Abstract
Although the spread of sushi restaurants in the European Union and United States is a relatively new phenomenon, they have rapidly become among the most popular food services globally. Recent studies indicate that they can be associated with very high levels (>70%) of fish species substitution. Based on indications that the European seafood retail sector may currently be under better control than its North American counterpart, here we investigated levels of seafood labelling accuracy in sushi bars and restaurants across England. We used the COI barcoding gene to screen samples of tuna, eel, and a variety of other products characterised by less visually distinctive ‘white flesh’. Moderate levels of substitution were found (10%), significantly lower than observed in North America, which lends support to the argument that public awareness, policy and governance of seafood labels is more effective in the European Union. Nevertheless, the results highlight that current labelling practice in UK restaurants lags behind the level of detail implemented in the retail sector, which hinders consumer choice, with potentially damaging economic, health and environmental consequences. Specifically, critically endangered species of tuna and eel continue to be sold without adequate information to consumers.
Keywords: Sushi restaurants, UK, COI barcoding, Traceability, Fish, Species substitution, Mislabelling
Introduction
Seafood is a popular and healthy food choice and, therefore, one of the most commonly traded food commodities in the world (FAO, 2014). Regardless of the growing demand, studies on seafood mislabelling have identified that consumers are still too often given insufficient, confusing or misleading information about the seafood they purchase (Warner et al., 2013; Pramod et al., 2014; Cawthorn et al., 2015; Di Pinto et al., 2015). Due to increasingly complex supply chains, it is often unclear where and when seafood fraud is actually taking place, but restaurants and take-aways have been identified as the worst point of consumption for species substitution (Jacquet & Pauly, 2008; Warner et al., 2013; Bénard-Capelle et al., 2015). For example, large studies across North America illustrate that sushi venues have the highest level of mislabelling (74%–16%), followed by restaurants (38%) and grocery stores (18%) (Warner et al., 2013; Pramod et al., 2014; Khaksar et al., 2015). Such findings suggest that, as restaurants often represent the end-point of these long and intricate supply chains, without needing to comply with the standardised labelling practices of the retail sector, they could be consistently associated with the highest levels of substitution.
Seafood fraud encompasses any illegal activity that misrepresents the fish being purchased. Although some mislabelling may result from unintended human errors in identifying fish or their origin, often it is driven by economic gain, where cheaper or more readily available species are sold instead of expensive, desirable or supply-limited species e.g., farmed tilapia, Oreochromis sp., sold as snapper, Lutjanus sp., (Jacquet & Pauly, 2008; Warner et al., 2013). Mislabelling can also provide cover and profit for illegal and unregulated fishing and seafood (Watson et al., 2015), which could have damaging implications for fisheries management and conservation, e.g., Atlantic halibut Hippoglossus hippoglossus sold as Pacific halibut Hippoglossus stenolepis (Warner et al., 2013). Seafood fraud can also have serious health consequences when mislabelled seafood masks undeclared allergens, contaminants or toxins. This is exemplified by escolar, Lepidocybium flavobrunneum, sold as “white tuna” (Lowenstein, Burger & Jeitner, 2010; Warner et al., 2013); escolar can naturally contain a toxin, gempylotoxin, which can cause mild to severe gastrointestinal problems, meaning this species is banned from the market in Italy and Japan.
The European Union (EU) is the largest single market for imported fish and fishery products, representing about 23% of world imports, and continuing to grow (FAO, 2014). As such, the EU has a great responsibility to demonstrate legal and sustainable seafood supply chains to consumers. Its illegal fishing regulation (EC No 1005/2008) is an innovative and pioneering legal tool that has placed the EU at the forefront of global efforts to address illegal, unreported and unregulated (IUU) fishing. Part of the ongoing legal framework is the new European regulation (EC No 1379/2013), enacted in December 2014, which places an onus on anybody selling seafood to label it clearly and accurately, providing consumers with highly transparent information. This new EU labelling legislation applies to all pre-packed and non-packed fishery and aquaculture products (excluding preserved and prepared meals) at all stages in the retail supply chain, but excludes restaurants, which only have to provide mandatory information on allergens. In other words, restaurants are not obliged to mention on their menu what species is being sold but they are obliged to keep and give this information to the consumer if asked for. Additionally, EU Member States have to draw up a list of the commercial designations accepted in their territory, together with their scientific names. However, for some groups, like eels or tunas, the authorized commercial names cover a large number of species, including those with serious conservation concern. In such cases, there is no way for knowledgeable consumers to choose according to sustainability criteria.
Given recent indication that the European seafood retail sector may have significantly lower levels of fraudulent substitutions than its North American counterpart (Bénard-Capelle et al., 2015; Helyar et al., 2014; Mariani et al., 2015), we set out to investigate the levels of seafood mislabelling in Britain’s raw seafood restaurants. Since sushi venues were so susceptible to fraud in the American seafood trade (Lowenstein, Amato & Kolokotronis, 2009; Warner et al., 2013), we focussed on this specific part of the supply chain. Sampling was spread across six different cities, focussing on tuna, eel and opportunistic samples of less distinguishable white-fleshed fish.
Materials and Methods
Sampling
A total of 115 fish samples were collected in 31 sushi restaurants in Manchester, London, Bristol, Liverpool, Exeter and Newcastle, between September 2014 and 2015. Two independent sets of samples were collected in restaurants in Manchester, Liverpool, and Newcastle, with a minimum of two weeks between sampling. In all cases the individuals involved in the collection of tissue posed as normal customers and sampled in an as unobtrusive way as possible.
Samples were placed in pre-numbered tubes and stored in 95% ethanol at −20 °C until extraction. Data were recorded, including commercial name, date, price, location, restaurant name, as well as photographs of samples when possible. Sampling focused on tuna (Thunnus sp.) and eel (Anguilla sp.) samples; these two product types are highly sought-after and include critically endangered species. A selection of less distinguishable white-fleshed fish available in each restaurant was also collected (Table 1) as these can comprise hundreds of fish species whose flesh is virtually unrecognisable by consumers and hence easily susceptible to substitution.
Table 1. Summary of the samples collected in sushi venues across the UK.
Identification represented in this table is obtained by using the BOLD ‘Public Record Barcode’ database. Samples marked by (∗) represent samples which were identified using cyt b sequencing and the Genbank public database, the (a) characterises samples identified by the COI mini-barcodes. Results by using other database can be found in Table S1. The conservation status of the species can by assessed by their IUCN Red List of Threatened Species status.
| City | Sold as | BOLD Public Record Barcode database (% match) | Accepted common name | Mislabelled | IUCN status | Accession number |
|---|---|---|---|---|---|---|
| Bristol | Tuna (Albacore) | Thunnus alalunga 100%, Thunnus obesus 100%, Thunnus orientalis 99.81%, Thunnus thynnus 99.61%, Thunnus atlanticus 99.03% | Albacore | NO | Near threatened | KU168615 |
| Exeter | Tuna (Albacore) | Thunnus alalunga 100%, Thunnus obesus 100%, Thunnus orientalis 99.81%, Thunnus maccoyii 99.81%, Thunnus atlanticus 99.04% | Albacore | NO | Near threatened | KU168616 |
| London | Tuna (Albacore) | Thunnus alalunga 99.79%, Thunnus obesus 99.38%, Thunnus orientalis 99.17%, Thunnus maccoyii 99.17%, Thunnus thynnus 98.96%, Thunnus albacares 98.33% | Albacore | NO | Near threatened | KU168617 |
| Bristol | Tuna (Bluefin) | Thunnus thynnus 100% | Atlantic Bluefin tuna | NO | Endangered | KU168618 |
| Liverpool | Tuna (Bluefin) | Thunnus albacares 100%, Thunnus atlanticus 100%, Thunnus obesus 100%, Thunnus maccoyii 99.85% | Yellowfin tuna | YES | Near threatened | KU168619 |
| Bristol | Tuna (Yellowfin) | Thunnus albacares 100%, Thunnus obesus 100%, Thunnus maccoyii 99.83%, Thunnus tonggol 99.83% | Yellowfin tuna | NO | Near threatened | KU168620 |
| Bristol | Tuna (Yellowfin) | Thunnus albacares 100%, Thunnus atlanticus 100%, Thunnus obesus 100%, Thunnus maccoyii 99.83%, Thunnus tonggol 99.83% | Yellowfin tuna | NO | Near threatened | KU168621 |
| Exeter | Tuna (Yellowfin) | Thunnus albacares 100%, | Yellowfin tuna | NO | Near threatened | KU168622 |
| London | Tuna (Yellowfin) | Thunnus albacares 100%, Thunnus atlanticus 99.79%, Thunnus obesus 99.79%, Thunnus maccoyii 99.79% | Yellowfin tuna | NO | Near threatened | KU168623 |
| Manchester | Tuna (Yellowfin) | Thunnus obesus 100%, Thunnus albacares 99.69%, Thunnus atlanticus 99.62%, Thunnus tonggol 99.52%, Thunnus maccoyii 99.4% | Bigeye tuna | YES | Vulnerable | KU168624 |
| Manchester | Tuna (Yellowfin) | Thunnus albacares 100%, Thunnus atlanticus 100%, Thunnus maccoyii 100%, Thunnus obesus 100% | Yellowfin tuna | NO | Near threatened | KU168625 |
| Bristol | Tuna | Thunnus albacares 100%, Thunnus obesus 99.82%, Thunnus maccoyii 99.67%, Thunnus tonggol 99.67% | Yellowfin tuna | NO | Near threatened | KU168627 |
| Bristol | Tuna | Thunnus albacares 100%, Thunnus obesus 99.82%, Thunnus maccoyii 99.67%, Thunnus tonggol 99.67% | Yellowfin tuna | NO | Near threatened | KU168628 |
| Bristol | Tuna | Thunnus albacares 100%, Thunnus atlanticus 100%, Thunnus obesus 100%, Thunnus maccoyii 99.83%, Thunnus tonggol 99.83% | Yellowfin tuna | NO | Near threatened | KU168629 |
| Bristol | Tuna | Thunnus albacares 100%, Thunnus atlanticus 100%, Thunnus obesus 100%, Thunnus maccoyii 99.83%, Thunnus tonggol 99.83% | Yellowfin tuna | NO | Near threatened | KU168630 |
| Bristol | Tuna | Thunnus obesus 100%, Thunnus albacares 99.34% | Bigeye tuna | NO | Vulnerable | KU168631 |
| Bristol | Tuna | Thunnus albacares 100%, Thunnus obesus 99.83% | Yellowfin tuna | NO | Near threatened | KU168632 |
| Bristol | Tuna | Thunnus albacares 100%, Thunnus atlanticus 100%, Thunnus maccoyii 100%, Thunnus obesus 100%, Thunnus tonggol 99.84% | Yellowfin tuna | NO | Near threatened | KU168633 |
| Bristol | Tuna | Thunnus albacares 100%, Thunnus atlanticus 100%, Thunnus obesus 100%, Thunnus maccoyii 99.84%, Thunnus tonggol 99.83% | Yellowfin tuna | NO | Near threatened | KU168634 |
| Exeter | Tuna | Thunnus albacares 100%, Thunnus atlanticus 99.49%, Thunnus obesus 99.49%, Thunnus maccoyii 99.48% | Yellowfin tuna | NO | Near threatened | KU168635 |
| Liverpool | Tuna* | Thunnus albacares 100%, Thunnus obesus 100%, Thunnus atlanticus 99.81%, Thunnus maccoyii 99.68% | Yellowfin tuna | NO | Near threatened | KU168636 |
| Liverpool | Tuna | Thunnus albacares 100% | Yellowfin tuna | NO | Near threatened | KU168637 |
| London | Tuna | Thunnus albacares 100%, Thunnus maccoyii 100%, Thunnus obesus 100% | Yellowfin tuna | NO | Near threatened | KU168638 |
| London | Tuna | Thunnus albacares 100%, Thunnus atlanticus 100%, Thunnus maccoyii 100%, Thunnus obesus 100% | Yellowfin tuna | NO | Near threatened | KU168639 |
| London | Tuna | Seriola lalandi 100%, Seriola zonata 99.36% | Yellowtail amberjack | YES | Not assessed | KU168640 |
| London | Tuna | Seriola lalandi 100%, Seriola zonata 99.36% | Yellowtail amberjack | YES | Not assessed | KU168641 |
| London | Tuna | Thunnus albacares 99.82%, Thunnus atlanticus 99.82%, Thunnus maccoyii 99.82%, Thunnus obesus 99.81% | Yellowfin tuna | NO | Near threatened | KU168642 |
| London | Tuna | Thunnus albacares 100%, Thunnus atlanticus 99.79%, Thunnus obesus 99.79%, Thunnus maccoyii 99.79% | Yellowfin tuna | NO | Near threatened | KU168643 |
| London | Tuna | Thunnus albacares 99.79%, Thunnus atlanticus 99.79%, Thunnus obesus 99.79%, Thunnus maccoyii 99.79% | Yellowfin tuna | NO | Near threatened | KU168644 |
| London | Tuna | Thunnus albacares 100%, Thunnus atlanticus 100%, Thunnus maccoyii 100%, Thunnus obesus 100% | Yellowfin tuna | NO | Near threatened | KU168645 |
| London | Tunaa | Thunnus thynnus 100% | Atlantic Bluefin tuna | NO | Endangered | KU168646 |
| London | Tuna | Thunnus albacares 100%, Thunnus atlanticus 100%, Thunnus maccoyii 100%, Thunnus obesus 100% | Yellowfin tuna | NO | Near threatened | KU168647 |
| London | Tuna | Thunnus albacares 100% | Yellowfin tuna | NO | Near threatened | KU168648 |
| London | Tuna | Thunnus albacares 100%, Thunnus atlanticus 100%, Thunnus maccoyii 100%, Thunnus obesus 100% | Yellowfin tuna | NO | Near threatened | KU168649 |
| Manchester | Tuna* | Seriola quinqueradiata 99.85%, Seriola lalandi 94.97% | Japanese amberjack | YES | Not assessed | KU168650 |
| Manchester | Tuna | Thunnus obesus 100%, Thunnus albacares 99.69% | Bigeye tuna | NO | Vulnerable | KU168651 |
| Manchester | Tuna* | Thunnus albacares 100%, Thunnus atlanticus 100%, Thunnus obesus 100%, Thunnus maccoyii 99.85% | Yellowfin tuna | NO | Near threatened | KU168652 |
| Manchester | Tuna (Spicy) | Thunnus albacares 100%, Thunnus atlanticus 100%, Thunnus obesus 100%, Thunnus maccoyii 99.85% | Yellowfin tuna | NO | Near threatened | KU168653 |
| Manchester | Tuna | Thunnus albacares 100%, Thunnus atlanticus 100%, Thunnus obesus 100%, Thunnus maccoyii 99.85% | Yellowfin tuna | NO | Near threatened | KU168654 |
| Manchester | Tuna | Thunnus thynnus 100%, Thunnus orientalis 99.69%, Thunnus atlanticus 99.69%, Thunnus maccoyii 99.54%, Thunnus albacares 99.53% | Atlantic Bluefin tuna | NO | Endangered | KU168655 |
| Manchester | Tuna | Thunnus albacares 100% | Bigeye tuna | NO | Vulnerable | KU168656 |
| Manchester | Tuna | Thunnus albacares 100% | Yellowfin tuna | NO | Near threatened | KU168657 |
| Manchester | Tuna | Thunnus thynnus 100%, Thunnus orientalis 99.84%, Thunnus maccoyii 99.84%, Thunnus alalunga 99.69%, Thunnus obesus 99.68%, Thunnus atlanticus 99.19%, Thunnus albacares 99% | Atlantic Bluefin tuna | NO | Endangered | KU168658 |
| Manchester | Tuna | Thunnus albacares 100%, Thunnus maccoyii 99.84%, Thunnus obesus 99.82%, Thunnus atlanticus 99.8% | Yellowfin tuna | NO | Near threatened | KU168659 |
| Manchester | Tuna* | Thunnus albacares 100%, Thunnus atlanticus 100%, Thunnus maccoyii 100%, Thunnus obesus 100%, Thunnus tonggol 99.84% | Yellowfin tuna | NO | Near threatened | KU168660 |
| Manchester | Tuna | Thunnus albacares 100%, Thunnus atlanticus 100%, Thunnus maccoyii 100%, Thunnus obesus 100% | Yellowfin tuna | NO | Near threatened | KU168661 |
| Newcastle | Tuna | Thunnus albacares 100%, Thunnus atlanticus 100%, Thunnus obesus 100%, Thunnus maccoyii 99.85%, | Yellowfin tuna | NO | Near threatened | KU168662 |
| Newcastle | Tuna | Thunnus albacares 100%, Thunnus atlanticus 99.84%, Thunnus maccoyii 99.84%, Thunnus obesus 99.82% | Yellowfin tuna | NO | Near threatened | KU168663 |
| Bristol | Eel | Anguilla anguilla 100% | European eel | NO | Critically endangered | KU168664 |
| Bristol | Eel | Anguilla anguilla 100% | European eel | NO | Critically endangered | KU168665 |
| Bristol | Eel | Anguilla marmorata 99.84% | Giant mottled eel | NO | Least concern | KU168666 |
| Exeter | Eel | Anguilla japonica 99.36% | Japanese eel | NO | Endangered | KU168667 |
| Liverpool | Eel | Anguilla anguilla 99.84% | European eel | NO | Critically endangered | KU168668 |
| Liverpool | Eel | Anguilla rostrata 99.84% | American eel | NO | Endangered | KU168669 |
| Liverpool | Eel | Anguilla japonica 100% | Japanese eel | NO | Endangere d | KU168670 |
| London | Eel (Freshwater)a | Anguilla japonica 100% | Japanese eel | NO | Endangered | KU168671 |
| London | Eel (grilled) | Anguilla anguilla 100% | European eel | NO | Critically endangered | KU168672 |
| London | Eela | Anguilla japonica 99.49% | Japanese eel | NO | Endangered | KU168673 |
| Manchester | Eel | Anguilla anguilla 99.84% | European eel | NO | Critically endangered | KU168674 |
| Manchester | Eel (Freshwater) | Anguilla anguilla 100% | European eel | NO | Critically endangered | KU168675 |
| Manchester | Eel | Anguilla anguilla 99.84% | European eel | NO | Critically endangered | KU168676 |
| Manchester | Eel | Anguilla japonica 99.54%, Anguilla marmorata 94.74% | Japanese eel | NO | Endangered | KU168677 |
| Manchester | Eel | Anguilla rostrata 99.84% | American eel | NO | Endangered | KU168678 |
| Manchester | Eel | Anguilla anguilla 100% | European eel | NO | Critically endangered | KU168679 |
| Manchester | Eel | Anguilla anguilla 100% | European eel | NO | Critically endangered | KU168680 |
| Manchester | Eel* | Anguilla anguilla 90% | European eel | NO | Critically endangered | KU168681 |
| Newcastle | Eel | Anguilla anguilla 100% | European eel | NO | Critically endangered | KU168683 |
| Newcastle | Eel | Anguilla anguilla 99.37% | European eel | NO | Critically endangered | KU168684 |
| Liverpool | Seabass* | Dicentrarchus labrax 99% | European seabass | NO | Least concern | KU168685 |
| Liverpool | Seabass* | Dicentrarchus labrax 100% | European seabass | NO | Least concern | KU168686 |
| Liverpool | Seabassa | Dicentrarchus labrax 100% | European seabass | NO | Least concern | KU168687 |
| London | Seabass* | Dicentrarchus labrax 100% | European seabass | NO | Least concern | KU168688 |
| London | Seabass | Lateolabrax japonicus 100%, Lateolabrax maculatus 99.63% | Japanese seabass | YES | Not assessed | KU168689 |
| London | Seabass | Lateolabrax japonicus 100%, Lateolabrax maculatus 99.49% | Japanese seabass | YES | Not assessed | KU168690 |
| London | Seabass* | Dicentrarchus labrax 100% | European seabass | NO | Least concern | KU168691 |
| London | Seabass* | Dicentrarchus labrax 100% | European seabass | NO | Least concern | KU168692 |
| London | Seabass* | Dicentrarchus labrax 100% | European seabass | NO | Least concern | KU168693 |
| Manchester | Seabass* | Dicentrarchus labrax 99% | European seabass | NO | Least concern | KU168694 |
| Manchester | Seabass* | Dicentrarchus labrax 99% | European seabass | NO | Least concern | KU168695 |
| Manchester | Seabass* | Dicentrarchus labrax 99% | European seabass | NO | Least concern | KU168696 |
| Manchester | Seabass* | Dicentrarchus labrax 100% | European seabass | NO | Least concern | KU168697 |
| Manchester | Seabass* | Dicentrarchus labrax 100% | European seabass | NO | Least concern | KU168698 |
| Manchester | Seabass* | Dicentrarchus labrax 100% | European seabass | NO | Least concern | KU168699 |
| Manchester | Seabass* | Dicentrarchus labrax 100% | European seabass | NO | Least concern | KU168700 |
| Bristol | Yellowtail | Seriola quinqueradiata 99.34%, Seriola lalandi 94.53% | Japanese amberjack | NO | Not assessed | KU168701 |
| Bristol | Yellowtail | Seriola quinqueradiata 99.51%, Seriola lalandi 94.75% | Japanese amberjack | NO | Not assessed | KU168702 |
| Bristol | Yellowtail | Seriola quinqueradiata 99.84%, Seriola lalandi 94.9% | Japanese amberjack | NO | Not assessed | KU168703 |
| Liverpool | Yellowtail | Seriola quinqueradiata 99.63%, Seriola lalandi 93.85% | Japanese amberjack | NO | Not assessed | KU168704 |
| London | Yellowtail | Seriola quinqueradiata 99.69% | Japanese amberjack | NO | Not assessed | KU168705 |
| London | Yellowtail | Seriola lalandi 100%, Seriola zonata 99.34% | Yellowtail amberjack | NO | Not assessed | KU168706 |
| London | Yellowtail | Seriola quinqueradiata 99.80% | Japanese amberjack | NO | Not assessed | KU168707 |
| London | Yellowtail | Seriola quinqueradiata 99.79% | Japanese amberjack | NO | Not assessed | KU168708 |
| London | Yellowtail | Seriola quinqueradiata 99.79% | Japanese amberjack | NO | Not assessed | KU168709 |
| London | Yellowtail | Seriola quinqueradiata 99.77% | Japanese amberjack | NO | Not assessed | KU168710 |
| Manchester | Yellowtail | Seriola quinqueradiata 99.55%, Seriola lalandi 94.97% | Japanese amberjack | NO | Not assessed | KU168711 |
| Manchester | Yellowtail | Seriola quinqueradiata 99.7%, Seriola lalandi 94.9% | Japanese amberjack | NO | Not assessed | KU168712 |
| London | Mackerel | Scomber scombrus 100% | Mackerel | NO | Least concern | KU168713 |
| London | Mackerel | Scomber scombrus 99.80% | Mackerel | NO | Least concern | KU168714 |
| London | Mackerel | Scomber scombrus 100% | Mackerel | NO | Least concern | KU168715 |
| London | Mackerel | Scomber scombrus 100% | Mackerel | NO | Least concern | KU168716 |
| London | Mackerel | Scomber scombrus 100% | Mackerel | NO | Least concern | KU168717 |
| London | Mackerel | Scomber scombrus 100% | Mackerel | NO | Least concern | KU168718 |
| London | Mackerel | Scomber scombrus 100% | Mackerel | NO | Least concern | KU168719 |
| London | Mackerel | Scomber scombrus 100% | Mackerel | NO | Least concern | KU168720 |
| Manchester | Seabream | Sparus aurata 100% | Gilthead bream | NO | Least concern | KU168721 |
| Manchester | Seabream | Sparus aurata 100% | Gilthead bream | NO | Least concern | KU168722 |
| Manchester | Seabream | Sparus aurata 100% | Gilthead bream | NO | Least concern | KU168723 |
| Liverpool | Swordfish | Makaira nigricans 99.52% | Blue marlin | YES | Data deficient | KU168724 |
| Newcastle | Swordfish | Xiphias gladius 100% | Swordfish | NO | Least concern | KU168725 |
| London | King Fish | Seriola lalandi 100%, Seriola zonata 99.38% | Yellowtail amberjack | YES | Not assessed | KU168726 |
| Manchester | King Fish (Tasmanian) | Seriola lalandi 100%, Seriola zonata 99.43% | Yellowtail amberjack | YES | Not assessed | KU168727 |
| Manchester | Barramundia | Lates calcarifer 100% | Barramundi | NO | Not assessed | KU168728 |
| Manchester | Black Cod | Anoplopoma fimbria 100% | Sablefish | NO | Not assessed | KU168729 |
| Liverpool | Flying Fish eggs | Clupea harengus 100% | Herring | YES | Least concern | KU168731 |
| London | Snapper | Sparus aurata 100% | Gilthead bream | YES | Least concern | KU168732 |
DNA extraction and sequencing
Genomic DNA was extracted from muscle tissue according to a Chelex resin protocol (Estoup et al., 1996). The partial cytochromoxidase 1 (COI) was amplified using the FishF2 and FishR2 from Ward et al. (2005), following the PCR amplifications by Serra-Pereira et al. (2010). If samples could not be successfully amplified, the COI mini-barcode primers (mICOIintF and jgHCO2198) following Leray et al. (2013) or the L14735 and H15149 cytochrome b (cytb) primers as described by Burgener (1997) were used. In the case of cytb amplification, 2 µl 10 × reaction buffer, 1.6 µl MgCl2 (50 mM), 1 µl of each primer (0.01 mM), 0.5 Units of DNA Taq Polymerase (PROMEGA, Madison, WI, USA) and 0.2 µl of each dNTP (10 µM) were used in a total volume of 20 µL. PCR conditions entailed 5 min at 94 °C, following a cycle of 40 s at 94 °C, 80 s at 55 °C, 80 s at 72 °C, which is repeated 35 times, finalized by 7 min at 72 °C, until the PCR was held at 10 °C.
DNA sequencing was carried out by Source Bioscience (Cambridge, UK) and all sequences were obtained with the forward primer. Sequences were checked manually against their chromatogram and edited in BioEdit (Hall, 1999). Each sequence was then used to BLAST-search both the GenBank reference database (www.ncbi.nlm.nih.gov/) and the Barcode of Life Data system (BOLD, http://www.boldsystems.org/, see Ratnasingham & Hebert, 2007), using the “Public Record Barcode Database”, which restricts the search to sequences that have been published. In the Supplemental Information, results are presented for the alternative BOLD reference databases: the default “Species Level Barcode Records” database and the “Full Length Record Barcode Database”, which is recommended to use with short sequences as it provides a maximum overlap. Identification was determined by sequence similarity to the reference dataset (Wong & Hanner, 2008), and checked by “Tree based identification” (i.e., distance trees in BOLD; Costa et al., 2012). With the NCBI database a minimum similarity of 90% was required. The match with the highest expectation value (E-value) of the BLAST program was retained as potential species identification. The E-value is a parameter that describes the number of hits one can expect to see just by chance when searching a database of a particular size.
For each sample, the list of admissible species that can be sold under the commercial name indicated on the menu was determined by consulting the UK governmental list with commercial designations of fish (DEFRA, 2013). The sample was declared mislabelled if the species name determined through molecular identification did not match the commercially accepted names in this list. Species or commercial names obtained orally from waiting staff in restaurants were not utilised in calculations of substitution rates, but this information is available in Table S1.
Results and Discussion
This study represents the largest sampling of UK sushi venues to date. A relatively intensive effort was made to collect samples across multiple time-points and regions, going beyond the sampling of only the most commonly consumed species like tuna, eel and salmon. The inherently high cost of sampling raw fish restaurants as consumers represents a limitation to the collection of huge sample sizes. However, the final sample size (N = 115) is of the same order of magnitude as recent comparable investigations and the sample design that was spread over 31 restaurants and a 12-month span, strove to avoid high levels of repeated sampling from any one location or restaurant, giving a degree of independence to the data.
Interpretable sequences were obtained for a total of 115 samples, ranging between 166 and 674 base pairs (bp) (average length 531 bp). These include 48 ‘tuna’, 20 ‘eel’, 16 ‘seabass’, 12 ‘yellowtail’, 8 ‘mackerel’, 3 ‘seabream’, 2 ‘swordfish’, 2 ‘kingfish’, and single samples of ‘black cod’, ‘barramundi’, ‘snapper’ and ‘flying fish’ (Table 1). Searches on BOLD and GenBank generally produced clear matches allowing for confident assignment of species and there was good agreement between databases (Table S1). In fact, all searches yielded matches that were within the 98% similarity to database records. For all sea bass samples and one eel sample, no successful COI amplifications could be produced, and the cytb primers were utilised instead. A BOLD search could not be made in these instances, as this database only contains COI sequences, so the GenBank identification was used.
In the case of certain Thunnus species, little interspecific divergence can limit the power of COI to discriminate among species pair, owing to the short evolutionary history and/or introgression among them (Tseng et al., 2012; Vinas & Tudela, 2009). However, in the current study this would not generally cause issues in assessing the levels of substitution as the commercial designation by DEFRA (2013) allows restaurants to sell all Thunnus species under the umbrella term “tuna” . Despite the limitation in Thunnus identification, in some instances there is the potential to go down to species level identification. We can distinguish T. thynnus from the other Thunnus species by following a set of criteria. First, when there is 100% sequence match criterion alongside the reduced similarity between the unknown sequences and any other matching species record. Second, the phylogenetic tree option in the BOLD reference database provides further evidence of the origin of the species. Finally, comparison of results of different/more stringent sets of reference data in BOLD further provides an unambiguous identification. Therefore, it was possible with some samples to assign the sequence obtained to either the yellowfin or bluefin tuna group, providing evidence of mislabelling.
The overall level of mislabelling and substitution was moderate (10.4%, Table 2). In the case of tuna, three samples were sold as tuna, but identified as Yellowtail and Japanese Amberjack (Seriola lalandi and Seriola quinqueradiata, respectively). In two other cases, the restaurant deliberately advertised a specific Thunnus species: one restaurant claimed to sell Yellowfin tuna (Thunnus albacares) while highest similarity scores by COI barcoding suggested potential substitution with Big-eye tuna (Thunnus obesus). Another restaurant claimed to serve Bluefin tuna, but COI barcoding revealed matches with Big-eye and Yellowfin tuna. Although the common name Bluefin tuna encompasses Atlantic Bluefin (Thunnus thynnus), Pacific Bluefin (Thunnus orientalis) and Southern Bluefin (Thunnus maccoyii), none of them matched the COI barcoding results. Kingfish was sampled in London and Manchester. According to the official list on commercial designation of fish in the United Kingdom (DEFRA, 2013) this common name represents all species of Scomberomorus. However, both samples were identified as Seriola lalandi and hence regarded as mislabelled. Among the 16 samples of seabass, two samples were identified as Lateolabrax maculaus also known as the Japanese seabass. In the case of one “swordfish” sample, the reference database inquiry identified the species Makaira nigricans (Atlantic blue marlin), with additional matches from closely related sister taxa belonging to other marlin species (Family: Istiophoridae). Although it is difficult to pinpoint the exact species ID, it is evident that the sample did not match with swordfish (Xiphias gladius). Further mislabelling was found for a sample of snapper (Family: Lutjanidae) which was identified as Sparus aurata (gilt-head sea bream) and the sample of the flying fish eggs (representing all species of the family Exocoetidae) were identified as herring (Clupea harengus) eggs. The sample of Black cod was identified as Anoplopoma fimbria. According to Fishbase, both Black cod and Sablefish are accepted common names for Anoplopoma fimbria; however, the official list on commercial designation of fish in the United Kingdom (DEFRA, 2013) only accepts ‘sablefish’. As both common names are accepted by the scientific community, this particular example was not deemed to be mislabelled, as the restaurant business aimed to serve a rather unfamiliar species to the UK public and used a scientifically correct name. Rather than mislabelling, this example can be seen as a misapplied market nomenclature, which shows how, in a context of increasingly global and diverse seafood market, regular communication between governments, fisheries managers and scientific advisors should be improved in order to guarantee an updated and accurate list of valid names. Yet, the new labelling regulations (EC 1379/2013, article 37) requiring the use of scientific names, may offer the necessary level of universality to commercial designations.
Table 2. Samples collected across the UK per species and per city.
| City | “Tuna” | “Eel” | Seabass | Yellowtail | Seabream | Mackerel | Swordfish | Black cod | Barramundi | Kingfish | Snapper | Flying fish eggs | TOTAL |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Manchester | 14 | 8 | 7 | 2 | 3 | 1 | 1 | 1 | 37 | ||||
| London | 14 | 3 | 6 | 6 | 8 | 1 | 1 | 39 | |||||
| Bristol | 12 | 3 | 3 | 18 | |||||||||
| Liverpool | 3 | 3 | 3 | 1 | 1 | 1 | 12 | ||||||
| Newcastle | 2 | 2 | 1 | 5 | |||||||||
| Exeter | 3 | 1 | 4 | ||||||||||
| TOTAL mislabelled | 5 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 2 | 1 | 1 | 12 |
| TOTAL | 48 | 20 | 16 | 12 | 3 | 8 | 2 | 1 | 1 | 2 | 1 | 1 | 115 |
When compared to recent studies on sushi labelling in North America, which returned 74% (Warner et al., 2013) and 16.3% (Khaksar et al., 2015) in the level of substitution, the UK food service sector comes under a more positive light (Table 1 and Fig. 1). Similarly, Bénard-Capelle et al. (2015) found only 3% substitution in French restaurants, which suggests lower levels of mislabelling in restaurants across Europe. In contrast to North America, mislabelling of tuna is less pronounced (10.2%). Generally in Europe substitution occurred between tuna species (Bénard-Capelle et al., 2015), or with amberjack, unlike in the US where a large portion of the tuna is substituted with escolar (Lepidocybium flavobrunneum, Warner et al., 2013). Comparisons between mislabelling in North America and the EU are valid as labelling regulation for the FDA (2016) and the EU are similar as to allowing umbrella term to be used for the sale of product in restaurants. Interestingly, in one case where oral enquiry about which tuna species was being sold was made to the waiting staff, the response was Bluefin tuna, which was not supported by the results of DNA barcoding. In this study, it was not included as a case of mislabelling, as the menu did not explicitly mention “Bluefin tuna”, but it does illustrate an absence of care or knowledge in the usage of this commercial name. Given that consumers are not expected to know every possible regional name, and the need to standardise labels across a large region with many different languages, the EU’s policy to require scientific names on display appears inevitable. The lowest level of mislabelling among the most studies detected only 16.3% of mislabelling in North America (Khaksar et al., 2015). In spite of the short sampling time and moderate samples size, their result is in sharp contrast to the study by Warner et al. (2013) who detected 74% mislabelling, suggesting a decreasing trends in mislabelling and illustrating that the role of media, environmental Non-governmental Organisations and scientific outputs in increasing public awareness is undeniable, which in turn raises the demand for enforcement of more rigorous inspection and audit processes in the food supply chain. Surveillance studies like this can help further refine the scope of such efforts and identify existing knowledge gaps.
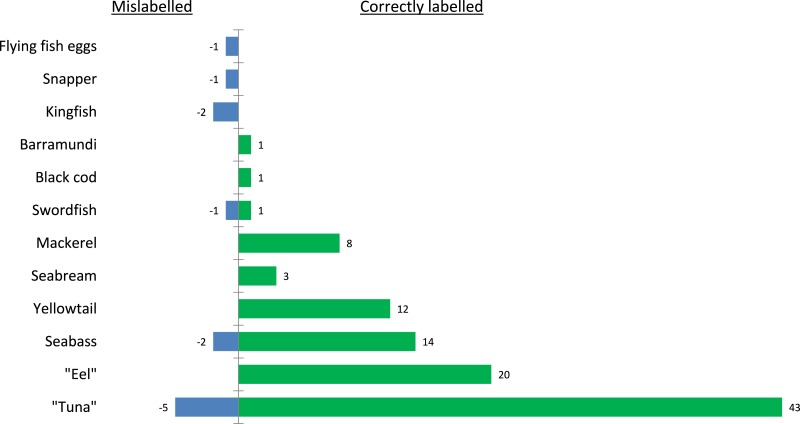
Figure 1. Level of mislabelling per species.
For the two ‘Swordfish’ samples, one sample was found correctly labelled, where the other was substituted with Marlin. Both the Marlin and Swordfish are depicted on either side of the diagram. Furthermore, substitution was recorded in tuna, seabass, kingfish, snapper and flying fish eggs samples.
Conservation issues
Concerns over the conservation and sustainable management of large oceanic fish are well established and the Big-eye and Yellowfin tunas identified in this study are listed as vulnerable and near-threatened by the International Union for Conservation of Nature and Natural Resources (IUCN) Red List (IUCN, 2015). Somewhat surprisingly, given the high conservation concern of Bluefin tuna species with the red listing of many species as endangered or critically endangered (IUCN, 2015) and its inclusion as a product to avoid due to sustainability issues in the Good Fish Guide (MSC, 2013), this product was listed on the menus of two restaurants. Bluefin tuna is particularly highly valued for its quality and taste. This would also make it an obvious target for economic fraud, with substitution for a lower value tuna species, as was identified in one case. In another instance, a product labelled with the umbrella term of “tuna” was also identified as Bluefin, which given its premium would appear as a missed promotion opportunity. Perhaps, due to the conservation issues around Bluefin tuna selling this meat under higher anonymity may help conceal that the species or individual was caught illegally (Jacquet & Pauly, 2008).
Mercury levels have been highlighted as a concern in some species. Species like Skipjack (Katsuwonus pelamis) and Yellowfin, often have lower mercury levels than other tuna species, such as Big-eye and Bluefin, and capture location in certain ocean basins can also be related to differing mercury levels (Lowenstein, Burger & Jeitner, 2010; Burger et al., 2014). Therefore, knowing what tuna species are being served and where they are caught is not only critical to making conservation informed consumer choices, but is also helpful in minimizing the health concerns of mercury exposure (Khaksar et al., 2015). This sort of crucial information is not easily accessible for consumers in restaurants, including sushi bars, and oral enquiries for this type of information appear to be unreliable.
Perhaps less well-known to the general public than conservation issues surrounding tuna, is the fact that most eel species are also of very poor conservation status. The European eel (Anguilla anguilla) is regarded as critically endangered (IUCN, 2015), and made up 62% of the eel products analysed. American (Anguilla rostrata) and Japanese (Anguilla japonica) eels were also found among the samples, and these are classified as endangered (IUCN, 2015). Although 90% of the freshwater eel consumed are farm-raised, they are not bred in captivity in economically relevant numbers (Mordenti et al., 2014; Okamura et al., 2014), young eels are still collected in the wild, further threatening wild populations (Okamura et al., 2014). The critical status of eels might explain why such a high diversity of species (4) is being found among the total of 21 samples analysed in this study. A worrying pattern of exploitation has already been noticed with eels; when one Anguilla species or population becomes over-exploited or fisheries restrictions are imposed, the industry moves to the next in order to fulfil demand (Crook & Nakamura, 2013). This may explain the occurrence of ‘new’ species, such as the Giant mottled eel (Anguilla marmorata), identified in the UK market for the first time.
Conclusion
This study detected a relatively low percentage of substitution, which could be an indicator that many restaurants have a positive attitude towards labeling accuracy due to heightened consumer awareness (Miller, Jessel & Mariani, 2012; Mariani et al., 2014). Even products, such as tuna, that are typically known to exhibit high levels of mislabeling, showed a remarkable level of compliance, corroborating the idea that seafood trade in the EU is addressing issues concerning mislabeling and food authenticity (Mariani et al., 2015). Although the substitutions appear infrequent compared to studies in other territories, or those conducted some years ago, improvements can be made to increase the reliability of the market. The legislation on labelling differs between restaurants, fresh sales and deep-frozen fish. For some groups, such as tuna, snapper or eel, the authorized commercial names cover a large number of species, including species with serious conservation and management issues. In such cases, consumers are unable to choose according to sustainability criteria. Additionally, because our study was restricted to seafood sold in a specific type of food service, at the end of a complex supply chain, it is difficult to determine if fraud is occurring at the landing site, during processing, at the wholesale level, at the retail counter or somewhere else along the way (Cawthorn, Steinman & Witthuhn, 2012). Therefore, in such a complex landscape, where restaurants may be just as much victims of mislabelling practices as consumers, more interdisciplinary research will be necessary to identify the mechanisms that still pose a threat to a transparent seafood supply chain.
Supplemental Information
Samples of Seabass are not included as identification was only possible with GenBank and is presented in Table 1. Samples marked with an astrics are the samples for which detailed species names were given from the waiting staff, however this information was not upheld in the analysis.
Sample names, genbank accession numbers and the sequences
Acknowledgments
We are grateful to the Academic Editor and three reviewers for their comments on earlier versions.
Funding Statement
This work was funded by the European Union INTERREG Atlantic Area Program (‘LabelFish’, project 2011-1/163). The UK Department for Environment, Food and Rural Affairs (DEFRA) grant FA0116, the University of Bristol and the University of Salford. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Sara G. Vandamme, Email: vandammesara@hotmail.com.
Stefano Mariani, Email: s.mariani@salford.ac.uk.
Additional Information and Declarations
Competing Interests
The authors declare there are no competing interests.
Author Contributions
Sara G. Vandamme conceived and designed the experiments, performed the experiments, analyzed the data, wrote the paper, prepared figures and/or tables, reviewed drafts of the paper.
Andrew M. Griffiths conceived and designed the experiments, performed the experiments, analyzed the data, contributed reagents/materials/analysis tools, wrote the paper, reviewed drafts of the paper.
Sasha-Ann Taylor, Jessica A. Towne and Mhairi Watson performed the experiments.
Cristina Di Muri prepared figures and/or tables, reviewed drafts of the paper.
Elizabeth A. Hankard performed the experiments, analyzed the data.
Stefano Mariani conceived and designed the experiments, contributed reagents/materials/analysis tools, wrote the paper, reviewed drafts of the paper.
Animal Ethics
The following information was supplied relating to ethical approvals (i.e., approving body and any reference numbers):
The work presented in this paper did not involve the use of live animals and no animals were killed expressly for this work. In fact, all samples we collected from food for human consumption at restaurants. Therefore, ethical oversight is not required in this case.
Data Availability
References
- Bénard-Capelle et al. (2015).Bénard-Capelle J, Guillonneau V, Nouvian C, Fournier N, Le Loët K, Dettai A. Fish mislabelling in France: substitution rates and retail types. PeerJ. 2015;2:e1891. doi: 10.7717/peerj.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgener (1997).Burgener M. PhD Thesis. 1997. Molecular species differentiation of fish and mammals. [Google Scholar]
- Burger et al. (2014).Burger J, Gochfeld M, Jeitner C, Donio M, Pittfield T. Sushi consumption rates and mercury levels in sushi: ethnic and demographic differences in exposure. Journal of Risk Research. 2014;17(8):981–997. doi: 10.1080/13669877.2013.822925. [DOI] [Google Scholar]
- Cawthorn, Steinman & Witthuhn (2012).Cawthorn D-M, Steinman HA, Witthuhn RC. DNA barcoding reveals a high incidence of fish species misrepresentation and substitution on the South African market. Food Research International. 2012;46(1):30–40. doi: 10.1016/j.foodres.2011.11.011. [DOI] [Google Scholar]
- Cawthorn et al. (2015).Cawthorn DM, Duncan J, Kastern C, Francis J, Hoffman LC. Fish species substitution and misnaming in South Africa: an economic, safety and sustainability conundrum revisited. Food Chemistry. 2015;185:165–181. doi: 10.1016/j.foodchem.2015.03.113. [DOI] [PubMed] [Google Scholar]
- Crook & Nakamura (2013).Crook V, Nakamura M. Glass eels: assessing supply chain and market impacts of a CITES listing on Anguilla species. TRAFFIC Bulletin. 2013;24(i–iv):9–24. [Google Scholar]
- Costa et al. (2012).Costa FO, Landi M, Martins R, Costa MH, Costa ME, Carneiro M, Alves MJ, Steinke D, Carvalho GR. A ranking system for reference libraries of DNA barcodes: application to marine fish species from Portugal. PLoS ONE. 2012;7(4):e1891. doi: 10.1371/journal.pone.0035858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pinto et al. (2015).Di Pinto A, Marchetti P, Mottola A, Bozzo G, Bonerba E, Ceci E, Bottaro M, Tantillo G. Species identification in fish fillet products using DNA barcoding. Fisheries Research. 2015;170:9–13. doi: 10.1016/j.fishres.2015.05.006. [DOI] [Google Scholar]
- DEFRA (2013).DEFRA . 2013. Commercial designations of fish: United Kingdom. Available at https://www.gov.uk/government/publications/commercial-designations-of-fish-united-kingdom . [Google Scholar]
- Estoup et al. (1996).Estoup A, Largiadèr CR, Perrot E, Chourrout D. Rapid one-tube DNA extraction for reliable PCR detection of fish polymorphic markers and transgenes. Molecular Marine Biology and Biotechnology. 1996;3(4):295–298. [Google Scholar]
- FAO (2014).FAO . Rome: Food and Agriculture Organization of the United Nations; 2014. (The state of world fisheries and aquaculture: opportunities and challenges). [Google Scholar]
- FDA (2016).FDA . 2016. Guidance for Industry: the Seafood List—FDA’s guide to acceptable market names for seafood sold in interstate commerce. Available at http://www.fda.gov/Food/GuidanceRegulation/GuidanceDocumentsRegulatoryInformation/Seafood/ucm113260.htm (accessed 07 March 2016) [Google Scholar]
- Hall (1999).Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Oxford University Press. Nucleic Acid Symposium Series. 1999;41:95–98. [Google Scholar]
- Helyar et al. (2014).Helyar SJ, Lloyd HAD, De Bruyn M, Leake J, Bennett N, Carvalho GR. Fish product mislabelling: failings of traceability in the production chain and implications for illegal, unreported and unregulated (IUU) fishing. PLoS ONE. 2014;9(6):e1891. doi: 10.1371/journal.pone.0098691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IUCN (2015).IUCN The IUCN Red List of Threatened Species. Version 2015.2. 2015. Available at http://www.iucnredlist.org (accessed 19 May 2015)
- Jacquet & Pauly (2008).Jacquet JL, Pauly D. Trade secrets: renaming and mislabelling of seafood. Marine Policy. 2008;32:309–318. doi: 10.1016/j.marpol.2007.06.007. [DOI] [Google Scholar]
- Khaksar et al. (2015).Khaksar R, Carlson T, Schaffner DW, Ghorashi M, Best D, Jandhyala S, Traverso J, Amini S. Unmasking seafood mislabelling in U.S. markets: DNA barcoding as a unique technology for food authentication and quality control. Food Control. 2015;56:71–76. doi: 10.1016/j.foodcont.2015.03.007. [DOI] [Google Scholar]
- Leray et al. (2013).Leray M, Yang JY, Meyer CP, Mills SC, Agudelo N, Ranwez V, Boehm JT, Machida RJ. A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents. Frontiers in Zoology. 2013;10:34. doi: 10.1186/1742-9994-10-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenstein, Amato & Kolokotronis (2009).Lowenstein JH, Amato G, Kolokotronis S-O. The real maccoyii: identifying Tuna Sushi with DNA barcodes—contrasting characteristic attributes and genetic distances. PLoS ONE. 2009;4:e1891. doi: 10.1371/JOURNAL.PONE.0007866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowenstein, Burger & Jeitner (2010).Lowenstein JH, Burger J, Jeitner CW. DNA barcodes reveal species-specific mercury levels in tuna sushi that pose a health risk to consumers. Biology Letters. 2010;6(5):692–695. doi: 10.1098/rsbl.2010.0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariani et al. (2014).Mariani S, Ellis J, O’Reilly A, Bréchon AL, Sacchi C, Miller DD. Mass media influence and the regulation of illegal practices in the seafood market. Conservation Letters. 2014;7(5):478–483. doi: 10.1111/conl.12085. [DOI] [Google Scholar]
- Mariani et al. (2015).Mariani S, Griffiths AM, Velasco A, Kappel K, Jérôme M, Perez-Martin RI, Schröder U, Verrez-Bagnis V, Silva H, Vandamme SG, Boufana B. Low mislabeling rates indicate marked improvements in European seafood market operations. Frontiers in Ecology and the Environment. 2015;13(10):536–540. doi: 10.1890/150119. [DOI] [Google Scholar]
- Miller, Jessel & Mariani (2012).Miller D, Jessel A, Mariani S. Seafood mislabelling: comparisons of two western European case studies assist in defining influencing factors, mechanisms and motives. Fish and Fisheries. 2012;13(3):345–358. doi: 10.1111/j.1467-2979.2011.00426.x. [DOI] [Google Scholar]
- Mordenti et al. (2014).Mordenti O, Casalini A, Mandelli M, Di Biase A. A closed recirculating aquaculture system for artificial seed production of the European eel (Anguilla anguilla): technology development for spontaneous spawning and eggs incubation. Aquaculture Engineering. 2014;58:88–94. doi: 10.1016/j.aquaeng.2013.12.002. [DOI] [Google Scholar]
- MSC (2013).MSC . 2013. Good fish guide—a guide to choosing sustainable seafood. Available at http://catering.southampton.ac.uk/sites/catering.southampton.ac.uk/files/PocketGoodFishGuide22January2013 (accessed 15 July 2015) [Google Scholar]
- Okamura et al. (2014).Okamura A, Horie N, Mikawa N, Yamada Y, Tsukamoto K. Recent advances in artificial production of glass eels for conservation of anguillid eel populations. Ecology of Freshwater Fish. 2014;23(1):95–110. doi: 10.1111/eff.12086. [DOI] [Google Scholar]
- Pramod et al. (2014).Pramod G, Nakamura K, Pitcher TJ, Delagran L. Estimates of illegal and unreported fish in seafood imports to the USA. Marine Policy. 2014;48:102–113. [Google Scholar]
- Ratnasingham & Hebert (2007).Ratnasingham S, Hebert PDN. Bold: the barcode of life data system (http://www.barcodinglife.org. ) Molecular Ecology Notes. 2007;7(3):355–364. doi: 10.1111/j.1471-8286.2007.01678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra-Pereira et al. (2010).Serra-Pereira B, Moura T, Griffiths AM, Gordo LS, Figueiredo I. Molecular barcoding of skates (Chondrichthyes: Rajidae) from the southern Northeast Atlantic. Zoologica Scripta. 2010;40(1):76–84. doi: 10.1111/j.1463-6409.2010.00461.x. [DOI] [Google Scholar]
- Tseng et al. (2012).Tseng MC, Jean CT, Smith PJ, Hung YH. Interspecific and intraspecific genetic diversity of Thunnus species. INTECH Open Access Publisher; 2012. [Google Scholar]
- Vinas & Tudela (2009).Vinas J, Tudela S. A Validated methodology for genetic identification of tuna species (Genus Thunnus) PLoS ONE. 2009;4:e1891. doi: 10.1371/journal.pone.0007606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward et al. (2005).Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PDN. DNA barcoding Australia’s fish species. Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences. 2005;360(1462):1847–1857. doi: 10.1098/rstb.2005.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner et al. (2013).Warner K, Timme W, Lowell B, Hirshfield M. Oceana study reveals seafood fraud nationwide. 2013. Available at http://oceana.org/reports/oceana-study-reveals-seafood-fraud-nationwide .
- Watson et al. (2015).Watson RA, Green BS, Tracey SR, Farmery A, Pitcher TJ. Provenance of global seafood. Fish and Fisheries. 2015 doi: 10.1111/faf.12129. [DOI] [Google Scholar]
- Wong & Hanner (2008).Wong EH-K, Hanner RH. DNA barcoding detects market substitution in North American seafood. Food Research International. 2008;41(8):828–837. doi: 10.1016/j.foodres.2008.07.005. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Samples of Seabass are not included as identification was only possible with GenBank and is presented in Table 1. Samples marked with an astrics are the samples for which detailed species names were given from the waiting staff, however this information was not upheld in the analysis.
Sample names, genbank accession numbers and the sequences
Data Availability Statement
The following information was supplied regarding data availability:
All 118 sequences generated in this study have been made publicly available on Genbank (accession numbers KU168615– KU168732) and also appear in Table 1.