Abstract
Purpose
Magnetic Resonance Spectroscopy (MRS) typically requires averaging of multiple acquisitions to achieve adequate signal to noise (SNR). In systems undergoing dynamic changes this can compromise the temporal resolution of the measurement. One such example is 31P MRS of exercising skeletal muscle. Spectral Improvement by Fourier Thresholding (SIFT) offers a way of suppressing noise without averaging. In this study we evaluate the performance of SIFT in healthy subjects and clinical cases.
Methods
31P MRS of the calf or thigh muscle of subjects (n = 12) was measured continuously before, during, and after exercise. The data was processed conventionally and with the addition of SIFT before quantifying peak amplitudes and frequencies. The post-exercise increase in the amplitude of phosphocreatine was also characterized by fitting with an exponential function to obtain the recovery time constant.
Results
Substantial reductions in the uncertainty of peak fitting for phosphocreatine (73%) and inorganic phosphate (60%) were observed when using SIFT relative to conventional processing alone. SIFT also reduced the phosphocreatine recovery time constant uncertainty by 38%.
Conclusion
SIFT considerably improves SNR, which improved quantification and parameter estimation. It is suitable for any type of time varying MRS and is both straightforward and fast to apply.
Keywords: 31P spectroscopy, exercise, phosphocreatine, energy metabolism, signal processing
Introduction
Magnetic Resonance Spectroscopy (MRS) is a powerful, non-invasive technique which permits the study of many metabolic processes in vivo. Some of the most interesting applications of MRS involve perturbing a particular metabolic process while acquiring a sequence of spectra to observe the temporal response of the system to the perturbation. One example is the depletion of phosphocreatine (PCr) in exercising muscle tissue, and the subsequent recovery of the peak.
31P MRS is widely accepted as the “gold standard” method for noninvasive metabolic measurements in exercising muscle (1,2). During exercise, consumption of Adenosine TriPhosphate (ATP) is stoichiometrically correlated to the hydrolysis of PCr to creatine and inorganic phosphate (Pi). Post-exercise, the recovery of PCr reflects ATP synthesis. The change in PCr peak height and the frequency difference between Pi and PCr directly measure the intracellular energy status and intracellular pH respectively. This method has been used to study therapeutic interventions and training-induced metabolic changes in children and adults as well as in patients with pulmonary, cardiac, and renal disorders (3). The dynamics of Pi/PCr have been used to evaluate mitochondrial respiration in skeletal muscle and the rate constant of post-exercise PCr recovery used as an index of mitochondrial function, i.e. oxidative ATP synthesis (4,5). Recently, 31P MRS has also been used in clinical trials to evaluate the effect of drug-based intervention on muscle energetics (6–8). 31P MRS is of particular interest in investigating the underlying mechanisms causing functional impairment in patients with peripheral artery disease (PAD) (9). PAD is a manifestation of atherosclerosis that affects more than 7 million adults in the US(10). Studies have shown reduced PCr recovery rates in PAD (11).
The numerous and diverse applications of 31P MRS depend upon accurate and robust measurement. Perhaps the most challenging aspect of in vivo 31P MRS is the low concentration of the observed metabolites and the correspondingly low Signal-to-Noise Ratio (SNR). This problem is typically addressed by averaging blocks of consecutive spectra, which improves the SNR by the square root of the number of averaged spectra. In most protocols this approach involves no loss of information and is only limited by considerations concerning the total length of the exam. However for a series involving variations over time, data averaging degrades the temporal resolution of the data and the averaged spectra will be blurred by the spectral changes across its components.
Goluch et al. recently published details of a new calf muscle coil with a three channel 31P phased array for localized spectroscopy at 7T (12). Despite the improved coil design, the spectra they present use five temporal averages per spectrum to achieve acceptable SNR, for a temporal resolution of one spectrum every 30s. In another report Meyerspeer et al. used a semi-LASER sequence for localized calf spectroscopy at 7T (13). They were able to quantify PCr in individual spectra (TR = 6s), but found that for the Pi it was necessary to average spectra from multiple subjects to obtain a reliable quantification.
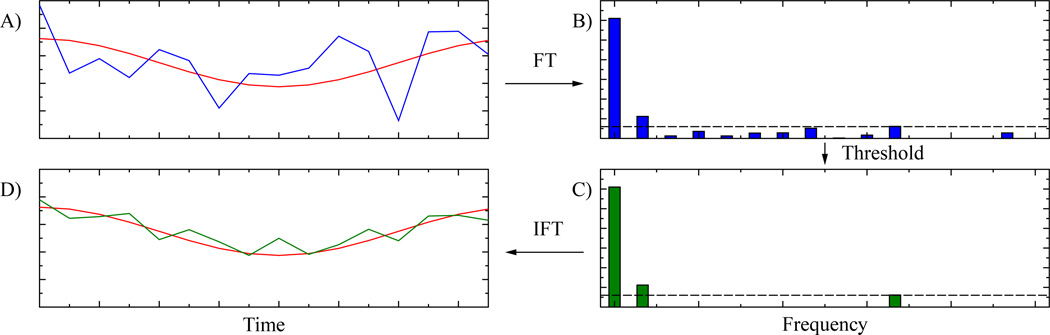
The SIFT method (Spectral Improvement by Fourier Thresholding) (14) provides improved SNR without compromising temporal resolution. SIFT entails: (i) Fourier transforming serially acquired spectra along the indirect (as opposed to direct) temporal dimension; (ii) performing a thresholding operation; and (iii) inverse Fourier transforming the MR data for the conventional analysis. Figure 1 diagrams the concept and sequence of operations in the SIFT method. The original SIFT report showed significant improvement for simulated data, and for a limited set of in vivo canine skeletal muscle 31P MR spectra (14).
Figure 1.
A diagram of the concept and components steps in the SIFT method, illustrated for a single spectral frequency. Panel A) the exercise time domain signal (red) is affected by white Gaussian noise (blue). Panel B) taking the Fourier transform of this noisy signal shows that the majority of the signal is concentrated in a small number of components. Panel C) removing all components below the threshold (dashed line) removes most of the noise but retains the signal components. Panel D) after the inverse Fourier transform, the exercise time domain signal is much less noisy (green).
In this study, we apply the SIFT method to 31P MRS data acquired from exercising muscle and evaluate its effectiveness in both healthy volunteers and PAD patients. This broad range of subjects provides a real-world evaluation of the performance of SIFT, particularly in those individuals with poor PCr recovery rates.
Methods
Two different forms of exercise were performed for this study: plantar flexion exercises observing the gastrocnemius muscle, and leg lifting exercises observing the quadriceps muscle. In both cases, healthy volunteers and patients suffering from PAD were scanned. For the volunteers, care was taken to select a diverse range of ages and physical fitness levels to provide a realistic evaluation of the methodology. All studies were conducted with the approval of the Institutional Review Board (IRB).
For the plantar flexion exercises, 5 subjects were scanned: 3 healthy volunteers (ages 21–24, three females) and 2 PAD patients (ages 77 and 84, both male). Subjects were scanned in a Siemens Skyra 3T whole-body MR system (Siemens Medical Solutions, Erlangen, Germany) using a single channel 10-cm diameter 31P tuned (51.8MHz) transmit/receive surface coil (Mirtech, Boston, MA), fixed to the subject's calf. Subjects lay supine, feet first with their lower legs resting on the base plate of the plantar exercise device (11). A second “exercise” plate is attached to the base plate by a hinge. The subject’s feet push against the upper plate to complete the exercise. Elastic bands are stretched between the two plates to provide resistance and increase the work-load of the exercise. The subject's legs were immobilized using a table strap across the ankles to minimize movement and thus changes in B0.
For the leg lift exercises, 7 subjects were scanned: 6 healthy volunteers (ages 24–65 years; 5 males, 1 female) and 1 PAD patient (70 year old female). Subjects were scanned in a Siemens Verio 3T whole-body MR system using a similar single channel 31P tuned transmit/receive surface coil strapped to the thigh at the vastus medialis muscle of the quadriceps. The subjects were positioned in the scanner supine on a padded bed with a custom-built dynamic knee extension apparatus (15). This apparatus has a bar that rests across the shins of the subjects, which attaches to a lever connected by rope to weights via a pulley. The subjects extend their knee, pushing up the bar to lift the weights. Maximum work capacity was determined by a 3 second knee extension with different weights to determine each subject’s maximum work-load. The load for the exercise was set to 30% of maximum work-load.
In both types of exercise the same MR protocol was followed. First the transmitter gain of a 500 μs hard excitation pulse was adjusted to determine the value giving the highest PCr peak. A simple pulse-acquire sequence (TR = 2s, spectral width = 6kHz) was then performed continuously during an exercise protocol consisting of two minutes of initial rest (baseline), three minutes of exercise at a rate of approximately one lift every two seconds, and five minutes of recovery also resting. A total of 300 Free Induction Decays (FIDs), with 4096 complex data points per FID, were acquired per subject. Subjects were given audio cues for each section of the exercise routine and an investigator would stay with the subject in the scanner room to provide cues for each lift and to ensure compliance with the exercise.
Data was exported from the Siemens scanner in the raw data format and the individual FIDs were extracted. Our analysis procedure consisted of three sequential steps. In the first step standard data processing including phasing and baseline correction was performed using in-house software written in Python. The SIFT method was then applied to the processed data. Finally metabolite concentrations were estimated using the AMARES plugin option in jMRUI (16), and the recovery parameters calculated.
Data Processing
The first five FIDs were discarded to ensure that steady state excitation had been reached. On initial loading of the data each FID was apodized with a 50Hz Gaussian filter, 50Hz matching the average Full Width at Half Maximum (FWHM) of the PCr peak.
During initial data exploration it was observed that substantial baseline distortion was present in the phased data. Such distortion is typically attributable to corruption of the first few points of the FID, which can be caused by several effects including: probe ringing, where the residual electrical currents from the excitation pulse introduce distortion in the first few points of the FID and gating artifacts where the receiver channel is turned off for a few ms at the start of the acquisition to avoid ringing.
Due to the low SNR of the spectra, fitting a baseline to each spectrum individually actually increases the deviation between spectra, leading to a noisier time course. To avoid this problem, the first few points of each FID were set to zero so that the baseline was identical in every spectrum. The 55 spectra from the pre-exercise period, where no temporal variation occurred, were averaged to create a high SNR spectrum. This spectrum was then used to phase the data and determine a universal baseline. These global parameters were then used to phase and baseline correct each individual spectrum.
The shape of the baseline in the frequency domain may be calculated by performing the Discrete Fourier Transform (DFT) of the distortion profile in the time domain. The DFT is defined in Eq. 1.
| Eq. 1 |
Where the coefficients cn describe the shape of the distortion term. In this case we are zeroing the first few points of the FID, providing a “top hat” distortion profile where cn is 1 for the first M points and 0 for all other points (Eq. 2.).
| Eq. 2 |
Note that this term is closely related to, but subtly different from, the sinc function obtained in the continuous case. For our data we found that zeroing the first two points of the FID and setting M=2 gave the best results.
A measured spectrum may be expressed as a linear combination of contributions from a basis set comprising the individual spectra of the observable metabolites and the baseline contribution. If the individual basis spectra are known, the contributions of each to the measured spectrum can be easily obtained using a linear least squares regression. In this study the leastsq function from the scipy.optimize library was applied in the frequency domain to calculate the magnitude of the baseline and to phase the spectrum.
The metabolite basis spectra can be constructed from Gaussian lineshapes, but the exact frequencies and linewidths of the peaks will vary slightly from exam to exam, so an additional iterative optimization must be performed to find the values for these parameters which best match the measured data. At each iteration the measured spectrum was phased with the current estimates of zeroth and first order terms, while a new basis set of simulated metabolite peaks was created based on the current estimates of frequency shift and linewidth. The calculated baseline spectrum was added to this basis set and leastsq used to obtain a fit to the phased measured spectrum.
When the root-mean-square difference between measured and simulated spectra reaches the minimum, the optimal values of the phase parameters and baseline can be extracted. These phase and baseline values were then used to correct the individual spectra in the exercise and recovery stages of the exam. Figure 2 shows an example of a pre-exercise composite FID with baseline distortion and the achieved fit of the simulated basis set, including the baseline contribution.
Figure 2.
Typical composite (55 averages from pre-exercise period) 31P MR spectrum (black) showing baseline distortion produced by coil ringing; the fit including the additional corrective basis vector (red). The residual from the fitting (blue) is shown offset below the spectrum. The labelled resonances are: phosphomonoesters (PME), inorganic phosphate (Pi), phosphodiesters (PDE), phosphocreatine (PCr), and the 3 resonances of ATP.
SIFT
The SIFT method applies the Fourier transform to the indirect time dimension of the data, here denoted the “exercise domain”. Just as the conventional FT in MRS concentrates the temporal variation of the FID into a few sharp peaks in the spectrum, the FT in the exercise domain concentrates the time course of the exercise into a small number of significant signal coefficients, with the rest containing solely noise. By application of a judicious threshold to suppress the noise while maintaining the signal terms, a significant noise reduction is achieved when the inverse FT is used to return the data to the time domain (Fig. 1).
In the limit of data with no temporal variation, the entire signal lies in the zeroth coefficient of the exercise spectrum (the average of all points in the time domain) so that suppressing all other coefficients is identically equivalent to the standard temporal averaging. However, with time varying data there will be other coefficients containing signal and a threshold-based approach must be used to try to distinguish noise from signal.
Care must be taken when choosing the threshold, as those noise coefficients that exceed it and are not removed are necessarily the largest. For a random variable X such as the receiver noise, the expected value E[X]in the spectrum is the first moment of the probability distribution. For Gaussian noise, this is expressed in Eq. 3.
| Eq. 3 |
Zeroing coefficients below a certain threshold changes the lower limit of the integral from 0 to the threshold t. Dividing by the total noise from the unmodified signal we obtain the fractional magnitude of the residual noise as shown in Eq 4.
| Eq. 4 |
where t is the threshold in standard deviations of the noise. For example, choosing a threshold of 2 standard deviations will suppress 95% of the pure noise coefficients, but the expected noise value decreases by only 86%.
Preliminary testing indicated that a threshold value of 2 standard deviations would be suitable for our datasets: this value was utilized for all results presented in this study. Generally, thresholds > 2 cause unacceptable deformation of the time domain trajectory (signal coefficients are removed with the noise), whilst thresholds that are lower lead to suboptimal improvements in SNR.
Fitting
After global phasing and baseline correction, the FID data, both with and without SIFT filtering, was exported to jMRUI, and the metabolite amplitudes computed using AMARES. Gaussian lineshapes were used for each metabolite as they were found to give better agreement with the data than Lorentzian lineshapes. AMARES reports both the fitted metabolite amplitudes and their standard deviations for each spectrum. These standard deviations were used to assess whether the SIFT method reduces the fitting uncertainty on a per spectrum basis. The post-exercise PCr amplitudes of each subject, for both raw and SIFTed data, were fitted to a monoexponential recovery function A + B exp(-t/τ) using the curve_fit routine of the scipy.optimize Python package to determine the recovery time constant τ. The curve_fit routine returns both the estimated values of the fitted parameters and their covariances, from which the standard deviation of the fit was calculated by taking the square root. This standard deviation was used to assess whether the SIFT method reduced the fitting uncertainty for the time course
Results
In both plantar flexion and leg lift exercises depletion of PCr was observed, along with an increase in Pi and a drop of pH. As expected, the wide range of ages and physical conditions of the subjects led to a corresponding range of recovery times, which ran from τ =21.22 +/− 2.36s to τ =107.76 +/− 11.39s.
Applying the SIFT method to the data removed most of the coefficients in the time domain. Across the entire spectrum, the average number of coefficients removed by the thresholding was 279 +/− 6. Within the region of the PCr peak fewer coefficients were removed (265 +/− 9).
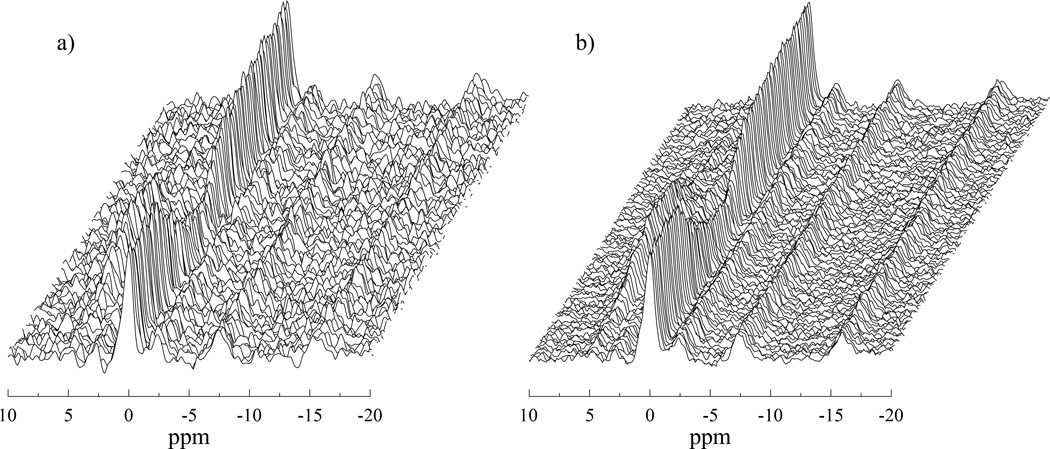
Figure 3 shows an example dataset both before and after application of the SIFT method. The reduction in noise across the frequency range of interest is clearly visible, as well as the improved smoothness of the time domain trajectory of the peaks. Single spectra from this dataset are shown in figure 4 to give a clearer idea of the improved spectral quality achieved by SIFT.
Figure 3.
Stack plots of 300 muscle 31P MR spectra from the entire exercise protocol. Panel (a) displays the data processed with the standard methods and, panel (b) the same data with the SIFT method applied. There is a great improvement in the SNR and the temporal trajectories of the peaks are much smoother in the SIFTed data.
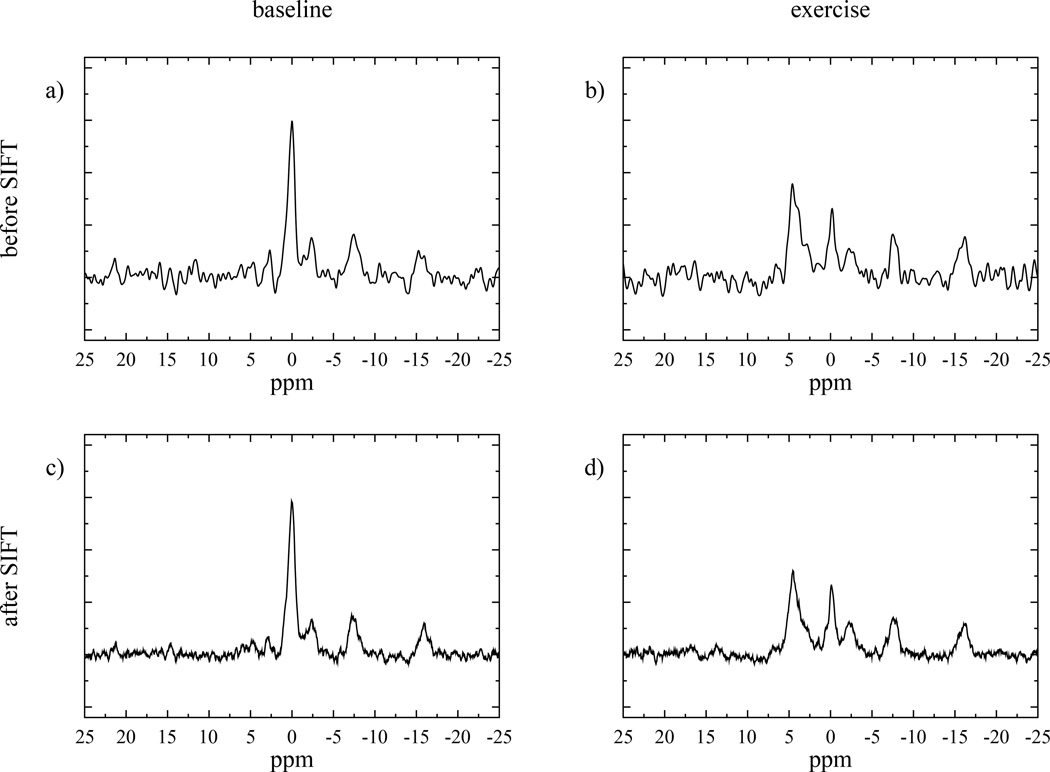
Figure 4.
Example spectra with standard processing (a, b) and, with the SIFT process (c, d). Spectra a) and c) are from the pre-exercise period, while b) and d) were acquired mid-exercise, showing depleted PCr and elevated Pi.
The improved SNR evident in figures 3 and 4 allows more accurate fitting of the metabolite peaks by AMARES. The standard deviation in the PCr amplitude fit was reduced by an average of 72.5% +/− 5.82% and that for the Pi resonance was reduced by 60.26% +/− 9.49%. The mean improvements per subject are shown in columns 1 and 2 of table 1.
Table 1.
| SD of PCr fit % reduction |
SD of PI fit % reduction |
Std τ / s |
Std τ % SD |
SIFT τ / s |
SIFT τ % SD |
SD of τ fit % reduction |
||
|---|---|---|---|---|---|---|---|---|
| Calf | VOL1 | 66.3 | 49.9 | 35.2 | 7 | 36.6 | 2.8 | 59.1 |
| VOL2 | 56.9 | 47.3 | 76.6 | 6.9 | 82.4 | 5.1 | 26.3 | |
| VOL3 | 72.2 | 53.3 | 21.2 | 11.1 | 21 | 6.7 | 39.6 | |
| PAD1 | 77.5 | 71.4 | 107.8 | 10.6 | 107.1 | 5.7 | 45.7 | |
| PAD2 | 73.9 | 52.3 | 49.5 | 10.7 | 51.6 | 4.1 | 61.5 | |
| Thigh | VOL4 | 75.1 | 69.1 | 36.5 | 3.9 | 39.5 | 3 | 24.3 |
| VOL5 | 75.7 | 67.3 | 62 | 3.6 | 60.6 | 2 | 45.1 | |
| VOL6 | 73.4 | 65.5 | 78.1 | 6.5 | 75.1 | 5.7 | 11.3 | |
| VOL7 | 73.1 | 65.3 | 63.1 | 4.8 | 62.9 | 3.5 | 28.7 | |
| VOL8 | 74.8 | 64.2 | 72.7 | 12.6 | 65.4 | 10.2 | 18.9 | |
| VOL9 | 72.5 | 70.6 | 22.4 | 12.3 | 24.4 | 7.5 | 39.1 | |
| PAD3 | 78.9 | 47.0 | 62.4 | 9 | 62.9 | 3.6 | 60.4 | |
| Mean | 72.5 | 60.3 | 8.3 | 5 | 38.4 | |||
VOL = volunteer; PAD = patient with Peripheral Artery Disease
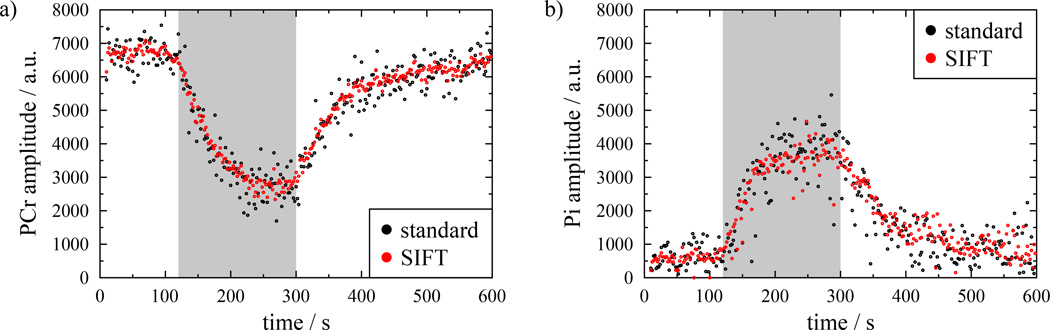
The combination of more accurate fitting at each time point with the exercise time domain smoothing introduced by SIFT improved the fit of the exponential fit to the PCr recovery. Figure 5 shows time courses for both the PCr and Pi fitted amplitudes from a sample dataset both before and after SIFT. The spread of amplitudes for both metabolites is considerably reduced in the SIFT case, but overall the shape of the temporal trajectory is preserved, with no distortion or smoothing.
Figure 5.
Time course of (a) PCr and (b) Pi amplitudes for both standard (black circles) and SIFTed data (red circles). The SIFTed data shows a reduced level of noise while following the same shape as the original data. The period of exercise is shaded.
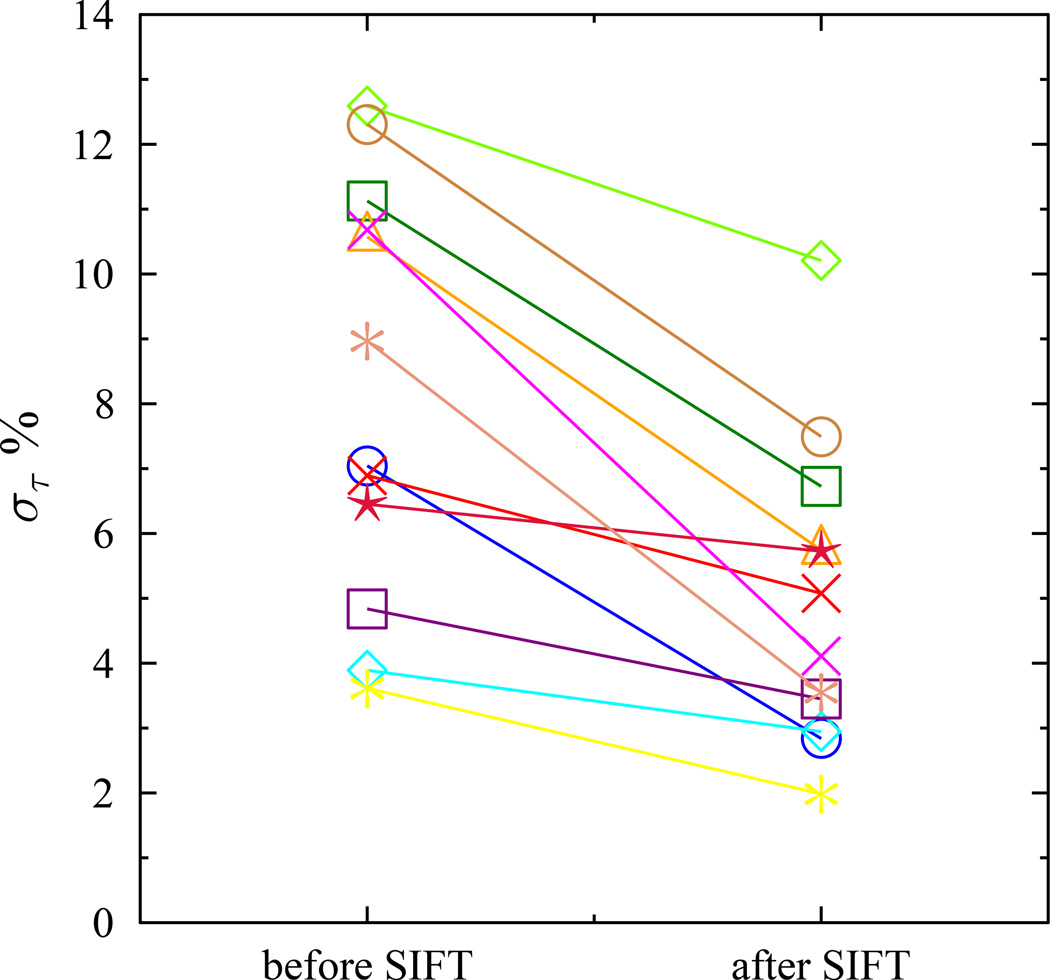
For every subject the standard deviation in τ for the SIFTed data was reduced relative that obtained for the uncorrected data (Figure 6). The size of the fitting error is correlated with the recovery time constant. To allow comparison across the range of values from the subjects, the standard deviation as a percentage of the mean is used. The mean improvement in standard deviation was 38.4%. The estimated values and standard deviations for τ in the standard and SIFTed case for each subject are shown in Table 1.
Figure 6.
Difference in τ fitting accuracy between standard and SIFTed data for each subject. In every case the error, here reported as the standard deviation (στ) of the τ (as a percentage) is reduced with SIFT, the largest improvements mostly occur for the subjects with the highest error in the standard case.
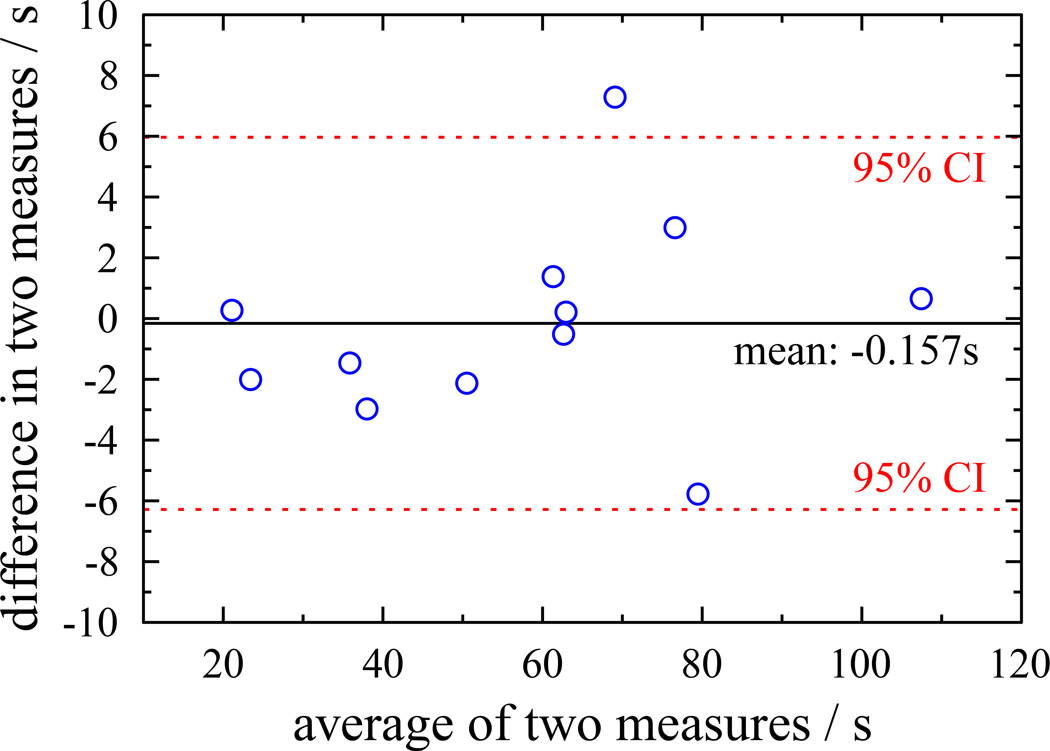
The evaluation of any new processing method must consider whether a bias or systematic error is introduced relative to the original data. One method for visually evaluating the differences between the results from two methods is the Bland-Altman plot, shown in figure 7. This plots the mean value of the two methods against the difference between them for each subject. The mean shift in τ from applying SIFT to the data is only −0.16s, showing that no bias is introduced, SIFT does not systematically lengthen or shorten τ. The points are also uniformly distributed either side of the mean for all values of τ, indicating that SIFT performs equally well across a wide range of recovery times.
Figure 7.
Bland-Altman plot showing difference in τ between standard and SIFTed results as a function of τ. Points are uniformly distributed on either side of zero and there are no apparent trends in the distribution showing that SIFT does not introduce any systematic bias.
Discussion
Across a diverse range of clinical scenarios, MR spectroscopy is a widely used tool in the study of time varying systems. However, the technique’s SNR is limited by (i) low concentrations of target molecules (particularly when observing spins other than protons) and (ii) its inability to apply the temporal averaging method used in static MRS.
The results demonstrate that SIFT substantially reduces measurement uncertainties in dynamic studies of healthy and PAD subjects. Both fast and slow PCr recovery curves can be equally improved by applying SIFT to the data. One of the strengths of the SIFT method is that it is independent of the shape of the temporal trajectory it is fitting. Although in this study it only quantified the monoexponential recovery for PCr, it qualitatively reduced the temporal noise throughout the entire exercise time domain for both PCr and Pi. SIFT should be applicable with non- exponential time courses.
SIFT is a very straightforward and fast operation to perform on even a large dataset. The method comprises only a Fourier transform in the exercise domain, thresholding of the coefficients and the inverse Fourier transform. In our implementation this takes less than a second for 295 spectra.
One characteristic of SIFT is that the noise suppression varies with spectral frequency, depending on the degree of temporal variation in the exercise time domain. Although the quantification algorithm used in AMARES is not itself sensitive to such variations in the character of the noise, the estimation of concentration uncertainties using the Cramer-Rao lower bound does depend on the noise level, which is normally assessed from a region of the spectrum far from the peak regions. If the noise level varies across the spectrum then this might affect the uncertainty estimates. In order to investigate this issue, the degree of noise suppression as a function of frequency was calculated.
Across the spectrum SIFT thresholding suppressed an average of 279 out of the 295 exercise frequency domain coefficients. A threshold of two standard deviations includes 95% of the pure noise coefficients. Therefore, the mean total number of pure noise coefficients should be 279 / 0.95 = 294. This is the expected value for a time invariant signal, which describes most of the spectrum. In this case the signal in the exercise frequency domain is concentrated in the first coefficient while the remaining 294 contain only noise. As noted in the methods section, although thresholding suppresses 95% of the noise coefficients the remaining 5% of coefficients contain 14% of the noise energy. Thus, SIFT only removes 86% of the noise energy.
For the 100 spectral points making up the PCr peak an average of 265 exercise frequency domain coefficients were suppressed. Since the mean total number of pure noise coefficients is estimated at 279 (265 / 0.95), PCr has 16 signal coefficients in the exercise frequency domain. The mean residual noise energy in this region is approximately 19%, calculated as (14% × 279 + 100% × 16) / 295 ≈ 19%. While the noise energy is higher in the PCr peak region, it is only ~ 5 % above the theoretical optimum found elsewhere in the spectrum. We therefore feel that this relatively small variation in noise level across the spectrum will not significantly impact the quantification process.
The degree to which SIFT is able to remove noise is dependent on the sparseness of the temporal trajectory’s representation in the exercise frequency domain. We chose to use SIFT in this case because of its simplicity and the previously reported effectiveness of the method, and our results show that for data of this kind, the signal is indeed sparse and SIFT is an effective denoising method. However for some alternative dynamic MRS studies, the temporal trajectory of the metabolite peaks might have a different form so that the indirect spectral domain has a larger number of signal coefficients, which would have a negative impact on the efficacy of SIFT. In such cases it would be interesting to explore other transforms that might improve sparsity in these cases, or alternate denoising techniques such as wavelet denoising or methods based on singular value decomposition.
In this study SIFT was applied to 31P MR muscle data. The method is, however, generally applicable to any MRS study acquiring time series spectra. A possible application would be improving 31P MR SNR obtained from localized sequences that isolate signal from specific muscles (30, 31). A typical 31P surface coil may have a total sensitive volume of around 300 cm3, while a localized muscle voxel might be only 30 cm3. In a typical localized study, the use of a PRESS echo sequence rather than a simple pulse acquire introduces T2 decay losses that also reduce SNR.
Serial spectra following labeled molecules e.g. 13C and 17O through metabolism would benefit from improved temporal resolution or improved SNR by making use of SIFT. For example, in dynamic 13C MR studies that track the transfer of labeled 13C from injected glucose or acetate to glutamate and glutamine through the TCA cycle by serial acquisition of spectra with a TR=1 sec for more than 2 hours (17). Due to the low SNR, a sliding window of 20 min (1200 acquisitions) was used to adequately measure the time course of the 13C metabolites (18). More recently, 1H MRS has also been used for dynamic measures of lactate, gamma-amino butyric acid and glutamate in functional tasks such as visual stimulation (19,20), pain (21,22), and cognitive tests (23,24), which also utilize serial acquisitions of spectra. These would also benefit from SIFT.
Conclusion
This study shows that the SIFT method produces a significant improvement in the SNR of time varying spectroscopy acquisitions, without compromising the temporal resolution in a realistic evaluation of healthy and diseased subjects. This improvement in the SNR translates to more precise quantification of peak amplitudes, with an average reduction in fit standard deviation of 73% for PCr. Better metabolite quantification leads to a reduced error in the fitting of the recovery curves. In this case a reduction in the standard deviation of 62% was observed. No evidence of bias in the SIFTed recovery curves relative to the original data was found.
Acknowledgements
Grants from the Osher Center for Integrative Medicine to JAB and APL and NIH EB 014414 to JAB supported this work. We thank Ms. Jessica Milian and Ms. Caitlin Parmer of the Cardiovascular Division, Brigham and Women’s Hospital for the recruitment of the PAD patients.
References
- 1.Chance B, Im J, Nioka S, Kushmerick M. Skeletal muscle energetics with PNMR: personal views and historic perspectives. NMR Biomed. 2006;19(7):904–926. doi: 10.1002/nbm.1109. Review PubMed PMID: 17075955. [DOI] [PubMed] [Google Scholar]
- 2.Layec G, Bringard A, Le Fur Y, Vilmen C, Micallef JP, Perrey S, Cozzone PJ, Bendahan D. Reproducibility assessment of metabolic variables characterizing muscle energetics in vivo: A 31P-MRS study. Magnetic resonance in med. 2009;62(4):840–854. doi: 10.1002/mrm.22085. PubMed PMID: 19725136. [DOI] [PubMed] [Google Scholar]
- 3.Kemp GJ, Radda GK. Quantitative interpretation of bioenergetic data from 31P and 1H magnetic resonance spectroscopic studies of skeletal muscle: an analytical review. Magnetic resonance quarterly. 1994;10(1):43–63. [PubMed] [Google Scholar]
- 4.Haseler LJ, Lin A, Hoff J, Richardson RS. Oxygen availability and PCr recovery rate in untrained human calf muscle: evidence of metabolic limitation in normoxia. Am J Physiol Regul Integr Comp Physiol. 2007;293(5):R2046–R2051. doi: 10.1152/ajpregu.00039.2007. [DOI] [PubMed] [Google Scholar]
- 5.Haseler LJ, Lin AP, Richardson RS. Skeletal muscle oxidative metabolism in sedentary humans: 31P-MRS assessment of O2 supply and demand limitations. J Appl Physiol. 2004;97(3):1077–1081. doi: 10.1152/japplphysiol.01321.2003. [DOI] [PubMed] [Google Scholar]
- 6.Makimura H, Murphy CA, Feldpausch MN, Grinspoon SK. The effects of tesamorelin on phosphocreatine recovery in obese subjects with reduced GH. The Journal of clinical endocrinology and metabolism. 2014;99(1):338–343. doi: 10.1210/jc.2013-3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Libri V, Brown AP, Gambarota G, Haddad J, Shields GS, Dawes H, Pinato DJ, Hoffman E, Elliot PJ, Vlasuk GP, Jacobson E, Wilkins MR, Matthews PM. A pilot randomized, placebo controlled, double blind phase I trial of the novel SIRT1 activator SRT2104 in elderly volunteers. PloS one. 2012;7(12):e51395. doi: 10.1371/journal.pone.0051395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.West AM, Anderson JD, Epstein FH, Meyer CH, Wang H, Hagspiel KD, Berr SS, Harthun NL, Weltman AL, Dimaria JM, Hunter JR, Christopher JM, Kramer CM. Low-density lipoprotein lowering does not improve calf muscle perfusion, energetics, or exercise performance in peripheral arterial disease. Journal of the American College of Cardiology. 2011;58(10):1068–1076. doi: 10.1016/j.jacc.2011.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDermott MM, Guralnik JM, Albay M, Bandinelli S, Miniati B, Ferrucci L. Impairments of muscles and nerves associated with peripheral arterial disease and their relationship with lower extremity functioning: the InCHIANTI Study. Journal of the American Geriatrics Society. 2004;52(3):405–410. doi: 10.1111/j.1532-5415.2004.52113.x. [DOI] [PubMed] [Google Scholar]
- 10.Pande RL, Perlstein TS, Beckman JA, Creager MA. Secondary prevention and mortality in peripheral artery disease: National Health and Nutrition Examination Study, 1999 to 2004. Circulation. 2011;124(1):17–23. doi: 10.1161/CIRCULATIONAHA.110.003954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isbell DC, Berr SS, Toledano AY, Epstein FH, Meyer CH, Rogers WJ, Harthun NL, Hagspiel KD, Weltman A, Kramer CM. Delayed calf muscle phosphocreatine recovery after exercise identifies peripheral arterial disease. Journal of the American College of Cardiology. 2006;47(11):2289–2295. doi: 10.1016/j.jacc.2005.12.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goluch S, Kuehne A, Meyerspeer M, Kriegl R, Schmid AI, Fiedler GB, Herrmann T, Mallow J, Hong SM, Cho ZH, Bernarding J, Moser E, Laistler E. A form-fitted three channel P, two channel H transceiver coil array for calf muscle studies at 7 T. Magnetic resonance in medicine. 2015;73(6):2376–2389. doi: 10.1002/mrm.25339. [DOI] [PubMed] [Google Scholar]
- 13.Meyerspeer M, Robinson S, Nabuurs CI, Scheenen T, Schoisengeier A, Unger E, Kemp GJ, Moser E. Comparing localized and nonlocalized dynamic 31P magnetic resonance spectroscopy in exercising muscle at 7 T. Magnetic resonance in medicine. 2012;68(6):1713–1723. doi: 10.1002/mrm.24205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doyle M, Chapman BLW, Balschi JA, Pohost GM. SIFT, a Postprocessing Method That Increases the Signal-to-Noise Ratio of Spectra Which Vary in Time. Journal of Magnetic Resonance, Series B. 1994;103(2):128–133. [Google Scholar]
- 15.Larson DE, Hesslink RL, Hrovat MI, Fishman RS, Systrom DM. Dietary effects on exercising muscle metabolism and performance by 31P-MRS. Journal of applied physiology. 1994;77(3):1108–1115. doi: 10.1152/jappl.1994.77.3.1108. [DOI] [PubMed] [Google Scholar]
- 16.Naressi A, Couturier C, Castang I, de Beer R, Graveron-Demilly D. Java-based graphical user interface for MRUI, a software package for quantitation of in vivo/medical magnetic resonance spectroscopy signals. Computers in biology and medicine. 2001;31(4):269–286. doi: 10.1016/s0010-4825(01)00006-3. [DOI] [PubMed] [Google Scholar]
- 17.Ross B, Lin A, Harris K, Bhattacharya P, Schweinsburg B. Clinical experience with 13C MRS in vivo. NMR Biomed. 2003;16(6–7):358–369. doi: 10.1002/nbm.852. [DOI] [PubMed] [Google Scholar]
- 18.Lin AP, Shic F, Enriquez C, Ross BD. Reduced glutamate neurotransmission in patients with Alzheimer's disease -- an in vivo (13)C magnetic resonance spectroscopy study. MAGMA. 2003;16(1):29–42. doi: 10.1007/s10334-003-0004-x. [DOI] [PubMed] [Google Scholar]
- 19.Lin Y, Stephenson MC, Xin L, Napolitano A, Morris PG. Investigating the metabolic changes due to visual stimulation using functional proton magnetic resonance spectroscopy at 7 T. Journal of cerebral blood flow and metabolism. 2012;32(8):1484–1495. doi: 10.1038/jcbfm.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reyngoudt H, Paemeleire K, Dierickx A, Descamps B, Vandemaele P, De Deene Y, Achten E. Does visual cortex lactate increase following photic stimulation in migraine without aura patients? A functional (1)H-MRS study. The journal of headache and pain. 2011;12(3):295–302. doi: 10.1007/s10194-011-0295-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gussew A, Rzanny R, Erdtel M, Scholle HC, Kaiser WA, Mentzel HJ, Reichenbach JR. Time-resolved functional 1H MR spectroscopic detection of glutamate concentration changes in the brain during acute heat pain stimulation. NeuroImage. 2010;49(2):1895–1902. doi: 10.1016/j.neuroimage.2009.09.007. [DOI] [PubMed] [Google Scholar]
- 22.Kupers R, Danielsen ER, Kehlet H, Christensen R, Thomsen C. Painful tonic heat stimulation induces GABA accumulation in the prefrontal cortex in man. Pain. 2009;142(1–2):89–93. doi: 10.1016/j.pain.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 23.Michels L, Martin E, Klaver P, Edden R, Zelaya F, Lythgoe DJ, Luchinger R, Brandeis D, O'Gorman RL. Frontal GABA levels change during working memory. PloS one. 2012;7(4):e31933. doi: 10.1371/journal.pone.0031933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lally N, Mullins PG, Roberts MV, Price D, Gruber T, Haenschel C. Glutamatergic correlates of gamma-band oscillatory activity during cognition: a concurrent ER-MRS and EEG study. NeuroImage. 2014;85(Pt 2):823–833. doi: 10.1016/j.neuroimage.2013.07.049. [DOI] [PubMed] [Google Scholar]