Abstract
Although the bioactive sphingolipid ceramide is an important cell signaling molecule, relatively few direct ceramide-interacting proteins are known. We used an approach combining yeast surface cDNA display and deep sequencing technology to identify novel proteins binding directly to ceramide. We identified 234 candidate ceramide-binding protein fragments and validated binding for 20. Most (17) bound selectively to ceramide, although a few (3) bound to other lipids as well. Several novel ceramide-binding domains were discovered, including the EF-hand calcium-binding motif, the heat shock chaperonin-binding motif STI1, the SCP2 sterol-binding domain, and the tetratricopeptide repeat region motif. Interestingly, four of the verified ceramide-binding proteins (HPCA, HPCAL1, NCS1, and VSNL1) and an additional three candidate ceramide-binding proteins (NCALD, HPCAL4, and KCNIP3) belong to the neuronal calcium sensor family of EF hand-containing proteins. We used mutagenesis to map the ceramide-binding site in HPCA and to create a mutant HPCA that does not bind to ceramide. We demonstrated selective binding to ceramide by mammalian cell-produced wild type but not mutant HPCA. Intriguingly, we also identified a fragment from prostaglandin D2 synthase that binds preferentially to ceramide 1-phosphate. The wide variety of proteins and domains capable of binding to ceramide suggests that many of the signaling functions of ceramide may be regulated by direct binding to these proteins. Based on the deep sequencing data, we estimate that our yeast surface cDNA display library covers ∼60% of the human proteome and our selection/deep sequencing protocol can identify target-interacting protein fragments that are present at extremely low frequency in the starting library. Thus, the yeast surface cDNA display/deep sequencing approach is a rapid, comprehensive, and flexible method for the analysis of protein-ligand interactions, particularly for the study of non-protein ligands.
The bioactive sphingolipid ceramide is involved in the regulation of a wide variety of cellular processes, including apoptosis, autophagy, and cell cycle progression in cancer (1–3). Ceramide has also been implicated in a number of disease states, including inflammation and inflammatory disorders (4) and neurodegenerative diseases (5).
Despite the wide range of processes regulated by ceramide, the precise molecular mechanisms by which ceramide acts as a signaling molecule are not clear. It has been suggested that plasma membrane ceramide acts to stabilize lipid rafts, which act as platforms for the concentration of signaling molecules (6, 7). Another possible mechanism of ceramide signaling is through direct interaction with target proteins. However, relatively few direct protein interactions with ceramide have been described. Examples of proteins that are regulated by direct ceramide binding include KSR (8), Raf-1 (9), protein kinase C-ζ (10), PP2A inhibitor SET (11), and cathepsin D (12). Thus, the identification of additional ceramide-binding proteins could lead to a better mechanistic understanding of how ceramide functions as a signaling molecule.
Although various techniques have been used previously, in general, efforts to systematically screen for protein-lipid interactions have proved challenging (13–15). The commonly used yeast two-hybrid system is ineffective when the bait cannot be expressed inside the yeast cell and phage and bacterial display is limited due to prokaryotic expression of eukaryotic proteins. Column-based affinity purification (16, 17) and protein chip methods (18, 19) have been utilized, but they also have drawbacks, including the difficulty in recovering low abundance proteins and cost of setup and quality control (14).
We have previously described the generation and application of yeast surface cDNA display libraries to novel protein-ligand discovery (13, 15, 20–24). Here, we describe their application for proteome-wide identification of human ceramide-binding proteins. Utilizing deep sequencing to comprehensively interrogate enriched selection outputs, we have identified a large number of ceramide-binding proteins, many of which represent novel interactions. For example, we have identified and validated EF-hand and STI1 domain-containing proteins as ceramide-specific binding proteins, suggesting that ceramide may regulate cellular pathways by interacting directly with those proteins.
EXPERIMENTAL PROCEDURES
Generation of Polyclonal Populations of Yeast Enriched with Clones Displaying Ceramide-binding Protein Fragments
The construction of yeast displayed human cDNA libraries and protocols, and media recipes for their use have been described previously (13, 20–25). Briefly, the yeast display cDNA library was inoculated into 100 ml of SR-CAA media (2% raffinose, 0.67% yeast nitrogen base, 0.5% casamino acids) and grown with shaking overnight at 25 °C. After adding 100 ml of additional SR-CAA, library expression was induced by the addition of galactose to a final concentration of 2%, and the yeast were grown overnight with shaking at 25 °C. Two selection methods were used, one based on pulldown using ligand-coated beads (26) and the other based on FACS (13, 20, 21, 27). For the bead-based pulldown method, ceramide-coated beads were generated by adding 10 μl of 200 μm ceramide-biotin (dissolved in DMSO) (Echelon Biosciences, Salt Lake City, UT) to 200 μl of magnetic Dynabeads M-280 streptavidin (Invitrogen/Life Technologies, Inc.), mixing, and incubating with rotation for 10 min at 25 °C. The ceramide-coated beads were washed twice with PBS with the aid of an Eppendorf-type microtube DYNAL MPC-E magnetic particle concentrator. Yeast cells from 3 ml of the overnight galactose-induced cDNA display library culture were collected by centrifugation, washed twice with PBS, resuspended in 1 ml of PBS, added to the ceramide-coated Dynabeads, and incubated for 2 h with rotation at 25 °C. To remove unbound yeast, the ceramide-coated Dynabeads were washed three times with PBS using the DYNAL MPC-E concentrator. The Dynabeads and associated yeast were plated on SD-CAA (2% dextrose, 0.67% yeast nitrogen base, 0.5% casamino acids) agar plates and incubated for 3 days at 30 °C to allow colonies to grow. Colonies were recovered by scraping, re-induced using the same protocol, and subjected to an additional round of selection on ceramide-coated beads using the same protocol and conditions to generate the second round output. Monoclonal screening revealed that ∼70% of the yeast clones in the second round selection output exhibit detectable binding to ceramide when tested by FACS at a concentration of 200 nm (data not shown).
For the FACS-based enrichment of ceramide-binding yeast clones, the yeast cDNA display library was induced as described above, and 3 ml of induced library was washed twice with PBS and resuspended in 1 ml of PBS. Ceramide-biotin was added at a final concentration of 1 μm, and the mixture was incubated with rotation at 25 °C. Cells were washed twice with PBS, incubated in 1 ml of PBS with 1:500 streptavidin-phycoerythrin (SA-PE)1 (Thermo Fisher/Life Technologies, Inc./Invitrogen) with rotation for 20 min at 25 °C and washed twice with PBS, and the PE-positive yeast were sorted using a Propel Labs/Bio-Rad Avalon/S3e cell sorter (Propel Labs, Fort Collins, CO). Sorted yeast were recovered on SD-CAA plates and incubated for 3 days at 30 °C to allow colonies to grow. Colonies were recovered by scraping, re-induced using the same protocol, and subjected to an additional round of sorting using the same protocol but with 200 nm ceramide-biotin and the substitution of streptavidin-FITC (Thermo Fisher/Life Technologies, Inc./Invitrogen) for SA-PA to generate the second round output. A third round of competitive sorting was performed using PE-labeled ceramide and FITC-labeled ceramide 1-phosphate (C1P). PE-labeled ceramide and FITC-labeled C1P were generated by mixing ceramide-biotin and C1P-biotin (Echelon Biosciences) with SA-PE and streptavidin-FITC, respectively, at a 1:2 molar ratio and incubating at 25 °C for 5 min. Induced yeast from the second round FACS-sorted output were incubated with 200 nm PE-labeled ceramide and FITC-labeled C1P in PBS with rotation for 1 h at 25 °C, washed twice with PBS, and analyzed by FACS. Yeast that were PE-positive, FITC-negative were sorted out, plated on SD-CAA plates, and incubated for 3 days at 30 °C to allow colonies to grow.
For testing of the starting library and polyclonal selection outputs, induced yeast were washed with PBS and incubated with 200 nm ceramide-biotin or C1P-biotin for 30 min at 25 °C with rotation, washed twice with PBS, and then incubated with SA-PE at 1:500 dilution in PBS for 10 min at 25 °C with rotation. Cells were then washed twice with PBS and analyzed by FACS using a BD Accuri C6 (BD Biosciences).
Deep Sequencing Analysis of Polyclonal Selection Outputs
Plasmids were recovered from the cDNA display library, and the ceramide-binding bead-enriched and FACS-enriched polyclonal yeast selection outputs using a modified QIAprep Spin Miniprep protocol (Qiagen, Hilden, Germany) incorporated a glass bead cell lysis step as described previously (13). To generate cDNA insert samples for the deep sequencing, the plasmid preparations were used as templates for PCRs using the primers 5-Illumina-2 (5′-cgacgatgacgataaggtaccag-3′) and PYD1 Illumina R (5′-ccgccactgtgctggatatc-3′) and Phusion high fidelity DNA polymerase (New England Biolabs, Ipswich, MA). The PCR products were isolated using the QIAquick PCR purification kit (Qiagen. Hilden, Germany). The PCR products representing the cDNA inserts from the starting yeast cDNA display library and the ceramide-binding enriched bead and FACS-selected polyclonal outputs were deeply sequenced by paired end 150 cycle Illumina MiSeq runs at the Gladstone Genomics Core. The deep sequencing data sets were analyzed using the web-based Galaxy server (28–30). Briefly, the paired FASTQ data files were processed within Galaxy by FASTQ Groomer (31), followed by TopHat for Illumina (32) to generate an accepted hits bam file, which was then processed by Cufflinks (33) using the iGenomes UCSC hg19 gene annotation reference file (hg19_genes.gtf) to generate gene-level expression values expressed in fragment/kb of exon per million fragments mapped (FPKM). The FPKM data were analyzed using Excel (Microsoft, Redmond, WA). To generate a candidate list of ceramide-binding protein fragments, frequency (FPKM >13) and enrichment (enrichment relative to library >3 for FPKM >100 and >7 for FPKM <100) cutoffs were applied to the data. To generate a list of unique cDNA insert sequences ranked by abundance for use in the recovery of candidate nucleotide sequences, the FASTQ Groomer outputs were processed (one file from each paired end read) using the reverse-complement tool, followed by the FASTQ joiner tool to join the paired end reads. The joined reads were then processed by the Collapse tool to generate a ranked list of nucleotide sequences. The top 3000 ranked sequences were BLAST searched using Blast2GO (BioBam, Valencia, Spain), and this list was used as a reference for recovering the nucleotide sequences of candidate gene cDNA inserts. In cases where candidate nucleotide sequences were not found among the BLAST results, the TopHat accepted bam file was viewed with the Integrated Genome Browser (Affymetrix, Santa Clara, CA), and predicted nucleotide cDNA insert sequences for candidate genes were recovered.
Cloning and Testing of Putative Ceramide-binding Protein Fragments
The cDNAs encoding the 20 chosen candidate ceramide-binding protein fragments were cloned into the pYD1 yeast display vector (20) by PCR using primers designed from the recovered nucleotide sequences of candidate gene cDNA inserts and the ceramide-binding enriched selection output plasmid preparations as a template. The plasmids were then transformed into the yeast strain EBY100 by a lithium acetate heat shock method, and individual colonies were verified by colony PCR as described previously (27).
Yeast clones were grown overnight in SRG-CAA (2% galactose, 2% raffinose, 0.67% yeast nitrogen base, 0.5% casamino acids) to induce surface display of the candidate ceramide-binding protein fragments. Biotinylated lipids were resuspended according to manufacturer's recommendations. Induced yeast clones were washed twice with PBS and incubated with rotation for 1 h at room temperature with 200 nm biotinylated ceramide, C1P, sphingomyelin (SM), phosphatidylcholine (PC), or phosphatidylethanolamine (PE) (Echelon Biosciences) and 1:1000 mouse anti-Xpress antibody (Thermo Fisher/Life Technologies, Inc./Invitrogen) in PBS to monitor surface display levels. The yeast were washed twice with PBS and incubated with 1:500 dilutions of SA-PE and Alexa Fluor 647-conjugated anti-mouse secondary antibody (Jackson ImmunoResearch, West Grove, PA) for 30 min. After two washes with PBS, the yeast were analyzed by FACS using a BD Accuri C6. For additional lipid binding specificity tests, induced yeast clones were washed twice with PBS and incubated with rotation for 1 h at room temperature with 800 nm biotinylated ceramide, C1P, lactosylceramide (Echelon Biosciences), or diacylglycerol (Avanti Polar Lipids, Alabaster, AL) and 1:1000 mouse anti-Xpress antibody. The yeast were washed twice with PBS and incubated with 1:500 dilutions of streptavidin-Alexa Fluor 647 (Thermo Fisher/Life Technologies, Inc./Invitrogen) and FITC-conjugated anti-mouse secondary antibody (Jackson ImmunoResearch) for 30 min. After two washes with PBS, the yeast were analyzed by FACS using a BD Accuri C6.
Isolation and Testing of Non-ceramide-binding HPCA Mutant
Nucleotide sequences encoding the first and second EF-hands (HPCA 1 + 2, amino acids 14–103) and the third and fourth EF-hands (HPCA 3 + 4, amino acids 96–193) were PCR-cloned into pYD1 and transformed into EBY100 as described previously. Yeast clones displaying HPCA 1 + 2 and HPCA 3 + 4 were tested for ceramide binding by FACS as described above. To generate libraries displaying randomly mutagenized versions of HPCA 1 + 2 and HPCA 3 + 4, a PCR-based gap repair method was employed. Briefly, primers PYD5 (5′-aaggtaccaggatccagtgtg-3′) and PYD3 (5′-gttagggataggcttaccttc-3′) were used along with either pYD1-HPCA 1 + 2 or pYD1 HPCA 3 + 4 as template for mutagenic PCRs. Mutagenic PCRs were carried out using Lucigen 1× Colony PCR Master Mix (Lucigen, Middleton, WI) with an additional 5 mm magnesium chloride added (total 6.5 mm) to increase the error rate. Mutagenized PCR products were gel-purified and transformed along with EcoRI-linearized pYD1 vector into EBY100 cells via electroporation (Benatuil), and transformants were selected on SD-CAA agar plates. The HPCA 1 + 2 and HPCA 3 + 4 mutagenized yeast display libraries were induced as described above, and 2 ml of the induced cultures were washed twice with PBS and resuspended in 1 ml of PBS with 1 μm ceramide-biotin and 1:500 mouse anti-V5 antibody (Pierce/Thermo Scientific, Waltham, MA) and incubated for 1 h with rotation at 25 °C. Cells were washed twice with PBS, incubated in 1 ml of PBS with 1:500 SA-PE and 1:500 FITC-conjugated anti-mouse secondary antibody (Jackson ImmunoResearch) with rotation for 20 min at 25 °C, washed twice with PBS, and PE-negative, FITC-positive yeast were sorted using a Propel Labs/Bio-Rad Avalon/S3e cell sorter. Yeast were recovered as described above, induced, and subjected to a second round of negative sorting. Individual yeast clones from the second round output were tested for binding to 200 nm ceramide by FACS as described above. Yeast clones with dramatically reduced ceramide binding were sequenced by colony PCR as described previously (27).
A QuikChange lightning site-directed mutagenesis kit (Agilent, Santa Clara, CA) was used to introduce L43A and I128A amino acid substitutions into pYD1-HPCA, and the plasmid was transformed into EBY100 yeast. The affinity of yeast displayed HPCA, and HPCA L43A/I128A for ceramide-biotin was measured by incubating induced yeast with varying concentrations of ceramide at 4 °C with rotation for 3 h. Yeast were washed twice with ice-cold PBS and incubated with 1:1000 SA-PE at 4 °C with rotation for 20 min, washed twice with ice-cold PBS, and analyzed by FACS using a BD Accuri C6. Mean fluorescence intensity data were plotted, and affinities were calculated using GraphPad Prism software (GraphPad software, La Jolla, CA).
Pulldown Experiments
The coding sequences for HPCA and HPCA L43A/I128A were cloned via PCR into the pCMV6-Myc-DDK mammalian expression vector with a C-terminal Myc epitope tag. HeLa cells were transfected with the HPCA and HPCA L43A/I128A mammalian expression vector plasmids with Lipofectamine 2000 (Thermo Fisher/Life Technologies, Inc./Invitrogen) according to the manufacturer's instructions. After 72 h, transfected cells were harvested by scraping into ice-cold HEPES-buffered saline and homogenized on ice using a Dounce homogenizer. Soluble extracts were recovered by centrifugation at 4 °C. Ceramide, C1P, and SM-coated magnetic Dynabeads were prepared (20 μl per pulldown) as described above and washed twice with HEPES-buffered saline using the DYNAL MPC-E concentrator. Pulldown experiments were carried out by incubating the coated Dynabeads with 500 μl of the HPCA and HPCA L43A/I128A cell extracts for 2 h at 4 °C with rotation. The beads were washed three times with ice-cold HEPES-buffered saline using the DYNAL MPC-E concentrator, boiled in SDS sample buffer, and analyzed by SDS-PAGE. After semi-dry transfer to Immobilon-P membrane (Millipore, Hayward, CA), Western blotting was performed using a mouse anti-Myc antibody (9B11, Cell Signaling Technology, Danvers, MA) followed by anti-mouse HRP (Pierce/Thermo Fisher Scientific) and detected by chemiluminescence with Pierce ECL Western blotting substrate (Pierce/Thermo Fisher Scientific) according to the manufacturer's instructions. Images were captured using a C-DiGit blot scanner (LI-COR Biosciences, Lincoln, NE).
RESULTS
Generation of Polyclonal Yeast Display Selection Outputs Enriched for Ceramide Binding
We have previously described the construction and application of large (>30 million clones) yeast surface display libraries expressing human protein fragments derived from cDNA from multiple human tissue sources (13–15, 20–24). To identify human proteins with affinity for ceramide, we used these previously generated libraries by employing two different methods to create polyclonal selection outputs enriched for clones that display ceramide-binding protein fragments (outlined in Fig. 1).
Fig. 1.
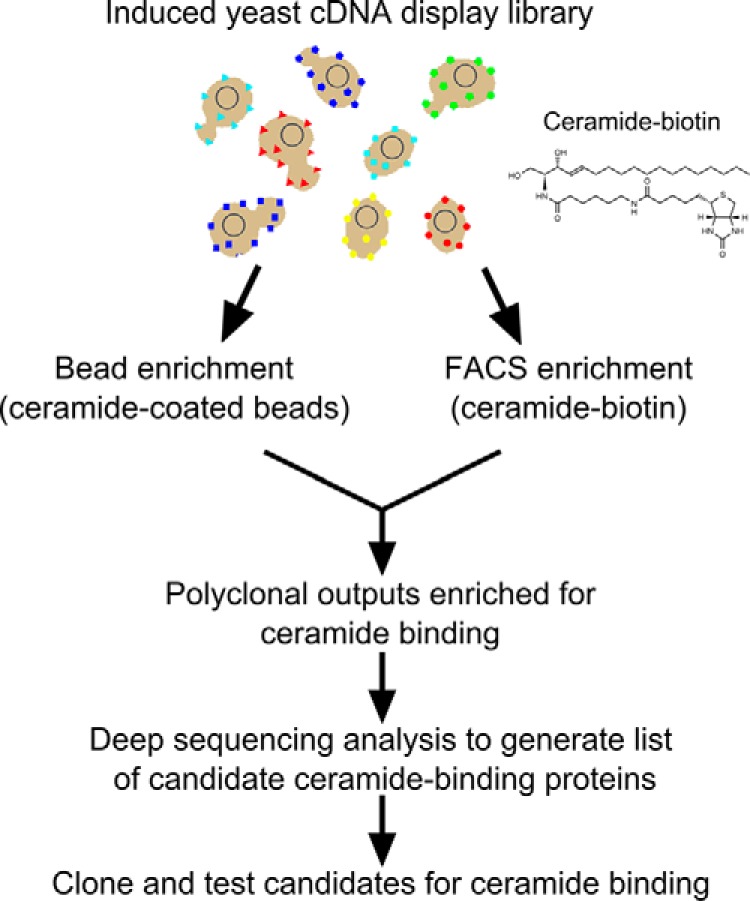
Flow chart of strategy for identification of ceramide-binding protein fragments.
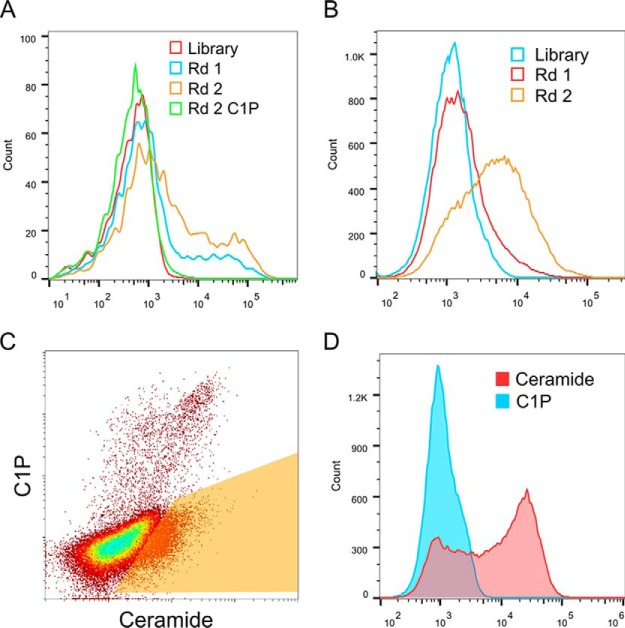
In the first method, we immobilized ceramide onto magnetic beads and used the ceramide-coated beads to capture and enrich yeast clones displaying protein fragments with affinity for ceramide. Two rounds of selection were performed, and the binding of the starting library and the polyclonal selection outputs from the first and second rounds were tested for binding to ceramide by FACS (Fig. 2A). The polyclonal output from the second round of selection exhibited significant binding to ceramide but very little binding to C1P (Fig. 2A).
Fig. 2.
Generation of polyclonal yeast display selection outputs enriched for ceramide binding. A, FACS analysis of ceramide-coated bead enrichment outputs. The starting library and polyclonal selection outputs from the first and second rounds were tested for binding to 200 nm ceramide by FACS. The second round output was also tested for binding to 200 nm C1P. B, FACS analysis of FACS-sorted enrichment outputs. The starting library and polyclonal selection outputs from the first and second rounds were tested for binding to 200 nm ceramide by FACS. C, two-color competitive selection with both ceramide and C1P. The FACS-enriched second round polyclonal output was incubated with 200 nm labeled ceramide and C1P, and the ceramide-specific binding population was gated and sorted. D, FACS analysis of the polyclonal competitive selection output. The polyclonal output of the third round competitive selection was tested by FACS using 200 nm ceramide and C1P.
In the second method, we utilized FACS-based selection to enrich ceramide-binding clones for two rounds. The first round of selection was done with 1 μm biotinylated ceramide. To increase stringency, the concentration of biotinylated ceramide was reduced to 200 nm in the second round. Binding of the first and second round sorting outputs was tested by FACS, and significant binding to 200 nm biotinylated ceramide was observed in the second round output (Fig. 2B). To enrich for clones that bind more specifically to ceramide, we performed an additional round of competitive two-color selection with both ceramide and C1P and gated and sorted the ceramide-specific population (Fig. 2C). The output of the competitive selection round was tested by FACS using 200 nm ceramide and C1P, and significant binding to ceramide was observed, although C1P binding was minimal (Fig. 2D).
Deep Sequencing of Ceramide-binding Polyclonal Selection Outputs
To comprehensively identify putative ceramide-binding protein fragments from the two polyclonal selection outputs, we amplified the cDNA inserts by PCR and subjected them to deep sequencing analysis. To estimate coverage of the starting library and provide a baseline for determining enrichment in the polyclonal selection outputs, we simultaneously performed deep sequencing on the starting library. The bead-selected polyclonal, FACS-sorted polyclonal output, and starting library yielded 1,788,470, 4,886,406, and 1,892,540 paired end reads, respectively. Data were processed and analyzed using the Galaxy web-based server (28–30) to determine abundance at the gene level (FPKM). Out of 24,897 genes in the human genome build used for analysis, 13,865 had a non-zero FPKM value in the processed data for the starting cDNA library. The bead-enriched and FACS-sorted ceramide-binding outputs had 5971 and 628 genes, respectively, with non-zero FPKM values, suggesting that the FACS-based selection was more stringent. Combined, the three datasets have 14,267 unique genes with non-zero FPKM values, indicating that a minimum of 57% of genes annotated by this human genome build are represented in our yeast display human cDNA library. A graphical representation of the starting library, bead-enriched, and FACS-enriched non-zero FPKM values is shown in Fig. 3A.
Fig. 3.
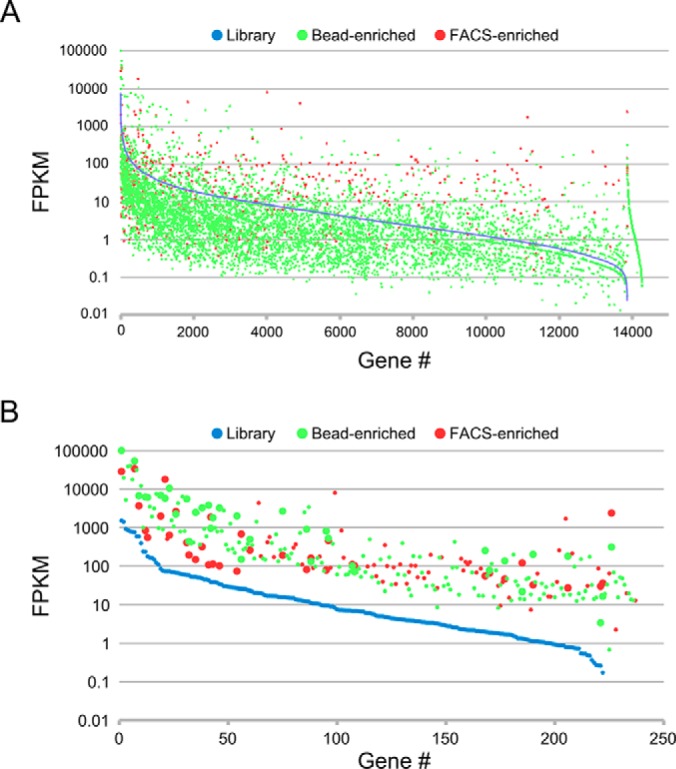
Graphical representations of starting library, bead-enriched, and FACS-enriched gene-level FPKM values calculated from deep sequencing data sets. The FPKM data for the individual genes is arranged by starting library FPKM (high to low from left to right). A, FPKM values for the 14,267 unique non-zero genes from the combined starting library, bead-enriched, and FACS-enriched analyzed data sets. B, FPKM values of the 234 unique genes that meet frequency and enrichment cutoffs and whose recovered nucleotide sequences encode protein fragments derived from the corresponding genes. Larger data points indicate candidates that meet the frequency and enrichment cutoffs in both the bead-enriched and FACS-enriched data sets.
To generate a candidate list of ceramide-binding protein fragments, we applied frequency and enrichment (relative to library) cutoffs to the data (see “Experimental Procedures”). Using these cutoffs yielded 206 candidates for the bead-enriched data set and 138 candidates for the FACS-enriched data set. We next recovered predicted nucleotide sequences for the cDNA inserts of the candidates from matched paired end reads from both the bead-enriched and FACS-enriched data sets and translated them to generate putative protein sequences that were subjected to a protein database search. From the bead-enriched and FACS-enriched candidates, we identified 182 and 91 protein fragment candidates, respectively (234 total unique protein fragment candidates), that match the predicted protein products of the corresponding genes (supplemental Table S1). A graphical representation of FPKM values for the 234 candidates is shown in Fig. 3B. Of the 234 candidates, 36 were identified in both the bead-enriched and FACS-enriched analyzed data sets (Table I and Fig. 3B).
Table I. Candidate ceramide-binding protein fragments that meet FPKM values and enrichment cutoffs in both the bead-enriched and FACS-enriched sequencing data.
FPKM values for the starting library, bead-enriched, and FACS-enriched are shown, and candidates are ranked by highest FPKM value of the enriched populations. The maximum enrichment was calculated by dividing the highest FPKM-enriched population value by the starting library value. Displayed amino acids predicted from the sequencing data, and the total amino acid counts of the full-length protein are shown. The names and accession numbers (in parentheses) of domains identified by searches of the NCBI conserved domain database, SMART domain database, and Pfam domain database are shown (RPN13-C, UCH-binding domain; EFh, EF-hand; SCP2, SCP-2 sterol transfer family; CX9C, CHCH-CHCH-like; SapB-1, saposin-like type B, region 1; SapB-2, saposin-like type B, region 2; STI1, heat shock chaperonin-binding motif; PAH, paired amphipathic helix repeat; PAM2, ataxin-2 C-terminal region; TPR, tetratricopeptide repeat; CRIB, Cdc42/Rac interactive binding; TAFH, NHR1 homology to TAF; PH-like, pleckstrin homology-like domain; UBQ, ubiquitin-like; Rod-C, rough deal protein C-terminal region).
| Gene | Library | Bead-enriched | FACS-enriched | Maximum enrichment | Amino acids | Total amino acids | Domain |
|---|---|---|---|---|---|---|---|
| ADRM1 | 1508 | 100,907 | 28,834 | 67 | 262–407 | 407 | RPN13-C (PF16550.1) |
| HPCA | 759 | 54,158 | 34,201 | 71 | 1–193 | 193 | 3× EFh (cl08302) |
| HSD17B4 | 73 | 5747 | 17,975 | 245 | 445–596 | 596 | SCP2 (cl01225) |
| CHCHD5 | 73 | 10,487 | 633 | 144 | 2–110 | 110 | CX9C (PF16860.1) |
| NCS1 | 85 | 6872 | 1977 | 81 | 2–190 | 190 | 3× EFh |
| RPLP2 | 587 | 6690 | 3673 | 11 | 1–115 | 115 | Ribosomal-P2 (cl21508) |
| PSAP | 236 | 6232 | 837 | 26 | 164–318 | 524 | SapB-1 (cl04972), SapB-2 (cl08395) |
| RAD23A | 177 | 6091 | 553 | 34 | 202–363 | 363 | XPC-binding (cl18177), UBA-like (cl21463), STI1 (SM00727) |
| CABP1 | 58 | 5543 | 406 | 95 | 237–370 | 370 | 3× EFh |
| PDCD6 | 43 | 3798 | 108 | 88 | 1–191 | 191 | 5× EFh |
| MYL6B | 48 | 3240 | 316 | 68 | 121–208 | 208 | 3× EFh |
| SIN3A | 34 | 3230 | 101 | 95 | 109–244 | 1273 | PAH (cl07842) |
| PAIP1 | 16 | 2661 | 190 | 168 | 92–233 | 479 | PAM2 (cl06258), MIF4G (cl02652) |
| SGTA | 67 | 2227 | 2616 | 39 | 88–313 | 313 | 3× TPR (cl22897) |
| CHP1 | 52 | 2474 | 148 | 48 | 1–91 | 195 | EFh |
| HBZ | 0.0 | 311 | 2378 | ∞ | 1–142 | 142 | Globin-like (cl21461) |
| NCALD | 27 | 1996 | 74 | 74 | 1–85 | 193 | 2× EFh |
| PDE6D | 38 | 952 | 1871 | 49 | 1–88 | 150 | GMP-PDE-δ (cl12307) |
| RPS29 | 38 | 1790 | 113 | 47 | 1–56 | 56 | |
| WASL | 12 | 906 | 81 | 74 | 217–337 | 505 | CRIB (cl00113) |
| IKBIP | 9.5 | 813 | 79 | 86 | 1–87 | 377 | |
| PDIA6 | 26 | 149 | 678 | 27 | 192–326 | 492 | Thioredoxin-like (cl00388) |
| ST13 | 9.4 | 525 | 462 | 56 | 301–369 | 369 | STI1 |
| RBP1 | 23 | 493 | 258 | 21 | 18–197 | 197 | Lipocalin (cl21528) |
| HNRNPUL1 | 58 | 434 | 193 | 8 | 1–64 | 285 | |
| GDAP1L1 | 2.0 | 251 | 55 | 127 | 1–179 | 367 | Thioredoxin-like |
| MDM1 | 1.1 | 200 | 32 | 176 | 1–92 | 714 | MDM1 (cl21312) |
| TAF4 | 0.8 | 180 | 27 | 227 | 590–717 | 1085 | TAFH (cl02658) |
| GIGYF1 | 11 | 131 | 160 | 14 | 479–701 | 754 | |
| ST13P4 | 1.7 | 136 | 46 | 79 | 97–369 | 369 | 3× TPR, STI1 |
| DAB2 | 1.3 | 22 | 121 | 91 | 35–221 | 235 | PH-like (cl17171) |
| UBQLN1 | 6.9 | 100 | 110 | 16 | 95–226 | 449 | UBQ (cl00155), STI1 |
| UBQLN2 | 6.9 | 71 | 102 | 15 | 1–310 | 624 | UBQ, 2× STI1 |
| SCP2 | 1.9 | 77 | 65 | 41 | 132–289 | 289 | SCP2 |
| KNTC1 | 0.2 | 17 | 36 | 208 | 2094–2209 | 2209 | Rod-C (cl20400) |
| BTG1 | 0.3 | 3.4 | 30 | 114 | 16–140 | 171 | BTG (cl15439) |
Next, we performed a search for known domains in the candidate ceramide-binding protein sequences. A total of 104 unique annotated protein domains were identified in the 234 candidate protein sequences (supplemental Table S1). Several of the domains are present in multiple candidates. Strikingly, 30 of the 234 (13%) candidate ceramide-binding protein fragments and 7 of the 36 (19%) candidates that are common to both selections have one or more EF-hand domains. Protein domains that are represented more than once among the candidate ceramide-binding protein fragments are shown in supplemental Table S2.
Testing Candidate Protein Fragments for Ceramide Binding
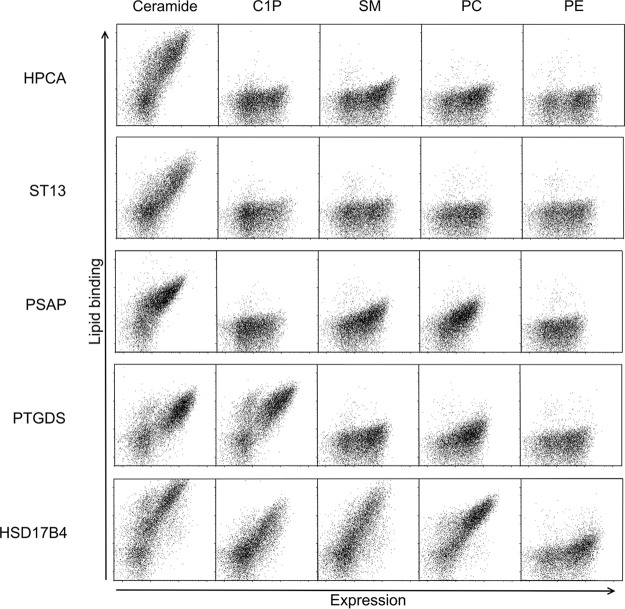
Based on the analyzed data, we chose 20 of the candidate ceramide-binding protein fragments for testing. We regenerated yeast clones displaying the 20 chosen candidate ceramide-binding protein fragments and tested them for binding to 200 nm ceramide, C1P, SM, PC, and PE by FACS (Fig. 4 and Table II). All of the protein fragments exhibited a detectable level of binding to ceramide, and most (17) of the protein fragments, including all of the EF-hand and STI1 domain containing fragments, bound only to ceramide among the tested lipids. Only fragments from HSD17B4 (SCP2 domain), PSAP (SapB-1, SapB-2 domains), and PTGDS (lipocalin domain) exhibited binding to lipids other than ceramide. Interestingly, the PTGDS protein fragment bound more strongly to C1P than ceramide or any of the other tested lipids (Fig. 4 and Table II). The protein fragments exhibited varying degrees of binding to ceramide, with the HSD17B4, HPCA, and ST13 protein fragments containing SCP2, EF-hands, and STI1 domains, respectively, exhibiting the strongest interaction (Fig. 4 and Table II). The 12 unique protein domains found in the 20 verified ceramide-binding protein fragments are described in Table III.
Fig. 4.
FACS plots depicting binding of ceramide, C1P, SM, phosphatidylcholine (PC), and PE by selected candidate ceramide-binding protein fragments displayed on yeast. All the lipids were tested at a concentration of 200 nm. Lipid binding is represented in the y axis, and surface expression level (Xpress epitope) is represented in the x axis.
Table II. Quantitative binding, FPKM, and domain data for the 20 tested candidates.
Binding values for ceramide, C1P, SM, phosphatidylcholine (PC), and PE are expressed as a ratio of the lipid-binding mean fluorescence intensity I to the Xpress epitope (surface expression) mean fluorescence intensity. Candidates are ranked top to bottom by ceramide binding, and data shading intensity increases according to value. FPKM values that meet frequency and enrichment cutoffs are shaded gray. The amino acids and domains present in the candidate protein fragments are indicated. Lib, representation in the unselected library. Bead, representation in the bead-based selection output. FACS, representation in the FACS-based selection output.
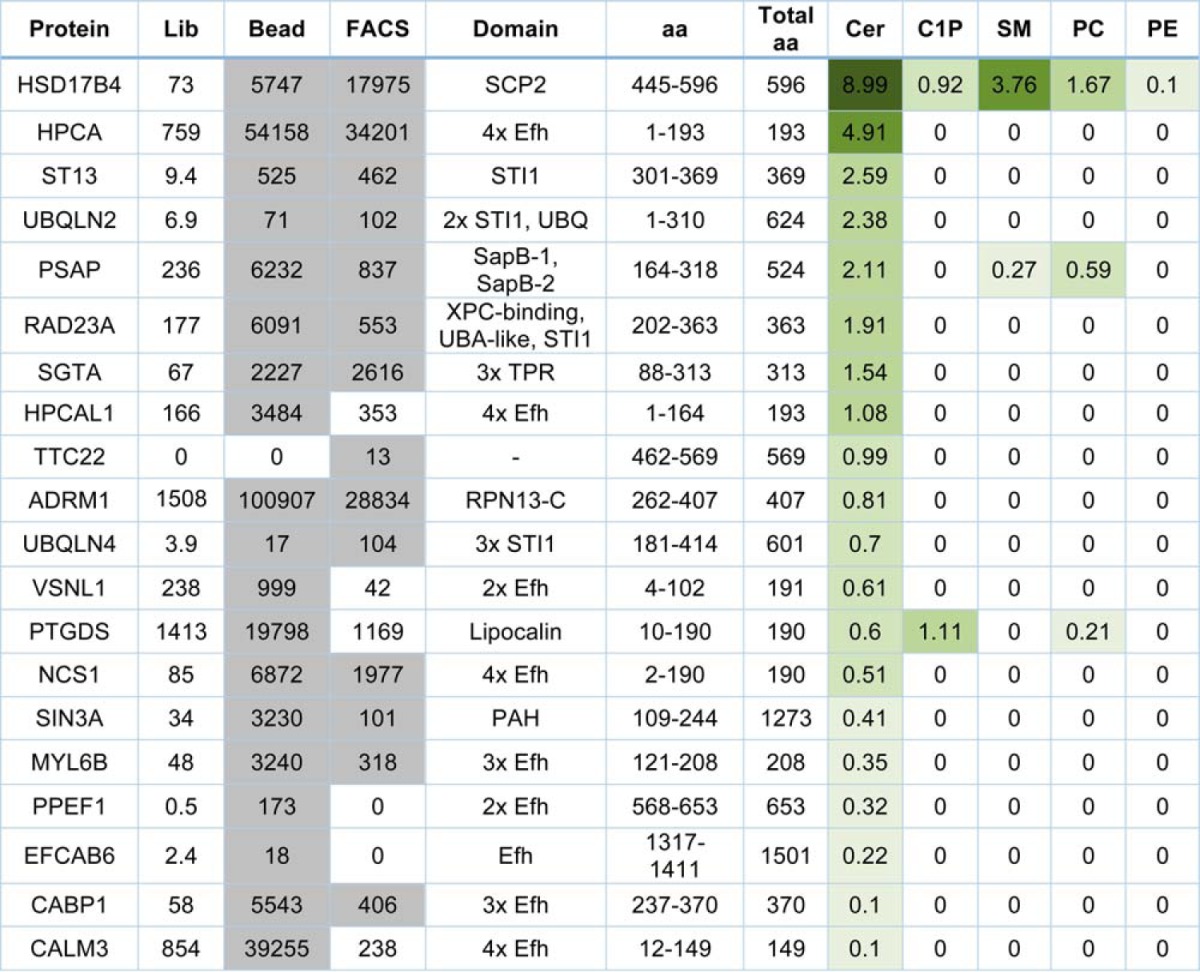
Table III. Annotated protein domains present in the 20 verified ceramide-binding protein fragments.
The names and accession numbers (in parentheses) of protein domains identified by database searches of the 20 verified ceramide-binding protein fragments. Names of the verified and candidate ceramide-binding proteins containing each domain are shown as well as a brief functional description.
| Domain | Verified | Candidates | Description |
|---|---|---|---|
| Efh (cl08302) | HPCA, HPCAL1, VSNL1, NCS1, MYL6B, PPEF1, EFCAB6, CABP1, CALM3 | ACTN1, ACTN4, C17orf57, CALB2, CALM1, CALM2, CALML3, CALN1, CETN1, CETN2, CETN3, CHP, CIB1, HPCAL4, KCNIP3, MYL12A, MYL9, NCALD, PDCD6, PPP3R2, S100A11, TNNC1 | EF-hand calcium-binding helix-loop-helix domain |
| STI1 (SM00727, cl17974) | ST13, UBQLN2, RAD23A, UBQLN4 | ST13P4, UBQLN1 | Heat shock chaperonin-binding motif |
| SCP2 (cl01225) | HSD17B4 | HSDL2, SCP2 | Sterol-binding domain; intracellular transport of lipids and cholesterol |
| UBQ (cl00155) | UBQLN2 | MAP1LC3B2, MYO9A, RAD23B, UBL4A, UBQLN1 | Domain characteristic of ubiquitin and ubiquitin-like proteins |
| SapB-1 (cl04972) | PSAP | Saposin-like type B, region 1; isolates lipid substrates | |
| SapB-2 (cl08395) | PSAP | Saposin-like type B, region 2; isolates lipid substrates | |
| XPC-binding (cl18177) | RAD23A | RAD23B | Xeroderma pigmentosum group C protein-binding; initiates nucleotide excision repair |
| UBA-like (cl21463) | RAD23A | RAD23B | Ubiquitin-associated-like domain; hydrophobic surface patch for protein-protein interactions |
| TPR (cl22897) | SGTA | ST13P4, TTC8 | Tetratricopeptide repeat; mediate protein-protein interactions |
| RPN13-C (PF16550) | ADRM1 (RPN13) | UCH-binding domain; mediates ubiquitin-receptor Rpn13 binding to de-ubiquitinating enzyme UCH37 | |
| Lipocalin (cl21528) | PTGDS | RBP1 | Cytosolic fatty-acid binding; transport of small hydrophobic molecules |
| PAH (cl07842) | SIN3A | SIN3B | Paired amphipathic helix repeat; mediates protein-protein interactions |
To further explore lipid binding specificity, we tested two additional lipids, lactosylceramide, which has a large polar headgroup, and diacylglycerol, which has a small non-polar headgroup. A diagram depicting the structures of all biotinylated lipids used in this study is shown in supplemental Fig. S1. Relative to ceramide binding, moderate binding to lactosylceramide was observed for several clones, while binding to diacylglycerol was very weak in comparison with ceramide binding for all clones with the exception of PTGDS (Figs. S2 and S3).
Identification of Non-ceramide-binding HPCA Mutant
Four of the verified ceramide-binding (HPCA, HPCAL1, NCS1, and VSNL1) and an additional three candidate ceramide-binding (NCALD, HPCAL4, and KCNIP3) protein fragments belong to the neuronal calcium sensor (NCS) family of EF-hand-containing proteins. NCS proteins have several functional roles in neurons and retinal photoreceptor cells and are composed of a conserved sequence of about 200 amino acids containing four EF-hand domains (34–37). We chose the NCS protein with the highest observed binding to ceramide, HPCA, as a model for the identification of amino acids important for interaction with ceramide.
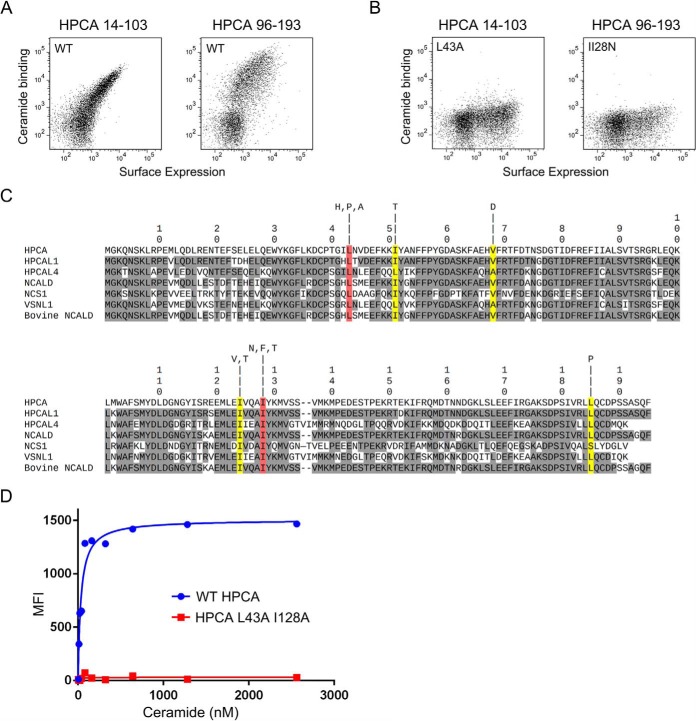
We first generated a library of yeast displaying randomly mutagenized full-length HPCA (amino acids 1–193) and attempted to isolate HPCA mutants that had lost the ability to bind ceramide by FACS selection. However, this attempt proved unsuccessful (data not shown). An examination of the molecular structure of the bovine neurocalcin protein (human version encoded by NCALD) (38), which is 88% identical to human HPCA, suggested that hydrophobic pockets formed by the first and second EF-hands and the third and fourth EF-hands might be capable of binding ceramide independently. We reasoned that this may have caused the difficulty in identifying HPCA mutants unable to bind ceramide because two independent mutations would be required. To test this possibility, we generated yeast displaying the HPCA domains consisting of the first and second EF-hands (HPCA 1 + 2, amino acids 14–103) and the third and fourth EF-hands (HPCA 3 + 4, amino acids 96–193) and tested them for binding to ceramide. Both HPCA 1 + 2 and HPCA 3 + 4 bound strongly to 200 nm ceramide (Fig. 5A).
Fig. 5.
Isolation of non-ceramide binding HPCA mutants. A, binding of HPCA 1 + 2 (amino acids 14–103) and HPCA 3 + 4 (amino acids 96–193) to 200 nm ceramide. Ceramide binding is represented by the y axis, and surface expression (Xpress epitope) is represented by the x axis. B, binding of HPCA 1 + 2 L43A and HPCA 3 + 4 I128N to 200 nm ceramide. C, alignment of candidate and verified ceramide-binding NCS family proteins showing the position of mutations in HPCA that reduce ceramide binding. Amino acids identical to HPCA are shaded gray. Mutations in amino acid residues shaded yellow or red dramatically reduce ceramide binding in the context of HPCA 1 + 2 or HPCA 3 + 4. Amino acid residues shaded red were additionally tested in combination in full-length HPCA. One-letter amino acid codes above each shaded mutation indicate the amino acid substitutions recovered that significantly reduce ceramide binding. D, measurement of ceramide affinity for yeast-displayed full-length HPCA and HPCA L43A/I128A mutant.
We generated two more libraries of yeast displaying randomly mutagenized versions of HPCA 1 + 2 and HPCA 3 + 4 and once again attempted to isolate mutants that had lost the ability to bind ceramide by FACS selection. This time we were successful in isolating non-ceramide-binding mutants. Amino acid mutations at several positions in HPCA 1 + 2 and HPCA 3 + 4 were found to greatly reduce binding to ceramide (Fig. 5, B and C). We sought to use this information to generate a full-length HPCA mutant incapable of ceramide binding. Based on sequence conservation and structural similarities among NCS family members (Figs. 5C and supplemental Fig. S1), we focused on mutations at positions Leu43 and Ile128 in HPCA 1 + 2 and HPCA 3 + 4, respectively, which are single-site mutations that were found to drastically reduce ceramide binding (Fig. 5B). To test whether the combination of these two mutations is able to eliminate ceramide binding in the context of full-length HPCA, we generated a yeast clone displaying full-length HPCA with L43A and I128A mutations (HPCA L43A/I128A) and measured its affinity for ceramide along with wild-type HPCA (Fig. 5D). Yeast-displayed wild-type HPCA bound to ceramide with an affinity of 33.7 ± 7.5 nm, whereas HPCA L43A/I128A binding to ceramide was undetectable (Fig. 5D). The Leu43 and Ile128 amino acid residues critical for ceramide binding are conserved in the NCS family (Fig. 5C). In addition, crystal structures of bovine NCALD (38) and human NCS1 (39) reveal that Leu43 and Ile128 are structurally conserved as well (supplemental Fig. S4).
HPCA but Not HPCA L43A/I128A Expressed by HeLa Cells Binds to Ceramide
To verify the ability of mammalian cell-produced HPCA to bind to ceramide, we made extracts from HeLa cells overexpressing epitope-tagged HPCA and HPCA L43A/I128A and used them for pulldown experiments with ceramide, C1P, and SM-coated beads. Wild-type HPCA was captured by the ceramide-coated beads but not the C1P or SM-coated beads, whereas the HPCA L43A/I128A mutant was not pulled down by any of the lipid-coated beads (Fig. 6). Thus, HPCA produced in mammalian cells is capable of binding to ceramide, and amino acid residues that we identified (L43A and I128A) are critical for mediating this interaction.
Fig. 6.
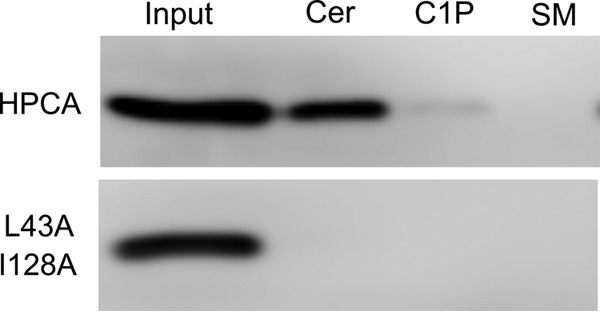
Pulldown of epitope-tagged HPCA and HPCA L43A/I128A mutant from transfected HeLa extracts. Beads used for capture were coated with ceramide (Cer), C1P, and SM. Extract equivalent to the total amount included in the pulldown experiments is shown in the 1st lane.
DISCUSSION
Despite the importance of ceramide as a signaling molecule, relatively few studies have attempted to systematically identify ceramide-binding proteins. Previous efforts utilized ceramide affinity chromatography followed by mass spectrometry analysis to identify ceramide-binding proteins (11, 40, 41). Only one of the studies identified a large number (97) of ceramide-binding protein candidates (41), and none of the candidates were verified directly by ceramide binding assays. Although the study by Mukhopadhyay et al. (11) identified only one ceramide-binding protein, inhibitor 2 of protein phosphatase 2A (I2PP2A), ceramide was demonstrated to directly bind I2PP2A and decrease its interaction with PP2A, thereby blocking inhibition of phosphatase activity. Several additional ceramide-binding proteins have been identified by direct assay approaches, including c-Raf (9), PKC-α and -δ (42), cathepsin D (12), MEKK1 (43), and kinase suppressor of Ras (KSR1) (44). Overall, the number of ceramide-interacting proteins with validated direct binding is limited. In addition, with the exception of cathepsin D, KSR1, and I2PP2A, the specificity of those ceramide-binding proteins was not analyzed in the context of binding to other related lipids. To understand how ceramide regulates diverse cellular functions, new approaches are needed to systematically identify cellular proteins that bind to ceramide directly. In this study, we continued our previously described yeast surface cDNA library selection approach to identify protein-ligand interaction on a proteome-wide scale (20, 21, 24). We used FACS or bead-based affinity capture to enrich for ceramide-binding clones from the yeast surface human proteome display library and analyzed the outputs by deep sequencing, which identified 234 candidate ceramide-binding protein fragments. Through direct testing of individual clones, 20 ceramide-binding protein fragments were confirmed, many of which represent novel interactions.
Of the 20 confirmed ceramide-binding protein fragments, 17 (including all the EF-hand-containing protein fragments) bound only ceramide and not any of the other lipids when tested at a concentration of 200 nm (Table II). When tested at a higher concentration of 800 nm, weak binding to C1P and lactosylceramide was observed for some protein fragments, although binding to diacylglycerol was negligible for all proteins with the exception of PTGDS (supplemental Fig. S3). For all proteins except PTGDS, the strongest interaction was observed with ceramide. The degree of binding specificity is notable and suggests that there may be some functional significance to the observed lipid-binding specificity of these proteins. Three of the verified ceramide-binding protein fragments bound to one or more of tested lipids in addition to ceramide when tested at a concentration of 200 nm. The HSD17B4 protein fragment additionally bound to C1P, SM, and PC, although the binding was less strong than binding to ceramide. The PSAP protein fragment also bound more weakly to SM and PC. Interestingly, the PTGDS protein fragment was unique in that it bound more strongly to C1P. PTGDS functions downstream of cytosolic phospholipase A2α in the prostaglandin biosynthesis pathway, and translocation of cytosolic phospholipase A2α to internal membranes is known to be mediated by C1P binding (45). Thus, it is possible that C1P binding by cytosolic phospholipase A2α and PTGDS facilitates prostaglandin biosynthesis by concentrating the enzymes in localized C1P-rich membrane domains. The domains in these protein fragments (SCP2, SapB-1 and SapB-2, and lipocalin, respectively) are known to be involved in isolating or transporting lipids or other small hydrophobic molecules so their observed nonspecific lipid binding is not surprising (Table III).
Based on the binding data for the different lipids, some speculation can be made concerning the structural features of the lipids that influence binding. The only difference between the biotinylated ceramide and C1P molecules used in this study is a headgroup phosphate. Because C1P binding is significantly weaker to all the protein fragments except PTGDS, it is likely that the presence of the phosphate headgroup interferes with binding. Indeed, binding to the lipids with large polar headgroups (SM, PC, PE, and lactosylceramide) is also weaker than binding to ceramide. Although the diacylglycerol headgroup is small and non-polar like ceramide, no binding was observed to any of the tested protein fragments with the exception of PTGDS. The ceramide-specific binding of the ceramide transporter CERT has been shown to be mediated by a hydrogen bond network between the amide and hydroxyl groups of ceramide and specific amino acid side chains in CERT (46). In contrast, fewer hydrogen bonds were observed in CERT complexed with diacylglycerol, resulting in a much weaker interaction (46). Thus, the dramatic preference for ceramide over diacylglycerol that we observed for all of the ceramide-binding protein fragments except PTGDS may be the result of a binding mechanism similar to that described for CERT. Alternatively, because the biotinylated diacylglycerol used in this study has longer acyl chains than the other tested lipids (supplemental Fig. S1), steric hindrance in the binding pocket could be a factor. Taken together, our data suggest that the polarity and/or size of the lipid headgroup is an important factor that influences binding efficiency, although structural differences that affect hydrogen bonding capacity and acyl chain length may also play important roles. As additional labeled lipid reagents become available, the factors important for binding specificity can be further investigated.
It should be noted that the biotinylated lipids used in this study are not identical to native cellular ceramide, which exists naturally in many forms in the cell. They have a shortened acyl chain modified by a linker with biotin, which may affect their biological properties, including solubility. Many of our assays are performed in aqueous solution at lipid concentrations up to the micromolar range. It is likely that the biotinylated lipids may exist as micelles at this concentration in aqueous solution, although this has not been investigated. Labeled lipids such as the biotinylated lipids used in this study are frequently used for binding or localization assays. The biotinylated ceramide used in this study has been previously successfully utilized in pulldown assays (47).
Database searches revealed the presence of 104 unique annotated protein domains in the 234 candidate ceramide-binding protein fragments (supplemental Table S1), including 12 unique domains in the 20 verified ceramide-binding protein fragments (Table III). The most highly represented domain is the calcium-binding EF-hand helix-loop-helix structural domain, which was found in one or more copies in 30 of the 234 candidates, including 9 of the 20 verified ceramide-binding protein fragments. Strikingly, four of the EF-hand-containing ceramide-binding protein fragments HPCA, HPCAL1, NCS1, and VSNL1, as well as three additional unverified candidates NCALD, KCNIP3, and HPCAL4, belong to the NCS family of EF-hand-containing proteins. There are 14 mammalian genes encoding NCS proteins (34, 36), and they are expressed in the brain and retina where they are involved in several cellular processes, including phototransduction (48), long term depression (49), slow after hyperpolarization (50, 51), receptor trafficking (52), channel regulation (53–55), axon growth and regeneration (56–58), and exocytosis (59–65). NCS proteins are highly conserved, composed of about 200 amino acids containing four EF-hand domains, the first of which does not bind calcium. A conserved feature of NCS proteins is a hydrophobic groove that has been shown to be directly involved in target interaction for multiple NCS proteins (34, 36). Interestingly, the HPCA mutations we identified that abolish ceramide binding reside in this groove, suggesting that binding of ceramide to NCS proteins could regulate their interaction with other targets.
Future experiments will be required to determine the functional significance of the identified ceramide-protein interactions. Many of the identified ceramide-binding protein fragments are known to mediate protein-protein interactions. In addition to the aforementioned NCS proteins, the STI1, XPC-binding, UBA-like, tetratricopeptide repeat region, RPN13-C, and PAH domains are all known to be involved in protein interactions (Table III). Thus, it is possible that ceramide binding by these domains may act to modify these protein interactions and thereby regulate function. Another potential mechanism of regulation is through lipid-mediated protein localization. The ability of ceramide to potentially interact with a broad range of protein motifs could be the physical basis for its observed regulatory activities in diverse cellular process.
Deep sequencing is an effective method to comprehensively analyze library diversity and selection outputs (66, 67). The combination of yeast surface human proteomes display affinity-based selections with deep sequencing of the outputs is a powerful method for the rapid identification of protein-ligand interactions. We used this method to identify 234 candidate ceramide-binding protein fragments, of which 20 were validated. Given the number of candidates tested, it is likely that additional testing of candidates would validate even more ceramide-binding proteins. This method allows the use of non-protein targets, such as lipids and small molecules, which would require complicated setups with traditional yeast hybrid methods. The most commonly used method for identifying lipid and small molecule-interacting proteins is affinity chromatography of extracts followed by mass spectrometry analysis. Although this method is powerful, the output tends to be dominated by proteins that are highly abundant in the extract. Protein chip-based experiments are rapid and straightforward to analyze, but the cost and expertise required to generate high quality comprehensive protein arrays has limited their application. In contrast, yeast cDNA display-based selection coupled with deep sequencing allow the rapid identification of even low abundance interactions. For example, we identified ceramide-binding protein fragments that were present at very low or even undetectable levels in the starting library (PPEF1, starting library FPKM = 0.5, and TTC22 starting library FPKM = 0). It is doubtful that these interactions would be identified using affinity chromatography-based methods. Deep sequencing data for the yeast cDNA display library and selection outputs contained reads for 14,267 of the 24,897 genes in the human genome build used for analysis, indicating good coverage of the human proteome. It is likely that the number of reads (1,892,540) acquired by the library's deep sequencing run was not sufficient to recover the full diversity of the library. This is supported by the fact that many genes with an FPKM score of zero in the library were nonetheless found to have non-zero FPKM scores in the selection outputs. Thus, the actual library coverage is likely higher than indicated by the deep sequencing library data analysis. In addition, the flexibility of the yeast display platform allows the rapid elucidation of amino acid residues important for mediating binding interactions, as demonstrated by our successful identification of non-ceramide-binding HPCA mutants.
Future experiments will be directed at understanding the functional significance of the identified ceramide-protein interactions. In particular, the generation of the HPCA non-ceramide-binding mutant will allow experiments directed at understanding the role of ceramide binding in HPCA function. Because the NCS family proteins are highly conserved, particularly the residues identified that are critical for ceramide binding, we expect that mutation of the equivalent amino acids in other ceramide-binding NCS proteins will abolish ceramide binding as well, making it possible to do functional experiments with additional NCS family proteins. Given the power of the yeast surface cDNA display/deep sequencing approach, future studies could apply the approach to other lipids, ultimately leading to a comprehensive map of protein-lipid interactions.
Supplementary Material
Acknowledgments
We thank Dr. Christopher R. Behrens for help with HPLC lipid analysis. We thank Drs. Daniel W. Sherbenou, Christopher R. Behrens, and Yafeng Zhang for helpful discussions.
Footnotes
Author contributions: S.B. and B.L. designed research; S.B., K.H., N.L., and Y.S. performed research; S.B., K.H., N.L., and Y.S. contributed new reagents or analytic tools; S.B. and B.L. analyzed data; S.B. and B.L. wrote the paper.
* This work was supported by National Institutes of Health Grants R01 CA171315, R01 CA118919, and R01 CA129491. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
 This article contains supplemental Figs. S1 to S4 and Tables S1 and S2.
This article contains supplemental Figs. S1 to S4 and Tables S1 and S2.
The authors declare that they have no conflicts of interest with the contents of this article.
1 The abbreviations used are:
- SA-PE
- streptavidin-phycoerythrin
- SA-647
- Alexa fluor 647-conjugated streptavidin
- FPKM
- fragments/kb of exon per million fragments mapped
- C1P
- ceramide 1-phosphate
- SM
- sphingomyelin
- PE
- phosphatidylethanolamine
- NCS
- neuronal calcium sensor
- SM
- sphingomyelin.
REFERENCES
- 1.Galadari S., Rahman A., Pallichankandy S., and Thayyullathil F. (2015) Tumor suppressive functions of ceramide: evidence and mechanisms. Apoptosis 20, 689–711 [DOI] [PubMed] [Google Scholar]
- 2.Morad S. A., and Cabot M. C. (2013) Ceramide-orchestrated signalling in cancer cells. Nat. Rev. Cancer 13, 51–65 [DOI] [PubMed] [Google Scholar]
- 3.Saddoughi S. A., and Ogretmen B. (2013) Diverse functions of ceramide in cancer cell death and proliferation. Adv. Cancer Res. 117, 37–58 [DOI] [PubMed] [Google Scholar]
- 4.Maceyka M., and Spiegel S. (2014) Sphingolipid metabolites in inflammatory disease. Nature 510, 58–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mencarelli C., and Martinez-Martinez P. (2013) Ceramide function in the brain: when a slight tilt is enough. Cell. Mol. Life Sci. 70, 181–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grassme H., Jekle A., Riehle A., Schwarz H., Berger J., Sandhoff K., Kolesnick R., and Gulbins E. (2001) CD95 signaling via ceramide-rich membrane rafts. J. Biol. Chem. 276, 20589–20596 [DOI] [PubMed] [Google Scholar]
- 7.Grassmé H., Jendrossek V., Bock J., Riehle A., and Gulbins E. (2002) Ceramide-rich membrane rafts mediate CD40 clustering. J. Immunol. 168, 298–307 [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y., Yao B., Delikat S., Bayoumy S., Lin X.-H., Basu S., McGinley M., Chan-Hui P.-Y., Lichenstein H., and Kolesnick R. (1997) Kinase suppressor of Ras is ceramide-activated protein kinase. Cell 89, 63–72 [DOI] [PubMed] [Google Scholar]
- 9.Huwiler A., Brunner J., Hummel R., Vervoordeldonk M., Stabel S., van den Bosch H., and Pfeilschifter J. (1996) Ceramide-binding and activation defines protein kinase c-Raf as a ceramide-activated protein kinase. Proc. Natl. Acad. Sci. U.S.A. 93, 6959–6963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bourbon N. A., Yun J., and Kester M. (2000) Ceramide directly activates protein kinase C ζ to regulate a stress-activated protein kinase signaling complex. J. Biol. Chem. 275, 35617–35623 [DOI] [PubMed] [Google Scholar]
- 11.Mukhopadhyay A., Saddoughi S. A., Song P., Sultan I., Ponnusamy S., Senkal C. E., Snook C. F., Arnold H. K., Sears R. C., Hannun Y. A., and Ogretmen B. (2009) Direct interaction between the inhibitor 2 and ceramide via sphingolipid-protein binding is involved in the regulation of protein phosphatase 2A activity and signaling. FASEB J. 23, 751–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heinrich M., Wickel M., Schneider-Brachert W., Sandberg C., Gahr J., Schwandner R., Weber T., Saftig P., Peters C., Brunner J., Krönke M., and Schütze S. (1999) Cathepsin D targeted by acid sphingomyelinase-derived ceramide. EMBO J. 18, 5252–5263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bidlingmaier S., and Liu B. (2011) Identification of protein/target molecule interactions using yeast surface-displayed cDNA libraries. Methods Mol. Biol. 729, 211–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bidlingmaier S., and Liu B. (2015) Utilizing yeast surface human proteome display libraries to identify small molecule-protein interactions. Methods Mol. Biol. 1319, 203–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bidlingmaier S., and Liu B. (2015) Identification of posttranslational modification-dependent protein interactions using yeast surface displayed human proteome libraries. Methods Mol. Biol. 1319, 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lomenick B., Olsen R. W., and Huang J. (2011) Identification of direct protein targets of small molecules. ACS Chem. Biol. 6, 34–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ziegler S., Pries V., Hedberg C., and Waldmann H. (2013) Target identification for small bioactive molecules: finding the needle in the haystack. Angew. Chem. Int. Ed. Engl. 52, 2744–2792 [DOI] [PubMed] [Google Scholar]
- 18.Chen R., and Snyder M. (2010) Yeast proteomics and protein microarrays. J. Proteomics. 73, 2147–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reymond Sutandy F. X., Qian J., Chen C. S., and Zhu H. (2013) Overview of protein microarrays. Curr Protoc Protein Sci. 2013. April; Chapter 27:Unit 27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bidlingmaier S., and Liu B. (2006) Construction and application of a yeast surface-displayed human cDNA library to identify post-translational modification-dependent protein-protein interactions. Mol. Cell. Proteomics 5, 533–540 [DOI] [PubMed] [Google Scholar]
- 21.Bidlingmaier S., and Liu B. (2007) Interrogating yeast surface-displayed human proteome to identify small molecule-binding proteins. Mol. Cell. Proteomics 6, 2012–2020 [DOI] [PubMed] [Google Scholar]
- 22.Bidlingmaier S., and Liu B. (2011) Construction of yeast surface-displayed cDNA libraries. Methods Mol. Biol. 729, 199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bidlingmaier S., and Liu B. (2015) Identification of novel protein-ligand interactions by exon microarray analysis of yeast surface displayed cDNA library selection outputs. Methods Mol. Biol. 1319, 179–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bidlingmaier S., Wang Y., Liu Y., Zhang N., and Liu B. (2011) Comprehensive analysis of yeast surface displayed cDNA library selection outputs by exon microarray to identify novel protein-ligand interactions. Mol. Cell. Proteomics 10.1074/mcp.M110.005116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boder E. T., and Wittrup K. D. (1997) Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15, 553–557 [DOI] [PubMed] [Google Scholar]
- 26.Angelini A., Chen T. F., de Picciotto S., Yang N. J., Tzeng A., Santos M. S., Van Deventer J. A., Traxlmayr M. W., and Wittrup K. D. (2015) Protein engineering and selection using yeast surface display. Methods Mol. Biol. 1319, 3–36 [DOI] [PubMed] [Google Scholar]
- 27.Bidlingmaier S., Su Y., and Liu B. (2015) Combining phage and yeast cell surface antibody display to identify novel cell type-selective internalizing human monoclonal antibodies. Methods Mol. Biol. 1319, 51–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blankenberg D., Kuster G. V., Coraor N., Ananda G., Lazarus R., Mangan M., Nekrutenko A., and Taylor J. (2010) Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. Chapter 19, Unit 19.10.1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giardine B., Riemer C., Hardison R. C., Burhans R., Elnitski L., Shah P., Zhang Y., Blankenberg D., Albert I., Taylor J., Miller W., Kent W. J., and Nekrutenko A. (2005) Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 15, 1451–1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goecks J., Nekrutenko A., Taylor J., and Galaxy Team (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11, R86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blankenberg D., Gordon A., Von Kuster G., Coraor N., Taylor J., Nekrutenko A., and Galaxy Team (2010) Manipulation of FASTQ data with Galaxy. Bioinformatics 26, 1783–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S. L. (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., Salzberg S. L., Wold B. J., and Pachter L. (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ames J. B., and Lim S. (2012) Molecular structure and target recognition of neuronal calcium sensor proteins. Biochim. Biophys. Acta 1820, 1205–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burgoyne R. D., and Haynes L. P. (2012) Understanding the physiological roles of the neuronal calcium sensor proteins. Mol. Brain. 5, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burgoyne R. D., and Haynes L. P. (2015) Sense and specificity in neuronal calcium signalling. Biochim. Biophys. Acta 1853, 1921–1932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim S., Dizhoor A. M., and Ames J. B. (2014) Structural diversity of neuronal calcium sensor proteins and insights for activation of retinal guanylyl cyclase by GCAP1. Front. Mol. Neurosci. 7, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vijay-Kumar S., and Kumar V. D. (1999) Crystal structure of recombinant bovine neurocalcin. Nat. Struct. Biol. 6, 80–88 [DOI] [PubMed] [Google Scholar]
- 39.Bourne Y., Dannenberg J., Pollmann V., Marchot P., and Pongs O. (2001) Immunocytochemical localization and crystal structure of human frequenin (Neuronal Calcium Sensor 1). J. Biol. Chem. 276, 11949–11955 [DOI] [PubMed] [Google Scholar]
- 40.Elsen L., Betz R., Schwarzmann G., Sandhoff K., and van Echten-Deckert G. (2002) Identification of ceramide binding proteins in neuronal cells: a critical point of view. Neurochem. Res. 27, 717–727 [DOI] [PubMed] [Google Scholar]
- 41.Kota V., Szulc Z. M., and Hama H. (2012) Identification of C6-ceramide-interacting proteins in D6P2T Schwannoma cells. Proteomics 12, 2179–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huwiler A., Fabbro D., and Pfeilschifter J. (1998) Selective ceramide binding to protein kinase C-α and -δ isoenzymes in renal mesangial cells. Biochemistry 37, 14556–14562 [DOI] [PubMed] [Google Scholar]
- 43.Huwiler A., Xin C., Brust A.-K., Briner V. A., and Pfeilschifter J. (2004) Differential binding of ceramide to MEKK1 in glomerular endothelial and mesangial cells. Biochim. Biophys. Acta 1636, 159–168 [DOI] [PubMed] [Google Scholar]
- 44.Yin X., Zafrullah M., Lee H., Haimovitz-Friedman A., Fuks Z., and Kolesnick R. (2009) A ceramide-binding C1 domain mediates kinase suppressor of ras membrane translocation. Cell. Physiol. Biochem. 24, 219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lamour N. F., Subramanian P., Wijesinghe D. S., Stahelin R. V., Bonventre J. V., and Chalfant C. E. (2009) Ceramide 1-phosphate is required for the translocation of group IVA cytosolic phospholipase A2 and prostaglandin synthesis. J. Biol. Chem. 284, 26897–26907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kudo N., Kumagai K., Tomishige N., Yamaji T., Wakatsuki S., Nishijima M., Hanada K., and Kato R. (2008) Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide. Proc. Natl. Acad. Sci. U.S.A. 105, 488–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tani H., Shiokawa M., Kaname Y., Kambara H., Mori Y., Abe T., Moriishi K., and Matsuura Y. (2010) Involvement of ceramide in the propagation of Japanese encephalitis virus. J. Virol. 84, 2798–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stephen R., Filipek S., Palczewski K., and Sousa M. C. (2008) Ca2+-dependent regulation of phototransduction. Photochem. Photobiol. 84, 903–910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Amici M., Doherty A., Jo J., Jane D., Cho K., Collingridge G., and Dargan S. (2009) Neuronal calcium sensors and synaptic plasticity. Biochem. Soc. Trans. 37, 1359–1363 [DOI] [PubMed] [Google Scholar]
- 50.Tzingounis A. V., and Nicoll R. A. (2008) Contribution of KCNQ2 and KCNQ3 to the medium and slow after hyperpolarization currents. Proc. Natl. Acad. Sci. 105, 19974–19979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim K. S., Kobayashi M., Takamatsu K., and Tzingounis A. V. (2012) Hippocalcin and KCNQ channels contribute to the kinetics of the slow after hyperpolarization. Biophys. J. 103, 2446–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kabbani N., Woll M. P., Nordman J. C., and Levenson R. (2012) Dopamine receptor interacting proteins: targeting neuronal calcium sensor-1/D2 dopamine receptor interaction for antipsychotic drug development. Curr. Drug Targets 13, 72–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiss J. L., Archer D. A., and Burgoyne R. D. (2000) Neuronal Ca2+ sensor-1/frequenin functions in an autocrine pathway regulating Ca2+ channels in bovine adrenal chromaffin cells. J. Biol. Chem. 275, 40082–40087 [DOI] [PubMed] [Google Scholar]
- 54.Yan J., Leal K., Magupalli V. G., Nanou E., Martinez G. Q., Scheuer T., and Catterall W. A. (2014) Modulation of CaV2.1 channels by neuronal calcium sensor-1 induces short-term synaptic facilitation. Mol. Cell. Neurosci. 63, 124–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lian L.-Y., Pandalaneni S. R., Todd P. A., Martin V. M., Burgoyne R. D., and Haynes L. P. (2014) Demonstration of binding of neuronal calcium sensor-1 to the cav2.1 p/q-type calcium channel. Biochemistry 53, 6052–6062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hui K., Fei G.-H., Saab B. J., Su J., Roder J. C., and Feng Z.-P. (2007) Neuronal calcium sensor-1 modulation of optimal calcium level for neurite outgrowth. Development 134, 4479–4489 [DOI] [PubMed] [Google Scholar]
- 57.Iketani M., Imaizumi C., Nakamura F., Jeromin A., Mikoshiba K., Goshima Y., and Takei K. (2009) Regulation of neurite outgrowth mediated by neuronal calcium sensor-1 and inositol 1,4,5-trisphosphate receptor in nerve growth cones. Neuroscience 161, 743–752 [DOI] [PubMed] [Google Scholar]
- 58.Dason J. S., Romero-Pozuelo J., Atwood H. L., and Ferrús A. (2012) Multiple roles for frequenin/NCS-1 in synaptic function and development. Mol. Neurobiol. 45, 388–402 [DOI] [PubMed] [Google Scholar]
- 59.McFerran B. W., Graham M. E., and Burgoyne R. D. (1998) Neuronal Ca2+ sensor 1, the mammalian homologue of frequenin, is expressed in chromaffin and PC12 cells and regulates neurosecretion from dense-core granules. J. Biol. Chem. 273, 22768–22772 [DOI] [PubMed] [Google Scholar]
- 60.Scalettar B. A., Rosa P., Taverna E., Francolini M., Tsuboi T., Terakawa S., Koizumi S., Roder J., and Jeromin A. (2002) Neuronal calcium sensor-1 binds to regulated secretory organelles and functions in basal and stimulated exocytosis in PC12 cells. J. Cell Sci. 115, 2399–2412 [DOI] [PubMed] [Google Scholar]
- 61.Pan C.-Y., Jeromin A., Lundstrom K., Yoo S. H., Roder J., and Fox A. P. (2002) Alterations in exocytosis induced by neuronal Ca2+ sensor-1 in bovine chromaffin cells. J. Neurosci. 22, 2427–2433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rajebhosale M., Greenwood S., Vidugiriene J., Jeromin A., and Hilfiker S. (2003) Phosphatidylinositol 4-OH kinase is a downstream target of neuronal calcium sensor-1 in enhancing exocytosis in neuroendocrine cells. J. Biol. Chem. 278, 6075–6084 [DOI] [PubMed] [Google Scholar]
- 63.Zheng Q., Bobich J. A., Vidugiriene J., McFadden S. C., Thomas F., Roder J., and Jeromin A. (2005) Neuronal calcium sensor-1 facilitates neuronal exocytosis through phosphatidylinositol 4-kinase. J. Neurochem. 92, 442–451 [DOI] [PubMed] [Google Scholar]
- 64.Gromada J., Bark C., Smidt K., Efanov A. M., Janson J., Mandic S. A., Webb D.-L., Zhang W., Meister B., Jeromin A., and Berggren P.-O. (2005) Neuronal calcium sensor-1 potentiates glucose-dependent exocytosis in pancreatic beta cells through activation of phosphatidylinositol 4-kinase β. Proc. Natl. Acad. Sci. U.S.A. 102, 10303–10308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de Barry J., Janoshazi A., Dupont J. L., Procksch O., Chasserot-Golaz S., Jeromin A., and Vitale N. (2006) Functional implication of neuronal calcium sensor-1 and phosphoinositol 4-kinase-β interaction in regulated exocytosis of PC12 cells. J. Biol. Chem. 281, 18098–18111 [DOI] [PubMed] [Google Scholar]
- 66.D'Angelo S., Kumar S., Naranjo L., Ferrara F., Kiss C., and Bradbury A. R. (2014) From deep sequencing to actual clones. Protein Eng. Des. Sel. 27, 301–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Blarcom T., Rossi A., Foletti D., Sundar P., Pitts S., Bee C., Melton Witt J., Melton Z., Hasa-Moreno A., Shaughnessy L., Telman D., Zhao L., Cheung W. L., Berka J., Zhai W., Strop P., Chaparro-Riggers J., Shelton D. L., Pons J., and Rajpal A. (2015) Precise and efficient antibody epitope determination through library design, yeast display and next-generation sequencing. J. Mol. Biol. 427, 1513–1534 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.