Abstract
In the classical view, the formation of a primary tumor is the consequence of a mutational event that first affects a single cell that subsequently passes through a multitude of consecutive hyperplastic and dysplastic stages. At the end of this pathogenetic sequence a cell arises that is potentially able to expanse infinitely having capacity to form a homogenous tumor mass. In contrary to this clonal expansion concept, the majority of primary human tumors display already a startling heterogeneity that can be reflected in different morphological features, physiological activities, and genetic diversity. In the past it was speculated that this cancer cell plasticity within a tumor is the result of an adaptive process that is induced by specific inhibiting therapies. In regard to the formation of hepatocellular carcinoma (HCC) this dogma was once challenged in a recent study that analysed tumor areas that were taken from HCC patients without medical pretreatment. Most of the analyzed samples showed highly significant intratumor heterogeneity. This affected morphological attributes, immunohistochemical stainability of five tumor-associated markers [α-fetoprotein (AFP), EpCAM, CK7, CD44 and glutamine synthetase], and integrity of genes (β-catenin and p53) that are critically involved in the pathogenesis of HCC. Altogether, this study showed that intratumor heterogeneity is a frequent finding in HCC that may contribute to treatment failure and drug resistance in HCC patients.
Keywords: Cancer stem cell model, clonal evolution model, stochastic model, hepatocellular carcinoma (HCC), subclone, tumor diversification
Hepatocellular carcinoma (HCC) is the most frequent primary liver cancer diagnosed worldwide and a prominent source of mortality (1). It develops on the background of many etiologies (chronic hepatitis B and C, non-alcoholic steatohepatitis, gene mutations) that either trigger hepatocytes to replicate at higher rate or by inducing a cellular phenotype that is resistant to apoptosis. Current pre-clinical research is focussed on genes that are deregulated during HCC development and predictive biomarkers that may lead to the identification of novel pharmacological relevant target structures. Prototypically, somatic mutations of the β-catenin gene (CTNNB1) leading to aberrant nuclear expression of β-catenin and activation of the Wnt/β-catenin pathway in HCC promote tumor progression by stimulating tumor cell proliferation (2). Likewise, there is a large mutational spectrum within the TP53 gene encoding the tumor suppressor p53. Several p53 mutations have profound effects on its protective activities towards DNA-damaging agents, chronic hepatitis virus infection, and during the molecular pathogenesis of HCC (3). Therefore, genetic testing for respective alterations is diagnostically widely applied. In addition, elevated expression of biliary/progenitor cell markers (e.g., cytokeratin 7, CK7; cytokeratin 19, CK19), cancer stem cell surface markers (CD34, CD44, EpCAM), α-fetoprotein (AFP), and other proteins that become differentially expressed in the tumor were introduced in diagnosing HCC. These immunohistochemical markers are widely used to classify HCC into different prognostic subclasses sharing similar characteristics or to guide therapeutic decision-making for personalized treatment in HCC. However, on the observed lack of consistent therapeutic outcome it was recognized during the last years that the histology-based definition of the morphological heterogeneity of HCC needs critical refinement (4).
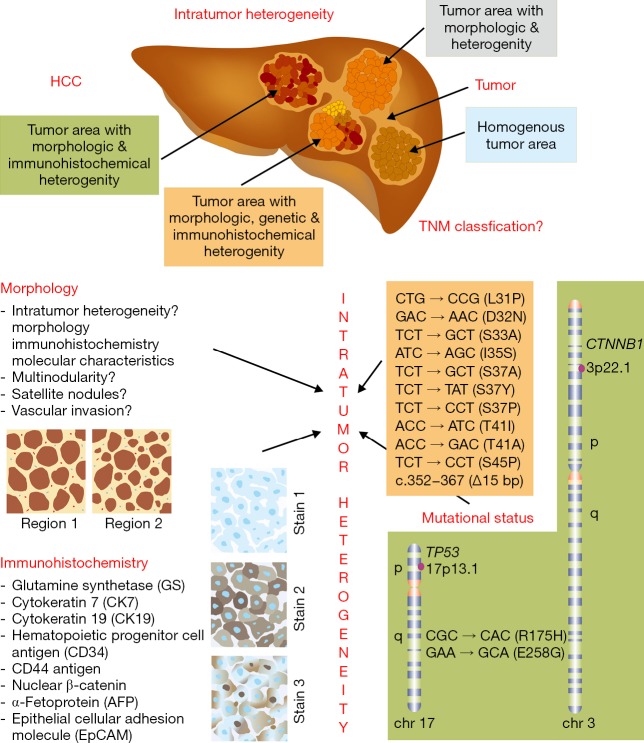
Beside the observed variability among patients, a recent study systematically characterized intratumor heterogeneity in HCC in regard to morphology, immune phenotype, and mutational status within the CTNNB1 and TP53 genes (5). In the mentioned study, the authors analyzed 120 tumor areas taken from 23 patients suffering from HCC without medical pretreatment. In particular, the samples were analysed for cell and tissue morphologies, expression of tumor-associated markers (CK7, CD44, AFP, EpCAM and glutamine synthetase) and for gene mutations affecting the TP53 or CTNNB1 genes. In most of the cases, the authors noticed intratumor heterogeneity that either affected the morphology alone, the morphology and immunohistochemical characteristics, or pertained morphology, exposed antigens and mutational status of the CTNNB1 and TP53 genes (Figure 1). Only three patients showed homogenous tumors lacking the morphologic and immunohistochemical intratumor heterogeneity.
Figure 1.
Intratumor heterogeneity. (A) Friemel and coworkers analysed 23 HCC patients without medical pretreatment. In most cases (n=20), intratumor heterogeneity was observed solely on the level of morphology (n=6), on the level of morphology combined with immunohistochemical heterogeneity (n=9), and heterogeneity in regard to morphology, immunohistochemistry and mutational status of the tumor suppressor p53 (TP53) and β-catenin (CTNNB1) (n=5). Only three tumors were phenotypically homogenous, meaning that there was no morphological or immunohistochemical variation observed. The authors concluded that this intratumor heterogeneity is a challenge for the establishment of a robust HCC classification and a critical factor that contributes to treatment failure and the development of drug resistance; (B) the analyzed morphological characteristics, immunohistochemical parameters as well as the detected TP53 and CTNNB1 gene mutations are depicted. More details on this study are given elsewhere (5). HCC, hepatocellular carcinoma.
Although the analyzed patient cohort in this study is rather small, the study unequivocally shows that intratumor heterogeneity is a frequent finding in HCC. Furthermore, the morphological and immunophenotypical heterogeneity within the tissue was associated with variable somatic TP53 and CTNNB1 gene mutations suggesting that the observed endogenous tumor cell plasticity and tumor cell subclonality in the affected liver tissue is crucially triggered by genetic factors.
The observed intratumor heterogeneity in the tumorigenic livers has major implications for diagnosis and therapy of HCC. In light of the present study, actual classification criteria and scoring systems that are presently used in prognostic staging of hepatic tumors are challenged by the finding of intratumor heterogeneity. The TNM system for example that is maintained by the American Joint Committee on Cancer (AJCC) and the International Union for Cancer Control (UICC) is widely used among clinicians for tumor classification, determination of a targeted therapy and assessment of the chance of a successful treatment outcome (6). However, criteria of intratumor heterogeneity are not included in this scoring system.
Since the study by Friemel and colleagues enrolled patients without medical pretreatment, the findings further confirm previous results that have shown that intratumor heterogeneity is an intrinsic property of primary tumors in which chemotherapy only promotes the dominance of existing previously minor or dormant lineages (7). Therefore, the imprinted heterogeneity of a primary tumor might be one of the driving forces predicting clonal evolution, tumor progression, and resistance to chemotherapy.
There is clear evidence from many other tumors that the phenotypic and functional heterogeneity hierarchically arise among cancer cells as a consequence of genetic drift and epigenetic environment differences (8). Based on this assumption, HCC tumor diversification is a highly dynamic process that might offer some new diagnostic avenues with prognostic value. It also implies that in the development of novel drugs or definition of therapeutic targets, the occurrence of intratumor heterogeneity in HCC has to be considered. As discussed above, well established HCC staging systems such as the TNM classification (6) incorporates only information about the characteristics of the original primary tumor (T), the involved regional lymph nodes (N), and the occurrence of distant metastasis (M). Data on intratumorigenic heterogeneity might on long-term added to these scoring systems to better support the requested personalization in HCC therapy and outcome prediction. In this regard, the development of novel single-cell Western blotting techniques (9), innovative mass spectrometric imaging techniques designed for detection of tumor heterogeneity (10) and single-cell imaging techniques that have diagnostic capacity to unravel different cell populations in a tumor (11) might offer new diagnostic options to early track down such imprinted intratumorigenic heterogeneities at single cell resolution.
During the last years several models were discussed that should explain tumor heterogeneity (12). Currently there are two models that are favoured (Figure 2). In the “cancer stem cell model”, it is supposed that within a population of tumor cells, there is a distinct subset of cells with self-renewal capacity that are potentially tumorigenic (13). These cells can drive tumor growth and intratumor heterogeneity might result from differences in the stem cells which contributed to the pathogenetic event. In the “clonal evolution model” that was already proposed in 1976 it is assumed that the primary tumor arises from a single mutated cell that accumulates additional mutations during its uncontrolled multiplication (14). The resulting heterogenic subclones in turn have also potential to form further subclones that have reproductive or survival advantages in the tumor environment. This hypothesis is also compatible with the establishment of a mosaic tumor that has the observed variations in genotype and phenotype. Certainly, these two models are not mutually exclusive and it is not excluded that they both cooperate or synergistically act in establishing intratumor heterogeneity during neoplastic transformation and HCC.
Figure 2.
Models of tumor growth. The cancer stem cell model (A) suggests a hierarchy of cells in which only a small subset of tumorigenic cells exists. These tumor-forming cancer stem cells (CSC) have self-renewal capacity (SR) and potential to differentiate into non-tumorigenic cells. As a consequence, a neoplasm contains cancer stem cells that feed the abnormal growth of the tissue, cells that divide a few times before they differentiate into specialized tumor cells, and inactive tumor cells. The clonal evolution theory (B) that is a stochastic model suggests that a tumor is the result of a single mutated somatic cell that acquires a highly proliferative phenotype and accumulates additional mutations during repeated divisions. There is no hierarchy during tumorigenesis and the resulting subpopulations have different potential to grow and divide. The resulting subclones can independently choose between self-renewal and differentiation and during time the tumor environment create dominant cell variants that have acquired growth advantages. While in the cancer stem cell model individual CSCs are therapeutic targets, individual somatic cells with unwanted reproductive or survival properties must be tackled therapeutically according to the clonal evolution model.
To emphasize it again, the observed intratumorigenic heterogeneity has wide implication in HCC therapy. It is obvious that the different clonal subpopulation within the tumor may exhibit different sensitivities to drugs and causative involved in mediating drug resistance. Moreover, since the epigenetic and genetic factors that provoke the formation of different tumor cell subclones is nearly infinitely, it can be assumed that each patient acquires a highly individually mixture of subtumors that is unique in regard to genetic, immunologic and clinico-pathological phenotype. This diversity is further modulated by patient's specific tumor microenvironment consisting of different numbers and amounts of soluble factors, signalling molecules, extracellular matrix components and many other factors.
Consequently, each patient needs a highly personalized therapy targeting its individual divergent cancer entity. The complexity in elaborating such sophisticated treatment regimens is a scary clinical challenge that will require new diagnostic approaches for definition of intratumorigenic diversity. It was recently proposed that a computationally predictive combination therapy in the context of intratumoral diversity is a chance to maximize tumor cell death and to minimize the outgrowth of clonal subpopulations (15).
In regard to HCC, it is now first necessary to estimate the potential relevance of intratumorigenic diversity for the pathogenesis and outcome prediction in larger patient cohorts. It is also required to dissect if the observed spatial and temporal alterations during the initiation and progression of HCC are dependent on the etiology of the tumor and to dissect the genetic or epigenetic factors that influence generation of intratumor heterogeneity. Unravelling of inter-individual differences in susceptibility for intratumor heterogeneity will possibly allow on long-term to establish novel personalized treatments designed for specific subsets of HCC patients that carry similar combinations of heterogenic morphological, immunohistochemical, immunologic or mutations.
Acknowledgements
The author is grateful to Sabine Weiskirchen for help in preparing the final figures.
Funding: Author’s laboratory is sponsored by the German Research Foundation (DFG, SFB TRR57 P13) and the Interdisciplinary Centre for Clinical Research within the Faculty of Medicine at the RWTH Aachen University (IZKF, project E7-6).
Footnotes
Provenance: This is a Guest Editorial commissioned by the Deputy Editor-in-Chief Haitao Zhao (Department of Liver Surgery, Peking Union Medical College Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China).
Conflicts of Interest: The author has no conflicts of interest to declare.
References
- 1.Waller LP, Deshpande V, Pyrsopoulos N. Hepatocellular carcinoma: A comprehensive review. World J Hepatol 2015;7:2648-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nhieu JT, Renard CA, Wei Y, et al. Nuclear accumulation of mutated beta-catenin in hepatocellular carcinoma is associated with increased cell proliferation. Am J Pathol 1999;155:703-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hussain SP, Schwank J, Staib F, et al. TP53 mutations and hepatocellular carcinoma: insights into the etiology and pathogenesis of liver cancer. Oncogene 2007;26:2166-76. [DOI] [PubMed] [Google Scholar]
- 4.Li L, Wang H. Heterogeneity of liver cancer and personalized therapy. Cancer Lett 2015. [Epub ahead of print]. doi: . 10.1016/j.canlet.2015.07.018 [DOI] [PubMed] [Google Scholar]
- 5.Friemel J, Rechsteiner M, Frick L, et al. Intratumor heterogeneity in hepatocellular carcinoma. Clin Cancer Res 2015;21:1951-61. [DOI] [PubMed] [Google Scholar]
- 6.Edge SB, Compton CC. The American Joint Committee on Cancer: the 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol 2010;17:1471-4. [DOI] [PubMed] [Google Scholar]
- 7.Kreso A, O'Brien CA, van Galen P, et al. Variable clonal repopulation dynamics influence chemotherapy response in colorectal cancer. Science 2013;339:543-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature 2013;501:328-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang CC, Lin JM, Xu Z, et al. Single-cell Western blotting after whole-cell imaging to assess cancer chemotherapeutic response. Anal Chem 2014;86:10429-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abramowski P, Kraus O, Rohn S, et al. Combined Application of RGB Marking and Mass Spectrometric Imaging Facilitates Detection of Tumor Heterogeneity. Cancer Genomics Proteomics 2015;12:179-87. [PubMed] [Google Scholar]
- 11.Di Palma S, Bodenmiller B. Unraveling cell populations in tumors by single-cell mass cytometry. Curr Opin Biotechnol 2015;31:122-9. [DOI] [PubMed] [Google Scholar]
- 12.Shackleton M, Quintana E, Fearon ER, et al. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell 2009;138:822-9. [DOI] [PubMed] [Google Scholar]
- 13.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell 2014;14:275-91. [DOI] [PubMed] [Google Scholar]
- 14.Nowell PC. The clonal evolution of tumor cell populations. Science 1976;194:23-8. [DOI] [PubMed] [Google Scholar]
- 15.Zhao B, Pritchard JR, Lauffenburger DA, et al. Addressing genetic tumor heterogeneity through computationally predictive combination therapy. Cancer Discov 2014;4:166-74. [DOI] [PMC free article] [PubMed] [Google Scholar]