Abstract
Rationale: Rhinoviruses (RVs) are a major cause of symptomatic respiratory tract infection in all age groups. However, RVs can frequently be detected in asymptomatic individuals.
Objectives: To evaluate the ability of host transcriptional profiling to differentiate between symptomatic RV infection and incidental detection in children.
Methods: Previously healthy children younger than 2 years old (n = 151) were enrolled at four study sites and classified into four clinical groups: RV− healthy control subjects (n = 37), RV+ asymptomatic subjects (n = 14), RV+ outpatients (n = 30), and RV+ inpatients (n = 70). Host responses were analyzed using whole-blood RNA transcriptional profiles.
Measurements and Main Results: RV infection induced a robust transcriptional signature, which was validated in three independent cohorts and by quantitative real-time polymerase chain reaction with high prediction accuracy. The immune profile of symptomatic RV infection was characterized by overexpression of innate immunity and underexpression of adaptive immunity genes, whereas negligible changes were observed in asymptomatic RV+ subjects. Unsupervised hierarchical clustering identified two main clusters of subjects. The first included 93% of healthy control subjects and 100% of asymptomatic RV+ subjects, and the second comprised 98% of RV+ inpatients and 88% of RV+ outpatients. Genomic scores of healthy control subjects and asymptomatic RV+ children were similar and significantly lower than those of RV+ inpatients and outpatients (P < 0.0001).
Conclusions: Symptomatic RV infection induced a robust and reproducible transcriptional signature, whereas identification of RV in asymptomatic children was not associated with significant systemic transcriptional immune responses. Transcriptional profiling represents a useful tool to discriminate between active infection and incidental virus detection.
Keywords: asymptomatic, children, rhinovirus, transcriptional profiling, viral detection
At a Glance Commentary
Scientific Knowledge on the Subject
Rhinoviruses (RVs) are the most frequent cause of respiratory tract infection in children and adults. However, they are also frequently detected in the absence of symptoms. The significance of detecting RVs in respiratory samples obtained from asymptomatic individuals remains unclear, and there are no standard diagnostic tools currently available to differentiate between viral detection and active infection.
What This Study Adds to the Field
Analysis of host whole-blood transcriptional profiles demonstrates that symptomatic RV infection in children is associated with a robust and reproducible transcriptional signature with significant changes in expression of immune-related genes, whereas negligible transcriptional host responses were observed in asymptomatic children in whom RV was incidentally detected. These findings suggest that transcriptional profiling may provide a useful tool to differentiate between incidental pathogen detection and active infection.
Rhinoviruses (RVs) are the most frequent etiologic agents of respiratory tract infections, both in children and in adults. In children, the clinical spectrum of RV infection ranges from fever or mild upper respiratory tract infections, such as the common cold and acute otitis media, to severe lower respiratory tract infections, including bronchiolitis, asthma exacerbations, and pneumonia (1–5). In addition, RV-induced wheezing during infancy has been linked to an increased risk for recurrent wheezing and asthma later in childhood (6, 7).
The use of molecular diagnostics has broadened the understanding of RV-associated illnesses. It has also shown that RV can frequently be detected in asymptomatic subjects. Detection rates of RV in asymptomatic subjects are highest in young children, with frequencies ranging from 14% to 50% (8–13). The high detection rates of RV in asymptomatic subjects and the common codetection with other viruses have raised questions about the etiological role of RV in certain clinical situations. Furthermore, it is unclear whether RV detection in asymptomatic subjects represents an active infection or whether it merely indicates the presence of viral RNA in the respiratory tract mucosa without detectable immune responses.
Whole-blood transcriptome analysis is a comprehensive tool to gain understanding of the pathogenesis of disease processes. It has also provided new insights into the diagnosis of autoimmune and infectious diseases (14–22). In this study, we evaluated the value of gene expression profiles to differentiate asymptomatic viral detection from symptomatic RV infection in young children. Some of the results of these studies have been reported previously in abstract form (23, 24).
Methods
Study Design
We conducted a prospective study involving a convenience sample of previously healthy children younger than 2 years of age recruited over six respiratory seasons (2007–2011 and 2013–2015). Children with respiratory symptoms were enrolled at the emergency department (outpatients) or within 48 hours of hospitalization (inpatients) at four study sites: Children’s Medical Center (Dallas, TX), Turku University Hospital (Turku, Finland), Regional University Hospital of Malaga (Malaga, Spain), and Nationwide Children’s Hospital (Columbus, OH). Healthy control subjects were enrolled during well-child visits or minor elective surgical procedures not involving the respiratory tract. Blood and nasopharyngeal samples were collected at enrollment from all subjects and analyzed for transcriptional profiles and respiratory viruses, respectively. Study subjects were classified into four clinical groups: (1) RV+ inpatients and (2) RV+ outpatients with respiratory tract infections, (3) asymptomatic subjects in whom RV was detected, and (4) RV− healthy control subjects. Exclusion criteria included documented bacterial or viral coinfections, systemic corticosteroid treatment in the preceding 2 weeks, prematurity (<36 wk of gestation), immunodeficiency, or chronic medical conditions. Healthy control subjects with a history of a respiratory tract infection within 2 weeks of enrollment were also excluded. This study was conducted in accordance with the Declaration of Helsinki and was approved by the institutional review boards at Nationwide Children’s Hospital (IRB 10-00028), the University of Texas Southwestern Medical Center (0802-447), Baylor Research Institute (002-141), Turku University Hospital (21.11.2006-492), and Malaga Maternal and Child Hospital (IP-17_271114). It was classified as a level 1 risk clinical study, indicating no greater than minimal risk (pursuant to 45 C.F.R. § 46.404 and 21 C.F.R. § 50.51). Informed consent procedures were followed in compliance with each institution’s guidelines for responsible conduct of research.
Sample and Data Collection
Nasal specimens from patients and control subjects were tested for respiratory viruses using either a multiplex polymerase chain reaction (PCR) panel or an individual real-time (RT)-PCR assay. Blood samples were collected for microarray analyses and stored at −20°C or −80°C within 2–4 hours of collection, and analyzed for white blood cell (WBC) counts and serum C-reactive protein (CRP) concentrations. Demographic and clinical parameters were collected using a standardized questionnaire, and disease severity was assessed according to the need for hospitalization (inpatients [moderate or severe disease] vs. outpatients [mild disease]) and using a standardized clinical disease severity score (CDSS) (20, 25). Blood and urine cultures were obtained according to individual physicians’ criteria. Details on virological testing, sample storage and processing, and CDSS can be found in the online supplement.
Microarray Data, Quantitative RT-PCR Validation, and Statistical Analyses
RNA samples were hybridized into Illumina Human WG-6 v3 or HT-12 v4 BeadChips (Illumina, San Diego, CA). Transcriptional signatures were identified and validated in four independent datasets (20, 21). We performed several analyses to account for the nonbiological and biological variability within the study, as explained in the online supplement. Briefly, to adjust for the experimental variation between batches (batch effect) and to assess the performance of the batch correction, we used the ComBat tool (26) followed by principal component analysis (see Figure E1 in the online supplement). The platform and chip information of the samples is included in Table E1. Except for modular analyses (27, 28), molecular distance to health (MDTH) scores (29), and Ingenuity Pathway Analysis (QIAGEN, Redwood City, CA), downstream analyses were performed in R statistical software. We used the limma package and applied stringent statistical filtering (Benjamini-Hochberg false discovery rate <0.01, ≥1.25-fold change) along with linear models to adjust for known biological factors, specifically age, race, and WBC composition (30). The variability in WBC composition between study groups was adjusted by deriving lineage-specific neutrophil, lymphocyte, and monocyte scores as proposed by Zhai and coworkers (22). Monocyte percentages were comparable between groups, and lymphocyte and neutrophil percentages were negatively correlated; therefore, to avoid overfitting, only neutrophil scores were left in the model (Figure E2, Table 1, and Table E2). For validation and to determine the accuracy of the RV signature to differentiate between groups, we used the support vector machine (SVM) algorithm and also applied unsupervised hierarchical clustering. The fit and stability of the clusters were assessed using average silhouette indices (Figure E3). Last, by performing quantitative RT-PCR (qRT-PCR), we validated the top representative transcripts identified in the RV signature (Table E3). The study data are deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (accession number GEO:GSE67059).
Table 1.
Demographic and Clinical Characteristics of the Study Subjects, by Clinical Group
| RV+ Inpatients (n = 70) | RV+ Outpatients (n = 30) | RV+ Asymptomatic Subjects (n = 14) | RV− Healthy Control Subjects (n = 37) | P Value | |
|---|---|---|---|---|---|
| Age, mo | 7.7* (2.2–15.4) | 9.3* (5.2–14.4) | 6.3 (2.6–9.3) | 4.3 (2.1–6.5) | 0.003 |
| Sex, male:female, n | 49:21 | 17:13 | 10:4 | 21:16 | 0.396 |
| Race/ethnicity, n (%) | 0.015 | ||||
| White | 44 (63) | 19 (63) | 7 (50) | 18 (49) | |
| Hispanic | 17 (24) | 1 (3) | 2 (14) | 13 (35) | |
| Black/other | 9 (13) | 10 (33) | 5 (36) | 6 (16) | |
| WBC | |||||
| Count, 103/mm3 | 11.3 (8.1–14.7) | 10.0 (8.2–12.1) | 9.2 (7.0–9.8) | 9.1 (6.2–10.8) | 0.061 |
| Neutrophils, % | 30* (24–43) | 30 (21–37) | 18 (15–22) | 21 (17–25) | <0.001 |
| Lymphocytes, % | 53* (39–60) | 57* (42–69) | 73 (66–77) | 70 (66–73) | <0.001 |
| Monocytes, % | 9 (5–13) | 8 (7–12) | 8 (5–10) | 6 (3–8) | 0.139 |
| CDSS | 46.7 (26.7–60.0) | 25.83 (6.7–40.4) | N/A | N/A | <0.001 |
| Duration of symptoms, d† | 4 (2–7) | 4 (3–7) | N/A | N/A | 0.872 |
Definition of abbreviations: CDSS = clinical disease severity score scaled from 0 to 100; N/A = not applicable; RV = rhinovirus; WBC = white blood cells.
Data are reported as median (interquartile range) unless otherwise indicated. WBC data were available in 129 (85%) of 151 children, and complete differential was available in 74 (49%) of 151 children. Categorical data were analyzed using a χ2 test. For continuous variables, the Mann-Whitney U test or the Kruskal-Wallis test followed by Dunn’s test for multiple comparisons was used.
Parameters that are significantly different compared with healthy control subjects.
Duration of symptoms at the time of sample collection.
Results
Characteristics of the Study Subjects
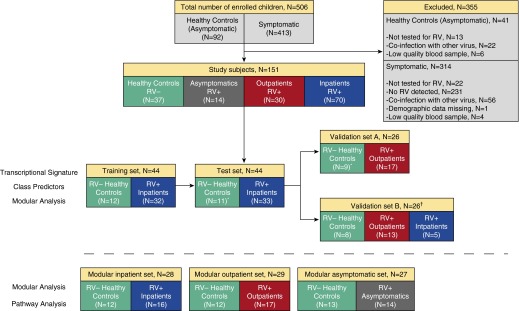
During the study period, 151 of 506 children screened fulfilled the inclusion criteria and were included in the analysis (Figure 1). Subjects were classified into four clinical groups according to disease severity and RV detection: (1) RV+ inpatients (n = 70), (2) RV+ outpatients (n = 30), (3) RV+ asymptomatic subjects (n = 14), and (4) RV− healthy control subjects (n = 37). Demographic, laboratory, and clinical parameters of study subjects according to clinical group are shown in Table 1. Overall, RV+ inpatients (median age, 7.7 mo) and RV+ outpatients (median age, 9.3 mo) were older than healthy control subjects (median age, 4.3 mo). WBC counts, which were obtained in 85% of the study subjects, were not different between the four clinical groups. However, neutrophil and lymphocyte percentages were significantly different in symptomatic patients compared with healthy control subjects (Table 1). CRP values were evaluated in 79 (79%) of the 100 symptomatic RV+ children and were low (median, 1.2 mg/dl; interquartile range, 0.4–2.3 mg/dl). In addition, blood and urine cultures were performed at the discretion of the attending physician in 23 (23%) and 14 (14%) of the 100 RV+ symptomatic children, respectively, and all results were negative. Overall, 83% of symptomatic RV+ children underwent either bacterial culture or CRP testing.
Figure 1.
Flowchart of study subjects. The upper panels indicate the number of patients and control subjects enrolled and the reasons for exclusion of others. Subjects included in the study (n = 151) were classified into four clinical groups according to disease severity and rhinovirus (RV) detection: RV+ inpatients, RV+ outpatients, RV+ asymptomatic subjects, and RV− healthy control subjects. Transcriptional signatures were identified and validated in four datasets (middle panels): training set, test set, validation set A, and validation set B. The first three groups underwent batch correction, and the fourth group was used as an additional, external validation cohort. A random selection of patients was used to derive the modular signatures (lower panels) for RV+ inpatients, RV+ outpatients, and RV+ asymptomatic subjects. *Three healthy control subjects were shared between the test set and validation set A. †Samples in validation set B were not included in the batch correction.
Symptomatic RV Infection Induces a Robust and Reproducible Transcriptional Signature
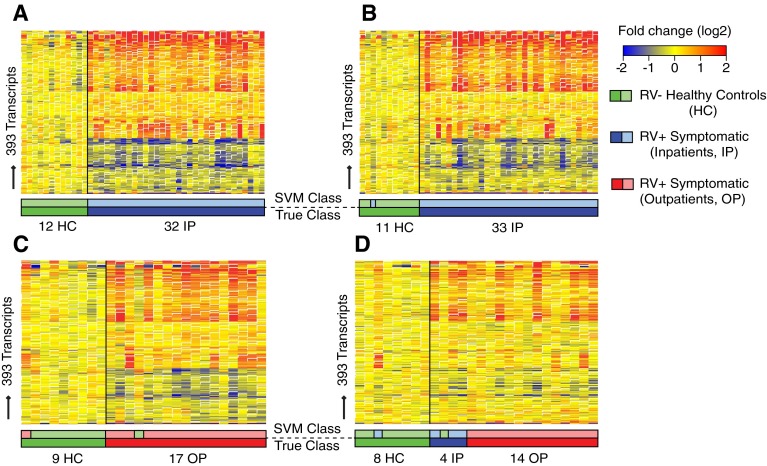
To first identify and then validate the host systemic response to RV infection (RV transcriptional signature), the 151 study subjects were divided into four distinct cohorts (training set, test set, validation set A, and validation set B) (Figure 1, Table 2). Using statistical group comparisons (Benjamini-Hochberg corrected false discovery rate <0.01 and ≥1.25-fold change) using linear models adjusted for age, race, and WBC composition (neutrophil score), we identified 393 transcripts differentially expressed between 32 RV+ inpatients and 12 healthy control subjects included in the training set (Figure 2A). Sixty-six percent of transcripts were overexpressed, and 34% were underexpressed. The top 20 overexpressed transcripts included genes related to IFN (IFI27, IFITM3), neutrophil function (DEFA1, DEFA3, DEFA4, BPI, GPR84, FCGR1), and apoptosis (MMP9), among others (Table E4). Representative transcripts among these top overexpressed genes were validated by qRT-PCR (Figure E4 and Table E3).
Table 2.
Demographic and Clinical Characteristics of the Study Subjects, Divided into Four Distinct Sets for Data Analysis
|
Training Set (n = 44) |
Test Set (n = 44) |
Validation Set A (n = 26) |
Validation Set B (n = 26) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inpatients (n = 32) | Healthy Control Subjects (n = 12) | P Value | Inpatients (n = 33) | Healthy Control Subjects (n = 11) | P Value | Outpatients (n = 17) | Healthy Control Subjects (n = 9) | P Value | Inpatients (n = 5) | Outpatients (n = 13) | Healthy Control Subjects (n = 8) | P Value | |
| Age, mo | 7.6 (2.3–16.1) | 6 (2.3–7.6) | 0.207 | 9.9 (2.7–15.1) | 6.0 (4.0–6.2) | 0.073 | 14.1 (6.5–15.4) | 2.3 (1.2–9.5) | 0.018 | 2* (1.6–4.0) | 6.7* (5.0–10.0) | 5.1 (2.9–6.7) | 0.007 |
| Sex, male:female, n | 23:9 | 7:5 | 0.476 | 22:11 | 5:4 | 0.289 | 9:8 | 3:6 | 0.429 | 4:1 | 8:5 | 7:1 | 0.400 |
| Race, n (%) | 0.143 | 0.017 | 0.161 | 0.014 | |||||||||
| White | 18 (56) | 3 (25) | 22 (67) | 3 (27) | 17 (100) | 8 (89) | 4 (80) | 2 (15) | 7 (88) | ||||
| Hispanic | 11 (34) | 6 (50) | 6 (18) | 7 (64) | 0 | 0 | 0 | 1 (8) | 0 | ||||
| Black/other | 3 (9) | 3 (25) | 5 (15) | 1 (9) | 0 | 1 (11) | 1 (20) | 10 (77) | 1 (13) | ||||
| WBC | |||||||||||||
| Count, 103/mm3 | 11.1 (7.6–15.4) | 9.6 (8.3–10.43) | 0.266 | 12.5 (9.1–13.8) | 10.8 (8.1–15.7) | 0.951 | 10.1 (8.8–12.4) | 8.9 (6.6–10.4) | 0.25 | 8.7 (6.7–13.6) | 9 (7.0–12.1) | 7.9 (6.6–9.4) | 0.520 |
| Neutrophils, % | 29 (25–43) | 26 (21–30) | 0.559 | 32 (17–44) | 22 (21–26) | 0.140 | 46 (24–69) | N/A | N/A | 28 (22–63) | 30 (20–37) | 18 (14–20) | 0.033 |
| Lymphocytes, % | 53 (36–60) | 66 (63–69) | 0.089 | 51 (39–62) | 70 (64–73) | 0.012 | 34 (15–52) | N/A | N/A | 59 (33–69) | 57* (44–69) | 72* (70–75) | 0.026 |
| Monocytes, % | 9 (7–21) | 6 (3–9) | 0.368 | 9 (5–13) | 5 (2–8) | 0.099 | 12 (10–14) | N/A | N/A | 7 (3–11) | 8 (7–11) | 7 (5–9) | 0.629 |
| CDSS | 46.7 (26.7–65.0) | N/A | N/A | 53.3 (26.7–62.5) | N/A | 0.563† | 33.3 (33.3–50.0) | N/A | 0.399† | 40.0 (26.7–56.7) | 6.7 (0–10.0) | N/A | <0.001 |
| LOS, d | 2.4 (0.9–3.9) | N/A | N/A | 1.6 (0.9–3.5) | N/A | 0.763† | N/A | N/A | N/A | 1.3 (0.6–3.3) | N/A | N/A | N/A |
Definition of abbreviations: CDSS = clinical disease severity score scaled from 0 to 100; LOS = length of stay; N/A = not applicable; WBC = white blood cells.
Data are reported as median (interquartile range) unless otherwise indicated. Three healthy control subjects were used in both the test set and the validation set A. WBC and complete differential were available in 119 (87%) and 63 (46%) of 137 of children, respectively. Categorical data were analyzed using a χ2 test. For continuous variables, we used the Mann-Whitney U test or the Kruskal-Wallis test followed by Dunn’s post hoc test to adjust for multiple comparisons.
Groups that were significantly different in the adjusted Kruskal-Wallis test.
Comparison between inpatients in the training and test sets.
Figure 2.
Symptomatic rhinovirus (RV) infection induces a robust and reproducible transcriptional signature. (A) Training set (n = 44). Class comparisons (Benjamini-Hochberg corrected false discovery rate <0.01 and ≥1.25-fold change) using linear models adjusted for age, race, and white blood cell composition (neutrophil score) between RV− healthy control subjects (HC) and RV+ inpatients (IP) identified 393 differentially expressed transcripts (transcriptional RV signature) (Table E4). Transcripts are organized in a heat map format in which each row represents a single transcript and each column represents a patient sample. Red indicates overexpression, and blue underexpression, of a transcript compared with the median expression of HC (yellow). The transcriptional signature was validated in three independent patient sets using the support vector machine (SVM) algorithm. The SVM algorithm predicted the condition (symptomatic RV infection or RV− HC) in (B) the test set (n = 44) with 98% accuracy and in (C) validation set A (n = 26) and (D) validation set B (n = 26) with 92% accuracy. Color bars below the heat maps indicate the true sample class below (darker colors) and above (lighter colors) the predicted SVM class. OP = outpatient.
This transcriptional signature was validated in three independent patient groups using the SVM learning algorithm: a test set with 33 RV+ inpatients and 11 healthy control subjects; a validation set A with 26 subjects comprising 17 RV+ outpatients and 9 healthy control subjects; and a validation set B (n = 26) comprising 5 RV+ inpatients, 13 RV+ outpatients, and 8 healthy control subjects. Samples in validation set B underwent quality control and normalization procedures separate from others in the whole cohort and were not included in the batch correction procedure, representing a separate external validation cohort. SVM predicted the sample’s condition in the test set with 98% accuracy and with 92% accuracy in both validation sets A and B (Figures 2B–2D).
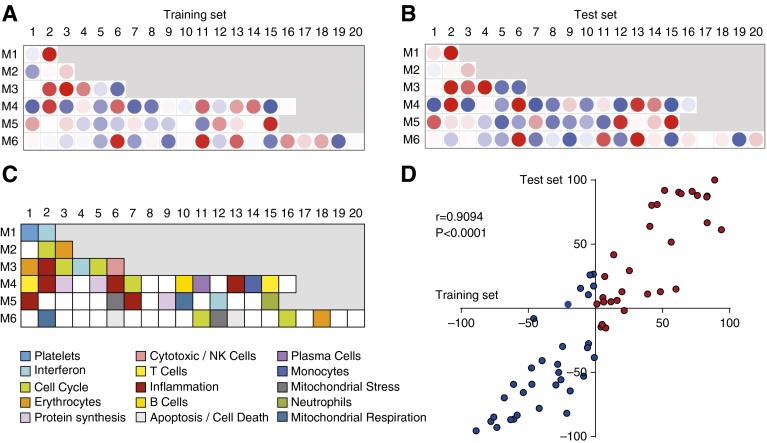
Next, to characterize the biological significance of the RV signature and the function of the differentially expressed genes, we applied a modular approach (27, 28) that was derived independently for the training and test sets (Figures 3A–3C). Inpatients with RV+ infection demonstrated overexpression of innate immunity–associated modules (IFN, inflammation, neutrophils, and monocytes) and underexpression of T-cell–, cytotoxic/natural killer (NK)-cell–, and B-cell–related modules. These findings were validated in the test set, as demonstrated by a significant correlation between both cohorts (Spearman’s r = 0.91; P < 0.0001) (Figure 3D). Thus, this initial analysis identified and validated the RV transcriptional signature at the gene and modular levels in independent sets of patients.
Figure 3.
Modular fingerprints of rhinovirus-induced acute respiratory infection. To characterize biological functions of the differentially expressed genes, we used modular analysis. The analysis was performed separately for (A) the training set and (B) the test set. The color intensity of the modules (dots) indicates the proportion of overexpressed (red) or underexpressed (blue) transcripts within each module. (C) Key to the functional annotations of modular sets 1–6. (D) Scatterplot representing the modular correlation between the training set (x-axis) and the test set (y-axis). The axes indicate the percentage of differentially expressed genes in each module. Correlations were assessed using Spearman’s correlation coefficient. NK = natural killer.
The RV Signature Discriminates between RV-induced Respiratory Tract Infection and Asymptomatic RV Detection
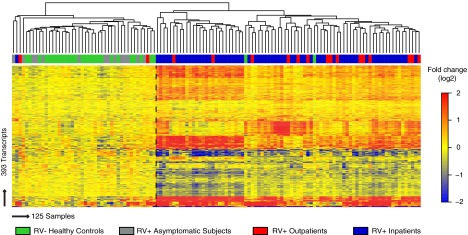
To determine the accuracy of the 393-transcript RV signature in grouping patients with similar clinical conditions, we combined samples from the training set (n = 44), the test set (n = 44), and validation set A (n = 26), together with 14 RV+ asymptomatic subjects (all 125 samples that were batch corrected). We then applied unsupervised hierarchical clustering with euclidean distance and average linkage (Figure 4) and calculated silhouette indices to determine the optimal number and the strength of the clusters (Figure E3). These 125 samples were divided into two main clusters. The first cluster comprised 93% of healthy control subjects (27 of 29), as well as all RV+ asymptomatic subjects (14 [100%] of 14) and 3 symptomatic subjects, 1 RV+ inpatient and 2 RV+ outpatients, all three with low CDSSs. The second cluster included 96% of children with symptomatic RV+ respiratory tract infections, including RV+ outpatients (15 [88.2%] of 17) and RV+ inpatients (64 [98.5%] of 65), as well as 2 healthy control subjects. Overall, these results indicate that the transcriptional profile of RV+ asymptomatic subjects closely resembles that of healthy control subjects and that this profile is significantly different from that of patients with RV+ symptomatic infection.
Figure 4.
Transcriptional signatures accurately discriminate between symptomatic and asymptomatic rhinovirus (RV) detection. Unsupervised hierarchical clustering (euclidean distance, average linkage) of samples included in the training set (n = 44), the test set (n = 44), validation set A (n = 26), and an additional cohort of 14 RV+ asymptomatic children (total N = 125) grouped the samples in two main clusters (separated by dashed vertical line). The first cluster included the majority of RV− healthy control subjects (27 [93%] of 29) and all RV+ asymptomatic subjects (14 [100%] of 14), and the second cluster comprised the majority of RV+ inpatients (64 [98%] of 65) and RV+ outpatients (15 [88%] of 17). The optimal number and the stability of the clusters were determined by calculating average silhouette indices (Figure E3).
Asymptomatic Children with RV Detection Lack Activation of the Systemic Immune Response
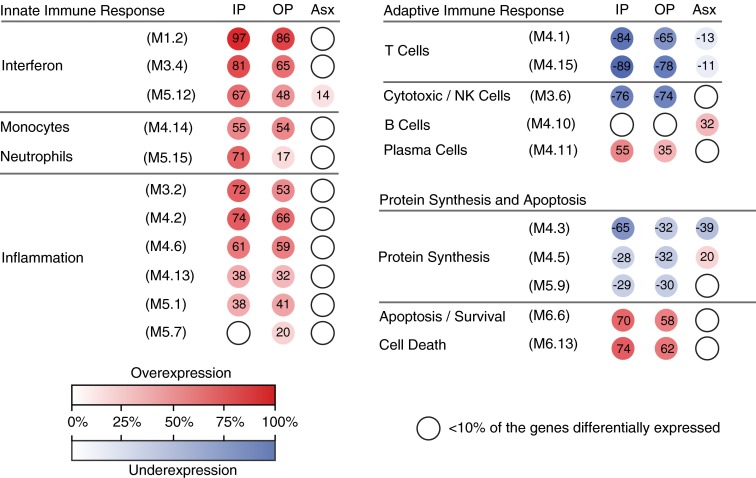
To determine whether the systemic host immune response to RV was influenced by the presence and severity of clinical symptoms, we compared all three RV+ clinical groups (inpatients, outpatients, and asymptomatic children) using modular analysis. We randomly selected children from each clinical group to create three independent sets of comparable size composed of RV+ children (RV+ inpatient set [n = 28], RV+ outpatient set [n = 29], and RV+ asymptomatic set [n = 27]) who were matched for age and sex with RV− healthy control subjects used as a reference group (Table E5). Children with RV+ respiratory tract infections (inpatients and outpatients) demonstrated overexpression of IFN, neutrophils, monocytes, inflammation, and plasma cell modules, whereas modules associated with T cells and cytotoxic/NK cells were underexpressed (Figure 5). The expression of IFN-, neutrophil-, T-cell–, plasma cell–, and apoptosis-related modules was stronger among inpatients (thus with a more severe disease phenotype) than in outpatients with milder symptoms. In contrast, asymptomatic children with RV detection exhibited mild overexpression of B-cell–related genes with mostly absent or low-level expression in the remaining modules. The findings derived from modular analyses were confirmed using Ingenuity Pathway Analysis, which showed marked differences in the IFN pathway between the RV+ symptomatic groups and RV+ asymptomatic children (Figure E5).
Figure 5.
Differences in modular expression according to disease severity. Comparison of modular expression between the clinical groups showed significant overexpression of innate immunity–associated modules and underexpression of adaptive immunity–associated modules in the rhinovirus-positive (RV+) inpatients (IP) and outpatients (OP), whereas only minor changes were detected in RV+ asymptomatic subjects (Asx). Each group was compared with matched healthy control subjects (modular inpatient set, modular outpatient set, and modular asymptomatic set) (Table E5). The color intensity of the modules (colored circles) indicates the proportion of overexpressed (red) or underexpressed (blue) transcripts within each module. Numeric values indicate the exact percentage of transcripts expressed in each specific module. Open circles indicate that less than 10% of the genes in the module were differentially expressed. NK = natural killer.
Molecular Distance to Health Scores Categorize RV+ Children according to Disease Severity
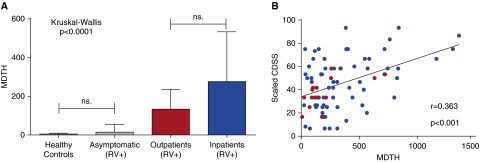
Last, we calculated the MDTH score, which measures the magnitude of transcriptional perturbation of each individual patient compared with the healthy control baseline values (20, 29). MDTH scores in RV+ asymptomatic children were comparable to those of healthy control subjects and significantly lower than those of both RV+ inpatients and RV+ outpatients (P < 0.0001) (Figure 6A). Although MDTH scores in RV+ inpatients (severe disease) were higher than in RV+ outpatients (mild disease), the difference did not reach statistical significance. Nevertheless, in RV+ symptomatic children (inpatients and outpatients combined [n = 82]), MDTH scores were significantly correlated with CDSS (r = 0.36; P < 0.001) (Figure 6B).
Figure 6.
Molecular distance to health (MDTH) score correlates with disease severity. (A) MDTH score discriminates between the human rhinovirus–positive (RV+) asymptomatic subjects and RV+ symptomatic subjects (inpatients and outpatients) (P < 0.0001 by Kruskal-Wallis test with Dunn’s test for multiple comparisons). (B) In the symptomatic subjects (inpatients and outpatients; n = 82), MDTH score correlated with the clinical disease severity score (CDSS) (Spearman’s correlation coefficient). Outpatients are represented by red dots and inpatients by blue dots. ns = nonsignificant.
Discussion
We have previously shown that host transcriptional profiles can discriminate between infants with severe respiratory syncytial virus, influenza A, or RV infection with great sensitivity and specificity (20, 31). In this study, we went one step further and applied transcriptional profiling to define and compare the systemic host response elicited by RV in different clinical scenarios. We found that symptomatic RV infection, after adjustment for age, race, and blood cell composition, induced a robust transcriptional signature, a finding that was validated in three additional patient cohorts of diverse genetic backgrounds. By contrast, transcriptional profiles of asymptomatic children in whom RV was incidentally detected mostly resembled the profiles of RV− healthy control children. These initial findings were confirmed by qRT-PCR and validated by applying a modular approach that clearly differentiated symptomatic RV infection from RV detection in asymptomatic children. The MDTH genomic score further confirmed the differences among the groups. To our knowledge, this is the first study in which transcriptional profiling has been applied to provide a comprehensive analysis of the systemic host response to RV in children representative of the whole disease spectrum.
The introduction of sensitive molecular assays has revolutionized the understanding of the etiology of respiratory viral infections. This has been particularly relevant for RV, whose identification in the past exclusively relied on viral culture (32, 33). With the use of these diagnostic tools, respiratory viruses are also frequently detected in asymptomatic subjects. However, the significance of RV detection in asymptomatic children and whether RV induces a detectable systemic host immune response remains unclear. In children, RVs are the most commonly detected respiratory viruses during symptomatic respiratory infections, including asthma exacerbations (34, 35), but they are also the most common respiratory viruses identified in the absence of symptoms (8–13). The rates of RV detection during symptomatic infection vary by condition. On one hand, RVs have been detected in 47–71% of children with the common cold (1, 36), in 26–76% of children with bronchiolitis or acute wheezing (4, 37), and in 64% of children with acute otitis media (38). On the other hand, studies have shown rates of RV detection in asymptomatic children varying from 14% to 35% (5, 8–13). Even higher asymptomatic RV detection rates were recently reported in a study conducted in the context of a pneumococcal vaccine trial where RVs were detected in 31–50% of asymptomatic children (39). Thus, high detection rates in asymptomatic subjects, together with common codetection with other respiratory viruses and the reported lack of clear correlation between RV loads and disease severity (40), have raised questions about the causality of RV detection in clinical illness.
Whole-blood transcriptional profiling can accurately discriminate between viral and bacterial infections as well as between infections caused by different viruses, both in children and in adults (14–20). Studies in which researchers have used these tools to better understand the role of asymptomatic viral detection are limited. Hu and colleagues analyzed a cohort of children with febrile viral and bacterial infections and afebrile control subjects. Of the 35 afebrile control subjects, 13 were positive for different viruses (RV, adenovirus, human herpesvirus 6). They found that afebrile, virus-positive, and virus-negative control subjects had indistinguishable gene expression profiles that were significantly different from those of febrile children (18). A recent study in which healthy adults were experimentally challenged with influenza virus showed that gene expression profiles of subjects who developed symptomatic illness significantly differed from the profiles of those who remained asymptomatic (41). Our study was focused specifically on children naturally exposed to RV. Using a large patient cohort, we analyzed the significance of RV detection by including groups of patients with different levels of clinical severity. We validated the results in independent patient populations of different genetic and/or ethnic backgrounds with great accuracy and also by qRT-PCR, thus providing a comprehensive analysis of transcriptome profiles in the context of RV identification.
Modular analyses showed that the RV transcriptional signature was characterized by overexpression of innate immunity, including IFN-, inflammation-, neutrophil-, and monocyte-related genes. Specifically, IFN responses including IFI27 (an IFN type I–related gene and the most overexpressed transcript) were strongly activated. The activation was more pronounced in RV+ inpatients, which corresponded to children with severe disease. This is plausible, as IFNs are known to play an important role in orchestrating antiviral host responses and activation of IFN genes has been described in vitro, in animal models, and in humans in response to different respiratory viral infections (15, 20, 31, 42). In contrast, we found marked underexpression of T-cell– and cytotoxic/NK-cell–related genes. Again, this suppression was more profound in RV+ inpatients than in RV+ outpatients. A study in adult volunteers showed a reduction in peripheral blood T-cell counts after challenge with RV. This decrease in T-cell numbers also correlated with increased disease severity (43). In previous studies in children, investigators have also found decreased expression of adaptive immunity genes in the blood during severe respiratory syncytial virus, influenza, or RV lower respiratory tract infection (20). Whether downregulation of T-cell–associated genes reflects a failure to mount an adequate host response that leads to a more severe illness, or whether it represents a well-controlled early step in the host response that balances the excessive inflammatory responses during viral infections, remains unclear.
There are different hypotheses to explain the mechanisms associated with viral detection in asymptomatic subjects. It could represent an active asymptomatic infection or the prodromal phase of an acute symptomatic infection, or it may indicate a past infection in which the PCR identifies remnants of viral RNA (13). While chronic RV infections have been described in subjects with hypogammaglobulinemia, cystic fibrosis, and lung transplantation, immunocompetent children usually clear RV from the respiratory tract within 2–4 weeks of the acute infection. Thus, long-term viral persistence would be unusual in otherwise healthy children (44, 45). In the present study, except for mild overexpression of B-cell–related genes, the host systemic immune response of children with no symptoms in whom RV was incidentally detected was similar to that of RV− asymptomatic healthy control subjects, suggesting that in most cases asymptomatic RV detection in children represents the presence of viral RNA in the respiratory tract mucosa without robust host responses. The observed low-level overexpression of B-cell–related genes in asymptomatic children might reflect the development of immunologic memory associated with either a past symptomatic RV infection or an ongoing asymptomatic RV infection.
Tools for use in discriminating between incidental pathogen detection and true infection are lacking. WBC counts and CRP levels are of limited value, and the significance of RV loads as a surrogate marker of disease severity is still inconclusive (10, 46–48). Our study introduces a proof of concept: The combination of molecular microbiological data and systemic host responses could help in assessment of the significance of pathogen detection and the causality of clinical illness. As this methodology is implemented in the future, it could help in situations where incidental pathogen detection could mislead clinical decision making. The clinical scenarios where differentiating between asymptomatic RV detection from active RV infection might be useful include situations in which scheduled surgical interventions (e.g., cardiac surgery) would need to be either postponed or performed as planned based on the significance of RV detection. In febrile children or in patients with pneumonia, the presence or absence of an RV biosignature could provide useful information for clinical decision making regarding the need for additional testing and/or antibiotic use. Last, if these findings are confirmed in high-risk groups (e.g., patients with cystic fibrosis, those with lung transplants, and other immunocompromised patients), the clinical utility of biosignatures could be further expanded in monitoring the activity of chronic infections, timing of interventions, and even assessing response to treatment (49).
Our study has limitations. To include a representative cohort of patients, we combined data from different datasets that were hybridized in different microarray platforms. This created a challenge for the analysis because of the technical variability between datasets (batch effect). However, we adjusted for this potential batch effect and validated the results in three different patient populations and also by qRT-PCR with high accuracy. Clinical samples were collected at a single time point, and therefore our observations provide only a snapshot of a dynamic process. Longitudinal studies with sequential samples would also have allowed us to characterize the temporal changes affecting the transcriptional profiles over time, and those studies will be important in the future. Although patients with confirmed bacterial coinfections were excluded from the study, systematic blood or urine cultures were not available for all patients, as those tests were not routinely performed in the participating institutions according to the standard of care. Nevertheless, 83% of patients with symptomatic RV infection underwent blood or urine culture testing with negative results and/or had serum CRP concentrations measured with median levels less than 1.5 mg/dl, suggesting a low likelihood of bacterial infections. Last, we characterized the host response to RV by analyzing the transcriptional changes in the systemic compartment. We did not measure the host–pathogen interaction in the respiratory epithelium, the primary site of infection, which prevents us from making conclusions about the presence or absence of mucosal immune responses in asymptomatic RV+ children. Nonetheless, blood transcriptional profiling provides a valuable tool to better understand the complex processes that take place during the infection by using clinical samples that are easy to obtain in a standardized manner across different clinical sites, as shown in previous studies (14–20).
In summary, we found that symptomatic RV infection induced a robust and reproducible host response characterized by overexpression of innate immunity and underexpression of adaptive immunity–associated genes. However, no significant changes were detected in the transcriptional profiles of children with asymptomatic RV detection. These data suggest that, in otherwise healthy children, asymptomatic RV detection likely represents detection of viral RNA without significant activation of systemic host responses. Our study suggests that whole-blood transcriptional profiling may become a useful diagnostic tool to discriminate between incidental pathogen detection and active infection.
Acknowledgments
Acknowledgment
The authors thank Evelyn Torres, R.N., Juanita Lozano, M.D., and Maria Rocha, R.N., at Children’s Medical Center (Dallas, TX); and Gail Arthur, R.N., Michael Lawson, R.N., Paula Davies, and Grace Wentzel at Nationwide Children’s Hospital (Columbus, OH) for their extraordinary efforts in enrolling patients and control subjects. The authors also thank Bennett Smith at the Research Institute at Nationwide Children’s Hospital for technical assistance; Phuong Nguyen, Jeanine Baisch, and Nicole Baldwin of the microarray core facility at the Baylor Institute for Immunology Research (Dallas, TX) for their help with RNA processing and hybridization; and especially our patients and their families for agreeing to participate in the study.
Footnotes
This work was supported in part by National Institute of Allergy and Infectious Diseases grant U19AI089987 (O. Ramilo and A.M.); Nationwide Children’s Hospital intramural funds (A.M.); Academy of Finland grants 132595 and 114034 and Sigrid Juselius Foundation grants (T.J.); and nonfederal grants from the European Society for Paediatric Infectious Diseases (ESPID Fellowship Award), the Finnish Medical Foundation, the Foundation for Pediatric Research, and the Maud Kuistila Memorial Foundation (S.H.). The sponsors had no role in the study design, data collection, data analysis, data interpretation, decision to publish, or preparation of the manuscript.
Author Contributions: S.H., O. Ramilo, and A.M. conceived and designed the study; T.J., S.O., C.G., and O. Ruuskanen acquired the clinical data; T.V. performed part of the virological analyses; S.H., B.D., N.M.S., and W.A.A.d.S.P. analyzed the gene expression data; C.S., V.P., and E.A. performed quantitative real-time polymerase chain reaction validation of the gene expression data; S.H. and A.M. interpreted the results and wrote the first and final drafts of the manuscript; and all authors were involved in critical revision of the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org
Originally Published in Press as 10.1164/rccm.201504-0749OC on November 16, 2015
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Ruohola A, Waris M, Allander T, Ziegler T, Heikkinen T, Ruuskanen O. Viral etiology of common cold in children, Finland. Emerg Infect Dis. 2009;15:344–346. doi: 10.3201/eid1502.081468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruuskanen O, Lahti E, Jennings LC, Murdoch DR. Viral pneumonia. Lancet. 2011;377:1264–1275. doi: 10.1016/S0140-6736(10)61459-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwane MK, Prill MM, Lu X, Miller EK, Edwards KM, Hall CB, Griffin MR, Staat MA, Anderson LJ, Williams JV, et al. Human rhinovirus species associated with hospitalizations for acute respiratory illness in young US children. J Infect Dis. 2011;204:1702–1710. doi: 10.1093/infdis/jir634. [DOI] [PubMed] [Google Scholar]
- 4.Mansbach JM, Piedra PA, Teach SJ, Sullivan AF, Forgey T, Clark S, Espinola JA, Camargo CA, Jr MARC-30 Investigators. Prospective multicenter study of viral etiology and hospital length of stay in children with severe bronchiolitis. Arch Pediatr Adolesc Med. 2012;166:700–706. doi: 10.1001/archpediatrics.2011.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rakes GP, Arruda E, Ingram JM, Hoover GE, Zambrano JC, Hayden FG, Platts-Mills TA, Heymann PW. Rhinovirus and respiratory syncytial virus in wheezing children requiring emergency care: IgE and eosinophil analyses. Am J Respir Crit Care Med. 1999;159:785–790. doi: 10.1164/ajrccm.159.3.9801052. [DOI] [PubMed] [Google Scholar]
- 6.Calışkan M, Bochkov YA, Kreiner-Møller E, Bønnelykke K, Stein MM, Du G, Bisgaard H, Jackson DJ, Gern JE, Lemanske RF, Jr, et al. Rhinovirus wheezing illness and genetic risk of childhood-onset asthma. N Engl J Med. 2013;368:1398–1407. doi: 10.1056/NEJMoa1211592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson DJ, Gangnon RE, Evans MD, Roberg KA, Anderson EL, Pappas TE, Printz MC, Lee WM, Shult PA, Reisdorf E, et al. Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am J Respir Crit Care Med. 2008;178:667–672. doi: 10.1164/rccm.200802-309OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Benten I, Koopman L, Niesters B, Hop W, van Middelkoop B, de Waal L, van Drunen K, Osterhaus A, Neijens H, Fokkens W. Predominance of rhinovirus in the nose of symptomatic and asymptomatic infants. Pediatr Allergy Immunol. 2003;14:363–370. doi: 10.1034/j.1399-3038.2003.00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van der Zalm MM, van Ewijk BE, Wilbrink B, Uiterwaal CS, Wolfs TFW, van der Ent CK. Respiratory pathogens in children with and without respiratory symptoms. J Pediatr. 2009;154:396–400.e1. doi: 10.1016/j.jpeds.2008.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jansen RR, Wieringa J, Koekkoek SM, Visser CE, Pajkrt D, Molenkamp R, de Jong MD, Schinkel J. Frequent detection of respiratory viruses without symptoms: toward defining clinically relevant cutoff values. J Clin Microbiol. 2011;49:2631–2636. doi: 10.1128/JCM.02094-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Advani S, Sengupta A, Forman M, Valsamakis A, Milstone AM. Detecting respiratory viruses in asymptomatic children. Pediatr Infect Dis J. 2012;31:1221–1226. doi: 10.1097/INF.0b013e318265a804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rhedin S, Lindstrand A, Rotzén-Östlund M, Tolfvenstam T, Ohrmalm L, Rinder MR, Zweygberg-Wirgart B, Ortqvist A, Henriques-Normark B, Broliden K, et al. Clinical utility of PCR for common viruses in acute respiratory illness. Pediatrics. 2014;133:e538–e545. doi: 10.1542/peds.2013-3042. [DOI] [PubMed] [Google Scholar]
- 13.Jartti T, Jartti L, Peltola V, Waris M, Ruuskanen O. Identification of respiratory viruses in asymptomatic subjects: asymptomatic respiratory viral infections. Pediatr Infect Dis J. 2008;27:1103–1107. doi: 10.1097/INF.0b013e31817e695d. [DOI] [PubMed] [Google Scholar]
- 14.Ramilo O, Allman W, Chung W, Mejias A, Ardura M, Glaser C, Wittkowski KM, Piqueras B, Banchereau J, Palucka AK, et al. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 2007;109:2066–2077. doi: 10.1182/blood-2006-02-002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zaas AK, Chen M, Varkey J, Veldman T, Hero AO, III, Lucas J, Huang Y, Turner R, Gilbert A, Lambkin-Williams R, et al. Gene expression signatures diagnose influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe. 2009;6:207–217. doi: 10.1016/j.chom.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berry MPR, Graham CM, McNab FW, Xu Z, Bloch SAA, Oni T, Wilkinson KA, Banchereau R, Skinner J, Wilkinson RJ, et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466:973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zaas AK, Burke T, Chen M, McClain M, Nicholson B, Veldman T, Tsalik EL, Fowler V, Rivers EP, Otero R, et al. A host-based RT-PCR gene expression signature to identify acute respiratory viral infection. Sci Transl Med. 2013;5:203ra126. doi: 10.1126/scitranslmed.3006280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu X, Yu J, Crosby SD, Storch GA. Gene expression profiles in febrile children with defined viral and bacterial infection. Proc Natl Acad Sci USA. 2013;110:12792–12797. doi: 10.1073/pnas.1302968110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anderson ST, Kaforou M, Brent AJ, Wright VJ, Banwell CM, Chagaluka G, Crampin AC, Dockrell HM, French N, Hamilton MS, et al. ILULU Consortium; KIDS TB Study Group. Diagnosis of childhood tuberculosis and host RNA expression in Africa. N Engl J Med. 2014;370:1712–1723. doi: 10.1056/NEJMoa1303657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mejias A, Dimo B, Suarez NM, Garcia C, Suarez-Arrabal MC, Jartti T, Blankenship D, Jordan-Villegas A, Ardura MI, Xu Z, et al. Whole blood gene expression profiles to assess pathogenesis and disease severity in infants with respiratory syncytial virus infection. PLoS Med. 2013;10:e1001549. doi: 10.1371/journal.pmed.1001549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suarez NM, Bunsow E, Falsey AR, Walsh EE, Mejias A, Ramilo O. Superiority of transcriptional profiling over procalcitonin for distinguishing bacterial from viral lower respiratory tract infections in hospitalized adults. J Infect Dis. 2015;212:213–222. doi: 10.1093/infdis/jiv047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhai Y, Franco LM, Atmar RL, Quarles JM, Arden N, Bucasas KL, Wells JM, Niño D, Wang X, Zapata GE, et al. Host transcriptional response to influenza and other acute respiratory viral infections – a prospective cohort study. PLoS Pathog. 2015;11:e1004869. doi: 10.1371/journal.ppat.1004869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heinonen S, Suarez NM, Jartti T, Oliva S, Garcia C, Ramilo O, Mejias A.Gene expression profiles discriminate between young children with human rhinovirus (HRV) symptomatic infection vs asymptomatic detection. Open Forum Infect Dis 2014;1(Suppl 1):S353 [Google Scholar]
- 24.Heinonen S, Suarez NM, Jartti T, Oliva S, Garcia C, Ramilo O, Mejias A.The molecular distance to health (MDTH) genomic score classifies infants with human rhinovirus infections according to disease severity. Presented at the 32nd annual meeting of the European Society for Paediatric Infectious Diseases (ESPID 2014). May 6–10, 2014, Dublin, Ireland. Abstract 718 [Google Scholar]
- 25.Jartti T, Lehtinen P, Vanto T, Hartiala J, Vuorinen T, Mäkelä MJ, Ruuskanen O. Evaluation of the efficacy of prednisolone in early wheezing induced by rhinovirus or respiratory syncytial virus. Pediatr Infect Dis J. 2006;25:482–488. doi: 10.1097/01.inf.0000215226.69696.0c. [DOI] [PubMed] [Google Scholar]
- 26.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 27.Chaussabel D, Quinn C, Shen J, Patel P, Glaser C, Baldwin N, Stichweh D, Blankenship D, Li L, Munagala I, et al. A modular analysis framework for blood genomics studies: application to systemic lupus erythematosus. Immunity. 2008;29:150–164. doi: 10.1016/j.immuni.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chaussabel D, Baldwin N. Democratizing systems immunology with modular transcriptional repertoire analyses. Nat Rev Immunol. 2014;14:271–280. doi: 10.1038/nri3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pankla R, Buddhisa S, Berry M, Blankenship DM, Bancroft GJ, Banchereau J, Lertmemongkolchai G, Chaussabel D. Genomic transcriptional profiling identifies a candidate blood biomarker signature for the diagnosis of septicemic melioidosis. Genome Biol. 2009;10:R127. doi: 10.1186/gb-2009-10-11-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ioannidis I, McNally B, Willette M, Peeples ME, Chaussabel D, Durbin JE, Ramilo O, Mejias A, Flaño E. Plasticity and virus specificity of the airway epithelial cell immune response during respiratory virus infection. J Virol. 2012;86:5422–5436. doi: 10.1128/JVI.06757-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jartti T, Söderlund-Venermo M, Hedman K, Ruuskanen O, Mäkelä MJ. New molecular virus detection methods and their clinical value in lower respiratory tract infections in children. Paediatr Respir Rev. 2013;14:38–45. doi: 10.1016/j.prrv.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arens MQ, Buller RS, Rankin A, Mason S, Whetsell A, Agapov E, Lee WM, Storch GA. Comparison of the Eragen Multi-Code Respiratory Virus Panel with conventional viral testing and real-time multiplex PCR assays for detection of respiratory viruses. J Clin Microbiol. 2010;48:2387–2395. doi: 10.1128/JCM.00220-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee WM, Lemanske RF, Jr, Evans MD, Vang F, Pappas T, Gangnon R, Jackson DJ, Gern JE. Human rhinovirus species and season of infection determine illness severity. Am J Respir Crit Care Med. 2012;186:886–891. doi: 10.1164/rccm.201202-0330OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller EK, Gebretsadik T, Carroll KN, Dupont WD, Mohamed YA, Morin LL, Heil L, Minton PA, Woodward K, Liu Z, et al. Viral etiologies of infant bronchiolitis, croup and upper respiratory illness during 4 consecutive years. Pediatr Infect Dis J. 2013;32:950–955. doi: 10.1097/INF.0b013e31829b7e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miller EK, Williams JV, Gebretsadik T, Carroll KN, Dupont WD, Mohamed YA, Morin LL, Heil L, Minton PA, Woodward K, et al. Host and viral factors associated with severity of human rhinovirus-associated infant respiratory tract illness. J Allergy Clin Immunol. 2011;127:883–891. doi: 10.1016/j.jaci.2010.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh AM, Moore PE, Gern JE, Lemanske RF, Jr, Hartert TV. Bronchiolitis to asthma: a review and call for studies of gene–virus interactions in asthma causation. Am J Respir Crit Care Med. 2007;175:108–119. doi: 10.1164/rccm.200603-435PP. [DOI] [PubMed] [Google Scholar]
- 38.Ruohola A, Pettigrew MM, Lindholm L, Jalava J, Räisänen KS, Vainionpää R, Waris M, Tähtinen PA, Laine MK, Lahti E, et al. Bacterial and viral interactions within the nasopharynx contribute to the risk of acute otitis media. J Infect. 2013;66:247–254. doi: 10.1016/j.jinf.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van den Bergh MR, Biesbroek G, Rossen JWA, de Steenhuijsen Piters WAA, Bosch AATM, van Gils EJM, Wang X, Boonacker CWB, Veenhoven RH, Bruin JP, et al. Associations between pathogens in the upper respiratory tract of young children: interplay between viruses and bacteria. PLoS One. 2012;7:e47711. doi: 10.1371/journal.pone.0047711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kennedy JL, Shaker M, McMeen V, Gern J, Carper H, Murphy D, Lee WM, Bochkov YA, Vrtis RF, Platts-Mills T, et al. Comparison of viral load in individuals with and without asthma during infections with rhinovirus. Am J Respir Crit Care Med. 2014;189:532–539. doi: 10.1164/rccm.201310-1767OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang Y, Zaas AK, Rao A, Dobigeon N, Woolf PJ, Veldman T, Øien NC, McClain MT, Varkey JB, Nicholson B, et al. Temporal dynamics of host molecular responses differentiate symptomatic and asymptomatic influenza A infection. PLoS Genet. 2011;7:e1002234. doi: 10.1371/journal.pgen.1002234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mallia P, Message SD, Gielen V, Contoli M, Gray K, Kebadze T, Aniscenko J, Laza-Stanca V, Edwards MR, Slater L, et al. Experimental rhinovirus infection as a human model of chronic obstructive pulmonary disease exacerbation. Am J Respir Crit Care Med. 2011;183:734–742. doi: 10.1164/rccm.201006-0833OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Levandowski RA, Ou DW, Jackson GG. Acute-phase decrease of T lymphocyte subsets in rhinovirus infection. J Infect Dis. 1986;153:743–748. doi: 10.1093/infdis/153.4.743. [DOI] [PubMed] [Google Scholar]
- 44.Peltola V, Waris M, Kainulainen L, Kero J, Ruuskanen O. Virus shedding after human rhinovirus infection in children, adults and patients with hypogammaglobulinaemia. Clin Microbiol Infect. 2013;19:E322–E327. doi: 10.1111/1469-0691.12193. [DOI] [PubMed] [Google Scholar]
- 45.Loeffelholz MJ, Trujillo R, Pyles RB, Miller AL, Alvarez-Fernandez P, Pong DL, Chonmaitree T. Duration of rhinovirus shedding in the upper respiratory tract in the first year of life. Pediatrics. 2014;134:1144–1150. doi: 10.1542/peds.2014-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chonmaitree T, Alvarez-Fernandez P, Jennings K, Trujillo R, Marom T, Loeffelholz MJ, Miller AL, McCormick DP, Patel JA, Pyles RB. Symptomatic and asymptomatic respiratory viral infections in the first year of life: association with acute otitis media development. Clin Infect Dis. 2015;60:1–9. doi: 10.1093/cid/ciu714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peltola V, Waris M, Osterback R, Susi P, Ruuskanen O, Hyypiä T. Rhinovirus transmission within families with children: incidence of symptomatic and asymptomatic infections. J Infect Dis. 2008;197:382–389. doi: 10.1086/525542. [DOI] [PubMed] [Google Scholar]
- 48.Jartti T, Hasegawa K, Mansbach JM, Piedra PA, Camargo CA., Jr Rhinovirus-induced bronchiolitis: lack of association between virus genomic load and short-term outcomes. J Allergy Clin Immunol. 2015;136:509–512.e11. doi: 10.1016/j.jaci.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruuskanen O, Waris M, Kainulainen L. Treatment of persistent rhinovirus infection with pegylated interferon α2a and ribavirin in patients with hypogammaglobulinemia. Clin Infect Dis. 2014;58:1784–1786. doi: 10.1093/cid/ciu169. [DOI] [PMC free article] [PubMed] [Google Scholar]