the end of scanning, the isoflurane and ventilator were shut off and the animals breathed a mixture of oxygen and room air during recovery from anesthesia. Animals were then extubated, provided pipeline oxygen until they recovered to sit in an upright position, returned to their home cages, and monitored until they were fully alert.
Imaging was performed on either a 1.5T or a 3T GE scanner with a circularly-polarized, single channel dedicated RF coil with an internal diameter of 18.4 cm (Litzcage, Doty Scientific, Columbia, SC), using a 3D SPGR sequence (TI 600ms, TE 3.432ms, TR 8.16ms; flip angle 15°; matrix 256×256 matrix; FOV 12.8 cm; 0.5 mm isotropic resolution).
SST Library
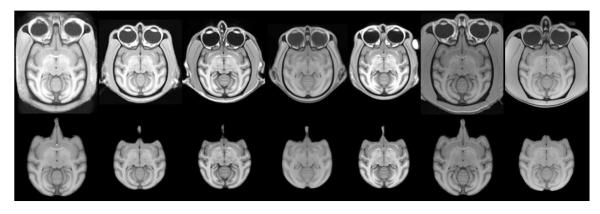
A library of multiple SSTs was created using our previously described methods (Maldjian et al. 2014). One SST was created for each study cohort for which the imaging data was acquired. These included 1 rhesus SST (6 subjects), 2 vervet SST (30 subjects and 15 subjects), and 3 cynomolgus SST (16, 41, and 42 subjects) (Figure 2). Creation of each SST involves generation of an unbiased population average template from the existing study-specific MRI scans, skull-stripping, tissue segmentation, and propagation of atlas labels. We make use of the INIA19 rhesus template to facilitate tissue segmentation and for propagation of the atlas labels, as it provides one of the most complete label segmentations currently available (Rohlfing et al. 2012). Additional label atlases can easily be incorporated. We have recently added the UNC subcortical and cortical label atlases (https://www.nitrc.org/projects/primate_atlas/), as well as manual probabilistic atlases into the automated SST generation procedure. The final atlas labels are generated in rigidly aligned population template space to avoid introducing any morphologic differences in species. For the template creation and normalization procedures we use symmetric diffeomorphic registration. Symmetric diffeomorphic registration (SyN) captures both large deformations and small shape changes (B. B. Avants et al. 2008) and provides consistently high accuracy across subjects and label sets (Klein et al. 2009). For each SST, we also generate a 6-class TPM and a 6-level Dartel multi-resolution template for use with SPM and VBM8 (http://dbm.neuro.uni-jena.de/vbm.html).
Fig. 2.
Multi-Atlas Library. Representative axial image (top) and skull-stripped image (bottom) from multi-atlas library including INIA template and 6 study-specific templates. From left to right: rhesus (Macaca mulatta) (INIA19), cynomolgus (M. fascicularis), vervet (Chlorocebus aethiops), cynomolgus, cynomolgus, rhesus, vervet.
Population Average (subject-space strategy)
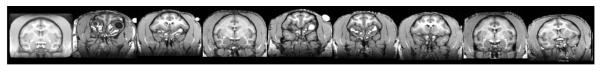
The population template is initially built using a diffeomorphic shape and intensity averaging technique (B. Avants and Gee 2004; B. B. Avants et al. 2008) constructed from all the subject scans for that cohort. The procedure accepts a list of native subjects to use in constructing the population average. The output space of the population average is in the space of the first subject on the list. In order to generate the SST, segmentation, and label maps, the population template is then registered to the INIA19 template. This step is a potential source of normalization error for the SST. We have adopted several strategies to eliminate normalization errors, and specifically, affine related errors, in generating this SST. The first strategy is at the level of choice of subject space for the initial population template. Each individual subject is registered to the INIA19 template, and the best performer is determined on the basis of an image similarity metric (Figure 3). In this study, we used the maximum of the pairwise image covariance, as implemented in the VBM8 toolbox (cg_check_cov) for the similarity metric. The input list to the population averaging procedure is then re-ordered with the best performer first. The output space of the unbiased population template is thus closer to that of that of the target template.
Fig. 3.
Selection of Template Output Space based on cross-correlation metric. Representative coronal images from a series of individual subject normalizations in a cohort to the INIA template demonstrates the third subject (fourth from the left) to be the best performer. From left to right: INIA 19 template, then individual subjects. Cross correlation values were 0.54, 0.57, 0.65, 0.55, 0.56, 0.53,0.59 and 0.6, respectively).
SST and TPM (multi-atlas library strategy)
The SST and corresponding TPM are created by registering the population template to the INIA19 Template and segmentation priors (Rohlfing et al. 2012). This is facilitated by a registration-based skull stripping, matching the population template to the INIA19 Template. This step is a critical potential area of normalization error from a poor affine estimate. This can be improved somewhat by using serial affine registrations as initializations, but this strategy alone is ineffective. We make use of our multi-atlas library of SSTs to facilitate accurate registration-based skull stripping. The population template is normalized to each SST using the diffeomorphic procedure and the best performer is selected on the basis of the pairwise image covariance similarity metric using the output masked and normalized skull-stripped images. The inverse transformation is used to generate a native skull-stripped population template. This native skull-stripped population template is used with the INIA skull-stripped image and priors to generate the final population SST, brain mask, TPM, and labels as previously described (Maldjian et al. 2014).
Individual Subjects
Each native space NHP undergoes an initial registration-based skull-stripping using the SST followed by an affine registration to the SST. The mask from the skull-stripping is dilated, and used as an initial estimate, with the final accurate skull stripping, normalization and tissue segmentation performed as part of the SPM8 new segment procedure as implemented in the VBM8 toolbox. The initial steps in this process are potential sources of registration error resulting in inadequate skull stripping, and poor affine registration, which are then propagated to the SPM8 procedure resulting in segmentation and normalization failure. The multi-atlas library of SSTs is used with each individual subject to facilitate the initial registration based skull-stripping. Images from individual subjects are normalized to each SST using the diffeomorphic procedure and the best performer is selected on the basis of the pairwise image similarity metric using the output masked and normalized skull-stripped images. The inverse transformation is used to generate a native skull-stripped individual image. This skull-stripped image is also used to generate the initial affine transformation to the actual SST. The skull-stripped mask is dilated and then carried forward with the affine transformation to SPM and VBM8 for the final segmentation, skull-stripping and normalization.
Subject Evaluation
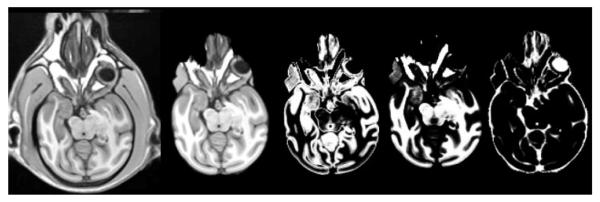
The original automated SST-generation and subject segmentation procedure was performed in 150 subjects for generation of 6 SSTs and corresponding individual subject segmentations. This was then repeated using the multi-atlas framework. When evaluating a cohort with the multi-atlas approach, the SST for that cohort was excluded from the multi-atlas library. Performance of each method was evaluated on the basis of visual assessment for accuracy of normalization, skull stripping, and segmentation. Evaluation was performed by a board certified neuroradiologist (JAM) with over 15 years of experience in the evaluation of image registration performance, clinical image interpretation, and 3 years of experience in the evaluation of NHP image segmentation and normalization algorithms. The failures for this evaluation were defined as obvious failures in image orientation and/or segmentation of grey matter, white matter, and CSF as shown in figure 4.
Fig. 4.
SST Failure. Example of a SST failure with poor normalization, skull-stripping and segmentation using the single atlas methodology. Left to right: normalized SST with skull, skull-stripped template, grey matter segmentation, white matter segmentation, and CSF segmentation.
Results
Using the original SST approach, only 1 of the 6 SSTs demonstrated a normalization failure at the level of the SST generation. Figure 4 demonstrates the results of the SST creation procedure for the failed vervet cohort. Notably, this particular cohort was from a group of 15 elderly vervets. The 15 individual subjects corresponding to this cohort were thus not included in the assessment of individual subject performance. The remaining 135 subjects for the other 5 SSTs demonstrated 1 segmentation/normalization failure. This provided a failure rate of 17% at the SST generation level, and approximately 1% at the individual subject level for the original method. Using the multi-atlas approach, there were no normalization failures for SST generation, and no normalization/segmentation failures for individual subjects providing a failure rate of 0%. This included the 15 individual subjects that could not be processed with the original method. Figure 5 demonstrates the results of the automated multi-atlas labeling procedure for the previously failed vervet SST. Note that the optimum template selected from the multi-atlas library for this particular cohort was for an inter-species normalization using a cynomolgus SST. Excellent skull stripping, segmentation, and label mapping to a variety of atlases is demonstrated. The subsequent processing performance of the 15 individual vervets in this cohort using the corresponding vervet SST was similarly excellent.
Fig. 5.
SST Multi-Atlas Segmentation. Same vervet cohort as in Figure 4, processed using fully automated multi-atlas approach demonstrates excellent normalization, skull stripping, segmentation and label mapping. From left to right: SST with skull, skull-stripped template, grey matter segmentation, white matter segmentation, CSF segmentation, NeuroMap labeling, UNC subcortical labeling, UNC cortical atlas labeling, and WFU probabilistic atlas labeling.
Discussion
A great deal of research has been directed at improving normalization approaches for brain imaging studies. There are many normalization approaches currently available, and several large-scale evaluations of these approaches (Tustison et al. 2014; B. B. Avants et al. 2011; Klein et al. 2009). A topic which has recently garnered some attention is the performance of the initial affine component of these registration approaches (B. B. Avants et al. 2011). Image registration begins with a linear transformation followed by the nonlinear registration steps. The initial quality of the affine registration is in fact critical to the performance of the final deformable registration and accuracy of label mapping (B. B. Avants et al. 2011). Normalization failures, when not due to bad or corrupted image data, can typically be traced to a poor affine registration. This problem can be compounded for NHPs in which image similarity between object and template is often driven by the dominating non-interesting features of the image, specifically the skull and massive extracranial soft tissues. The approach we have previously described provides a marked improvement over existing tools in terms of normalization performance, as well as providing study-specific templates, an area which has been largely ignored for NHP studies. In order to obtain a quality normalization and segmentation, our method relies on a reasonably accurate skull stripping. However, in order to obtain an accurate skull-stripping, our approach relies on an accurate normalization. Our solution to this image-equivalent conundrum of the chicken and egg problem is to use multiple eggs of different varieties. Similar to human studies, the failure of these normalizations can be traced to a poor initial affine estimate. The multi-atlas library can effectively drive the normalization error rate to zero. There can be a great deal of variability in the size and shape of the extracranial structures in NHP. These driving image features can be problematic even for within-species normalization algorithms. For example, an elderly vervet head may have more similarity in terms of the extracranial soft tissues to a young cynomolgus, than to a middle-aged vervet. The multi-atlas library takes advantage of this variability by providing a range of templates to accomplish a clean initial skull-stripping. Note that this approach does not introduce morphologic alterations into the normalization pipeline. The multi-atlas library is used strictly for the purpose of effecting a high quality image-based native-space skull stripping. Once the extracranial tissues have been cleanly removed, the existing normalization and segmentation algorithms perform very effectively using the skull-stripped brains. The failed vervet SST (acquired at 3T) provides a striking example of this approach. The automated normalization selected a cynomolgus template (acquired at 1.5 T) from the library to accomplish the skull-stripping. Examining the representative images from the multi-atlas library in Figure 2 demonstrates this concept well. The third image from the left is the SST eventually created for this elderly vervet cohort. The last image in Figure 2 is an SST for another vervet cohort, but this is for a group of young vervets. Note the tremendous differences in the bulk of the extracranial soft tissues between these cohorts. The cynomolgus templates in the library provide more similarity to the elderly vervets for the normalization-based skull stripping algorithms to perform well. Also note that the multi-atlas approach allows the use of SSTs acquired at different field strengths.
The concept of a multi-atlas approach has recently gained traction for label mapping (Wu et al. 2015; Wang et al. 2014). Specifically, more accurate individual subject labeling can be achieved by mapping to multiple templates. A similar approach has recently been suggested for achieving segmentation as well as for skull stripping (Doshi et al. 2013). These approaches are designed to provide accurate segmentation on the basis of averaging the results of the labeling, or introducing a hierarchy and weighting of performance based on evaluation of a similarity metric. These approaches, however, do not address the problem of eliminating normalization errors altogether. A multi-atlas labeling approach can in fact be combined with the multi-atlas library to eliminate any affine-related registration errors in the labeling.
The approach outlined here is not limited to NHP. This can be useful in a variety of difficult normalization settings, such as in neonatal brains and in the elderly. Performing normalizations to a series of atlases in a library can add considerable time to the analysis of a subject. This can be minimized by reducing the number of nonlinear registration steps, and focusing on the affine component, which is typically fast, in order to quickly determine the best performer. A more accurate normalization can then be performed using the best performer. Alternatively, this approach can be reserved for use in normalization failures only, with automatic detection on the basis of a minimum similarity metric.
Limitations
The multi-atlas approach is designed to create an excellent native space skull stripping and thereby eliminate downstream affine registration and normalization failures. The quality of this skull-stripping was determined on the basis of visual assessment. This is a common method employed across image-based studies for quality control, and currently, is still the gold standard. Failed normalizations and segmentations are quite obvious on visual inspection and well-known to both human and NHP neuroimaging researchers (similar to Figure 4). Additionally, our use of the pairwise covariance similarity metric quantitatively demonstrates the superiority of the use of the multi-atlas library. Differences in the resulting segmentations on the basis of choice of multi-atlas template for the initial skull stripping were not assessed. Any differences introduced on the basis of an improved native-space skull stripping are likely to be similar to those in existing analysis data flows in which visual inspection and manual intervention is frequently used to edit the final skull stripping, or where the use of additional tuning parameters on an individual subject basis are employed to effect a better “clean-up” of extraneous tissues. The final skull stripping, normalization, and segmentation in our procedure uses the appropriate SST and is performed within VBM8. The multi-atlas framework is used strictly for the native-space skull-stripping without introducing morphologic changes in the brain image data. While we have proposed additional steps for improving the affine registration (e.g. re-ordering the population template subject list), for the data in this study, use of the multi-atlas approach alone for the native space skull-stripping is sufficient to eliminate normalization failures. Although we have demonstrated a zero error rate for this method using our available data, the actual error rate is likely to be higher. This error rate can be further mitigated as more SSTs are incorporated into the multi-atlas library. Additionally, the several hundred individual skull-stripped and normalized NHP can each serve as part of the library, effectively expanding the range of variation for improving the success of the normalization approach and further driving the error rate to zero.
Conclusion
We describe a novel multi-atlas library framework for eliminating normalization and segmentation errors in NHP brain imaging analyses. The multi-atlas library allows for a high quality native space skull-stripping and initial affine estimate across a variety of species, effectively removing the problem of extracranial tissue that adversely affects normalization and segmentation performance. This approach can have a tremendous impact on the performance of automated NHP image analysis tools that rely on accurate matching to a template. It is also an important advance towards developing fully automated high-throughput processing pipelines that are critical for future high volume multi-center NHP imaging studies for studies of drug abuse and brain health.
Acknowledgements
This study was supported in part by R01DA025120, R37DA010584, (PI: MAN), R01HL087103 (PI: CAS), and AA014106 (PI: DPF). Support for this research was also provided by NIH grants NS0075107 and NS082453 (PI:JAM). The authors would also like to thank Ben Wagner for programming assistance and the Center for Biomolecular Imaging.
Footnotes
Information Sharing Statement
All the software used or mentioned in this article can be accessed by using the bibliographic references or URLs provided. These include SPM (http://www.fil.ion.ucl.ac.uk/spm/) , VBM8 (http://dbm.neuro.uni-jena.de/vbm.html) , ANTS (https://www.nitrc.org/projects/ants/), and the INIA19 Primate Brain Atlas (https://www.nitrc.org/projects/inia19/). The multi-atlas library will be available through NITRC (https://www.nitrc.org).
Conflict of Interest
The authors declare that they have no conflict of interest.
References
- Avants B, Gee JC. Geodesic estimation for large deformation anatomical shape averaging and interpolation. Neuroimage. 2004;23(Suppl 1):S139–150. doi: 10.1016/j.neuroimage.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med Image Anal. 2008;12(1):26–41. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC. A reproducible evaluation of ANTs similarity metric performance in brain image registration. [Evaluation Studies Research Support, N.I.H., Extramural] Neuroimage. 2011;54(3):2033–2044. doi: 10.1016/j.neuroimage.2010.09.025. doi:10.1016/j.neuroimage.2010.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czoty PW, Gage HD, Garg PK, Garg S, Nader MA. Effects of repeated treatment with the dopamine D2/D3 receptor partial agonist aripiprazole on striatal D2/D3 receptor availability in monkeys. Psychopharmacology (Berl) 2013 doi: 10.1007/s00213-013-3274-7. doi:10.1007/s00213-013-3274-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czoty PW, Nader MA. Individual differences in the effects of environmental stimuli on cocaine choice in socially housed male cynomolgus monkeys. [Research Support, N.I.H., Extramural] Psychopharmacology (Berl) 2012;224(1):69–79. doi: 10.1007/s00213-011-2562-3. doi:10.1007/s00213-011-2562-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czoty PW, Nader MA. Effects of oral and intravenous administration of buspirone on food-cocaine choice in socially housed male cynomolgus monkeys. [Research Support, N.I.H., Extramural] Neuropsychopharmacology. 2015;40(5):1072–1083. doi: 10.1038/npp.2014.300. doi:10.1038/npp.2014.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doshi J, Erus G, Ou Y, Gaonkar B, Davatzikos C. Multi-atlas skull-stripping. Acad Radiol. 2013;20(12):1566–1576. doi: 10.1016/j.acra.2013.09.010. doi:10.1016/j.acra.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorov A, Li X, Pohl KM, Bouix S, Styner M, Addicott M, et al. Atlas-guided segmentation of vervet monkey brain MRI. Open Neuroimag J. 2011;5:186–197. doi: 10.2174/1874440001105010186. doi:10.2174/1874440001105010186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang MC, et al. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. Neuroimage. 2009;46(3):786–802. doi: 10.1016/j.neuroimage.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldjian JA, Daunais JB, Friedman DP, Whitlow CT. Vervet MRI atlas and label map for fully automated morphometric analyses. Neuroinformatics. 2014;12(4):543–550. doi: 10.1007/s12021-014-9231-8. doi:10.1007/s12021-014-9231-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren DG, Kosmatka KJ, Kastman EK, Bendlin BB, Johnson SC. Rhesus macaque brain morphometry: a methodological comparison of voxel-wise approaches. Methods. 2010;50(3):157–165. doi: 10.1016/j.ymeth.2009.10.003. doi:10.1016/j.ymeth.2009.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren DG, Kosmatka KJ, Oakes TR, Kroenke CD, Kohama SG, Matochik JA, et al. A population-average MRI-based atlas collection of the rhesus macaque. Neuroimage. 2009;45(1):52–59. doi: 10.1016/j.neuroimage.2008.10.058. doi:10.1016/j.neuroimage.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohlfing T, Kroenke CD, Sullivan EV, Dubach MF, Bowden DM, Grant KA, et al. The INIA19 Template and NeuroMaps Atlas for Primate Brain Image Parcellation and Spatial Normalization. Front Neuroinform. 2012;6:27. doi: 10.3389/fninf.2012.00027. doi:10.3389/fninf.2012.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tustison NJ, Cook PA, Klein A, Song G, Das SR, Duda JT, et al. Large-scale evaluation of ANTs and FreeSurfer cortical thickness measurements. [Comparative Study Research Support, U.S. Gov't, Non-P.H.S.] Neuroimage. 2014;99:166–179. doi: 10.1016/j.neuroimage.2014.05.044. doi:10.1016/j.neuroimage.2014.05.044. [DOI] [PubMed] [Google Scholar]
- Wang J, Vachet C, Rumple A, Gouttard S, Ouziel C, Perrot E, et al. Multi-atlas segmentation of subcortical brain structures via the AutoSeg software pipeline. Front Neuroinform. 2014;8:7. doi: 10.3389/fninf.2014.00007. doi:10.3389/fninf.2014.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Kim M, Sanroma G, Wang Q, Munsell BC, Shen D, et al. Hierarchical multi-atlas label fusion with multi-scale feature representation and label-specific patch partition. Neuroimage. 2015;106:34–46. doi: 10.1016/j.neuroimage.2014.11.025. doi:10.1016/j.neuroimage.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]