Abstract
Tobacco mature pollen has extremely desiccated cytoplasm, and is metabolically quiescent. Upon re-hydration it becomes metabolically active and that results in later emergence of rapidly growing pollen tube. These changes in cytoplasm hydration and metabolic activity are accompanied by protein phosphorylation. In this study, we subjected mature pollen, 5-min-activated pollen, and 30-min-activated pollen to TCA/acetone protein extraction, trypsin digestion and phosphopeptide enrichment by titanium dioxide. The enriched fraction was subjected to nLC-MS/MS. We identified 471 phosphopeptides that carried 432 phosphorylation sites, position of which was exactly matched by mass spectrometry. These 471 phosphopeptides were assigned to 301 phosphoproteins, because some proteins carried more phosphorylation sites. Of the 13 functional groups, the majority of proteins were put into these categories: transcription, protein synthesis, protein destination and storage, and signal transduction. Many proteins were of unknown function, reflecting the fact that male gametophyte contains many specific proteins that have not been fully functionally annotated. The quantitative data highlighted the dynamics of protein phosphorylation during pollen activation; the identified phosphopeptides were divided into seven groups based on the regulatory trends. The major group comprised mature pollen-specific phosphopeptides that were dephosphorylated during pollen activation. Several phosphopeptides representing the same phosphoprotein had different regulation, which pinpointed the complexity of protein phosphorylation and its clear functional context. Collectively, we showed the first phosphoproteomics data on activated pollen where the position of phosphorylation sites was clearly demonstrated and regulatory kinetics was resolved.
Tobacco mature pollen represents an extremely resistant structure filled with a desiccated cytoplasm that is surrounded by an extremely tough cell wall. This metabolically quiescent stage of male gametophyte has to reach stigma tissue in a viable state. After pollination, the rehydration and metabolic activation of a pollen grain starts. The pollen activation is represented by a time period when there is no pollen tube growth, and only metabolic processes within the original volume of cytoplasm occur together with cytoplasm hydration (1). Within this period, the pollen aperture later used for pollen tube outgrowth is selected. After that, a rapid pollen tube tip growth starts in order to deliver the genetic information carried by two sperm cells to the ovaries. Desiccated mature pollen of many angiosperm species can be also rehydrated and activated in vitro (2). Here we aim to elucidate the regulation processes of pollen grain re-hydration and activation mediated by protein phosphorylation.
Protein phosphorylation, representing one of the most frequent regulatory mechanisms, was shown to control a number of cellular processes, such as signal transduction, regulation of transcription and translation, regulation of cytoskeleton dynamics, cell cycle regulation, metabolism regulation, regulation of protein stability, and protein targeting (3–5). Similar to pollen activation, the rehydration of African xerophyte Craterostigma plantagineum was accompanied by changes in protein phosphorylation (6). Attachment of a phosphate group to the polypeptide chain shifts the pI of a protein to more acidic range (7). Such pI shift usually causes changes of protein conformation within a single domain (8) or even influences domain-domain interactions (9). In case of enzymes, phosphorylation sometimes inhibits activity by occupying the active site of the protein, as was documented for instance for isocitrate dehydrogenase (10).
In order to be able to identify phosphorylated proteins, it is inevitable to apply some of the various enrichment protocols (11, 12) because of several reasons: (i) Phosphoproteins are mostly low abundant so they are overwhelmed by the excess of nonphosphorylated proteins. (ii) A given protein is expressed in many copies and contains many potential phosphorylation sites (Ser/Thr/Tyr residues), but individual phosphorylation sites are usually only partly phosphorylated— i.e. only a small share of the present protein molecules will be phosphorylated at a given position whereas the majority will be nonphosphorylated. (iii) The identification of phosphopeptides by mass spectrometry is still challenging from the technical point of view. The enrichment can be performed at two levels. The first possibility is to fish the intact phosphoproteins out of a protein mixture whereas the second approach relies on the enrichment of phosphorylated peptides of the protease-digested protein sample. A plethora of protocols are meanwhile available for both approaches, whereas for both advantages as well as disadvantages have been reported (11). In order to broaden the phosphoproteome coverage, a tandem procedure applying first the former approach and then after protease cleavage also the latter one was suggested (13, 14).
The first angiosperm pollen phosphoproteome published was that of Arabidopsis thaliana (15), which completed the pollen proteomic data because before that, three Arabidopsis pollen proteomic data sets achieved by the conventional in-gel approach (16–18) and one high-throughput proteomic study (19) were published. Mayank and colleagues identified many phosphopeptides, notable number of which played their roles in regulation of metabolism and protein function, metabolism, protein fate, binding other proteins, signal transduction, and cellular transport. Many kinases were identified in the data set, showing that these were indeed subject to phosphorylation, for instance AGC protein kinases, calcium-dependent protein kinases, and sucrose non-fermenting 1-related protein kinases (15).
The tobacco pollen proteome was studied directly by a high-throughput approach but appeared only recently (20). In this study, Ischebeck and colleagues compared the proteome of eight male gametophyte stages ranging from diploid microsporocytes to pollen tubes. Interestingly, the first tobacco pollen phosphoproteomic paper appeared earlier than the whole proteome was published (21). In order to identify phosphoproteins in tobacco mature pollen and pollen activated in vitro for 30 min, metal oxide/hydroxide affinity chromatography phosphoprotein enrichment employing an aluminum hydroxide matrix (Al(OH)3) was carried out (22). This approach led to the identification of only one phosphorylation site, so that additionally titanium dioxide (TiO2)1 enrichment was applied, identifying 51 more phosphorylation sites in the already-identified proteins from mature pollen. Among those proteins were for instance various translation initiation and elongation factors, metabolic proteins (for instance fructose bisphosphate-aldolase, glyceraldehyde-3-phosphate dehydrogenase, and alcohol dehydrogenase), Rho guanine nucleotide dissociation inhibitor, and several ribosomal proteins. However, not many signaling proteins were identified in this study. The third male gametophyte phosphoproteome revealed to date was that of a gymnosperm Picea wilsonii. However, the proteome of this species was studied from the perspective of deficient growth media, and several phosphoproteins linked to Ca2+ and sucrose deficiency were identified (23).
The present study is a continuation of our male gametophyte phosphoproteomic studies. Herein, we employed phosphopeptide enrichment by metal oxide/hydroxide affinity chromatography with TiO2 matrix (24) on three stages of male gametophyte, this time including two stages of activated pollen (5 min and 30 min) as well as mature pollen. Collectively, 471 phosphopeptides carrying 432 phosphorylation sites (phosphoRS probabilities >90%) have been identified in the three stages of male gametophyte. These phosphorylation sites belonged to 301 phosphoproteins that were classified into 13 functional categories; with transcription, protein synthesis, destination and storage, as well as signal transduction being the dominant functional groups. A phosphorylation motif search revealed 5 motifs with a central phosphoserine and one motif with a central phosphothreonine. Quantitative data led to the discovery of regulated phosphopeptides, which were grouped into seven categories based on their regulatory trends throughout the studied developmental stages.
EXPERIMENTAL PROCEDURES
Plant Material and Pollen Activation In Vitro
Tobacco plants (Nicotiana tabacum cv. Samsun) were grown in a greenhouse from April to September. Flower buds shortly before anthesis were collected between June and September. Anthers were removed from the buds and let dehisce at room temperature on a filtration paper overnight. Then, mature pollen was sieved by a stocking and stored at −20 °C (25) until it was further used. The collected pollen represented bulk samples originating from three groups of 15 plants that were grown in separate parts of the greenhouse. These bulk samples were further referred to as the three biological replicates.
Mature pollen was activated in vitro as a shaken suspension for 5 min, and 30 min, respectively, each stage in three biological replicates as mentioned above, at 27 °C in sucrose-mineral medium buffered with MES (SMM-MES; 175 mm sucrose, 1.6 mm boric acid, 3 mm Ca(NO3)2·4H2O, 0.8 mm MgSO4·H2O, 1 mm KNO3, 25 mm MES, pH 5.9) (26). The activated pollen was then harvested by filtration on a vacuum pump-driven apparatus, and immediately frozen in liquid nitrogen. The three stages differ from each other as follows: mature pollen represents an oval-shaped structure with a desiccated cytoplasm. Upon re-hydration, 5-min activated pollen becomes round-shaped with a hydrated cytoplasm. Furthermore, one pollen aperture is usually chosen for pollen tube outgrowth after 30-min imbibition (see supplemental Fig. S1).
Protein Extraction and Phosphopeptide Enrichment
The total proteins were extracted from all the above stages by TCA/acetone precipitation (27) with slight modifications (21) in three biological replicates as mentioned above (see Fig. 1 for workflow overview). In detail, mature or activated pollen was homogenized by a pestle in a mortar. The acquired fine powder was resuspended in 10 volumes of 10% w/v TCA in acetone supplemented with 1% w/v DTT. After 5 min sonication in an ultrasonic bath, the samples were briefly frozen in liquid nitrogen, incubated at −20 °C for 45 min, and centrifuged (23,000 × g, 15 min, 4 °C). After the removal of the supernatant, the samples were washed by 1.5 ml acetone with 1% w/v DTT, sonicated for 5 min, briefly frozen in liquid nitrogen, and kept at −20 °C for 30 min. After the centrifugation under the above conditions, the washing step was repeated. Finally, the pellet was dried and stored at −20 °C.
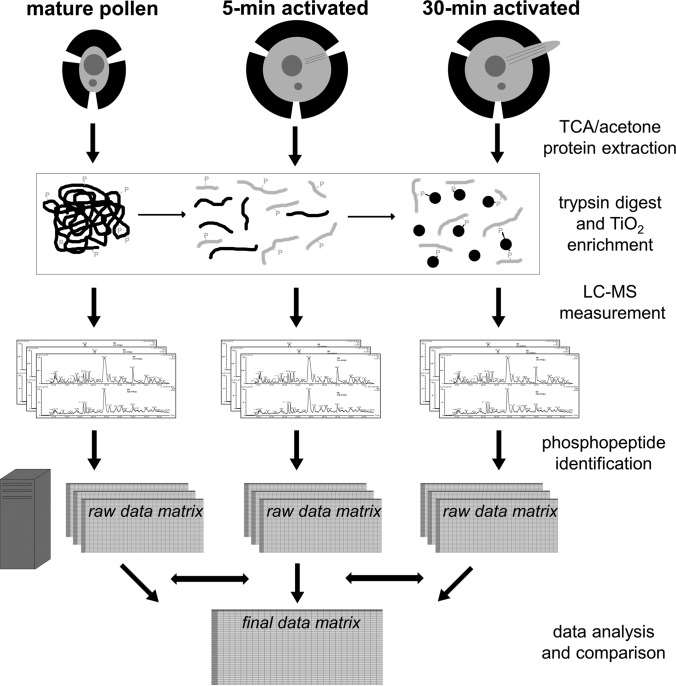
Fig. 1.
A schematic workflow of the performed experiments. The three stages of tobacco male gametophyte (particularly mature pollen, pollen activated in vitro for 5 min and pollen activated in vitro for 30 min) were subjected to TCA/acetone protein extraction, trypsin digest and phosphopeptide enrichment by TiO2. The obtained phosphopeptide-enriched eluate was fractionated by nLC and measured by MS/MS. The present phosphopeptides were identified (if possible with the unambiguous position of the phosphosite) and the results further analyzed.
The total protein extracts from all stages were resuspended in 0.2 m guanidinium chloride and 50 mm ammonium bicarbonate supplemented with PhosStop phosphatase inhibitor mixture (Roche, Penzberg, Germany), carbamidomethylated as described elsewhere (28) and subsequently trypsin-digested (trypsin-to-protein ratio 1:50; 37 °C, 12 h).
There were three biological replicates for each studied stage. For each of the triplicates, 500 μg peptides were dissolved in loading buffer (80% v/v ACN, 6% v/v TFA, saturated with phthalic acid) and subjected to phosphopeptide enrichment by TiO2. Seven synthetic peptides were spiked to the peptide mixture in order to check the reproducibility of the replicates. The phosphopeptides bound to TiO2 beads were washed and eluted as described previously (21, 29).
nLC-MS/MS Measurement and Phosphopeptide Identification
The phosphopeptide-enriched samples were analyzed by nLC-MS/MS on an LTQ Orbitrap Elite (Thermo Fisher Scientific, Bremen, Germany) mass spectrometer coupled to an Ultimate 3000 nLC (Thermo Fisher Scientific). Peptides were pre-concentrated on a self-packed Synergi HydroRP trapping column (100 μm ID,4 μm particle size, 10 nm pore size, 2 cm length) and separated on a self-packed Synergi HydroRP main column (75 μm ID, 2.5 μm particle size, 10 nm pore size, 30 cm length) at 60 °C and a flow rate of 270 nl·min−1 using a binary gradient (A: 0.1% formic acid, B: 0.1% formic acid, 84% ACN) ranging from 3% to 45% B in 240 min.
MS survey scans were acquired from 350–2000 m/z in the Orbitrap at a resolution of 60,000 using the polysiloxane m/z 445.120030 as lock mass. The ten most intense ions were subjected to collision-induced dissociation and MS/MS using normalized collision energy of 35% and an activation time of 30 ms and MS/MS were acquired in the LTQ. AGC values were set to 106 for MS and 104 for MS/MS scans.
The acquired spectra were searched against the TIGR EST sequence database for Tobacco (ftp://occams.dfci.harvard.edu/pub/bio/tgi/data/; release version 10/04/2011, 48961 entries) using Proteome Discoverer 1.3 with Mascot. Quantification, false discovery assessment and phosphorylation site localization were performed using the following nodes: Precursor Ions Area Detector, Peptide Validator, and phoshoRS (30). Searches were conducted with the following settings: 10 ppm MS tolerance, 0.5 Da MS/MS tolerance, trypsin as a cleaving enzyme with max. two missed cleavage sites, carbamidomethylation (Cys) as fixed, and oxidation (Met) together with phosphorylation (Ser, Thr, Tyr) as variable modifications. Finally, the results were subjected to the filtering criteria of mass deviation ≤ 4 ppm and high confidence (corresponding to a false discovery rate <1% on the peptide-spectrum match level). The standard deviation of the peak areas of the synthetic peptides was below 25% so the results were considered reproducible. Peak areas were considered per peptide, i.e. different charge states were combined. Of all identified phosphopeptides, only the ones that showed a standard deviation <30% of the abundance between the biological replicates of the same stage, and that were identified in all of the replicates were listed in the result tables. Moreover, only phosphopeptides with an unambiguously assigned phosphorylation site with a probability higher than 90% (phoshoRS) were considered. All raw data and search results have been deposited in proteomeXchange (31) with the accession PXD003042.
nLC-MS/MS of the Trypsinized Crude Protein Extract
For each sample ∼1 μg of the trypsin digest was analyzed by nLC-MS/MS prior to TiO2 enrichment, using the same conditions as above. Data analysis was also conducted as above, however, omitting phosphorylation as variable modification. Only proteins meeting the following criteria were quantified: (1) at least 2 unique peptides quantified in at least 2 out of 3 biological replicates, (2) for all conditions standard deviations between biological replicates had to be <40%. Proteins that differed among any of two studied stages at least twofold in abundance were considered as regulated.
Protein Categories and Motif Search
The gene ontology (GO) and enzyme codes were originally acquired by Blast2GO ver 2.7.2 (https://www.blast2go.com); the identified tobacco ESTs translated in the longest reading frame were searched against the Arabidopsis proteome. For many of the sequences, the GO terms (divided into three groups: molecular function, biological process, and cellular compartment) together with the EC enzyme codes were assigned according to the homologous Arabidopsis sequences. However, some of the tobacco sequences lacked their Arabidopsis homologue in the proteome database and/or the gene ontology was not informative enough. So finally, the acquired GO terms were manually converted to protein categories and subcategories according to Bevan et al. (32) to enable better categorization of the data. In case a protein had more functions, it was catalogued according to the prevailing function.
All unambiguous phosphopeptides (supplemental Table S2) were analyzed for the significant phosphorylation motifs by Motif-X software (33, 34). Two searches were performed, one looking up phosphorylated serine and the other one searching for phosphorylated threonine (phosphotyrosine motifs were not searched because there was only one phosphorylated tyrosine in the phosphopeptide data set). The width of a phosphorylation motif was set to 13 (where the phosphoamino acid was placed into the central position), number of occurrences to 15, and significance score to 0.000001. As a background, data set of tobacco Uniprot sequences was uploaded.
The regulated phosphopeptides were manually divided into seven categories according to their regulatory trends. The motif search was not performed on the regulated phosphopeptide data set because it contained only a limited number of phosphopeptides. The graphical representation of the peptide abundances in the various stages was performed by the VANTED software package (http://www.vanted.org, ref. 35).
RESULTS
Phosphopeptide Enrichment and Identification
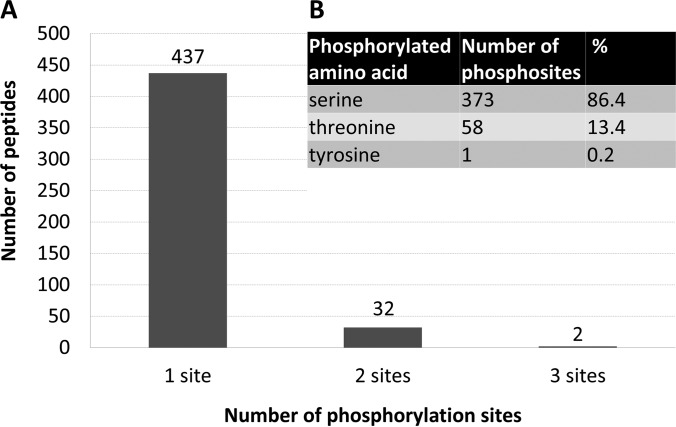
In this study, 471 phosphopeptides were identified with an unambiguously assigned position of the phosphorylation site (supplemental Tables S1 and S2). The vast majority of the identified phosphopeptides was singly phosphorylated (437), whereas only a minority was doubly (32) or triply phosphorylated (2), see Fig. 2A. These 471 identified phosphopeptides contained collectively 432 unique unambiguous phosphorylation sites. The number of unique phosphorylation sites is lower than the number of phosphosites identified in all phosphopeptides because some of the identified phosphorylation sites were redundant. Such a redundancy was observed for instance in case of couples of peptides, where one of which was completely cleaved whereas the other carried one missed-cleaved trypsin site (e.g. represented by the peptides KQLVSVAS*AVK and QLVSVAS*AVK from adenine nucleotide α hydrolases-like protein or peptides S*WDDADLK and S*WDDADLKLPGK from eukaryotic translation initiation factor 5B-like protein; an asterisk represents the phosphorylation site) or alternatively in case of two peptides, one of which was oxidized on a methionine whereas the other was not modified in that way (e.g. peptides KENVGPMVNLENPTS*PK and KENVGPmVNLENPTS*PK from low-temperature-induced 65 kDa protein or peptides EES*DDDMGFSLFD and EES*DDDmGFSLFD from acidic ribosomal protein P1a-like protein; an asterisk represents phosphorylation site, and lowercase “m” represents an oxidized methionine).
Fig. 2.
The statistics of identified phosphopeptides. A, Column diagram showing the number of phosphopeptides according to the number of unambiguously identified phosphorylation sites in a single peptide. If a phosphorylation site was identified in more than one peptide, it was counted repeatedly every time on every peptide. B, Number of phosphorylation sites according to the phosphorylated amino acid (serine, threonine, or tyrosine) identified. If a phosphorylation site was identified in more than one peptide, it was counted only once.
Because conventional phosphoproteomic workflows were applied, only O-phosphorylated amino acids were identified, particularly serine, threonine, and tyrosine (Fig. 2B). The dominant phosphorylated amino acid was phosphoserine with 373 phosphorylation sites (86.4%), followed by phosphothreonine represented by 58 phosphorylation sites (13.4%). Only one phosphorylation site (corresponding to 0.2%) was detected on a tyrosine making it the rarest phosphorylated amino acid in the data set.
Identified Phosphoprotein Categories
The 471 identified phosphopeptides revealing 432 unique phosphorylation sites were assigned to 301 proteins as several proteins contained more than one phosphorylation site. A protein was defined throughout the article as a sequence identified either with a single accession number or with a unique combination of accession numbers. The combination of accession numbers was applied in case of one peptide being assigned to two or more identifiers, e.g. the couple NT_TC85822_1 and NT_TC87771_1 or the pair NT_TC82971_1 and NT_TC77872_1). Also, some accession numbers were assigned to more than one peptide, either as an exclusive number (e.g. NT_TC95936_1 or NT_TC83486_1), or in combination with another accession (e.g. NT_TC95936_1 and NT_FG166442_1, or NT_TC83486_1 and NT_FG175056_1). Thus, a row in supplemental Table S1 represents a single protein; the phosphopeptides belonging to one phosphoprotein are put together into one cell. The proteins were annotated according to the original TIGR protein descriptions. However, many of these annotations were not explanatory enough so in case of some phosphoproteins, the annotation was improved using the homologues found by tblastx in the GenBank database (http://blast.ncbi.nlm.nih.gov).
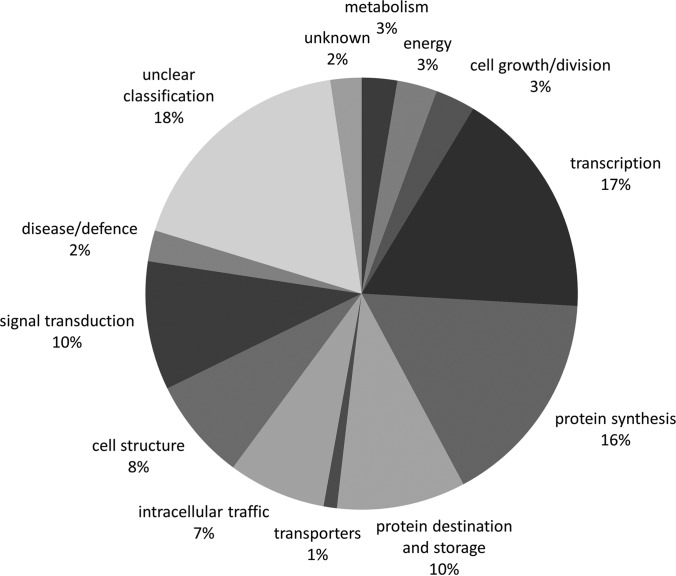
The annotated proteins were sorted according to their prevailing function. The GO search was performed by blast2GO software (https://www.blast2go.com). Because the obtained results did not allow an easy categorization according to protein function, the gene ontology assignment was further performed manually into the categories according to Bevan et al. (32). Every protein was catalogued into just one category. In case one protein had more distinct functions, it was sorted into the category with the dominant function. The difference between “unclear classification” and “unknown” was as follows: the proteins with a known homologue and/or annotation (characterizing them only to some extent) with an unclear function were catalogued as “unclear classification” whereas the proteins without a known homologue and/or functional annotation were classified as “unknown.” The protein categories are summarized by a pie chart in Fig. 3. The main category cataloguing almost one fifth of the phosphoproteins was represented by species with “unclear classification.” Over one quarter of proteins (falling into two separate categories, protein synthesis, and protein destination and storage) was connected with translation. More than 10% belonged also to transcription (17%), and exactly 10% to signal transduction. Cell structure and intracellular traffic reached 8% or 7%, respectively. The other categories were represented only by a few percent: metabolism, energy, cell growth/division, disease/defense, unknown, and transporters. For enzymes, the EC numbers are given in supplemental Table S2.
Fig. 3.
Pie chart showing the percentage of the individual phosphoprotein categories.
Motif Analysis
Protein phosphorylation occurs usually on particular short amino acid motifs rather than on random sequences. Some of these motifs can be kinase-specific so their knowledge can reveal cellular regulatory networks in more detail. The dominant phosphorylation motifs in our data set compared with the random background based on the tobacco sequences from Uniprot protein (http://www.uniprot.org) database were identified by Motif-X software (supplemental Fig. S2). Two independent searches were performed - one focused on phosphoserine motifs and the other one looking up phosphothreonine motifs. The phosphotyrosine was not subjected to this analysis because only one phosphorylated tyrosine was present in the entire data set. The phosphorylation motifs had to be found at least 15 times in the experimental data set to be considered. The most abundant phosphoserine motif and the only phosphothreonine motif were represented by the phosphorylation site that was followed by a prolin: xxxxxxS*Pxxxxx, and xxxxxxT*Pxxxxx (where phosphorylation site is marked by an asterisk and the position that can be occupied by any amino acid is shown as “x”). The proline motif with a serine was detected 118 times, whereas proline following a threonine was found only 31 times. The remaining phosphoserine motifs were two basic and two acidic ones. The basic motifs were represented by a phosphorylated serine, preceded by a lysine or an arginine followed by any two amino acids. In particular, the motif xxxRxxS*xxxxxx was detected 37 times, whereas the slightly less abundant xxxKxxS*xxxxxx was carried by 30 phosphorylated peptides. The acidic motifs were composed of a serine followed either by a glutamic acid, one any amino acid, and a glutamic acid or by one any amino acid with two glutamic acids. The motif xxxxxxS*DxExxx was found 23 times whereas the second acidic motif xxxxxxS*xDDxxx was present in 15 phosphopeptides (supplemental Fig. S2). The kinase families that usually recognize such motifs are referred to more in detail in the discussion.
Regulated Phosphopeptides
In order to determine whether substantial changes on the level of protein expression occurred between the different time points, additional nLC-MS/MS measurements were performed on the complex peptide mixture without prior TiO2 enrichment.
The concentration of the nonphosphorylated peptides from this analysis served as a reference (protein abundance), and the abundance of the phosphopeptides was compared with this reference. Some of the phosphorylated peptides changed their concentration in accordance with the abundance changes of the whole protein. The global abundance ratios of these phosphoproteins are shown in supplemental Table S3 in red. The concentration of such phosphorylated peptides changed likely because of the synthesis or degradation of the whole proteins rather than as a consequence of the sole phosphorylation or dephosphorylation. On the other hand, other phosphorylated peptides did not reflect the concentration changes of the corresponding proteins and showed either opposite abundance change or showed a changed abundance exclusively at the phosphopeptide level (and not on the level of the whole protein). The concentration ratio of such proteins is shown in supplemental Table S3 in black. Such changes in phosphopeptide abundance that were not reflected by the concentration of the whole protein are likely to be caused exclusively by protein phosphorylation or dephosphorylation processes.
Because proteins were quantified based on at least two unique peptides leading to a reduced precision compared with single peptide-based phosphopeptide quantification, here the maximum standard deviation allowed among biological replicates was 40%, compared to 30% for phosphopeptides. Moreover, because of the increased complexity of the samples the identification in two out of three candidates was considered sufficient.
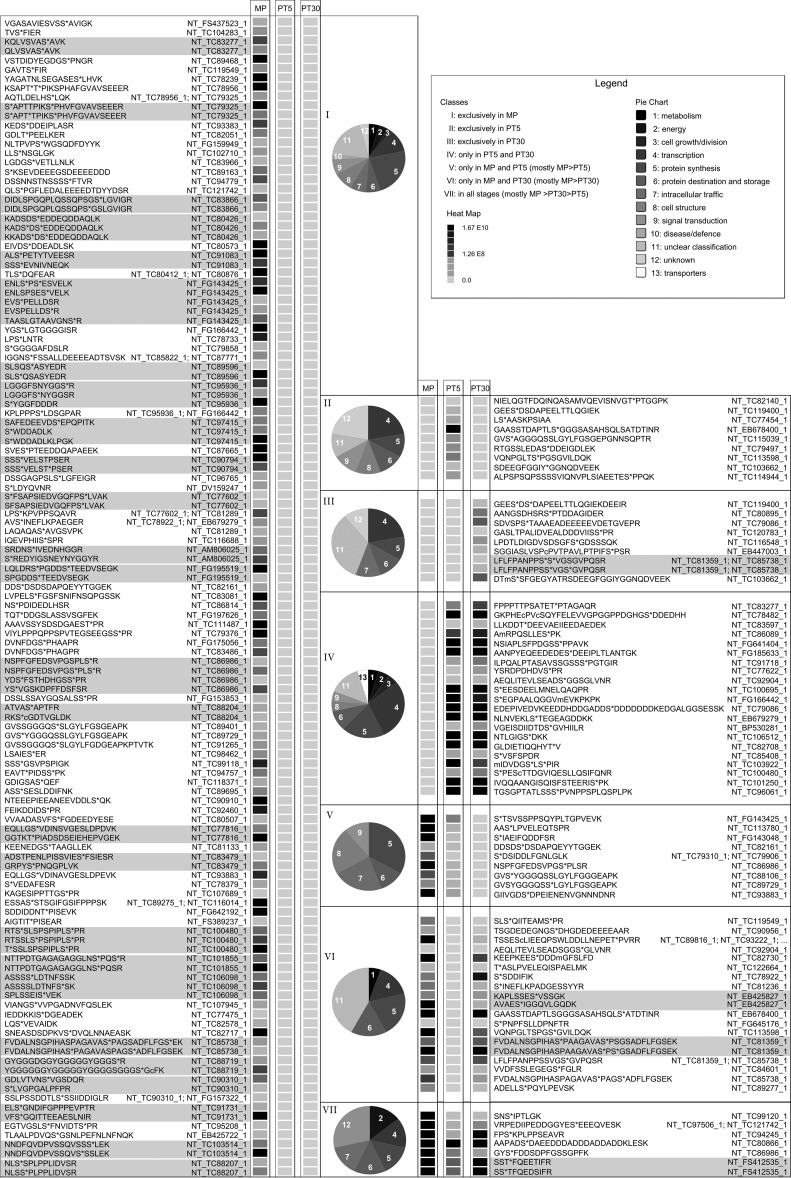
The proteins considered to be of a different abundance had to show twofold difference between at least two stages. If we consider these proteins and the phosphopeptides that belonged to them, we counted 209 phosphopeptides, which were sorted into seven regulatory groups (see Fig. 4 and supplemental Fig. S3). The first three groups presented phosphopeptides that were identified exclusively in one of the three studied stages. The highest number of phosphorylated peptides fell into the category unique for mature pollen—135 phosphopeptides (group I). Nine phosphopeptides were identified exclusively in both 5-min (group II), and 30-min activated pollen (group III). The other three groups contained phosphorylated peptides that were detected in two stages out of three. Twenty-one common phosphorylated peptides were detected in 5-min and 30-min activated pollen (group IV), and 19 common phosphopeptides were detected in mature pollen and 30-min activated pollen (group VI). Only nine phosphopeptides fell into the group that was missing in 30-min activated pollen (group V). The last regulation group was represented by the phosphopeptides common to all three stages, represented by seven phosphopeptides (group VII).
Fig. 4.
Expression profiles of the selected phosphopeptides with a different abundance in the studied male gametophyte stages. The phosphopeptides were sorted into seven regulation groups based on their abundance differences in the three analyzed male gametophyte stages (group I - left panel; groups II-VII - right panel). The relative peptide abundance in each group is shown based on a gray scale (light gray - not detected; black - the highest concentration). Each column represents the average peptide abundance of the three independent LC-MS experiments. In the rows, the normalized abundance of peptides as extracted from the Proteome discoverer LC-MS software is presented. Peptides assigned to one and the same identifier are highlighted in gray. Gene ontology (GO) categories are presented for each group as a pie chart. The full presentation of the data set is provided in the supplemental Table S3.
The main protein categories where the regulated phosphopeptides belong to were transcription, translation, and protein synthesis and storage (please refer to the pie charts in Fig. 4). These categories collectively accounted for one third to one half of phosphopeptides in the respective regulation group. Quite common were also phosphopeptides with “unclear classification” that accounted for about a quarter of the phosphopeptides in the regulatory groups exclusive to any of the stages (group I, II, and III), and in the group IV with the common regulation to the 5-min and the 30-min activated stage. Furthermore, it represented almost a half in the group VI (i.e. peptides that were absent from 5-min activated pollen). In the regulation group VII (common to all studied stages), the functional category “unclear classification” was supplemented with “unknown,” which represented over a quarter of identified phosphopeptides.
Group I, and group V represented the phosphopeptides that were phosphorylated in mature pollen, and then were dephosphorylated upon pollen activation. Exclusive phosphosites in mature pollen, concentration changes of which were not reflected by changes on the protein level were represented for instance by eukaryotic initiation factor 4B, various RNA binding proteins, mini zinc finger protein, C2 domain-containing protein, ubiquitin-activating enzyme 2, vesicle-associated protein 25, MODIFIER OF SNC1 (SUPRESSOR OF NPR1–1, CONSTITUTIVE 1) 1-like protein, and a variety of proteins with unclear classification, such as muscle M-line assembly protein UNCOORDINATED-89-like (UNC-89-like), dentin sialophosphoprotein-like protein, and glycine-rich protein 2. The phosphosites that were shared by 5-min activated pollen and mature pollen were found for example in eukaryotic translation initiation factor 4γ-like protein, nucleic acid binding protein, UBA (ubiquitin associating) and UBX (ubiquitin-like) domain-containing protein At4g15410-like, auxilin-related protein 2-like, and pollen tube Rho guanine nucleotide dissociation inhibitor 2 (Rho GDI2). The groups II, III, and IV were composed of phosphorylation sites that appeared only upon pollen activation. There were detected for instance zinc finger CCCH domain-containing protein 31-like protein, ribosomal protein S6-like, protein phosphatase inhibitor 2-like protein, serine/arginine-rich splicing factor RS2Z32-like, E3 ubiquitin-protein ligase RING FINGER PROTEIN 4 (RNF4)-like, WD repeat-containing protein 24 homolog, cytochrome c oxidase subunit 5b-1 protein, methyl-CpG-binding domain 10 protein, histone deacetylase 1 (HDT 1). The most dynamic regulatory trend was documented by group VI, peptides of which showed phosphorylation in mature pollen, then temporary dephosphorylation immediately upon pollen activation and a re-phosphorylation later during pollen activation (30 min). Such a dynamic regulation was detected in these phosphoproteins: phospholipase A2/esterase, bZIP transcription factor bZIP100, acidic ribosomal protein P1a-like protein, late embryogenesis abundant (LEA) proteins, RNA binding proteins and transcription initiation factor IIF subunit α-like protein. Finally, group VII collected the proteins that were present in all stages and showed significant abundance changes throughout the development. These species were for example represented by serine/threonine-protein kinase DST2-like, calreticulin precursor, 2-phosphoglycerate kinase-related family protein, and RNA polymerase-associated protein LEO1-like.
Multiple Phosphorylation
A single protein can carry several phosphorylation sites that show different regulatory trends (36, 37). Examples of such proteins identified in our study were actin cytoskeleton-regulatory complex PAN1-like protein, and octicosapeptide/PHOX/BEM1p-domain-containing protein (PB1-containing protein). The former is characterized by six phosphorylation sites that showed three regulatory trends (Fig. 5A, and supplemental Table S4). The phosphopeptides NSPFGFEDSVPGS*PLS*R and NSPFGFEDSVPGSPLS*R were identified exclusively in mature pollen whereas the phosphopeptide NSPFGFEDSVPGS*PLSR was present in mature pollen and 5-min activated pollen. We can speculate that the first serine became phosphorylated in mature pollen, peaked in 5-min activated pollen, and in 30-min activated pollen remained undetectable. On the other hand, the second serine dominated in mature pollen whereas later on was undetectable. Furthermore, it is likely that in mature pollen both phosphorylation forms (a singly and a doubly phosphorylated) coexisted possibly each showing a different regulatory activity. The latter, PB1-containing protein is characterized by seven phosphorylation sites showing three regulatory trends (Fig. 5B, and supplemental Table S4). The phosphopeptides FVDALNSGPIHASPAGAVAS*PAGSADFLFGS*EK, and FVDALNSGPIHAS*PAGAVASPAGS*ADFLFGSEK coexisted exclusively in mature pollen, whereas the phosphopeptide FVDALNSGPIHASPAGAVAS*PAGS*ADFLFGSEK was present only in mature pollen and 30-min activated pollen. It is likely that phosphorylation of all these four serines occurs in mature pollen, it vanishes in 5-min activated pollen, and that two of the phosphorylation sites re-appear after 30 min of pollen activation. The dephosphorylation after 5 min of activation might be directly related to pollen activation/hydration. On the other hand, the phosphopeptides LFLFPANPPS*S*VGSGVPQSR, and LFLFPANPPSS*VGS*GVPQSR were identified exclusively in 30-min activated pollen, whereas the phosphopeptide LFLFPANPPSSVGS*GVPQSR was found also in mature pollen. Possibly, the third phosphorylated serine appeared in mature pollen, vanished and re-appeared in 30-min activated pollen whereas the other two phosphoserines appeared only after 30 min of pollen activation. Collectively our results showed differential phosphorylation patterns for a large number of proteins likely involved in the early processes during tobacco pollen activation.
Fig. 5.
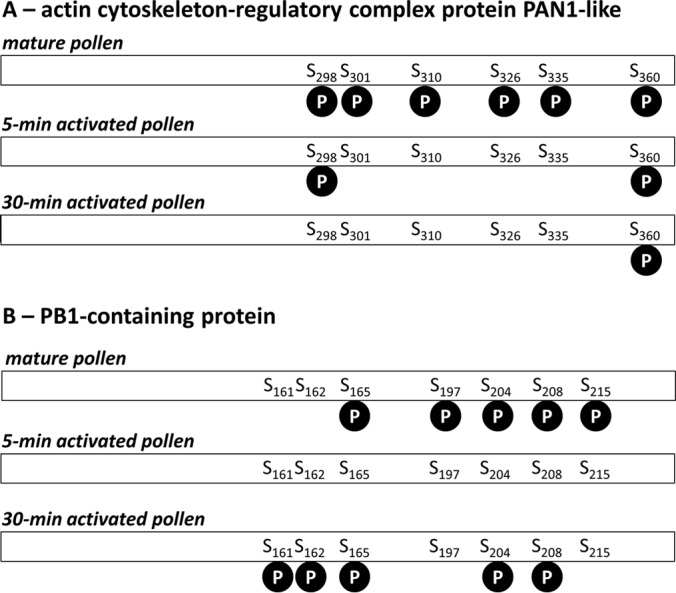
Phosphorylation patterns of two selected phosphoproteins identified in our data set in three stages of male gametophyte. A, Actin cytoskeleton-regulatory complex protein PAN1-like. B, PB1-containing protein. “S” stands for serine, the number indicates the position of the amino acid in the polypeptide chain, and phosphorylation site is depicted as a “P” in the black circle.
DISCUSSION
In the presented data set, 471 phosphopeptides have been identified in three stages of male gametophyte (mature pollen, pollen activated for 5 min and pollen activated for 30 min), which carried 432 unambiguous phosphorylation sites. The observed redundancy was caused by couples of phosphopeptides, one of which was “normal”, and the other one was either missed-cleaved by trypsin or came from chemical modifications, such as methionine oxidation. These 432 unique phosphorylation sites have been assigned to 301 individual proteins. The number of phosphorylation sites identified represents a great improvement in comparison to our previous tobacco male gametophyte phosphoproteomic study that identified 52 unambiguous phosphorylation sites (21). In that study we applied Al(OH)3- metal oxide/hydroxide affinity chromatography for phosphoprotein enrichment (allowing the annotation of only one phosphorylation site), and TiO2 phosphopeptide enrichment for the analysis of phosphorylation sites of selected candidate proteins from mature pollen. Such improvement in the number of phosphorylation sites after phosphopeptide enrichment in the actual study compared with the phosphoprotein enrichment in the previous study is in accordance with several previous studies where phosphoprotein enrichment revealed only a limited number of phosphorylation sites (6, 38, 39). Moreover, a tandem approach enriching first for phosphoproteins and then after trypsin digest also for phosphopeptides was shown beneficial (13, 14).
The Proportion of the Phosphorylated Amino Acids in the Presented Phosphoproteome
The conventional phosphoproteomic techniques lead to the identification of O-phosphorylated amino acids: serine, threonine, and tyrosine because phosphorylated histidine (that carries phosphate attached to a nitrogen atom in its imidazole ring) is labile under acidic pH (that is usually applied during the conventional enrichment protocols and during conventional LC-MS; ref. 40). In most phosphoproteomics studies, the dominant phosphoamino acid is serine with 80–90%, followed by threonine occupying around 10–15%, and tyrosine reaching few percent. In our study, we observed the pSer/pThr/pTyr ratio of 86.4:13.4:0.2 that was astonishingly similar to the Arabidopsis mature pollen phosphoproteome with a ratio of 86:14:0.16 (15). In case of various human cell cultures, around 2–4% of phosphotyrosine were reported—particularly 1.8% (41), 2.3% (42), or 3.8% (43). Usually, there was less phosphotyrosine (<1%) observed in plants than in human cell cultures, although the human phosphoproteomic research was often conducted on cancer cell lines that have a huge phosphorylation level. The pSer/pThr/pTyr ratios in various Arabidopsis cell cultures ranged from 91.8:7.5:0.7 (44) to 83.81:16.18:0.01 (45). On the contrary, other studies reported a phosphotyrosine content comparable to the animal phosphoproteomes, such as 85:10.7:4.3 (46), and 82.7:13.1:4.2 (47) in Arabidopsis cell cultures, and 84.8:12.3:2.9 in a rice cell culture (47). From the differing contents of phosphotyrosine in the presented data sets, it is obvious that it still remains speculative how abundant phosphotyrosine phosphorylation in plants actually is (48, 49). Furthermore, the inhibitors of tyrosine phosphorylation (phenylarsine oxide and genistein) applied to the lily cultivated pollen strongly affected its growth rate, likely influencing the dynamics of actin cytoskeleton (50). However, the exact position of phosphotyrosine phosphorylation in pollen proteins remains to be elucidated as well as any further possible role of tyrosine phosphorylation during pollen tube growth. Phosphotyrosine was shown to be carried by proteins playing an essential role throughout the life of a plant, such as brassinosteroid receptor BRI1 (51), or proteins involved in phytochrome signaling (52). The only phosphorylated tyrosine in our data set was identified in the peptide GVSY*GGGQSSLGYLFGGGEAPK of the SPIRAL1-like 1 protein.
Phosphoproteins with Unknown Function
Among the identified phosphoproteins, the dominant functional category was “unclear classification.” Collectively with “unknown,” it counted for one fifth of the identified phosphoproteome (Fig. 3). It is likely that some of these “unknown” proteins are pollen-specific or pollen-enriched compared with sporophyte tissues, and that their role is still unknown. In tobacco proteome, it was clearly shown that gametophytic tissues contained specific proteins (837 out of 2135 proteins) that were not shared by sporophyte tissues, particularly leaves and roots (or were at least not as abundant as in gametophyte, and so remained below the detection limit of the proteomic techniques; ref. 20). Out of these 837 proteins, 120 fell into the GO category “not assigned”, that represented approx. 14% of all pollen-specific proteins reported by Ischebeck and colleagues (20). From this point of view, our phosphoproteomic data set is consistent with the published tobacco male gametophyte proteome.
Phosphoproteins Involved in Translation and Protein Fate
Almost a quarter of the identified phosphoproteins have a likely role in translation; either in protein synthesis, or in protein destination and storage. Tobacco pollen activation and subsequent pollen tube growth was originally shown to be vitally dependent on translation but almost independent of transcription (53). Although our recent microarray transcriptomic analyses revealed a number of mRNAs being synthesized during pollen tube growth even after 24 h of cultivation (54, 55), many of the transcripts in the desiccated mature pollen are stored in EDTA/puromycine-resistant particles (EPPs). These particles contain parts of ribosomes and translation apparatus together with mRNAs (56, 57) and the translation of EPP-stored mRNAs starts after pollen activation. Translation initiation was shown to be regulated by protein phosphorylation of initiation factors and other regulatory proteins (5) so the presence of the translation initiation factors such as various forms of eukaryotic translation initiation factor 2, and eukaryotic translation initiation factors 3B, 4A-9, 4B, 4G, iso4F, 5B-like in our data set indicates ongoing translation regulation. The fate of proteins during cellular processes is also determined by their degradation via proteasome pathway, to which proteins labeled by polyubiquitine chain are subjected. Protein degradation is likely to have a key role during male gametophyte development. Recently, it was demonstrated that defective in cullin neddylation protein 1 (DCN1) was crucial for proper pollen tube development (58).
Phosphoproteins Role During Transcription
We detected a remarkable proportion of phosphoproteins involved in transcription (17% in particular). On the contrary, there was no phosphoprotein candidate connected to transcription in our previous phosphoprotein-enriched data set (21), probably because of their generally low abundance and the limited dynamic range of protein visualization techniques used. Such a fact was already demonstrated for Arabidopsis mitochondrial phosphoproteome where phosphopeptide enrichment led to the identification of novel phosphorylation sites that were not previously identified by the alternative approaches (38). As mentioned above, active transcription in activated pollen grain as long as 24 h of pollen tube growth has been shown (54, 55). Here, we identified several transcription factors, most of which contained a zinc finger motif. One of them, ZAT10, was shown to be phosphorylated by two mitogen-activated protein kinases (MAPK3 and MAPK6) (59). Interestingly, most of our zinc finger transcription factors showed also prolyl-directed phosphorylation motif making them likely substrates of MAP kinases (60). Some MAP kinases were already identified in tobacco male gametophyte (20, 61, 62). However, experimental data directly linking these MAP kinases to their targets have yet to be established.
Signaling Phosphoproteins
Compared with the data achieved before (21), herein, we identified almost a twofold number of proteins connected with signaling (10% in this study compared to 6% after phosphoprotein enrichment). Some of the signaling molecules are of a low abundance and therefore likely below the detection limits of the phosphoprotein enrichment. From our data the pollen-specific Rho guanine nucleotide dissociation inhibitors (Rho GDIs) should be mentioned. Small GTPases from the Rho family play an essential role in a polarized tip cell growth of pollen tubes, and their activity is regulated by other interacting proteins, including Rho GDI among others. Rho GDI removes the prenylated Rho GTPase from the membrane and helps to maintain the cytoplasmic pool of this protein. Its activity was shown to be essential for pollen tube growth (63). The other signaling proteins from our data set were various protein kinases and phosphatases. Their presence was expected because the precise regulation accompanying the switch from the metabolically quiescent pollen grain to the rapidly-growing pollen tube is likely to involve the activity of kinases and phosphatases, phosphorylation of which was shown to regulate their activity (64). Many of the identified kinases showed low homology to the known sequences in the database making the specification of the appropriate kinase family hard or even impossible. This might be caused by the fact that they represented pollen-specific and/or tobacco-specific proteins, homologues of which were absent in recent databases.
Kinase Motifs
Many protein kinases show phosphorylation motif specificity or at least phosphorylation motif preference. In order to find any possible up-regulated kinase motifs, we searched the presented data set using Motif-X algorithm. It should be noted that the information about linking a particular kinase to a phosphorylation motif is limited in plants and consequently the information is often extrapolated from other model organisms, mostly human (65). Two searches were performed looking up either for phosphoserine or for phosphothreonine (supplemental Fig. S2). Phosphorylated serine was shown to be present in five phosphorylation motifs whereas phosphothreonine occupied the central position of only one phosphorylation motif.
The first motif to be discussed is the prolyl-directed phosphorylation, i.e. a phosphorylated amino acid followed by a proline, regardless of the presence of phosphoserine (xxxxxxS*Pxxxxx) or phosphothreonine (xxxxxxT*Pxxxxx). The prolyl-directed phosphorylation is typical for two big groups of protein kinases - mitogen-activated protein kinases (MAPK), and cyclin-dependent protein kinases (CDK; ref. 60). Both these large kinase families were identified in the tobacco male gametophyte proteome (20), supplemental Table S5. MAPKs play a key regulatory role in many physiological processes including stress reactions and pollen hydration (62). CDKs were originally shown to regulate cell cycle and their activity in male gametophyte was expected because both pollen mitoses are precisely regulated (54). The alternative function of CDKs is for example the regulation of pre-mRNA splicing of callose synthase in pollen tube that influences cell wall formation (66). Alkaline phosphorylation motifs xxxRxxS*xxxxxx, and xxxKxxS*xxxxxx are recognized by Ca2+/calmodulin-dependent protein kinase (CAMK2; ref. 60). A chimeric CAMK with two distinct domains, one of which reacts to free Ca2+ and the other to Ca2+/calmodulin, was shown to be expressed in male gametophyte of lily and tobacco (67). Its expression started in pollen mother cell and then continued to peak in the tetrad stage. Such an expression profile tends us to speculate that the expression of this kinase reacts to Ca2+ oscillations, and that precisely regulates the synchronous events during microsporogenesis. Besides, this alkaline motif is in plants also recognized by the Ca2+-dependent protein kinase-sucrose-nonfermenting-related kinase (CDPK-SnRK) superfamily of protein kinases (68). Two kinases of this family were actually identified in tobacco male gametophyte proteome (Supplemental Table 5, and ref. 20). Last but not least, we identified two acidic kinase motifs with a central phosphoserine - xxxxxxS*DxExxx, and xxxxxxS*xDDxxx - corresponding in principle to the motif xxxxxxS*(D/E)(D/E)(D/E)xxx that is recognized by casein kinase 2 (CK2; ref. 60). Casein kinase 2 was shown to be activated by salicylic acid in tobacco (69), and two casein kinases were identified in tobacco male gametophyte proteome (supplemental Table S4, and ref. 20). We have to point out that although the corresponding kinases were identified in our data set, it still remains unproven whether they really interact with the phosphoproteins containing the corresponding motifs, and which of the kinases is actually responsible for a particular phosphorylation event.
Regulated Phosphopeptides and Their Function
There were established seven groups collecting the regulated phosphopeptides according to their regulatory trends. As mentioned above the groups I and V collected phosphopeptides that were phosphorylated in mature pollen. Because the phosphopeptides included in both group I and group V decreased in abundance after pollen activation, we assumed that the role of their phosphorylation is mainly required in dry mature pollen and/or their dephosphorylation represents the actual activation/de-repression. The dominant categories were protein synthesis and protein destination and storage, represented by many proteins for example by various translation initiation factors, LA-related protein like, among others. There was also identified protein Rho GDI that regulates the activity of Rho GTPases that are essential for tip growth of pollen tube (63). However, to our knowledge, the role of its phosphorylation site was not reported yet. According to our results, it can be assumed that its activity is switched on by dephosphorylation (at least of the particular phosphopeptide found in this regulatory group V) because the phosphates were attached to the protein exclusively in mature pollen and the concentration of the only phosphopeptide in group V decreased in pollen activated in vitro for 5 min. The other candidate specific to mature pollen was MAP kinase. MAP kinases were reported to play their roles upon pollen rehydration (62) so this phosphorylation might again be switching off the MAP kinase ready for pollen grain activation.
The groups II, III, and IV collected proteins phosphorylated strictly upon pollen activation. There appeared for example E3 ubiquitin-protein ligase RING FINGER 4 (RNF4)-like, the α-subunit of a nascent polypeptide-associated complex, protein phosphatase inhibitor 2, cytochrome oxidase c, histone deacetylase HDT1, villin and peptidyl-prolyl cis-trans isomerase 1 (PPI1), among others. Protein ubiquitination is likely to be initiated upon pollen activation in order to degrade the present proteins and to replace them with the newly synthesized species. Another E3 ubiquitin-protein ligase in Arabidopsis was reported to bind its target 14–3-3-proteins only upon phosphorylation of its particular amino acids (70). If the E3 ligase identified in our data set acts also after phosphorylation, we might speculate that this phosphorylation event represents an activation phosphorylation. The phosphorylated peptides from phosphatase inhibitor 2 appeared only upon protein phosphorylation. However, we might only speculate whether their phosphorylation promotes their activity or rather blocks it. Villin plays a role in actin cytoskeleton dynamics and it was shown to be phosphorylated on a tyrosine (71). The role of tyrosine phosphorylation during pollen tube growth was deduced from the pollen tube treatment by drugs influencing tyrosine phosphorylation that caused lower pollen germination rate and shorter pollen tubes (50). Because the treated pollen tubes showed a different arrangement of actin filaments, it might be possible that not only actin itself but also actin-binding proteins (such as villin) have to be precisely tyrosine-phosphorylated.
The most dynamic regulation was shown for the group VI phosphopeptides. This category grouped phosphopeptides that were phosphorylated in mature pollen, then dephosphorylated in 5-min activated pollen and later after 30-min activation re-appeared again among phosphopeptides. Phosphopeptides of the following proteins were put exclusively to this category (i.e. they did not show any other phosphopeptides belonging to any other group of regulated phosphopeptides): transcription initiation factor IIF, acidic ribosomal protein P1a-like, LEA protein D34, and ARA4-interacting protein. The other phosphoproteins had their corresponding phosphopeptides also in other regulation groups. These phosphorylation sites might represent phosphoproteins that reflect with their phosphorylation/dephosphorylation cycles the ion signal pulses during pollen tube growth (72). However, we do not have the phosphoproteomics data regarding longer periods of pollen tube growth in vitro, so making any bold conclusion is beyond the scope of this article.
Group VII comprised the regulated phosphopeptides that appeared in all studied stages. There were only three proteins that fell with their phosphopeptides exclusively into this category—2-phosphoglycerate kinase-related family protein, nuclear RNA binding protein-like, and calreticulin precursor. The other proteins were identified by peptides that fell not only into this group but also in at least another one (mostly group I, see supplemental Table S3, and Fig. 4).
CONCLUSION
Collectively, we purified and identified phosphopeptides from mature pollen, 5-min activated pollen, and 30-min activated pollen, the three stages covering an early phase of male gametophyte activation. This study presents the first developmental phosphoproteomics data from angiosperm activated pollen including the dynamics of very early phosphorylation events during pollen re-hydration and activation (i.e. 5-min activated pollen). The only other studied pollen tubes were these of Picea wilsonii, a gymnosperm (23). We identified 471 phosphopeptides carrying 432 phosphorylation sites that were assigned to 301 phosphoproteins. Moreover, the quantitative data highlighted the dynamics of protein phosphorylation during pollen activation and the differential regulation of several phosphopeptides of the same phosphoprotein pinpointed the complexity of protein phosphorylation in its functional context. Such list of phosphorylated proteins also represents a good starting point for the selection of the most interesting candidates for subsequent studies revealing the function of their phosphorylation and its integration into the molecular processes underlying pollen tube growth and development. Thus, this study brought new insights into the activation of pollen because highlighted the phosphorylated proteins that are very likely candidates, which would take part in the regulation and processes of pollen tube activation.
Supplementary Material
Footnotes
Author contributions: J. Fila, H.M., V.C., and D.H. designed research; J. Fila, S.R., A.M., J. Fecikova, and V.C. performed research; S.R., A.M., A.H., U.S., R.P.Z., and D.H. analyzed data; J. Fila, A.M., and D.H. wrote the paper.
* This work was supported by the Czech Science Foundation (15–16050S, 15–22720S, and P305/12/2611), and Czech Ministry of Education, Youth and Sports (LD13049). RPZ and SR thank the Ministry for Innovation, Science and Research of the Federal State of North Rhine-Westphalia for funding.
 This article contains supplemental Figs. S1 to S3 and Tables S1 to S5.
This article contains supplemental Figs. S1 to S3 and Tables S1 to S5.
1 The abbreviations used are:
- page: TiO2
- titanium dioxide
- Al(OH)3
- aluminium hydroxide
- bZIP
- basic leucine zipper
- CAMK2
- Ca2+/calmodulin-dependent protein kinase
- CDK
- cyclin-dependent protein kinase
- CDPK—SnRK
- Ca2+-dependent protein kinase–sucrose-nonfermenting-related kinase
- CK2
- casein kinase 2
- Cys
- cysteine
- DCN1
- defective in cullin neddylation protein 1
- DHB
- 2,5–dihydroxybenzoic acid
- EPP
- EDTA/puromycine-resistant particle
- GO
- gene ontology
- IMAC
- immobilized metal affinity chromatography
- LEA
- late embryogenesis abundant
- MAPK
- mitogen-activated protein kinase
- Met
- methionine
- MS/MS
- tandem mass spectrometry
- nLC
- nano liquid chromatography
- PB1
- octicosapeptide/PHOX/BEM1p
- PPI1
- peptidyl-prolyl cis-trans isomerase 1
- Rho GAP
- Rho GTPase activation protein
- Rho GDI2
- Rho guanine nucleotide dissociation inhibitor 2
- RNF4
- RING FINGER PROTEIN 4
- Ser
- serine
- SIMAC
- sequential elution from IMAC
- SMM-MES
- sucrose-mineral medium buffered with MES
- SNC1
- SUPRESSOR OF NPR1–1, CONSTITUTIVE 1
- Thr
- threonine
- Tyr
- tyrosine
- UBA
- ubiquitin-associated
- UBX
- ubiquitin-like
- UNC-89
- UNCOORDINATED-89
- WVD2
- WAVE-DAMPENED 2.
REFERENCES
- 1.Vogler F., Konrad S. S. A., and Sprunck S. (2015) Knockin' on pollen's door: live cell imaging of early polarization events in germinating Arabidopsis pollen. Front. Plant Sci. 6, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mascarenhas J. P. (1993) Molecular mechanisms of pollen tube growth and differentiation. Plant Cell 5, 1303–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mishra N. S., Tuteja R., and Tuteja N. (2006) Signaling through MAP kinase networks in plants. Arch. Biochem. Biophys. 452, 55–68 [DOI] [PubMed] [Google Scholar]
- 4.Francis D., and Halford N. G. (1995) The plant cell cycle. Physiol. Plant. 93, 365–374 [Google Scholar]
- 5.van der Kelen K., Beyaert R., Inze D., and de Veylder L. (2009) Translational control of eukaryotic gene expression. Crit. Rev. Biochem. Mol. Biol. 44, 143–168 [DOI] [PubMed] [Google Scholar]
- 6.Röhrig H., Colby T., Schmidt J., Harzen A., Facchinelli F., and Bartels D. (2008) Analysis of desiccation-induced candidate phosphoproteins from Craterostigma plantagineum isolated with a modified metal oxide affinity chromatography procedure. Proteomics 8, 3548–3560 [DOI] [PubMed] [Google Scholar]
- 7.Darewicz M., Dziuba J., and Minkiewicz P. (2005) Some properties of beta-casein modified via phosphatase. Acta Alimentaria 34, 403–415 [Google Scholar]
- 8.Fletterick R. J., and Sprang S. R. (1982) Glycogen-phosphorylase structures and function. Acc. Chem. Res. 15, 361–369 [Google Scholar]
- 9.Kim J., Shen Y., Han Y. J., Park J. E., Kirchenbauer D., Soh M. S., Nagy F., Schafer E., and Song P. S. (2004) Phytochrome phosphorylation modulates light signaling by influencing the protein-protein interaction. Plant Cell 16, 2629–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garnak M., and Reeves H. C. (1979) Phosphorylation of isocitrate dehydrogenase of Escherichia coli. Science 203, 1111–1112 [DOI] [PubMed] [Google Scholar]
- 11.Fíla J., and Honys D. (2012) Enrichment techniques employed in phosphoproteomics. Amino Acids 43, 1025–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunn J. D., Reid G. E., and Bruening M. L. (2010) Techniques for phosphopeptide enrichment prior to analysis by mass spectrometry. Mass Spectrom. Rev. 29, 29–54 [DOI] [PubMed] [Google Scholar]
- 13.Hoehenwarter W., Thomas M., Nukarinen E., Egelhofer V., Röhrig H., Weckwerth W., Conrath U., and Beckers G. J. M. (2013) Identification of novel in vivo MAP kinase substrates in Arabidopsis thaliana through use of tandem metal oxide affinity chromatography. Mol. Cell. Proteomics 12, 369–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beckers G. J., Hoehenwarter W., Röhrig H., Conrath U., and Weckwerth W. (2014) Tandem metal-oxide affinity chromatography for enhanced depth of phosphoproteome analysis. Methods Mol. Biol. 1072, 621–632 [DOI] [PubMed] [Google Scholar]
- 15.Mayank P., Grossman J., Wuest S., Boisson-Dernier A., Roschitzki B., Nanni P., Nuehse T., and Grossniklaus U. (2012) Characterization of the phosphoproteome of mature Arabidopsis pollen. Plant J. 72, 89–101 [DOI] [PubMed] [Google Scholar]
- 16.Holmes-Davis R., Tanaka C. K., Vensel W. H., Hurkman W. J., and McCormick S. (2005) Proteome mapping of mature pollen of Arabidopsis thaliana. Proteomics 5, 4864–4884 [DOI] [PubMed] [Google Scholar]
- 17.Noir S., Bräutigam A., Colby T., Schmidt J., and Panstruga R. (2005) A reference map of the Arabidopsis thaliana mature pollen proteome. Biochem. Biophys. Res. Commun. 337, 1257–1266 [DOI] [PubMed] [Google Scholar]
- 18.Sheoran I. S., Sproule K. A., Olson D. J. H., Ross A. R. S., and Sawhney V. K. (2006) Proteome profile and functional classification of proteins in Arabidopsis thaliana (Landsberg erecta) mature pollen. Sex. Plant Reprod. 19, 185–196 [Google Scholar]
- 19.Grobei M. A., Qeli E., Brunner E., Rehrauer H., Zhang R., Roschitzki B., Basler K., Ahrens C. H., and Grossniklaus U. (2009) Deterministic protein inference for shotgun proteomics data provides new insights into Arabidopsis pollen development and function. Genome Res. 19, 1786–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ischebeck T., Valledor L., Lyon D., Gingl S., Nagler M., Meijon M., Egelhofer V., and Weckwerth W. (2014) Comprehensive cell-specific protein analysis in early and late pollen development from diploid microsporocytes to pollen tube growth. Mol. Cell. Proteomics 13, 295–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fíla J., Matros A., Radau S., Zahedi R. P., Čapková V., Mock H.-P., and Honys D. (2012) Revealing phosphoproteins playing role in tobacco pollen activated in vitro. Proteomics 12, 3229–3250 [DOI] [PubMed] [Google Scholar]
- 22.Wolschin F., Wienkoop S., and Weckwerth W. (2005) Enrichment of phosphorylated proteins and peptides from complex mixtures using metal oxide/hydroxide affinity chromatography (MOAC). Proteomics 5, 4389–4397 [DOI] [PubMed] [Google Scholar]
- 23.Chen Y., Liu P., Hoehenwarter W., and Lin J. (2012) Proteomic and phosphoproteomic analysis of Picea wilsonii pollen development under nutrient limitation. J. Proteome Res. 11, 4180–4190 [DOI] [PubMed] [Google Scholar]
- 24.Pinkse M. W. H., Uitto P. M., Hilhorst M. J., Ooms B., and Heck A. J. R. (2004) Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-nanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Chem. 76, 3935–3943 [DOI] [PubMed] [Google Scholar]
- 25.Petrů E., Hrabětová E., and Tupý J. (1964) The technique of obtaining germinating pollen without microbial contamination. Biologia Plantarum 6, 68–69 [Google Scholar]
- 26.Tupý J., and Říhová L. (1984) Changes and growth effect of pH in pollen tube culture. J. Plant Physiol. 115, 1–10 [DOI] [PubMed] [Google Scholar]
- 27.Méchin V., Damerval C., and Zivy M. (2006) Total protein extraction with TCA-acetone. In: Thiellement H., Zivy M., Damerval C., and Méchin V., eds. Methods in Mol. Biol., pp. 1–8, Springer; [DOI] [PubMed] [Google Scholar]
- 28.Kollipara L., and Zahedi R. P. (2013) Protein carbamylation: In vivo modification or in vitro artefact? Proteomics 13, 941–944 [DOI] [PubMed] [Google Scholar]
- 29.Beck F., Lewandrowski U., Wiltfang M., Feldmann I., Geiger J., Sickmann A., and Zahedi R. P. (2011) The good, the bad, the ugly: Validating the mass spectrometric analysis of modified peptides. Proteomics 11, 1099–1109 [DOI] [PubMed] [Google Scholar]
- 30.Taus T., Koecher T., Pichler P., Paschke C., Schmidt A., Henrich C., and Mechtler K. (2011) Universal and confident phosphorylation site localization using phosphoRS. J. Proteome Res. 10, 5354–5362 [DOI] [PubMed] [Google Scholar]
- 31.Vizcaino J. A., Deutsch E. W., Wang R., Csordas A., Reisinger F., Rios D., Dianes J. A., Sun Z., Farrah T., Bandeira N., Binz P. A., Xenarios I., Eisenacher M., Mayer G., Gatto L., Campos A., Chalkley R. J., Kraus H. J., Albar J. P., Martinez-Bartolome S., Apweiler R., Omenn G. S., Martens L., Jones A. R., and Hermjakob H. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nature Biotechnol. 32, 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bevan M., Bancroft I., Bent E., Love K., Goodman H., Dean C., Bergkamp R., Dirkse W., Van Staveren M., Stiekema W., Drost L., Ridley P., Hudson S. A., Patel K., Murphy G., Piffanelli P., Wedler H., Wedler E., Wambutt R., Weitzenegger T., Pohl T. M., Terryn N., Gielen J., Villarroel R., De Clerck R., Van Montagu M., Lecharny A., Auborg S., Gy I., Kreis M., Lao N., Kavanagh T., Hempel S., Kotter P., Entian K. D., Rieger M., Schaeffer M., Funk B., Mueller-Auer S., Silvey M., James R., Montfort A., Pons A., Puigdomenech P., Douka A., Voukelatou E., Milioni D., Hatzopoulos P., Piravandi E., Obermaier B., Hilbert H., Dusterhoft A., Moores T., Jones J. D. G., Eneva T., Palme K., Benes V., Rechman S., Ansorge W., Cooke R., Berger C., Delseny M., Voet M., Volckaert G., Mewes H. W., Klosterman S., Schueller C., Chalwatzis N., and Project E. U. A. G. (1998) Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 391, 485–488 [DOI] [PubMed] [Google Scholar]
- 33.Chou M. F., and Schwartz D. (2011) Biological sequence motif discovery using motif-x. Curr. Protoc. Bioinformatics Chapter 13, Unit 13.15–24 [DOI] [PubMed] [Google Scholar]
- 34.Schwartz D., and Gygi S. P. (2005) An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nature Biotechnol. 23, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 35.Rohn H., Junker A., Hartmann A., Grafahrend-Belau E., Treutler H., Klapperstueck M., Czauderna T., Klukas C., and Schreiber F. (2012) VANTED v2: a framework for systems biology applications. BMC Syst. Biol. 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCoy C. E., Campbell D. G., Deak M., Bloomberg G. B., and Arthur J. S. C. (2005) MSK1 activity is controlled by multiple phosphorylation sites. Biochem. J. 387, 507–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X. M., Paulin F. E. M., Campbell L. E., Gomez E., O'Brien K., Morrice N., and Proud C. G. (2001) Eukaryotic initiation factor 2B: identification of multiple phosphorylation sites in the epsilon-subunit and their functions in vivo. EMBO J. 20, 4349–4359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ito J., Taylor N. L., Castleden I., Weckwerth W., Millar A. H., and Heazlewood J. L. (2009) A survey of the Arabidopsis thaliana mitochondrial phosphoproteome. Proteomics 9, 4229–4240 [DOI] [PubMed] [Google Scholar]
- 39.Luis Carrasco J., Jose Castello M., Naumann K., Lassowskat I., Navarrete-Gomez M., Scheel D., and Vera P. (2014) Arabidopsis protein phosphatase DBP1 nucleates a protein network with a role in regulating plant defense. Plos One 9, e90734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hultquist D. E. (1968) The preparation and characterization of phosphorylated derivatives of histidine. Biochim Biophys Acta 153, 329–340 [DOI] [PubMed] [Google Scholar]
- 41.Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., and Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 42.Molina H., Horn D. M., Tang N., Mathivanan S., and Pandey A. (2007) Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 104, 2199–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beausoleil S. A., Jedrychowski M., Schwartz D., Elias J. E., Villén J., Li J. X., Cohn M. A., Cantley L. C., and Gygi S. P. (2004) Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. U.S.A. 101, 12130–12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Bentem S. D., Anrather D., Dohnal I., Roitinger E., Csaszar E., Joore J., Buijnink J., Carreri A., Forzani C., Lorkovic Z. J., Barta A., Lecourieux D., Verhounig A., Jonak C., and Hirt H. (2008) Site-specific phosphorylation profiling of Arabidopsis proteins by mass spectrometry and peptide chip analysis. J. Prot. Res. 7, 2458–2470 [DOI] [PubMed] [Google Scholar]
- 45.Benschop J. J., Mohammed S., O'Flaherty M., Heck A. J. R., Slijper M., and Menke F. L. H. (2007) Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteomics 6, 1198–1214 [DOI] [PubMed] [Google Scholar]
- 46.Sugiyama N., Nakagami H., Mochida K., Daudi A., Tomita M., Shirasu K., and Ishihama Y. (2008) Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol. Syst. Biol. 4, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nakagami H., Sugiyama N., Mochida K., Daudi A., Yoshida Y., Toyoda T., Tomita M., Ishihama Y., and Shirasu K. (2010) Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 153, 1161–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Bentem S. D., and Hirt H. (2009) Protein tyrosine phosphorylation in plants: more abundant than expected? Trends Plant Sci. 14, 71–76 [DOI] [PubMed] [Google Scholar]
- 49.Mithoe S. C., and Menke F. L. H. (2011) Phosphoproteomics perspective on plant signal transduction and tyrosine phosphorylation. Phytochem. 72, 997–1006 [DOI] [PubMed] [Google Scholar]
- 50.Zi H. J., Xiang Y., Li M., Wang T., and Ren H. Y. (2007) Reversible protein tyrosine phosphorylation affects pollen germination and pollen tube growth via the actin cytoskeleton. Protoplasma 230, 183–191 [DOI] [PubMed] [Google Scholar]
- 51.Oh M. H., Wang X. F., Kota U., Goshe M. B., Clouse S. D., and Huber S. C. (2009) Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 658–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nito K., Wong C. C. L., Yates J. R., and Chory J. (2013) Tyrosine phosphorylation regulates the activity of phytochrome photoreceptors. Cell Rep. 3, 1970–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Čapková V., Hrabětová E., and Tupý J. (1988) Protein synthesis in pollen tubes: preferential formation of new species independent of transcription. Sex. Plant Reprod. 1, 150–155 [Google Scholar]
- 54.Hafidh S., Breznenová K., Růžička P., Feciková J., Čapková V., and Honys D. (2012) Comprehensive analysis of tobacco pollen transcriptome unveils common pathways in polar cell expansion and underlying heterochronic shift during spermatogenesis. BMC Plant Biol. 12, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hafidh S., Breznenová K., and Honys D. (2012) De novo post-pollen mitosis II tobacco pollen tube transcriptome. Plant Signal. Behav. 7, 918–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Honys D., Combe J. P., Twell D., and Čapková V. (2000) The translationally repressed pollen-specific ntp303 mRNA is stored in nonpolysomal mRNPs during pollen maturation. Sex. Plant Reprod. 13, 135–144 [Google Scholar]
- 57.Honys D., Reňák D., Feciková J., Jedelský P. L., Nebesářová J., Dobrev P., and Čapková V. (2009) Cytoskeleton-associated large RNP complexes in tobacco male gametophyte (EPPs) are associated with ribosomes and are involved in protein synthesis, processing, and localization. J. Proteome Res. 8, 2015–2031 [DOI] [PubMed] [Google Scholar]
- 58.Hosp J., Ribarits A., Retzer K., Jin Y. F., Tashpulatov A., Resch T., Friedmann C., Ankele E., Voronin V., Palme K., Heberle-Bors E., and Touraev A. (2014) A tobacco homolog of DCN1 is involved in pollen development and embryogenesis. Plant Cell Rep. 33, 1187–1202 [DOI] [PubMed] [Google Scholar]
- 59.Nguyen X. C., Kim S. H., Lee K., Kim K. E., Liu X. M., Han H. J., My H. T. H., Lee S. W., Hong J. C., Moon Y. H., and Chung W. S. (2012) Identification of a C2H2-type zinc finger transcription factor (ZAT10) from Arabidopsis as a substrate of MAP kinase. Plant Cell Rep. 31, 737–745 [DOI] [PubMed] [Google Scholar]
- 60.Lee T. Y., Bretana N. A., and Lu C. T. (2011) PlantPhos: using maximal dependence decomposition to identify plant phosphorylation sites with substrate site specificity. BMC Bioinformatics 12, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heberle-Bors E., Voronin V., Touraev A., Testillano P. S., Risueno M. C., and Wilson C. (2001) MAP kinase signaling during pollen development. Sex. Plant Reprod. 14, 15–19 [Google Scholar]
- 62.Wilson C., Voronin V., Touraev A., Vicente O., and Heberle-Bors E. (1997) A developmentally regulated MAP kinase activated by hydration in tobacco pollen. Plant Cell 9, 2093–2100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klahre U., Becker C., Schmitt A. C., and Kost B. (2006) Nt-RhoGDI2 regulates Rac/Rop signaling and polar cell growth in tobacco pollen tubes. Plant J. 46, 1018–1031 [DOI] [PubMed] [Google Scholar]
- 64.Payne D. M., Rossomando A. J., Martino P., Erickson A. K., Her J. H., Shabanowitz J., Hunt D. F., Weber M. J., and Sturgill T. W. (1991) Identification of the regulatory phosphorylation sites in PP42/mitogen-activated protein kinase (MAP kinase). EMBO J. 10, 885–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee T.-Y., Lin Z.-Q., Hsieh S.-J., Bretana N. A., and Lu C.-T. (2011) Exploiting maximal dependence decomposition to identify conserved motifs from a group of aligned signal sequences. Bioinformatics 27, 1780–1787 [DOI] [PubMed] [Google Scholar]
- 66.Huang X.-Y., Niu J., Sun M.-X., Zhu J., Gao J.-F., Yang J., Zhou Q., and Yang Z.-N. (2013) Cyclin-dependent kinase G1 is associated with the spliceosome to regulate callose synthase 5 splicing and pollen wall formation in Arabidopsis. Plant Cell 25, 637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Poovaiah B. W., Xia M., Liu Z. H., Wang W. Y., Yang T. B., Sathyanarayanan P. V., and Franceschi V. R. (1999) Developmental regulation of the gene for chimeric calcium/calmodulin-dependent protein kinase in anthers. Planta 209, 161–171 [DOI] [PubMed] [Google Scholar]
- 68.Hrabak E. M., Chan C. W. M., Gribskov M., Harper J. F., Choi J. H., Halford N., Kudla J., Luan S., Nimmo H. G., Sussman M. R., Thomas M., Walker-Simmons K., Zhu J. K., and Harmon A. C. (2003) The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 132, 666–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hidalgo P., Garreton V., Berrios C. G., Ojeda H., Jordana X., and Holuigue L. (2001) A nuclear casein kinase 2 activity is involved in early events of transcriptional activation induced by salicylic acid in tobacco. Plant Physiol. 125, 396–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yasuda S., Sato T., Maekawa S., Aoyama S., Fukao Y., and Yamaguchi J. (2014) Phosphorylation of Arabidopsis ubiquitin ligase ATL31 is critical for plant carbon/nitrogen nutrient balance response and controls the stability of 14–3-3 proteins. J. Biol. Chem. 289, 15179–15193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhai L. W., Zhao P. L., Panebra A., Guerrerio A. L., and Khurana S. (2001) Tyrosine phosphorylation of villin regulates the organization of the actin cytoskeleton. J. Biol. Chem. 276, 36163–36167 [DOI] [PubMed] [Google Scholar]
- 72.Geitmann A., and Cresti M. (1998) Ca2+ channels control the rapid expansions in pulsating growth of Petunia hybrida pollen tubes. J. Plant Physiol. 152, 439–447 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.