Abstract
Plants are sessile organisms that have evolved a complex immune system which helps them cope with pathogen attacks. However, the capacity of a plant to mobilize different defense responses is strongly affected by its physiological status. Nitrogen (N) is a major nutrient that can play an important role in plant immunity by increasing or decreasing plant resistance to pathogens. Although no general rule can be drawn about the effect of N availability and quality on the fate of plant/pathogen interactions, plants’ capacity to acquire, assimilate, allocate N, and maintain amino acid homeostasis appears to partly mediate the effects of N on plant defense. Nitric oxide (NO), one of the products of N metabolism, plays an important role in plant immunity signaling. NO is generated in part through Nitrate Reductase (NR), a key enzyme involved in nitrate assimilation, and its production depends on levels of nitrate/nitrite, NR substrate/product, as well as on L-arginine and polyamine levels. Cross-regulation between NO signaling and N supply/metabolism has been evidenced. NO production can be affected by N supply, and conversely NO appears to regulate nitrate transport and assimilation. Based on this knowledge, we hypothesized that N availability partly controls plant resistance to pathogens by controlling NO homeostasis. Using the Medicago truncatula/Aphanomyces euteiches pathosystem, we showed that NO homeostasis is important for resistance to this oomycete and that N availability impacts NO homeostasis by affecting S-nitrosothiol (SNO) levels and S-nitrosoglutathione reductase activity in roots. These results could therefore explain the increased resistance we noted in N-deprived as compared to N-replete M. truncatula seedlings. They open onto new perspectives for the studies of N/plant defense interactions.
Keywords: nitrogen metabolism, plant immunity, Aphanomyces euteiches, Medicago truncatula, nitric oxide homeostasis
Nitrogen and the Plant Immune Response
Plants are under the constant threat of pathogen attacks that limit their survival and are major yield-limiting factors. In response to these attacks, plants activate multiple defense reactions both at the site of infection and systemically, which in many cases lead to resistance. These reactions include massive transcriptional reprogramming, cell wall reinforcement, synthesis of antimicrobial metabolites, and production of pathogenesis-related (PR) proteins. These events are mediated by a variety of rapidly mobilized molecules, such as second messengers, e.g., Ca2+, protein kinases, reactive oxygen species (ROS), or reactive nitrogen species (RNSs), including nitric oxide (NO). Although these defense responses have been widely studied, it has become increasingly obvious over the past years that a plant’s capacity to mobilize them is greatly affected by its physiological status (Snoeijers et al., 2000) and its development (Develey-Riviere and Galiana, 2007).
Nutrients are important for the growth and development of plants and microorganisms. Among them, nitrogen (N) can affect the fate of an interaction between a plant and a pathogen (Dordas, 2008). No general rules can be drawn about modification of resistance by N. Although we know that N lack or excess, along with the nature of available N in soil, can modulate plant resistance (Huber and Watson, 1974), the underlying mechanisms remain unclear. Recent works indicate that plants’ capacity to acquire and assimilate N could partly explain nutrition effects on plant defense. N is taken up by the roots mostly in the form of nitrate (NO3-) in aerobic soils and ammonium (NH4+) in flooded wetlands or acidic soils. Ammonium taken up directly from the soil or resulting from the reduction of NO3- and nitrite (NO2-) by nitrate reductase (NR) and nitrite reductase (NiR), respectively, is assimilated via the glutamine synthetase (GS)/glutamate synthase cycle (Xu et al., 2012). The uptake of mineral N from the soil and the subsequent distribution to the whole plant is driven by nitrate transporters from the multigenic NRT2 and NPF families and by ammonium transporters from the AMT family (Krapp, 2015). The contribution of several of these transporters to plant defense has recently been highlighted in Arabidopsis thaliana. For instance, induction of AMT1.1 expression was evidenced upon infection by the bacterium Pseudomonas syringae or the fungus Erysiphe cichoracearum (Liu et al., 2010). The role of specific transporters was demonstrated using plant mutants: nrt2 (deficient in the expression of the NRT2.1 and NRT2.2 genes) and nrt2.6-1 mutants displayed altered sensitivity to the bacterial phytopathogens P. syringae and E. amylovora (Camanes et al., 2012; Dechorgnat et al., 2012). Besides N uptake into plants and its subsequent allocation, several results indicate that N assimilation and particularly amino acid homeostasis can impact plant-pathogen interactions (Zeier, 2013; Luna et al., 2014). Conversely, pathogen attacks are correlated with modulation of the expression of genes or of the activity of enzymes involved in N assimilation such as NR or GS2, in N remobilization such as GS1, and in amino acid metabolism [reviewed by Fagard et al. (2014)]. Whether these changes in N metabolism reflect the manipulation of host metabolism by the pathogen or result from the modulation of plant defenses is not always clear. Interestingly, some members of the GLR glutamate receptor family were recently proposed to play a role as amino acid sensors during plant defense, perhaps by sensing changes in extracellular amino acids caused by pathogen infection (Forde and Roberts, 2014). Crosstalk between N metabolism and phytohormones can also interfere with plant stress responses and could be considered as a mechanism involved in the partitioning of available resources between defense and growth. For instance, N limitation induced the accumulation of salicylic acid (SA) in A. thaliana leaves (Yaeno and Iba, 2008). Conversely, ethylene/jasmonic acid signaling coordinated the upregulation of the nitrate transporter NRT1.8 (AtNPF7.2) and the downregulation of NRT1.5 (AtNPF7.3) genes to tune NO3- reallocation in plants from the shoot to the roots under stress conditions (Zhang et al., 2014). Finally, experiments on rice showed that N-induced susceptibility to Magnaporthe oryzae is genotype-dependent, and may be linked to N use efficiency (Ballini et al., 2013). These interesting data raise the question of the genetic control of N effects on plant immunity. The identification of the corresponding QTLs will permit to uncover new molecular actors of N-controlled resistance to pathogens.
Nitric Oxide and N Metabolism
The role of NO in plant defense is widely accepted. NO is involved in transcriptional regulation of defense genes encoding PR proteins or proteins involved in phytoalexin synthesis, SA accumulation, and post-translational protein modifications (Wendehenne et al., 2014). NO is a nitrogen species produced via a variety of pathways in plants (reviewed by Gupta et al., 2011c). Briefly, these pathways can be classified into two groups according to nitrogen-containing precursors: the L-arginine-dependent pathway (oxidative pathway), and the NO2--dependent pathway (reductive pathway). NO2--dependent NO synthesis involves NR which reduces NO2- to NO both in vitro and in vivo in specific physiological contexts (Yamasaki and Sakihama, 2000); alternatively, formation of NO through the reduction of NO2- by the mitochondrial respiratory chain can also be observed, particularly in roots (Gupta et al., 2011a; Horchani et al., 2011). Finally, NO can be produced by an apoplastic non-enzymatic conversion of NO2- to NO at acidic pH, in the presence of reductants such as ascorbic acid (Bethke et al., 2004).
Several pathways involved in NO transformation and turnover and balancing the bioavailability of this molecule have been identified (Leitner et al., 2009). Firstly, NO can react with reduced glutathione to produce S-nitrosoglutathione (GSNO), a low-molecular-weight S-nitrosothiol (SNO) that is more stable than NO and considered to be a mobile reservoir of NO. The cellular level of GSNO is enzymatically regulated primarily by GSNO reductase (GSNOR), which catalyzes the reduction of GSNO to oxidized glutathione and ammonium. Importantly, Yun et al. (2016) recently reported that NO and GSNO have additive functions in plant immunity but also in plant development. NO and GSNO might have distinct or overlapping molecular targets, thus allowing differential control of key cellular processes belonging to both defense and development. Secondly, besides their O2 binding properties, hemoglobins (Hbs) can metabolize NO into NO3- and therefore are also considered as NO and NO2- concentration modulators (Gupta et al., 2011b). Finally, NO rapidly reacts with superoxide (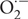 ) to form peroxynitrite (ONOO-), an oxidizing and nitrating RNS produced for instance in plant cells during immune responses (Vandelle and Delledonne, 2011). These molecules associated with NO turnover also play a role in the plant immune response. For instance, GSNO plays a key role in mediating the structural and functional changes of NPR1, a key transcription coactivator of plant immunity (Tada et al., 2008).
) to form peroxynitrite (ONOO-), an oxidizing and nitrating RNS produced for instance in plant cells during immune responses (Vandelle and Delledonne, 2011). These molecules associated with NO turnover also play a role in the plant immune response. For instance, GSNO plays a key role in mediating the structural and functional changes of NPR1, a key transcription coactivator of plant immunity (Tada et al., 2008).
Nitric oxide is partly produced through NR, dependent on its substrate/product NO3-/ NO2- as well as on L-arginine and polyamines. As a result, cross-regulation between NO signaling and N supply/metabolism is expected. Several lines of evidence show that NO production is likely to be affected by N supply. In a physio-pathological context, plant NO production is dependent on the form of N supply. Besson-Bard et al. (2008) and Gupta et al. (2013) showed that tobacco cell suspensions or leaves from plants grown on ammonium instead of nitrate as an N source emitted less NO when elicited by cryptogein or P. syringae. Thus these data highlight the determining role of the N source on the rate of NO synthesis. Modifications of the intracellular concentration of diverse intermediates of N metabolism such as amino acids or polyamines also result in the modulation of NO production. For instance, exogenously added polyamines induced rapid NO biosynthesis in A. thaliana (Tun et al., 2006). In the same manner, overexpression of the Asparagine synthetase 1 gene significantly enhanced the NO burst (Hwang et al., 2011). Finally, N nutrition could also impact important redox molecules associated with NO homeostasis. Nitrate deprivation led to altered levels of ROSs in A. thaliana and tobacco (Schachtman and Shin, 2007; Besson-Bard et al., 2008). Pathogen-induced expression of the nitrate transporter NRT2.6 was also correlated with ROS accumulation (Dechorgnat et al., 2012). Concentrations of antioxidant molecules such as glutathione (GSH) were altered (decreased in shoots and increased in roots) in A. thaliana and barley plants exposed to N deficiency (Kandlbinder et al., 2004; Kovacik et al., 2014).
Reciprocally, NO and derived RNS could participate in the regulation of N metabolism. NO can control physiological processes by modifying gene transcription. By analyzing available literature and databases, we identified interesting candidates likely to contribute to the crosstalk between N metabolism and NO among the numerous NO-regulated genes. Transcriptomic studies highlighted the up- or down-regulation of transcripts encoding N transporters (Ahlfors et al., 2009; Corti Monzon et al., 2014; Trevisan et al., 2015) or N assimilation/remobilization genes (Ferrarini et al., 2008; Ahlfors et al., 2009; Xu et al., 2013; Begara-Morales et al., 2014; Corti Monzon et al., 2014; Zeng et al., 2014; Trevisan et al., 2015) and amino acid metabolism-related genes (Ferrarini et al., 2008; Xu et al., 2013) upon modulation of NO homeostasis by treatment with NO donors, NO scavengers, or using mutants affected in NO homeostasis. Physiological studies identified NO as a regulator of N uptake in Chlamydomonas reinhardtii, possibly through the control of the expression of the nitrate or ammonium (AMT1.1 and AMT2.2) transporters. In A. thaliana, the expression of the high affinity nitrate transporter NRT2.1 was down-regulated by NO donors and in a GSNOR knock-out mutant, but the expression of the low-affinity nitrate transporter NRT1.1 remained unaltered (Frungillo et al., 2014), suggesting a switch from high- to low-affinity nitrate transport. By contrast, the expression of NRT2.1 was up-regulated through an NO-dependent process in A. thaliana roots exposed to cadmium (Besson-Bard et al., 2009). In addition to NO-mediated transcriptional regulation, many of NO biological functions arise as a direct consequence of chemical reactions between proteins and NO/RNS. Metal-nitrosylation, S-nitrosylation, and tyrosine nitration are notably emerging as main NO-dependent post-translational protein modifications (Astier and Lindermayr, 2012). Among the soluble proteins identified as S-nitrosylated or Tyr-nitrated, possible candidates contributing to the NO/N metabolism interplay are mainly involved in both N assimilation/remobilization and amino acid metabolism (Table 1). Post-translational inhibition of high-affinity ammonium and high-affinity NO3-/ NO2- transporters by NO was highlighted in C. reinhardtii (Sanz-Luque et al., 2013). However, whether the reversible effect of NO was linked to S-nitrosylation of the transporters or to an indirect effect of NO leading to other post-translational modifications of the transporters remains to be determined (Sanz-Luque et al., 2013). In that same study, NO also inhibited NR activity reversibly, but not NiR or GS activity. This post-translational effect of NO on N transporters and NR might mediate the fast inhibition of N uptake and assimilation by ammonium in C. reinhardtii. More recently, inhibition of NR activity by NO was proposed to be partly mediated by a truncated hemoglobin THB1 whose gene expression is highly induced by NO (Sanz-Luque et al., 2015).
Table 1.
Examples of S-nitrosylated or Tyr-nitrated proteins involved in N and amino acid metabolism.
| Functions | Post-translational modifications | Identified Proteins | Conditions | Reference |
|---|---|---|---|---|
| Amino acid metabolism | Tyrosine nitration | Methionine synthase | – | Lozano-Juste et al., 2011 |
| S-nitrosylation | Asparagine synthase 3 | Biotic stress | Maldonado-Alconada et al., 2011 | |
| Glutamate decarboxylase | Biotic stress | Maldonado-Alconada et al., 2011 | ||
| EPSP synthase | Biotic stress | Astier et al., 2012 | ||
| Acetohydroxy acid isomeroreductase (Val and Ile synthesis) | Biotic stress | Astier et al., 2012 | ||
| Aspartate aminotransferase | Biotic stress | Astier et al., 2012 | ||
| Cysteine synthase | Abiotic stress | Puyaubert et al., 2014 | ||
| Alanine glyoxylate aminotransferase | Abiotic stress | Puyaubert et al., 2014 | ||
| Glutamate glyoxylate aminotransferase | Abiotic stress | Puyaubert et al., 2014 | ||
| Nitrogen metabolism | Tyrosine nitration | Glutamine synthetase 2 | Biotic stress | Cecconi et al., 2009; Lozano-Juste et al., 2011 |
| Glutamine synthetase 1 | Rhizobium-legume symbiosis | Melo et al., 2011 | ||
| S-nitrosylation | Argininosuccinate synthase | Biotic stress | Maldonado-Alconada et al., 2011 | |
| Nitrite reductase | atgsnor1–3 | Hu et al., 2015 | ||
| Glutamate synthase | Abiotic stress | Puyaubert et al., 2014 | ||
| Glutamate dehydrogenase 1 | Biotic stress | Maldonado-Alconada et al., 2011 | ||
| Glutamate dehydrogenase 2 | Biotic stress | Maldonado-Alconada et al., 2011 | ||
In higher plants, NO produced by denitrification in the rhizosphere of forest soils impacts N uptake without affecting gene expression patterns of putative N transporters, suggesting post-translational modification of these transporters (Dong et al., 2015). NR is also highly regulated by complex transcriptional and post-translational mechanisms. Studies on different models using NO donors, NO synthase inhibitors, or the scavenger cPTIO indicate that NO modulates NR activity. Results are sometimes contradictory. NR activity in leaves was inhibited under high NO concentrations (Rosales et al., 2011, 2012; Frungillo et al., 2014), but was enhanced in cabbage (Du et al., 2008). Moreover, the inhibition or activation of NR by NO in tomato roots could depend on the NO3- concentration (Jin et al., 2009). The mechanisms explaining these effects of NO on NR are poorly understood. Regulation of NR by NO could occur through transcriptional downregulation of the NR NIA genes in Chlamydomonas and A. thaliana (de Montaigu et al., 2010). A direct interaction of NO with NR is possible, as S-nitrosylation of NR was evidenced in poplar exposed to cold stress (Cheng et al., 2015). Glutamine synthetase 2 is a second key enzyme of plant N metabolism involved in the synthesis of essentially nitrogenous compounds via Gln production. Interestingly, GS1 and GS2 were identified as molecular targets of NO (Table 1). GS activity was inhibited by Tyr nitration in root nodules of Medicago truncatula. This post-translational modification may mediate channeling of glutamate to boost plant antioxidant defenses (Melo et al., 2011) in response to NO. This interesting feature does not seem to be shared across the plant kingdom since GS activity was not affected by the NO donor DEA-NONOate in the alga Chlamydomonas (Sanz-Luque et al., 2013).
Role of NO/RNS in the Modulation of the Immune Response by N Nutrition: First Experimental Evidence
Altogether, these data indicate that N supply has an impact on plant immunity and NO/RNS signaling and lead us to wonder about the role of NO/RNS in the modulation of the immune response by N nutrition. In the present work, we used an in vitro pathosystem composed of the legume M. truncatula challenged with the soil-borne root pathogen Aphanomyces euteiches. This oomycete is considered as the most limiting factor for legume production. Resistance of M. truncatula roots includes protection of the central cylinder against pathogen invasion, associated with frequent pericycle cell divisions, lignin deposition, and accumulation of soluble phenolic compounds (Djébali et al., 2009). First investigations of the biochemical processes underlying the expression of this resistance showed modulation of H2O2 levels and of the activity of antioxidant enzymes (Djébali et al., 2009, 2011). Interestingly, in the M. truncatula A17 genotype, resistance against A. euteiches was significantly enhanced in response to NO3- starvation as compared to sufficient N conditions (Thalineau et al., unpublished). Based on the current literature, we hypothesized that NO could play a role in this N-induced modulation of M. truncatula defense responses against A. euteiches. We therefore first assessed whether changes in NO homeostasis could indeed affect M. truncatula resistance to A. euteiches. Secondly, we determined whether NO homeostasis could be modulated by N nutrition during the M. truncatula-A. euteiches interaction. We considered NO homeostasis as the maintenance of a functional NO concentration in a specific condition, through a balance between its biosynthesis (e.g., NR activity) and turnover pathways (e.g., interactions with GSH or O2⋅- to form GSNO or ONOO-, respectively).
Materials and Methods
Plant Growth and Inoculation by A. euteiches
We used the M. truncatula Jemalong-A17 genotype. M. truncatula seeds were scarified according to Djébali et al. (2009). After stratification overnight at 4°C, they were germinated in phytochambers with 16 h light under 350 μmol m-2 s-1 photons at 23°C/8 h night at 21°C. One day after germination, the seedlings were transferred to 12 cm × 12 cm square Petri dishes containing modified M medium (Bécard and Fortin, 1988). This modified medium was sugar-free, enriched in phosphate (1.3 mM final concentration), and contained either 3.2 mM nitrate (complete medium) or no nitrate (NØ medium). The Petri dishes were sealed with parafilm and the roots were protected from light with aluminum foil, and then placed vertically in the culture chamber (16 h light under 350 μmol m-2 s-1 photons at 23°C/8 h night at 21°C) for 7 days. The strain Aphanomyces euteiches Drechs ATCC 201684 was used to inoculate the seeds one day after germination. Zoospores were produced as described in Rey et al. (2013), and each root was inoculated with 500 zoospores.
Agrobacterium rhizogenes Root Transformation
The pENTR4 vector carrying the MtNR1 or the MtNR2 fragment (Horchani et al., 2011) was recombined with the pK7GWIWG2d vector using LR clonase II enzyme mix (Invitrogen, France) to create RNA interference expression vectors. The MtGSNOR gene (M. truncatula Gene code Medtr7g099040) (1,143 bp) was amplified using M. truncatula cDNA as a template and the specific primers GSNOR-F 5′-AAAAAGCAGGCTTCACCATGGCATCGTCGACTGAAGGT-3′ and GSNOR-R 5′- AGAAAGCTGGGTGTCAATGCAATGCAAGCACAC containing the corresponding attB recombination sites. The PCR product was recombined into the pDONR entry vector (Invitrogen) and checked by sequencing. The pDONR vector carrying the MtGSNOR gene was recombined with pK7WG2d plasmids1 to create the overexpression vector. The constructs pK7GWIGW2d-MtNR1-2/GFP (RNAi::MtNIA1/2) and pK7WG2d-MtGSNOR/GFP (35S::GSNOR) were introduced into A. rhizogenes strain Arqua1 (Quandt et al., 1993). M. truncatula plants were transformed with A. rhizogenes according to Boisson-Dernier et al. (2001). Control plants were transformed with A. rhizogenes containing the pK7GWIGW5D or the pK7WG2d empty vectors. Hairy roots were selected based on the fluorescent marker GFP 21 days after transformation.
RNA Extraction, Reverse Transcription, and Quantitative PCR on Transformed Roots
Total RNA was extracted from transformed roots using TRIzol® Reagent (Life Technologies) according to the manufacturer’s recommendations. To carry out the qPCR reaction, RNAs (0.5–1 μg) were reverse-transcribed in a final volume of 20 μL in the presence of RNasin (Promega, Charbonnières, France), and oligo(dT)15, with M-MLV reverse transcriptase (Promega, Charbonnières, France), as recommended by the manufacturer.
Quantitative PCR was performed on reverse-transcribed RNAs from four independent biological replicates per condition and from two independent plant cultures. Quantitative PCR reactions were performed in an ABI PRISM 7900 sequence detection system (Applied Biosystems®, Saint-Aubin, France), in a final volume of 15 μL containing Absolute SYBR green ROX Mix (Thermo Scientific, Surrey, UK), 0,3 μM of gene-specific primers, and 5 μL of cDNA template diluted 60-fold. The reference gene used for normalization was MtEF1α. Relative expression was expressed as 2-ΔCt test genes-reference gene. The primers used for the qPCR all displayed a high amplification efficiency (90–100%). They were the following:
MtGSNORforward 5′-GTGACTGGGCGTGTATGGAA-3′
MtGSNORreverse 5′-TGCAAGCACACAACGAAGAC-3′
MtNIA1forward 5′-TGTTCCACAGGCTTCTCCAGATACA-3′
MtNIA1reverse 5′-CATACAGCGTCGTACTCAGCGACA-3′
MtNIA2forward 5′GCAAACCGGACGGAGGATGA-3′
MtNIA2reverse 5′CCGTGATGAATCCCACACTATATTCC-3′
MtEF1αforward 5′-AAGCTAGGAGGTATTGACAAG-3′
MtEF1αreverse 5′-ACTGTGCAGTAGTACTTGGTG-3′
Inoculation of Transformed Root Cultures with A. euteiches
Roots were cultured on Shb10 medium (Boisson-Dernier et al., 2001) and transferred on modified Fahraeus medium enriched in ammonitrate (1 mM NH4NO3 final) one day before inoculation. Inoculation of the root cultures with A. euteiches strain ATCC 201684 was carried out by adding 10 mL of an A. euteiches zoospore suspension containing 80,000 zoospores.mL-1 in sterilized Volvic (Colditz et al., 2007) water. Zoospore production was initiated as described in Rey et al. (2013). Control root cultures were inoculated with 10 mL of sterile Volvic water. After 4 h of incubation in the dark, the zoospore solution was drained off the roots, and the Petri dishes were placed back into the growth room and left there for 7 days in the dark.
Assessing Infection Levels by Enzyme-Linked Immunosorbent Assay (ELISA)
Assessment of A. euteiches development in roots was performed by ELISA, using rabbit polyclonal serum raised against A. euteiches, and a mouse anti-rabbit IgG alkaline phosphatase conjugate as described by Slezack et al. (1999), on protein extracts from roots from pooled plants. Alkaline phosphatase activity was monitored by recording the increase in absorbance at 405 nm for 2–3 h, and was expressed as the slope of the resulting curve per mg of root fresh weight.
Hydrogen Peroxide Quantification
H2O2 concentration was measured using an Amplex Red®/peroxidase-coupled fluorescence assay adapted from Ashtamker et al. (2007). Roots were ground on ice and in the dark, in 1 mL of KRPG buffer (145 mM NaCl; 5.7 mM K2HPO4; 4.86 mM KCl; 0.54 mM CaCl2; 1.22 mM MgSO4; 5.5 mM glucose; pH 7.35) with 10 μM Amplex Red® and 0.2 U/mL of Horse Radish Peroxidase (HRP) per 100 mg of fresh weight. Catalase, an H2O2 scavenger, was used as a control. After 10 min of incubation at 4°C with catalase (1 unit/μL), 10 μM Amplex Red® and 0.2 U/mL of HRP were added to the samples. After centrifugation (10,000×g, 15 min, 4°C), 100 μL of supernatant were used to quantify resorufin (λex = 560 nm; λem = 584 nm) by spectrofluorimetry (Mithras, Berthold Technology). The relative fluorescence units were converted into μmol of H2O2 mg-1 root fresh weight on the basis of a standard curve established from known concentrations of H2O2.
Nitric Oxide and Peroxynitrite Quantification
ONOO- and NO concentrations were determined using A17 or transformed roots ground on ice and in the dark, with 1 mL of Tris-HCl (10 mM, pH 7.5), KCl (10 mM) buffer with 5 μM aminophenyl fluorescein (APF) or 10 μM 4,5-diaminofluorescein (DAF), respectively, per 100 mg of fresh weight. Epicatechin, an ONOO- scavenger, was used as a control. After 10 min of incubation at 4°C with epicatechin (1 mM), APF was added to the samples at a final concentration of 5 μM. cPTIO, an NO scavenger, was used as a control. After 10 min of incubation at 4°C with cPTIO (500 μM), DAF was added to the samples at a final concentration of 10 μM.
After centrifugation (10,000×g, 15 min, 4°C), 100 μL of supernatant were used to quantify ONOO- or NO (λex = 485 nm; λem = 535 nm) by spectrofluorimetry (Mithras, Berthold Technology).
S-nitrosothiol Quantification
S-nitrosothiol quantification was performed using the Saville–Griess assay (Gow et al., 2007). A17 roots or transformed roots were ground, on ice and in the dark, in extraction buffer (1 mL/100 mg of fresh weight, 0.1 M Tris-HCl, pH 7.5; 1 mM PMSF). After centrifugation (10,000×g, 15 min, 4°C), 100 μL of supernatant were incubated with 100 μL of buffer A (0.5 M HCl; 1% sulfanilamide) or 100 μL of buffer B (0.5 M HCl; 1% sulfanilamide; 0.2% HgCl2). After incubation (15 min at room temperature), 100 μL of Griess reagent[(0.5 M HCl; 0.02% N-(1-naphtyl)-ethylenediamine dihydrochloride] were added. After 15 min, SNOs were quantified by measuring absorbance at 540 nm. A standard curve was obtained using different concentrations of GSNO.
Nitrate Determination
Nitrate determination was performed according to Miranda et al. (2001), based on the reduction of nitrate to nitrite by vanadium and colorimetric detection at 540 nm of nitrite in the presence of sulfanilamide and N-1-naphthylethylenediamine. Approximately 100 mg of 7-day-old plant roots were collected, flash-frozen in liquid N2, and ground into powder. Three hundred micro liter of ultra-pure water were added to 20 mg of frozen sample, thoroughly vortexed, and incubated with occasional mixing for 15 min on ice. After centrifugation 15 min at 13,000×g and 4°C, the supernatant was recovered and used for nitrate determination.
Nitrate Reductase Activity Measurements
Transformed root samples were frozen in liquid nitrogen and ground using pestle and mortar. Extraction was performed in MOPS buffer (1 mL per 100 mg of fresh weight, 50 mM MOPS-KOH buffer, pH 7.6; 5 mM NaF; 1 μM Na2MoO4; 10 μM FAD; 1 μM leupeptin, 0.2 g/g FW polyvinylpolypyrrolidone; 2 mM β-mercaptoethanol; 5 mM EDTA). After centrifugation (20,000×g, 5 min, 4°C), the supernatant was used to measure NR activity. The reaction mixture consisted of 50 mM MOPS-KOH buffer, pH 7.6, containing 1 mM NaF, 10 mM KNO3, 0.17 mM NADH, and 5 mM EDTA. After incubation 15 min at 30°C, the reaction mixture was stopped by adding an equal volume of sulfanilamide (1% w/v in 3 N HCl) followed by N-naphtylethylenediamine dihydrochloride (0.02%, w/v), and the A540 was measured. A standard curve was obtained based on different concentrations of nitrite.
GSNOR Activity Measurements
To measure GSNOR activity, roots were ground in liquid nitrogen and proteins were extracted in 50 mM Tris-HCl buffer, pH 8, 0.5 mM EDTA, and 1 mg/mL of a protease inhibitor cocktail (1 mL of buffer per 100 mg of fresh weight). GSNOR activity was assayed from the rate of NADH oxidation by measuring the decrease in absorbance at 340 nm at 25°C using 25 μg of proteins in a total volume of 200 μL of extraction buffer containing 350 μM NADH with or without 350 μM GSNO. GSNO reductase activity was determined by subtracting NADH oxidation values in the absence of GSNO from values in the presence of GSNO. All samples were protected from light during the assay and tested for linearity. A standard curve was obtained using different concentrations of NADH.
Statistical Analyses
Statistical analyses were performed using one- or two-way analysis of variance (ANOVA) followed by Fisher’s test. Data were considered as significantly different when p < 0.05.
Results and Discussion
NO Homeostasis Participates in the M. truncatula Immune Response
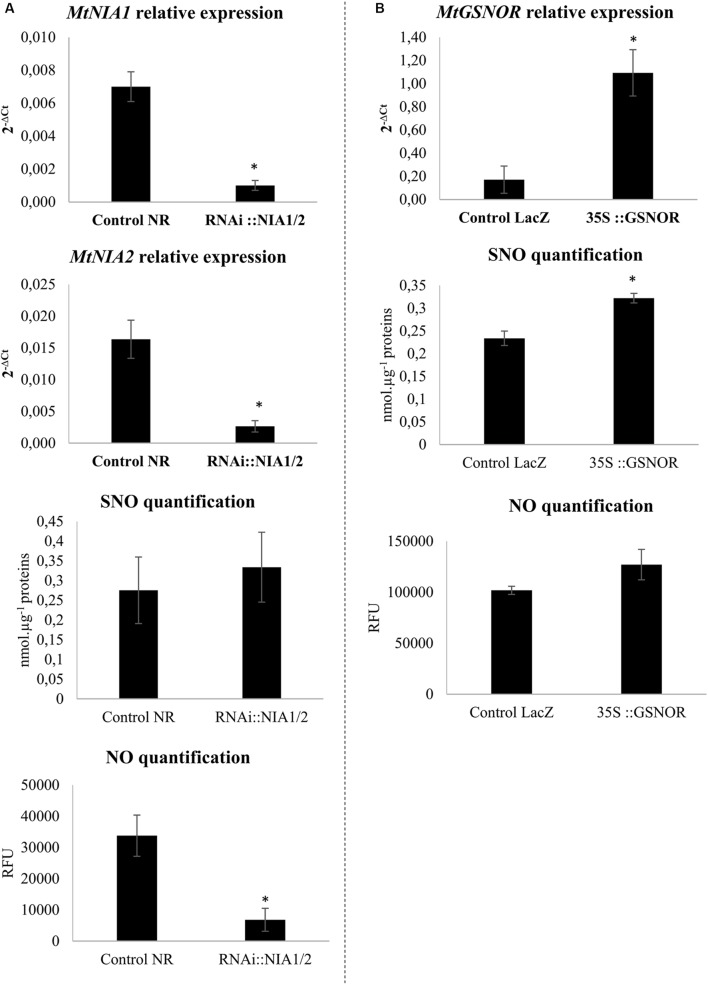
To investigate the putative role of NO homeostasis in the M. truncatula/A. euteiches interaction, roots were transformed to inactivate the NR-encoding MtNIA1/2 genes or to overexpress GSNOR-encoding genes. Quantification of gene transcripts in transformed roots using RT-qPCR confirmed that the two NIA genes were repressed (Figure 1A) while GSNOR was overexpressed (Figure 1B). To perform functional validation of the different constructs, we quantified NO and SNO levels in transformed roots. The two genetic manipulations modulated NO or SNO levels (Figure 1). SNO levels remained unchanged in RNAi::MtNIA1/2 roots as compared to the controls, whereas NO levels clearly decreased (Figure 1A). This was in accordance with the downregulation of NR, a major enzymatic source of NO. Conversely, NO levels in the 35S::GSNOR roots did not significantly change, but SNO significantly increased as compared to control roots (Figure 1B). This was surprising because in most previous experiments a negative correlation was described between SNO levels and GSNOR activity (Feechan et al., 2005; Rusterucci et al., 2007; Yun et al., 2011). However, it is interesting to note that in pea (a legume closely related to M. truncatula), higher SNO levels induced by wounding were correlated with higher GSNOR activity (Corpas et al., 2008).
FIGURE 1.
Transformed root validation. (A) Transcript levels of MtNIA1 and MtNIA2 in RNAi::NIA1/2-transformed roots were compared to control transformed roots (control NR). SNO quantification using the Saville–Griess assay and NO quantification using the fluorophore DAF (10 μM). Control NR and RNAi::NIA1/2-transformed roots extracts were pre-incubated or not with 500 μM cPTIO as an NO scavenger. (B) Transcript levels of MtGSNOR in 35S::GSNOR-transformed roots were compared to control transformed roots (control LacZ). SNO quantification using the Saville–Griess assay and NO quantification using the fluorophore DAF (10 μM). Control LacZ and 35S::GSNOR-transformed roots extracts were pre-incubated or not with 500 μM cPTIO as an NO scavenger. Error bars indicate standard errors (n = 4 for transcripts and NO levels; n = 8 for SNO levels), and ∗ indicates significant differences (p < 0.05).
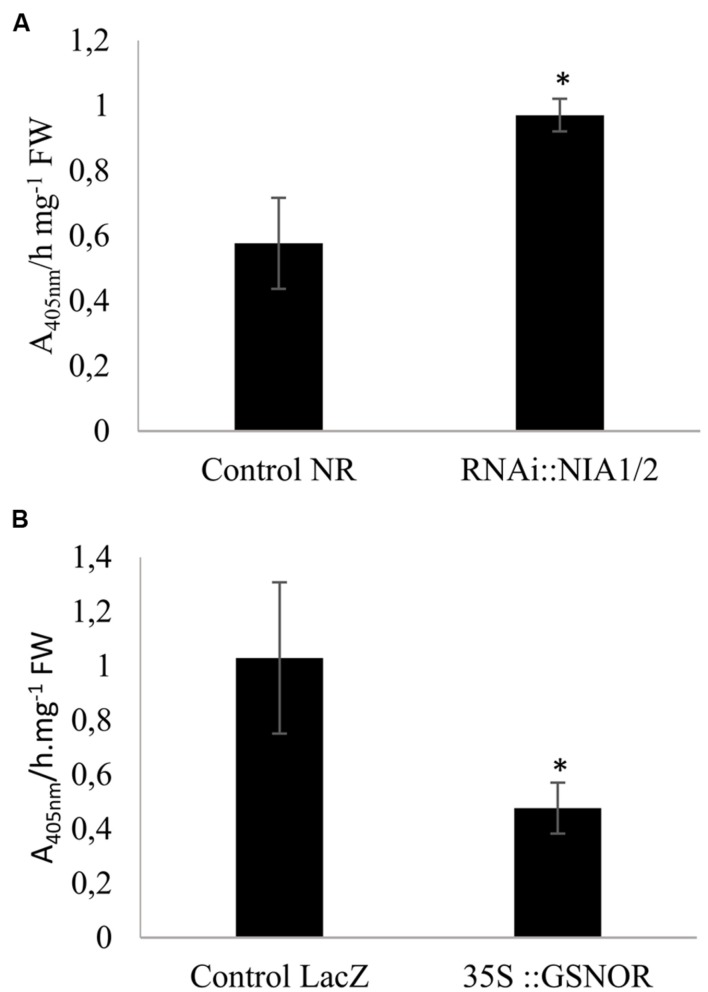
We studied the impact of these genetic transformations on the M. truncatula/A. euteiches interaction. ELISA tests using antibodies raised against A. euteiches (Slezack et al., 1999) were performed to quantify the presence of the pathogen in roots. In RNAi::MtNIA1/2 roots (Figure 2A), A. euteiches colonization was significantly greater than in control transformed roots (Control NR roots). These data reaffirm the role of the NR enzyme in the plant immune response. In A. thaliana, the NR-deficient double mutant (nia1 nia2) failed to exhibit a hypersensitive response and was hyper-susceptible to P. syringae (Modolo et al., 2006; Oliveira et al., 2009) and to the necrotrophic fungal pathogens Sclerotinia sclerotiorum or Botrytis cinerea (Perchepied et al., 2010; Rasul et al., 2012). Although these effects were attributed to the substantially reduced NO levels in this mutant, a side effect of N metabolism on plant defense cannot be excluded as NR stands at the crossroads between N metabolism and NO production.
FIGURE 2.
Quantification of Aphanomyces euteiches in extracts from inoculated transformed roots. RNAi::NIA1/2-transformed roots (A) and 35S::GSNOR-transformed roots (B) were extracted for ELISA tests. Roots were cultivated in vitro for 7 days on Fahraeus medium and then inoculated with A. euteiches. The background signal in non-inoculated roots was subtracted from the signal detected in inoculated roots. Error bars indicate standard errors (n = 4), and ∗ indicates significant differences (p < 0.05). Data from one representative experiment out of four independent experiments.
Our results using GSNOR-transformed roots showed that pathogen levels were lower in GSNOR-overexpressing roots (Figure 2B) than in control transformed roots (Control LacZ roots). GSNOR could therefore be considered as a positive regulator of M. truncatula resistance to A. euteiches. Previous works already investigated the physiological roles of GSNOR in plant-pathogen interactions, using transgenic A. thaliana plants (Feechan et al., 2005; Rusterucci et al., 2007; Yun et al., 2011). Results are sometimes contradictory, as modulation of AtGSNOR expression enhanced or decreased plant disease resistance depending on the pathosystem. GSNOR could play a significant role in plant immunity because GSNO is considered as a mobile reservoir of NO, is more stable than NO, and is a transnitrosylation agent of proteins. The contrasted results obtained in our study with NR and GSNOR constructs could be attributed to the specific roles of the corresponding proteins in NO homeostasis. NR is involved in NO synthesis, whereas the primary role of GSNOR is to regulate GSNO contents. The recent results from Yun et al. (2016) confirm that GSNO and NO may play distinct roles in plant immunity by acting on different molecular targets. In addition, GSNOR indirectly affects NO, GSH, ROS, and total intracellular nitrosothiol (SNO) levels, indicating that GSNOR might be more globally involved in the regulation of the cell redox state (Espunya et al., 2006; Yun et al., 2011).
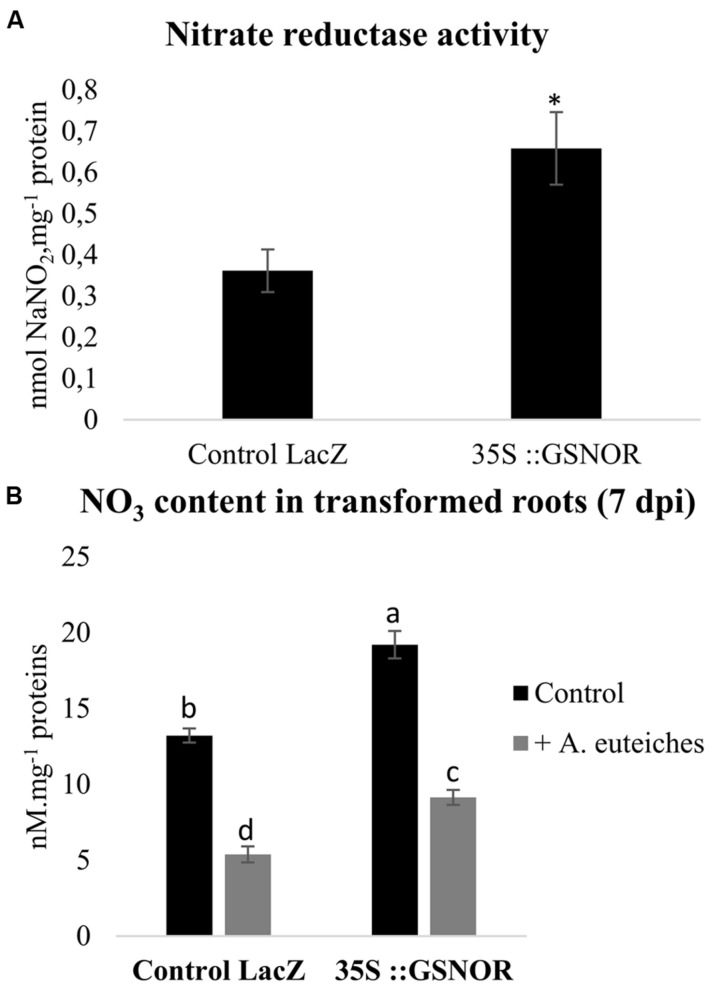
Nitric Oxide partly regulates N metabolism. Therefore we also investigated the effects of GSNOR overexpression on root NO3- contents and NR activity in transformed roots. GSNOR overexpression increased basal NO3- content and NR activity (Figures 3A,B). Modulation of N metabolism by GSNO and NO in A. thaliana has been described (Frungillo et al., 2014), and was explained by the effect of NO and GSNO on NR activity and on the expression of the AtNRT2.1 high-affinity NO3- transporter gene. Similarly to our data, that study shows that GSNOR overexpression is correlated with higher NR activity and NO3- content. Interestingly, we noted that pathogen colonization reduced NO3- concentrations in roots by approximately 65%, suggesting an effect of A. euteiches on nitrate transport and/or NO3- assimilation. Although we found a higher NO3- content in 35S::GSNOR-infected roots than in control infected roots, the amplitude of the pathogen-induced decrease in NO3- level was not impacted in 35S::GSNOR roots, suggesting that this process is independent of GSNO homeostasis. This reduced level of NO3- is unlikely to result from consumption of NO3- by the pathogen: data mining of the A. euteiches database revealed that no homologs of the NR, NIR, and NO3- transporter (NRT2) genes were detected in the genome of this pathogen2, confirming earlier observations that NO3- is unfavorable for A. euteiches development (Huber and Watson, 1974). Alternatively, we cannot exclude that the decreased NO3- content in infected roots could be due to nitrate leakage from the roots related to developing necrosis.
FIGURE 3.
Nitrate reductase (NR) activity and NO3- contents in transformed roots. (A) NR activity in control transformed roots (Control LacZ roots transformed with pK7GWG2D-GFP) and in transformed roots overexpressing GSNOR (35S::GSNOR). Transformed roots were cultivated in vitro on Shb10 medium. (B) NO3- concentrations in control transformed roots (Control LacZ) and GSNOR-overexpressing roots. Transformed roots were cultivated in vitro for 7 days on Fahraeus medium, and inoculated with A. euteiches. Error bars indicate standard errors (n = 4), and letters or ∗ indicate significant differences (p < 0.05). Data from one representative experiment out of three independent experiments for both NR activity and NO3- contents (n = 12).
Effect of N Nutrition on NO/ H2O2/ONOO- Accumulation and SNO Contents
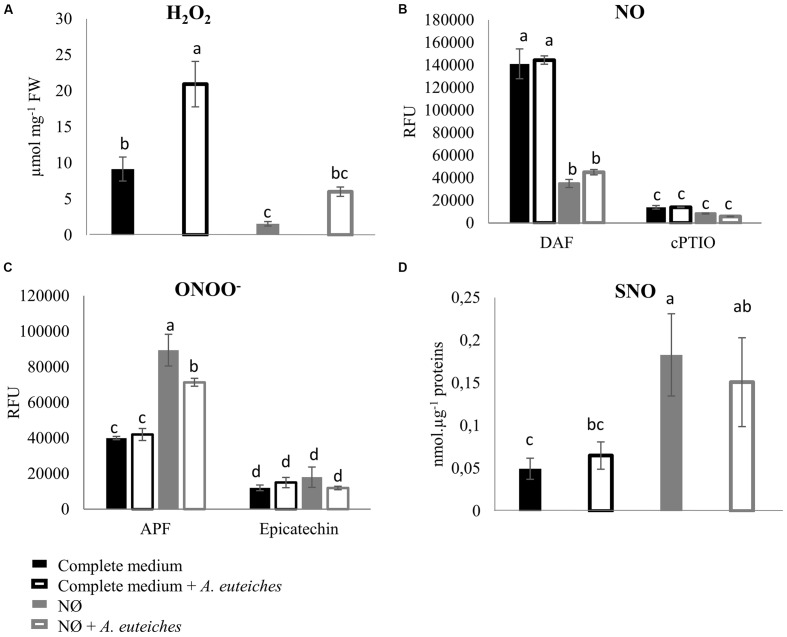
To analyze the role of N availability on NO, H2O2, and ONOO- accumulation, M. truncatula plants were cultivated in complete medium or NO3--deficient medium (NØ), and inoculated or not with A. euteiches. The NO scavenger cPTIO and the ONOO- scavenger epicatechin were used as controls to check the specificity of the fluorescence probes. We observed that NO3- deficiency caused a significant increase in ONOO- content on NØ medium (Figure 4C), whereas NO and H2O2 levels decreased (Figures 4A,B), highlighting a link between NO3- content and production of these reactive species. A clear effect of pathogen colonization was only evidenced for H2O2 contents (Figure 4A), and this increase was abolished on NØ. Surprisingly, although NO production is considered as a common response to pathogens, no increase in NO levels was detected in response to A. euteiches (Figure 4B). More generally, whereas NO, ROS, or ONOO- production has been widely described in response to pathogens, the literature does not give a clear picture of the cross-talks between these molecules. For instance, we observed a negative correlation between NO and ONOO- contents in response to NO3- deficiency, but in other models high NO levels are often correlated with high ONOO- levels (Abramowski et al., 2015; Kulik et al., 2015). These conflicting observations raise some questions. Are these discrepancies due to plant models or due to the difficulty in measuring and precisely localizing these molecules? Differences in the stability of these molecules or their specific scavenging by plants during pathogen attack could explain why we did not detect changes in ONOO- or NO levels in response to A. euteiches. Moreover, NO could also be used by the pathogen to activate its own metabolism, an important step in plant infection by fungi (Sedlářová et al., 2016).
FIGURE 4.
H2O2, NO, ONOO-, and SNO quantification in Medicago truncatula roots 7 dpi. M. truncatula was cultivated on complete medium or NØ medium, and roots were harvested 7 days after inoculation with A. euteiches and used to detect H2O2, NO, and ONOO- concentrations using fluorescent probes, and SNO concentrations using the Saville–Griess assay. (A) H2O2 quantification using 10 μM Amplex Red® fluorophore and 0.2 U/mL of peroxidase. Catalase (1 U/μL), used as an H2O2 scavenger, abolished Amplex Red® fluorescence. (B) NO quantification using the fluorophore DAF (10 μM). Root extracts were pre-incubated or not with 500 μM cPTIO as an NO scavenger. (C) ONOO- quantification using the fluorophore APF (5 μM). Root extracts were pre-incubated or not with 1 mM of the ONOO- scavenger epicatechin. (D) SNO quantification by the Saville–Griess assay. Error bars indicate standard errors (n = 4 for A–C; n = 14 for D), and letters indicate significant differences (p < 0.05). Data from one representative experiment out of three independent experiments for H2O2, NO, and ONOO-concentrations, and data corresponding to two independent experiments pooled together for SNO concentrations. RFU, relative fluorescence units.
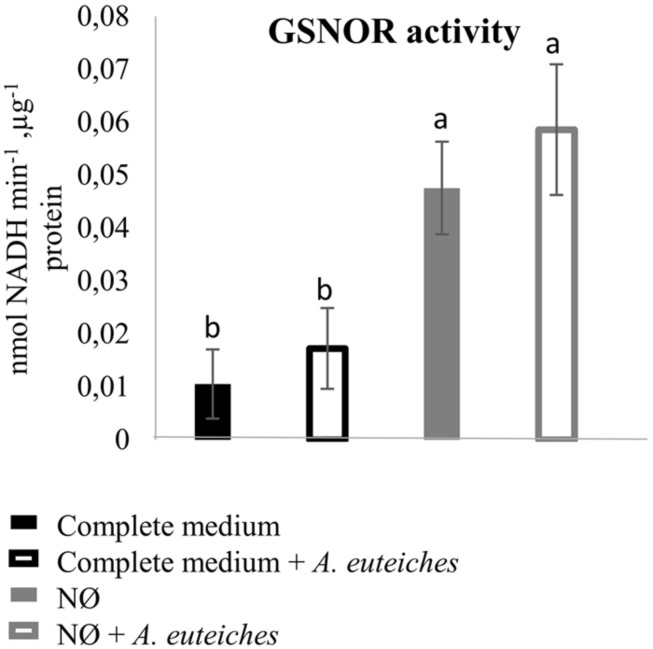
We also measured root SNO levels and GSNOR activity in the biological conditions of interest. Root SNO contents, determined using the Saville–Griess method, significantly increased on NØ medium as compared to the complete medium (Figure 4D). In response to A. euteiches, no significant change in SNO levels was highlighted (Figure 4D). Therefore, on NØ medium, the SNO content evolved in an opposite way to the NO content, similarly to the ONOO- content. This result is in accordance with results reported in Helianthus annuus (Chaki et al., 2011), and can be attributed to the fact that NO is the source for ONOO- and SNO. By contrast, a high NO content can be correlated with a high SNO content when plants are grown on culture medium containing NO3- (Abramowski et al., 2015; Pietrowska et al., 2015). Our data also suggest that NO3- nutrition impacts the overall balance between NO, ONOO-, and SNO. Regarding GSNOR, no changes in its activity was detected upon inoculation, in line with the absence of change in SNO levels in infected roots. In the roots of plants cultivated on NØ (Figure 5), higher GSNOR activity was correlated with higher SNO levels, confirming the positive correlation between GSNOR activity and SNO levels observed in 35S::GSNOR-transformed Medicago roots (Figure 1) and in pea, a closely related legume (Corpas et al., 2008). The positive or negative correlation between GSNOR activity and SNO levels or between NO and SNO levels depending on plant species and experimental conditions can be explained by several hypotheses. The SNO level is regulated through nitrosylation and denitrosylation; GSNOR, by controlling the level of GSNO, indirectly affects the level of S-nitrosylation. However, the TRX (thioredoxin)/NTR (NADPH-dependent TRX reductase) enzymatic system also controls S-nitrosylation (Kneeshaw et al., 2014). Interestingly, these activities were also identified in roots and activated by NO, leading to denitrosylation of specific proteins (Correa-Aragunde et al., 2015). Thus, these results, together with our study, illustrate the complex relationships between NO production/GSNOR activity and total SNO levels. Abiotic stresses also increase GSNOR activity (Kubienova et al., 2014), and this appears to be the case for M. truncatula plants under NO3- deficiency. Higher GSNOR activity in N-deprived roots (Figure 5) could lead to a physiological state inducing higher resistance to A. euteiches, as observed in the 35S::GSNOR-transformed roots (Figure 2B). This could partly explain the enhanced resistance to this oomycete on NØ medium despite the low levels of NO in the roots. Thus, altogether our data highlight the possible positive and non-redundant roles of NO (Figures 1A and 2A) and SNO (Figures 1B, 2B, and 4D) in mediating M. truncatula resistance to A. euteiches.
FIGURE 5.
GSNO reductase activity in M. truncatula roots 7 dpi. M. truncatula seedlings were inoculated or not with A. euteiches, and cultivated on complete medium or NØ medium for 7 days. Root extracts were used to measure GSNOR activity. Error bars indicate standard errors (n = 4), and letters indicate significant differences (p < 0.05). Data from one representative experiment out of four independent experiments.
Conclusion and New Hypotheses
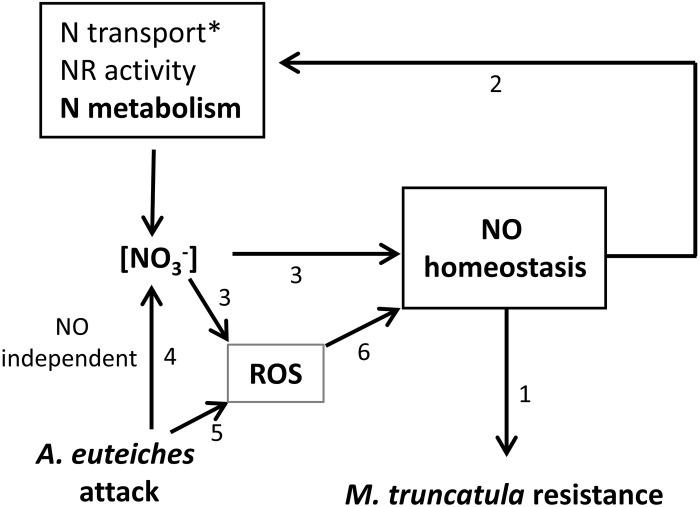
The results obtained in the present study are summarized in Figure 6. We have demonstrated, using transformed roots affected in genes involved in NO synthesis (NIA genes) and turnover (GSNOR gene), that deregulation of NO homeostasis has an effect on M. truncatula resistance against A. euteiches, as observed in other pathosystems (1). In addition, it appears that the modulation of NO homeostasis (through GSNOR overexpression) impacts NR activity and NO3-content, indicating possibly an effect of GSNOR (or GSNO) on basal NO3- transport/assimilation and confirming the results of Frungillo et al. (2014) (2). In return, NO3- availability in the medium can affect NO homeostasis by modulating ROS/RNS/NO contents and their balance (3). Finally, infection by A. euteiches decreases root NO3- content (4) and induces higher ROS levels (5). Altogether these results highlight the close interplay occurring between N nutrition and NO homeostasis as well as the involvement of NO in the modulation of plant resistance by N nutrition.
FIGURE 6.
Working model. Results from the present work indicate that RNAi::MtNIA1/2 and 35S::GSNOR transformed roots are, respectively, more susceptible and more resistant to A. euteiches (1). NR activity and NO3- content were impacted by GSNOR overexpression, indicating a possible effect of GSNOR on basal NO3- transport/assimilation (2). NO3- availability in the medium causes quantitative modulation of ROS/RNS/NO content and affects their balance (3). Infection by A. euteiches decreases root NO3- content (4) and induces higher ROS levels (5). According to the literature superoxyde (O2⋅-), by reacting with NO to form peroxynitrite, can influence the concentration of NO available for signaling (6). ∗: GSNO was shown to regulate NO3- uptake through transcriptional regulation of NRT2.1 (Frungillo et al., 2014).
Future work should take into account the role of N availability on NO-mediated plant molecular responses. Thus, the study of the specific role of GSNO in this process through the identification S-nitrosylated/denitrosylated proteins under different N availability conditions and N sources seems promising. A focus will be made on proteins involved in the plant immune response (1), but also on the feedback regulation of N metabolism by NO because NO could control NO3- availability and therefore plant resistance (2) (Figure 6). Investigations using foliar pathogens and other plant models will lead to a possible generalization of this phenomenon. More generally, plant N use efficiency can be affected by NO since NO controls not only N metabolism but also plant root growth and architecture changes in response to NO3- (Manoli et al., 2014; Sun et al., 2015). Recent data show that plant N use efficiency and N-induced susceptibility to pathogens may be linked (Ballini et al., 2013). Consequently future studies should also focus on candidate proteins involved in root development. Finally, experiments conducted with plant genotypes differing in their resistance levels will permit to study the quantitative effect of NO/ROS production on plant defense.
Author Contributions
ET, H-NT, ABo, and SJ conceived and designed the research; ET, H-NT, CF, ABo, and ABe carried out the experiments and analysis/interpretation of data; ET, H-NT, DW, and SJ wrote the manuscript. All authors contributed to the discussion and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank N. Pauly for practical advice on M. truncatula root transformation, E. Gaulin, A. Nars, and C. Jacquet for providing the A. euteiches strain Drechs. ATCC 201684, and advice on the pathosystem, and E. Dumas-Gaudot for gift of the polyclonal antibody directed against A. euteiches.
Funding. ET was funded by a Ph.D. contract (2013–2032) from the Ministère de l’Enseignement Supérieur et de la Recherche. This work was supported by a grant from AgroSup Dijon and the Conseil Régional de Bourgogne (PARI8).
References
- Abramowski D., Arasimowicz-Jelonek M., Izbiańska K., Billert H., Floryszak-Wieczorek J. (2015). Nitric oxide modulates redox-mediated defense in potato challenged with Phytophthora infestans. Eur. J. Plant Pathol. 143 237–260. 10.1007/s10658-015-0677-9 [DOI] [Google Scholar]
- Ahlfors R., Brosche M., Kollist H., Kangasjarvi J. (2009). Nitric oxide modulates ozone-induced cell death, hormone biosynthesis and gene expression in Arabidopsis thaliana. Plant J. 58 1–12. 10.1111/j.1365-313X.2008.03756.x [DOI] [PubMed] [Google Scholar]
- Ashtamker C., Kiss V., Sagi M., Davydov O., Fluhr R. (2007). Diverse subcellular locations of cryptogein-induced reactive oxygen species production in tobacco bright Yellow-2 cells. Plant Physiol. 143 1817–1826. 10.1104/pp.106.090902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astier J., Besson-Bard A., Lamotte O., Bertoldo J., Bourque S., Terenzi H., et al. (2012). Nitric oxide inhibits the ATPase activity of the chaperone-like AAA+ ATPase CDC48, a target for S-nitrosylation in cryptogein signalling in tobacco cells. Biochem. J. 447 249–260. 10.1042/BJ20120257 [DOI] [PubMed] [Google Scholar]
- Astier J., Lindermayr C. (2012). Nitric oxide-dependent posttranslational modification in plants: an update. Int. J. Mol. Sci. 13 15193–15208. 10.3390/ijms131115193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballini E., Nguyen T. T., Morel J. B. (2013). Diversity and genetics of nitrogen-induced susceptibility to the blast fungus in rice and wheat. Rice 6 32 10.1186/1939-8433-6-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bécard G., Fortin J. A. (1988). Early events of vesicular-arbuscular mycorrhiza formation on Ri T-DNA transformed roots. New Phytol. 108 211–218. 10.1111/j.1469-8137.1988.tb03698.x [DOI] [PubMed] [Google Scholar]
- Begara-Morales J. C., Sanchez-Calvo B., Luque F., Leyva-Perez M. O., Leterrier M., Corpas F. J., et al. (2014). Differential transcriptomic analysis by RNA-seq of GSNO-responsive genes between Arabidopsis roots and leaves. Plant Cell Physiol. 55 1080–1095. 10.1093/pcp/pcu044 [DOI] [PubMed] [Google Scholar]
- Besson-Bard A., Courtois C., Gauthier A., Dahan J., Dobrowolska G., Jeandroz S., et al. (2008). Nitric oxide in plants: production and cross-talk with Ca2+ signaling. Mol. Plant 1 218–228. 10.1093/mp/ssm016 [DOI] [PubMed] [Google Scholar]
- Besson-Bard A., Gravot A., Richaud P., Auroy P., Duc C., Gaymard F., et al. (2009). Nitric oxide contributes to cadmium toxicity in Arabidopsis by promoting cadmium accumulation in roots and by up-regulating genes related to iron uptake. Plant Physiol. 149 1302–1315. 10.1104/pp.108.133348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke P. C., Badger M. R., Jones R. L. (2004). Apoplastic synthesis of nitric oxide by plant tissues. Plant Cell 16 332–341. 10.1105/tpc.017822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A., Chabaud M., Garcia F., Becard G., Rosenberg C., Barker D. G. (2001). Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant-Microbe Interact. 14 695–700. 10.1094/MPMI.2001.14.6.695 [DOI] [PubMed] [Google Scholar]
- Camanes G., Pastor V., Cerezo M., Garcia-Andrade J., Vicedo B., Garcia-Agustin P., et al. (2012). A deletion in NRT2.1 attenuates Pseudomonas syringae-induced hormonal perturbation, resulting in primed plant defenses. Plant Physiol. 158 1054–1066. 10.1104/pp.111.184424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi D., Orzetti S., Vandelle E., Rinalducci S., Zolla L., Delledonne M. (2009). Protein nitration during defense response in Arabidopsis thaliana. Electrophoresis 30 2460–2468. 10.1002/elps.200800826 [DOI] [PubMed] [Google Scholar]
- Chaki M., Valderrama R., Fernandez-Ocana A. M., Carreras A., Gomez-Rodriguez M. V., Pedrajas J. R., et al. (2011). Mechanical wounding induces a nitrosative stress by down-regulation of GSNO reductase and an increase in S-nitrosothiols in sunflower (Helianthus annuus) seedlings. J. Exp. Bot. 62 1803–1813. 10.1093/jxb/erq358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T., Chen J., Ef A. A., Wang P., Wang G., Hu X., et al. (2015). Quantitative proteomics analysis reveals that S-nitrosoglutathione reductase (GSNOR) and nitric oxide signaling enhance poplar defense against chilling stress. Planta 242 1361–1390. 10.1007/s00425-015-2374-5 [DOI] [PubMed] [Google Scholar]
- Colditz F., Niehaus K., Krajinski F. (2007). Silencing of PR-10-like proteins in Medicago truncatula results in an antagonistic induction of other PR proteins and in an increased tolerance upon infection with the oomycete Aphanomyces euteiches. Planta 226 57–71. 10.1007/s00425-006-0466-y [DOI] [PubMed] [Google Scholar]
- Corpas F. J., Chaki M., Fernandez-Ocana A., Valderrama R., Palma J. M., Carreras A., et al. (2008). Metabolism of reactive nitrogen species in pea plants under abiotic stress conditions. Plant Cell Physiol. 49 1711–1722. 10.1093/pcp/pcn144 [DOI] [PubMed] [Google Scholar]
- Correa-Aragunde N., Cejudo F. J., Lamattina L. (2015). Nitric oxide is required for the auxin-induced activation of NADPH-dependent thioredoxin reductase and protein denitrosylation during root growth responses in Arabidopsis. Ann. Bot. 116 695–702. 10.1093/aob/mcv116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti Monzon G., Pinedo M., Di Rienzo J., Novo-Uzal E., Pomar F., Lamattina L., et al. (2014). Nitric oxide is required for determining root architecture and lignin composition in sunflower. Supporting evidence from microarray analyses. Nitric Oxide 39 20–28. 10.1016/j.niox.2014.04.004 [DOI] [PubMed] [Google Scholar]
- de Montaigu A., Sanz-Luque E., Galvan A., Fernandez E. (2010). A soluble guanylate cyclase mediates negative signaling by ammonium on expression of nitrate reductase in Chlamydomonas. Plant Cell 22 1532–1548. 10.1105/tpc.108.062380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechorgnat J., Patrit O., Krapp A., Fagard M., Daniel-Vedele F. (2012). Characterization of the Nrt2.6 gene in Arabidopsis thaliana: a link with plant response to biotic and abiotic stress. PLoS ONE 7:e42491 10.1371/journal.pone.0042491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Develey-Riviere M. P., Galiana E. (2007). Resistance to pathogens and host developmental stage: a multifaceted relationship within the plant kingdom. New Phytol. 175 405–416. 10.1111/j.1469-8137.2007.02130.x [DOI] [PubMed] [Google Scholar]
- Djébali N., Jauneau A., Ameline-Torregrosa C., Chardon F., Jaulneau V., Mathe C., et al. (2009). Partial resistance of Medicago truncatula to Aphanomyces euteiches is associated with protection of the root stele and is controlled by a major QTL rich in proteasome-related genes. Mol. Plant-Microbe Interact. 22 1043–1055. 10.1094/MPMI-22-9-1043 [DOI] [PubMed] [Google Scholar]
- Djébali N., Mhadhbi H., Lafitte C., Dumas B., Esquerré-Tugayé M.-T., Aouani M. E., et al. (2011). Hydrogen peroxide scavenging mechanisms are components of Medicago truncatula partial resistance to Aphanomyces euteiches. Eur. J. Plant Pathol. 131 559–571. 10.1007/s10658-011-9831-1 [DOI] [Google Scholar]
- Dong F., Simon J., Rienks M., Lindermayr C., Rennenberg H. (2015). Effects of rhizopheric nitric oxide (NO) on N uptake in Fagus sylvatica seedlings depend on soil CO2 concentration, soil N availability and N source. Tree Physiol. 35 910–920. 10.1093/treephys/tpv051 [DOI] [PubMed] [Google Scholar]
- Dordas C. (2008). Role of nutrients in controlling plant diseases in sustainable agriculture, A review. Agron. Sustain. Dev. 28 33–46. 10.1051/agro:2007051 [DOI] [Google Scholar]
- Du S., Zhang Y., Lin X., Wang Y., Tang C. (2008). Regulation of nitrate reductase by nitric oxide in Chinese cabbage pakchoi (Brassica chinensis L.). Plant Cell Environ. 31 195–204. 10.1111/j.1365-3040.2007.01750.x [DOI] [PubMed] [Google Scholar]
- Espunya M. C., Diaz M., Moreno-Romero J., Martinez M. C. (2006). Modification of intracellular levels of glutathione-dependent formaldehyde dehydrogenase alters glutathione homeostasis and root development. Plant Cell Environ. 29 1002–1011. 10.1111/j.1365-3040.2006.01497.x [DOI] [PubMed] [Google Scholar]
- Fagard M., Launay A., Clement G., Courtial J., Dellagi A., Farjad M., et al. (2014). Nitrogen metabolism meets phytopathology. J. Exp. Bot. 65 5643–5656. 10.1093/jxb/eru323 [DOI] [PubMed] [Google Scholar]
- Feechan A., Kwon E., Yun B.-W., Wang Y., Pallas J. A., Loake G. J. (2005). A central role for S-nitrosothiols in plant disease resistance. Proc. Natl. Acad. Sci. U.S.A. 102 8054–8059. 10.1073/pnas.0501456102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrarini A., De Stefano M., Baudouin E., Pucciariello C., Polverari A., Puppo A., et al. (2008). Expression of Medicago truncatula genes responsive to nitric oxide in pathogenic and symbiotic conditions. Mol. Plant-Microbe Interact. 21 781–790. 10.1094/mpmi-21-6-0781 [DOI] [PubMed] [Google Scholar]
- Forde B. G., Roberts M. R. (2014). Glutamate receptor-like channels in plants: a role as amino acid sensors in plant defence? F1000Prime Rep. 6:37 10.12703/p6-37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frungillo L., Skelly M. J., Loake G. J., Spoel S. H., Salgado I. (2014). S-nitrosothiols regulate nitric oxide production and storage in plants through the nitrogen assimilation pathway. Nat. Commun. 5:5401 10.1038/ncomms6401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gow A., Doctor A., Mannick J., Gaston B. (2007). S-Nitrosothiol measurements in biological systems. J. Chromatogr. B 851 140–151. 10.1016/j.jchromb.2007.01.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K. J., Brotman Y., Segu S., Zeier T., Zeier J., Persijn S. T., et al. (2013). The form of nitrogen nutrition affects resistance against Pseudomonas syringae pv. phaseolicola in tobacco. J. Exp. Bot. 64 553–568. 10.1093/jxb/ers348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta K. J., Fernie A. R., Kaiser W. M., Van Dongen J. T. (2011a). On the origins of nitric oxide. Trends Plant Sci. 16 160–168. 10.1016/j.tplants.2010.11.007 [DOI] [PubMed] [Google Scholar]
- Gupta K. J., Hebelstrup K. H., Mur L. A., Igamberdiev A. U. (2011b). Plant hemoglobins: important players at the crossroads between oxygen and nitric oxide. FEBS Lett. 585 3843–3849. 10.1016/j.febslet.2011.10.036 [DOI] [PubMed] [Google Scholar]
- Gupta K. J., Igamberdiev A. U., Manjunatha G., Segu S., Moran J. F., Neelawarne B., et al. (2011c). The emerging roles of nitric oxide (NO) in plant mitochondria. Plant Sci. 181 520–526. 10.1016/j.plantsci.2011.03.018 [DOI] [PubMed] [Google Scholar]
- Horchani F., Prevot M., Boscari A., Evangelisti E., Meilhoc E., Bruand C., et al. (2011). Both plant and bacterial nitrate reductases contribute to nitric oxide production in Medicago truncatula nitrogen-fixing nodules. Plant Physiol. 155 1023–1036. 10.1104/pp.110.166140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Huang X., Chen L., Sun X., Lu C., Zhang L., et al. (2015). Site-specific nitrosoproteomic identification of endogenously S-nitrosylated proteins in Arabidopsis. Plant Physiol. 167 1731–1746. 10.1104/pp.15.00026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber D. M., Watson R. D. (1974). Nitrogen form and plant disease. Annu. Rev. Phytopathol. 12 139–165. 10.1146/annurev.py.12.090174.001035 [DOI] [PubMed] [Google Scholar]
- Hwang I. S., An S. H., Hwang B. K. (2011). Pepper asparagine synthetase 1 (CaAS1) is required for plant nitrogen assimilation and defense responses to microbial pathogens. Plant J. 67 749–762. 10.1111/j.1365-313X.2011.04622.x [DOI] [PubMed] [Google Scholar]
- Jin C. W., Du S. T., Zhang Y. S., Lin X. Y., Tang C. X. (2009). Differential regulatory role of nitric oxide in mediating nitrate reductase activity in roots of tomato (Solanum lycocarpum). Ann. Bot. 104 9–17. 10.1093/aob/mcp087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandlbinder A., Finkemeier I., Wormuth D., Hanitzsch M., Dietz K. J. (2004). The antioxidant status of photosynthesizing leaves under nutrient deficiency: redox regulation, gene expression and antioxidant activity in Arabidopsis thaliana. Physiol. Plant. 120 63–73. 10.1111/j.0031-9317.2004.0272.x [DOI] [PubMed] [Google Scholar]
- Kneeshaw S., Gelineau S., Tada Y., Loake G. J., Spoel S. H. (2014). Selective protein denitrosylation activity of Thioredoxin-h5 modulates plant immunity. Mol. Cell. 56 153–162. 10.1016/j.molcel.2014.08.003 [DOI] [PubMed] [Google Scholar]
- Kovacik J., Klejdus B., Babula P., Jarosova M. (2014). Variation of antioxidants and secondary metabolites in nitrogen-deficient barley plants. J. Plant Physiol. 171 260–268. 10.1016/j.jplph.2013.08.004 [DOI] [PubMed] [Google Scholar]
- Krapp A. (2015). Plant nitrogen assimilation and its regulation: a complex puzzle with missing pieces. Curr. Opin. Plant Biol. 25 115–122. 10.1016/j.pbi.2015.05.010 [DOI] [PubMed] [Google Scholar]
- Kubienova L., Ticha T., Jahnova J., Luhova L., Mieslerova B., Petrivalsky M. (2014). Effect of abiotic stress stimuli on S-nitrosoglutathione reductase in plants. Planta 239 139–146. 10.1007/s00425-013-1970-5 [DOI] [PubMed] [Google Scholar]
- Kulik A., Noirot E., Grandperret V., Bourque S., Fromentin J., Salloignon P., et al. (2015). Interplays between nitric oxide and reactive oxygen species in cryptogein signalling. Plant Cell Environ. 38 331–348. 10.1111/pce.12295 [DOI] [PubMed] [Google Scholar]
- Leitner M., Vandelle E., Gaupels F., Bellin D., Delledonne M. (2009). NO signals in the haze: nitric oxide signalling in plant defence. Curr. Opin. Plant Biol. 12 451–458. 10.1016/j.pbi.2009.05.012 [DOI] [PubMed] [Google Scholar]
- Liu G., Ji Y., Bhuiyan N. H., Pilot G., Selvaraj G., Zou J., et al. (2010). Amino acid homeostasis modulates salicylic acid-associated redox status and defense responses in Arabidopsis. Plant Cell 22 3845–3863. 10.1105/tpc.110.079392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Juste J., Colom-Moreno R., León J. (2011). In vivo protein tyrosine nitration in Arabidopsis thaliana. J. Exp. Bot. 62 3501–3517. 10.1093/jxb/err042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna E., Van Hulten M., Zhang Y., Berkowitz O., Lopez A., Petriacq P., et al. (2014). Plant perception of beta-aminobutyric acid is mediated by an aspartyl-tRNA synthetase. Nat. Chem. Biol. 10 450–456. 10.1038/nchembio.1520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado-Alconada A., Echevarría-Zomeńo S., Lindermayr C., Redondo-López I., Durner J., Jorrín-Novo J. (2011). Proteomic analysis of Arabidopsis protein S-nitrosylation in response to inoculation with Pseudomonas syringae. Acta Physiol. Plant 33 1493–1514. 10.1007/s11738-010-0688-2 [DOI] [Google Scholar]
- Manoli A., Begheldo M., Genre A., Lanfranco L., Trevisan S., Quaggiotti S. (2014). NO homeostasis is a key regulator of early nitrate perception and root elongation in maize. J. Exp. Bot. 65 185–200. 10.1093/jxb/ert358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo P. M., Silva L. S., Ribeiro I., Seabra A. R., Carvalho H. G. (2011). Glutamine synthetase is a molecular target of nitric oxide in root nodules of Medicago truncatula and is regulated by tyrosine nitration. Plant Physiol. 157 1505–1517. 10.1104/pp.111.186056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda K. M., Espey M. G., Wink D. A. (2001). A rapid, simple spectrophotometric method for simultaneous detection of nitrate and nitrite. Nitric Oxide 5 62–71. 10.1006/niox.2000.0319 [DOI] [PubMed] [Google Scholar]
- Modolo L. V., Augusto O., Almeida I. M. G., Pinto-Maglio C. A. F., Oliveira H. C., Seligman K., et al. (2006). Decreased arginine and nitrite levels in nitrate reductase-deficient Arabidopsis thaliana plants impair nitric oxide synthesis and the hypersensitive response to Pseudomonas syringae. Plant Sci. 171 34–40. 10.1016/j.plantsci.2006.02.010 [DOI] [Google Scholar]
- Oliveira H. C., Justino G. C., Sodek L., Salgado I. (2009). Amino acid recovery does not prevent susceptibility to Pseudomonas syringae in nitrate reductase double-deficient Arabidopsis thaliana plants. Plant Sci. 176 105–111. 10.1016/j.plantsci.2008.09.017 [DOI] [Google Scholar]
- Perchepied L., Balague C., Riou C., Claudel-Renard C., Riviere N., Grezes-Besset B., et al. (2010). Nitric oxide participates in the complex interplay of defense-related signaling pathways controlling disease resistance to Sclerotinia sclerotiorum in Arabidopsis thaliana. Mol. Plant-Microbe Interact. 23 846–860. 10.1094/mpmi-23-7-0846 [DOI] [PubMed] [Google Scholar]
- Pietrowska E., Różalska S., Kaźmierczak A., Nawrocka J., Małolepsza U. (2015). Reactive oxygen and nitrogen (ROS and RNS) species generation and cell death in tomato suspension cultures—Botrytis cinerea interaction. Protoplasma 252 307–319. 10.1007/s00709-014-0680-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puyaubert J., Fares A., Reze N., Peltier J. B., Baudouin E. (2014). Identification of endogenously S-nitrosylated proteins in Arabidopsis plantlets: effect of cold stress on cysteine nitrosylation level. Plant Sci. 21 150–156. 10.1016/j.plantsci.2013.10.014 [DOI] [PubMed] [Google Scholar]
- Quandt H. J., Puhler A., Broer I. (1993). Transgenic root-nodules of Vicia-hirsuta - a fast and efficient system for the study of gene-expression in indeterminate-type nodules. Mol. Plant-Microbe Interact. 6 699–706. 10.1094/mpmi-6-699 [DOI] [Google Scholar]
- Rasul S., Dubreuil-Maurizi C., Lamotte O., Koen E., Poinssot B., Alcaraz G., et al. (2012). Nitric oxide production mediates oligogalacturonide-triggered immunity and resistance to Botrytis cinerea in Arabidopsis thaliana. Plant Cell Environ. 35 1483–1499. 10.1111/j.1365-3040.2012.02505.x [DOI] [PubMed] [Google Scholar]
- Rey T., Nars A., Bonhomme M., Bottin A., Huguet S., Balzergue S., et al. (2013). NFP, a LysM protein controlling Nod factor perception, also intervenes in Medicago truncatula resistance to pathogens. New Phytol. 198 875–886. 10.1111/nph.12198 [DOI] [PubMed] [Google Scholar]
- Rosales E. P., Iannone M. F., Groppa M. D., Benavides M. P. (2011). Nitric oxide inhibits nitrate reductase activity in wheat leaves. Plant Physiol. Biochem. 49 124–130. 10.1016/j.plaphy.2010.10.009 [DOI] [PubMed] [Google Scholar]
- Rosales E. P., Iannone M. F., Groppa M. D., Benavides M. P. (2012). Polyamines modulate nitrate reductase activity in wheat leaves: involvement of nitric oxide. Amino Acids 42 857–865. 10.1007/s00726-011-1001-4 [DOI] [PubMed] [Google Scholar]
- Rusterucci C., Espunya M. C., Diaz M., Chabannes M., Martinez M. C. (2007). S-nitrosoglutathione reductase affords protection against pathogens in Arabidopsis, both locally and systemically. Plant Physiol. 143 1282–1292. 10.1104/pp.106.091686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Luque E., Ocana-Calahorro F., Galvan A., Fernandez E. (2015). THB1 regulates nitrate reductase activity and THB1 and THB2 transcription differentially respond to NO and the nitrate/ammonium balance in Chlamydomonas. Plant Signal. Behav. 10:e1042638 10.1080/15592324.2015.1042638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Luque E., Ocana-Calahorro F., Llamas A., Galvan A., Fernandez E. (2013). Nitric oxide controls nitrate and ammonium assimilation in Chlamydomonas reinhardtii. J. Exp. Bot. 64 3373–3383. 10.1093/jxb/ert175 [DOI] [PubMed] [Google Scholar]
- Schachtman D. P., Shin R. (2007). Nutrient sensing and signaling: NPKS. Annu. Rev. Plant Biol. 58 47–69. 10.1146/annurev.arplant.58.032806.103750 [DOI] [PubMed] [Google Scholar]
- Sedlářová M., Kubienová L., Drábková Trojanová Z., Luhová L., Lebeda A., Petřivalský M. (2016). “Chapter Thirteen - the role of nitric oxide in development and pathogenesis of biotrophic phytopathogens – downy and powdery mildews,” in Advances Botnical Research, ed. David W. (Cambridge, MA: Academic Press; ), 263–283. [Google Scholar]
- Slezack S., Dumas-Gaudot E., Rosendahl S., Kjøller R., Paynot M., Negrel J., et al. (1999). Endoproteolytic activities in pea roots inoculated with the arbuscular mycorrhizal fungus Glomus mosseae and/or Aphanomyces euteiches in relation to bioprotection. New Phytol. 142 517–529. 10.1046/j.1469-8137.1999.00421.x [DOI] [Google Scholar]
- Snoeijers S., Pérez-García A., Joosten M. A. J., De Wit P. G. M. (2000). The effect of nitrogen on disease dvelopment and gene expression in bacterial and fungal pathogens. Eur. J. Plant Pathol. 106 493–506. 10.1023/a:1008720704105 [DOI] [Google Scholar]
- Sun H., Li J., Song W., Tao J., Huang S., Chen S., et al. (2015). Nitric oxide generated by nitrate reductase increases nitrogen uptake capacity by inducing lateral root formation and inorganic nitrogen uptake under partial nitrate nutrition in rice. J. Exp. Bot. 66 2449–2459. 10.1093/jxb/erv030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y., Spoel S. H., Pajerowska-Mukhtar K., Mou Z., Song J., Wang C., et al. (2008). Plant immunity requires conformational changes [corrected] of NPR1 via S-nitrosylation and thioredoxins. Science 321 952–956. 10.1126/science.1156970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevisan S., Manoli A., Ravazzolo L., Botton A., Pivato M., Masi A., et al. (2015). Nitrate sensing by the maize root apex transition zone: a merged transcriptomic and proteomic survey. J. Exp. Bot. 66 3699–3715. 10.1093/jxb/erv165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tun N. N., Santa-Catarina C., Begum T., Silveira V., Handro W., Floh E. I., et al. (2006). Polyamines induce rapid biosynthesis of nitric oxide (NO) in Arabidopsis thaliana seedlings. Plant Cell Physiol. 47 346–354. 10.1093/pcp/pci252 [DOI] [PubMed] [Google Scholar]
- Vandelle E., Delledonne M. (2011). Peroxynitrite formation and function in plants. Plant Sci. 181 534–539. 10.1016/j.plantsci.2011.05.002 [DOI] [PubMed] [Google Scholar]
- Wendehenne D., Gao Q. M., Kachroo A., Kachroo P. (2014). Free radical-mediated systemic immunity in plants. Curr. Opin. Plant Biol. 20C, 127–134. 10.1016/j.pbi.2014.05.012 [DOI] [PubMed] [Google Scholar]
- Xu G., Fan X., Miller A. J. (2012). Plant nitrogen assimilation and use efficiency. Annu. Rev. Plant Biol. 63 153–182. 10.1146/annurev-arplant-042811-105532 [DOI] [PubMed] [Google Scholar]
- Xu S., Guerra D., Lee U., Vierling E. (2013). S-nitrosoglutathione reductases are low-copy number, cysteine-rich proteins in plants that control multiple developmental and defense responses in Arabidopsis. Front. Plant Sci. 4:430 10.3389/fpls.2013.00430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaeno T., Iba K. (2008). BAH1/NLA, a RING-type ubiquitin E3 ligase, regulates the accumulation of salicylic acid and immune responses to Pseudomonas syringae DC3000. Plant Physiol. 148 1032–1041. 10.1104/pp.108.124529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki H., Sakihama Y. (2000). Simultaneous production of nitric oxide and peroxynitrite by plant nitrate reductase: in vitro evidence for the NR-dependent formation of active nitrogen species. FEBS Lett. 468 89–92. 10.1016/S0014-5793(00)01203-5 [DOI] [PubMed] [Google Scholar]
- Yun B. W., Feechan A., Yin M., Saidi N. B., Le Bihan T., Yu M., et al. (2011). S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature 478 264–268. 10.1038/nature10427 [DOI] [PubMed] [Google Scholar]
- Yun B. W., Skelly M. J., Yin M., Yu M., Mun B. G., Lee S. U., et al. (2016). Nitric oxide and S-nitrosoglutathione function additively during plant immunity. New Phytol. 10.1111/nph.13903 [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Zeier J. (2013). New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant Cell Environ. 36 2085–2103. 10.1111/pce.12122 [DOI] [PubMed] [Google Scholar]
- Zeng F., Sun F., Li L., Liu K., Zhan Y. (2014). Genome-scale transcriptome analysis in response to nitric oxide in birch cells: implications of the triterpene biosynthetic pathway. PLoS ONE 9:e116157 10.1371/journal.pone.0116157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G. B., Yi H. Y., Gong J. M. (2014). The Arabidopsis ethylene/jasmonic Acid-NRT signaling module coordinates nitrate reallocation and the trade-off between growth and environmental adaptation. Plant Cell 26 3984–3994. 10.1105/tpc.114.129296 [DOI] [PMC free article] [PubMed] [Google Scholar]