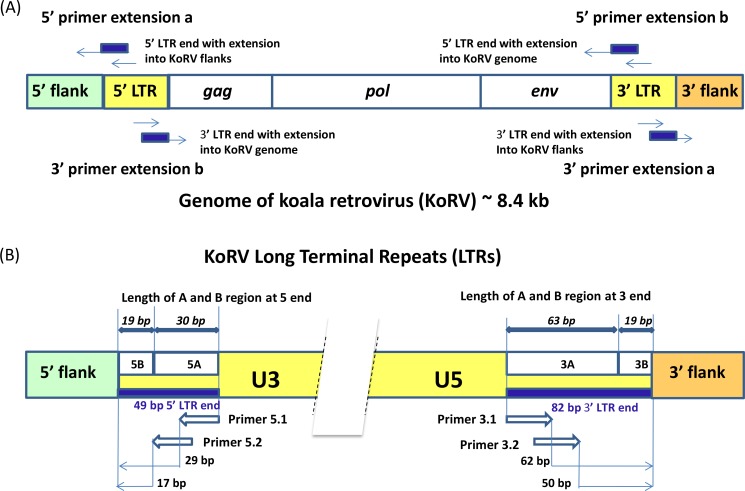
Figure 2. Experimental design for the identification of KoRV integration sites.
A illustrates that the genome of the koala retrovirus (KoRV) has two identical long terminal repeats (LTRs) on both ends. The primers or baits can bind to both LTRs, so two categories of products exist: (A) products extending into the flanks from primer extension a; (B) products extending into the middle of KoRV genome from primer extension b. In principle, there should be an equal number of sequences for the two categories. B indicates that the KoRV LTRs contain three components, U3, R and U5. For SPEX, primers were partially nested. All primers are 20 bp long, and there is a 8 bp-overlap between the inner primers (3.1 and 5.1) and outer primers (3.2 and 5.2) respectively. To avoid known polymorphisms in the LTR, the 3′ end of outer primers are 17 bp from the 5′ end of LTR and 50 bp from the 3′ end of LTR. Since the 5′ LTR and 3′ LTR of the same KoRV are identical products can also extend into the KoRV genome. The 5′ and 3′ flanks can be distinguished by their linked LTR end, with the 5′ flank linked to 5′ LTR and 3′ flank linked to 3′ LTR. Considering the longest deletion found at the end of LTR is 19 bp, the LTR end was divided into two segments for subsequent computational identification: the B region representing the last 19 bp of the LTR, and the A region representing the rest of LTR end.