Abstract
Gene therapy with genetically modified human CD34+ hematopoietic stem cells (HSCs) may be safer using targeted integration (TI) of transgenes into a genomic ‘safe harbor’ site than random viral integration. We demonstrate that temporally optimized delivery of zinc finger nuclease mRNA via electroporation and adeno associated virus (AAV) 6 delivery of donor constructs in human HSCs approaches clinically relevant levels of TI into the AAVS1 safe harbor locus. Up to 58% Venus-positive HSCs with 6–16% human cell marking were observed following engraftment into mice. In HSCs from patients with X-linked chronic granulomatous disease (X-CGD), caused by mutations in the gp91phox subunit of the NADPH oxidase, TI of a gp91phox transgene into AAVS1 in resulted in ~15% gp91phox expression and increased NADPH oxidase activity in ex vivo–derived neutrophils. In mice transplanted with corrected HSCs, 4–11% of human cells in the bone marrow expressed gp91phox. This method for TI into AAVS1 may be broadly applicable to correction of other monogenic diseases.
Ex vivo transfer of therapeutic transgenes into autologous HSCs using viral vectors has benefitted patients with rare genetic conditions, such as the leukodystrophies1 and immunodeficiencies2, 3, but has been associated with oncogenesis4, 5due to random genomic integration. Genome editing with engineered nucleases6 permits targeted rather than random genetic manipulation and is being tested in clinical trials of autologous genome-edited T cells7. For treatment of monogenic recessive diseases, insertion of transgenes at a safe harbor locus could substantially reduce the risk of insertional mutagenesis. The AAVS1 locus8 on chromosome 19 has been shown to be a safe harbor for several transgenes in a broad range of primary and transformed cell types9. Here we describe gene addition to the AAVS1 locus in human HSCs using a zinc finger nuclease (ZFN) pair and AAV6 donor. Genome editing with engineered nucleases begins with a sequence-specific double strand DNA break (DSB) that is most commonly repaired via non-homologous end-joining (NHEJ), which yields either an error-free repair or small insertions and deletions (indels) at the break site10. Alternatively, the DSB undergoes homology-directed repair (HDR) using the sister chromatid or donor sequence as a repair template, enabling targeted gene correction11 or gene addition12 if a DNA substrate that bears a small genetic change or a longer fragment flanked by homology arms to the nuclease target site is provided.
Gene correction and gene addition are usually less efficient than gene disruption, particularly in quiescent human CD34+ HSCs, in part due to cell-cycle-imposed control on the DSB-repair pathway choice9. With the goal of developing a clinical relevant strategy for gene addition at a safe harbor locus in human CD34+ HSCs, we focused first on identifying the optimal ZFN design. We used a ZFN pair8 targeting the AAVS1 locus combined with a promoterless donor construct in which the Venus fluorescent marker cDNA is preceded by a splice acceptor and a ribosome stuttering signal, and flanked by regions homologous to the AAVS1 locus (SA-2A-Venus) (Fig. 1a). In this genome editing approach, following HDR-driven targeted addition of cDNA to the AAVS1 locus, the cell becomes marker-positive because the nuclease target site at the AAVS1 locus lies within the first intron downstream of exon 1 of the PPP1R12C gene. PPP1R12C gene promoter capture is required to drive expression of the Venus coding sequence using the promoterless SA-2A-Venus donor. Based on a previously described and characterized ZFN heterodimer for targeted addition to the AAVS1 safe harbor region with a composite 24 bp-recognition site8, we generated and screened a panel of ZFN variants with longer composite recognition sites. A heterodimer of 6-finger ZFNs with the same cut-site but a longer (36 bp) recognition site was selected and used for all subsequent experiments, delivered to cells in the form of in vitro transcribed, capped, polyadenylated mRNAs13. To enhance HDR efficiency and minimize cytotoxicity of repair construct delivery to the CD34 cells, we explored a recombinant adeno-associated virus (AAV), a non-integrating vector that exists transiently in various episomal single- and double-stranded forms—in principle, optimal substrates for HDR. Recombinant vectors based on this non-pathogenic human virus have been used effectively and safely in recent clinical trials, with the AAV6 serotype showing the highest efficiency for transducing human HSCs14, 15.
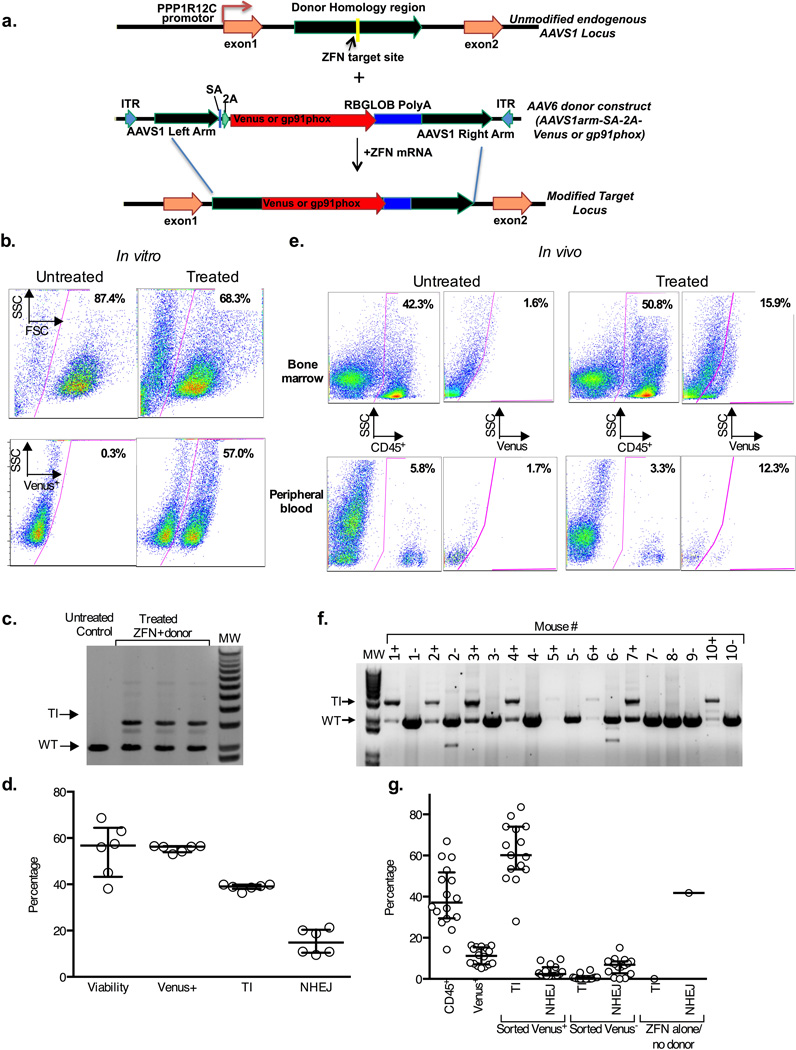
Figure 1. AAVS1 specific ZFN and AAV6 donor mediated targeted insertion of Venus fluorescent marker into human CD34+ HSCs.
a) A schematic of the AAVS1 locus (top) indicating the ZFN target site within PPP1R12C and the AAV6 donor homology regions (green). The AAV6 donor construct (middle) contains homology sequences flanking the Venus marker or gp91phox therapeutic cDNA (red) preceded by a splice-acceptor (SA) and a 2A cleavage element, and followed by a rabbit globin polyA signal. The TI modified locus is depicted below. Not shown is the gp91phox donor construct in which SA-2A is replaced by the MND promoter.
b) Flow cytometry analysis of Venus expression in 10 day cultured human HSCs with untreated or gene targeted cells in left and right panels, respectively. Treated cells were targeted by electroporation of ZFN mRNA (25µg/ml each) and transduction with AAV6-SA-2A-Venus (1×106 MOI) on day 2 of culture. Top panels show gating of viable cells analyzed for Venus expression in the bottom panels.
c) Electrophoresis gel molecular analysis of TI at the AAVS1 locus in HSCs treated as in Fig 1b. PCR with primers outside the homology region (HDR-F4 and HDR-R5 in Suppl. Fig. 5a) result in a 3.1kb product of targeted Venus insertion (TI) only seen in treated HSCs (n=3), or a 2kb product representing endogenous locus (wild type; WT).
d) Bar graph summary of in vitro efficiency of Venus TI (n=6) showing flow cytometry analysis as analyzed in Fig 1b for viability and Venus expression or showing molecular analysis using MiSeq of TI versus NHEJ at the AAVS1 site in treated HSCs as described in detail in the Methods.
e) Flow cytometry analysis of Venus expression in bone marrow (top panels) or peripheral blood (bottom panels) from NSG mice 17 weeks after transplant of human HSCs treated as in Fig 1b, but with average Venus expression of 30% before transplant. Shown are results from mice transplanted with untreated human HSCs (left panel pairs) or Venus TI treated HSCs (right panel pairs). The left panel of each pair shows gating of human CD45+ cells analyzed for Venus expression in the right panel of each pair.
f) Electrophoresis gel molecular analysis of human HSCs harvested from bone marrow of 17-week post-transplant NSG mice treated as described in Fig 1e and using primers for analysis of TI at the AAVS1 locus as described in Fig 1c. Marrow from each mouse was sorted into a human CD45+ Venus+ fraction (“+” after each mouse # at top of gel lane) and human CD45+ Venus− fractions “−“) for TI analysis. 16 mice were transplanted with human HSCs treated with Venus TI of which analysis of 8 is shown in this gel (Mice 1–7 and 10). Two mice were transplanted with control human HSCs, where mouse #8 received HSCs electroporated with AAVS1 ZFNs without AAV6 Venus donor, while mouse #9 received HSCs treated with AAV6 Venus donor alone (#9). No Venus expression was detected by flow cytometry in marrow cells from the controls, so only TI analysis of the human CD45+ Venus− sorted fractions were analyzed.
g) Bar graph summary of flow cytometry analysis and molecular analysis of Venus transgene in bone marrow from the NSG mice 17 weeks after transplant of human HSCs treated and analyzed as in Fig 1e and 1F. The first 2 bars from the left show, respectively, the level of human cell engraftment in mice transplanted with Venus TI treated human HSCs and percent of Venus expression in the human cell graft (n=16). Following cell sorting of marrow as described in Fig 1f the Venus+ (sorted V+) and Venus− (sorted V−) fractions (middle 4 bars) were analyzed by MiSeq for presence of Venus transgene (T) and for NHEJ (n=14). The two bars at the right side of the graph show analysis of the control mouse transplanted with human HSCs treated with ZFN without AAV6 Venus donor, leading to high NHEJ without any detectable transgene. The quantification of Venus transgene and NHEJ was assessed using only the MiSeq primer set (Mi-F,Mi-R, plus 2A-R), without first amplifying with an Out/Out primer set (see Suppl. Fig 5a and as described in detail in the Methods). Thus, this primer set detects donor both within AAVS1 and off-target anywhere in the genome (T).
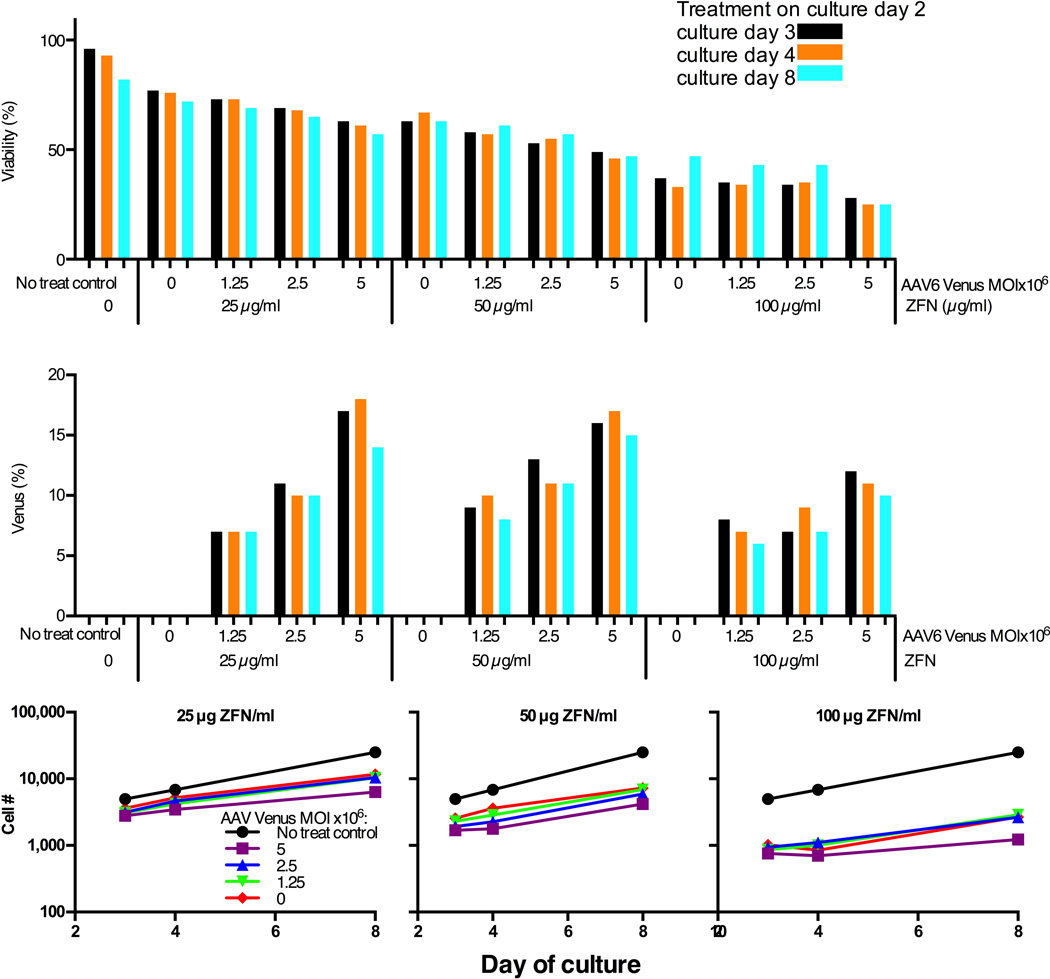
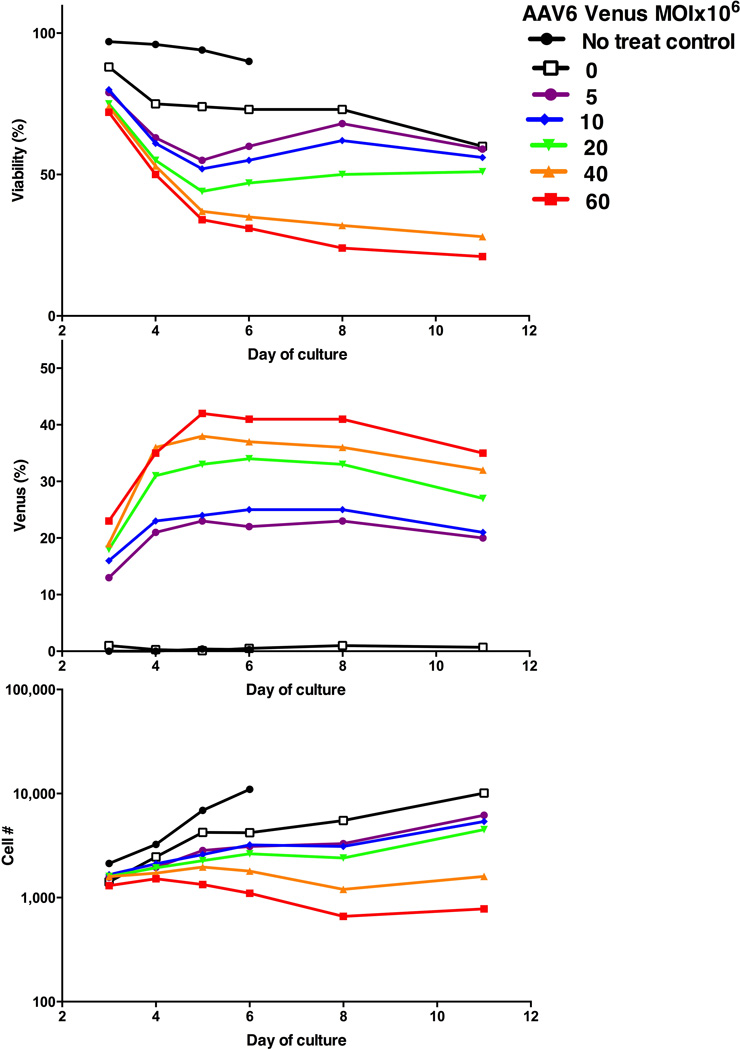
Next, we optimized parameters for ZFN mRNA electroporation and AAV6 delivery, using healthy donor (HD) peripheral blood mobilized CD34+ HSCs obtained by apheresis and immunoselection. Delivery of the ZFN mRNAs was performed in a clinical-grade electroporator (MaxCyte GT) which has a Device Master File with the FDA and is scalable for processing of clinically relevant numbers of HSCs. To determine the amount of ZFN mRNA required to promote integration of a transgene delivered by AAV6, HSCs were electroporated with ZFN mRNA concentrations ranging from 6.25 µg/mL to 100 µg/mL for each ZFN, each titrated against incremental amounts of AAV6-SA-2A-Venus. As gauged by cell viability, % of live cells expressing Venus, and cell proliferation (Fig. 2 & Suppl. Fig. 1), 25 µg/mL ZFN mRNA was selected for subsequent experiments. Cells were then infected with varying amounts of AAV6 bearing a promoterless Venus construct and electroporated with the optimal (25 µg/ml) ZFN mRNA dose. Increased Venus expression in transfected HSCs was associated with reduced cell viability and proliferation (Fig. 3), illustrating the importance of balancing transduction efficiency from increasing AAV6-SA-2A-Venus virus input with cytotoxicity. Finally, because the donor-delivering AAV is replication-incompetent and diluted upon mitosis, and because donor-dependent HDR-directed targeted addition occurs preferentially in S/G2 phase, we reasoned that the timing of the delivery of the virus bearing a repair donor template is critical. We evaluated the timing of ZFN mRNA transfection by electroporating HSCs with ZFN mRNA following 1, 2, or 3 days of in vitro culture, followed by AAV6 SA-2A-Venus delivery. As shown in Suppl. Fig. 2, optimal integration of promoterless Venus into the AAVS1 site occurred if electroporation is performed following 2 days of in vitro culture. The timing of AAV6 addition relative to the timing of electroporation of HSCs with ZFN mRNA was also investigated, and robust rates of TI were consistently achieved when the cells were transduced with the AAV6 donor immediately after ZFN electroporation (data not shown).
Figure 2. Optimization of ratios of AAVS1 ZFN mRNA to AAV6 Venus donor.
On day two of culture healthy donor CD34+ HSCs (2×105 cells/10 µl per sample) were electroporated (MaxCyte GT instrument) with a series of indicated AAVS1 ZFN mRNA pair concentrations immediately followed by exposure to indicated AAV6 Venus donor MOIs. This figure shows cell viability (top), percent of live cells expressing Venus marker (middle) and relative viable cell numbers at each day of culture after treatment in each sample following treatment to reflect cell proliferative capability after treatment (bottom). Values were determined for each analysis at culture day 3, 4 and 8 (corresponding to 1, 2 and 6 days post treatment).
Figure 3. Optimization of AAV6 Venus donor MOI.
Healthy donor CD34+ HSCs (2×105 cells/10 µl per sample) were electroporated with AAVS1 ZFN mRNAs at 25g/mL, followed by transduction with the indicated dilutions of AAV6 Venus. This figure shows cell viability (top), percent of live cells expressing Venus marker (middle) and relative viable cell numbers (bottom) at indicated days of culture after treatment in each sample.
Under optimal ZFN electroporation and AAV6 donor delivery conditions, we observed high levels of Venus marker-positive cells in HSCs with robust viability as assessed by flow cytometry after 10 days of culture (Fig. 1b), averaging 55.4% Venus positive, 54.7% viability (Fig. 1d, n=6 experiments). The AAV6-Venus construct contains a splice acceptor (SA) and a 2A ribosome stuttering signal preceding the Venus cDNA but no promoter, thus ensuring that marker expression in the target cell is driven by activity of the PPP1R12C gene promoter following HR-driven TI into the AAVS1 locus. To determine the efficiency of TI into the AAVS1 locus in human HSCs, we used PCR employing two primers located in the AAVS1 locus outside the region of homology present in the donor16,12(Fig. 1c, Schematic in Suppl. Fig. 3a), and a semi-quantitative TI deep sequencing assay (MiSeq) to determine the ratio of the AAVS1 alleles containing the integrated transgene to that of alleles containing the wild-type locus (Fig 1d, 38.8% TI and 15.2% NHEJ-derived indels). Because AAVS1 is on an autosome, TI into just one of the two alleles of the target locus is sufficient to yield a marker-positive cell. Thus, if all cells bore an editing-driven transgene at only one AAVS1 allele, a TI efficiency of 38.8% TI would yield 77.6% Venus positive cells. We observed 55.4% Venus positive cells (Fig. 1d); our previous data and Poisson distribution statistics indicate that a fraction of cells bear a transgene at both alleles of the nuclease-targeted locus8, 11, 12.
We next used long-read next-generation sequencing (PacBio, Suppl. Fig. 3b) to characterize the integration junctions between the chromosomal and donor-specified sequences. This analysis showed that 1083 out of 1085 (>99.7%) AAVS1 integration events resulted from bona fide HDR (Suppl. Fig. 3c) rather than NHEJ. Further, a junction PCR assay failed to reveal end-capture based integration of the repair construct into the two most common off-target sites for the ZFNs used in these experiments (data not shown).
We also assessed ZFN-driven targeted integration of a Venus transgene into the subset of CD34+ HSCs capable of long-term engraftment in immune-deficient NOD-scid IL2Rγnull (NSG) mice preconditioned with busulfan. For our initial mouse transplant experiments, in which in vitro Venus expression averaged 30% of the human HSCs injected into NSG mice, engraftment of human cells at 8 weeks was 15% in mouse bone marrow, and 12.4% of these human cells were Venus positive (Suppl. Fig. 4a, b). Significant Venus expression was observed in the human cells present in spleen and peripheral blood of these mice (Suppl. Fig. 4a ii, 4a iii), demonstrating efficient differentiation of TI-targeted HSCs into mature blood cells.
NSG transplant experiments to assess engrafted human HSCs at 17 weeks (4 months) showed similar in vitro Venus targeting efficiency. In the representative flow cytometry analysis shown in Fig. 1e, engraftment of human cells in the NSG bone marrow was similar for untreated and treated human HSCs, where 15.9% of human CD45+ cells were Venus positive (Fig. 1e). Long term marking of human cells in NSG mouse peripheral blood was also observed, where the comparable levels of Venus expression in peripheral blood and bone marrow suggest efficient engraftment and differentiation into multiple peripheral blood lineages of the human HSCs bearing the transgene at the safe harbor locus (Fig. 1e). Overall, average CD45+ human cell engraftment in the bone marrow (n=16) was 40.1 ±14.6% (mean ±SD), of which 10.8±4.2% (mean ±SD) were Venus+ (Fig. 1g first two bars on the left).
We further investigated the efficiency and molecular structure of targeted and, potentially, random integration events in the long-term NSG transplants by sorting for human CD45+ and Venus expression (positive and negative). Targeted integration into the AAVS1 locus was measured on DNA from all sorted fractions from each animal using a conventional PCR-based (Fig. 1f) as well as a deep sequencing–based assay (Fig. 1g). As shown in Fig.1f, Venus+, but not the Venus−, cells bear a substantial fraction of TI in the AAVS1 alleles; where the size of the band is consistent with HDR repair. These data indicate that Venus TI in AAVS1 maintains expression during long-term engraftment in immune-deficient mice. Further, these data argue that, at least under these experimental conditions, endcapture–based integration of the donor construct into the targeted locus (and, by inference, into potential off-target sites that are cut at significantly lower efficiency) occurs at a level below the limit of detection of the assay shown in Fig. 1f.
Deep sequence analysis of the same fractions was performed to assess frequency of TI versus NHEJ-induced indels (Fig. 1g 3rd to 8th bars from the left). For all 16 mice, sorted Venus+ CD45+ bone marrow cells averaged 61.5% versus 3.7% of AAVS1 alleles with TI versus NHEJ, respectively, with the remaining alleles retaining wild-type sequence. For 14 of the 16 mice (2 failed PCR amplification), the MiSeq analysis showed that sorted Venus− CD45+ bone marrow cells had an average of 0.7% versus 6.2% of AAVS1 alleles with TI versus NHEJ, respectively, with the remaining alleles retaining wild-type sequence. This confirms that there is minimal, if any, functional silencing of the Venus TI over 4 months in vivo. In further support of our notion that endcapture–based integration into the AAVS1 target as well as potential off-target sites occurs at a level below the limit of detection of our assays, a deep-sequencing assay for the transgene itself in the marker-negative cell fraction yielded signal indistinguishable from background levels (data not shown).
As noted above, when human CD34+ HSCs that are 30% Venus positive from gene targeting were transplanted into NSG mice, the average marking of 4 month engrafted human cells from the mouse marrow was 10.8%. Our deep sequence data show that the observed decrease of marking in vivo represents real differences in initial TI marking of short-term, medium-term and long-term repopulating HSCs initially transplanted, rather than functional silencing of TI at the AAVS1 site. Further, our data argue against the likelihood that the genome of the edited cells bear a substantial burden of randomly integrated, non-expressing Venus transgenes. In sum, this protocol achieved efficient TI of a repair-construct-specified transgene into the AAVS1 locus in primary human HSPCs.
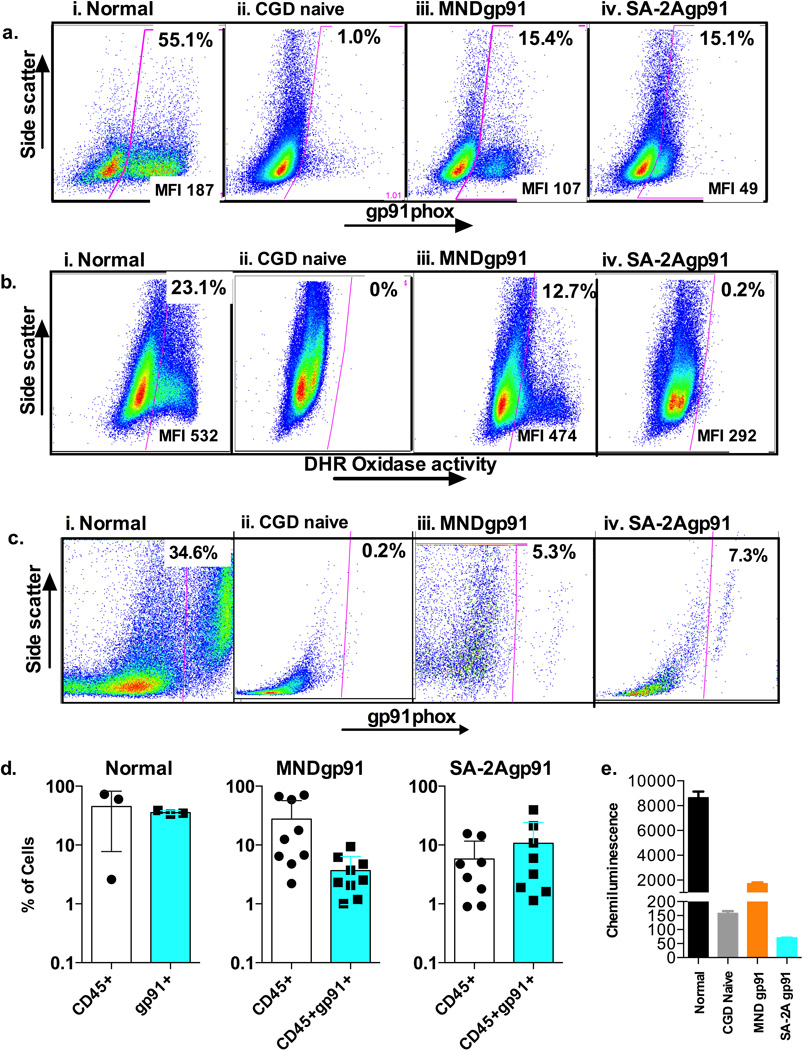
X-linked chronic granulomatous disease (X-CGD) is caused by mutations in the CYBB gene that encodes the gp91phox subunit of the phagocyte NADPH oxidase, resulting in impaired production of antimicrobial reactive oxidative species17. Consequently, patients with X-CGD suffer from severe bacterial and fungal infections18 with excessive inflammation. To date, 3 of the reported 13 X-CGD patients who have undergone gene therapy have developed myelodysplasia related to integrations in MDS-EVI15, thus prompting us to test whether TI of functional gp91phox into the AAVS1 safe harbor locus of HSCs from patients with X-CGD could repair the defect in reactive oxidative species generation. The optimized delivery protocol for ZFN mRNA electroporation and AAV6 delivery described above was used to test TI of the therapeutic cassette in CD34+ HSCs obtained from X-CGD patients using AAV6 donors that either contained an MND-promoter-driving gp91phox cDNA or a promoterless construct with a splice acceptor and a 2A ribosomal stutter sequence preceding gp91phox cDNA. The MND (Myeloproliferative sarcoma virus MPSV enhancer, negative control region NCR deleted, d1587rev primer-binging site substituted) promoter has been shown to be suitable to drive high levels of transgene expression in HSCs19, 20. Further optimization of AAV6-donor delivery into HSCs from CGD patients to balance effects on cell growth and transfection efficiency achieved 15% gp91phox protein expression in vitro (Fig. 4a) from both the MND-gp91 donor and the SA-2A-gp91 donor. Deep sequencing assays (not shown) demonstrated a TI efficiency of 7.1% TI (a result consistent with most or all of the gp91 positive cells having gene targeting of only one of the two AAVS1 alleles), with NHEJ rate of 20.5%. Despite similar TI efficiencies with both MND-91phox and SA-2A-gp91 donors, corrected cells produce significantly more gp91 from the MND promoter (MFI 107) versus from the captured PPP1R12C promoter (MFI 49), though both are less than the native production of gp91 in the normal control (MFI 187). DHR assay (Fig. 4b) showed that MND-gp91 TI corrected CGD HSCs at this stage of differentiation have NADPH oxidase activity approaching normal levels on a per cell basis. When the culture was allowed to mature further and a chemiluminescence assay performed (Fig. 4e) the MND-gp91 TI corrected CGD HSCs generated reactive oxidase species that was approximately 20% of the normal control. The low amount of gp91 produced per cell following SA-2A-gp91 donor correction (Fig. 4a, iv) was associated with very low oxidase activity, whether by DHR analysis (Fig 4b, iv) or by chemiluminescence (Fig. 4e).
Figure 4. AAVS1 specific ZFN and AAV6 donor mediated targeted insertion of gp91phox corrective gene into human CD34+ HSCs from patients with X-linked chronic granulomatous disease (X-CGD).
a) Flow cytometry analysis of gp91phox expression in 7 day cultured untreated healthy human donor HSCs (normal), untreated XCGD patient HSCs (CGD naïve), ZFN plus AAV6-MND-gp91 TI treated XCGD patient HSCs (MNDgp91), and ZFN plus AAV6-SA-2A-gp91 TI treated XCGD patient HSCs (SA-2Agp91). Indicated at the top of the gated area of each panel is the percent of gp91phox positive cells, where the mean fluorescence intensity (MFI) indicates the average per cell expression of gp91phox positive cell.
b) Flow cytometry dihydrorhodamine (DHR) analysis of NADPH oxidase activity in 14 day cultured untreated healthy human donor HSCs (normal), untreated XCGD patient HSCs (CGD naïve), ZFN plus AAV6-MND-gp91 TI treated XCGD patient HSCs (MNDgp91), and ZFN plus AAV6-SA-2A-gp91 TI treated XCGD patient HSCs (SA-2Agp91). Indicated at the top of the gated area of each panel is the percent of oxidase positive cells, where the mean fluorescence intensity (MFI) indicates the average per cell oxidase activity per positive cell.
c) Flow cytometry analysis of gp91phox expression of the human CD45+ cells in bone marrow from NSG mice 8 weeks after transplant of untreated healthy human donor HSCs (normal), untreated XCGD patient HSCs (CGD naïve), ZFN plus AAV6-MND-gp91 TI treated XCGD patient HSCs (MNDgp91), and ZFN plus AAV6-SA-2A-gp91 TI treated XCGD patient HSCs (SA-2Agp91). The gp91phox expression in culture cells before transplant is as shown in Fig 2a. Indicated at the top of the gated area of each panel is the percent of gp91phox positive cells.
d) Bar graph summary of flow cytometry analysis of the level of human cell engraftment (CD45+) and percent of the human CD45+ cells expressing gp91phox (CD45+gp91+) in bone marrow of mice transplanted with untreated healthy human donor HSCs (normal) (n=3), ZFN plus AAV6-MND-gp91 TI treated XCGD patient HSCs (MNDgp91) (n=9), and ZFN plus AAV6-SA-2A-gp91 TI treated XCGD patient HSCs (SA-2Agp91) (n=9). The average % is shown at the top of each bar.
e) Functional correction in X-CGD patient CD34+ progenitors. Bar graph summary of phorbol myristate acetate stimulated chemiluminescence detection of reactive oxidative species production from similarly in vitro cultured CD34+ progenitors differentiated into myeloid cells over × days in culture from a normal donor (N), untreated XCGD patient HSCs (CGD naïve), ZFN plus AAV6-MND-gp91 TI treated XCGD patient HSCs (MNDgp91), and ZFN plus AAV6-SA-2A-gp91 TI treated XCGD patient HSCs (SA-2Agp91). The mean chemiluminescence intensity units are shown at the top of each bar.
CGD patient CD34+ HSCs transfected with the AAVS1-targeting ZFNs and transduced with AAV6-MNDgp91 or AAV6-SA-2A-gp91 were transplanted into NSG mice, and following 8-week engraftment were analyzed for expression of gp91 in the human CD45+ cells from NSG mouse bone marrow (Figs. 2c and d). A representative set of flow cytometry analyses of gp91phox expression in human CD45+ cells engrafted in NSG marrow is shown in Fig 2c following transplant of normal, CGD non-treated, and CGD TI corrected HSCs. In this example, there is negligible expression of gp91 from the untreated CGD HSCs transplant, but the TI corrected CGD transplants result in rates of gp91 positive cells that are >20% of the normal control. The aggregate data for these studies from the bone marrow of transplanted NSG mice are shown in Fig. 2d where it can be appreciated that healthy volunteer HSCs transplanted into NSG mice result in 35.6±2.8% (mean ±SD) of human CD45+ cells from marrow expressing gp91. The MND-91 TI and 2A-2A-gp91 corrected HSC grafts average 3.7±4.2% (mean ±SD)(Fig. 2d ii), and 10.7±4.2% (mean ±SD) (Fig.2d iii) of human CD45+ cells from marrow expressing gp91, or >10% of the normal control (Fig. 2d i).
A fundamental challenge in the clinical management of genetic diseases is the gap between recognition of the causative genetic lesion and the ability to functionally repair cells harboring the mutation. Using optimized delivery of ZFN and AAV6 to repair the genetic defect in HSCs, we demonstrate >50% targeted genomic incorporation of a marker transgene in vitro. We then applied this method to repair of HSCs from patients with X-CGD and observed persistent gp91 expression and restoration of NADPH activity in neutrophils derived from these cells following engraftment into NSG mice, which approaches clinically relevant levels. Because all genome editing reagents used here can be deployed in a GMP setting, this approach may enable therapy for X-CGD and other monogenic diseases.
Methods
Approvals for human blood and animal use
Human CD34+ hematopoietic stem cells (HSC) from healthy volunteer donors and patients with chronic granulomatous disease (CGD) purified from G-CSF mobilized peripheral blood were obtained after written informed consent under the auspices of National Institute of Allergy and Infectious Diseases (NIAID) Institutional Review Board–approved protocols 05-I-0213 and 94-I-0073. The conduct of these studies conforms to the Declaration of Helsinki protocols and all US federal regulations required for protection of human subjects.
Use of immune deficient NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice (from The Jackson Laboratory) for xenotransplant studies was approved by the NIAID Institutional Animal Care and Use Committee under animal use protocol LHD 3E. The conduct of these studies conforms to AAALAC International guidelines and all US federal regulations required for protection of research animals.
ZFN reagents
ZFNs targeting the AAVS1 locus have been described previously8. An optimized pair of 423 the AAVS1-targeting ZFNs with ELD:KKR obligate heterodimeric FokI variants were used in this study.
| AAVS1 ZFN SBS#30035 (NELD): | |||||||
| 1 | MDYKDHDGDY | KDHDIDYKDD | DDKMAPKKKR | KVGIHGVPAA | MAERPFQCRI | CMRNFSRSDH | |
| 61 | LSRHIRTHTG | EKPFACDICG | RKFATSGHLS | RHTKIHTGSQ | KPFQCRICMR | NFSYNWHLQR | |
| 121 | HIRTHTGEKP | FACDICGRKF | ARSDHLTTHT | KIHTGSQKPF | QCRICMRNFS | HNYARDCHIR | |
| 181 | THTGEKPFAC | DICGRKFAQN | STRIGHTKIH | LRGSQLVKSE | LEEKKSELRH | KLKYVPHEYI | |
| 241 | ELIEIARNST | QDRILEMKVM | EFFMKVYGYR | GKHLGGSRKP | DGAIYTVGSP | IDYGVIVDTK | |
| 301 | AYSGGYNLPI | GQADEMERYV | EENQTRDKHL | NPNEWWKVYP | SSVTEFKFLF | VSGHFKGNYK | |
| 361 | AQLTRLNHIT | NCNGAVLSVE | ELLIGGEMIK | AGTLTLEEVR | RKFNNGEINF | RS* | |
| AAVS1 ZFN SBS#30054 (CKKR): | |||||||
| 1 | MDYKDHDGDY | KDHDIDYKDD | DDKMAPKKKR | KVGIHGVPAA | MAERPFQCRI | CMRNFSDRSN | |
| 61 | LSRHIRTHTG | EKPFACDICG | RKFALKQHLT | RHTKIHTHPR | APIPKPFQCR | ICMRNFSTSG | |
| 121 | NLTRHIRTHT | GEKPFACDIC | GRKFARRDWR | RDHTKIHTGS | QKPFQQCRICM | ||
| RNFSQSSHLT | |||||||
| 181 | RHIRTHTGEK | PFACDICGRK | FARLDNRTAH | TKIHLRGSQL | VKSELEEKKS | ELRHKLKYVP | |
| 241 | HEYIELIEIA | RNSTQDRILE | MKVMEFFMKV | YGYRGKHLGG | SRKPDGAIYT | VGSPIDYGVI | |
| 301 | VDTKAYSGGY | NLPIGQADEM | QRYVKENQTR | NKHINPNEWW | KVYPSSVTEF | KFLFVSGHFK | |
| 361 | GNYKAQLTRL | NRKTNCNGAV | LSVEELLIGG | EMIKAGTLTL | EEVRRKFNNG | EINF* | |
mRNA production
Plasmids encoding the ZFNs with 64 alanines 3’ of the coding sequence were linearized with SpeI (New England Biolabs) digestion and purified by phenol:chloroform before use as a template for mRNA synthesis by in vitro transcription. mRNA was prepared using the mMESSAGE mMACHINE® T7 ULTRA Kit (Life Technologies, Carlsbad, CA) according to the manufacturer’s protocol and was cleaned up with the RNeasy MinElute Cleanup Kit (Qiagen). Alternatively, mRNA was purchased from TriLink Biotechnologies (San Diego, CA).
AAV6 Donor Construction and Virus Production
The AAV donor plasmid construct for the Venus fluorescent marker has the cDNA preceded by a splice acceptor (SA) sequence and 2A peptide sequences, and flanked by homology arms to the human AAVS1 gene locus8. Two versions of the AAV donor construct for gp91 were designed; the first one resembles the AAV Venus construct with a SA and 2A elements preceding the gp91phox cDNA; and a second one that contains a MND promoter driving the gp91 cDNA, both constructs are flanked by AAVS1 homology arms. AAV viruses were produced using a previously described triple plasmid transfection method13. Briefly, HEK 293 cells plated in 10-layer CellSTACK chambers (Corning, Acton, MA) were cultured for 3 days until cells were 80% confluent before calcium phosphate transfection of an AAV helper plasmid expressing AAV2 Rep and AAV6 serotype specific Cap genes, an adenovirus helper plasmid, and the ITR-containing AAV vector genome donor plasmid. 3 days later, the cells were harvested, lysed by repeat freeze/thaw, and the virus precipitated using polyethylene glycol, followed by overnight ultracentrifugation on a cesium chloride gradient, and final formulation by dialysis and filter sterilization. For some experiments (the Venus targeting experiments in the 17 week engrafted NSG mice), the virus preps were generated by Virovek (Hayward, Ca) using their commercial proprietary methodology that is similar to the details provided above.
Human CD34+ HSC culture, electroporation and transduction for in vitro analysis, and transplant into NSG mice
Cryopreserved human CD34+ HSC were thawed at 37°C, washed in phosphate-buffered saline (Life Technologies) and prestimulated in complete culture medium (Stemspan supplemented with stem cell factor (SCF), fms-related tyrosine kinase 3 ligand (Flt3-L), and thrombopoietic (TPO), each at 100ng/mL (Peprotech, NJ). For electroporation, 3–6 ×106 cells were spun at 100–300 × g for 10 minutes, followed by resuspension in 100 µL of MaxCyte electroporation buffer. The cell suspension was mixed with ZFN mRNA and pulsed with HSC-CL1 program (MaxCyte Systems, Gaithersburg, MD). Following electroporation, the cells were transduced with AAV6-donor (MOI 1–3 × 106 vector genomes/cell) and washed after 3 hours. Treated cells were kept in culture and either transplanted into NSG mice the following day (day 3 of culture), or maintained in vitro for analyses.
For transplant studies, following overnight recovery, treated cells (1–2 × 106 per mouse) were transplanted into 6- to 8-week old NSG mice (Jackson Laboratory) pre-conditioned with intraperitoneal busulfan (20mg/kg) 24 hours prior. The number of mice transplanted depended on amount of material available, particularly with patient CD34+ HSC. Following 6–8 weeks, the mice were euthanized for analysis.
Analysis of human cell engraftment and transgene expression by flow cytometry
Fluorochrome-conjugated anti-human monoclonal antibodies were used to identify human hematopoietic cells (CD45-PE (phycoerythrin), human myeloid cells (CD13-APC (allophycocyanin). Human gp91phox expression was determined by indirect staining with murine monoclonal antibody 7D5 followed by FITC-conjugated goat anti-mouse immunoglobulin G antibody. For flow cytometry, a FACSsort (Argon laser; Becton Dickinson, San Jose, CA) was used and the data analyzed using Flowjo version 9.7.6 software.
Dihydrorhodamine flow cytometric assay
We measured phorbol 12-myristate 13-acetate (PMA)–stimulated ROS production either in NSG mice bone marrow cells, or in human myeloid cells differentiated in ex vivo culture from naive and gene-edited X-CGD CD34+ HSC or normal donor CD34+ HSC.
Analysis of gene modification
Several molecular assays were used to detect integration at the AAVS1 locus. Out/Out PCR used primers that recognize human AAVS1 genomic sequences outside of homology regions present in the donor. Targeted integration in the target AAVS1 locus will result in PCR product that is larger than the wild type product, and the relative integration frequency can be estimated using densitometry based on the relative intensity of the bands. The primers used are listed in Supplemental Table 1 and the approximate location of each of these primers is shown in Supplemental Figure 5a For the analyses shown in Figures 1c and 1f the Out/Out PCR primers HDR-F4 and HDR-R5 were used. For a more robust semi-quantitative estimate of the frequency of targeted integration, we developed an assay based on Illumina MiSeq deep sequencing. For the molecular analysis shown in Figure 1d DNA was first amplified using the Out/Out PCR as described above in order to avoid amplification analysis of contaminating AAV6 donor, then amplified using a target-specific MiSeq adaptor primer pair containing a set of MiSeq primers recognizing the AAVS1 locus sequences flanking the ZFN binding site (Mi-F and Mi-R) as well as a third primer recognizing sequence from within the transgene (2A-R or PolyA-F) (the size of TI PCR product generated by the Mi-F/2A-R pair or PolyA-F/Mi-R pair is very similar in size to the NHEJ or wild-type product generated by the Mi-F/Mi-R pair, Supplemental Table 1). Barcodes were then added in this secondary PCR reaction. For the molecular analyses of the 17 week engrafted NSG mouse bone marrow fractions shown in Fig 1g an Out/Out primary PCR was not needed because it was presumed there was no contaminating AAV6 donor. Only the MiSeq primers Mi-F, 2A-R and Mi-R were used. Also as a result of not first using Out/Out primary PCR, the molecular analysis shown in Figure 1f would detect not only TI at the AAVS1 site, but also any non-homologous insertion of Venus donor throughout the genome. The final PCR products were cleaned and sequenced in an Illumina MiSeq sequencer using manufacturer’s protocols (Illumina, San Diego, CA). For analysis of gene modification levels, a custom computer script was used to merge paired-end 150bp sequences, and adapter trimmed via SeqPrep (John St. John, https://github.com/jstjohn/SeqPrep, unpublished) and aligned to the wild-type template sequence. Merged reads were filtered using the following criteria: the 5’ and 3’ ends (23bp) must match the expected amplicon exactly, the read must not map to a different locus in the target genome as determined by Bowtie222 with default settings, and deletions must be <70% of the amplicon size or <70bp long. Indel events in aligned sequences were defined as described previously23, with the exceptions that indels of 1bp in length were also considered true indels to avoid undercounting real events, and true indels must include deletions occurring at or near the predicted ZFN cut site. Targeted integration TI events were defined based on perfect alignment with the DNA sequence containing the expected integrated sequences (2a or gp91 polyA).
Extended sequencing of AAVS1 locus
We investigated the integration mechanism at the AAVS1 site using PacBio single molecule sequencing. PCR primers were designed with one inside the vector specific region and the other outside the vector in the non-overlapping genomic region of the AAVS1 locus. This allowed us to amplify the junctions of the integrated vector. Following HDR-mediated integration, the ITR is lost due to homologous recombination since it is located outside the homologous region. Therefore the NHEJ junction sequence did not contain any ITR sequence. In contrast, NHEJ mediated integration retains an ITR in the junction sequence. The expected amplicon sizes range from 1.1 kb to 2.5 kb depending on whether the ITR is retained. We amplified both AAVS1L junction and AAVS1R junction. PacBio long single DNA molecule sequencing identified 1082 integration events without ITR sequence and 3 integration events retaining ITR sequence. The results suggest that the vast majority of AAV integration at the AAVS1 locus occurred through homologous recombination between vector bearing-AAVS1 sequence flanking the transgene and host AAVS1 sequence.
Primers for Pac-Bio sequencing
| AAVS1-L | CTGCCGTCTCTCTCCTGAGT |
| Venus_AAVS1-L | GCTGAACTTGTGGCCGTTTA |
| Venus_AAVS1-R | GAGCAAAGACCCCAACGAG |
| AAVS1-R | AAAAGGCAGCCTGGTAGACA |
Superoxide measurement
A quantiative ferricytochrome C reduction assay was used to measure superoxide production at 10 and 60 minutes after stimulation.β
Supplementary Material
Acknowledgments
These studies were supported in part by project 1 ZIA AI000644 of the intramural program of NIAID, NIH. Human CD34+ HSCs were obtained under NIAID IRB approved Protocol 94-I-0073 after written informed consent. Murine animal studies were performed under NIAID ACUC approved protocol LHD 3E.
Footnotes
Author contributions:
SDR, AR, P-QL, LL, XW performed most of the experiments, NT, UC, JL, SK, CR, HN, LS, SW, AHS, AC, JRP, DEP, DAS developed reagent, ran assays and analyzed samples, MVP, PDG provided support, SDR, KZ, AR, FDU, HLM designed experiments, wrote manuscript.
Competing Financail Intersests:
- Sangamo BioSciences, Inc.; AR, P-Q L, CR,AHS,AC, JRP, DEP, DAS, PDG (now at Bluebird Bio), FU; and
- MaxCyte lnc.: LL, MVP; and
- Leidos Biomedical Research, lnc.-XW and LS.
References
- 1.Biffi A, et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science. 2013;341:1233158. doi: 10.1126/science.1233158. [DOI] [PubMed] [Google Scholar]
- 2.Hacein-Bey-Abina S, et al. Efficacy of gene therapy for X-linked severe combined immunodeficiency. N Engl J Med. 2010;363:355–364. doi: 10.1056/NEJMoa1000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavazzana-Calvo M, et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288:669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- 4.Hacein-Bey-Abina S, et al. A serious adverse event after successful gene therapy for X-linked severe combined immunodeficiency. The New England journal of medicine. 2003;348:255–256. doi: 10.1056/NEJM200301163480314. [DOI] [PubMed] [Google Scholar]
- 5.Stein S, et al. Genomic instability and myelodysplasia with monosomy 7 consequent to EVI1 activation after gene therapy for chronic granulomatous disease. Nature medicine. 2010;16:198–204. doi: 10.1038/nm.2088. [DOI] [PubMed] [Google Scholar]
- 6.Carroll D. Genome Engineering with Targetable Nucleases. Annu Rev Biochem. 2014 doi: 10.1146/annurev-biochem-060713-035418. [DOI] [PubMed] [Google Scholar]
- 7.Tebas P, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370:901–910. doi: 10.1056/NEJMoa1300662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeKelver RC, et al. Functional Genomics, Proteomics, and Regulatory DNA Analysis in Isogenic Settings Using Zinc Finger Nuclease-Driven Transgenesis Into a Safe Harbor Locus in the Human Genome. Genome Research. 2010;20:1133–1142. doi: 10.1101/gr.106773.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Genovese P, et al. Targeted genome editing in human repopulating haematopoietic stem cells. Nature. 2014;510:235–240. doi: 10.1038/nature13420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carroll D, Beumer KJ. Genome engineering with TALENs and ZFNs: repair pathways and donor design. Methods. 2014;69:137–141. doi: 10.1016/j.ymeth.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urnov FD, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 12.Moehle EA, et al. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc Natl Acad Sci U S A. 2007;104:3055–3060. doi: 10.1073/pnas.0611478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vierstra J, et al. Functional footprinting of regulatory DNA. Nat Methods. 2015 doi: 10.1038/nmeth.3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song L, et al. Optimizing the transduction efficiency of capsid-modified AAV6 serotype vectors in primary human hematopoietic stem cells in vitro and in a xenograft mouse model in vivo. Cytotherapy. 2013;15:986–998. doi: 10.1016/j.jcyt.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Song L, et al. High-efficiency transduction of primary human hematopoietic stem cells and erythroid lineage-restricted expression by optimized AAV6 serotype vectors in vitro and in a murine xenograft model in vivo. PLoS One. 2013;8:e58757. doi: 10.1371/journal.pone.0058757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orlando SJ, et al. Zinc-finger nuclease-driven targeted integration into mammalian genomes using donors with limited chromosomal homology. Nucleic Acids Res. 2010;38:e152. doi: 10.1093/nar/gkq512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhns DB, et al. Residual NADPH oxidase and survival in chronic granulomatous disease. The New England journal of medicine. 2010;363:2600–2610. doi: 10.1056/NEJMoa1007097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marciano BE, et al. Common severe infections in chronic granulomatous disease. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2015;60:1176–1183. doi: 10.1093/cid/ciu1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Challita PM, et al. Multiple modifications in cis elements of the long terminal repeat of retroviral vectors lead to increased expression and decreased DNA methylation in embryonic carcinoma cells. J Virol. 1995;69:748–755. doi: 10.1128/jvi.69.2.748-755.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Astrakhan A, et al. Ubiquitous high-level gene expression in hematopoietic lineages provides effective lentiviral gene therapy of murine Wiskott-Aldrich syndrome. Blood. 2012;119:4395–4407. doi: 10.1182/blood-2011-03-340711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xiao X, Li J, Samulski RJ. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. Journal of virology. 1998;72:2224–2232. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nature methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gabriel R, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nature biotechnology. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- 24.Malech HL, et al. Prolonged production of NADPH oxidase-corrected granulocytes after gene therapy of chronic granulomatous disease. Proc Natl Acad Sci U S A. 1997;94:12133–12138. doi: 10.1073/pnas.94.22.12133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.