Abstract
Intrathecal immunoglobulin G (IgG) synthesis and oligoclonal IgG bands in cerebrospinal fluid (CSF) are hallmarks of multiple sclerosis (MS), but the antigen specificities remain enigmatic. Our study is the first investigating the autoantibody repertoire in paired serum and CSF samples from patients with relapsing-remitting MS (RRMS), primary progressive MS (PPMS), and other neurological diseases by the use of high-density peptide microarrays. Protein sequences of 45 presumed MS autoantigens (e.g. MOG, MBP, and MAG) were represented on the microarrays by overlapping 15mer peptides. IgG reactivities were screened against a total of 3991 peptides, including also selected viral epitopes. The measured antibody reactivities were highly individual but correlated for matched serum and CSF samples. We found 54 peptides to be recognized significantly more often by serum or CSF antibodies from MS patients compared with controls (p values <0.05). The results for RRMS and PPMS clearly overlapped. However, PPMS patients presented a broader peptide-antibody signature. The highest signals were detected for a peptide mapping to a region of the Epstein-Barr virus protein EBNA1 (amino acids 392–411), which is homologous to the N-terminal part of human crystallin alpha-B. Our data confirmed several known MS-associated antigens and epitopes, and they delivered additional potential linear epitopes, which await further validation. The peripheral and intrathecal humoral immune response in MS is polyspecific and includes antibodies that are also found in serum of patients with other diseases. Further studies are required to assess the pathogenic relevance of autoreactive and anti-EBNA1 antibodies as well as their combinatorial value as biomarkers for MS.
Multiple sclerosis (MS)1 is a chronic disease of the central nervous system (CNS) that typically affects young adults, especially women. The disease is characterized by discrete areas of inflammation (lesions), demyelination, axonal loss, and astrogliosis in the brain and spinal cord. The clinical correlate of these processes is a wide range of neurological signs and symptoms involving mobility problems, vision problems, cognitive dysfunction, fatigue, and pain (1, 2). This variability is also reflected in the disease courses observed, which include relapsing-remitting MS (RRMS), usually with secondary progression (SPMS) in later stages, and primary progressive MS (PPMS) (3). The majority of patients (∼85%) are diagnosed with RRMS, in which a first clinical attack heralds the onset of the disease (clinically isolated syndrome, CIS). In RRMS, episodes with new or recurrent neurological deficits (relapses) are followed by phases of partial or complete recovery (remission). The remaining ∼15% of the patients have PPMS, which is from the beginning gradually progressive without relapses. Current therapeutics predominantly target the inflammatory component of the disease in order to reduce the frequency and severity of relapses and to prevent the accumulation of disability. However, although several disease-modifying treatments have shown to be efficacious in RRMS, none have yet been approved to alleviate PPMS (4). Moreover, the course of disease is largely unpredictable on an individual level, and there is no single clinical feature or diagnostic test that is sufficient to diagnose MS or to distinguish RRMS from PPMS. Therefore, ongoing research efforts are dedicated to better understand the disease and to identify biomarkers for an improved diagnosis and prognosis of MS.
The primary cause of MS is unknown, and the molecular mechanisms of inflammation and neurodegeneration are still elusive. However, it is generally accepted that MS involves an immune response to self-antigens in genetically predisposed individuals exposed to environmental risk factors. A fundamental step in the development of demyelinating lesions is the recruitment and migration of activated leukocytes into the CNS through a deficient blood-brain barrier (5). Irreversible neuroaxonal damage within the lesions is associated with accumulating neurological disability. Episodes of inflammatory activity are mainly characteristic for RRMS patients, whereas PPMS patients present less prominent inflammation and more neurodegenerative pathology. T cells are assumed to be critical drivers of the disease, but B-cells and other immune cells play significant roles as well (5, 6). Autoreactive T- and B cells may be activated in the periphery and then reactivated after entering the CNS. On the other hand, enhanced T- and B-cell reactivities may represent merely secondary responses to neurodegeneration. As of today, the complex interplay of neuronal dysfunction and immune responses (innate and adaptive, humoral and cell-mediated) is far from being understood.
The immune dysregulation in MS is partly driven by B cells. B cells have regulatory and antigen-presenting functions, and the activation of antigen-specific B cells (usually dependent on helper T cells) results in their proliferation and differentiation, culminating in the generation of memory B cells and antibody-secreting plasmablasts and plasma cells. A role of B cells in MS is supported by studies, in which effector B cells as well as antibodies were found in CNS lesions and in cerebrospinal fluid (CSF) of MS patients (7, 8), and by clinical trials, which showed that B-cell depletion is effective in the treatment of this disease (9). Recently, a genetic fine-mapping revealed that active cis-regulatory elements of B cells are enriched of MS-associated single-nucleotide polymorphisms (10). Moreover, intrathecal immunoglobulin G (IgG) synthesis and oligoclonal IgG bands (OCB) in the CSF (not mirrored in serum) are hallmarks of MS and are therefore useful diagnostic markers (11, 12). This oligoclonal antibody pattern is generated by clonal expansion of a limited repertoire of activated B cells, but it is still a mystery whether it derives from a small but random sample of peripheral B cells or from a pathogenic response. Interestingly, there is evidence for ongoing B-cell stimulation and maturation to antibody-expressing cells locally inside the CNS (13). It is assumed that ectopic B-cell follicles resembling a germinal center reaction occur in the brain of MS patients (14, 15). A point of contention is whether B cells that are latently infected with Epstein-Barr virus (EBV) may contribute to such B-cell aggregations and sustain the compartmentalized immunopathological response (16, 17).
Despite extensive research on intrathecally produced antibodies in MS, their major antigen specificities remain enigmatic. Myelin antigens such as MBP, PLP, MAG, and MOG were first investigated as potential targets of antibodies to substantiate the concept of MS as an autoimmune disease. However, the pathogenetic relevance of myelin antibodies (IgG and other isotypes) has so far not been established unequivocally. Within the last years, it became evident that the B-cell response is not restricted to myelin but is much more widespread. Antibodies against several other CNS components (proteins, lipids, and glycans) have been described (reviewed in (6, 18)). Additionally, apparently nonspecific immune responses in the serum and CSF of MS patients involve nonbrain targets and common infectious agents. Antibodies to measles, rubella, and varicella zoster virus have a >90% seroprevalence in the population (19), but their intrathecal synthesis is a marker for MS (called MRZ reaction) (20). The antiviral response further includes EBV (also called human herpesvirus 4, HHV-4). The seropositivity for EBV appears to be 100% in MS patients (21). However, although several antibody targets have been defined, there is no single antibody pattern, which can distinguish MS patients from healthy individuals or patients with other diseases with high sensitivity and high specificity. It has been argued that this may indicate different (immuno)pathologies, thereby explaining that MS is a heterogeneous disease with variable clinical presentation.
Disease-specific antibodies are difficult to identify, because of experimental limitations and biological variation. The antibodyome, that is, the complete set of antibodies existing in an organism at a given time, is extremely individual. It is influenced by many kinds of environmental stimuli, it is relatively stable but also changes over time, and it varies with age, diet, and health status. Autoimmune responses can be initiated by diverse mechanisms such as molecular mimicry, epitope spreading, and viral support of autoreactive cell survival (e.g. by EBV) (22). It is also possible that the presence of autoantibodies in MS represents just a bystander effect during inflammatory responses secondary to demyelination and CNS injury. Accordingly, normal immune system processes, which include naturally occurring autoreactive antibodies involved in removing cellular debris and maintaining homeostasis (23, 24), may be generally raised as an epiphenomenon. To which extent autoreactive antibodies are pathogenic in vivo remains largely unclear. Detrimental processes within the MS brain may influence whether self-antigens are sufficiently abundant, accessible, and in the specific conformation to be recognized by such antibodies.
To better understand the role of antibodies in MS, it is also important to define the antigen epitopes (the specific parts of antigens to which antibodies bind). Multiple epitopes of different type can be contained in a single protein antigen. Conformational epitopes depend on the protein folding and are thus lost on denaturation of the protein. Linear epitopes consist of a sequence of adjacent amino acids that is recognized by antibodies binding either the denatured protein only (epitope is inaccessible in the folded protein) or both the native and the denatured protein (epitope is accessible in the folded protein). A significant amount of literature already addressed the study of epitopes. When searching the Immune Epitope Database (IEDB) (25) using the parameters “Linear Peptide,” “B Cell Response,” and “multiple sclerosis,” more than 50 publications are listed. However, the results differ across the studies because of different experimental settings and assay systems used.
Peptide microarrays are an attractive novel tool for screening and validation of linear epitopes by measuring antibody reactivities against many peptides simultaneously (26). When using microarrays, a tiling strategy is usually employed to represent a protein sequence by a set of overlapping short peptides. An advantage of this screening approach is that patients can be better stratified because antibodies of different patients may bind to different epitopes of the same protein, possibly implying pathophysiological differences. Information about the precise epitopes is thus potentially relevant for MS diagnosis, prognosis, and monitoring (27). However, it should be noted that peptide microarrays are in general not suited to detect conformational epitopes, even if secondary structure conformations such as an alpha-helix can be formed by some short peptides (28). Different research groups have already applied microarrays to profile the autoantibody repertoire of MS patients against selected sets of peptides. They used serum samples (29–31), CSF samples (32–34), or both (35). By this means, MS-specific antibody signatures (IgG and IgM) were defined, comprising panels of antigenic peptides derived from CNS proteins. However, all these studies were limited to <370 peptides. High-density peptide microarrays containing thousands of peptides were so far not used for examining autoantibodies in MS.
Here, we screened for potential autoantibody reactivities in serum and CSF of MS patients using custom high-density microarrays containing almost 4000 peptides. The analysis of both compartments facilitated the study of the relatedness between intrathecal and peripheral immune responses and how they are linked with MS alone and in combination. The peptides were derived from 45 human proteins described in the literature to be MS autoantigens. Additional antigenic peptides were included as well (e.g. known EBV epitopes). IgG reactivities against these peptides were measured for 10 RRMS patients, 10 PPMS patients, and 10 controls with other neurological diseases (CTR). This is the first peptide microarray study that compares the antibody binding profiles of RRMS and PPMS patients for serum and CSF in parallel. The antibody repertoire against the peptides differed markedly between the individual subjects but was qualitatively similar in paired CSF and serum samples. In general, much higher peptide-antibody reactivities were measured for serum, but some peptides showed higher signals for CSF. Antibody responses against dozens of peptides were significantly elevated in MS patients compared with controls. Unexpectedly, the antibody signature was particularly increased in PPMS. The potential pathogenic relevance of selected epitopes is discussed.
EXPERIMENTAL PROCEDURES
Study Population
Paired CSF and serum samples were collected at the Department of Neurology of the University of Rostock from 10 patients with RRMS (RRMS_01–10), 10 patients with PPMS (PPMS_01–10), and 10 controls with other noninflammatory neurological diseases (CTR_01–10). The samples of each individual were always obtained at the same day and stored at −80 °C.
The patient cohorts were matched for age and gender, with an average age of 43 years and a balanced sex ratio in each group (Table I). All patients and controls were of Western European descent, and they did not suffer from any other autoimmune disorder. The MS patients' diagnosis was confirmed according to the revised McDonald criteria (36). They were given routine care following the consensus treatment guidelines and recommendations of the German Society of Neurology. Two RRMS patients were treated with glatiramer acetate, whereas the remaining MS patients received no immunomodulatory or immunosuppressive treatment at the time of sampling of serum and CSF, respectively (hereinafter abbreviated to “serum/CSF”). The patients were assessed neurologically, monitored for relapses and rated using the Expanded Disability Status Scale (EDSS) for a clinical follow-up period of 12 months. At sample collection, the degree of disability was, on average, significantly higher in the PPMS group than in the RRMS group (t test p value = 0.017). CSF-specific OCB were found for 17 of the 20 MS patients, and an elevated CSF IgG index was detected for five RRMS patients and four PPMS patients, consistent with the abnormal production of IgG in the CNS that is characteristic of MS. The demographic and clinical data of the individual patients and controls are given in supplementary File S1.
Table I. Clinical and demographic parameters of the three cohorts. Mean ± standard deviation (S.D.) are shown for age and EDSS at the sampling time point. Absolute numbers of patients are given for gender and markers of intrathecal IgG synthesis. For full patient information (supplemental File S1) n.d. = not determined in general.
| RRMS (n = 10) | PPMS (n = 10) | CTR (n = 10) | |
|---|---|---|---|
| Age in years (mean ± S.D.) | 43.7 ± 8.4 | 43.4 ± 7.4 | 43.9 ± 7.7 |
| Gender (female/male) | 5/5 | 5/5 | 5/5 |
| CSF IgG index (elevated/normal) | 5/5 | 4/6 | 0/10 |
| Oligoclonal bands (positive/negative) | 9/1 | 8/2 | n.d. |
| Expanded Disability Status Scale (mean ± S.D.) | 2.4 ± 0.8 | 4.1 ± 1.9 | n.d. |
The study was approved by the local ethics committee and carried out according to the Declaration of Helsinki. All patients gave written informed consent to participate in this study.
Peptide Microarray Design and Production
High-density peptide microarrays were used to compare the reactivities of IgG in serum and CSF of MS patients and controls. A decisive factor in such an experiment is the choice of peptides.
Initially, we searched the literature and identified 45 different human proteins that were described as potential autoantigens in MS (supplemental File S2). The majority of peptides spotted on the microarrays were derived from the sequences of these proteins and all their isoforms (generated, e.g. by alternative splicing) listed in the UniProt Knowledgebase (37). The peptides were selected in a manner that each protein sequence was completely represented by 15mer peptides with at least nine amino acids overlap (linear epitope mapping approach). According to this tiling strategy, 3747 different peptides were deduced for the 45 candidate proteins.
Moreover, we used the manually curated IEDB (http://www.iedb.org) (25) to gather previously reported MS-specific epitopes. With the search parameters “Linear Peptide,” “B Cell Response,” and “multiple sclerosis,” a set of 137 “positive” up to 20 amino acids long peptide sequences were obtained. Some of these peptides belonged to the already selected 45 proteins (in particular MOG and MBP), and others were potential MS mimotopes identified by researchers who used phage-displayed random peptide libraries. We further used IEDB to extract peptides described as epitopes of viral proteins from HHV-4 (n = 103) and HHV-6 (n = 4). In sum, 244 additional peptides, 10–20 amino acids in length, were included to this study, and 10 of these peptides contained the nonstandard amino acid citrulline (Z). Taken together, 3991 different peptides were selected for the design of the custom peptide microarrays to investigate the peripheral and intrathecal antibody profiles in MS.
Peptide synthesis and production of the high-density peptide microarrays were performed by JPT Peptide Technologies (Berlin, Germany) (26). For technical reasons, all synthesized peptides contained an additional C-terminal glycine (G). A linker at the N terminus of the peptides facilitated the immobilization of the peptides in a directed fashion on the glass slides (standard microscope slides, 76 × 26 mm). The peptides were suspended in printing solution (45% dimethyl sulfoxide, 50% 0.2 m sodium acetate pH 4.5, 5% glycerol) and spotted on each microarray in triplicate as three subarrays. Each microarray thus contained ∼12,000 spots with a diameter of 150 μm. The microarrays were blocked after spotting with bovine serum albumin (BSA). Further details on peptide synthesis and spotting can be found elsewhere (38).
Peptide Microarray Experiment
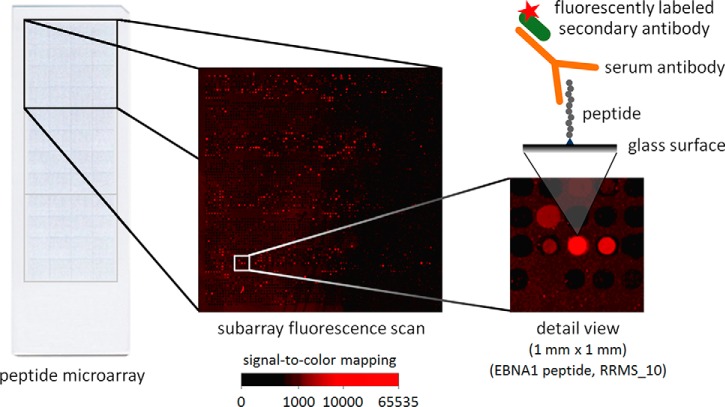
Microarray incubation was done following previously described protocols (38). Briefly, 100 μl of diluted patient serum/CSF were added to the arrays. Serum samples were always diluted 1:100, and CSF samples were always diluted 1:5 in wash buffer (50 mm Tris-HCl pH 7.4, 150 mm NaCl, 0.05% Tween 20, 0.1% BSA). After 4 h of incubation at room temperature, the microarrays were washed, and residual fluid was removed by centrifugation. Secondary incubation was performed at room temperature in the dark for 2 h using as secondary (anti-)antibody 100 μl of polyclonal Zenon goat anti-human IgG (Fab fragments, final concentration of 0.8 μg/ml in wash buffer) conjugated to Alexa Fluor 647 (Thermo Fisher Scientific, Waltham, MA, USA, a single batch of catalog number Z-25408). After another round of washing, fluorescence signals were acquired using a G2565 scanner from Agilent (Santa Clara, CA, USA). The scans were performed at 10 μm resolution with 16-bit depth. The Alexa Fluor 647 fluorescence in the red channel (635 nm) was used to assess whether the spotted peptides were bound by IgG (Fig. 1). All steps from incubation to scanning were realized at the same day and by the same person (KF) for all serum/CSF samples to prevent potential sources of bias. In the end, 60 peptide microarrays were used to profile the antibody reactivities in serum and CSF of the 20 MS patients and 10 controls. Two additional microarrays were incubated with wash buffer plus secondary antibody only. They were used as negative controls.
Fig. 1.
Multiplex analysis of antibody reactivities with peptide microarrays. Each peptide microarray glass slide consisted of three replicate subarrays (top, middle, bottom). In each subarray, antibody reactivities against 3991 different peptides were measured. The Alexa Fluor 647 fluorescence image and a detail view (one square millimeter) are presented for an exemplary microarray incubated with serum of a patient with RRMS. The region on the right shows a spot with high signal intensity. It contains a 20mer peptide derived from the EBNA1 protein that is bound by patient IgG (“GRRPFFHPVGEADYFEYHQE,” EBNA1 401–420).
Data Preprocessing
The following summarizes the preprocessing of the peptide microarray data. For a more complete description of the bioinformatic workflow, the reader is referred to our previous publication (39).
First of all, each microarray image was processed with the GenePix Pro software version 6.1 (Molecular Devices, Sunnyvale, CA, USA) to calculate the median Alexa Fluor 647 fluorescence intensity for each peptide spot and its respective local background (that is, the area surrounding a spot). The software also automatically assigned flags to mark abnormal spots. For each peptide spot, we subtracted the background intensity from the spot intensity. If the fluorescence was higher in the background than in the spot, we set the signal intensity to 1 (truncation). Outliers in the data of each microarray were detected by computing the coefficient of variation (CV) for each triplicate. Spot triplicates with CV >1.685 indicated outliers. Each outlier spot was flagged and corrected by setting its spot intensity to the mean intensity of the two respective spots from the other two subarrays. Finally, to end up with one signal value for each peptide and each microarray, we used the average of the triplicate spot intensities after background subtraction, truncation and outlier correction (Fig. 2).
Fig. 2.
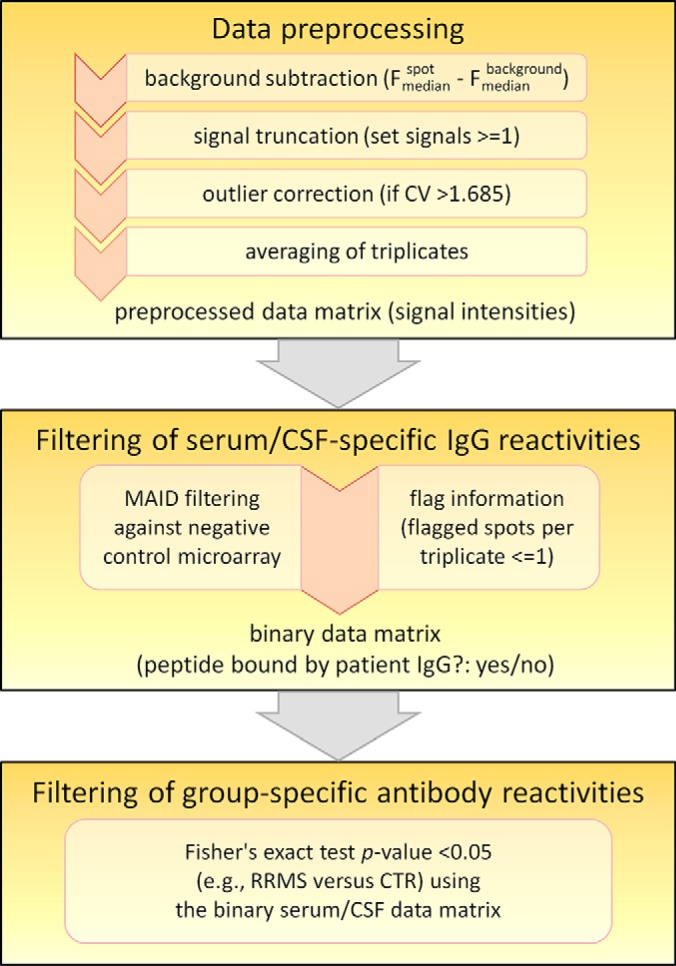
Data analysis workflow. This diagram depicts the bioinformatic steps after microarray image analysis. The GenePix Pro software delivered the fluorescence (F) of each spot as well as the local background around each spot (median pixel intensities) and assigned flags to mark irregular spots. The subsequent preprocessing of the data included a correction of outliers, which were detected by calculating the coefficient of variation (CV) for each triplicate. A two-step filtering strategy was applied to obtain lists of peptides recognized significantly more often by serum or CSF antibodies from one group of patients compared with another group.
Filtering of Specific Antibody Reactivities
Because antibody reactivities depend on both affinity and titer, the signal intensities measured for a peptide can vary greatly among individuals, even if they have, in principle, antibodies against the same epitope. Moreover, the specificity of the secondary anti-IgG antibody affects the data as well. Therefore, a negative control is needed to evaluate if the secondary antibody directly binds to some of the spotted peptides. We previously implemented a two-step filtering method to identify peptides specifically bound by patient IgG (39). Firstly, we select the peptides for which the signal of at least one microarray incubated with serum/CSF is notably higher compared with the respective signal of the negative control microarray. Secondly, the remaining peptides are filtered according to their capacity to distinguish the groups of patients.
In the first step, a MA plot-based signal intensity-dependent (MAID) filtering method was applied to extract for each microarray the set of peptides that showed serum/CSF-specific IgG reactivities. The MAID method calculates signal intensity-dependent fold-changes (MAID-scores) based on the MA plot visualization as described previously (Fig. 3) (39, 40). MAID-scores realize a smooth combination of two requirements, high signal intensity, and high fold-change (that is, the ratio comparing the peptide signal of a microarray incubated with serum/CSF to the respective signal of a negative control microarray). The MAID-score cut-off was chosen so that a filtered peptide had to have a signal > = 400 even if the negative control showed no reactivity (preprocessed signal = 1). For peptides with much higher signal intensities, a lower fold-change was sufficient to survive the filtering. This relatively stringent filtering was further restricted by rejecting peptides for which >1 of the triplicate spots were flagged.
Fig. 3.
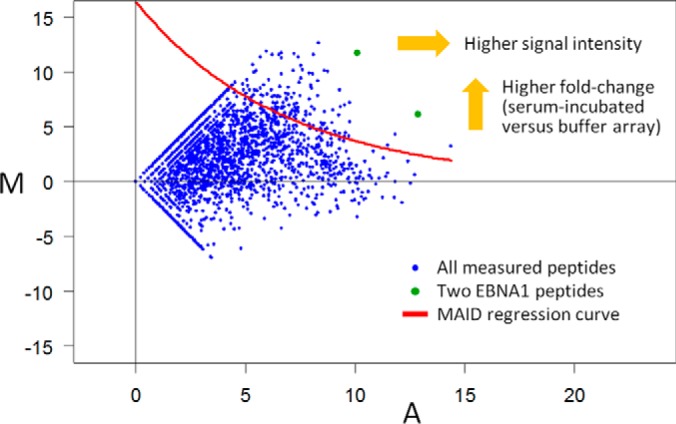
Exemplary MA plot visualizing the filtering method. This MA plot compares the signal intensities of a negative control microarray (Buffer_2) with those of a microarray incubated with serum of patient RRMS_10. For each peptide spotted in triplicate, A is the mean log2 signal of both microarrays and M is the log2 fold-change. An M value of 10 thus means that the peptide signal was 1024-fold higher for the array incubated with serum than for the buffer control. To identify peptides specifically bound by patient-derived IgG, an exponential function (MAID regression curve (40)) was fitted to the signal intensity-dependent variability in the data. In this case, 138 peptides survived this filtering (blue dots above the red line), including two overlapping peptides of the viral protein EBNA1 (shown in green, “GSPPRRPPPGRRPFFHPVGE” and “GRRPFFHPVGEADYFEYHQE,” EBNA1 392–420).
In the second step, peptides with group-specific antibody reactivities were filtered. For this purpose, we used the binary categorization of the first filtering step, which provided for each array the information whether or not a peptide signal can be regarded as serum/CSF-specific. We then used the one-tailed Fisher's exact test to evaluate for each peptide if either the RRMS patients or the PPMS patients had more often a serum/CSF-specific signal than the control cohort (Fig. 2). Raw p values <0.05 were considered statistically significant and indicated peptides recognized by MS-specific antibodies. This significance threshold seems somewhat permissive in the context of multiple testing (with an achieved power of >0.60 per test). However, we did not want to miss potentially discriminating peptides. As outlined in the introduction, usually only a subpopulation of the patients shows IgG reactivities against a certain antigen, and different patients may have antibodies binding to different epitopes of the same target protein.
All analyses were performed in the R environment for statistical computing accessed through a local webserver maintained at the Institute of Immunology of the University of Rostock. Protein structures were obtained from the Protein Data Bank Japan (PDBj, http://www.pdbj.org) (41) and visualized with Chimera version 1.10.1 (42).
Verification of Peptide–Antibody Reactivities
Antibody reactivities against four selected peptides were re-evaluated using enzyme-linked immunosorbent assays (ELISA). We used the same serum/CSF samples for the ELISA as for the peptide microarray experiment. The peptides were prepared with an additional C-terminal glycine and with biotin coupled at the N termini via a double PEG2 (8-amino-3,6-dioxaoctanoic acid) linker. They were commercially synthesized, purified to >95% purity and delivered as a lyophilized powder by Biosyntan (Berlin, Germany). Serum/CSF IgG binding to these peptides was detected with polyclonal Zenon goat horseradish peroxidase (HRP)-conjugated anti-human IgG Fab fragments (Thermo Fisher Scientific, catalog number Z-25454, diluted 1:3000 in wash buffer). This secondary anti-antibody is similar to the one used for the peptide microarrays. Moreover, peptide-antibody reactivities in serum (but not in CSF because of limited amounts of samples) were also tested with three other secondary antibodies that were HRP-conjugated and anti-human IgG Fc fragment-specific, namely polyclonal mouse antibodies (Jackson ImmunoResearch, West Grove, PA catalog number 209–035-098, 1:6500 dilution) and two monoclonal mouse antibody preparations (clone HP6017: Thermo Fisher Scientific, catalog number 05–4220, 1:250 dilution; clone H2: Abcam, Cambridge, UK, catalog number ab99765, 1:8000 dilution). Different secondary antibodies were used, because their specificities (possibly including cross-reactivities against the peptide sequences) differ among different preparations dependent on, e.g. the host animal, clonality, and antibody structure (whole IgG or Fab fragments).
Following our standard ELISA protocol, Pierce NeutrAvidin Coated High Capacity Plates (clear, 96-well) (Thermo Fisher Scientific) were used to evaluate the individual antibody responses in serum/CSF of all patients and controls. All plates were first washed with Tris-buffered saline (TBS) (50 mm Tris-HCl pH 7.4 and 150 mm NaCl). The lyophilized peptides were dissolved to 0.5 mm concentration in TBS containing 5% dimethyl sulfoxide, diluted in TBS, dispensed into each well (50 pmol in 100 μl) and incubated for 2 h at room temperature. Each well was then washed with wash buffer (TBS, 0.05% Tween 20, 0.1% BSA). Subsequently, 100 μl of serum samples (diluted 1:100 in wash buffer) and CSF samples (1:2.75 dilution) were added to each well, respectively. After 1 h of incubation at room temperature, the plates were washed, incubated for 1 h with diluted secondary antibodies (100 μl/well) and washed again with wash buffer to remove unbound antibodies. In all ELISA reactions, 2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS) (Sigma-Aldrich, St. Louis, MO, USA) was used as a chromogenic substrate for HRP. Each well was filled with 100 μl of 1.74 mm ABTS in buffer (39.8 mm citric acid, 3.25 mm NaBO3, 60 mm Na2HPO4, pH 4.4). Finally, the absorbance was read after 10 min at 405 nm using an Anthos 2010 ELISA reader with ADAP Basic software (Anthos Labtec Instruments, Wals-Siezenheim, Austria). In the same way, we also measured the reactivities of a pooled mixture of all 30 sera as well as of negative controls at least in triplicates.
RESULTS
Overview on the Peptide Microarray Data
The peptide microarray antibody profiling was performed in parallel for serum and CSF samples from RRMS patients (n = 10), PPMS patients (n = 10), and controls (n = 10). Each custom peptide microarray facilitated the investigation of antibody reactivities against 3991 different peptides spotted in triplicate.
An initial quality control included visual inspections of the microarray fluorescence scans and comparisons of the data of the three subarrays of each microarray. Data consistency and similarity between microarrays were in general good, and only minor issues were observed in case of four CSF-incubated microarrays (supplemental File S3). Therefore, all 62 microarrays were used for the further analysis without an additional across-array normalization.
As a typical finding of peptide microarray experiments, the signal distribution was highly skewed. For the vast majority of peptides, the measured signal intensities were very low. In the serum data set and in the CSF data set 90.8% and 95.8% of the data points had a preprocessed peptide signal of <400, respectively. Only few peptides showed strong antibody responses. On the other hand, 35 peptides showed a signal of >400 in both negative control microarrays because of reactivities of the secondary anti-antibody.
In general, lower IgG responses were observed in CSF than in serum. For 82.7% of the peptides, the average signal was higher for the sera than for the CSF samples. The preprocessed data of all peptide microarrays are available in supplementary File S4.
Filtering of Peptides Bound by IgG from Serum and CSF
In the first filtering step, we identified IgG-reactive peptides. For this purpose, we compared the signals of serum/CSF-incubated microarrays with those of a negative control microarray using the MAID method (39) and flag information. The microarray labeled as “Buffer_2” served as negative control (“buffer array”) in all comparisons. This microarray had somewhat higher but overall similar signal intensities compared with “Buffer_1” (Pearson correlation coefficient r = 0.828). As can be seen in Fig. 3, the MAID filtering approach considers that the relative stochastic variation is higher for low signal intensities. Therefore, MAID implements a signal intensity-dependent fold-change to differentiate patient-derived IgG reactivities from secondary antibody effects.
Relatively few peptides survived this first filtering. For most peptides, the signal was always either low or just reflecting the reactivities of the secondary antibody. Fewer peptides were IgG-reactive in CSF than in serum. In the serum data set, 42–312 peptides (on average 140 peptides) were filtered per individual sample. In the CSF data set, 2–131 peptides (on average 30 peptides) were filtered per sample. The number of peptides filtered as bound by IgG for each of the 30 subjects correlated with the amount of total IgG in serum and CSF, respectively (Pearson correlation p values <0.05).
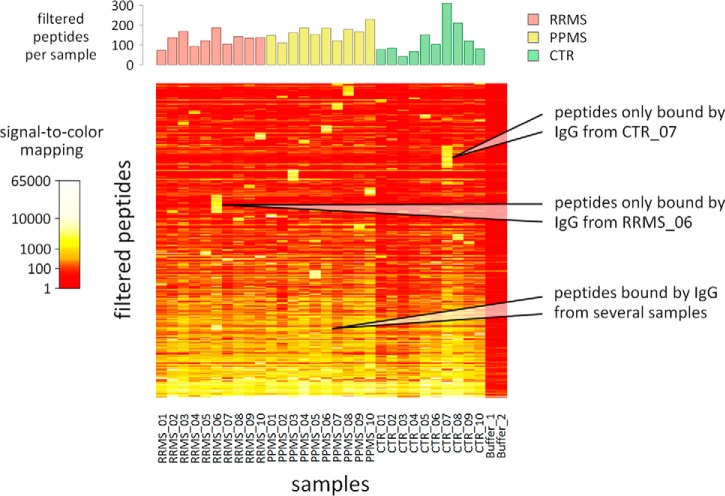
In consequence, only some of the 3991 spotted peptides showed serum/CSF-specific reactivities for at least onesample. In total, signals of 852 peptides were filtered as serum-specific, and signals of 255 peptides were filtered as CSF-specific (supplemental File S4). These two subsets overlapped significantly with 218 peptides, thus 889 peptides were filtered altogether. The data of these peptides are visualized as heatmaps in Fig. 4 (serum samples) and supplemental File S5 (CSF samples). These heatmaps display the comparability of the microarray measurements but also the strong individual differences. Different samples were usually reactive against different peptides. In fact, many of the filtered peptides were recognized by only one of the 30 serum samples (n = 412) or only one of the 30 CSF samples (n = 119). In contrast, high signals across several samples were observed for only few peptides. There was no peptide with IgG-specific signals in all CSF samples, but 16 peptides were bound by IgG from all 30 sera (supplemental File S4).
Fig. 4.
Heatmap visualization of serum antibody reactivities. This heatmap shows the signal intensities in the serum data set for 852 of the 3991 spotted peptides. These peptides were filtered as recognized by IgG from at least one of the 30 serum samples. Individually distinct antibody reactivities could be seen. Many peptides showed a serum-specific signal only for one sample (upper half). On the other hand, many peptides were detected with high signals for several samples, with no obvious difference between MS patients and controls (lower half). This is a typical finding in peptide microarray studies (39). The number of peptides filtered for each sample is displayed on top.
The antibody signatures of the serum samples were in general similar to those of the matched CSF samples. Correlation of the data for the sera and CSF samples was significant (Pearson correlation p value <0.05) for 425 of the 889 filtered peptides.
None of the four HHV-6 epitopes obtained from IEDB (25) was filtered. In contrast, serum/CSF-specific peptide signals were observed for 61 of the 103 epitopes that were listed for HHV-4.
Filtering of Antibody Reactivities Specific for Multiple Sclerosis
In the second filtering step, MS-specific antibody reactivities were identified by applying Fisher's exact test on the data of each peptide that survived the first filtering. This filtering did not require a peptide to be recognized by MS samples exclusively. Instead, we searched for peptides that showed significantly more often a serum/CSF-specific signal for samples of MS patients than for samples of controls. Four main comparisons were made, namely RRMS>CTR and PPMS>CTR for both the serum profiles and the CSF profiles.
In the serum data set, we filtered 13 peptides (RRMS>CTR) and 26 peptides (PPMS>CTR) as MS-specific. In the CSF data set, we filtered two peptides (RRMS>CTR) and 23 peptides (PPMS>CTR). The respective recognition frequencies (that is, the number of samples with a much higher peptide signal than the negative control array) are presented in Table II and Table III. Overlapping results were obtained for RRMS and PPMS patients. Serum IgG reactivities against six peptides and CSF IgG reactivities against two peptides were found to be more frequent in both types of MS than in controls (RRMS>CTR and PPMS>CTR). In total, 33 and 23 different peptides were thus filtered as MS-specific in serum and CSF, respectively (Table II and Table III). Two peptides in turn appeared in both of these peptide sets, ultimately resulting in 54 different peptides. The Venn diagram in Fig. 5 summarizes all filtering results and depicts the intersections of the sets for both MS groups as well as for serum/CSF.
Table II. MS-specific antibody reactivities in serum. List of 33 peptides which were significantly more often bound by serum IgG of either RRMS or PPMS patients compared to controls. Three groups of peptides were distinguished: peptides of human origin, peptides from EBV protein sequences and peptides from random peptide libraries described in the literature to be recognized by MS antibodies. UniProt protein names in bold indicate that an overlapping peptide was filtered as well. The region within the respective protein is given for the main isoform if not otherwise indicated. In case of EBV peptides, sequences were mapped to proteins of strain B95–8. For each peptide, the number of patients with serum/CSF-specific antibody reactivity is presented. The numbers are bold and underlined if they are significantly higher for MS patients according to Fisher's exact test. Mean signal intensities are shown in the rightmost columns for each group and the negative control microarrays (“Buffer”).
| Peptide | Source | Serum |
CSF |
Average signal (serum) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| RRMS | PPMS | CTR | RRMS | PPMS | CTR | RRMS | PPMS | CTR | Buffer | ||
| Peptides derived from human proteins | |||||||||||
| KRGILTLKYPIEHGI | ACTB/ACTG (61–75) | 4 | 8 | 2 | 0 | 0 | 0 | 451 | 862 | 451 | 2 |
| LKYPIEHGIVTNWDD | ACTB/ACTG (67–81) | 1 | 7 | 0 | 0 | 0 | 0 | 244 | 440 | 169 | 2 |
| PIYEGYALPHAILRL | ACTB/ACTG (164–178) | 3 | 8 | 2 | 0 | 4 | 0 | 1478 | 8479 | 1457 | 48 |
| SLSTFQQMWISKQEY | ACTB/ACTG (348–362) | 2 | 6 | 1 | 0 | 0 | 0 | 876 | 1135 | 703 | 21 |
| AVLAGGLYEYVFCPD | AQP4 (241–255) | 10 | 10 | 4 | 1 | 4 | 1 | 1329 | 2057 | 975 | 8 |
| AGQVFLEELGNHKAF | CN37 (205–219) | 0 | 6 | 1 | 0 | 0 | 0 | 218 | 861 | 271 | 3 |
| MDIAIHHPWIRRPFF | CRYAB (1–15) | 3 | 7 | 0 | 1 | 1 | 0 | 25948 | 40751 | 10885 | 4451 |
| IDFLIEEIERLGQDL | DCE2 (571–585) | 1 | 4 | 0 | 0 | 1 | 0 | 430 | 697 | 326 | 5 |
| DQWREWADSKSCCDY | DPYL2 (121–135) | 0 | 4 | 0 | 0 | 0 | 0 | 278 | 560 | 89 | 10 |
| SWYDNEFGYSNRVVD | G3P (312–326) | 9 | 8 | 4 | 0 | 3 | 0 | 1207 | 1257 | 595 | 6 |
| MHEAEEWYRSKFADL | GFAP (250–264) | 4 | 6 | 0 | 0 | 0 | 0 | 710 | 1148 | 398 | 6 |
| DEMARHLQEYQDLLN | GFAP (340–354) | 6 | 4 | 1 | 0 | 0 | 0 | 929 | 659 | 207 | 5 |
| RGQYSRASWEGHWSP | GFAP isoform 2 (390–404) | 5 | 7 | 1 | 0 | 1 | 0 | 1593 | 1852 | 877 | 48 |
| ELRPAVVHGVWYFNS | MAG (49–63) | 9 | 10 | 6 | 0 | 1 | 0 | 1371 | 1790 | 1152 | 15 |
| LLSNVSPELGGKYYF | MAG (103–117) | 2 | 5 | 0 | 0 | 0 | 0 | 414 | 600 | 399 | 3 |
| CQASFPNTTLQFEGY | MAG (217–231) | 2 | 8 | 3 | 0 | 1 | 0 | 707 | 1391 | 857 | 9 |
| YEDGFSQPRGWNPGF | PERT (181–195) | 7 | 9 | 4 | 3 | 4 | 0 | 1570 | 2122 | 1607 | 37 |
| YSNISKDRRYADLTE | PGAM1 (133–147) | 4 | 7 | 1 | 0 | 0 | 0 | 534 | 1089 | 337 | 7 |
| ELDENGDGEVDFQEY | S10A1 (61–75) | 5 | 3 | 0 | 1 | 0 | 0 | 2931 | 1017 | 176 | 7 |
| VAALTVACNNFFWEN | S10A1 (79–93) | 2 | 5 | 0 | 0 | 0 | 0 | 480 | 717 | 225 | 9 |
| AALTVACNNFFWENS | S10A1 (80–94) | 4 | 8 | 0 | 0 | 1 | 0 | 637 | 1313 | 400 | 10 |
| QQQQFNRNVEDIELW | SPTA2 (679–693) | 4 | 2 | 0 | 0 | 0 | 0 | 1048 | 429 | 54 | 5 |
| FFMPGFAPLTAQGSQ | TBB1 (265–279) | 1 | 8 | 3 | 0 | 1 | 0 | 496 | 1219 | 679 | 10 |
| Peptides derived from EBV proteins | |||||||||||
| ETFTETWNRFITHTE | EAR (63–77) | 3 | 7 | 0 | 0 | 0 | 0 | 657 | 804 | 239 | 10 |
| GRPGAPGGSGSGPRHRDGVR | EBNA1 (52–71) | 6 | 4 | 1 | 1 | 2 | 0 | 3551 | 1938 | 2991 | 44 |
| GSGPRHRDGVRRPQKRPSCI | EBNA1 (61–80) | 3 | 7 | 2 | 1 | 1 | 0 | 4149 | 8767 | 2020 | 22 |
| RPQKRPSCIGCKGTHGGTGA | EBNA1 (72–91) | 6 | 7 | 2 | 0 | 0 | 0 | 5604 | 2428 | 852 | 36 |
| GCKGTHGGTGAGAGAGGAGA | EBNA1 (81–100) | 4 | 0 | 0 | 1 | 0 | 0 | 772 | 288 | 174 | 18 |
| SGSPPRRPPPGRRPFFHPVG | EBNA1 (391–410) | 5 | 6 | 0 | 0 | 0 | 0 | 11092 | 11408 | 1964 | 565 |
| PPAAGPPAAGPPAAGPPAAG | EBNA6 (566–595) | 8 | 5 | 3 | 1 | 0 | 0 | 9645 | 1616 | 1351 | 26 |
| QVSLESVDVYFQDVFGTMWC | GP350 (122–141) | 10 | 10 | 6 | 4 | 3 | 3 | 8552 | 7649 | 7556 | 13 |
| Peptides not derived from known proteins | |||||||||||
| HTQPYAYEARDH | - | 8 | 8 | 1 | 1 | 2 | 0 | 5688 | 1382 | 434 | 13 |
| VPQKYWWLSDHT | - | 6 | 9 | 3 | 1 | 3 | 1 | 915 | 1559 | 1058 | 17 |
Table III. MS-specific antibody reactivities in CSF. List of 23 peptides which were significantly more often bound by CSF IgG of either RRMS or PPMS patients compared to controls. CSF antibody reactivities against 17 peptides from human proteins, 1 EBV-derived peptide and 5 mimotopes previously identified using phage-displayed random peptide libraries were significantly associated with MS (p values <0.05). The structure of the table is as for Table II. Note that due to high sequence homology, ACTB and ACTG share the same potential epitopes. For 5 peptides (indicated in bold), the average signal intensities were higher for CSF samples than for serum samples, suggesting intrathecal IgG synthesis.
| Peptide | Source | Serum |
CSF |
Average signal (CSF) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| RRMS | PPMS | CTR | RRMS | PPMS | CTR | RRMS | PPMS | CTR | Buffer | ||
| Peptides derived from human proteins | |||||||||||
| PIYEGYALPHAILRL | ACTB/ACTG (164–178) | 3 | 8 | 2 | 0 | 4 | 0 | 692 | 2539 | 397 | 48 |
| PLYFGWFLTKKSSET | CN37 (187–201) | 4 | 7 | 5 | 0 | 4 | 0 | 370 | 842 | 235 | 21 |
| LFPTSTSLSPFYLRP | CRYAB (37–51) | 0 | 1 | 3 | 2 | 4 | 0 | 4118 | 7240 | 2761 | 593 |
| SLSPFYLRPPSFLRA | CRYAB (43–57) | 0 | 0 | 0 | 0 | 4 | 0 | 4585 | 7860 | 2469 | 1367 |
| EGSGRYIPRKPFPDF | DPYL2 (463–477) | 1 | 0 | 0 | 0 | 4 | 0 | 2512 | 4700 | 1143 | 573 |
| VHGVWYFNSPYPKNY | MAG (55–69) | 1 | 4 | 4 | 1 | 4 | 0 | 1354 | 1977 | 814 | 126 |
| YYFRGDLGGYNQYTF | MAG (115–129) | 1 | 1 | 1 | 3 | 4 | 0 | 2395 | 2418 | 860 | 134 |
| PGFGYGGRASDYKSAHKGFK | MBP (257–276) | 1 | 1 | 1 | 0 | 4 | 0 | 597 | 1564 | 523 | 144 |
| RNVRFSDEGGFTCFF | MOG (115–129) | 0 | 0 | 0 | 0 | 4 | 0 | 140 | 392 | 49 | 2 |
| ELLVLRQKHSEPSRF | NFL (121–135) | 0 | 2 | 1 | 0 | 4 | 0 | 809 | 1514 | 336 | 94 |
| FFPFISRGKELLWGK | PERT (19–33) | 2 | 5 | 4 | 1 | 4 | 0 | 637 | 1640 | 558 | 123 |
| YEDGFSQPRGWNPGF | PERT (181–195) | 7 | 9 | 4 | 3 | 4 | 0 | 905 | 1305 | 414 | 37 |
| DDRYSDLLMAWGQYI | PERT (223–237) | 9 | 10 | 10 | 4 | 4 | 0 | 431 | 704 | 67 | 3 |
| LTSTVICRWRVLGWK | PERT isoform 3 (865–879) | 0 | 0 | 1 | 0 | 4 | 0 | 260 | 716 | 117 | 75 |
| QALRDAGYEFDICFT | PGAM1 (43–57) | 1 | 1 | 1 | 1 | 4 | 0 | 252 | 472 | 143 | 7 |
| RQKLEDSYRFQFFQR | SPTA2 (37–51) | 0 | 1 | 0 | 0 | 4 | 0 | 1868 | 3854 | 1131 | 576 |
| YNYYKKFSYKTIVMG | TALDO (221–235) | 0 | 0 | 0 | 0 | 4 | 0 | 683 | 1567 | 547 | 138 |
| Peptides derived from EBV proteins | |||||||||||
| GSPPRRPPPGRRPFFHPVGE | EBNA1 (392–411) | 8 | 10 | 8 | 5 | 8 | 2 | 14490 | 15898 | 4074 | 543 |
| Peptides not derived from known proteins | |||||||||||
| ARWELNLEPLYGPY | - | 10 | 10 | 10 | 8 | 10 | 3 | 3805 | 4726 | 1627 | 34 |
| EPMTPHQWITLYRSY | - | 1 | 0 | 1 | 1 | 4 | 0 | 1483 | 1976 | 588 | 163 |
| SCNYHGRTLTCW | - | 4 | 5 | 4 | 2 | 4 | 0 | 1468 | 2115 | 741 | 119 |
| WHAPQTPYWASM | - | 10 | 10 | 10 | 5 | 6 | 1 | 4637 | 2137 | 426 | 27 |
| WHIPPNIGRTFS | - | 4 | 7 | 5 | 1 | 4 | 0 | 1602 | 2025 | 496 | 97 |
Fig. 5.
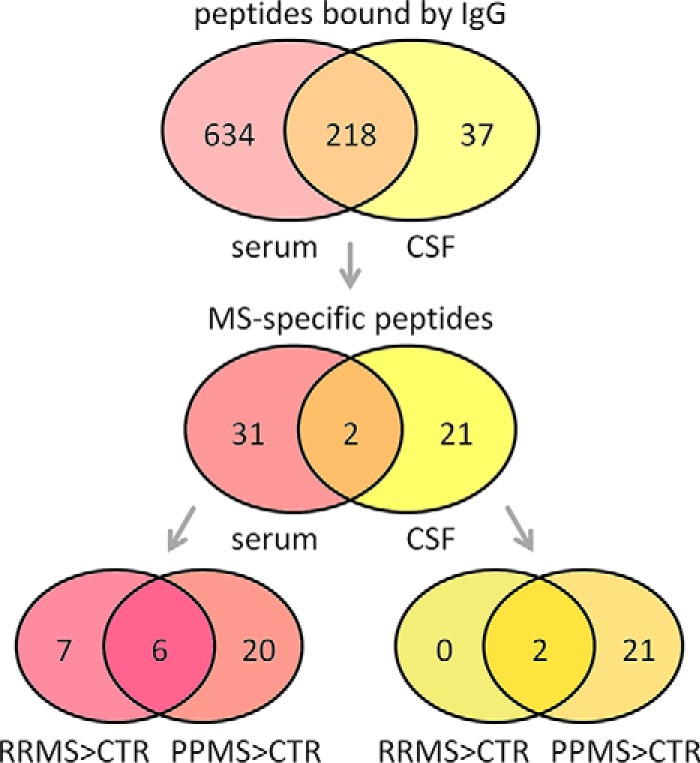
Summary of the results from profiling (auto)antibodies in multiple sclerosis. Of the 3991 peptides analyzed, 852 were found to be bound by IgG of at least one serum sample, and 255 peptides were found to be bound by IgG of at least one CSF sample (first filtering step based on MAID-scores and flag information). The upper Venn diagram depicts the intersection of these two sets with 218 peptides. In the second filtering step, we used Fisher's exact test to identify peptides that were IgG-reactive more often either in the RRMS patient group or in the PPMS patient group than in the control group. In the serum data set, 13 peptides were preferentially bound by sera of RRMS patients (“RRMS>CTR”). Antibody reactivities against six of these peptides were also characteristic for sera of PPMS patients (intersection with “PPMS>CTR” in the lower left Venn diagram). In total, 33 and 23 peptides were filtered as MS-specific for serum and CSF, respectively.
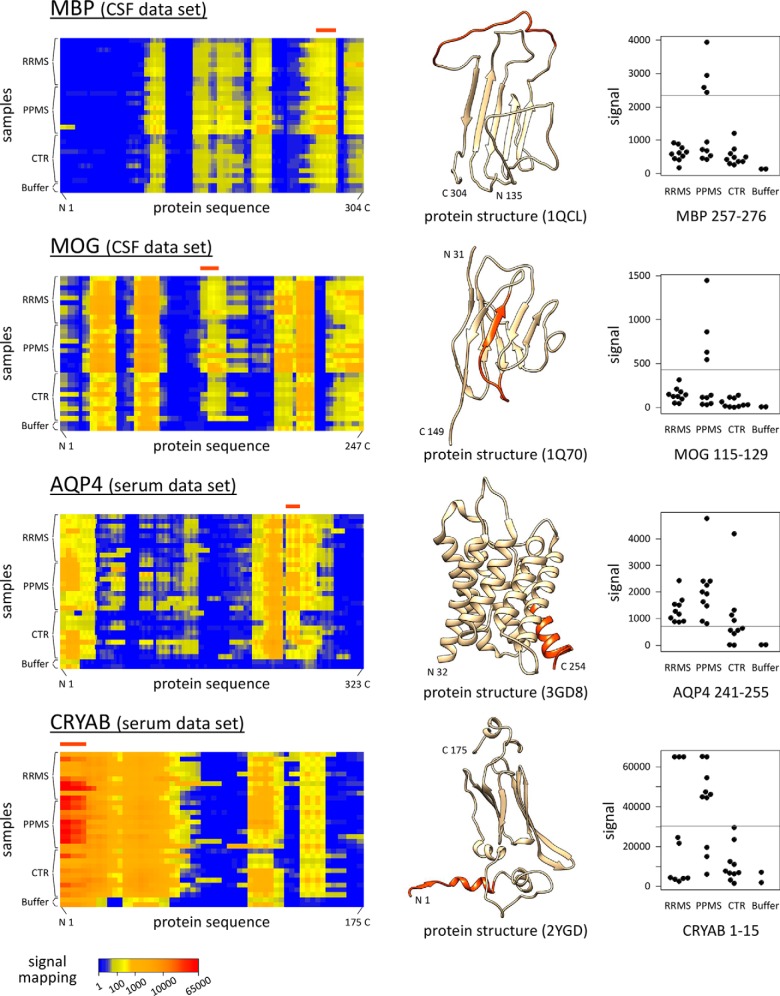
A subset of 38 of the 54 filtered peptides was derived from 19 human proteins (and their isoforms) that were selected for the peptide microarray design. The data for peptides from MBP, MOG, AQP4, and CRYAB are exemplarily shown in Fig. 6. The remaining 16 peptides represent EBV protein sequences (n = 9) and potential MS-specific mimotopes determined by others using random peptide libraries (n = 7). No HHV-6 epitope and no peptide with citrulline (Z) was found to present increased IgG reactivities in serum/CSF of MS patients.
Fig. 6.
Potential autoantigenic regions of selected proteins. IgG reactivities against 15mer peptides derived from the amino acid sequences of MBP, MOG, AQP4, and CRYAB were measured in CSF and serum. The figures on the left show compact heatmap visualizations (VisualMaps (39)) of the data for the serum/CSF-incubated microarrays and the two negative control microarrays (Buffer_1–2). Each row corresponds to a microarray, and the antibody reactivities are visualized from the N- to the C terminus as calculated from the signals of overlapping peptides. Bands with elevated signals (yellow to red) and “epitope silent” regions (blue) are seen for MS patients as well as controls. If the signals are as high as for the negative control microarrays, the bands are caused by cross-reactivities of the secondary anti-IgG antibody. The MAID filtering method (40) followed by Fisher's exact test identified linear epitopes that were significantly more frequently recognized by MS patients (Table II and Table III). Filtered peptides were colored in orange in the three-dimensional protein structures, which were downloaded from PDBj (41). Known or predicted structures were usually available only for truncated protein fragments, and monomers are shown even if the proteins may form higher-order assemblies. MS-specific antibodies were found to bind peptides matching, e.g. a loop within MBP (CSF data set) and the N-terminal region of CRYAB (serum data set). The respective peptide signal intensities are plotted in the figures on the right for each individual subject. Gray horizontal lines indicate the cut-off for serum/CSF-specific signals. For instance, IgG to CRYAB 1–15 (“MDIAIHHPWIRRPFF”) were detected for three RRMS, seven PPMS, and zero CTR serum samples.
Although reactivities against only 54 peptides were filtered as MS-specific, their relevance is clearly supported by the fact that the results for RRMS and PPMS overlapped partially. Moreover, when we applied exactly the same criteria to screen for peptides that were less often bound by IgG from MS patients (CTR>RRMS and CTR>PPMS), only four peptides from four different proteins were filtered (supplemental File S6). The results also illustrate that antibody reactivities against the analyzed peptides are sometimes elevated or more common in MS, but they are usually not limited to MS. For instance, each of the 10 control sera showed IgG responses against at least three of the 23 peptides filtered as MS-specific in the CSF data set. Only 18 of the 54 peptides filtered in total showed consistently no serum/CSF-specific signal for all 10 controls. Therefore, the majority of these peptides was recognized by at least one of the control samples.
The CSF samples of four PPMS patients (PPMS_05, PPMS_07, PPMS_09, and PPMS_10) each showed IgG-specific signals for all 23 peptides filtered for CSF (Table III). This dominant signature was already evident in the heatmap (supplemental File S5). All of these four PPMS patients had a particularly high CSF/serum quotient of total IgG (supplemental File S1), indicative of intrathecal IgG synthesis or blood-CSF barrier dysfunction.
For the human proteins ACTB, ACTG, S100A1, and CRYAB, antibody reactivities against overlapping peptides were filtered to be MS-specific. In these cases, the linear epitope can be confined to the amino acid sequence that is shared by both 15mer peptides. One of the two overlapping peptides always tended to show higher signals than the other one. Therefore, all samples with an IgG response against one of the peptides also showed an IgG response to the other peptide, thus confirming the measurement. For instance, the peptides “KRGILTLKYPIEHGI” and “LKYPIEHGIVTNWDD” showed serum-specific signals for 12 and eight MS patients, respectively. They represent the amino acid positions 61–75 and 67–81 from β-actin (ACTB) and gamma-actin (ACTG). ACTB and ACTG have 98.9% sequence identity and they differ only at the N-terminal end. Their protein sequences were screened for autoantibody binding patterns by 129 different peptides on the microarrays (linear epitope mapping). The region 67–75 (“LKYPIEHGI”), that is covered by both filtered peptides, is likely to be an epitope of these proteins. However, on closer inspection, the absolute peptide signals were only moderately elevated (<2000) in MS samples compared with controls (Table II, supplemental File S4).
Marked IgG reactivities were observed for the EBV protein EBNA1. Peptides matching this protein were selected from the immune epitope repository IEDB (25). Hence, the EBNA1 protein was not represented in full length on the arrays. All 30 patients had serum antibodies binding to at least one peptide derived from EBNA1. The peptide “SGSPPRRPPPGRRPFFHPVG” (EBNA1 391–410) was filtered as MS-specific in the serum data set (Table II), whereas the peptide “GSPPRRPPPGRRPFFHPVGE” (EBNA1 392–411) was filtered as MS-specific in the CSF data set (Table III). The sequences of the two peptides are shifted only by a single amino acid. However, much stronger IgG reactivities were always observed for the latter peptide, which had a higher average signal for CSF than any other tested peptide and even saturated signal intensities (>60,000) for 13 of the 30 sera (supplementary File S4). Moreover, four overlapping peptides spanning the region EBNA1 52–100 were found to be bound more often by IgG from serum samples of MS patients (Table II). Because these peptides were recognized differently by the samples, one can combine the results to better discriminate MS patients and controls. With the criterion that a sample must show IgG effects for at least two of the four peptides, which was the case for one CTR, seven RRMS, and eight PPMS serum samples, the differences between the groups were more significant (Fisher's exact test p values <0.01) than for each peptide alone.
Association of Antibody Responses to Clinical Parameters
After the sampling of serum/CSF, the individual course of disease was followed for 12 months for each MS patient. Six of the RRMS patients were relapse-free in this observation period, and three RRMS patients (RRMS_01, RRMS_03, and RRMS_08) experienced a relapse (RRMS_09 left the study during follow-up). In general, the degree of disability remained stable. Only three female patients (RRMS_07, PPMS_01, and PPMS_06) showed disease progression with an increase in the EDSS of at least one point (supplemental File S1). In the next step, we evaluated whether the peptide microarray data correlated with the clinical and demographic characteristics of the patients. More specifically, we tested whether the antibody profiles were associated with age, gender, type of MS, disease duration, disease activity (occurrence of relapses), level of disability (change in EDSS), treatment status, presence of OCB, and CSF IgG index.
When we used the same filtering criteria to directly compare both groups of MS patients (RRMS<>PPMS), largely the same sets of peptides were filtered as in the comparisons against the control group (Table II and Table III). The RRMS<>PPMS comparisons yielded only 11 additional nonoverlapping peptides (supplemental File S6). In general, stronger antibody responses were seen for PPMS patients than for RRMS patients (Fig. 5). Four PPMS patients (PPMS_05, PPMS_07, PPMS_09, and PPMS_10) showed a dominant CSF antibody profile. High signals were observed exclusively for the CSF samples of these four patients in case of 10 peptides (Table III). Six of the MS-specific peptides filtered from the serum data set (Table II) were preferentially bound by serum IgG of PPMS patients (PPMS>RRMS). For instance, “LKYPIEHGIVTNWDD” (ACTB/ACTG 67–81) showed a serum-specific signal for seven PPMS patients but only for one RRMS patient. On the other hand, antibody reactivities against “GCKGTHGGTGAGAGAGGAGA” (EBNA1 81–100) were measured for four patients with RRMS but no patient with PPMS.
We further noted that serum antibodies against some peptides derived from the EBV protein EBNA1 were less frequent for OCB-negative patients and patients in clinical remission. For instance, serum-specific reactivities against “RPQKRPSCIGCKGTHGGTGA” (EBNA1 72–91) were detected for none of the three RRMS patients in remission (RRMS_01, RRMS_03, and RRMS_09) but for six out of seven RRMS patients that were recruited during a phase of relapse. Similarly, the three female MS patients without IgG OCB (RRMS_01, PPMS_04, and PPMS_06) each showed low signal intensities (<200) for “GRPGAPGGSGSGPRHRDGVR” (EBNA1 52–71), whereas 10 OCB-positive MS patients showed clear IgG effects (signals >2000).
Apart from that, there was no compelling evidence that the individual antibody binding profiles correlate with disease activity or even prognosticate disease progression. However, this study was not explicitly designed and powered to detect significant clinicodemographic differences between patients with different antibody signatures.
Validation of CRYAB and EBNA1 Epitopes
Four peptides (CRYAB 1–15 as well as EBNA1 52–71, 391–410, and 392–411), which passed the MS-specific filtering of the peptide microarray data (Table II and Table III), were selected for the ELISA analysis. Serum antibodies recognizing the peptides were detected in the ELISA experiment with four different HRP-conjugated anti-human IgG secondary antibodies. In case of CSF, only polyclonal Zenon goat anti-human IgG Fab fragments were used as secondary antibody. The full ELISA results for all 30 paired serum/CSF samples and all negative controls are given in supplemental File S7.
The ELISA data correlated with the microarray data for both the serum samples and the CSF samples and for all peptides and secondary antibodies used (Pearson correlation coefficients r>0.477 and respective p values <0.01). In both data sets, the measured serum IgG reactivities against the three peptides from CRYAB 1–15 and EBNA1 391–411 correlated with each other because of sequence similarities (Pearson correlation p values <0.01), but they did not correlate with the data for EBNA1 52–71 (p values >0.45). Higher optical densities (OD) were usually raised with Zenon Fab fragments, but it should be noted that the absolute values largely depend on the preparation and dilution of each secondary antibody. In contrast to the microarray results, the secondary antibodies did in general not bind to the peptides directly in the ELISA negative control experiments (OD ≤ 0.040). The only exception was a monoclonal anti-IgG antibody, which slightly cross-reacted with the peptide representing EBNA1 391–410 (supplemental File S7).
When comparing the groups of patients, ELISA confirmed in part the findings obtained with the peptide microarrays, despite the differences between both experimental approaches. Using an OD cut-off of ≥ 0.610 for the data obtained with Zenon anti-IgG Fab fragments, serum antibodies binding to “MDIAIHHPWIRRPFF” (CRYAB 1–15) and “SGSPPRRPPPGRRPFFHPVG” (EBNA1 391–410) were detected significantly more frequently in PPMS than in CTR (one-tailed Fisher's exact test p values <0.05). The OD for these two peptides are visualized in Fig. 7. For CSF samples, there was no ELISA reaction with OD≥ 0.610. However, with the cut-off set at OD≥ 0.065, all four peptides were bound significantly more often by CSF antibodies from MS patients compared with controls (supplementary File S7).
Fig. 7.
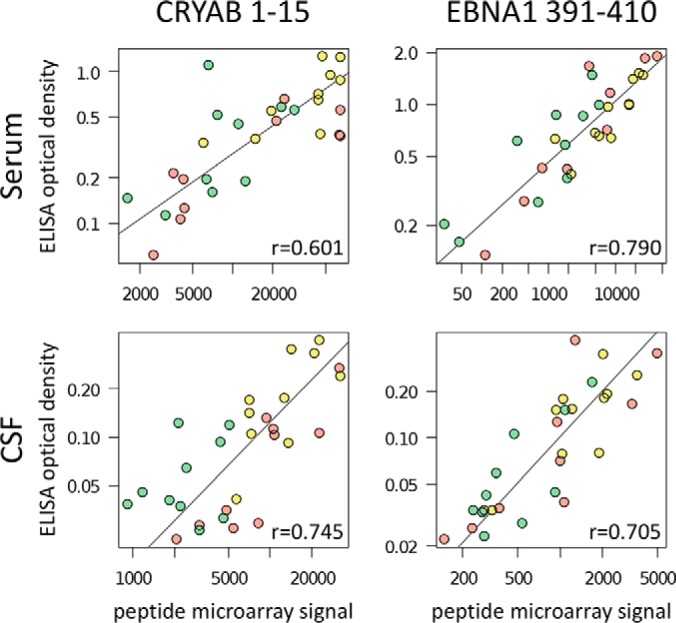
Verification of antibody responses to linear peptides by ELISA. ELISA were run with selected peptides to validate the measurement of serum/CSF antibody reactivities for all 20 MS patients and 10 controls. In this figure, the peptide microarray data (preprocessed signal intensities, supplementary File S4) and the ELISA data (optical density values, supplementary File S7) are compared for serum and CSF and the two peptides “MDIAIHHPWIRRPFF” (CRYAB 1–15) and “SGSPPRRPPPGRRPFFHPVG” (EBNA1 391–410). These peptides share the sequence motif “RRPFF,” and they were significantly more frequently detected by IgG in sera of MS patients in the microarray data (Table II). Patient IgG, which bound to the peptides, was measured using as secondary antibody polyclonal Zenon goat anti-human IgG Fab fragments conjugated with either a fluorescent dye (in case of peptide microarrays) or horseradish peroxidase (in case of ELISA) (Thermo Fisher Scientific). As indicated by the gray orthogonal linear regression lines, the data of both experiments correlated well (Pearson correlation coefficients r>0.6 and respective p values <0.001). Different colors designate the three patient cohorts RRMS (red), PPMS (yellow), and CTR (green).
DISCUSSION
Overview on Study Design, Analysis, and Results
Here, we examined the intrathecal and peripheral antibody repertoire against almost 4000 peptides in progressive and relapsing MS patients as well as controls by the use of custom high-density peptide microarrays. These microarrays allowed to screen for IgG against linear epitopes of many potential autoantigens (n = 45 proteins, supplementary File S2) and additional peptides previously described in the literature to be MS-specific or viral epitopes. The antibody signatures of the paired serum and CSF samples correlated, though signal intensities were in general lower for CSF-incubated arrays than for serum-incubated arrays. Still, ∼17% of the peptides had a higher mean signal intensity in the CSF data set, indicating that the antibody repertoire is somewhat different between the body fluids. The group-specific analysis revealed significantly increased serum/CSF antibody responses in MS patients compared with the control cohort. More peptides were filtered for serum, and a different and smaller set of peptides was filtered for CSF, suggesting both a peripheral and intrathecal immune dysregulation. A possible scenario is that the concentration of certain antibodies is reduced in the CSF because they are bound to tissue, as has been shown for anti-MOG antibodies (43). MS-associated antibody specificities identified in this study included peptides derived from 19 human proteins and four EBV proteins (Table II and Table III), which will be discussed in more detail later in this section.
The microarray data clearly showed that samples of different patients recognized different sets of peptides, illustrating that antibody reactivities are highly individual (Fig. 4). Environmental and genetic factors have an impact on adaptive immune responses, which can result in a diverse range of antibodies possibly targeting distinct epitopes of the same antigen. Therefore, a significant proportion of the immunosignature is personal. We used a two-step filtering approach to identify a disease signature over the personal ones (39). First, we determined for each serum/CSF sample, which peptides were bound by IgG. Secondly, we evaluated for each peptide whether MS patients had more often a serum/CSF-specific signal than the controls. By this means, antibody reactivities against 54 peptide sequences were filtered as more common in RRMS or PPMS, which may seem surprisingly few considering that 3991 peptides were tested, most of which were derived from proteins for which earlier studies provided evidence that they are autoantigens in MS (18, 39). However, one should note that large parts of an antigen are “epitope silent” and that the B-cell immunodominant regions usually consist of only few epitopes (44) (Fig. 6). Despite the relatively small sample size with 10 patients per group, there was a remarkable overlap in the results for RRMS and PPMS. Antibodies in serum/CSF against eight peptides were significantly more often found in both subtypes of MS than in controls (Fig. 5). Thus, more than half of the peptides filtered as RRMS-specific were also filtered as PPMS-specific. The robustness of the results is further shown by the fact that pairs of overlapping peptides were filtered. Nevertheless, MS is a heterogeneous disease, and if certain disease-specific antibodies are present in only a small percentage of patients, we could not detect them in the present study with the applied filtering criteria. Another critical point is that, although we filtered no MS-specific linear epitope for a subset of the examined proteins (supplemental File S2), our data should not be misinterpreted as disproving these proteins as autoantigens in MS. The antibody binding of epitopes depends on both the specific amino acid residues and their structure in three-dimensional space. Peptides presented on arrays represent just small parts of a protein sequence and may thus not be displayed in the correct conformation to be bound by IgG with high affinity. On the other hand, while conformational epitopes have been missed, peptides recognized by antibodies may not be accessible in the natively folded protein as a linear epitope. Therefore, the microarray results may contain peptide-antibody reactivities that are not pathologically relevant in vivo. These methodological issues are important to consider because the variety of technologies used for assaying antibodies also explains why sometimes divergent results are reported by different research groups (18).
None of the tested peptides was 100% specific and 100% sensitive for relapsing or progressive MS. In fact, many IgG responses were also seen in serum/CSF samples of the control group. Only 18 of the 54 filtered peptides showed signal intensities below the MAID-score cut-off in all control samples. This suggests that immune reactivities against these peptides are typically just more frequently seen in MS and possibly reflect an overabundance of naturally occurring autoreactive antibodies (23). Increased reactivities in control samples (CTR>RRMS or CTR>PPMS), on the contrary, were observed for only four peptides (supplementary File S6). Samples of RRMS and PPMS patients recognized to some degree the same peptides, but, unexpectedly, more peptides were filtered for PPMS>CTR than for RRMS>CTR (Fig. 5). Though, in the CSF data set, this pattern was dominated by four PPMS patients with relatively high total CSF IgG levels (PPMS_05, PPMS_07, PPMS_09, and PPMS_10), the serum antibody profiles pointed to a broadened immune response in the PPMS cohort as well. It may be speculated whether this reflects the PPMS patients' higher grade of disability (Table I) and/or diffuse rather than focal CNS inflammation (45). An earlier study by Quintana et al. with microarrays containing <370 peptides analyzed serum antibody signatures in RRMS and PPMS as well. Compared with healthy controls, they found five peptides with higher IgG reactivity in PPMS patients, but none in RRMS patients (29). These five peptides were not filtered as MS-specific in our study. However, our data confirmed several previously known antigens and epitopes, and, in addition, a number of hitherto unknown potential epitopes could be revealed.
MBP: Evidence of a Flexible Loop Epitope
Some researchers already used human samples to screen the MBP protein for linear epitopes. Belogurov et al. tested 12 peptides covering the MBP sequence (isoform 5) for serum IgG reactivities (46). Three of their peptides overlapped with the peptide MBP 257–276 (“PGFGYGGRASDYKSAHKGFK”), which was filtered as PPMS-specific in our CSF IgG data set (Table III). In their study, all three peptides showed significantly higher antibody levels in the PPMS/SPMS group compared with the healthy group. Moreover, we filtered “DTGILDSIGRFFGGD” (MBP 168–182) as frequently bound by sera of PPMS patients (supplemental File S6), which is also in accordance with the findings by Belogurov et al. (46). The region MBP 257–276 forms a flexible loop at the protein surface (Fig. 6), and it was previously shown to be the immunodominant epitope in a peptide microarray analysis with CSF of MS patients (33). However, serum antibodies against this region seem to be present in other (auto)immune diseases as well. Pediatric patients with acute disseminated encephalomyelitis (ADEM) presented even higher peripheral IgG reactivities for this epitope than RRMS patients (31). Moreover, roughly 20% of sera from patients with Sjögren's syndrome, systemic lupus erythematosus, and rheumatoid arthritis were found to be IgG-positive for the peptide “HKGFKGVD” (47), and serum antibodies against another peptide overlapping with this region were described for 29% of cases with Semple rabies vaccine-induced autoimmune encephalomyelitis (SAE) (48). Because of the immune response to MBP and pathological demyelination, SAE can be regarded as the human homolog of experimental autoimmune encephalomyelitis (EAE), a disease that can be induced in animals by immunization with myelin antigens from the CNS (49). EAE is widely used as a model for MS, and a considerable amount of literature has investigated the cellular and humoral response to MBP in this condition. However, differences in encephalitogenicity of MBP peptides have been observed between different animal strains, most likely because of differences in antigen presentation. For the same reason, it is difficult to compare the characteristics and the relevance of anti-MBP antibodies in EAE and MS. Some researchers have shown that autoantibodies against native human MBP have low binding affinities, that they are relatively infrequent in MS (50) and that they are found in sera of healthy individuals as well (51).
MOG: Peptides Mimic Parts of a Milk Protein
Several studies have aimed to establish a role for antibodies targeting MOG, a membrane protein expressed on the surface of myelin sheaths. MOG is homologous to the N-terminal domain of the bovine milk protein butyrophilin (BTN). It has therefore been postulated that the consumption of milk provides a source of BTN-derived peptides that cross the gut mucosa to stimulate cross-reactive immune responses (52). Our peptide microarray analysis revealed MS-specific CSF antibodies against the peptide “RNVRFSDEGGFTCFF” (MOG 115–129) (Table III, Fig. 6). This confirmed the results by Guggenmos et al., who measured IgG reactivities in paired serum and CSF samples against 25mer peptides derived from MOG and BTN and showed that antibodies binding to a peptide containing this region were present more frequently in CSF (60%) than in sera (9%) of MS patients (52). In our data, a higher average signal intensity was found for this peptide in CSF compared with serum, suggesting intrathecal IgG synthesis, but the signals were overall modest, indicating low concentration and/or low affinity. Another study has been reported by Mayer et al., who expressed mutants of MOG on human HeLa cells and analyzed sera from patients with different diseases, including ADEM, neuromyelitis optica (NMO) and pediatric MS (53). No association between epitope recognition and clinical presentation was found. However, the second most frequently recognized IgG epitope was located at the membrane-distal FG-loop (His132 and Ser133) and thus close to the region identified in our study. For the overlapping peptide “DEGGFTCFFRDHSYQ” (MOG 121–135), we measured a serum-specific and CSF-specific signal for 28 and seven subjects, respectively. Hence, antibody reactivities against this epitope were indeed common, but they were not filtered as MS-specific in our study as they were also often seen in controls. The most frequently recognized epitope described by Mayer et al. encompasses amino acid Pro71 (53). Others found significantly elevated levels of IgA and IgG binding to the respective region in serum and plasma of MS patients (54, 55), which correlated with anti-BTN antibody levels (55), again supporting the hypothesis that an autoimmune response is caused by molecular mimicry. However, in our data, the corresponding peptide “VGWYRPPFSRVV” (MOG 66–77) was detected with high signals (>1000) for the arrays incubated with the secondary antibody only. Because no serum/CSF-incubated microarray had a much higher signal, the peptide did not pass the first filtering step. The specificity of MOG antibodies thus remains controversial. Evidence is accumulating that MOG antibodies are in fact associated with a broad spectrum of CNS demyelinating diseases, that they are more often present in pediatric patients than in adults and that conformational MOG epitopes are more relevant in vivo than linear epitopes (56).
Other Brain Proteins: Common IgG Response to Linear AQP4-Derived Peptides
Another putative myelin autoantigen in MS is MAG (18). In our study, reactivities against five peptides from three different regions of this protein (MAG 49–69, 103–129, and 217–231) were filtered as PPMS-specific for serum/CSF IgG. Such distributed epitopes, which may result from intramolecular epitope spreading, were also identified for other proteins, e.g. ACTB/ACTG and PERT. Andersson et al. previously examined the B- and T-cell response of MS patients to five peptides representing 16% of the MAG protein sequence (57). However, there was no overlap with our results. Thus, the validation of linear MAG epitopes merits further research.
In contrast, much more is known about the astrocyte protein AQP4, which was included in the profiling, because AQP4-specific antibodies are important in the differential diagnosis of MS. Anti-AQP4 seropositivity was originally found in 73% of NMO patients, but 5–10% of MS patients also have such autoantibodies (58). The antibodies were reported to bind conformational epitopes formed by three extracellular loops (A, C, and E) when AQP4 is organized in membrane aggregates called orthogonal arrays of particles (59, 60). Especially loop E (AQP4 226–230) seems to contribute to an IgG epitope (61, 62). In our data set, the corresponding peptide “IMGNWENHWIYWVGP” (AQP4 223–237) was bound by serum antibodies of all 30 patients and by CSF antibodies of 14 patients (supplemental File S4). The group-specific filtering (PPMS>CTR and RRMS>CTR) revealed a sequence close to loop E (“AVLAGGLYEYVFCPD,” AQP4 241–255), which showed a serum-specific signal for all 20 MS patients but only four controls (Table II, Fig. 6). Another research group performed an epitope mapping using peptides spanning all intracellular and extracellular (but not transmembrane) parts of AQP4 (63, 64). They found that ∼20% of NMO patients and ∼10% of MS patients have increased serum IgG reactivities against an intracellular region (AQP4 252–275), which overlaps with the peptide filtered in our study. It can be speculated that this region contains a neoepitope, that is, a site normally not exposed to the immune system, except during pathological processes. However, whether such neoepitope antibodies contribute to disease or are simply secondary markers of CNS tissue damage remains to be determined.
Some Autoreactive Antibodies in MS Resemble Those of Other Diseases
Several of the 54 filtered peptides were derived from proteins that are not specifically expressed in brain and spinal cord. Antibodies targeting these proteins have been described for MS, but they are occasionally also present in serum of patients with other diseases (18, 39). For instance, about 10% of MS patients were shown to have serum antibodies specific for DCE2 (also known as GAD65) (65), which are markers for diabetes mellitus type 1. In our analysis, antibodies toward the C-terminal end of DCE2 (571–585, “IDFLIEEIERLGQDL”) were significantly more frequently found in sera of the PPMS group (Table II). The N- and C-terminal regions of proteins are typically nonstructured and exposed. Therefore, if there is an epitope, it is likely to be detected by linear peptides on microarrays. The C terminus of DCE2 was already located as the main linear epitope in earlier studies focusing on diabetes mellitus type 1 (66, 67) and stiff person syndrome (68, 69). In addition to DCE2 571–585, we filtered MS-specific reactivities to peptides from the N- and C terminus of CRYAB and S100A1 (Table II).
We also identified elevated IgG levels against distinct potential epitopes of PERT (also called TPO), which is a major autoantigen in autoimmune thyroid disease. Some antipeptide reactivities were present in most of the sera. For instance, 29 and 28 serum samples were found to contain antibodies binding to “DDRYSDLLMAWGQYI” (PERT 223–237) and “LYKHPDNIDVWLGGL” (PERT 625–639), respectively. For CSF samples, these two peptides showed high signals for more MS patients than controls (Table III, supplementary File S6). Thus, antibodies recognizing these peptides seem to belong to the natural human immune repertoire, and their presence in CSF of MS patients may be a consequence of blood-brain barrier defects and/or local bystander activation of B cells during ongoing neuroinflammation. Although the underlying processes deserve further investigation, there is evidence in the literature that the two filtered peptides (PERT 223–237 and 625–639) indeed reflect epitopes of the protein (70). Gora et al. determined Arg225 and Lys627 as key residues for the immunodominant epitopes of PERT by single amino acid mutagenesis (71). Moreover, anti-PERT IgG were shown to recognize the 589–633 fragment when expressed as recombinant truncated fusion protein (72).
Our data further highlighted two SPTA2-derived peptides. SPTA2 679–693 (“QQQQFNRNVEDIELW”) was filtered with MS-specific signals for serum (Table II), whereas SPTA2 37–51 (“RQKLEDSYRFQFFQR”) was filtered for CSF (Table III), again suggesting differential antibody responses in the periphery and within the CNS. The latter peptide overlaps with an epitope (“FQFFQRDAEELEKW”) described for autoantibodies in patients with Sjögren's syndrome (73).
CRYAB: IgG Binding the N-terminal Region May Result from EBV Infection
One of the most debated autoantigens in MS is crystallin alpha-B (CRYAB), a heat shock protein with suppressive effects on (neuro)inflammation through chaperone-like activities (74). CRYAB binds a spectrum of unfolded or aggregated proteins, and it is highly expressed as a 24-subunit multimer in the eye lens, where it is a target antigen in patients with intraocular inflammation (75). Similar autoimmune processes might take place in CNS inflammation. Our microarray data revealed two different MS-specific epitopes, CRYAB 1–15, which showed serum-specific signals for 10 MS patients but no control (Table II), and CRYAB 37–57, for which CSF-specific reactivities were observed for six MS patients but no control (Table III). Thus, different parts of the protein were preferentially recognized in the different compartments. The peptide representing the N-terminal epitope (“MDIAIHHPWIRRPFF,” CRYAB 1–15) showed higher average signals in the data than any other peptide derived from human proteins (supplemental File S4), and, remarkably, the respective signals were also relatively high (>1000) for both negative control microarrays. Because all negative controls in the ELISA analysis clearly showed no reactivity to this peptide (supplementary File S7), we speculate that the secondary antibody cross-reactivity in the microarray experiment is explained by the negatively charged fluorescence dye (Alexa Fluor 647), which may interact with basic side chains (e.g. of arginine). The second epitope was identified by two overlapping 15mer peptides (“LFPTSTSLSPFYLRP” and “SLSPFYLRPPSFLRA,” CRYAB 37–57), which showed CSF reactivities at levels suggestive of intrathecal IgG production. Interestingly, some MS patients had antibodies against only one of these two epitopes. Therefore, in combination, a better stratification of MS and CTR is possible. Earlier microarray studies by Steinman et al. already identified these two regions as immunogenic. The group found antibodies recognizing CRYAB 1–20 in the CSF of MS patients (33, 34), but when comparing sera of RRMS and ADEM patients, reactivities to this peptide were even increased in the ADEM cohort (31). In yet another study, they measured elevated CSF antibody responses to CRYAB 21–40, which partially overlaps with the second epitope from our study, in MS patients (32). T cells from immunized mice and rat strains also show proliferative responses specifically against these two regions (as reviewed by Starckx et al. (76)). Notably, the putatively MS-specific epitopes CRYAB 1–15 and 37–57 (in particular the amino acids Pro13 and Pro46) are very close to each other in the secondary structure of the protein (77). We measured antibodies binding to the region between these epitopes (CRYAB 13–27, “PFFPFHSPSRLFDQF”) in all 30 sera and in 24 of the 30 CSF samples (supplemental File S4). Thus, in principle, all subjects presented a peripheral anti-CRYAB response, but the IgG fine-specificity seems to be different between MS and CTR. This is in line with findings reported in a series of publications by van Noort et al. They concluded that in an adult human immune repertoire, selective reactivity against CRYAB at the level of serum antibodies, but also at the level of T cells, appears to be normal (78, 79). They further suggested that, rather than this repertoire as such, an accumulation of CRYAB and its local presentation as an antigen in the CNS distinguishes MS patients from healthy subjects (79). Indeed, CRYAB is expressed at elevated transcript and protein levels in active MS lesions (80, 81). This up-regulation of CRYAB is assumed to be part of an early reparative innate response, but the acquired immune response against CRYAB abrogates these immunosuppressive effects and thus promotes the progression of demyelination (82).
The autoimmune response to CRYAB has been discussed to involve cross-priming during viral infection. There is evidence that infection of B cells with EBV induces the expression of CRYAB and its presentation to T-cells (83). Moreover, CRYAB and a known epitope of the EBV protein EBNA1 share an amino acid sequence motif (“RRPFF,” CRYAB 11–15, and EBNA1 402–406) (84). In our peptide microarray and ELISA analyses, both epitopes were filtered as MS-specific (Table II, supplemental File S7), with IgG recognizing CRYAB 1–15 detected only in samples that also contained IgG recognizing the EBNA1 epitope. Molecular mimicry and altered B-cell activity can result in the loss of immunological tolerance. The appearance of autoreactive antibodies targeting CRYAB could thus be the side effect of an infection with EBV, but further research is needed to confirm this hypothesis.
Distinct Epitopes of EBV Protein EBNA1 are Preferentially Recognized in MS
Because of the strong epidemiological association, it is assumed that EBV may play a necessary but not sufficient role in the initiation of MS (85). However, the mechanisms have not been fully elucidated. The EBV-mediated transformation of infected B cells, which confers resistance to apoptosis, may pose a challenge. On the other hand, a dysregulated EBV-specific immune control in MS patients may contribute to autoimmune processes (86). A major antigen of EBV is EBNA1, a multifunctional DNA-binding protein maintaining the virus in latently infected proliferating cells (87). A total of 39 EBNA1-derived peptides, representing 70% of the entire protein sequence, were included on the arrays based on IEDB (25). The analysis of the data revealed that all 30 sera were IgG-positive for at least one of these peptides, that respective antibody reactivities were generally lower in CSF and that MS patients typically present a robust and qualitatively different anti-EBNA1 antibody signature. The six overlapping peptides that were actually filtered (Table II and Table III) resemble distinct known epitopes. For instance, IgG against EBNA1 391–411 (“SGSPPRRPPPGRRPFFHPVGE”) were significantly more often found in CSF of PPMS patients and in serum of both RRMS and PPMS patients. This is consistent with various previous studies, which showed increased IgG responses to EBNA1 385–420 (or fragments thereof) in serum, plasma and CSF of MS patients compared with controls (88–93). According to reports by Sundström et al., antibodies recognizing this region are more strongly associated to MS than antibodies against the full-length EBNA1 protein (88, 89), and, in combination with the genetic marker HLA-DRB1*1501, they implicate a 24-fold increased risk to develop MS (90). However, linear epitope mapping studies found serum IgG binding to EBNA1 398–411 also in patients with systemic lupus erythematosus (SLE) (94) and in patients with infectious mononucleosis (a disease resulting from primary infection with EBV) (95) at elevated levels in comparison to healthy controls. The region was shown to cross-react with the sequence “PPPGMRPP” of the lupus spliceosomal autoantigen SNRPB (95, 96). Moreover, as described above, the region contains the sequence motif “RRPFF” (EBNA1 402–406), which is also present in CRYAB (84).
The filtering further identified four overlapping peptides highlighting MS-specific antibody responses to EBNA1 52–100, a transition sequence at the end of a region with GA-rich repeats (EBNA1 90–328). The GA-rich region has been described as the primary target of the normal peripheral humoral immune response (92–95, 97). This is verified by our data. For instance, the GA-rich peptide “GGAGAGGGAGAGGGAG,” which occurs four times in EBNA1 (101–116, 122–137, 140–155, and 167–182), was bound by almost all sera (all except CTR_07 and RRMS_01). Therefore, this peptide did not pass the MS-specific filtering either for the serum data set or for the CSF data set, where less samples showed high signals (n = 12). Otherwise, in line with previous studies (88, 90), serum IgG to EBNA1 52–100 were significantly more frequently observed in the MS group (Table II). A study with nasopharyngeal carcinoma (NPC) patients, however, also showed increased IgG and IgA reactivities to this region in comparison to healthy subjects (97), and antibodies to an epitope overlapping with this region (EBNA1 35–58) were found at higher concentrations in sera of both SLE and MS patients (96). Thus, EBNA1 52–100 harbors distinct epitopes, which explains the differences between the individual patients in the data for the four filtered peptides (see “Results”). It is worth to mention that identical amino acid sequences are present in EBNA1 and MBP. EBNA1 64–68 (“PRHRD”) matches MBP 164–168, and EBNA1 74–78 (“QKRPS”) matches MBP 137–141. This potentially means that protective immune responses against the virus cause pathological immune responses against human proteins. For the respective MBP peptides, no serum/CSF-specific signal was measured for any MS patient, but cross-reactive conformational epitopes cannot be excluded based on our data.
Other EBV proteins were recognized as well. Eight RRMS patients had serum IgG to “PPAAGPPAAGPPAAGPPAAG,” a known linear epitope occurring repeatedly in EBNA6 566–595 (98), though few controls (n = 3) showed similar reactivities. Most PPMS patients had antibodies binding to EAR (also known as BHRF1) 63–77 (“ETFTETWNRFITHTE”). This peptide was found to be the dominant IgG epitope in patients with NPC and other EBV-associated tumors (99), again illustrating that the immune response to this common virus is a marker for a range of diseases. In a recent peptide microarray study by Ruprecht et al., serum IgG against a larger panel of EBV proteins were profiled more systematically. In accordance with our results, they concluded that the anti-EBV response is elevated and broadened in MS, with EBNA1 being the predominant target (100). However, future studies may compare the response to EBV in different phases of the disease (e.g. relapse and remission) and for different antibody isotypes (IgG, IgM, and IgA).
Mimotopes of CSF IgG from MS Patients
A subset of seven of the 54 filtered peptides were selected for the arrays based on four IEDB-linked studies (101–104), which have determined mimotopes of CSF IgG from MS patients using random peptide libraries displayed on phage. Our data confirmed that these seven peptides are more often recognized by CSF antibodies from MS patients than from controls. However, IgG against these peptides were also found in serum. The peptides “WHAPQTPYWASM” and “ARWELNLEPLYGPY” even showed high signals for all 30 serum samples. Because the native antigens of the respective antibodies have so far not been identified, their further study is warranted.
Outlook and Conclusion
In the present study, we explored antibody reactivities to almost 4000 peptides. However, this still represents a very incomplete antibody profiling as only a fraction of the “epitome” was addressed. So far, it was not possible to investigate the binding of antibodies in a proteome-wide manner, but new methods for in situ synthesis of peptides on ultra-dense arrays have made this now achievable. Using a photolithographic approach, about 2 million different peptides can be synthesized on a slide (105), which facilitates a linear epitope mapping of all human proteins (106). With such technologies, much broader MS-specific (auto)antibody patterns will be defined, and this may ultimately yield better biomarkers as well as important insights into the pathogenesis of MS. On the other hand, the antigens and epitopes that have been associated with MS in the past (e.g. the epitopes in EBNA1 and CRYAB) should be further scrutinized in focused studies. The true specificity of combinations of antigenic targets has yet to be established for different stages of MS (e.g. CIS, RRMS, and SPMS) and in comparison to MS-related diseases (e.g. NMO and ADEM) as well as other neurological and inflammatory diseases. Moreover, it is largely unclear whether the presence of certain antibodies distinguishes SPMS and PPMS or patients with a more aggressive course of disease, whether the (auto)immune response changes over time with disease progression and whether current treatments for MS modulate the antibody repertoire in the long term. The potential epitopes identified in our study (Table II and Table III) justify testing in larger patient cohorts according to these settings, but also recently proposed MS autoantigens not included in our analysis, e.g. Kir4.1 (KCNJ10) (107), alpha-actinin-1 (ACTN1) (108), and additional candidates that were identified by screening for IgG against proteins or protein fragments (109–112), should be further investigated. Complementing methods have to be exploited to validate whether respective antibodies are indeed pathogenetically or physiologically autoreactive by binding properly folded proteins and/or degraded proteins from damaged tissues. The reactivities of IgG in serum/CSF should be also compared more systematically to those of other immunoglobulin isotypes (e.g. IgM and IgA) in other body fluids. Furthermore, to confine putatively MS-specific epitopes, the binding of modified linear peptides (e.g. mutated, glycosylated, or citrullinated ones) might be studied. This may help to uncover cross-reactive protein sequences in infectious agents and thus provide clues about the initiation of autoreactive immune responses that are implicated with neurodegeneration in MS. However, a more complete picture of the disease will be only achieved by a better understanding of the formation of immunoglobulins (113), the presentation of antigens, the activation of B and T cells and the processes in the CNS that are altered in MS patients.
In summary, high-density peptide microarrays were used for the first time to profile autoantibody reactivities in paired serum and CSF samples of RRMS and PPMS patients. The measured antibody responses were highly individual but correlated to a large degree for CSF and serum. IgG reactivities against 54 peptides were significantly more frequently observed in MS patients compared with controls. Similar results were obtained for RRMS and PPMS, but the reactivities were increased and broader for PPMS patients. The filtered peptides match to sequences of 19 human proteins and four EBV proteins. The MS-specific anti-EBNA1 response, which is suspected to cross-react with CRYAB, was particularly robust. Despite the limited sample size bearing a risk of false-positive results, several known antigens and epitopes were confirmed by our data, and hitherto unknown potential linear epitopes were identified. However, as discussed, some of the respective antibodies were previously described to be present also in serum of patients with various other diseases, and autoantibodies that belong to normal immune repertoires seem to be synthesized intrathecally in MS. More research is clearly needed to understand the etiology and implications of (epitope-specific) autoreactive antibodies as well as anti-EBV antibodies and to translate this knowledge into clinical care by defining better diagnostic and prognostic biomarkers and novel therapeutic strategies.
Supplementary Material
Acknowledgments
We thank Nele Retzlaff for assisting in clinical documentation, Elke Schade for her help in collecting the microarray data and Yvonne Tillack for contributing to the interpretation of the data. We are also grateful to study nurse Christa Tiffert for her excellent support in this study.
Footnotes
Authors' contributions: UKZ and H-JT inspired and directed the study. IS and UKZ were responsible for patient care and clinical documentation. The lab experiments were performed by BF, MW, PL and KF. MH and FS carried out the analysis of the data. MH drafted the manuscript and prepared all tables and figures. BF, PL and UKZ contributed to the interpretation of the results and corrected and improved the paper. All authors read and approved the final manuscript.
* Support for the peptide microarray and ELISA experiments were funded by Novartis (grant MFTY720A_FVAA024) and Merck Serono (grant 4501218059). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
 This article contains supplemental Files S1 to S7.
This article contains supplemental Files S1 to S7.
Conflicts of interest: UKZ received speaking fees and financial support for research activities from Bayer HealthCare, Biogen, Merck Serono, Novartis, Sanofi, Almirall and Teva. MH received speaking fees from Bayer HealthCare, Biogen, Novartis and Teva. HJT is co-founder and MH and FS were employees of the Gesellschaft für Individualisierte Medizin mbH (IndyMED, Rostock, Germany), a company providing services in the collection and analysis of peptide microarray data. BF, MW, PL, KF and IS declare that they have no potential conflicts of interest in connection with this paper.
1 The abbreviations used are:
- MS
- multiple sclerosis
- ABTS
- 2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt
- ADEM
- acute disseminated encephalomyelitis
- AQP4
- aquaporin 4
- BSA
- bovine serum albumin
- CIS
- clinically isolated syndrome
- CNS
- central nervous system
- CRYAB
- crystallin alpha-B
- CSF
- cerebrospinal fluid
- CTR
- controls with other neurological diseases
- CV
- coefficient of variation
- EAE
- experimental autoimmune encephalomyelitis
- EBNA1
- Epstein-Barr nuclear antigen 1
- EBV
- Epstein-Barr virus
- EDSS
- Expanded Disability Status Scale
- ELISA
- enzyme-linked immunosorbent assay
- HHV
- human herpesvirus
- HRP
- horseradish peroxidase
- IEDB
- Immune Epitope Database
- IgG
- immunoglobulin G
- MAID
- MA plot-based signal intensity-dependent fold-change
- MBP
- myelin basic protein
- MOG
- myelin oligodendrocyte glycoprotein
- MRZ
- measles, rubella and varicella zoster
- n.d.
- not determined
- NMO
- neuromyelitis optica
- NPC
- nasopharyngeal carcinoma
- OCB
- oligoclonal bands
- OD
- optical density
- PDBj
- Protein Data Bank Japan
- PEG2
- 8-amino-3,6-dioxaoctanoic acid
- PMID
- PubMed identifier
- PPMS
- primary progressive MS
- RRMS
- relapsing-remitting MS
- SAE
- Semple rabies vaccine-induced autoimmune encephalomyelitis
- S.D.
- standard deviation
- SLE
- systemic lupus erythematosus
- SPMS
- secondary progressive MS
- TBS
- Tris-buffered saline.
REFERENCES
- 1.Compston A., and Coles A. (2008) Multiple sclerosis. Lancet 372, 1502–1517 [DOI] [PubMed] [Google Scholar]
- 2.Katz Sand I. B., and Lublin F. D. (2013) Diagnosis and differential diagnosis of multiple sclerosis. Continuum 19, 922–943 [DOI] [PubMed] [Google Scholar]
- 3.Lublin F. D., Reingold S. C., Cohen J. A., Cutter G. R., Sørensen P. S., Thompson A. J., Wolinsky J. S., Balcer L. J., Banwell B., Barkhof F., Bebo B. Jr., Calabresi P. A., Clanet M., Comi G., Fox R. J., Freedman M. S., Goodman A. D., Inglese M., Kappos L., Kieseier B. C., Lincoln J. A., Lubetzki C., Miller A. E., Montalban X., O'Connor P. W., Petkau J., Pozzilli C., Rudick R. A., Sormani M. P., Stüve O., Waubant E., and Polman C. H. (2014) Defining the clinical course of multiple sclerosis: the 2013 revisions. Neurology 83, 278–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wingerchuk D. M., and Carter J. L. (2014) Multiple sclerosis: current and emerging disease-modifying therapies and treatment strategies. Mayo Clin. Proc. 89, 225–240 [DOI] [PubMed] [Google Scholar]
- 5.Sospedra M., and Martin R. (2005) Immunology of multiple sclerosis. Annu. Rev. Immunol. 23, 683–747 [DOI] [PubMed] [Google Scholar]
- 6.Cross A. H., and Waubant E. (2011) MS and the B cell controversy. Biochim. Biophys. Acta 1812, 231–238 [DOI] [PubMed] [Google Scholar]
- 7.Henderson A. P., Barnett M. H., Parratt J. D., and Prineas J. W. (2009) Multiple sclerosis: distribution of inflammatory cells in newly forming lesions. Ann. Neurol. 66, 739–753 [DOI] [PubMed] [Google Scholar]
- 8.Cepok S., Rosche B., Grummel V., Vogel F., Zhou D., Sayn J., Sommer N., Hartung H. P., and Hemmer B. (2005) Short-lived plasma blasts are the main B cell effector subset during the course of multiple sclerosis. Brain 128, 1667–1676 [DOI] [PubMed] [Google Scholar]
- 9.von Büdingen H. C., Palanichamy A., Lehmann-Horn K., Michel B. A., and Zamvil S. S. (2015) Update on the autoimmune pathology of multiple sclerosis: B-cells as disease-drivers and therapeutic targets. Eur. Neurol. 73, 238–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farh K. K., Marson A., Zhu J., Kleinewietfeld M., Housley W. J., Beik S., Shoresh N., Whitton H., Ryan R. J., Shishkin A. A., Hatan M., Carrasco-Alfonso M. J., Mayer D., Luckey C. J., Patsopoulos N. A., De Jager P. L., Kuchroo V. K., Epstein C. B., Daly M. J., Hafler D. A., and Bernstein B. E. (2015) Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature 518, 337–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fitzner B., Hecker M., and Zettl U. K. (2015) Molecular biomarkers in cerebrospinal fluid of multiple sclerosis patients. Autoimmun. Rev. 14, 903–913 [DOI] [PubMed] [Google Scholar]
- 12.Tumani H., Deisenhammer F., Giovannoni G., Gold R., Hartung H. P., Hemmer B., Hohlfeld R., Otto M., Stangel M., Wildemann B., and Zettl U. K. (2011) Revised McDonald criteria: the persisting importance of cerebrospinal fluid analysis. Ann. Neurol. 70, 520. [DOI] [PubMed] [Google Scholar]
- 13.Monson N. L., Brezinschek H. P., Brezinschek R. I., Mobley A., Vaughan G. K., Frohman E. M., Racke M. K., and Lipsky P. E. (2005) Receptor revision and atypical mutational characteristics in clonally expanded B cells from the cerebrospinal fluid of recently diagnosed multiple sclerosis patients. J. Neuroimmunol. 158, 170–181 [DOI] [PubMed] [Google Scholar]
- 14.Magliozzi R., Howell O., Vora A., Serafini B., Nicholas R., Puopolo M., Reynolds R., and Aloisi F. (2007) Meningeal B-cell follicles in secondary progressive multiple sclerosis associate with early onset of disease and severe cortical pathology. Brain 130, 1089–1104 [DOI] [PubMed] [Google Scholar]
- 15.Corcione A., Casazza S., Ferretti E., Giunti D., Zappia E., Pistorio A., Gambini C., Mancardi G. L., Uccelli A., and Pistoia V. (2004) Recapitulation of B cell differentiation in the central nervous system of patients with multiple sclerosis. Proc. Natl. Acad. Sci. U.S.A. 101, 11064–11069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franciotta D., Salvetti M., Lolli F., Serafini B., and Aloisi F. (2008) B cells and multiple sclerosis. Lancet Neurol. 7, 852–858 [DOI] [PubMed] [Google Scholar]
- 17.Serafini B., Muzio L., Rosicarelli B., and Aloisi F. (2013) Radioactive in situ hybridization for Epstein-Barr virus-encoded small RNA supports presence of Epstein-Barr virus in the multiple sclerosis brain. Brain 136, e233. [DOI] [PubMed] [Google Scholar]
- 18.Fraussen J., Claes N., de Bock L., and Somers V. (2014) Targets of the humoral autoimmune response in multiple sclerosis. Autoimmun. Rev. 13, 1126–1137 [DOI] [PubMed] [Google Scholar]
- 19.González-Escalada A., García-García L., Viguera-Ester P., Marín-García P., García J., Gil-de-Miguel A., and Gil-Prieto R. (2013) Seroprevalence of antibodies against measles, rubella, mumps, varicella-zoster, and B. Pertussis in young adults of Madrid, Spain. Hum. Vaccin. Immunother. 9, 1918–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarius S., Eichhorn P., Jacobi C., Wildemann B., Wick M., and Voltz R. (2009) The intrathecal, polyspecific antiviral immune response: specific for MS or a general marker of CNS autoimmunity? J. Neurol. Sci. 280, 98–100 [DOI] [PubMed] [Google Scholar]
- 21.Deuschle K., Hofmann J., Otto C., Bellmann-Strobl J., Scherner O., Klumbies K., Schneider E., Broddack J., Paul F., and Ruprecht K. (2013) Are there Epstein-Barr virus seronegative patients with multiple sclerosis? Mult. Scler. 19, 1242–1243 [DOI] [PubMed] [Google Scholar]
- 22.Kakalacheva K., Münz C., and Lünemann J. D. (2011) Viral triggers of multiple sclerosis. Biochim. Biophys. Acta 1812, 132–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gold M., Pul R., Bach J. P., Stangel M., and Dodel R. (2012) Pathogenic and physiological autoantibodies in the central nervous system. Immunol. Rev. 248, 68–86 [DOI] [PubMed] [Google Scholar]
- 24.Nagele E. P., Han M., Acharya N. K., DeMarshall C., Kosciuk M. C., and Nagele R. G. (2013) Natural IgG autoantibodies are abundant and ubiquitous in human sera, and their number is influenced by age, gender, and disease. PLoS One 8, e60726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vita R., Overton J. A., Greenbaum J. A., Ponomarenko J., Clark J. D., Cantrell J. R., Wheeler D. K., Gabbard J. L., Hix D., Sette A., and Peters B. (2015) The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 43, D405-D412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Masch A., Zerweck J., Reimer U., Wenschuh H., and Schutkowski M. (2010) Antibody signatures defined by high-content peptide microarray analysis. Methods Mol. Biol. 669, 161–172 [DOI] [PubMed] [Google Scholar]
- 27.Andresen H., and Grötzinger C. (2009) Deciphering the antibodyome – peptide arrays for serum antibody biomarker diagnostics. Curr. Proteomics 6, 1–12 [Google Scholar]
- 28.Al-Majdoub M., Koy C., Lorenz P., Thiesen H.-J., and Glocker M. O. (2013) Mass spectrometric and peptide chip characterization of an assembled epitope: analysis of a polyclonal antibody model serum directed against the Sjøgren/systemic lupus erythematosus autoantigen TRIM21. J. Mass. Spectrom. 48, 651–659 [DOI] [PubMed] [Google Scholar]
- 29.Quintana F. J., Farez M. F., Viglietta V., Iglesias A. H., Merbl Y., Izquierdo G., Lucas M., Basso A. S., Khoury S. J., Lucchinetti C. F., Cohen I. R., and Weiner H. L. (2008) Antigen microarrays identify unique serum autoantibody signatures in clinical and pathologic subtypes of multiple sclerosis. Proc. Natl. Acad. Sci. U.S.A. 105, 18889–18894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quintana F. J., Patel B., Yeste A., Nyirenda M., Kenison J., Rahbari R., Fetco D., Hussain M., O'Mahony J., Magalhaes S., McGowan M., Johnson T., Rajasekharan S., Narayanan S., Arnold D. L., Weiner H. L., Banwell B., and Bar-Or A.; Canadian Pediatric Demyelinating Disease Network (2014) Epitope spreading as an early pathogenic event in pediatric multiple sclerosis. Neurology 83, 2219–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Haren K., Tomooka B. H., Kidd B. A., Banwell B., Bar-Or A., Chitnis T., Tenembaum S. N., Pohl D., Rostasy K., Dale R. C., O'Connor K. C., Hafler D. A., Steinman L., and Robinson W. H. (2013) Serum autoantibodies to myelin peptides distinguish acute disseminated encephalomyelitis from relapsing-remitting multiple sclerosis. Mult. Scler. 19, 1726–1733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ousman S. S., Tomooka B. H., van Noort J. M., Wawrousek E. F., O'Connor K. C., Hafler D. A., Sobel R. A., Robinson W. H., and Steinman L. (2007) Protective and therapeutic role for alphaB-crystallin in autoimmune demyelination. Nature 448, 474–479 [DOI] [PubMed] [Google Scholar]
- 33.Garren H., Robinson W. H., Krasulová E., Havrdová E., Nadj C., Selmaj K., Losy J., Nadj I., Radue E. W., Kidd B. A., Gianettoni J., Tersini K., Utz P. J., Valone F., and Steinman L.; BHT-3009 Study Group (2008) Phase 2 trial of a DNA vaccine encoding myelin basic protein for multiple sclerosis. Ann. Neurol. 63, 611–620 [DOI] [PubMed] [Google Scholar]
- 34.Bar-Or A., Vollmer T., Antel J., Arnold D. L., Bodner C. A., Campagnolo D., Gianettoni J., Jalili F., Kachuck N., Lapierre Y., Niino M., Oger J., Price M., Rhodes S., Robinson W. H., Shi F. D., Utz P. J., Valone F., Weiner L., Steinman L., and Garren H. (2007) Induction of antigen-specific tolerance in multiple sclerosis after immunization with DNA encoding myelin basic protein in a randomized, placebo-controlled phase 1/2 trial. Arch. Neurol. 64, 1407–1415 [DOI] [PubMed] [Google Scholar]
- 35.Quintana F. J., Farez M. F., Izquierdo G., Lucas M., Cohen I. R., and Weiner H. L. (2012) Antigen microarrays identify CNS-produced autoantibodies in RRMS. Neurology 78, 532–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Polman C. H., Reingold S. C., Banwell B., Clanet M., Cohen J. A., Filippi M., Fujihara K., Havrdova E., Hutchinson M., Kappos L., Lublin F. D., Montalban X., O'Connor P., Sandberg-Wollheim M., Thompson A. J., Waubant E., Weinshenker B., and Wolinsky J. S. (2011) Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann. Neurol. 69, 292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.UniProt Consortium (2015) UniProt: a hub for protein information. Nucleic Acids Res. 43, D204-D212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lorenz P., Kreutzer M., Zerweck J., Schutkowski M., and Thiesen H.-J. (2009) Probing the epitope signatures of IgG antibodies in human serum from patients with autoimmune disease. Methods Mol. Biol. 524, 247–258 [DOI] [PubMed] [Google Scholar]
- 39.Hecker M., Lorenz P., Steinbeck F., Hong L., Riemekasten G., Li Y., Zettl U.K., and Thiesen H.-J. (2012) Computational analysis of high-density peptide microarray data with application from systemic sclerosis to multiple sclerosis. Autoimmun. Rev. 11, 180–190 [DOI] [PubMed] [Google Scholar]
- 40.Hecker M., Goertsches R. H., Engelmann R., Thiesen H.-J., and Guthke R. (2009) Integrative modeling of transcriptional regulation in response to antirheumatic therapy. BMC Bioinformatics 10, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kinjo A. R., Suzuki H., Yamashita R., Ikegawa Y., Kudou T., Igarashi R., Kengaku Y., Cho H., Standley D. M., Nakagawa A., and Nakamura H. (2012) Protein Data Bank Japan (PDBj): maintaining a structural data archive and resource description framework format. Nucleic Acids Res. 40, D453-D460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang C. C., Meng E. C., Morris J. H., Pettersen E. F., and Ferrin T. E. (2014) Enhancing UCSF Chimera through web services. Nucleic Acids Res. 42, W478-W484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Connor K. C., Appel H., Bregoli L., Call M. E., Catz I., Chan J. A., Moore N. H., Warren K. G., Wong S. J., Hafler D. A., and Wucherpfennig K. W. (2005) Antibodies from inflamed central nervous system tissue recognize myelin oligodendrocyte glycoprotein. J. Immunol. 175, 1974–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hjelm B., Fernández C. D., Löfblom J., Ståhl S., Johannesson H., Rockberg J., and Uhlén M. (2010) Exploring epitopes of antibodies toward the human tryptophanyl-tRNA synthetase. N. Biotechnol. 27, 129–137 [DOI] [PubMed] [Google Scholar]
- 45.Pender M. P. (2004) The pathogenesis of primary progressive multiple sclerosis: antibody-mediated attack and no repair? J. Clin. Neurosci. 11, 689–692 [DOI] [PubMed] [Google Scholar]
- 46.Belogurov A. A. Jr., Kurkova I. N., Friboulet A., Thomas D., Misikov V. K., Zakharova M. Y., Suchkov S. V., Kotov S. V., Alehin A. I., Avalle B., Souslova E. A., Morse H. C. 3rd, Gabibov A. G., and Ponomarenko N. A. (2008) Recognition and degradation of myelin basic protein peptides by serum autoantibodies: novel biomarker for multiple sclerosis. J. Immunol. 180, 1258–1267 [DOI] [PubMed] [Google Scholar]
- 47.Terzoglou A. G., Routsias J. G., Sakarellos C., Sakarellos-Daitsiotis M., Moutsopoulos H. M., and Tzioufas A. G. (2003) Linear epitopes of two different autoantigens-La/SSB and myelin basic protein – with a high degree of molecular similarity, cause different humoral immune responses. J. Autoimmun. 21, 47–57 [DOI] [PubMed] [Google Scholar]
- 48.Piyasirisilp S., Hemachudha T., and Griffin D. E. (1999) B-cell responses to myelin basic protein and its epitopes in autoimmune encephalomyelitis induced by Semple rabies vaccine. J. Neuroimmunol. 98, 96–104 [DOI] [PubMed] [Google Scholar]
- 49.Robinson A. P., Harp C. T., Noronha A., and Miller S. D. (2014) The experimental autoimmune encephalomyelitis (EAE) model of MS: utility for understanding disease pathophysiology and treatment. Handb. Clin. Neurol. 122, 173–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.O'Connor K. C., Chitnis T., Griffin D. E., Piyasirisilp S., Bar-Or A., Khoury S., Wucherpfennig K. W., and Hafler D. A. (2003) Myelin basic protein-reactive autoantibodies in the serum and cerebrospinal fluid of multiple sclerosis patients are characterized by low-affinity interactions. J. Neuroimmunol. 136, 140–148 [DOI] [PubMed] [Google Scholar]
- 51.Hedegaard C. J., Chen N., Sellebjerg F., Sørensen P. S., Leslie R. G., Bendtzen K., and Nielsen C. H. (2009) Autoantibodies to myelin basic protein (MBP) in healthy individuals and in patients with multiple sclerosis: a role in regulating cytokine responses to MBP. Immunology 128, e451-e461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guggenmos J., Schubart A. S., Ogg S., Andersson M., Olsson T., Mather I. H., and Linington C. (2004) Antibody cross-reactivity between myelin oligodendrocyte glycoprotein and the milk protein butyrophilin in multiple sclerosis. J. Immunol. 172, 661–668 [DOI] [PubMed] [Google Scholar]
- 53.Mayer M. C., Breithaupt C., Reindl M., Schanda K., Rostásy K., Berger T., Dale R. C., Brilot F., Olsson T., Jenne D., Pröbstel A. K., Dornmair K., Wekerle H., Hohlfeld R., Banwell B., Bar-Or A., and Meinl E. (2013) Distinction and temporal stability of conformational epitopes on myelin oligodendrocyte glycoprotein recognized by patients with different inflammatory central nervous system diseases. J. Immunol. 191, 3594–3604 [DOI] [PubMed] [Google Scholar]
- 54.Khalil M., Reindl M., Lutterotti A., Kuenz B., Ehling R., Gneiss C., Lackner P., Deisenhammer F., and Berger T. (2006) Epitope specificity of serum antibodies directed against the extracellular domain of myelin oligodendrocyte glycoprotein: Influence of relapses and immunomodulatory treatments. J. Neuroimmunol. 174, 147–156 [DOI] [PubMed] [Google Scholar]
- 55.Kennel De March A., De Bouwerie M., Kolopp-Sarda M. N., Faure G. C., Béné M. C., and Bernard C. C. (2003) Anti-myelin oligodendrocyte glycoprotein B-cell responses in multiple sclerosis. J. Neuroimmunol. 135, 117–125 [DOI] [PubMed] [Google Scholar]
- 56.Reindl M., Di Pauli F., Rostásy K., and Berger T. (2013) The spectrum of MOG autoantibody-associated demyelinating diseases. Nat. Rev. Neurol. 9, 455–461 [DOI] [PubMed] [Google Scholar]
- 57.Andersson M., Yu M., Söderström M., Weerth S., Baig S., Solders G., and Link H. (2002) Multiple MAG peptides are recognized by circulating T and B lymphocytes in polyneuropathy and multiple sclerosis. Eur. J. Neurol. 9, 243–251 [DOI] [PubMed] [Google Scholar]
- 58.Kira J. (2011) Neuromyelitis optica and opticospinal multiple sclerosis: Mechanisms and pathogenesis. Pathophysiology 18, 69–79 [DOI] [PubMed] [Google Scholar]
- 59.Crane J.M., Lam C., Rossi A., Gupta T., Bennett J. L., and Verkman A. S. (2011) Binding affinity and specificity of neuromyelitis optica autoantibodies to aquaporin-4 M1/M23 isoforms and orthogonal arrays. J. Biol. Chem. 286, 16516–16524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pisani F., Mastrototaro M., Rossi A., Nicchia G. P., Tortorella C., Ruggieri M., Trojano M., Frigeri A., and Svelto M. (2011) Identification of two major conformational aquaporin-4 epitopes for neuromyelitis optica autoantibody binding. J. Biol. Chem. 286, 9216–9224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu X., Green M., Gilden D., Lam C., Bautista K., and Bennett J. L. (2011) Identification of peptide targets in neuromyelitis optica. J. Neuroimmunol. 236, 65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tani T., Sakimura K., Tsujita M., Nakada T., Tanaka M., Nishizawa M., and Tanaka K. (2009) Identification of binding sites for anti-aquaporin 4 antibodies in patients with neuromyelitis optica. J. Neuroimmunol. 211, 110–113 [DOI] [PubMed] [Google Scholar]
- 63.Alexopoulos H., Kampylafka E. I., Chatzi I., Travasarou M., Karageorgiou K. E., Dalakas M. C., and Tzioufas A. G. (2013) Reactivity to AQP4 epitopes in relapsing-remitting multiple sclerosis. J. Neuroimmunol. 260, 117–120 [DOI] [PubMed] [Google Scholar]
- 64.Kampylafka E. I., Routsias J. G., Alexopoulos H., Dalakas M. C., Moutsopoulos H. M., and Tzioufas A. G. (2011) Fine specificity of antibodies against AQP4: epitope mapping reveals intracellular epitopes. J. Autoimmun. 36, 221–227 [DOI] [PubMed] [Google Scholar]
- 65.Hermitte L., Martin-Moutot N., Boucraut J., Barone R., Atlan-Gepner C., Seagar M., Pouget J., Kleisbauer J. P., Couraud F., and Vialettes B. (2000) Humoral immunity against glutamic acid decarboxylase and tyrosine phosphatase IA-2 in Lambert-Eaton myasthenic syndrome. J. Clin. Immunol. 20, 287–293 [DOI] [PubMed] [Google Scholar]
- 66.Mauch L., Abney C. C., Berg H., Scherbaum W. A., Liedvogel B., and Northemann W. (1993) Characterization of a linear epitope within the human pancreatic 64-kDa glutamic acid decarboxylase and its autoimmune recognition by sera from insulin-dependent diabetes mellitus patients. Eur. J. Biochem. 212, 597–603 [DOI] [PubMed] [Google Scholar]
- 67.Tree T. I., Morgenthaler N. G., Duhindan N., Hicks K. E., Madec A. M., Scherbaum W. A., and Banga J. P. (2000) Two amino acids in glutamic acid decarboxylase act in concert for maintenance of conformational determinants recognised by Type I diabetic autoantibodies. Diabetologia 43, 881–889 [DOI] [PubMed] [Google Scholar]
- 68.Söhnlein P., Müller M., Syren K., Hartmann U., Böhm B. O., Meinck H. M., Knip M., Akerblom H. K., and Richter W. (2000) Epitope spreading and a varying but not disease-specific GAD65 antibody response in Type I diabetes. The Childhood Diabetes in Finland Study Group. Diabetologia 43, 210–217 [DOI] [PubMed] [Google Scholar]
- 69.Al-Bukhari T. A., Radford P. M., Bouras G., Davenport C., Trigwell S. M., Bottazzo G. F., Lai M., Schwartz H. L., Tighe P. J., and Todd I. (2002) Distinct antigenic features of linear epitopes at the N-terminus and C-terminus of 65 kDa glutamic acid decarboxylase (GAD65): implications for autoantigen modification during pathogenesis. Clin. Exp. Immunol. 130, 131–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.McLachlan S. M., and Rapoport B. (2007) Thyroid peroxidase as an autoantigen. Thyroid 17, 939–948 [DOI] [PubMed] [Google Scholar]
- 71.Gora M., Gardas A., Watson P. F., Hobby P., Weetman A. P., Sutton B. J., and Banga J. P. (2004) Key residues contributing to dominant conformational autoantigenic epitopes on thyroid peroxidase identified by mutagenesis. Biochem. Biophys. Res. Commun. 320, 795–801 [DOI] [PubMed] [Google Scholar]
- 72.Arscott P. L., Koenig R. J., Kaplan M. M., Glick G. D., and Baker J. R. Jr. (1996) Unique autoantibody epitopes in an immunodominant region of thyroid peroxidase. J. Biol. Chem. 271, 4966–4973 [DOI] [PubMed] [Google Scholar]
- 73.Chen Q., Li X., He W., Zhang H., Gao A., Cheng Y., Lei J., Li S., and Zeng L. (2007) The epitope study of alpha-fodrin autoantibody in primary Sjögren's syndrome. Clin. Exp. Immunol. 149, 497–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Masilamoni J. G., Jesudason E. P., Baben B., Jebaraj C. E., Dhandayuthapani S., and Jayakumar R. (2006) Molecular chaperone alpha-crystallin prevents detrimental effects of neuroinflammation. Biochim. Biophys. Acta 1762, 284–293 [DOI] [PubMed] [Google Scholar]
- 75.Doycheva D., Preuss B., Deuter C., Zierhut M., and Klein R. (2012) Identification of immunodominant epitopes of alpha-crystallins recognized by antibodies in sera of patients with uveitis. Graefes Arch. Clin. Exp. Ophthalmol. 250, 297–305 [DOI] [PubMed] [Google Scholar]
- 76.Starckx S., Van den Steen P. E., Verbeek R., van Noort J. M., and Opdenakker G. (2003) A novel rationale for inhibition of gelatinase B in multiple sclerosis: MMP-9 destroys alpha B-crystallin and generates a promiscuous T cell epitope. J. Neuroimmunol. 141, 47–57 [DOI] [PubMed] [Google Scholar]
- 77.Braun N., Zacharias M., Peschek J., Kastenmüller A., Zou J., Hanzlik M., Haslbeck M., Rappsilber J., Buchner J., and Weinkauf S. (2011) Multiple molecular architectures of the eye lens chaperone αB-crystallin elucidated by a triple hybrid approach. Proc. Natl. Acad. Sci. U.S.A. 108, 20491–20496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van Noort J. M., van Sechel A. C., Bajramovic J. J., el Ouagmiri M., Polman C. H., Lassmann H., and Ravid R. (1995) The small heat-shock protein alpha B-crystallin as candidate autoantigen in multiple sclerosis. Nature 375, 798–801 [DOI] [PubMed] [Google Scholar]
- 79.van Noort J. M., Verbeek R., Meilof J. F., Polman C. H., and Amor S. (2006) Autoantibodies against alpha B-crystallin, a candidate autoantigen in multiple sclerosis, are part of a normal human immune repertoire. Mult. Scler. 12, 287–293 [DOI] [PubMed] [Google Scholar]
- 80.Sinclair C., Mirakhur M., Kirk J., Farrell M., and McQuaid S. (2005) Up-regulation of osteopontin and alphaBeta-crystallin in the normal-appearing white matter of multiple sclerosis: an immunohistochemical study utilizing tissue microarrays. Neuropathol. Appl. Neurobiol. 31, 292–303 [DOI] [PubMed] [Google Scholar]
- 81.Chabas D., Baranzini S. E., Mitchell D., Bernard C. C., Rittling S. R., Denhardt D. T., Sobel R. A., Lock C., Karpuj M., Pedotti R., Heller R., Oksenberg J. R., and Steinman L. (2001) The influence of the proinflammatory cytokine, osteopontin, on autoimmune demyelinating disease. Science 294, 1731–1735 [DOI] [PubMed] [Google Scholar]
- 82.van Noort J. M., Bsibsi M., Gerritsen W. H., van der Valk P., Bajramovic J. J., Steinman L., and Amor S. (2010) Alphab-crystallin is a target for adaptive immune responses and a trigger of innate responses in preactive multiple sclerosis lesions. J. Neuropathol. Exp. Neurol. 69, 694–703 [DOI] [PubMed] [Google Scholar]
- 83.van Sechel A. C., Bajramovic J. J., van Stipdonk M. J., Persoon-Deen C., Geutskens S. B., and van Noort J. M. (1999) EBV-induced expression and HLA-DR-restricted presentation by human B cells of alpha B-crystallin, a candidate autoantigen in multiple sclerosis. J. Immunol. 162, 129–135 [PubMed] [Google Scholar]
- 84.Rand K. H., Houck H., Denslow N. D., and Heilman K. M. (1998) Molecular approach to find target(s) for oligoclonal bands in multiple sclerosis. J. Neurol. Neurosurg. Psychiatry 65, 48–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lucas R. M., Hughes A. M., Lay M. L., Ponsonby A. L., Dwyer D. E., Taylor B. V., and Pender M. P. (2011) Epstein-Barr virus and multiple sclerosis. J. Neurol. Neurosurg. Psychiatry 82, 1142–1148 [DOI] [PubMed] [Google Scholar]
- 86.Pender M. P., and Burrows S. R. (2014) Epstein-Barr virus and multiple sclerosis: potential opportunities for immunotherapy. Clin. Transl. Immunol. 3, e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Westhoff Smith D., and Sugden B. (2013) Potential cellular functions of Epstein-Barr Nuclear Antigen 1 (EBNA1) of Epstein-Barr Virus. Viruses 5, 226–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sundqvist E., Sundström P., Lindén M., Hedström A. K., Aloisi F., Hillert J., Kockum I., Alfredsson L., and Olsson T. (2012) Epstein-Barr virus and multiple sclerosis: interaction with HLA. Genes Immun. 13, 14–20 [DOI] [PubMed] [Google Scholar]
- 89.Salzer J., Nyström M., Hallmans G., Stenlund H., Wadell G., and Sundström P. (2013) Epstein-Barr virus antibodies and vitamin D in prospective multiple sclerosis biobank samples. Mult. Scler. 19, 1587–1591 [DOI] [PubMed] [Google Scholar]
- 90.Sundström P., Nyström M., Ruuth K., and Lundgren E. (2009) Antibodies to specific EBNA-1 domains and HLA DRB1*1501 interact as risk factors for multiple sclerosis. J. Neuroimmunol. 215, 102–107 [DOI] [PubMed] [Google Scholar]
- 91.Mameli G., Cossu D., Cocco E., Masala S., Frau J., Marrosu M. G., and Sechi L. A. (2014) Epstein-Barr virus and Mycobacterium avium subsp. paratuberculosis peptides are cross recognized by anti-myelin basic protein antibodies in multiple sclerosis patients. J. Neuroimmunol. 270, 51–55 [DOI] [PubMed] [Google Scholar]
- 92.Mechelli R., Anderson J., Vittori D., Coarelli G., Annibali V., Cannoni S., Aloisi F., Salvetti M., James J. A., and Ristori G. (2011) Epstein-Barr virus nuclear antigen-1 B-cell epitopes in multiple sclerosis twins. Mult. Scler. 17, 1290–1294 [DOI] [PubMed] [Google Scholar]
- 93.Jafari N., van Nierop G. P., Verjans G. M., Osterhaus A. D., Middeldorp J. M., and Hintzen R. Q. (2010) No evidence for intrathecal IgG synthesis to Epstein Barr virus nuclear antigen-1 in multiple sclerosis. J. Clin. Virol. 49, 26–31 [DOI] [PubMed] [Google Scholar]
- 94.McClain M. T., Poole B. D., Bruner B. F., Kaufman K. M., Harley J. B., and James J.A. (2006) An altered immune response to Epstein-Barr nuclear antigen 1 in pediatric systemic lupus erythematosus. Arthritis Rheum. 54, 360–368 [DOI] [PubMed] [Google Scholar]
- 95.McClain M. T., Rapp E. C., Harley J. B., and James J. A. (2003) Infectious mononucleosis patients temporarily recognize a unique, cross-reactive epitope of Epstein-Barr virus nuclear antigen-1. J. Med. Virol. 70, 253–257 [DOI] [PubMed] [Google Scholar]
- 96.Csuka D., Simon D., Hóbor R., Uray K., Prohászka Z., Bánlaki Z., Jani P. K., Szilágyi Á., Hudecz F., Rajczy K., Beke G., Boros Major A., Tordai A., Illés Z., Berki T., Czirják L., and Füst G. (2013) Serum concentration of immunoglobulin G-type antibodies against the whole Epstein-Barr nuclear antigen 1 and its aa35–58 or aa398–404 fragments in the sera of patients with systemic lupus erythematosus and multiple sclerosis. Clin. Exp. Immunol. 171, 255–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cheng H. M., Foong Y. T., Sam C. K., Prasad U., and Dillner J. (1991) Epstein-Barr virus nuclear antigen 1 linear epitopes that are reactive with immunoglobulin A (IgA) or IgG in sera from nasopharyngeal carcinoma patients or from healthy donors. J. Clin. Microbiol. 29, 2180–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Parkkonen P., Hyöty H., Ilonen J., Reijonen H., Ylä-Herttuala S., and Leinikki P. (1994) Antibody reactivity to an Epstein-Barr virus BERF4-encoded epitope occurring also in Asp-57 region of HLA-DQ8 beta chain. Childhood Diabetes in Finland Study Group. Clin. Exp. Immunol. 95, 287–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pothen S., Cao T., Smith R., Levine P. H., Levine A., and Pearson G. R. (1993) Identification of T- and B-cell epitopes associated with a restricted component of the Epstein-Barr virus-induced early antigen complex. Int. J. Cancer 53, 199–204 [DOI] [PubMed] [Google Scholar]
- 100.Ruprecht K., Wunderlich B., Gieβ R., Meyer P., Loebel M., Lenz K., Hofmann J., Rosche B., Wengert O., Paul F., Reimer U., and Scheibenbogen C. (2014) Multiple sclerosis: the elevated antibody response to Epstein-Barr virus primarily targets, but is not confined to, the glycine-alanine repeat of Epstein-Barr nuclear antigen-1. J. Neuroimmunol. 272, 56–61 [DOI] [PubMed] [Google Scholar]
- 101.Jolivet-Reynaud C., Perron H., Ferrante P., Becquart L., Dalbon P., and Mandrand B. (1999) Specificities of multiple sclerosis cerebrospinal fluid and serum antibodies against mimotopes. Clin. Immunol. 93, 283–293 [DOI] [PubMed] [Google Scholar]
- 102.Yu X., Gilden D. H., Ritchie A. M., Burgoon M. P., Keays K. M., and Owens G. P. (2006) Specificity of recombinant antibodies generated from multiple sclerosis cerebrospinal fluid probed with a random peptide library. J. Neuroimmunol. 172, 121–131 [DOI] [PubMed] [Google Scholar]
- 103.Yu X., Owens G. P., and Gilden D. H. (2006) Rapid and efficient identification of epitopes/mimotopes from random peptide libraries. J. Immunol. Methods 316, 67–74 [DOI] [PubMed] [Google Scholar]
- 104.Yu X., Gilden D., Schambers L., Barmina O., Burgoon M., Bennett J., and Owens G. (2011) Peptide reactivity between multiple sclerosis (MS) CSF IgG and recombinant antibodies generated from clonally expanded plasma cells in MS CSF. J. Neuroimmunol. 233, 192–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Buus S., Rockberg J., Forsström B., Nilsson P., Uhlen M., and Schafer-Nielsen C. (2012) High-resolution mapping of linear antibody epitopes using ultrahigh-density peptide microarrays. Mol. Cell. Proteomics 11, 1790–1800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Forsström B., Axnäs B. B., Stengele K. P., Bühler J., Albert T. J., Richmond T. A., Hu F. J., Nilsson P., Hudson E. P., Rockberg J., and Uhlen M. (2014) Proteome-wide epitope mapping of antibodies using ultra-dense peptide arrays. Mol. Cell. Proteomics 13, 1585–1597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Srivastava R., Aslam M., Kalluri S. R., Schirmer L., Buck D., Tackenberg B., Rothhammer V., Chan A., Gold R., Berthele A., Bennett J. L., Korn T., and Hemmer B. (2012) Potassium channel KIR4.1 as an immune target in multiple sclerosis. N. Engl. J. Med. 367, 115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pandey S., Dioni I., Lambardi D., Real-Fernandez F., Peroni E., Pacini G., Lolli F., Seraglia R., Papini A. M., and Rovero P. (2013) Alpha actinin is specifically recognized by Multiple Sclerosis autoantibodies isolated using an N-glucosylated peptide epitope. Mol. Cell. Proteomics 12, 277–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ayoglu B., Häggmark A., Khademi M., Olsson T., Uhlén M., Schwenk J. M., and Nilsson P. (2013) Autoantibody profiling in multiple sclerosis using arrays of human protein fragments. Mol. Cell. Proteomics 12, 2657–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Querol L., Clark P. L., Bailey M. A., Cotsapas C., Cross A. H., Hafler D. A., Kleinstein S. H., Lee J. Y., Yaari G., Willis S. N., and O'Connor K. C. (2013) Protein array-based profiling of CSF identifies RBPJ as an autoantigen in multiple sclerosis. Neurology 81, 956–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Beyer N. H., Lueking A., Kowald A., Frederiksen J. L., and Heegaard N. H. (2012) Investigation of autoantibody profiles for cerebrospinal fluid biomarker discovery in patients with relapsing-remitting multiple sclerosis. J. Neuroimmunol. 242, 26–32 [DOI] [PubMed] [Google Scholar]
- 112.Cepok S., Zhou D., Srivastava R., Nessler S., Stei S., Büssow K., Sommer N., and Hemmer B. (2005) Identification of Epstein-Barr virus proteins as putative targets of the immune response in multiple sclerosis. J. Clin. Invest. 115, 1352–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Singh V., Stoop M. P., Stingl C., Luitwieler R. L., Dekker L. J., van Duijn M. M., Kreft K. L., Luider T. M., and Hintzen R. Q. (2013) Cerebrospinal-fluid-derived immunoglobulin G of different multiple sclerosis patients shares mutated sequences in complementarity determining regions. Mol. Cell. Proteomics 12, 3924–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.