Abstract
Background
Neuroprotection for Parkinson Disease (PD) remains elusive. Biomarkers hold the promise of removing roadblocks to therapy development. The National Institute of Neurological Disorders and Stroke has therefore established the Parkinson’s Disease Biomarkers Program to promote discovery of PD biomarkers for use in phase II-III clinical trials.
Methods
Utilizing a novel consortium design, the Parkinson’s Disease Biomarker Program is focused on the development of clinical and laboratory-based biomarkers for PD diagnosis, progression, and prognosis. Standardized operating procedures and pooled reference samples were created to allow cross-project comparisons and assessment of batch effects. A web-based Data Management Resource facilitates rapid sharing of data and biosamples across the research community for additional biomarker projects.
Results
Eleven consortium projects are ongoing, seven of which recruit participants and obtain biosamples. As of October 2014, 1082 participants have enrolled (620 PD, 101 with other causes of parkinsonism, 23 essential tremor, and 338 controls), 1040 of whom have at least one biosample. There are 6898 total biosamples from baseline, 6, 12, and 18-month visits: 1006 DNA, 1661 RNA, 1419 whole blood, 1382 plasma, 1200 serum, and 230 cerebrospinal fluid (CSF). Quality control analysis of plasma, serum, and CSF samples indicates almost all samples are high quality (24 of 2812 samples exceed acceptable hemoglobin levels).
Conclusions
By making samples and data widely available, using stringent operating procedures based upon existing standards, hypothesis testing for biomarker discovery, and providing a resource which complements existing programs, the Parkinson’s Disease Biomarker Program will accelerate the pace of PD biomarker research.
Keywords: Parkinsonism, disease-modifying strategies, biofluids, data management
Background
Trial design and clinical management in Parkinson disease (PD) could be greatly improved with biomarkers. A biomarker is “a characteristic that is objectively measured and evaluated as an indicator of normal biologic processes, pathogenic processes, or pharmacologic response to a therapeutic intervention.” (1).
The Parkinson Disease Biomarkers Program (PDBP) is a consortium of laboratory and clinical biomarker discovery projects funded by the National Institute of Neurological Diseases and Stroke (NINDS, National Institutes of Health (NIH), United States). The PDBP was developed in the context of the Harvard Biomarker Study (HBS), the Michael J. Fox Foundation (MJFF) Parkinson’s Progression Markers Initiative (PPMI), and the MJFF BioFIND Project. The HBS has developed a longitudinal biobank for the discovery and replication of biomarkers. PPMI has created a biorepository for validation of markers. BioFIND is a cross-sectional case/control collection for biomarker discovery. PDBP is building a resource of longitudinally collected data and biosamples from well-characterized patients with PD of different stages and duration, parkinsonism, and control participants, and includes hypothesis-driven biomarker discovery projects. PDBP will in turn support replication of findings from BioFIND and HBS, and provide validation candidates for PPMI. In order to provide the necessary infrastructure, a PDBP Data Management Resource (DMR) was established (described further below). Data and samples are already available through the PDBP DMR and a funding announcement has been issued (see PAR-14-259 and https://pdbp.ninds.nih.gov/jsp/biospecimens.jsp).
Here we describe the design of the longitudinal cohort, the individual projects comprising the consortium, and the standardized biospecimen collection protocols and clinical assessments. We also summarize the characteristics of the 1082 participants enrolled as of October 2014, from whom 1040 unique samples have been collected and catalogued in the DMR.
Methods
PDBP Establishment
Established November 2012, the PDBP seeks to identify biomarkers to improve therapeutic development in PD. The PDBP serves both as a biomarker discovery engine and a resource for clinical information and biospecimens. The PDBP includes 1) biomarker hypothesis testing and collection of clinical data and biospecimens, 2) identification of novel PD biomarkers, 3) biospecimen banking and distribution, and 4) data management through the DMR.
NINDS established the PDBP based on the vision that biomarker discovery and replication was needed to improve the efficiency and outcome of therapeutic clinical trials. Funding opportunity announcements (see RFA-NS-12-010, and RFA-NS-12-011) solicited applications, which underwent peer review. NINDS Advisory Council input then led to selection of projects based on scientific merit (as judged in review) and uniqueness of the project (either scientifically or in terms of the cohort recruited, or both) and considered in the space of existing efforts. The NINDS invested approximately $5M per year in the PDBP; three projects (under the U18 mechanism) are three years; the others are five years, allowing longitudinal follow-up for clinical cohorts. Milestones (go-no-go decision points) are used by NINDS to maximize scientific progress and compliance with rapid sharing.
There are several key differences between PDBP and other programs. PPMI’s goal is validation of biomarker discovery projects and participants must not be on anti-PD medications at the time of enrollment. BioFIND is a cross-sectional study for the characterization and collection of samples for biomarkers discovery from participants with resting-tremor predominant PD and controls. BioFIND does not have hypothesis testing included as a core component of its activities. However, hypothesis-based discovery projects, funded separately, are expected to utilize the collected samples and associated data. PDBP fills the gap in between these two and complements both by including longitudinal collection and by creating a resource for replication of early discoveries made in BioFIND. PDBP is agnostic regarding the use of DaTScan’s for enrollment, resting tremor predominance, and treatment status, and thus represents the typical participant for a future clinical trial. Additionally, each funded PDBP project pursues a biomarkers discovery project.
Consortium model
PDBP uses a novel consortium model that is self-assembled and self-managed (with NINDS input). The 11 projects comprising the PDBP consortium were independently peer-reviewed. However, PDBP investigators engage in collaborative activities spanning the entire consortium. For example, the consortium is in the process of identifying biochemical markers that will be tested consortium-wide and has performed genome-wide genotyping of samples. There are standardized operating procedures for biospecimen collection and clinical assessments. Participating projects interact at least monthly through a steering committee call and at an annual meeting. Data is shared immediately within the consortium; it is also shared rapidly once a requesting investigator has requested and received approval for controlled access (following the database for Genotypes and Phenotypes (dbGaP) example). The data and sample request process are further discussed below.
Clinical standards and Biospecimen Collection
Standardized clinical and biospecimen collection procedures are used for all participants. Inclusion/exclusion criteria include the ability to sign an informed consent (or have consent signed by an appropriate surrogate) and diagnosis of a neurodegenerative disorder (patient) or no evidence of a clinically significant neurological disorder (control). Each participant is evaluated for these and other criteria that in the case of PD participants allows for a broad spectrum of disease severity and duration. Additional inclusion/exclusion criteria are contained in Supplemental Table 1.
Assessments were chosen based on both NINDS Common Data Elements and overlap with BioFIND and PPMI. All 7 recruiting sites collect whole blood, plasma/serum, and RNA (6 of the sites collect these biofluids every 6 months, 1 site collects this material annually for a total of 2 visits); CSF is collected annually at 5 sites. Each biospecimen collection follows the PDBP protocol, which intentionally mirrors the Alzheimer’s Disease Neuroimaging Initiative (ADNI), BioFIND and PPMI protocols. NINDS staff performs site visits to ensure protocol compliance. Collected biosamples are sent to the NINDS Repository (Coriell Laboratories), undergo quality control, and are cataloged.
Data management overview
The NINDS and NIH Center for Information Technology (CIT) developed the PDBP DMR under a contract mechanism using both existing tools developed for NINDS-associated projects (including the Federal Interagency Traumatic Brain Injury Research Informatics System (FITBIR)) and novel tools. The DMR is a web-based system that allows receipt, management, and rapid sharing of clinical and laboratory data compliant with federal sharing mandates. The DMR generates unique, anonymous identifiers (Global Unique Identifiers, GUIDs), which are assigned to individual participants for clinical data in the DMR and associated biospecimens. All data from the standard clinical assessments are either entered into the DMR in real time or within 2 weeks of participant visits. The data then undergoes quality control measures (typically over 2–4 days) and is available immediately thereafter.
Data on previously existing cohorts have also been added to the PDBP DMR in order to streamline the data request process for PD studies. To date, four legacy studies have been added including the National Brain and Tissue Resource for Parkinson’s Disease and Related Disorders, HBS, University of Washington Udall Center, and University of Pennsylvania Udall Center. In total, this legacy data consists of 11 clinical assessments on over 1700 unique participants, and is also available to approved requesters.
Access to the clinical data and biospecimens
The DMR query tool allows approved users to review data and request biospecimens, including all clinical elements as well as quality control results from biospecimens banked at the NINDS Repository. The PDBP Data Access Committee reviews requests for data access to ensure subject privacy and non-tertiary re-distribution, along with other requirements. The Biospecimen Review Access Committee (BRAC) reviews all requests for biospecimens (submitted via an NIH X01 (PAR-14-340)) using the following criteria: 1) the appropriateness of the sample for accomplishing the scientific goals, 2) statistical power/analysis plans and 3) overlap with existing projects. If approved, the NINDS Repository is directed to ship the requested samples. A further description of the activities and members of the BRAC are listed at https://pdbp.ninds.nih.gov/jsp/brac.jsp. In order to ensure sharing of results, the recipient is blinded to clinical status until raw data are returned to the DMR.
Laboratory testing
Quality control measures applied to collected biosamples are documented in DMR and include 1) micro-satellite “barcoding”, 2) gender checks of extracted DNA and RNA, 3) hemoglobin level measurement in the plasma, serum, and CSF, and 4) assessment of turbidity in plasma and serum (as an indication of dietary fat).
Pooled aliquots for plasma, serum, and CSF have been developed to provide standards to compare measures across different assays and/or across different sites or projects. The NINDS Repository provides 3 different reference sample pools of each biofluid to investigators. These pooled samples are included with all shipments and expected to be included in analyses with resultant data also submitted to the DMR prior to unblinding of data.
Genetic characterization of DNA samples has been performed using the NeuroX array (3) This array provides a cost-effective method for screening 16,000 GWAS derived or GWAS related variants across a number of neurological disorders including Alzheimer’s Disease, Frontotemporal Degeneration (FTD), Multiple System Atrophy (MSA), myasthenia gravis, Charcot Marie Tooth, Progressive Supranuclear Palsy (PSP), Amyotrophic Lateral Sclerosis, and PD. The NeuroX array also includes 7,485 rare sequenced based variants for the diseases listed above resulting in 242,901 variants (3). The NeuroX array has been used in a replication cohort of 5,353 PD cases and 5,551 controls (4) to validate 24 risk loci for PD as well as for genotyping the PPMI cohort. The NeuroX has also been used in a replication study for FTD (2,154 FTD cases, 1372 controls) (5) to validate genome wide association variants. It is expected that the NeuroX data from the PDBP cohort, available now, will further enrich the value of this collection.
Results
Description of individual projects
Perelman School of Medicine, University of Pennsylvania
The goal of this project is to discover and develop blood-based PD biomarkers for diagnosis and prognostication. Unbiased screens evaluate thousands of proteins simultaneously, using both multiplex immunoassays and aptamer-based platforms, to discover new PD biomarkers. Investigators will also move two previously nominated biomarkers -- epidermal growth factor as a correlate of cognition in PD (6), and Apolipoprotein A1 as a correlate of dopaminergic system integrity (7) -- forward in a pipeline towards clinical translation.
University of Texas Southwestern Medical Center
Clinical Project
Researchers will determine the suitability of an instrumented gait and balance device (APDM Mobility Lab) (8) for assessing PD severity and progression. The investigators have found that two of the tests from this system discriminated PD participants from controls and correlated with disease severity in the PD group (9). Participants will be followed longitudinally to evaluate the correlation between clinical disease progression and change in gait and balance parameters.
Laboratory-based Project
This project aims to identify a serum biomarker useful for tracking disease progression. Antibody biomarkers will be identified using a high-throughput screening approach with a combinatorial peptoid library using samples from PDBP, Alzheimer’s disease (AD), and cognitively normal participants.
Johns Hopkins University School of Medicine
This program is identifying biomarkers for both PD and PD dementia. In addition to the standard PDBP clinical assessments, participants undergo an 80-minute cognitive testing battery annually. Outcomes from this test battery are combined with informant information to determine the participants’ cognitive diagnosis. With support from other sources, researchers are using a mass spectrometry approach to compare differences between PD and controls in the concentration of post-translationally modified proteins in the CSF and to determine if these proteins also serve as markers for development and/or progression of PD dementia.
Penn State Milton S. Hershey Medical Center
This team is investigating whether multimodal magnetic resonance imaging (MRI) techniques in combination with fluid-based iron protein profiles serve as in vivo markers for diagnosing PD and predicting progression. Diffusion tensor imaging (DTI) and R2* MRI measures have been promoted as biomarker(s) for PD-related pathology in nigrostriatal pathways, but fall short by the lack of understanding of their clinical implications and biological/pathological underpinnings. The imaging modalities in this project address these concerns and will determine whether these specific images and iron related proteins are potential PD markers.
Harvard Medical School, Brigham and Women’s Hospital
This team focuses on transcriptional markers. To discover PD-linked transcripts, the team is developing a high-resolution encyclopedia of all non-coding and protein-coding transcribed elements in dopamine neurons. Cell type-specific, ultra deep RNA-sequencing techniques are being applied in situ to dopamine neurons of human postmortem brains from controls and individuals with Parkinson’s neuropathology. Transcripts linked to earliest disease processes are evaluated for translation into markers accessible in blood or CSF. A private-public partnership with the PDBP will also allow all US investigators to search and access the substantial HBS biobank (established in 2003 and funded by the Harvard NeuroDiscovery Center) through the PDBP web site.
University of Florida College of Medicine
This team is focused on developing innovative and non-invasive techniques to understand differences in the pathophysiology and structural degeneration of the brain at baseline and following one year of progression across different movement disorders. The focus is on motor task-based functional MRI using a reliable motor task, and diffusion MRI using novel analytic methods (10, 11). The goal is to develop unique candidate markers of progression in PD (in comparison to other movement disorders). The study is enrolling individuals with PD, other parkinsonisms, essential tremor, and controls.
University of Washington School of Medicine
In collaboration with the University of Miami and Somalogics, these researchers will study three types of biomarkers: proteins, aptamers, and RNAs (coding and non-coding). Mass spectrometry and immunoassay techniques will be used to evaluate differences in the post-translational protein modifications between individuals with PD and controls in both CSF and plasma. Aptamer and RNA sequencing technologies will be used to evaluate differences in RNA species, CSF, and plasma between individuals with PD and controls.
Columbia University Mailman School of Public Health
This project will develop statistical methods to determine multimodal imaging biomarkers for PD that are more accurate and robust than single modality markers and that deepen our understanding of PD neuropathophysiology. The project uses data from MRI, neuromelanin MRI, DTI, and resting-state functional MRI. Separately, the investigators will consider electronic medical records from a massive patient database arising from a large integrated healthcare system. The goal is to identify clinical risk factors for early stage PD and use these to develop a clinical risk score for use in future studies.
Columbia University School of Medicine
This project’s focus is on the role of glucorebrosidase (GBA) mutations and glucocerebrosidase enzymatic (GCase) activity in PD. PD risk in GBA heterozygotes was estimated by obtaining family history information from participants with Gaucher disease both in the USA and in Israel (12). The investigators will now determine whether GCase activity is a marker for PD generally by measuring GCase activity in blood spots from PD cases and controls without GBA mutations, and PD cases and controls who carry GBA mutations (13).
Pacific Northwest National Laboratory (PNNL)
This project is developing biofluid protein-based assays for Lewy body load by focusing on proteins, their post-translational modifications, and isoforms that are associated with Lewy bodies. By analyzing substantia nigra tissue the investigators have discovered 33 proteins that differ in abundance patterns between cases with high densities of Lewy bodies and matching controls. A CSF biofluid assay using ultra-sensitive mass spectrometry based on selected reaction monitoring (14) is currently being used to measure the abundances of 33 protein panel and determine whether they reflect the presence of Lewy bodies in the substantia nigra.
University of Alabama at Birmingham (UAB)
This team is determining whether the proteins inside microvesicles (“exosomes”) derived from both urine and serum have prognostic and/or diagnostic potential for PD. The team has recently discovered that these exosomes contain proteins linked to PD such as LRRK2. In their analysis, LRRK2 levels are being measured directly along with hundreds of other proteins using a high-throughput mass spectrometry approach.
Data overview
The PDBP has enrolled 1082 individuals, including 620 with PD, 101 with parkinsonism (46 PSP, 36 MSA, 19 parkinsonism with currently undetermined diagnosis), 23 with essential tremor, and 338 controls (72% of planned enrollment) as of 10/21/2014 (enrollment ends 8/31/2015). Table 2 summarizes demographic and clinical measures recorded in the DMR and compares their mean values across groups. Note that sites added to the consortium subsequent to its launch might not perform every assessment; therefore, the number of participants (N) varies depending on the measure analyzed.
Table 2. Demographic and clinical variables and associated statistics.
Mean and standard deviation are reported for continuous measures; percent and standard deviation are reported for dichotomous values. Participants were categorized as Healthy Controls (HC), Parkinson’s Disease patients (PD), or patients exhibiting parkinsonism (APD, i.e., “Atypical Parkinsonian Disorders”). Due to the low number of essential tremor participants, they were not included in the enrollment data analysis, but their biofluids are included in the biospecimen data analysis described in the text. Two main subsets of participants were identified based on site participation. The first (N = 1030) consists of participants who have values recorded for Demographics (Age, Sex, Education), MoCA Score, MDS-UPDRS (Part III), and a composite indicator for Depression. (For participants with valid HAM-D scores, depression is indicated by a HAM-D score equal to or greater than 18; for participants with valid BDI scores, depression is indicated by a BDI score greater than 16.) The second (N = 862) consists of participants who have values recorded for MDS-UPDRS (Total Score), HAM-D, HAM-A, ESS, and UPSIT rating scales. Also reported are BDI scores and Disease Duration for those participants for whom this is possible. Significant p-values (P) for the omnibus test (ANOVA for continuous variables, Chi-Square for dichotomous variables) merit post-hoc Welch t-tests (for continuous variables) to examine pairwise differences (Bonferroni-corrected) between groups. Abbreviations: PD, Parkinson’s disease; HC, healthy control; APD, Atypical Parkinsonian Disease; MoCA, Montreal Cognitive Assessment; MDS-UPDRS, Unified Parkinson’s Disease Rating Scale; HAM-D, Hamilton Depression Rating Scale; HAM-A, Hamilton Anxiety Rating Scale; ESS, Epworth Sleepiness Scale; BDI, Beck Depression Inventory; UPSIT, University of Pennsylvania Smell Identification Test.
| Mean (or Mean Percent) ± Standard Deviation | Omnibus Test | Pairwise Comparisons (p-values) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | HC (n=332) | PD (n=606) | APD (n=92) | P | PD vs HC | APD vs HC | APD vs PD | |||||||
| Age (Years) | 1030 | 62.218 | ± | 10.769 | 64.811 | ± | 9.170 | 68.880 | ± | 8.527 | <0.001 | <0.001 | <0.001 | <0.001 |
| Male (Percent) | 1030 | 48.193 | ± | 49.967 | 60.726 | ± | 48.836 | 52.174% | ± | 49.953 | <0.001 | <0.001 | 1 | 0.447 |
| Education (Years) | 1030 | 15.753 | ± | 2.765 | 15.790 | ± | 2.710 | 14.880 | ± | 2.847 | 0.011 | 1 | 0.030 | 0.014 |
| MoCA (Total) | 1030 | 26.340 | ± | 2.543 | 25.419 | ± | 3.506 | 21.304 | ± | 5.201 | <0.001 | <0.001 | <0.001 | <0.001 |
| MDS-UPDRS (Part III) | 1030 | 2.060 | ± | 3.616 | 26.193 | ± | 13.792 | 46.250 | ± | 16.301 | <0.001 | <0.001 | <0.001 | <0.001 |
| Depressed (Percent) | 1030 | 0.904 | ± | 9.463 | 1.320 | ± | 11.414 | 31.522 | ± | 46.460 | <0.001 | 1 | <0.001 | <0.001 |
| N | HC (n=287) | PD (n=559) | APD (n=16) | P | PD vs HC | APD vs HC | APD vs PD | |||||||
| MDS-UPDRS (Total) | 862 | 7.456 | ± | 7.365 | 49.530 | ± | 24.099 | 74.500 | ± | 28.664 | <0.001 | <0.001 | <0.001 | 0.010 |
| HAM-D (Total) | 862 | 2.341 | ± | 2.769 | 4.537 | ± | 4.019 | 6.250 | ± | 6.061 | <0.001 | <0.001 | 0.064 | 0.835 |
| HAM-D (Percent Total≥18) | 862 | 0.697 | ± | 8.319 | 0.894 | ± | 9.415 | 6.250 | ± | 24.206 | 0.078 | 1 | 1 | 1 |
| HAM-A (Total) | 862 | 2.495 | ± | 3.385 | 5.209 | ± | 4.983 | 8.938 | ± | 5.674 | <0.001 | <0.001 | 0.001 | 0.059 |
| HAM-A (Percent Total≥18) | 862 | 1.394 | ± | 11.723 | 2.862 | ± | 16.674 | 0.0% | ± | 0.0 | 0.334 | 0.824 | 1 | 1 |
| ESS (Total) | 862 | 5.415 | ± | 3.349 | 8.411 | ± | 4.919 | 6.750 | ± | 4.669 | <0.001 | <0.001 | 0.828 | 0.541 |
| UPSIT (Total) | 862 | 32.404 | ± | 6.029 | 19.751 | ± | 7.730 | 23.688 | ± | 10.203 | <0.001 | <0.001 | 0.012 | 0.438 |
| N | HC (n=42) | PD (n=43) | APD (n=75) | P | PD vs HC | APD vs HC | APD vs PD | |||||||
| BDI (Total) | 160 | 3.571 | ± | 3.933 | 8.000 | ± | 5.880 | 14.040 | ± | 9.059 | <0.001 | <0.001 | <0.001 | <0.001 |
| BDI (Percent Total > 16) | 160 | 2.381% | ± | 15.246 | 6.977 | ± | 25.475 | 37.333 | ± | 48.369 | <0.001 | 1 | <0.001 | 0.002 |
| N | HC (n=0) | PD (n=449) | APD (n=101) | P | PD vs HC | APD vs HC | APD vs PD | |||||||
| Disease Duration (Years) | 550 | 7.188 | ± | 7.483 | 3.128 | ± | 2.998 | <0.001 | <0.001 | |||||
Demographic data
Individuals with PD and other types of parkinsonism are older than controls and more likely to be male. Total years of education also differ between the three groups, with individuals with PD having greater years of education than individuals with parkinsonism and controls. We anticipate balancing these demographic variables between PD and controls by the end of enrollment.
Clinical Data
Individuals with parkinsonism were generally more impaired in their motor, cognitive, and psychiatric function than those with PD, who in turn were more impaired than controls. This trend was observed in the MDS-UPDRS, Montreal Cognitive Assessment, depression symptomatology and Hamilton Anxiety scales. Individuals with PD were more impaired than those with parkinsonism and controls on the University of Pennsylvania Smell Test and Epworth Sleepiness Scale.
The PD cohort generally suffers from mild-to-moderate disease. The average disease duration is approximately 7 years, and participants have a relatively low total MDS-UPDRS (Table 2), MDS-UPDRS III (mean 26.19, standard deviation 13.79) and Hoehn and Yahr (mean 2.023, standard deviation 0.69).
Biospecimen Data
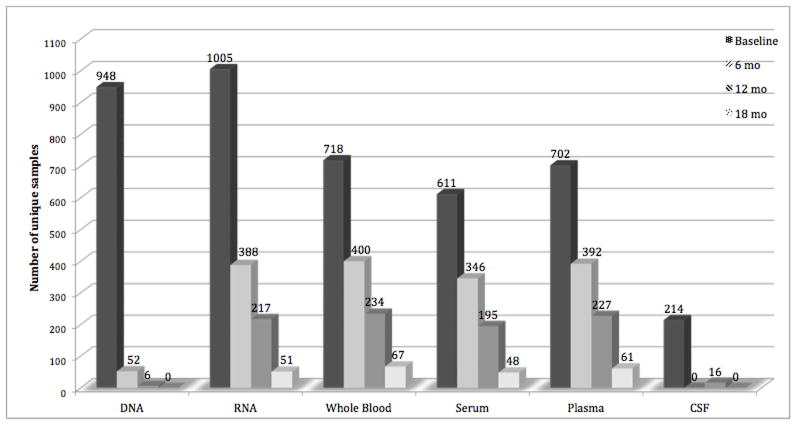
PDBP biorepository biospecimens as of 10/21/2014 are summarized in Figure 1. The biorepository includes 6898 samples from baseline, 6 month, 12 month, and 18 month visits. The full collection will include longitudinal samples from baseline to 36 months’ follow-up, with some participants followed for up to 60 months. A detailed schedule of measures for each clinical visit can be found on the PDBP website (see the Required Forms list).
Figure 1. Number of unique samples available in the NINDS bioropository as of 10/21/2014.
DNA is collected once, primarily at the baseline visit but in some cases at the 6 or 12 month visit RNA is drawn at least once on almost every participant and repeatedly for some participants. About two-thirds of the participants have baseline whole blood, serum and plasma available and about one quarter of the participants have baseline CSF available. As participants return for longitudinal assessments, there will be increased availability of biofluids from those visits.
Because elevated hemoglobin levels indicate erythrocyte lysis, we measured hemoglobin from biofluids. Samples with hemoglobin >200ng/mL were considered unacceptable (although not discarded). Overall, quality control analysis of plasma, serum, and CSF samples indicates only 24 of 2812 samples exceed acceptable hemoglobin levels.
At the baseline visit, only 0.3% (n=2) of 702 plasma samples and 0.2% (n=1) of 611 serum samples had hemoglobin concentrations >200ng/mL. Another 63.8% (n=448) of plasma and 76.8% (n=469) of serum samples demonstrated hemoglobin concentrations between 30 and 200 ng/mL, and the remaining 35.9% (n=252) of plasma and 23.1% (n=141) of serum samples demonstrated hemoglobin concentrations below the level of detection (<30 ng/mL). At the 6-month and 12-month visits, no plasma or serum samples had concentrations >200 ng/mL, and the percentages for the lower tiers were similar to those at baseline. There were statistically significant, but not clinically significant, differences (determined via one-way ANOVA) in mean hemoglobin concentration between some sites at baseline (p<0.01) and 6 months (p<0.01) for plasma, but only at baseline (p<0.01) for serum. There were no significant differences for plasma or serum at 12 months.
Among the 214 CSF samples assessed at the baseline visit, 9.8% (n=21) showed hemoglobin concentrations >200ng/mL. Another 15.4% (n=33) demonstrated hemoglobin concentrations between 30 and 200ng/mL, and the remaining 74.8% (n=160) had undetectable hemoglobin levels (<30ng/mL). At the 12-month visit, only 16 CSF samples have been collected; none exceeded hemoglobin concentrations of 200 ng/mL.
Discussion
The NINDS PDBP combines hypothesis-driven research, creation of a research resource, a novel collaborative consortium, and a flexible data management system. It has achieved 72% of target recruitment in less than 2 years. Quality control measures of plasma, serum, and CSF hemoglobin underscore the standardization of biospecimen collection and handling. The resource is further enhanced by NeuroX array-based genotyping.
The unique consortium arrangement of PDBP incorporates many of the strengths of a multi-site project while allowing for the flexibility and speed of investigator-initiated research. The individual sites are able to proceed with specific investigations, thereby capitalizing on each researcher’s unique line of research. Concurrently, the DMR and biorepository are available to researchers outside the PDBP consortium, further increasing the value of the cohort.
We previously noted that the PDBP complements other PD biomarker initiatives. The PDBP is a US-based longitudinal initiative comprising of PD patients of all stages, individuals with parkinsonism and controls. The PPMI is an international cohort that comprises PD individuals at early stages of the disease followed longitudinally, and also includes a control group. The BioFIND cohort is cross sectional and enrolls individuals in the mid-to-late stages of PD but is significantly smaller (~200 vs. ~1500 individuals). The HBS is a longitudinal case-control study of more than 700 patients with early-stage PD, >600 individuals with impaired memory, and >700 controls without neurologic disease. In the HBS, high-quality biosamples and high-resolution clinical phenotypes are tracked over a five-year period (15, 16). Importantly, while PDBP, HBS, and BioFIND provide samples for early-stage biomarker discovery, PPMI is aimed at replication of biomarker findings that have been observed in initial discovery studies. Together, PPMI, PDBP, HBS and BIOFIND provide the research community with access to samples across the spectrum of PD. This resource infrastructure allows new markers to be discovered and replicated within an accelerated time frame. The ability to assess a marker in multiple, large, well-characterized, independent patient populations should facilitate separating the “wheat from the chaff” and aid the development of replicable, robust and clinically useful biomarkers.
In addition to the four biorepository efforts aimed at biomarker development in PD described above, there are several other large biomarker efforts in neurodegenerative diseases that share samples and/or data widely. The Genetic Epidemiology of Parkinson’s Disease (GEO-PD) consortium comprises more than 60 sites in more than 30 countries on 6 continents, with a focus on PD genetics. ADNI follows individuals with AD and mild cognitive impairment (MCI), as well as control individuals, with the goal of developing imaging and biochemical biomarkers for AD. ADNI also features cohort-wide procedures and data are widely shared through a web-based download site; ADNI protocols have been widely adapted to PDBP, PPMI and BioFIND, in order to allow future analyses across projects.
The advent of large-scale efforts for biospecimen and data sharing is a relatively new development in the field of neurodegenerative disease research. We anticipate that much will be learned over the next decade about how best to design these efforts, which are emerging from government-funded, foundation-funded, and private-public-partnership-funded sources. Our hope is that PDBP will contribute to the widespread availability of well-characterized biospecimens for early-stage PD biomarker development, accelerating diagnostic and therapeutic discovery for the growing number of individuals suffering from PD worldwide.
Supplementary Material
Table 1. Description of the Parkinson’s Disease Biomarkers Program (PDBP) projects.
The PDBP consists of 11 projects that work together as a consortium to ensure standardized assessments and quality control of biofluid ascertainment and processing. Enrolled participants were counted from the start of the study through October 2014. Some of the sites have exceeded their planned enrollment, such that currently enrolled patients are greater than planned enrollment. *includes 16 Progressive Supranuclear Palsy (PSP) and 10 Multiple System Atrophy (MSA) participants. ^Includes 24 essential tremor (ET), 28 MSA, 30 PSP and 20 Atypical Parkinson’s Disorder participants. Abbreviations: EGF, Epidermal Growth Factor; FA. Fractional Anisotropy; LBs, Lewy Bodies; NR, Not Recruiting.
| Investigator (Last Name) | Institution | Project Title | PD participants enrolled / Total planned | Parkinsonism participants enrolled / Total planned | Control participants enrolled / Total planned |
|---|---|---|---|---|---|
| Chen-Plotkin | U. Pennsylvania | Unbiased approaches to novel biomarker discovery in Parkinson's disease | NR | NR | NR |
| Dewey; German | U. Texas Southwestern | Diagnostic and prognostic biomarkers for Parkinson's disease | 185 / 200 | NR | 26 / 25 |
| Dawson; Rosenthal | Johns Hopkins U. | Johns Hopkins medicine biomarker discovery in Parkinson's disease | 72 / 75 | NR | 23 / 75 |
| Huang | Penn State Hershey | Multimodal MRI markers of nigrostriatal pathology in Parkinson's disease | 116 / 108 | *26 / 42 | 77 / 72 |
| Scherzer | Brigham and Women's Hospital | Biomarkers for early intervention in Parkinson disease | 17 / 38 | NR | 27 / 37 |
| Vaillancourt | U. Florida | Non-invasive markers of Neurogeneration in Movement Disorders | 46 / 30 | ^102 / 90 | 45/ 30 |
| Zhang | U. Washington | Large scale biomarker discovery and validation for Parkinson's disease | 3/ 50 | NR | NR |
| Bowman | Columbia U. | Analytic methods for determining multimodal biomarkers for Parkinson's disease | NR | NR | NR |
| Alcalay | Columbia U. | The Role of Glucocerebrosidase in Parkinson’s Disease | 517/500 | NR | 252/250 |
| Petyuk | Pacific Northwest National Laboratory | Development of Lewy bodies biofluid signatures by targeted proteomics | NR | NR | NR |
| West | U. Alabama Birmingham | LRRK2 and Other Novel Exosome Proteins in Parkinson's Disease | 181 / 300 | NR | 139 / 300 |
Acknowledgments
The authors would like to thank Ms. Yekaterina A. Salnikova for her assistance with manuscript preparation and figure development.
Financial disclosure/Conflict of interest concerning research related to manuscript:
Liana Rosenthal: Dr. Rosenthal is supported for this project by NIH/NINDS U01 NS082133 and P50NS38377.
Daniel Drake: Dr. Drake is supported for this project by Emory University, Columbia University and NIH/NINDS U18 NS082143.
Roy Alcalay: Dr. Alcalay is supported for this project by NIH/NINDS K02 NS080915.
Debra Babcock: Dr. Babcock is an employee of the National Institutes of Health, which funded this project.
DuBois Bowman: Dr. Bowman is supported for this project by Columbia University and NIH/NINDS U18 NS082143.
Alice Chen-Plotkin: Dr. Chen-Plotkin is supported for this project by NIH/NINDS U01 NS082134.
Ted Dawson: Dr. Dawson is supported for this project by NIH/NINDS U01NS082133 and P50NS38377.
Richard Dewey: Dr. Dewey is supported for this project by NINDS U01 NS082148.
Dwight German: Dr. German is supported for this project by NINDS U01 NS082148.
Xuemei Huang: This project is supported by NINDS U01 NS082151. Dr. Huang’s work related to the project is also supported in part by UL1 RR033184 and UL1 TR00012, the Pennsylvania Department of Health Tobacco CURE Funds and intramural program of PSU- HMC. During the past three years, Dr. Huang also receives research funding from GE Healthcare. She is a consultant for the National Institute of Environmental Health Sciences.
Barry Landin: Mr. Landin is a contractor to the National Institutes of Health, which funded this project through contract ZIHCT000272.
Matthew McAuliffe: Dr. McAuliffe is an employee of the National Institutes of Health, which funded this project through contract ZIHCT000272.
Vladislav Petyuk: Dr. Petyuk is supported for this project by NIH/NINDS U18 NS082140.
Clemens Scherzer: Dr. Scherzer is supported for this project by NIH/NINDS U01 NS082157.
Coryse St. Hillaire-Clarke: Dr. St. Hillaire-Clarke is an employee of the National Institutes of Health, which funded this project.
Beth-Anne Sieber: Dr. Sieber is an employee of the National Institutes of Health, which funded this project.
Margaret Sutherland: Dr. Sutherland is an employee of the National Institutes of Health, which funded this project.
Chi Tarn: Dr. Tarn is an employee of the Coriell Institute for Medical Research, which receives funding from National Institutes of Health for this Project.
Andrew West: Dr. West is supported for this research by NIH/NINDS U18NS082132-01.
David Vaillancourt: Dr. Vaillancourt is supported by this project by NINDS R01 NS075012.
Jing Zhang: Dr. Zhang is supported for this project by NINDS U01 NS082137.
Katrina Gwinn: Dr. Gwinn is an employee of the National Institutes of Health, which funded this project.
Funding Sources for study:
This study is funded by the National Institute of Neurological Disorders and Stroke, National Institutes of Health, Bethesda, Maryland.
Documentation of author roles:
Liana S. Rosenthal: 1B, 1C, 2A, 3A, 3B
Daniel Drake: 2A, 2B, 2C, 3A, 3B
Roy Alcalay: 1B, 1C, 3A, 3B
Debra Babcock: 1A, 1B, 1C, 3A, 3B
F. DuBois Bowman: 1B, 1C, 2A, 2B, 2C, 3A, 3B
Alice Chen-Plotkin: 1B, 1C, 3A, 3B
Ted Dawson: 1B, 1C, 3A, 3B
Richard Dewey: 1B, 1C, 3A, 3B
Dwight German: 1B, 1C, 3A, 3B
Xuemei Huang: 1B, 1C, 3A, 3B
Matthew McAuliffe: 1C, 3B
Vladislav Petyuk: 1B, 1C, 3A, 3B
Clemens Scherzer: 1B, 1C, 3A, 3B
Coryse St. Hillaire-Clarke: 1C, 3B
Beth-Anne Sieber: 1A, 1B, 1C, 3A, 3B
Margaret Sutherland: 1A, 1B, 1C, 3A, 3B
Chi Tarn: 1C, 3B
Andrew West: 1B, 1C, 3A, 3B
David Vaillancourt: 1B, 1C, 3A, 3B
Jing Zhang: 1B, 1C, 3A, 3B
Katrina Gwinn: 1A, 1B, 1C, 3A, 3B
Financial disclosures from past 12 months:
Liana Rosenthal: Dr. Rosenthal also receives support in the previous 12 months from NIH/NINDS P50NS038377, Marilyn and Edward Macklin Foundation, and the Michael J. Fox Foundation. She also received an honorarium from the Edmond J. Safra Foundation and Functional Neuromodulation.
Daniel Drake: Dr. Drake also receives support in the previous 12 months from Georgia State University, Emory University, and Columbia University.
Roy Alcalay: Dr. Alcalay receives additional support from the Parkinson’s Disease Foundation, the Smart Foundation and the Michael J. Fox Foundation.
Debra Babcock: Dr. Babcock does not have any additional disclosures.
DuBois Bowman: Dr. Bowman’s receives additional support from Columbia University and Emory University.
Alice Chen-Plotkin: Dr. Chen-Plotkin also receives research support from the Burroughs Wellcome Fund, the Doris Duke Charitable Foundation, the Benaroya Fund, and the NIH/NINDS (P50 NS053488, RO1 NS082265).
Ted Dawson: Dr. Dawson also acknowledges the Adrienne Helis Malvin and Diana Henry Helis Medical Research Foundations and their direct engagement in the continuous active conduct of medical research in conjunction with The Johns Hopkins Hospital and The Johns Hopkins University School of Medicine and the Foundation’s Parkinson’s Disease Programs. Funding for a portion of Dr. Dawson’s research was provided by Merck KGAA. Under a licensing agreement between Merck KGAA and The Johns Hopkins University, Dr. Dawson and the University shared fees received by the University on licensing some of the reagents used in his research. Dr. Dawson also was a paid consultant to Merck KGAA. The terms of this arrangement are being managed by The Johns Hopkins University in accordance with its conflict of interest policies. Dr. Dawson is a founder of Valted, LLC and holds an ownership equity interest in the company. This arrangement has been reviewed and approved by the Johns Hopkins University in accordance with its conflict of interest policies.
His work is also supported by NIH/NINDS P50NS038377, NIH/NINDS R01NS067525, NIH/NIDA P50 DA00266, the JPB Foundation and the MDSCRF 2007-MSCRFI-0420-00, 2009-MSCRFII-0125-00, MDSCRF 2013-MSCRFII-0105-00. Dr. Dawson is the Leonard and Madlyn Abramson Professor in Neurodegenerative Diseases. Dr. Dawson is a member of the Board of Directors of the Bachmann Strauss Dystonia and Parkinson’s Disease Foundation. Dr. Dawson is a member of Scientific Advisory Board of CurePSP. Dr. Dawson is a member of American Gene Technologies International Inc., advisory board. The terms of this arrangement are being managed by The Johns Hopkins University in accordance with its conflict of interest policies.
Richard Dewey: Dr. Dewey has served as a consultant and speaker for Teva Neuroscience, Merz, Xenoport, UCB, Acadia, US WorldMeds, Lundbeck, and Impax Pharmaceuticals.
Dwight German: Dr. German has received additional support from the Dallas Foundation, Simons Foundation and the Department of Defense.
Xuemei Huang: During past 12 months, Dr. Huang is also receiving funding by NIH R01 NS060722, R01 ES019672. During the past three years, she has received reimbursement for travel expenses from Medtronics Inc. and a consulting fee related to a medical malpractice lawsuit. She also has an interest in intellectual property related to dopamine agonists through her husband (details available upon request), which activities are monitored by a standing conflict-of-interest committee at the Penn State College of Medicine.
Barry Landin: Mr. Landin has no other conflicts of interest to report.
Matthew McAuliffe: Dr. McAuliffe has no additional financial disclosures.
Vladislav Petyuk: Dr. Petyuk has no additional financial disclosures.
Clemens Scherzer: Dr. Scherzer has collaborated with Pfizer, Opko, and Proteome Sciences, and is funded by NIH grants R01 NS064155, R01 AG044113, U01 NS082080, U01 AT000613, the Department of Defense, the Harvard NeuroDiscovery Center, the Michael J. Fox Foundation, and the M.E.M.O. Hoffman Foundation.
Coryse St. Hillaire-Clark: Dr. St. Hillaire-Clark has no additional financial disclosures.
Beth-Anne Sieber: Dr. Sieber does not have any additional disclosures.
Margaret Sutherland: Dr. Sutherland does not have any additional disclosures.
Chi Tarn: Dr. Tarn does not have any additional disclosures.
Andrew West: Dr. West’s additional support includes a research grant from Pfizer Inc., NIH/NINDS R01-NS064934, NIH/NINDS Mentor for 1F31NS081963-01 Training Award to Mark Moehle, Michael J. Fox Foundation for Parkinson’s Disease Research, American Parkinson Disease Association Postdoctoral Fellowship Training Award to JP Daher, and the Alabama Drug Discovery Alliance.
David Vaillancourt: Dr. Vaillancourt receives grant support from NIH, Bachmann-Strauss Foundation Dystonia & Parkinson Foundation, Tyler’s Hope Foundation, and consults for projects at UT Southwestern Medical Center, University of Illinois at Chicago, and Great Lakes NeuroTechnologies. He is co-founder of Neuroimaging Solutions, LLC.
Jing Zhang: Dr. Zhang has also received support from NIH grants P42 ES004696. P30ES00703319, R01ES01687305, and R01ES01927704, as well as grants from the FOX Foundation.
Katrina Gwinn: Dr. Gwinn has no additional financial disclosures.
References
- 1.Atkinson AJ, Colburn WA, DeGruttola G, et al. Biomarkers and surrogate endpoints: Preferred definitions and conceptual framework. Clinical Pharmacology & Therapeutics. 2001;69:89–95. doi: 10.1067/mcp.2001.113989. [DOI] [PubMed] [Google Scholar]
- 2.Mohammadi D. The Harvard Biomarker Study’s big plan. The Lancet Neurology. 2013;12(8):739–740. doi: 10.1016/S1474-4422(13)70155-8. [DOI] [PubMed] [Google Scholar]
- 3.Nalls MA, Bras J, Hernandez DG, et al. NeuroX, a fast and efficient genotyping platform for investigation of neurodegenerative diseases. Neurobiology of Aging. 2015;36(3):1605. doi: 10.1016/j.neurobiolaging.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nalls Mike A, et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nature genetics. 2014;46(9):989–93. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrari R, Hernandez DG, Nalls MA, et al. Frontotemporal dementia and its subtypes: a genome-wide association study. The Lancet Neurology. 2014;13(7):686–699. doi: 10.1016/S1474-4422(14)70065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen-Plotkin Alice S, Hu William T, Andrew Siderowf, et al. Plasma epidermal growth factor levels predict cognitive decline in Parkinson disease. Annals of neurology. 2011;69(4):655–63. doi: 10.1002/ana.22271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qiang Judy K, Wong Yvette C, Andrew Siderowf, et al. Plasma apolipoprotein A1 as a biomarker for parkinson's disease. Annals of neurology. 2013;74(1):119–27. doi: 10.1002/ana.23872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mancini M, King L, Salarian A, Holmstrom L, McNames J, et al. Mobility Lab to Assess Balance and Gait with Synchronized Body-worn Sensors. J Bioeng Biomed Sci. 2011;S1:007. doi: 10.4172/2155-9538.S1-007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dewey DC, Miocinovic S, Bernstein I, et al. Automated gait and balance parameters diagnose and correlate with severity in Parkinson disease. Journal of Neurological Sciences. 2014;345(1–2):131–138. doi: 10.1016/j.jns.2014.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ofori E, Pasternak O, Planetta PJ, Li H, Burciu R, Snyder A, Lai S, Okun MS, Vaillancourt DE. Longitudinal changes in free-water within the substantia nigra of Parkinson’s disease. Brain. 2015;138(Pt 8):2322–2331. doi: 10.1093/brain/awv136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spraker MB, Prodoehl J, Corcos DM, Comella CL, Vaillancourt DE. Basal ganglia hypoactivity during grip force in drug naïve Parkinson’s disease. Human Brain Mapping. 2010;31(12):1928–1941. doi: 10.1002/hbm.20987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alcalay RN, Diner T, Quinn T, et al. Comparison of Parkinson risk in Ashkenazi Jewish patients with Gaucher disease and GBA heterozygotes. JAMA Neurology. 2014 Jun;71(6):752–7. doi: 10.1001/jamaneurol.2014.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alcalay RN, Levy O, Waters C, et al. Glucocerebrosidase Activity in Parkinson Disease with and Without GBA Mutations. Brain. 2015 doi: 10.1093/brain/awv179. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi T1, Fillmore TL, Sun X, et al. Antibody-free, targeted mass-spectrometric approach for quantification of proteins at low picogram per milliliter levels in human plasma/serum. Proc Natl Acad Sci USA. 2012;109(38):15395–400. doi: 10.1073/pnas.1204366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ziegler DA, Ashourian P, Wonderlick JS, et al. Motor impulsivity in Parkinson disease: Associations with COMT and DRD2 polymorphisms. Scandinavian Journal of Psychology. 2014;55(3):278–286. doi: 10.1111/sjop.12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ding H, Dhima K, Lockhart KC, et al. Unrecognized vitamin D3 deficiency is common in Parkinson disease. Neurology. 2013;81(17):1531–1537. doi: 10.1212/WNL.0b013e3182a95818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.