Abstract
Introduction
Rho GTPases are master regulators of actomyosin structure and dynamics and play pivotal roles in a variety of cellular processes including cell morphology, gene transcription, cell cycle progression and cell adhesion. Because aberrant Rho GTPase signaling activities are widely associated with human cancer, key components of Rho GTPase signaling pathways have attracted increasing interest as potential therapeutic targets. Similar to Ras, Rho GTPases themselves were, until recently, deemed “undruggable” because of structure-function considerations. Several approaches to interfere with Rho GTPase signaling have been explored and show promise as new ways for tackling cancer cells.
Areas covered
This review focuses on the recent progress in targeting the signaling activities of three prototypical Rho GTPases, i.e. RhoA, Rac1, and Cdc42. The authors describe the involvement of these Rho GTPases, their key regulators and effectors in cancer. Furthermore, the authors discuss the current approaches for rationally targeting aberrant Rho GTPases along their signaling cascades, upstream and downstream of Rho GTPases and posttranslational modifications at a molecular level.
Expert opinion
To date, while no clinically effective drugs targeting Rho GTPase signaling for cancer treatment are available, tool compounds and lead drugs that pharmacologically inhibit Rho GTPase pathways have shown promise. Small molecule inhibitors targeting Rho GTPase signaling may add new treatment options for future precision cancer therapy, particularly in combination with other anti-cancer agents.
Keywords: Cancer, Cdc42, Rac1, RhoA, RhoGEF, Rho GTPases, Small molecule inhibitor, Targeted therapy
1. Introduction
Rho GTPases are members of the Ras GTPase superfamily [1]. Since the first discovery in 1980s as Ras homologous small GTPases, over twenty mammalian Rho family members have been identified that can be divided into several subgroups [2]. Among Rho GTPases, RhoA, Rac1, and Cdc42 are the best characterized members, and have well-characterized roles in regulating actin cytoskeleton organization and dynamics [3]. RhoA, Rac1 and Cdc42 were first described to promote the formation of stress fibers, lamellipodia and filopodia in fibroblast respectively [4], and were subsequently appreciated for their roles in regulating signaling pathways affecting cell polarity, gene expression, cell-cycle progression, and cell survival [5].
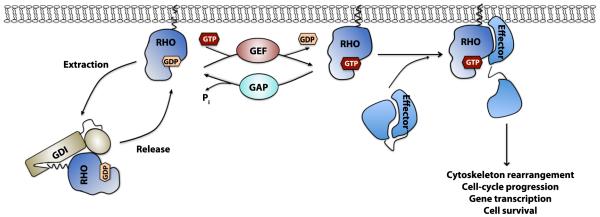
Like Ras, classical Rho GTPases such as RhoA, Rac1, and Cdc42 are molecular switches that cycle between an active GTP-bound form and an inactive GDP-bound form. They bind to GTP or GDP and can hydrolyze bound GTP. Three main families of regulatory proteins tightly control Rho GTPase activities: guanine-nucleotide exchange factors (GEFs), GTPase-activating proteins (GAPs), and guanine-nucleotide dissociation inhibitors (GDIs) (Figure 1). GEFs catalyze the exchange of GDP for GTP to activate Rho GTPases [6], while GAPs accelerate the intrinsic GTPase activity of Rho GTPases to inactivate them [7]. After translation, the Rho GTPases are geranylgeranylated, or less commonly farnesylated, in a C-terminal CAAX motif to promote their translocation to intracellular membranes where they are activated [8]. Rho GDIs bind to Rho GTPases in their inactive GDP bound form and sequesters them in the cytosol, thus acting as the inhibitor of Rho GTPases [9].
Figure 1. Biochemical model of the Rho GTPase regulatory mechanism.
Rho, Rac and Cdc42 cycle between an inactive GDP-bound and an active GTP-bound state. GEFs catalyze the GDP/GTP nucleotide exchange and activate the Rho GTPases, whereas GAPs enhance the intrinsic GTP-hydrolysis activity and inactivate them. GDIs can sequester Rho GTPases in the cytosol and prevent their activation. Activated Rho GTPases can interact with a variety of effector molecules to trigger downstream signaling events leading to diverse cellular responses.
The actions of Rho GTPases are mediated by effector proteins [10]. When bound to GTP, Rho GTPases undergo conformational changes and associate with a large number of potential effectors, including enzymes and scaffolding proteins, to mediate diverse yet specific cell behaviors. For example, during 2-dimensional cell migration, Rac1 and Cdc42 promote the actin-driven protrusions at the cell leading edge, whereas RhoA controls the actomyosin contraction at the cell body and rear. In 3-dimensional conditions, cells adapt different modes of migration dependent on spatial and temporal activation of Rho GTPases in response to the environmental cues [11,12]. RhoA mediates stress fiber formation mainly via Rho-kinase (ROCK) and mammalian diaphanous (mDia), while Rac1 and Cdc42 regulate actin re-organization by signaling via p21-activated kinases (Pak) or IQ-domain GTPase-activating protein (IQGAP). In addition, Cdc42 further signals via the Wiskott-Aldrich syndrome proteins (WASP) and Rac1 via the WASP-related WAVE to initiate actin rearrangement. In addition to actin-dependent activities, Rac and Cdc42 also activate the JNK and p38 MAP kinase cascade [13,14] that may affect tumor cell proliferation and metastasis.
In this review, we focus largely on the prototypical Rho GTPases, RhoA, Rac1, and Cdc42, and discuss their involvement in cancer and the ongoing strategies targeting their signaling pathways for future cancer therapy.
2. Rho GTPase signaling pathways and cancer
Rho GTPases, as well their regulators and effectors, have been implicated in multiple aspects of cancer [15-17] (Table 1). Unlike Ras, which is mutated in approximately 20-30% of human cancers, mutations in Rho GTPases are much less frequent and have just been appreciated. While emerging data suggest that mutations in various Rho proteins may occur in multiple cancer types, the main body of experimental evidence has focused on the regulatory mechanisms by which Rho GTPase activities are controlled. These mechanisms include altered expression levels of Rho GTPases or their regulators and altered localization patterns.
Table 1.
Selected Rho GTPases and regulators relevant to human cancer:
| Involvement in cancer | Reference | |
|---|---|---|
| Rho GTPase | ||
| RhoA | Mutated in gastric carcinoma, Burkitt lymphoma, peripheral and angioimmunoblastic T-cell lymphoma |
[21,143-152] |
| Over-expressed in gastric carcinoma, testicular germ cell tumor, head and neck squamous cell carcinoma, bladder, colon, breast and lung cancer |
[147-152] | |
| Rac1 | Mutated in melanoma, head and neck, and cutaneous squamous cell carcinoma |
[153-156] |
| Over-expressed in oral squamous cell carcinoma, gastric cell carcinoma, breast, lung, testicular and prostate cancer |
[147,148,151,152,157,158] | |
| Cdc42 | Over-expressed in breast cancer, melanoma, head and neck squamous cell carcinoma, colorectal, non-small cell lung and testicular cancer |
[148,149,151,152,159- 161] |
| RhoGEFs | ||
| P-Rex1 | Over-expressed in metastatic prostate tumor, breast cancer, and melanoma |
[162-164] |
| P-Rex2 | Over-expressed in PTEN wild type breast cancer | [165] |
| ECT2 | Over-expressed in lung, esophageal squamous cell carcinoma, glioblastoma, colorectal carcinoma, pancreatic, non-small cell lung cancer |
[166-170] |
| Trio | Over-expressed in glioblastoma, breast cancer | [171,172] |
| Activated in adult T-cell leukemia by alternative splicing | [173] | |
| Vav1 | Over-expressed in neuroblastoma, pancreatic carcinoma, metastatic melanoma and B-cell chronic lymphocytic leukemia |
[174-177] |
| Vav2 | Over-expressed in ovarian and breast cancer; Hyper-activated in head and neck squamous cell carcinoma |
[178-180] |
| Vav3 | Over-expressed in prostate, breast cancer and glioblastoma | [171,181] |
| Tiam1 | Over-expressed in melanoma, breast, colon, prostate and renal cell carcinoma |
[182-186] |
| LARG | Truncated and fusion to MLL in acute myelogenous leukemia | [49] |
| BCR-Abl1 | Fusion protein resulted from chromosome translocation associated most CML. BCR contains a GEF and a GAP domain |
[187] |
| DOCK1 | Over-expressed in glioblastoma | [188] |
| RhoGAPs | ||
| DLC-1 | Two missense mutations in pancreatic cancer | [189] |
| Loss-of-expression or down-regulation in lung, breast, liver, colon, pancreatic, ovarian, uterine, gastric, prostate, renal cancer, lymphoma, and ALL |
[52,53,190-192] | |
| p190-B | Gene amplification and protein overexpression in hepatocellular carcinoma cells |
[58] |
| β2- chimaerin |
Down-regulated in breast cancer | [96] |
| BCR-Abl1 | Fusion protein resulted from chromosome translocation associated with most CML. BCR contain a GEF and a GAP domain |
[187] |
| RhoGDIs | ||
| RhoGDI1 | Over-expressed in colorectal and ovarian cancers | [193,194] |
| Down-regulated in brain cancer and hepatocellular carcinoma | [195-197] | |
| Over-expressed or down-regulated in breast cancer in different studies |
[152,198] | |
| RhoGDI2 | Increased expression in pancreatic and ovarian cancer | [199,200] |
| Down-regulated in bladder cancer | [201,202] | |
| Over-expressed or down-regulated in breast cancer in different studies |
[203] | |
2.1 Rho GTPases in cancer
In addition to the notable case of RhoH [18], activating mutations in RhoA and Rac1 have been reported in several cancer types (Table 1). According to Catalogue of Somatic Mutations in Cancer (COSMIC) [19], somatic mutations of RhoA, Rac1, and Cdc42 are present in a variety of tumor tissues, most at low frequency (0.5% and below). Rac1 mutations are enriched in melanoma (5-6%) and RhoA mutations in both hematopoietic/lymphoid (4.6%) and stomach (7%) tissues. Cdc42 mutations, however, show no obvious tissue enrichment. The hotspot mutations of RhoA (G17V) and Rac1 ( P29S) account for 46% and 68% of all mutations identified, respectively. However, the biological and clinical significance of each hotspot mutation remains unclear. A recent study suggests that Rac1 P29S melanoma mutation may confer resistance to RAF inhibitors [20]. Although somatic mutations of Cdc42 are detected in many tumor tissues, these mutations are more sporadic. In a study to identify driver mutations for melanoma, G12V mutation of Cdc42, similar to an oncogenic Ras mutation in various cancers, was identified in one sample [21]. Whether such Cdc42 mutation can drive cancer needs to be verified.
There is ample experimental evidence that over-expression of Rho GTPases contributes directly to the proliferative and metastatic properties of cancer cells [22,23]. Most of these experiments involve manipulating the expression or activity of Rho GTPases in cancer cell lines by over-expressing constitutive active form or dominant negative form of Rho GTPases, their regulators or effectors, or using RNAi or small molecule inhibitors. However, those experiments are limited by approaches regarding specificity, dosage, and cell clonal variation. More recent genetic mouse models targeting individual Rho GTPases have provided convincing evidence for their physiological roles. Direct targeting of RhoA, Rac1, or Cdc42 is embryonic lethal, so a variety of tissue-specific conditional knockout mouse models have been generated (reviewed in [24-26]).
RhoA is highly over-expressed in a variety of cancer types (Table 1). In the case of breast cancer, CD44 interacts with a RhoA-specific GEF to induce RhoA and ROCK activation, which promotes cell growth and metastasis [27]. Inhibition of RhoA signaling by over-expression of a dominant negative form of ROCK blocks the metastatic activity by CD44. RhoA is also important for the metastasis of hepatocellular carcinoma cells. Inhibition of ROCK by expressing dominant negative ROCK or using small molecule inhibitor Y-27632 reduces the intrahepatic metastasis of those cells in SCID mice [28]. Similarly, knockdown of RhoA or RhoC in aggressive non-inflammatory or inflammatory breast cancer cells reduces invasion, motility and growth rate [29]. In colonic adenocarcinoma cells, leptin-induced invasion is potentiated by over-expression of the active form of RhoA and abrogated by the dominant negative form of RhoA or Rac1 [30]. In lung carcinoma A549 cells, ROCK inhibitor Y-27632 enhances the cisplatin-induced cytotoxicity through suppression of focal adhesion kinase (FAK)-independent mechanism [31]. These observations suggest that inhibition of RhoA signaling enhances the effect of anti-cancer agents. The role of RhoA in cancer is beginning to be revealed by genetic studies. RhoC deletion of RhoC shows no effect on tumor growth but decreases tumor metastasis in a breast cancer model [32]. Gene targeting of RhoA does not appear to affect K-Ras driven lung adenoma formation; rather, a combined RhoA and RhoC gene deletion is required to suppress lung cancer initiation [33].
Increased expression of Rac1 is reported for a variety of cancers (Table 1). Rac1 signaling is important for leptin-induced invasion in colonic adenocarcinoma cells [30]. Rac1 and Cdc42 are necessary for autotaxin-induced tumor motility in A2058 melanoma cells by a mechanism that may involve PAK and FAK [34]. In line with the observations obtained using dominant negative form of Rac1, RNA interference-mediated depletion of Rac1 strongly inhibits lamellipodia formation, cell migration and invasion of SNB19 glioblastoma cells and BT549 breast carcinoma cells [35]. Similarly, in human PC-3 prostate cancer cells, shRNA knockdown of Rac1 inhibits tumor cell diapedesis [36]. Inhibition of Rac1 by dominant-negative mutant inhibits aberrant cell proliferation of NF2-deficient cells [37]. Rac1 conditional knockout mouse studies have shown that Rac1 is required for K-Ras driven tumorigenesis in lung and skin cancer [38,39]. Such a Rac1 requirement may be cancer-type dependent as endothelial-specific Rac1 deletion shows no effect on tumor growth or angiogenesis [40].
Over-expression of Cdc42 has also been detected in some cancer types (Table 1). Rat mammary adenocarcinoma cells MTLn3 with over-expression of dominant negative form of RhoA or Cdc42 shown reduces number of focal contacts, inhibits colony formation in soft agar and affects cell growth in vivo [41]. In anaplastic large cell lymphoma (ALCL) driven by oncogenic fusion proteins, knockdown of Cdc42 by shRNA results in a cell cycle arrest and apoptosis of ALCL cells. In addition, Cdc42 is necessary for the growth and maintenance of established lymphomas in vivo [42]. Genetic deletion of Cdc42 in Ras-transformed cells inhibits proliferation, cell cycle progression and tumorigenicity [43]. Loss of Cdc42 also attenuats the tumorigenicity of mutant intestinal tumor cells [44]. Notably, the role of Cdc42 in cancer may be double-sided, as Cdc42 knockout in hematopoietic stem cells results in myeloproliferative disorder [45].
2.2 Rho regulators in cancer
The fact that Rho GTPases are often over-expressed or hyper-activated but less frequently mutated in many cancers suggests that regulatory proteins likely play a crucial role in dysregulating signaling pathways that promote cancer initiation and progression. Up-regulation of RhoGEFs or inhibition of RhoGAP and RhoGDI can lead to increased activation of Rho GTPases and promote aberrant signaling cascades. Mutations or aberrant expression of RhoGEFs, RhoGAPs, and RhoGDIs have all been detected in human cancer [16,17]. Because RhoGEFs are positive regulators, they represent good candidates for aberrant Rho GTPases activation in cancer.
There are two classes of RhoGEFs: the classical Dbl family RhoGEFs and the DOCK family RhoGEFs. The Dbl family of RhoGEFs includes over 70 members, many of which are conserved from yeast to human [46]. Dbl RhoGEFs share conserved tandem Dbl homology (DH)-pleckstrin homology (PH) domains for GEF activity and diverge significantly elsewhere [6]. The DOCK family of RhoGEFs includes 11 members and acts as GEFs for Rac and/or Cdc42, but not RhoA, and is structurally and mechanistically distinct from Dbl RhoGEFs [47]. Several RhoGEFs are mutated or up-regulated in human cancer. Among them, Dbl, Vav1/2/3, Ect2, Tiam1/2, P-Rex1/2 are best validated and extensively reviewed [46,48]. Compared to up-regulation of GEF activity in human cancer, gain-of-function mutations in RhoGEFs are less common. One example of activating mutation is the leukemia-associated RhoGEF (LARG), which was isolated as a fusion partner to the mixed lineage leukemia (MLL) gene in a patient with acute myeloid leukemia (AML) [49]. Later, it was shown to specifically activate RhoA in vivo, suggesting a role of LARG/RhoA signaling axis in leukemia [50]. Mutations in LARG are identified at low frequency in a variety of tumor tissues (COSMIC). LARG belongs to the subfamily of the regulator of G-protein signaling (RGS) domain-containing RhoGEFs and couples the signaling from G-protein coupled receptors through Gα12/13 to RhoA signaling. In childhood AML driven by the fusion gene AML1-ETO (AE), LARG is identified as a direct target for AE. As a transcription factor, AE significantly increases the expression level of LARG, resulting in hyper-activation of RhoA. Remarkably, LARG-specific shRNA targeting in human AE cells leads to increased apoptosis and inhibits the growth of AE+ leukemia without affecting control human blood progenitor cells or MLL-AF9 expressing leukemia cells. These findings, together with the ubiquitous expression of LARG, make this RhoGEF an exciting target for pharmaceutical intervention.
The negative Rho GTPase regulators, RhoGAPs and RhoGDIs, are relatively less well understood in the context of cancer. RhoGAPs constitute a large family with more than 70 members in eukaryotes [7]. All RhoGAPs contain a conserved ~150-amino-acid RhoGAP catalytic domain but share little other sequence homology. In both RhoGEFs and RhoGAPs, the diversity outside the catalytic domain allow them to be controlled in a spatial and temporal manner and serve as critical nodes to specifically mediate different signaling events. RhoGDIs, including RhoGDI1, 2 and 3, contain two domains: the C-terminal domain that contains the geranylgeranyl-binding pockets and binds the Rho GTPases, and the N-terminal regulatory domain that locks-in the Rho GTPases and prevents their nucleotide exchange and hydrolysis. They associate with Rho GTPases in their GDP-bound form and maintain a stable, soluble pool of inactivated Rho GTPases in the cytosol [8].
Mutations or dysregulation of RhoGAPs or RhoGDIs has been detected in human cancer [16,17], although less evident than that for RhoGEFs. One of the best characterized RhoGAPs, Deleted in liver cancer-1 (DLC-1), was originally discovered as a gene deleted in liver cancer [51]. Subsequently, it has been found to lose expression in many human cancers (Table 1). Reintroduce of DLC-1 into liver, lung or breast cancer cell lines results in reduced growth in vitro and in vivo [52-54]. These observations suggest an important role of DLC-1 in cancer as a tumor suppressor [55]. Another RhoGAP implicated in cancer is p190RhoGAP. p190-A is encoded on chromosome 19q13.3 that is often deleted in oligodendrogliomas [56]. There is evidence that p190-A can act to inhibit PDGF-induced gliomas in mice and serves as tumor suppressor [57]. Whereas P190-B, located on chromosome 14q12, is likely a target for amplification in hepatocellular carcinoma [58]. RhoGDIs have been associated with many cancers [9,16] (Table 1), but the specificity of each RhoGDI for each Rho GTPase is less understood. Since there are only three RhoGDIs that may act on more than twenty Rho GTPases, each RhoGDI may interact with multiple Rho GTPases and thus be subject to complex regulation. The involvement of RhoGDIs in cancer is likely complicated [9]. RhoGDI expression patterns in a single type of cancer, expression of a given RhoGDI among cancer types, and their activities toward particular Rho GTPases, vary significantly. For example, RhoGDI1 expression is upregulated in colorectal and ovarian cancer, but reduced in brain cancers. In breast cancers, both RhoGDI1 and GDI2 expression levels are reported to be increased or decreased in different studies.
2.3 Downstream Rho effectors and signaling in cancer
Rho GTPases utilize a wide variety of downstream effector proteins to regulate diverse cellular functions in response to various intracellular and extracellular stimuli [10]. There is a long-standing interest in understanding how each Rho GTPase, when hyper-activated by mutations or aberrant expression of upstream regulators, contributes to tumorigenesis through a unique set of effectors and signaling pathways.
Two of the best-characterized effector proteins downstream of RhoA are the Rho-associated protein kinases, ROCK1 and ROCK2 (also known as Rho kinases). They are serine/threonine kinases that phosphorylate multiple targets including myosin light chain (MLC), the myosin binding subunit (MYPT1) of the MLC phosphatase, and LIM kinases 1 and 2 (LIMK1 and LMK2). Activated ROCKs lead to increased myosin-driven contraction through MLC phosphorylation and promote actin filament stabilization through LIMK-mediated phosphorylation and inactivation of cofilin. ROCKs regulate cancer cell invasion and metastasis and have been studied extensively [59]. Recent studies have revealed additional functions of ROCK to their classic roles in cell motility, which include gene transcription, proliferation, differentiation, apoptosis and oncogenic transformation [60]. Notably, somatic ROCK1 activating mutations have been identified in human cancers [61], and large-scale sequencing has revealed mutations in both ROCK1 and ROCK2 that are associated with human cancers [62,63].
Another well known effector protein serine/threonine kinase family implicated in cancer is the P21-associated kinase (Pak) family effectors of active Rac1 and Cdc42 [64-66]. There are six mammalian members that can be divided into two subgroups based on their homology: group I (PAK1-3) and group II (PAK4-6). In mice, PAK1 is highly expressed in brain, muscle, and spleen, PAK2 and PAK4 is ubiquitously expressed, whereas PAK3, PAK5, and PAK6 are enriched in neuronal tissues. Genetic deletion of each Pak showed that they have both overlapping and distinct functions. Group I Paks are autoinhibited homodimers when inactive and binding of Rac/Cdc42 results in the dissociation of the dimer and subsequent activation, but the activation mechanism of group II Paks is less clear. In addition to dysregulated Rac/Cdc42 activities, over-expression or mutational activation of Pak isoforms contributes to the increased Pak activity found in various human cancers. Amplification of PAK1 on chromosome 11q13 has been reported in breast [67], ovarian cancer [68], and melanoma [69]. Similarly, amplification of PAK4 on chromosome 19q13 is commonly observed in pancreatic cancer [70,71] and oral squamous-cell carcinoma [72]. Recently, activating mutations in the PAK4 and PAK5 gene are associated with colon and lung cancers [73,74].
Activated Paks drive several oncogenic signaling pathways to impact tumor cell motility, survival and proliferation [66]. As the major effectors of Rac1 and Cdc42, Paks promote cell motility via several mechanisms. PAK1 facilitates actin stabilization through phosphorylation of MLC, LIMK, filamin A and dynein light chain 1 (DLC1) [75]. The PAK1/LIMK pathway is required for Rac1-induced actin reorganization at the cell leading edge during migration [76]. PAK1 also functions to induce rapid turnover of focal contacts at the cell leading edge via phosphorylation of paxillin [77]. Expression of dominant negative PAK1 in invasive breast carcinoma cells reduces invasion and metastasis [78]. Group II Paks seem to utilize different mechanisms to participate in cytoskeleton reorganization. Cdc42 recruits PAK4 to the Golgi and induces the formation of filopodia. Activated PAK4 leads to dissolution of stress fibers and loss of focal adhesions [79]. In addition to their role in tumor invasion and metastasis, most Paks promote cell cycle progression when over-expressed. Paks activate the Erk, PI3K/Akt, and Wnt signaling pathways that are tightly associated with cell proliferation. In the Erk pathway, PAK1 phosphorylates both MEK1 and Raf1 for efficient Erk activation. It has been shown that PAK1 drives anchorage-independent growth in human mammary epithelial cells through MAPK and MET signaling [80]. PAK1 and PAK4 also induce proliferation independent of RAF/MEK/ERK or PI3K/Akt pathways in mutant K-RAS or BRAF colon cancer cells by an unknown mechanism [81]. In the Wnt pathway, PAK1 and PAK4 directly interact and phosphorylate β-catenin, a key component of Wnt signaling [82,83]. Paks are also linked with the NF-κB signaling pathway, although a direct target in this pathway has yet to be identified. Other targets of Paks include nuclear hormone receptors such as estrogen receptor (ER) [84], androgen receptor (AR) [85], apoptosis signaling molecules such as BAD [86], and the E-cadherin repressor Snail [87].
There are many other Rho effectors in addition to ROCKs and Paks. Rac1 regulates components of the MAPK pathways, especially JNK and p38. Rac1 and Cdc42 both regulate cell polarity via PAR6. Rac1 also constitutes part of the phagocyte NADPH oxidase complex NOX2 that generates reactive oxygen species (ROS). This enzyme complex consists of at least six components: two membrane-bound subunits p22phox and gp91phox, and four cytosolic regulatory subunits Rac1/2, p47phox, p67phox and p40phox. Activated Rac1/2 binds to p67phox, leading to the translocation of the regulatory complex from cytosol to plasma membrane for full assembly and activation of the NOX2 complex. In addition as a host defense, ROS also activates various transcription factors such as NF-κB, AP-1, HIF-1α and STAT3 and plays critical roles in many signaling pathways including cell-cycle progression, apoptosis, and inflammation.
3. Approaches to target Rho GTPase signaling
Key components in the Rho GTPase pathways are attractive targets for therapeutic interventions in cancer. Rho GTPases themselves are difficult to target by small-molecule modulations. Given the micromolar GTP concentration in cells and the sub-nanomolar binding affinity of Rho GTPases for GTP or GDP, it is difficult to drug Rho GTPases by nucleotide analogs like that of protein kinases. Besides the nucleotide-binding pocket, there are few stable, tractable cavities present on Rho GTPases. Thus, the family of small GTPases, including Ras and Rho, are generally deemed “undruggable” in cancer research. A variety of bacterial toxins can modify the activity of Rho GTPases [88]. For example, the exoenzyme C3 transferase, an ADP-ribosyltransferase from Clostridium botulinum, inactivates RhoA/RhoB/RhoC, while Clostridium difficile toxin A and B glucosylate and inactivate multiple Rho GTPase subfamilies. These bacterial toxins have been widely used to dissect the biological functions of Rho GTPases. However, they are large enzymes that introduce covalent modifications to the substrates and are non-specific, therefore cannot be used clinically. Based on the biochemical mechanisms of Rho GTPase regulation and function, significant effort has been dedicated to developing small molecule inhibitors that act on various aspects of Rho GTPase signaling mechanisms (Figure 2). In this section, we discuss these strategies and representative inhibitors (Table 2).
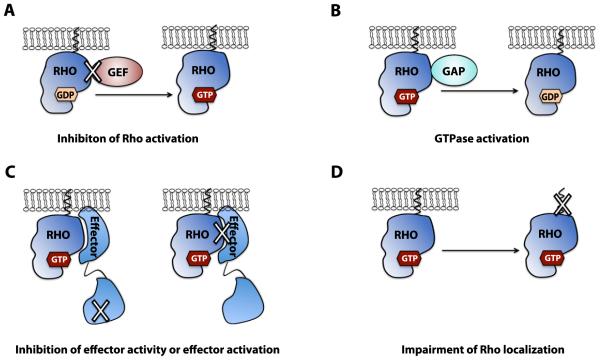
Figure 2. Approaches for rational targeting the Rho GTPase signaling module.
A: Inhibition of Rho GTPase activation by GEFs via disrupting Rho-GEF interactions. B: Enhancing the intrinsic GTPase activity or Rho-GAP interaction. There has been limited experimental evidence for this approach to date. C: Inhibition of effector activity or disrupting Rho-effector interactions. D: Impairment of a Rho GTPase intracellular localization by altering its post-translational lipid modifications.
Table 2.
Selected small molecule inhibitors for Rho GTPase signaling:
| Pathway | Target | Name of compound |
Mechanism | Reference |
|---|---|---|---|---|
| Inhibition of Rho activation | ||||
| Rho | RhoA | Rhosin | Block GEF binding | [204] |
| LARG | Y16 | Block RhoA binding | [92] | |
| MKL1 | CCG-1423 | Block RhoA-dependent gene transcription |
[205,206] | |
| PLD1 | CHS-111 | Block RhoA membrane recruitment |
[207] | |
| Rac | Rac1 | NSC23766 | Block GEF binding | [89] |
| Rac1 | EHop-016 | Derivative of NSC23766 | [208] | |
| Rac1/Rac2 | EHT-1864 | Lock in inactive state | [93] | |
| Cdc42 | Cdc42 | CASIN | Block GEF binding | [90] |
| Cdc42 | ZCL278 | Block GEF binding | [209] | |
| Cdc42 | AZA197 | Block GEF binding | [210] | |
| Cdc42/Rac1 | AZA1 | Block GEF binding | [211] | |
| Cdc42 | Secramine | Enhance interaction with RhoGDI |
[212] | |
| Cdc42 | ML141 (CID2950007) |
Block nucleotide binding | [94] | |
| Inhibition of effector activation | ||||
| Rac | p67phox | Phox-I1 | Block Rac1-GTP binding | [123] |
| Inhibition of effector activity | ||||
| Rho | ROCK | Fasudil | Compete with ATP | [102] |
| Dimethylfasudil | Also known as H-1152, derivative of Fasudil |
[105] | ||
| Y-27632 | Compete with ATP | [100] | ||
| WF-536 | Derivative of Y-27632 | [104] | ||
| Y-39983 | Also known as RKI-983, derivative of Y-27632 |
[213] | ||
| SNJ-1656 | Ophthalmic solution of RKI-983. In Phase II trials for glaucoma |
[107] | ||
| SB-772077-B | Compete with ATP | [214] | ||
| GSK269962A | Compete with ATP | [214] | ||
| K-115 | Compete with ATP; in Phase II trial for glaucoma and ocular hypertension |
[108,215] | ||
| SR-3677 | Compete with ATP | [216] | ||
| AR-12286 | Compete with ATP, in Phase II trial for glaucoma and ocular hypertension |
[109] | ||
| SAR407899 | Compete with ATP, in Phase II trial for erectile dysfunction |
[110] | ||
| PT-262 | Compete with ATP | [217] | ||
| Azaindole 1 | Compete with ATP | [218] | ||
| Rhodblock 6 | unknown | [219] | ||
| RKI-18 | Compete with ATP | [138,220] | ||
| RKI-1447 | Compete with ATP | [106] | ||
| AT13148 | Compete with ATP, multi- AGC kinase inhibitor, in Phase I trial for advanced solid tumors |
[111,112] | ||
| Rac/Cdc42 | Pak | K252 | Compete with ATP | [113] |
| KT-D606 | Derivative of K252 | [114] | ||
| CEP-1347 | Derivative of K252 | [114] | ||
| OSU-03012 | Compete with ATP | [115] | ||
| IPA-3 | Lock Paks in auto-inhibited conformation |
[141] | ||
| Λ-FL172/Λ-FL411 | With bulk scaffolds, compete with ATP |
[116] | ||
| PF-3758309 | Compete with ATP | [117] | ||
| LCH-7749944 | Compete with ATP | [118] | ||
| FRAX597 | Compete with ATP | [119] | ||
| MLK | CEP-1347 | Compete with ATP; pan- MLK inhibitor |
[221] | |
| CEP-5104 | Derivative of CEP-1347, more selective for MLK1 |
[222] | ||
| CEP-6331 | Derivative of CEP-1347, more selective for MLK3 |
[222] | ||
| Cdc42 | MRCK | Chelerythrine | Not compete with ATP | [223,224] |
| Cycloartane- 3,24,25-triol |
Compete with ATP | [225] | ||
| BDP5290 | Compete with ATP | [224] | ||
| N-Wasp | 187-1 | Lock N-Wasp in auto- inhibited conformation |
[226] | |
| Wiskostatin | Stabilize N-Wasp in its inactive state |
[227] | ||
| Inhibition of geranylgeranyltransferase I | ||||
| Rho/Rac/C dc42 |
GGTI | GGTI-286 | CAAL mimetic | [127] |
| GGTI-298 | CAAL mimetic | [128] | ||
| GGTI-2154 | CAAL mimetic | [129] | ||
| GGTI-2166 | CAAL mimetic | [130] | ||
| GGTI-DU40 | Nonpeptidomimetic, compete with protein substrate |
[131] | ||
| P61A6 | Nonpeptidomimetic, compete with protein substrate |
[132] | ||
| P61-E7 | Nonpeptidomimetic, compete with protein substrate |
[133] | ||
3.1 Inhibition of Rho GTPase activation by RhoGEFs or prevention of Rho-GTP formation
To activate Rho, Rac or Cdc42, RhoGEFs catalyze the GDP/GTP nucleotide exchange on the Rho GTPases. As Rho GTPases are activated by different RhoGEFs that response to different signals in a temporally and spatially manner, targeting specific RhoGEFs provide better selectivity compared to targeting Rho GTPases directly. To date, however, only limited success in targeting RhoGEF activity directly has been achieved. The diversity of the sequences and domain structures beyond the shared DH-PH domains indicates the existence of diverse mechanisms for the regulation and functions of RhoGEFs.
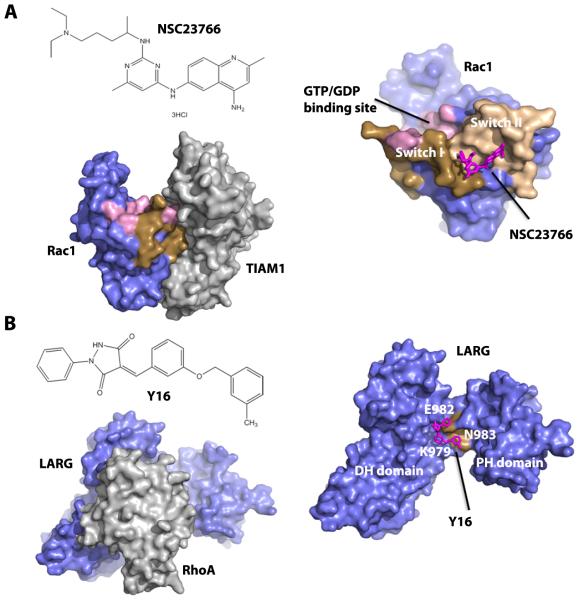
Attempts have been made to block the formation of Rho-GTP by interrupting the interactions between RhoGEFs and Rho. There are several small molecule inhibitors developed to bind Rho GTPases at the GEF-binding surface and inhibit RhoGEF function. For example, Rac1 inhibitor NSC23766 was discovered through a structure-based virtual screening of compounds to fit into a surface groove of Rac1 that interacts with GEFs [89] (Figure 3A). NSC23766 specifically inhibits Rac1 activation by the Rac-specific GEFs Trio or Tiam1 in a dose-dependent manner, but not Cdc42 or RhoA activation by their GEFs. Another small molecule inhibitor, CASIN, specifically inhibits Cdc42 by a similar mechanism. CASIN inhibits Cdc42 activation by its GEF intersectin in a specific, reversible and dose-dependent manner without affecting other Rho GTPases [90]. Since these Rho inhibitors target the surface of Rho GTPases that is required for activation by various GEFs, they are unlikely to be GEF-specific.
Figure 3. Structural basis for inhibition of Rac1-GEF and RhoA-GEF interactions.
A: chemical structure of the lead Rac inhibitor, NSC23766 (upper left), crystal structure of Rac1-TIAM1 complex (PDB code 1FOE, lower left), and a docking model of NSC23766 onto Rac1 surface (right). B: chemical structure of Y16, a lead Rho GEF inhibitor that binds to LARG (upper left), crystal structure of RhoA-LARG complex (PDB code 1X86, lower left) and a docking model of Y16 onto the LARG DH-PH domains hinge region (right). In both A and B, the docking model was generated by AutoDock Vina [142] and visualized by PyMOL (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC). Crystal structures used for docking were Rac1-TIAM1 complex and RhoA-LARG complex, respectively.
Another way to target Rho GTPases activity is through targeting specific RhoGEFs and preventing their binding to Rho GTPases. Conceivably, this strategy could lead to better selectivity conferred by individual GEFs. One example is the development of inhibitors for the RhoA-specific GEF, LARG. With different screening methods, two groups independently identified several structurally distinctive inhibitors for LARG. Using a fluorescence polarization guanine nucleotide-binding assay, five compounds were identified to selectively inhibit LARG-stimulated RhoA-GTP binding [91]. Subsequently, Y16 was identified to bind to the hinge region of LARG DH/PH domain through virtual screening coupled with high throughput screen [92] (Figure 3B). Y16 specifically inhibits LARG and other RGS-containing RhoGEFs by binding to RhoA without detectable effect on other Dbl family of RhoGEFs, Rho effectors, or RhoGAPs. Y16 selectively inhibits serum-induced RhoA signaling and mammary sphere formation in MCF7 breast cancer cells. Since this discovery, the inhibitors of the RhoGEF-Rho interaction are proven as useful research tools to provide experimental evidence that targeting RhoGEF to suppress Rho GTPase activation is feasible. Whether this approach will result in lead compounds with suitable potency and selectivity for clinical trials remains to be seen.
In addition to the discussed competitive inhibitors, non-competitive inhibitors have been developed to inhibit Rho signaling by promoting the loss of bound nucleotide and inhibiting nucleotide re-association. These include EHT 1864 for Rac subfamily [93] and ML141 (CID2950007) for Cdc42 [94]. Those compounds may function as conformation-dependent allosteric inhibitors that bind to guanine nucleotide-bound Rho GTPase, thus inducing dissociation of the bound nucleotide and locking the respective GTPase in an inactive conformation. ETH 1864 does not affect the interaction between Rac1 and its GEF Tiam1 in vitro, but inhibits nucleotide exchange induced by Tiam1. Similarly, ML141 acts independently of effects on GEF activity or interaction with Cdc42.
3.2 Stimulation of GTP-hydrolysis activity of Rho GTPases by RhoGAP
RhoGAPs stimulate the intrinsic GTPase activity of RhoA, Rac and Cdc42 by up to 105-fold. Our current knowledge suggests that involvement of RhoGAPs in cancer is often associated with loss-of-function mutations, and they often exhibit properties consistent with tumor suppressors. RhoGAPs are less attractive to researchers as potential targets for cancer therapy since it is difficult to develop small molecule agonists than antagonists. Further understanding of the mechanisms by which RhoGAPs are regulated in specific cellular contexts may reveal more possible approaches to target this important family of Rho GTPases regulators. Like RhoGEFs, RhoGAPs exist in large numbers and show tremendous diversity in their sequences beyond the conserved GAP catalytic domain. RhoGAPs typically contain multiple additional protein-protein and protein-lipid interacting domains, as well as many phosphorylation sites. Coordinately, RhoGEFs and RhoGAPs control Rho GTPase on/off status in a highly regulated temporal and spatial manner. It remains a challenge in the Rho GTPase field to uncover the signaling pathways by which RhoGAPs regulate specific aspects of Rho GTPase function. In particular, better understanding of which and how RhoGAPs are involved in cancer would promote the development of small molecule modulators of RhoGAPs.
There is limited but promising evidence that small molecules can be used to enhance the ability of RhoGAPs to suppress Rho GTPases. One class of RhoGAPs, the Rac-specific Chimaerins (CHN), has a C1-zinc finger domain that binds the lipid second messenger diacyglycerol, a cofactor for its activity [95]. Chimaerins are implicated in development, axon guidance, metabolism, cell migration, and T-cell activation, and deregulation of CHN genes is associated with cancer and many other diseases [96]. It is speculated that small molecules that bind C1 domains may activate the GAP activities which in turn down-regulate Rac signaling. Although there are other proteins with C1 domains [97] and off-target effect could be a potential problem, C1 binding molecules may still have some degree of selectivity for a subset of C1-containing proteins and would be a therapeutic option for Rac hyperactivation dependent cancers.
3.3 Inhibition of Rho-effector interaction or effector activity
Approaches to target upstream Rho signaling by either blocking Rho activation through RhoGEFs or stimulating Rho intrinsic GTPase activity through RhoGAPs, are still in the early stage of development with relatively limited success. The task to modulate RhoGEFs or GAPs is a particular challenge in part due to the diversity and complexity of those upstream regulators signaling and regulation, which are still largely unclear. Another approach to block Rho signaling is by interfering the activation of effectors by Rho GTPases or inhibiting specific effector activities directly. Similar to targeting upstream signaling of Rho, higher selectivity may be achieved targeting Rho effectors. In contrast to upstream signaling pathways, downstream effector signaling pathways are much better understood. As a result, development of small molecules inhibitors against effector signaling has been more successful.
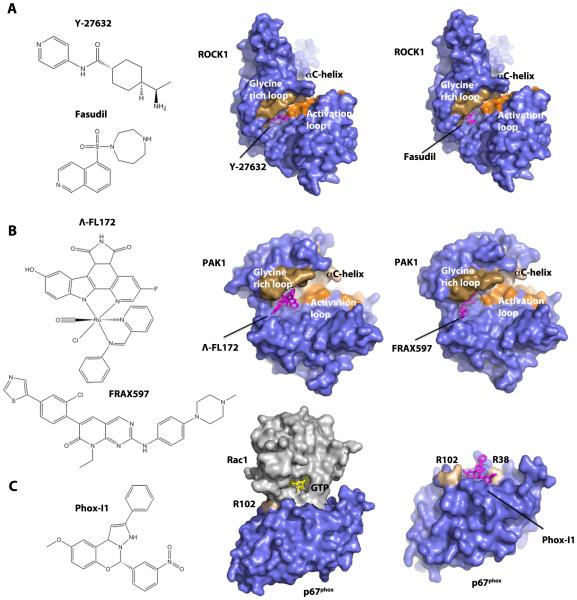
Some downstream effectors are classical kinases and, therefore, are more readily targeted by traditional pharmacologic approaches. In fact, inhibitors of ROCKs, the major effector family downstream of RhoA, are most promising among all therapeutic interventions of Rho signaling for cancer therapy. ROCK inhibitors are reviewed extensively elsewhere [98,99] and will only be briefly discussed here. One widely used ROCK inhibitor, Y-27632, is a pyridine-analog that competes with ATP for binding to ROCKs [100] (Figure 4A). Y-27632 treatment decreases invasion and alters cell survival of melanoma cells in vitro resulting in a reduction in melanoma tumor volume in vivo [101]. Another extensively studied inhibitor of ROCKs is Fasudil (also known as HA-1077), which is also ATP-competitive and inhibits a wide spectrum of kinases [102] (Figure 4A). Fasudil is the only clinically approved ROCK inhibitor, initially approved for the treatment of cerebral vasospasms and pulmonary hypertension. Various animal models of cancer have revealed that Fasudil can inhibit tumor invasion and metastasis [103]. Recently developed analogs of Fasudil and Y-27632, with improved selectivity and potency, such as WF-536 (Y-27632 derivative) [104], H1152 (Fasudil derivative) [105], and RKI-1447 [106], have been shown to reduce tumor progression. Despite the great interest in ROCK for cancer therapy, no ROCK inhibitor has been approved for cancer treatment. Newer generations of ROCK inhibitors have proven more potent and selective and several are currently under clinical trials for glaucoma and ocular hypertension [107-109] and erectile dysfunction [110]. The multi-AGC kinase inhibitor AT13148, which also inhibits ROCK, shows potent antitumor and anti-metastatic activities [111,112] and is in phase I clinical trial for advanced solid tumors.
Figure 4. Structural basis for the inhibition of a Rho GTPase effector activity and a Rho-effector interaction.
A: Chemical structures of the ROCK inhibitor Y-27632 and Fasudil (left), and crystal structures of ROCK1 with bound Y-27632 (PDB code 2ETR, middle) or Fasudil (PDB code 2ESM, right). B: Chemical structures of lead inhibitors of PAK, Λ-FL172 and FRAX597 (left), and crystal structures of PAK1 with bound Λ-FL172 (PDB code 3FXZ, middle) or FRAX597 (PDB code 4EQC, right). C: Chemical structure of the Rac-p67phox binding inhibitor of NOX2 enzyme, Phox-I1 (left), crystal structure of Rac1-p67phox complex (PDB code 1E96, middle), and the docking model of Phox-I1 on p67phox surface (right). This model was generated by AutoDock Vina [142] using the crystal structure of Rac1-p67phox complex. All crystal structures and models were visualized by PyMOL (The PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC).
Another family of effectors that are attractive targets for drug discovery is the Pak family, downstream of both Rac and Cdc42. Several ATP-competitive inhibitors have been identified for Paks. Earlier generations of Pak inhibitors include K252 [113], and K252 derivative KT-D606 and CEP-1347 [114]. Although they demonstrate potent Pak inhibition, these inhibitors are less selective and have limited utility. Later, more selective ATP-competitive Pak inhibitors were developed, such as OSU-03012 [115], Λ-FL172 [116], PF-3758309 [117], LCH-7749944 [118], and FRAX597 [119]. Of them, Λ-FL172 is of particular interest (Figure 4B). Λ-FL172 and related molecules were generated by modifying other compounds with bulky and rigid octahedral ruthenium scaffolds to selectively target the large ATP-binding site of PAK1. Among 264 kinases tested, only fifteen showed an inhibition similar to that of PAK1 [116]. However, such compounds usually have poor solubility and relatively high toxicity, and their potential as clinical drugs remains to be determined. FRAX597 was a potent inhibitor of group I Paks identified from high-throughput screening and shows beneficial effects on inhibiting cancer in animal models (Figure 4B). Non-ATP-competitive inhibitors have also been described. For example, IPA-3 binds to the regulatory domain of group I Paks, locking the kinases in their auto-inhibited conformation [120]. Thereby, IPA-3 is exceptionally selective. A few studies have shown that IPA-3 induces apoptosis in a number of cancer cell lines [121] and decreases cell spreading and adhesion in Schwannoma cell lines [122]. However, IPA-3 is unstable under physiological condition and unsuitable for further clinical development.
In addition to directly targeting effector activities, blocking Rho GTPase-effector interactions prevents effector activation and allows for greater selectivity through targeting unique Rho GTPase interactive sites. One example is the development of small molecule inhibitor phox-I1 for p67phox, a subunit of the NOX2 superoxide producing enzyme that is required for Rac1-GTP binding [123] (Figure 4C). Phox-I1 and derivatives recognize a surface pocket on p67phox, disrupt Rac1 binding and effectively inhibit NOX2-mediated superoxide production dose-dependently in human and mouse neutrophils. Although phox-I1 might not have sufficient efficacy to be used clinically, this study provides a proof of principle that inhibition of downstream effector activation by a specific pathway via rational targeting of a Rho GTPase-effector interface is a viable approach for drug development.
3.4 Inhibition of Rho posttranslational modification
To be effective as on/off switches, Rho GTPases require proper subcellular localization to function at defined intracellular sites. The recruitment of Rho GTPase to cell membranes is mediated by post-translational modification processes as well as signaling events that release Rho-GDP from bound RhoGDI. The first and crucial step is the progressive post-translational modification of a Rho GTPase at the C-terminal “CAAX box”. A geranylgeranyl, or less frequently, a farnesyl lipid group is attached to the cysteine residue of the CAAX sequence in the cytoplasm, catalyzed by geranylgeranyl or farnesyl transferases. This allows the Rho GTPase to translocate to the ER, where the –AAX tail is cleaved off and the newly exposed α-carboxyl group of the C-terminal cysteine residue is methylesterified. Incorporation of geranylgeranyl or farnesyl group is essential for the proper localization and biological activity for most small GTPases. In contrast, the latter methylesterification is only essential for farnesylated Ras, but not geranylgenranylated Rho [124]. However, this methylesterification step may have additional functions for Rho GTPases as isoprenylcysteine carboxyl methyltransferase (ICMT) knockout resulted in greatly reduced level of RhoA [125].
Inhibitors of prenyltransferases have long been proposed as potential therapeutic drugs. There are three related enzymes of prenyltransferases: farnesyltransferase (Ftase), geranylgeranyl transferase I (GGTase I) and GGTase II. Ftase and GGTase I recognize the CAAX motif and catalyze the lipidation of their protein substrates. GGTase II is specific for Rabs and its working mechanism is different from that of Ftase and GGTase I. Since Ras is well established as the most frequently mutated oncogene in human cancer and both normal and oncogenic Ras require farnesylation, Ftase has been one of the most attractive targets for anti-cancer drug discovery. A large number of Ftase inhibitors (FTIs) have been developed and tested in more than seventy clinical trials [126]. The results were largely disappointing, however, with the surprising finding that H-Ras and K-Ras can still be geranylgeranyled when Ftase is inhibited. Comparing to FTIs, less work has been done on GGTase I-specific inhibitors (GGTIs). To date, several GGTIs have been developed, including inhibitors that mimic the CAAL peptide such as GGTI-286 [127], GGTI-298 [128], GGTI-2154 [129], and GGTI-2166 [130], as well as inhibitors that are non-peptidomimetic such as GGTI-Du40 [131], P61A6 [132], P61-E7 [133]. P61A6 significantly suppresses tumor growth and cell proliferation in a human pancreatic cancer xenograft [134] and non-small cell lung cancer xenograft in nude mice [135]. However, PPTIs have failed to show clinical efficacy thus far [136].
4. Conclusions
Interference of signaling from upstream regulators like RhoGEFs and RhoGAPs can impact GDP/GTP exchange or GTP hydrolysis to prevent Rho GTPases activation. Several currently available inhibitors block upstream signaling by disrupting protein-protein interactions between Rho GTPases and RhoGEFs. While RhoGAP domains have been engineered to target specific members of Rho GTPases, it remains a challenge to design chemicals that may mimic or enhance RhoGAP activity to manipulate Rho-GTP level in cells. Although RhoGDIs are proposed as potential targets in drug discovery, it is not clear if the promiscuous nature of RhoGDIs might allow for specific Rho GTPase signaling modulation. Similarly, whether targeting Rho GTPase localization through a manipulation of FTase or GGTase may satisfy selectivity and toxicity considerations.
The approach of targeting Rho GTPase effectors, in particular kinases like ROCKs and PAKs, has been extensively investigated. A number of ROCK inhibitors have been developed and significant structure-activity relationship data for improving potency and selectivity are available [137,138]. ROCK inhibitors are currently in several clinical trials, although only AT13148 is developed for cancer therapy. The development of PAK inhibitors, however, are still in their early stage and only one has entered clinical trials [139]. Owing to their large and flexible ATP binding pocket, Paks, particular group I Paks, are difficult to target. Few Pak inhibitors with satisfactory selectivity and drug-like properties have been reported to date. More studies are needed to decipher the role of each Pak in cancer before pan-Pak or Pak isoform selective inhibitors can move to clinical trials. An alternative to inhibiting effector activity is to block effector activation by disrupting Rho/effector interaction. Proof-of-principle studies have suggested a few such inhibitors. It remains to be seen if these inhibitors meet the selectivity and efficacy requirements in animal models.
5. Expert’s Opinion
Relative to Ras, Rho GTPases are considered emerging anti-cancer targets. One reason is that, while oncogenic Ras is indisputably a driver for many cancers, mutations in Rho GTPases are relatively rare in human cancers. Partly due to setbacks in strategies directly drugging Ras and the increasing understanding of Rho GTPase involvement in cancer over the last two decades, Rho GTPases have gained considerable interests as potential cancer targets. Since aberrant Rho activities associated with cancer are often caused by over-expression or dysregulation of regulators or effectors, strategies targeting Rho GTPases either directly or indirectly through other signaling pathway components have been proposed. Parallel to the continuing effort to drug Ras signaling [140], approaches in advancing drug discovery of Rho GTPase signaling axes will enrich future cancer therapy portfolio.
One of the challenges remaining is to better understand the detailed picture of Rho GTPase signaling and regulation, particularly in the context of specific cancer types. With the wide application of genome-wide analyses of cancer cells, additional evidence for aberrant Rho, GEF, GAP, and effector expression and function is expected to grow at a rapid rate. Experimental validation of their functional significance in primary cancer cells and animal models will be a rate-limiting step. Careful target validation will be key for further development of Rho pathway inhibitors for therapy. In particular because RhoGEFs and RhoGAPs are multi-domain and multi-functional, caution must be taken to more rigorously validate their roles in cancer in the context of Rho GTPase signaling.
Another challenge is druggability. To date, there is no clinically available drug that targets Rho GTPase signaling for cancer treatment. Aside from a few kinase effectors which constitutes a small portion of effector portfolios, pharmacological intervention of Rho signaling by targeting Rho, Rac, Cdc42, GEFs, Rho GTPase isoprenylation and other non-kinase effectors is still in an early stage of development. Much effort so far is proof-of-concept in nature and most academic drug leads remain far from clinical applications. Nevertheless, advances in drug design technologies, particularly those related to the design of protein-protein interaction inhibitors and high-throughput experimental assays, will accelerate the drug development process that target Rho GTPases.
Rho GTPase signaling crosstalks with other cancer-associated pathways such as EGFR, PDGFR, VEGFR, G-protein coupled receptors, integrins, p53, NF1/NF2, FAK, Src kinases, and Ras, constitute a complex and delicate network with multiple feedback and compensatory mechanisms. It is now generally accepted that specific targeting of a particular signaling pathway may show less efficacy and encounter rapid resistance, but drug combinations that target multiple nodal points within the network are more likely to achieve substantial clinical benefit. Even if Rho signaling inhibitors by themselves are not sufficient for effective cancer cell suppression, they may provide an option in combinatory therapy to achieve the desired efficacy. For example, the group I Pak inhibitor IPA-3 can overcome resistance to PI3K inhibitors in lymphomas [141]. Thus, small molecule inhibitors targeting Rho GTPase signaling may add new treatment regimens in future precision cancer therapy, particularly in combination with other anti-cancer agents.
Acknowledgments
Financial and Competing Interests
Y Zheng is supported by Cincinnati Children's Hospital. He is also supported by the US National Institutes of Health with grant number NIH CA193350, NIH CA150547 and NIH CA141341.
Footnotes
Disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
References
- 1.Wennerberg K, Rossman KL, Der CJ. The Ras superfamily at a glance. J Cell Sci. 2005;118:843–846. doi: 10.1242/jcs.01660. [DOI] [PubMed] [Google Scholar]
- 2.Boureux A, Vignal E, Faure S, et al. Evolution of the Rho family of ras-like GTPases in eukaryotes. Mol Biol Evol. 2007;24:203–216. doi: 10.1093/molbev/msl145. * Evolutional study that established the 20 mammalian Rho GTPase members to be structured into 8 subfamilies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall A. Rho GTPases and the actin cytoskeleton. Science. 1998;279:509–514. doi: 10.1126/science.279.5350.509. [DOI] [PubMed] [Google Scholar]
- 4.Nobes CD, Hall A. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. * Showes different roles of Rho, Rac, and Cdc42 in actin organization. [DOI] [PubMed] [Google Scholar]
- 5.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 6.Rossman KL, Der CJ, Sondek J. GEF means go: Turning on Rho GTPases with guanine nucleotide-exchange factors. Nature Reviews Molecular Cell Biology. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 7.Tcherkezian J, Lamarche-Vane N. Current knowledge of the large RhoGAP family of proteins. Biology of the Cell. 2007;99:67–86. doi: 10.1042/BC20060086. [DOI] [PubMed] [Google Scholar]
- 8.Adamson P, Marshall CJ, Hall A, et al. Post-translational modifications of p21rho proteins. J Biol Chem. 1992;267:20033–20038. [PubMed] [Google Scholar]
- 9.Garcia-Mata R, Boulter E, Burridge K. The 'invisible hand': regulation of RHO GTPases by RHOGDIs. Nat Rev Mol Cell Biol. 2011;12:493–504. doi: 10.1038/nrm3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bishop AL, Hall A. Rho GTPases and their effector proteins. Biochem J. 2000;348(2):241–255. [PMC free article] [PubMed] [Google Scholar]
- 11.Doyle AD, Petrie RJ, Kutys ML, et al. Dimensions in cell migration. Curr Opin Cell Biol. 2013;25:642–649. doi: 10.1016/j.ceb.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riching KM, Keely PJ. Rho family GTPases: making it to the third dimension. Int J Biochem Cell Biol. 2015;59:111–115. doi: 10.1016/j.biocel.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coso OA, Chiariello M, Yu JC, et al. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 14.Minden A, Lin A, Claret FX, et al. Selective activation of the JNK signaling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- 15.Gomez del Pulgar T, Benitah SA, Valeron PF, et al. Rho GTPase expression in tumourigenesis: evidence for a significant link. Bioessays. 2005;27:602–613. doi: 10.1002/bies.20238. ** Reviews the evidence of dysregulation of Rho signaling by overexpression of different members of the Rho GTPases in human tumors. [DOI] [PubMed] [Google Scholar]
- 16.Harding MA, Theodorescu D. RhoGDI signaling provides targets for cancer therapy. Eur J Cancer. 2010;46:1252–1259. doi: 10.1016/j.ejca.2010.02.025. ** Reviews the role of RhoGDIs in a variety of cancers and discusses possible therapeutic strategies and potential complications arising from their implementation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vigil D, Cherfils J, Rossman KL, et al. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10:842–857. doi: 10.1038/nrc2960. ** Reviews the association of GEFs and GAPs with cancer and their druggability for cancer therapeutics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Preudhomme C, Roumier C, Hildebrand MP, et al. Nonrandom 4p13 rearrangements of the RhoH/TTF gene, encoding a GTP-binding protein, in non-Hodgkin's lymphoma and multiple myeloma. Oncogene. 2000;19:2023–2032. doi: 10.1038/sj.onc.1203521. * Identification of RhoH mutations in non-hodgkin's lymphoma and multiple myoloma. [DOI] [PubMed] [Google Scholar]
- 19.Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watson IR, Li L, Cabeceiras PK, et al. The RAC1 P29S hotspot mutation in melanoma confers resistance to pharmacological inhibition of RAF. Cancer Res. 2014;74:4845–4852. doi: 10.1158/0008-5472.CAN-14-1232-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hodis E, Watson IR, Kryukov GV, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–263. doi: 10.1016/j.cell.2012.06.024. ** Identification of mutations in Rac1, Rac2, and Cdc42 in melanoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaffe AB, Hall A. Rho GTPases in transformation and metastasis. Adv Cancer Res. 2002;84:57–80. doi: 10.1016/s0065-230x(02)84003-9. [DOI] [PubMed] [Google Scholar]
- 23.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 24.Melendez J, Grogg M, Zheng Y. Signaling role of Cdc42 in regulating mammalian physiology. J Biol Chem. 2011;286:2375–2381. doi: 10.1074/jbc.R110.200329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Zheng Y. Cell type-specific functions of Rho GTPases revealed by gene targeting in mice. Trends Cell Biol. 2007;17:58–64. doi: 10.1016/j.tcb.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 26.Zhou X, Zheng Y. Cell type-specific signaling function of RhoA GTPase: lessons from mouse gene targeting. J Biol Chem. 2013;288:36179–36188. doi: 10.1074/jbc.R113.515486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bourguignon LY, Singleton PA, Zhu H, et al. Hyaluronan-mediated CD44 interaction with RhoGEF and Rho kinase promotes Grb2-associated binder-1 phosphorylation and phosphatidylinositol 3-kinase signaling leading to cytokine (macrophage-colony stimulating factor) production and breast tumor progression. J Biol Chem. 2003;278:29420–29434. doi: 10.1074/jbc.M301885200. [DOI] [PubMed] [Google Scholar]
- 28.Takamura M, Sakamoto M, Genda T, et al. Inhibition of intrahepatic metastasis of human hepatocellular carcinoma by Rho-associated protein kinase inhibitor Y-27632. Hepatology. 2001;33:577–581. doi: 10.1053/jhep.2001.22652. [DOI] [PubMed] [Google Scholar]
- 29.Wu M, Wu ZF, Rosenthal DT, et al. Characterization of the roles of RHOC and RHOA GTPases in invasion, motility, and matrix adhesion in inflammatory and aggressive breast cancers. Cancer. 2010;116:2768–2782. doi: 10.1002/cncr.25181. [DOI] [PubMed] [Google Scholar]
- 30.Attoub S, Noe V, Pirola L, et al. Leptin promotes invasiveness of kidney and colonic epithelial cells via phosphoinositide 3-kinase-, rho-, and rac-dependent signaling pathways. FASEB J. 2000;14:2329–2338. doi: 10.1096/fj.00-0162. [DOI] [PubMed] [Google Scholar]
- 31.Igishi T, Mikami M, Murakami K, et al. Enhancement of cisplatin-induced cytotoxicity by ROCK inhibitor through suppression of focal adhesion kinase-independent mechanism in lung carcinoma cells. Int J Oncol. 2003;23:1079–1085. [PubMed] [Google Scholar]
- 32.Hakem A, Sanchez-Sweatman O, You-Ten A, et al. RhoC is dispensable for embryogenesis and tumor initiation but essential for metastasis. Genes Dev. 2005;19:1974–1979. doi: 10.1101/gad.1310805. * Genetic knockout model of RhoC suggests the role of RhoC in tumor cell metastasis in a breast cancer model. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuent IA, Hu G, Zheng Y. RhoA and RhoC combined are required for K-Ras induced lung adenoma initiation. PLoS One. 2015 doi: 10.1371/journal.pone.0127923. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jung ID, Lee J, Yun SY, et al. Cdc42 and Rac1 are necessary for autotaxin-induced tumor cell motility in A2058 melanoma cells. FEBS Lett. 2002;532:351–356. doi: 10.1016/s0014-5793(02)03698-0. [DOI] [PubMed] [Google Scholar]
- 35.Chan AY, Coniglio SJ, Chuang YY, et al. Roles of the Rac1 and Rac3 GTPases in human tumor cell invasion. Oncogene. 2005;24:7821–7829. doi: 10.1038/sj.onc.1208909. [DOI] [PubMed] [Google Scholar]
- 36.Sequeira L, Dubyk CW, Riesenberger TA, et al. Rho GTPases in PC-3 prostate cancer cell morphology, invasion and tumor cell diapedesis. Clinical & Experimental Metastasis. 2008;25:569–579. doi: 10.1007/s10585-008-9173-3. [DOI] [PubMed] [Google Scholar]
- 37.Bosco EE, Nakai Y, Hennigan RF, et al. NF2-deficient cells depend on the Rac1-canonical Wnt signaling pathway to promote the loss of contact inhibition of proliferation. Oncogene. 2010;29:2540–2549. doi: 10.1038/onc.2010.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kissil JL, Walmsley MJ, Hanlon L, et al. Requirement for Rac1 in a K-ras induced lung cancer in the mouse. Cancer Res. 2007;67:8089–8094. doi: 10.1158/0008-5472.CAN-07-2300. * Genetic model of Rac1 suggests Rac1 is required for tumorigenesis in K-Ras driven lung cancer. [DOI] [PubMed] [Google Scholar]
- 39.Wang Z, Pedersen E, Basse A, et al. Rac1 is crucial for Ras-dependent skin tumor formation by controlling Pak1-Mek-Erk hyperactivation and hyperproliferation in vivo. Oncogene. 2010;29:3362–3373. doi: 10.1038/onc.2010.95. * Genetic model of Rac1 suggests Rac1 is required for tumorigenesis in Ras-dependent skin cancer. [DOI] [PubMed] [Google Scholar]
- 40.D'Amico G, Robinson SD, Germain M, et al. Endothelial-Rac1 is not required for tumor angiogenesis unless alphavbeta3-integrin is absent. PLoS One. 2010;5:e9766. doi: 10.1371/journal.pone.0009766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bouzahzah B, Albanese C, Ahmed F, et al. Rho family GTPases regulate mammary epithelium cell growth and metastasis through distinguishable pathways. Mol Med. 2001;7:816–830. [PMC free article] [PubMed] [Google Scholar]
- 42.Ambrogio C, Voena C, Manazza AD, et al. The anaplastic lymphoma kinase controls cell shape and growth of anaplastic large cell lymphoma through Cdc42 activation. Cancer Res. 2008;68:8899–8907. doi: 10.1158/0008-5472.CAN-08-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stengel KR, Zheng Y. Essential role of Cdc42 in Ras-induced transformation revealed by gene targeting. PLoS One. 2012;7:e37317. doi: 10.1371/journal.pone.0037317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakamori R, Yu S, Zhang X, et al. CDC42 inhibition suppresses progression of incipient intestinal tumors. Cancer Res. 2014;74:5480–5492. doi: 10.1158/0008-5472.CAN-14-0267. * Genetic model of Cdc42 suggests Cdc42 function is required for the malignant progression of early-stage mutant intestinal epithelial cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang L, Wang L, Kalfa TA, et al. Cdc42 critically regulates the balance between myelopoiesis and erythropoiesis. Blood. 2007;110:3853–3861. doi: 10.1182/blood-2007-03-079582. * Conditional knockout of Cdc42 in hematopoietic stem cells results in myeloproliferative disorder. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cook DR, Rossman KL, Der CJ. Rho guanine nucleotide exchange factors: regulators of Rho GTPase activity in development and disease. Oncogene. 2014;33:4021–4035. doi: 10.1038/onc.2013.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang J, Zhang Z, Roe SM, et al. Activation of Rho GTPases by DOCK exchange factors is mediated by a nucleotide sensor. Science. 2009;325:1398–1402. doi: 10.1126/science.1174468. [DOI] [PubMed] [Google Scholar]
- 48.Lazer G, Katzav S. Guanine nucleotide exchange factors for RhoGTPases: good therapeutic targets for cancer therapy? Cell Signal. 2011;23:969–979. doi: 10.1016/j.cellsig.2010.10.022. [DOI] [PubMed] [Google Scholar]
- 49.Kourlas PJ, Strout MP, Becknell B, et al. Identification of a gene at 11q23 encoding a guanine nucleotide exchange factor: evidence for its fusion with MLL in acute myeloid leukemia. Proc Natl Acad Sci U S A. 2000;97:2145–2150. doi: 10.1073/pnas.040569197. * Original identification of LARG gene. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reuther GW, Lambert QT, Booden MA, et al. Leukemia-associated Rho guanine nucleotide exchange factor, a Dbl family protein found mutated in leukemia, causes transformation by activation of RhoA. J Biol Chem. 2001;276:27145–27151. doi: 10.1074/jbc.M103565200. [DOI] [PubMed] [Google Scholar]
- 51.Yuan BZ, Miller MJ, Keck CL, et al. Cloning, characterization, and chromosomal localization of a gene frequently deleted in human liver cancer (DLC-1) homologous to rat RhoGAP. Cancer Res. 1998;58:2196–2199. * Original identification of DLC-1 gene. [PubMed] [Google Scholar]
- 52.Yuan BZ, Jefferson AM, Baldwin KT, et al. DLC-1 operates as a tumor suppressor gene in human non-small cell lung carcinomas. Oncogene. 2004;23:1405–1411. doi: 10.1038/sj.onc.1207291. [DOI] [PubMed] [Google Scholar]
- 53.Yuan BZ, Zhou X, Durkin ME, et al. DLC-1 gene inhibits human breast cancer cell growth and in vivo tumorigenicity. Oncogene. 2003;22:445–450. doi: 10.1038/sj.onc.1206064. [DOI] [PubMed] [Google Scholar]
- 54.Zhou X, Thorgeirsson SS, Popescu NC. Restoration of DLC-1 gene expression induces apoptosis and inhibits both cell growth and tumorigenicity in human hepatocellular carcinoma cells. Oncogene. 2004;23:1308–1313. doi: 10.1038/sj.onc.1207246. [DOI] [PubMed] [Google Scholar]
- 55.Durkin ME, Yuan BZ, Zhou X, et al. DLC-1:a Rho GTPase-activating protein and tumour suppressor. J Cell Mol Med. 2007;11:1185–1207. doi: 10.1111/j.1582-4934.2007.00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tikoo A, Czekay S, Viars C, et al. p190-A, a human tumor suppressor gene, maps to the chromosomal region 19q13.3 that is reportedly deleted in some gliomas. Gene. 2000;257:23–31. doi: 10.1016/s0378-1119(00)00387-5. [DOI] [PubMed] [Google Scholar]
- 57.Wolf RM, Draghi N, Liang X, et al. p190RhoGAP can act to inhibit PDGF-induced gliomas in mice: a putative tumor suppressor encoded on human chromosome 19q13.3. Genes Dev. 2003;17:476–487. doi: 10.1101/gad.1040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gen Y, Yasui K, Zen K, et al. A novel amplification target, ARHGAP5, promotes cell spreading and migration by negatively regulating RhoA in Huh-7 hepatocellular carcinoma cells. Cancer Lett. 2009;275:27–34. doi: 10.1016/j.canlet.2008.09.036. [DOI] [PubMed] [Google Scholar]
- 59.Olson MF, Sahai E. The actin cytoskeleton in cancer cell motility. Clin Exp Metastasis. 2009;26:273–287. doi: 10.1007/s10585-008-9174-2. [DOI] [PubMed] [Google Scholar]
- 60.Rath N, Olson MF. Rho-associated kinases in tumorigenesis: re-considering ROCK inhibition for cancer therapy. EMBO Rep. 2012;13:900–908. doi: 10.1038/embor.2012.127. * Reviews ROCK, its roles in cancer and potential of ROCK inhibitors for cancer therapy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lochhead PA, Wickman G, Mezna M, et al. Activating ROCK1 somatic mutations in human cancer. Oncogene. 2010;29:2591–2598. doi: 10.1038/onc.2010.3. [DOI] [PubMed] [Google Scholar]
- 62.Liu P, Morrison C, Wang L, et al. Identification of somatic mutations in non-small cell lung carcinomas using whole-exome sequencing. Carcinogenesis. 2012;33:1270–1276. doi: 10.1093/carcin/bgs148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Holbrook JD, Parker JS, Gallagher KT, et al. Deep sequencing of gastric carcinoma reveals somatic mutations relevant to personalized medicine. J Transl Med. 2011;9:119. doi: 10.1186/1479-5876-9-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.King H, Nicholas NS, Wells CM. Role of p-21-activated kinases in cancer progression. Int Rev Cell Mol Biol. 2014;309:347–387. doi: 10.1016/B978-0-12-800255-1.00007-7. [DOI] [PubMed] [Google Scholar]
- 65.Kumar R, Gururaj AE, Barnes CJ. p21-activated kinases in cancer. Nat Rev Cancer. 2006;6:459–471. doi: 10.1038/nrc1892. [DOI] [PubMed] [Google Scholar]
- 66.Radu M, Semenova G, Kosoff R, et al. PAK signalling during the development and progression of cancer. Nat Rev Cancer. 2014;14:13–25. doi: 10.1038/nrc3645. * Reviews Pak and its role in cancer development and progression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lundgren K, Holm K, Nordenskjold B, et al. Gene products of chromosome 11q and their association with CCND1 gene amplification and tamoxifen resistance in premenopausal breast cancer. Breast Cancer Res. 2008;10:R81. doi: 10.1186/bcr2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brown LA, Kalloger SE, Miller MA, et al. Amplification of 11q13 in ovarian carcinoma. Genes Chromosomes Cancer. 2008;47:481–489. doi: 10.1002/gcc.20549. [DOI] [PubMed] [Google Scholar]
- 69.Ong CC, Jubb AM, Jakubiak D, et al. P21-activated kinase 1 (PAK1) as a therapeutic target in BRAF wild-type melanoma. J Natl Cancer Inst. 2013;105:606–607. doi: 10.1093/jnci/djt054. [DOI] [PubMed] [Google Scholar]
- 70.Chen S, Auletta T, Dovirak O, et al. Copy number alterations in pancreatic cancer identify recurrent PAK4 amplification. Cancer Biol Ther. 2008;7:1793–1802. doi: 10.4161/cbt.7.11.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mahlamaki EH, Kauraniemi P, Monni O, et al. High-resolution genomic and expression profiling reveals 105 putative amplification target genes in pancreatic cancer. Neoplasia. 2004;6:432–439. doi: 10.1593/neo.04130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Begum A, Imoto I, Kozaki K, et al. Identification of PAK4 as a putative target gene for amplification within 19q13.12-q13.2 in oral squamous-cell carcinoma. Cancer Sci. 2009;100:1908–1916. doi: 10.1111/j.1349-7006.2009.01252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fawdar S, Trotter EW, Li Y, et al. Targeted genetic dependency screen facilitates identification of actionable mutations in FGFR4, MAP3K9, and PAK5 in lung cancer. Proc Natl Acad Sci U S A. 2013;110:12426–12431. doi: 10.1073/pnas.1305207110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Whale AD, Dart A, Holt M, et al. PAK4 kinase activity and somatic mutation promote carcinoma cell motility and influence inhibitor sensitivity. Oncogene. 2013;32:2114–2120. doi: 10.1038/onc.2012.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bokoch GM. Biology of the p21-activated kinases. Annu Rev Biochem. 2003;72:743–781. doi: 10.1146/annurev.biochem.72.121801.161742. [DOI] [PubMed] [Google Scholar]
- 76.Yang N, Higuchi O, Ohashi K, et al. Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization. Nature. 1998;393:809–812. doi: 10.1038/31735. [DOI] [PubMed] [Google Scholar]
- 77.Delorme-Walker VD, Peterson JR, Chernoff J, et al. Pak1 regulates focal adhesion strength, myosin IIA distribution, and actin dynamics to optimize cell migration. J Cell Biol. 2011;193:1289–1303. doi: 10.1083/jcb.201010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Adam L, Vadlamudi R, Mandal M, et al. Regulation of microfilament reorganization and invasiveness of breast cancer cells by kinase dead p21-activated kinase-1. J Biol Chem. 2000;275:12041–12050. doi: 10.1074/jbc.275.16.12041. [DOI] [PubMed] [Google Scholar]
- 79.Qu J, Cammarano MS, Shi Q, et al. Activated PAK4 regulates cell adhesion and anchorage-independent growth. Mol Cell Biol. 2001;21:3523–3533. doi: 10.1128/MCB.21.10.3523-3533.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shrestha Y, Schafer EJ, Boehm JS, et al. PAK1 is a breast cancer oncogene that coordinately activates MAPK and MET signaling. Oncogene. 2012;31:3397–3408. doi: 10.1038/onc.2011.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tabusa H, Brooks T, Massey AJ. Knockdown of PAK4 or PAK1 inhibits the proliferation of mutant KRAS colon cancer cells independently of RAF/MEK/ERK and PI3K/AKT signaling. Mol Cancer Res. 2013;11:109–121. doi: 10.1158/1541-7786.MCR-12-0466. [DOI] [PubMed] [Google Scholar]
- 82.He H, Shulkes A, Baldwin GS. PAK1 interacts with beta-catenin and is required for the regulation of the beta-catenin signalling pathway by gastrins. Gastroenterology. 2008;134:A249–A250. doi: 10.1016/j.bbamcr.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 83.Wong LE, Reynolds AB, Dissanayaka NT, et al. p120-Catenin Is a Binding Partner and Substrate for Group B Pak Kinases. Journal of Cellular Biochemistry. 2010;110:1244–1254. doi: 10.1002/jcb.22639. [DOI] [PubMed] [Google Scholar]
- 84.Wang RA, Mazumdar A, Vadlamudi RK, et al. P21-activated kinase-1 phosphorylates and transactivates estrogen receptor-alpha and promotes hyperplasia in mammary epithelium. EMBO J. 2002;21:5437–5447. doi: 10.1093/emboj/cdf543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schrantz N, da Silva Correia J, Fowler B, et al. Mechanism of p21-activated kinase 6-mediated inhibition of androgen receptor signaling. J Biol Chem. 2004;279:1922–1931. doi: 10.1074/jbc.M311145200. [DOI] [PubMed] [Google Scholar]
- 86.Schurmann A, Mooney AF, Sanders LC, et al. p21-activated kinase 1 phosphorylates the death agonist bad and protects cells from apoptosis. Mol Cell Biol. 2000;20:453–461. doi: 10.1128/mcb.20.2.453-461.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang Z, Rayala S, Nguyen D, et al. Pak1 phosphorylation of snail, a master regulator of epithelial-to-mesenchyme transition, modulates snail's subcellular localization and functions. Cancer Res. 2005;65:3179–3184. doi: 10.1158/0008-5472.CAN-04-3480. [DOI] [PubMed] [Google Scholar]
- 88.Aktories K, Schmidt G, Just I. Rho GTPases as targets of bacterial protein toxins. Biol Chem. 2000;381:421–426. doi: 10.1515/BC.2000.054. [DOI] [PubMed] [Google Scholar]
- 89.Gao Y, Dickerson JB, Guo F, et al. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci U S A. 2004;101:7618–7623. doi: 10.1073/pnas.0307512101. * First-generation small molecule inhibitor of Rac activation by targeting GEF-binding pocket on Rac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Florian MC, Dorr K, Niebel A, et al. Cdc42 activity regulates hematopoietic stem cell aging and rejuvenation. Cell Stem Cell. 2012;10:520–530. doi: 10.1016/j.stem.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Evelyn CR, Ferng T, Rojas RJ, et al. High-throughput screening for small-molecule inhibitors of LARG-stimulated RhoA nucleotide binding via a novel fluorescence polarization assay. J Biomol Screen. 2009;14:161–172. doi: 10.1177/1087057108328761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Shang X, Marchioni F, Evelyn CR, et al. Small-molecule inhibitors targeting G-protein-coupled Rho guanine nucleotide exchange factors. Proc Natl Acad Sci U S A. 2013;110:3155–3160. doi: 10.1073/pnas.1212324110. * Small molecule inhibitor of RhoA activation by targeting RhoA-binding surface on RhoGEF LARG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shutes A, Onesto C, Picard V, et al. Specificity and mechanism of action of EHT 1864, a novel small molecule inhibitor of Rac family small GTPases. J Biol Chem. 2007;282:35666–35678. doi: 10.1074/jbc.M703571200. * Inhibitor of Rac subfamily that inhibits nucleotide association. [DOI] [PubMed] [Google Scholar]
- 94.Hong L, Kenney SR, Phillips GK, et al. Characterization of a Cdc42 protein inhibitor and its use as a molecular probe. J Biol Chem. 2013;288:8531–8543. doi: 10.1074/jbc.M112.435941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Caloca MJ, Garcia-Bermejo ML, Blumberg PM, et al. beta2-chimaerin is a novel target for diacylglycerol: binding properties and changes in subcellular localization mediated by ligand binding to its C1 domain. Proc Natl Acad Sci U S A. 1999;96:11854–11859. doi: 10.1073/pnas.96.21.11854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang C, Liu Y, Leskow FC, et al. Rac-GAP-dependent inhibition of breast cancer cell proliferation by {beta}2-chimerin. J Biol Chem. 2005;280:24363–24370. doi: 10.1074/jbc.M411629200. [DOI] [PubMed] [Google Scholar]
- 97.Colon-Gonzalez F, Kazanietz MG. C1 domains exposed: from diacylglycerol binding to protein-protein interactions. Biochim Biophys Acta. 2006;1761:827–837. doi: 10.1016/j.bbalip.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 98.Duan WG, Yuan ST, Liao H, et al. Advances in the study of Rho kinase and its inhibitors. Yao Xue Xue Bao. 2007;42:1013–1022. [PubMed] [Google Scholar]
- 99.Guan R, Xu X, Chen M, et al. Advances in the studies of roles of Rho/Rho-kinase in diseases and the development of its inhibitors. Eur J Med Chem. 2013;70:613–622. doi: 10.1016/j.ejmech.2013.10.048. [DOI] [PubMed] [Google Scholar]
- 100.Uehata M, Ishizaki T, Satoh H, et al. Calcium sensitization of smooth muscle mediated by a Rho-associated protein kinase in hypertension. Nature. 1997;389:990–994. doi: 10.1038/40187. * Identification of the first generation ROCK inhibitor. [DOI] [PubMed] [Google Scholar]
- 101.Routhier A, Astuccio M, Lahey D, et al. Pharmacological inhibition of Rho-kinase signaling with Y-27632 blocks melanoma tumor growth. Oncol Rep. 2010;23:861–867. [PubMed] [Google Scholar]
- 102.Nagumo H, Sasaki Y, Ono Y, et al. Rho kinase inhibitor HA-1077 prevents Rho-mediated myosin phosphatase inhibition in smooth muscle cells. Am J Physiol Cell Physiol. 2000;278:C57-65. doi: 10.1152/ajpcell.2000.278.1.C57. * Characterization of fasudil as ROCK inhibitor. [DOI] [PubMed] [Google Scholar]
- 103.Ying H, Biroc SL, Li WW, et al. The Rho kinase inhibitor fasudil inhibits tumor progression in human and rat tumor models. Mol Cancer Ther. 2006;5:2158–2164. doi: 10.1158/1535-7163.MCT-05-0440. [DOI] [PubMed] [Google Scholar]
- 104.Nakajima M, Hayashi K, Egi Y, et al. Effect of Wf-536, a novel ROCK inhibitor, against metastasis of B16 melanoma. Cancer Chemother Pharmacol. 2003;52:319–324. doi: 10.1007/s00280-003-0641-9. [DOI] [PubMed] [Google Scholar]
- 105.Sasaki Y, Suzuki M, Hidaka H. The novel and specific Rho-kinase inhibitor (S)-(+)-2-methyl-1-[(4-methyl-5-isoquinoline)sulfonyl]-homopiperazine as a probing molecule for Rho-kinase-involved pathway. Pharmacol Ther. 2002;93:225–232. doi: 10.1016/s0163-7258(02)00191-2. [DOI] [PubMed] [Google Scholar]
- 106.Patel RA, Forinash KD, Pireddu R, et al. RKI-1447 is a potent inhibitor of the Rho-associated ROCK kinases with anti-invasive and antitumor activities in breast cancer. Cancer Res. 2012;72:5025–5034. doi: 10.1158/0008-5472.CAN-12-0954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tanihara H, Inatani M, Honjo M, et al. Intraocular pressure-lowering effects and safety of topical administration of a selective ROCK inhibitor, SNJ-1656, in healthy volunteers. Arch Ophthalmol. 2008;126:309–315. doi: 10.1001/archophthalmol.2007.76. [DOI] [PubMed] [Google Scholar]
- 108.Tanihara H, Inoue T, Yamamoto T, et al. Phase 2 randomized clinical study of a Rho kinase inhibitor, K-115, in primary open-angle glaucoma and ocular hypertension. Am J Ophthalmol. 2013;156:731–736. doi: 10.1016/j.ajo.2013.05.016. [DOI] [PubMed] [Google Scholar]
- 109.Williams RD, Novack GD, van Haarlem T, et al. Ocular hypotensive effect of the Rho kinase inhibitor AR-12286 in patients with glaucoma and ocular hypertension. Am J Ophthalmol. 2011;152:834–841. doi: 10.1016/j.ajo.2011.04.012. e831. [DOI] [PubMed] [Google Scholar]
- 110.Lohn M, Plettenburg O, Ivashchenko Y, et al. Pharmacological characterization of SAR407899, a novel rho-kinase inhibitor. Hypertension. 2009;54:676–683. doi: 10.1161/HYPERTENSIONAHA.109.134353. [DOI] [PubMed] [Google Scholar]
- 111.Sadok A, McCarthy A, Caldwell J, et al. Rho kinase inhibitors block melanoma cell migration and inhibit metastasis. Cancer Res. 2015 doi: 10.1158/0008-5472.CAN-14-2156. [DOI] [PubMed] [Google Scholar]
- 112.Yap TA, Walton MI, Grimshaw KM, et al. AT13148 is a novel, oral multi-AGC kinase inhibitor with potent pharmacodynamic and antitumor activity. Clin Cancer Res. 2012;18:3912–3923. doi: 10.1158/1078-0432.CCR-11-3313. * A multi-AGC kinase inhibitor that also inhibits ROCK activity, is in Phase I clinical trial for cancer. [DOI] [PubMed] [Google Scholar]
- 113.Kaneko M, Saito Y, Saito H, et al. Neurotrophic 3,9-bis[(alkylthio)methyl]-and-bis(alkoxymethyl)-K-252a derivatives. J Med Chem. 1997;40:1863–1869. doi: 10.1021/jm970031d. * Identification of first generation inhibitor for PAK. [DOI] [PubMed] [Google Scholar]
- 114.Nheu TV, He H, Hirokawa Y, et al. The K252a derivatives, inhibitors for the PAK/MLK kinase family selectively block the growth of RAS transformants. Cancer J. 2002;8:328–336. doi: 10.1097/00130404-200207000-00009. [DOI] [PubMed] [Google Scholar]
- 115.Porchia LM, Guerra M, Wang YC, et al. 2-amino-N-{4-[5-(2-phenanthrenyl)-3-(trifluoromethyl)-1H-pyrazol-1-yl]-phenyl} acetamide (OSU-03012), a celecoxib derivative, directly targets p21-activated kinase. Mol Pharmacol. 2007;72:1124–1131. doi: 10.1124/mol.107.037556. [DOI] [PubMed] [Google Scholar]
- 116.Maksimoska J, Feng L, Harms K, et al. Targeting large kinase active site with rigid, bulky octahedral ruthenium complexes. J Am Chem Soc. 2008;130:15764–15765. doi: 10.1021/ja805555a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Murray BW, Guo C, Piraino J, et al. Small-molecule p21-activated kinase inhibitor PF-3758309 is a potent inhibitor of oncogenic signaling and tumor growth. Proc Natl Acad Sci U S A. 2010;107:9446–9451. doi: 10.1073/pnas.0911863107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang J, Wang J, Guo Q, et al. LCH-7749944, a novel and potent p21-activated kinase 4 inhibitor, suppresses proliferation and invasion in human gastric cancer cells. Cancer Lett. 2012;317:24–32. doi: 10.1016/j.canlet.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 119.Licciulli S, Maksimoska J, Zhou C, et al. FRAX597, a small molecule inhibitor of the p21-activated kinases, inhibits tumorigenesis of neurofibromatosis type 2 (NF2)-associated Schwannomas. J Biol Chem. 2013;288:29105–29114. doi: 10.1074/jbc.M113.510933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Viaud J, Peterson JR. An allosteric kinase inhibitor binds the p21-activated kinase autoregulatory domain covalently. Mol Cancer Ther. 2009;8:2559–2565. doi: 10.1158/1535-7163.MCT-09-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ong CC, Jubb AM, Haverty PM, et al. Targeting p21-activated kinase 1 (PAK1) to induce apoptosis of tumor cells. Proc Natl Acad Sci U S A. 2011;108:7177–7182. doi: 10.1073/pnas.1103350108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Flaiz C, Chernoff J, Ammoun S, et al. PAK kinase regulates Rac GTPase and is a potential target in human schwannomas. Exp Neurol. 2009;218:137–144. doi: 10.1016/j.expneurol.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bosco EE, Kumar S, Marchioni F, et al. Rational design of small molecule inhibitors targeting the Rac GTPase-p67(phox) signaling axis in inflammation. Chem Biol. 2012;19:228–242. doi: 10.1016/j.chembiol.2011.12.017. * Small molecule inhibitor of p67phox activation by targeting Rac/p67phox interaction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Michaelson D, Ali W, Chiu VK, et al. Postprenylation CAAX processing is required for proper localization of Ras but not Rho GTPases. Mol Biol Cell. 2005;16:1606–1616. doi: 10.1091/mbc.E04-11-0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bergo MO, Gavino BJ, Hong C, et al. Inactivation of Icmt inhibits transformation by oncogenic K-Ras and B-Raf. J Clin Invest. 2004;113:539–550. doi: 10.1172/JCI18829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Holstein SA, Hohl RJ. Is there a future for prenyltransferase inhibitors in cancer therapy? Curr Opin Pharmacol. 2012;12:704–709. doi: 10.1016/j.coph.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 127.Bredel M, Pollack IF, Freund JM, et al. Inhibition of Ras and related G-proteins as a therapeutic strategy for blocking malignant glioma growth. Neurosurgery. 1998;43:124–131. doi: 10.1097/00006123-199807000-00081. discussion 131-122. [DOI] [PubMed] [Google Scholar]
- 128.Miquel K, Pradines A, Sun J, et al. GGTI-298 induces G0-G1 block and apoptosis whereas FTI-277 causes G2-M enrichment in A549 cells. Cancer Res. 1997;57:1846–1850. [PubMed] [Google Scholar]
- 129.Sun J, Ohkanda J, Coppola D, et al. Geranylgeranyltransferase I inhibitor GGTI-2154 induces breast carcinoma apoptosis and tumor regression in H-Ras transgenic mice. Cancer Res. 2003;63:8922–8929. [PubMed] [Google Scholar]
- 130.Woo JT, Nakagawa H, Krecic AM, et al. Inhibitory effects of mevastatin and a geranylgeranyl transferase I inhibitor (GGTI-2166) on mononuclear osteoclast formation induced by receptor activator of NF kappa B ligand (RANKL) or tumor necrosis factor-alpha (TNF-alpha) Biochem Pharmacol. 2005;69:87–95. doi: 10.1016/j.bcp.2004.08.036. [DOI] [PubMed] [Google Scholar]
- 131.Peterson YK, Kelly P, Weinbaum CA, et al. A novel protein geranylgeranyltransferase-I inhibitor with high potency, selectivity, and cellular activity. J Biol Chem. 2006;281:12445–12450. doi: 10.1074/jbc.M600168200. [DOI] [PubMed] [Google Scholar]
- 132.Watanabe M, Fiji HD, Guo L, et al. Inhibitors of protein geranylgeranyltransferase I and Rab geranylgeranyltransferase identified from a library of allenoate-derived compounds. J Biol Chem. 2008;283:9571–9579. doi: 10.1074/jbc.M706229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chan LN, Fiji HD, Watanabe M, et al. Identification and characterization of mechanism of action of P61-E7, a novel phosphine catalysis-based inhibitor of geranylgeranyltransferase-I. PLoS One. 2011;6:e26135. doi: 10.1371/journal.pone.0026135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lu J, Chan L, Fiji HD, et al. In vivo antitumor effect of a novel inhibitor of protein geranylgeranyltransferase-I. Mol Cancer Ther. 2009;8:1218–1226. doi: 10.1158/1535-7163.MCT-08-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zimonjic DB, Chan LN, Tripathi V, et al. In vitro and in vivo effects of geranylgeranyltransferase I inhibitor P61A6 on non-small cell lung cancer cells. BMC Cancer. 2013;13:198. doi: 10.1186/1471-2407-13-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Berndt N, Hamilton AD, Sebti SM. Targeting protein prenylation for cancer therapy. Nat Rev Cancer. 2011;11:775–791. doi: 10.1038/nrc3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Jacobs M, Hayakawa K, Swenson L, et al. The structure of dimeric ROCK I reveals the mechanism for ligand selectivity. J Biol Chem. 2006;281:260–268. doi: 10.1074/jbc.M508847200. [DOI] [PubMed] [Google Scholar]
- 138.Li R, Martin MP, Liu Y, et al. Fragment-based and structure-guided discovery and optimization of Rho kinase inhibitors. J Med Chem. 2012;55:2474–2478. doi: 10.1021/jm201289r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rudolph J, Crawford JJ, Hoeflich KP, et al. Inhibitors of p21-Activated Kinases (PAKs) J Med Chem. 2015;58:111–129. doi: 10.1021/jm501613q. [DOI] [PubMed] [Google Scholar]
- 140.Baker NM, Der CJ. Cancer: Drug for an 'undruggable' protein. Nature. 2013;497:577–578. doi: 10.1038/nature12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Deacon SW, Beeser A, Fukui JA, et al. An isoform-selective, small-molecule inhibitor targets the autoregulatory mechanism of p21-activated kinase. Chem Biol. 2008;15:322–331. doi: 10.1016/j.chembiol.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kakiuchi M, Nishizawa T, Ueda H, et al. Recurrent gain-of-function mutations of RHOA in diffuse-type gastric carcinoma. Nat Genet. 2014;46:583–587. doi: 10.1038/ng.2984. * Identification of RhoA mutations in diffuse-type gastric carcinoma. [DOI] [PubMed] [Google Scholar]
- 144.Manso R, Sanchez-Beato M, Monsalvo S, et al. The RHOA G17V gene mutation occurs frequently in peripheral T-cell lymphoma and is associated with a characteristic molecular signature. Blood. 2014;123:2893–2894. doi: 10.1182/blood-2014-02-555946. * Identification of RhoA G17V mutation in peripheral T-cell lymphoma. [DOI] [PubMed] [Google Scholar]
- 145.Rohde M, Richter J, Schlesner M, et al. Recurrent RHOA Mutations in Pediatric Burkitt Lymphoma Treated According to the NHL-BFM Protocols. Genes Chromosomes & Cancer. 2014;53:911–916. doi: 10.1002/gcc.22202. * Identification of RhoA mutations in pediatric Burkitt lymphoma. [DOI] [PubMed] [Google Scholar]
- 146.Yoo HY, Sung MK, Lee SH, et al. A recurrent inactivating mutation in RHOA GTPase in angioimmunoblastic T cell lymphoma. Nat Genet. 2014;46:371–375. doi: 10.1038/ng.2916. * Identification of RhoA mutation in angioimmunoblastic T cell lymphoma. [DOI] [PubMed] [Google Scholar]
- 147.Pan Y, Bi F, Liu N, et al. Expression of seven main Rho family members in gastric carcinoma. Biochem Biophys Res Commun. 2004;315:686–691. doi: 10.1016/j.bbrc.2004.01.108. [DOI] [PubMed] [Google Scholar]
- 148.Kamai T, Yamanishi T, Shirataki H, et al. Overexpression of RhoA, Rac1, and Cdc42 GTPases is associated with progression in testicular cancer. Clin Cancer Res. 2004;10:4799–4805. doi: 10.1158/1078-0432.CCR-0436-03. [DOI] [PubMed] [Google Scholar]
- 149.Abraham MT, Kuriakose MA, Sacks PG, et al. Motility-related proteins as markers for head and neck squamous cell cancer. Laryngoscope. 2001;111:1285–1289. doi: 10.1097/00005537-200107000-00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kamai T, Tsujii T, Arai K, et al. Significant association of Rho/ROCK pathway with invasion and metastasis of bladder cancer. Clin Cancer Res. 2003;9:2632–2641. [PubMed] [Google Scholar]
- 151.Fritz G, Just I, Kaina B. Rho GTPases are over-expressed in human tumors. Int J Cancer. 1999;81:682–687. doi: 10.1002/(sici)1097-0215(19990531)81:5<682::aid-ijc2>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 152.Fritz G, Brachetti C, Bahlmann F, et al. Rho GTPases in human breast tumours: expression and mutation analyses and correlation with clinical parameters. Br J Cancer. 2002;87:635–644. doi: 10.1038/sj.bjc.6600510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Davis MJ, Ha BH, Holman EC, et al. RAC1P29S is a spontaneously activating cancer-associated GTPase. Proc Natl Acad Sci U S A. 2013;110:912–917. doi: 10.1073/pnas.1220895110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Krauthammer M, Kong Y, Ha BH, et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat Genet. 2012;44:1006–1014. doi: 10.1038/ng.2359. * Identification of Rac1 mutations in melanoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Stransky N, Egloff AM, Tward AD, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. * Identification of Rac1 mutations in head and neck squamous cell carcinoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Pickering CR, Zhou JH, Lee JJ, et al. Mutational landscape of aggressive cutaneous squamous cell carcinoma. Clin Cancer Res. 2014;20:6582–6592. doi: 10.1158/1078-0432.CCR-14-1768. * Identification of Rac1 mutations in cutaneous squamous cell carcinoma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Shieh DB, Godleski J, Herndon JE, et al. Cell motility as a prognostic factor in Stage I nonsmall cell lung carcinoma: the role of gelsolin expression. Cancer. (2nd) 1999;85:47–57. doi: 10.1002/(sici)1097-0142(19990101)85:1<47::aid-cncr7>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 158.Liu SY, Yen CY, Yang SC, et al. Overexpression of Rac-1 small GTPase binding protein in oral squamous cell carcinoma. J Oral Maxillofac Surg. 2004;62:702–707. doi: 10.1016/j.joms.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 159.Eisenmann KM, McCarthy JB, Simpson MA, et al. Melanoma chondroitin sulphate proteoglycan regulates cell spreading through Cdc42, Ack-1 and p130cas. Nat Cell Biol. 1999;1:507–513. doi: 10.1038/70302. [DOI] [PubMed] [Google Scholar]
- 160.Gomez Del Pulgar T, Valdes-Mora F, Bandres E, et al. Cdc42 is highly expressed in colorectal adenocarcinoma and downregulates ID4 through an epigenetic mechanism. Int J Oncol. 2008;33:185–193. [PubMed] [Google Scholar]
- 161.Liu Y, Wang Y, Zhang Y, et al. Abnormal expression of p120-catenin, E-cadherin, and small GTPases is significantly associated with malignant phenotype of human lung cancer. Lung Cancer. 2009;63:375–382. doi: 10.1016/j.lungcan.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 162.Qin J, Xie Y, Wang B, et al. Upregulation of PIP3-dependent Rac exchanger 1 (P-Rex1) promotes prostate cancer metastasis. Oncogene. 2009;28:1853–1863. doi: 10.1038/onc.2009.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sosa MS, Lopez-Haber C, Yang C, et al. Identification of the Rac-GEF P-Rex1 as an essential mediator of ErbB signaling in breast cancer. Mol Cell. 2010;40:877–892. doi: 10.1016/j.molcel.2010.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Shields JM, Thomas NE, Cregger M, et al. Lack of extracellular signal-regulated kinase mitogen-activated protein kinase signaling shows a new type of melanoma. Cancer Res. 2007;67:1502–1512. doi: 10.1158/0008-5472.CAN-06-3311. [DOI] [PubMed] [Google Scholar]
- 165.Fine B, Hodakoski C, Koujak S, et al. Activation of the PI3K pathway in cancer through inhibition of PTEN by exchange factor P-REX2a. Science. 2009;325:1261–1265. doi: 10.1126/science.1173569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Sano M, Genkai N, Yajima N, et al. Expression level of ECT2 proto-oncogene correlates with prognosis in glioma patients. Oncol Rep. 2006;16:1093–1098. [PubMed] [Google Scholar]
- 167.Hirata D, Yamabuki T, Miki D, et al. Involvement of epithelial cell transforming sequence-2 oncoantigen in lung and esophageal cancer progression. Clin Cancer Res. 2009;15:256–266. doi: 10.1158/1078-0432.CCR-08-1672. [DOI] [PubMed] [Google Scholar]
- 168.Jung Y, Lee S, Choi HS, et al. Clinical validation of colorectal cancer biomarkers identified from bioinformatics analysis of public expression data. Clin Cancer Res. 2011;17:700–709. doi: 10.1158/1078-0432.CCR-10-1300. [DOI] [PubMed] [Google Scholar]
- 169.Zhang ML, Lu S, Zhou L, et al. Correlation between ECT2 gene expression and methylation change of ECT2 promoter region in pancreatic cancer. Hepatobiliary Pancreat Dis Int. 2008;7:533–538. [PubMed] [Google Scholar]
- 170.Justilien V, Fields AP. Ect2 links the PKCiota-Par6alpha complex to Rac1 activation and cellular transformation. Oncogene. 2009;28:3597–3607. doi: 10.1038/onc.2009.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Salhia B, Tran NL, Chan A, et al. The guanine nucleotide exchange factors trio, Ect2, and Vav3 mediate the invasive behavior of glioblastoma. Am J Pathol. 2008;173:1828–1838. doi: 10.2353/ajpath.2008.080043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Lane J, Martin TA, Mansel RE, et al. The expression and prognostic value of the guanine nucleotide exchange factors (GEFs) Trio, Vav1 and TIAM-1 in human breast cancer. Int Semin Surg Oncol. 2008;5:23. doi: 10.1186/1477-7800-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yoshizuka N, Moriuchi R, Mori T, et al. An alternative transcript derived from the trio locus encodes a guanosine nucleotide exchange factor with mouse cell-transforming potential. J Biol Chem. 2004;279:43998–44004. doi: 10.1074/jbc.M406082200. [DOI] [PubMed] [Google Scholar]
- 174.Fernandez-Zapico ME, Gonzalez-Paz NC, Weiss E, et al. Ectopic expression of VAV1 reveals an unexpected role in pancreatic cancer tumorigenesis. Cancer Cell. 2005;7:39–49. doi: 10.1016/j.ccr.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 175.Hornstein I, Pikarsky E, Groysman M, et al. The haematopoietic specific signal transducer Vav1 is expressed in a subset of human neuroblastomas. J Pathol. 2003;199:526–533. doi: 10.1002/path.1314. [DOI] [PubMed] [Google Scholar]
- 176.Fujikawa K, Miletic AV, Alt FW, et al. Vav1/2/3-null mice define an essential role for Vav family proteins in lymphocyte development and activation but a differential requirement in MAPK signaling in T and B cells. J Exp Med. 2003;198:1595–1608. doi: 10.1084/jem.20030874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bartolome RA, Molina-Ortiz I, Samaniego R, et al. Activation of Vav/Rho GTPase signaling by CXCL12 controls membrane-type matrix metalloproteinase-dependent melanoma cell invasion. Cancer Res. 2006;66:248–258. doi: 10.1158/0008-5472.CAN-05-2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Patel V, Rosenfeldt HM, Lyons R, et al. Persistent activation of Rac1 in squamous carcinomas of the head and neck: evidence for an EGFR/Vav2 signaling axis involved in cell invasion. Carcinogenesis. 2007;28:1145–1152. doi: 10.1093/carcin/bgm008. [DOI] [PubMed] [Google Scholar]
- 179.Bourguignon LY, Zhu H, Zhou B, et al. Hyaluronan promotes CD44v3-Vav2 interaction with Grb2-p185(HER2) and induces Rac1 and Ras signaling during ovarian tumor cell migration and growth. J Biol Chem. 2001;276:48679–48692. doi: 10.1074/jbc.M106759200. [DOI] [PubMed] [Google Scholar]
- 180.Miller SL, DeMaria JE, Freier DO, et al. Novel association of Vav2 and Nek3 modulates signaling through the human prolactin receptor. Mol Endocrinol. 2005;19:939–949. doi: 10.1210/me.2004-0443. [DOI] [PubMed] [Google Scholar]
- 181.Dong Z, Liu Y, Lu S, et al. Vav3 oncogene is overexpressed and regulates cell growth and androgen receptor activity in human prostate cancer. Mol Endocrinol. 2006;20:2315–2325. doi: 10.1210/me.2006-0048. [DOI] [PubMed] [Google Scholar]
- 182.Adam L, Vadlamudi RK, McCrea P, et al. Tiam1 overexpression potentiates heregulin-induced lymphoid enhancer factor-1/beta -catenin nuclear signaling in breast cancer cells by modulating the intercellular stability. J Biol Chem. 2001;276:28443–28450. doi: 10.1074/jbc.M009769200. [DOI] [PubMed] [Google Scholar]
- 183.Jin H, Li T, Ding Y, et al. Methylation status of T-lymphoma invasion and metastasis 1 promoter and its overexpression in colorectal cancer. Hum Pathol. 2011;42:541–551. doi: 10.1016/j.humpath.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 184.Minard ME, Ellis LM, Gallick GE. Tiam1 regulates cell adhesion, migration and apoptosis in colon tumor cells. Clin Exp Metastasis. 2006;23:301–313. doi: 10.1007/s10585-006-9040-z. [DOI] [PubMed] [Google Scholar]
- 185.Stebel A, Brachetti C, Kunkel M, et al. Progression of breast tumors is accompanied by a decrease in expression of the Rho guanine exchange factor Tiam1. Oncol Rep. 2009;21:217–222. [PubMed] [Google Scholar]
- 186.Uhlenbrock K, Eberth A, Herbrand U, et al. The RacGEF Tiam1 inhibits migration and invasion of metastatic melanoma via a novel adhesive mechanism. J Cell Sci. 2004;117:4863–4871. doi: 10.1242/jcs.01367. [DOI] [PubMed] [Google Scholar]
- 187.Shtivelman E, Lifshitz B, Gale RP, et al. Fused transcript of abl and bcr genes in chronic myelogenous leukaemia. Nature. 1985;315:550–554. doi: 10.1038/315550a0. [DOI] [PubMed] [Google Scholar]
- 188.Jarzynka MJ, Hu B, Hui KM, et al. ELMO1 and Dock180, a bipartite Rac1 guanine nucleotide exchange factor, promote human glioma cell invasion. Cancer Res. 2007;67:7203–7211. doi: 10.1158/0008-5472.CAN-07-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Jones S, Zhang X, Parsons DW, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ng IO, Liang ZD, Cao L, et al. DLC-1 is deleted in primary hepatocellular carcinoma and exerts inhibitory effects on the proliferation of hepatoma cell lines with deleted DLC-1. Cancer Res. 2000;60:6581–6584. [PubMed] [Google Scholar]
- 191.Ullmannova V, Popescu NC. Expression profile of the tumor suppressor genes DLC-1 and DLC-2 in solid tumors. Int J Oncol. 2006;29:1127–1132. [PubMed] [Google Scholar]
- 192.Yuan BZ, Durkin ME, Popescu NC. Promoter hypermethylation of DLC-1, a candidate tumor suppressor gene, in several common human cancers. Cancer Genet Cytogenet. 2003;140:113–117. doi: 10.1016/s0165-4608(02)00674-x. [DOI] [PubMed] [Google Scholar]
- 193.Jones MB, Krutzsch H, Shu H, et al. Proteomic analysis and identification of new biomarkers and therapeutic targets for invasive ovarian cancer. Proteomics. 2002;2:76–84. [PubMed] [Google Scholar]
- 194.Zhao L, Wang H, Li J, et al. Overexpression of Rho GDP-dissociation inhibitor alpha is associated with tumor progression and poor prognosis of colorectal cancer. J Proteome Res. 2008;7:3994–4003. doi: 10.1021/pr800271b. [DOI] [PubMed] [Google Scholar]
- 195.Ding J, Huang S, Wu S, et al. Gain of miR-151 on chromosome 8q24.3 facilitates tumour cell migration and spreading through downregulating RhoGDIA. Nat Cell Biol. 2010;12:390–399. doi: 10.1038/ncb2039. [DOI] [PubMed] [Google Scholar]
- 196.Forget MA, Desrosiers RR, Del M, et al. The expression of rho proteins decreases with human brain tumor progression: potential tumor markers. Clin Exp Metastasis. 2002;19:9–15. doi: 10.1023/a:1013884426692. [DOI] [PubMed] [Google Scholar]
- 197.Liang L, Li Q, Huang LY, et al. Loss of ARHGDIA expression is associated with poor prognosis in HCC and promotes invasion and metastasis of HCC cells. Int J Oncol. 2014;45:659–666. doi: 10.3892/ijo.2014.2451. [DOI] [PubMed] [Google Scholar]
- 198.Jiang WG, Watkins G, Lane J, et al. Prognostic value of rho GTPases and rho guanine nucleotide dissociation inhibitors in human breast cancers. Clin Cancer Res. 2003;9:6432–6440. [PubMed] [Google Scholar]
- 199.Abiatari I, DeOliveira T, Kerkadze V, et al. Consensus transcriptome signature of perineural invasion in pancreatic carcinoma. Mol Cancer Ther. 2009;8:1494–1504. doi: 10.1158/1535-7163.MCT-08-0755. [DOI] [PubMed] [Google Scholar]
- 200.Tapper J, Kettunen E, El-Rifai W, et al. Changes in gene expression during progression of ovarian carcinoma. Cancer Genet Cytogenet. 2001;128:1–6. doi: 10.1016/s0165-4608(01)00386-7. [DOI] [PubMed] [Google Scholar]
- 201.Seraj MJ, Harding MA, Gildea JJ, et al. The relationship of BRMS1 and RhoGDI2 gene expression to metastatic potential in lineage related human bladder cancer cell lines. Clin Exp Metastasis. 2000;18:519–525. doi: 10.1023/a:1011819621859. [DOI] [PubMed] [Google Scholar]
- 202.Moissoglu K, McRoberts KS, Meier JA, et al. Rho GDP dissociation inhibitor 2 suppresses metastasis via unconventional regulation of RhoGTPases. Cancer Res. 2009;69:2838–2844. doi: 10.1158/0008-5472.CAN-08-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Hu LD, Zou HF, Zhan SX, et al. Biphasic expression of RhoGDI2 in the progression of breast cancer and its negative relation with lymph node metastasis. Oncol Rep. 2007;17:1383–1389. [PubMed] [Google Scholar]
- 204.Shang X, Marchioni F, Sipes N, et al. Rational design of small molecule inhibitors targeting RhoA subfamily Rho GTPases. Chem Biol. 2012;19:699–710. doi: 10.1016/j.chembiol.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Evelyn CR, Wade SM, Wang Q, et al. CCG-1423: a small-molecule inhibitor of RhoA transcriptional signaling. Mol Cancer Ther. 2007;6:2249–2260. doi: 10.1158/1535-7163.MCT-06-0782. [DOI] [PubMed] [Google Scholar]
- 206.Hayashi K, Watanabe B, Nakagawa Y, et al. RPEL proteins are the molecular targets for CCG-1423, an inhibitor of Rho signaling. PLoS One. 2014;9:e89016. doi: 10.1371/journal.pone.0089016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Chang LC, Huang TH, Chang CS, et al. Signaling mechanisms of inhibition of phospholipase D activation by CHS-111 in formyl peptide-stimulated neutrophils. Biochem Pharmacol. 2011;81:269–278. doi: 10.1016/j.bcp.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 208.Montalvo-Ortiz BL, Castillo-Pichardo L, Hernandez E, et al. Characterization of EHop-016, novel small molecule inhibitor of Rac GTPase. J Biol Chem. 2012;287:13228–13238. doi: 10.1074/jbc.M111.334524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Friesland A, Zhao Y, Chen YH, et al. Small molecule targeting Cdc42-intersectin interaction disrupts Golgi organization and suppresses cell motility. Proc Natl Acad Sci U S A. 2013;110:1261–1266. doi: 10.1073/pnas.1116051110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Zins K, Gunawardhana S, Lucas T, et al. Targeting Cdc42 with the small molecule drug AZA197 suppresses primary colon cancer growth and prolongs survival in a preclinical mouse xenograft model by downregulation of PAK1 activity. J Transl Med. 2013;11:295. doi: 10.1186/1479-5876-11-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Zins K, Lucas T, Reichl P, et al. A Rac1/Cdc42 GTPase-specific small molecule inhibitor suppresses growth of primary human prostate cancer xenografts and prolongs survival in mice. PLoS One. 2013;8:e74924. doi: 10.1371/journal.pone.0074924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Pelish HE, Peterson JR, Salvarezza SB, et al. Secramine inhibits Cdc42-dependent functions in cells and Cdc42 activation in vitro. Nat Chem Biol. 2006;2:39–46. doi: 10.1038/nchembio751. [DOI] [PubMed] [Google Scholar]
- 213.Sagawa H, Terasaki H, Nakamura M, et al. A novel ROCK inhibitor, Y-39983, promotes regeneration of crushed axons of retinal ganglion cells into the optic nerve of adult cats. Exp Neurol. 2007;205:230–240. doi: 10.1016/j.expneurol.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 214.Doe C, Bentley R, Behm DJ, et al. Novel Rho kinase inhibitors with anti-inflammatory and vasodilatory activities. J Pharmacol Exp Ther. 2007;320:89–98. doi: 10.1124/jpet.106.110635. [DOI] [PubMed] [Google Scholar]
- 215.Fang X, Yin Y, Chen YT, et al. Tetrahydroisoquinoline derivatives as highly selective and potent Rho kinase inhibitors. J Med Chem. 2010;53:5727–5737. doi: 10.1021/jm100579r. [DOI] [PubMed] [Google Scholar]
- 216.Feng Y, Yin Y, Weiser A, et al. Discovery of substituted 4-(pyrazol-4-yl)-phenylbenzodioxane-2-carboxamides as potent and highly selective Rho kinase (ROCK-II) inhibitors. J Med Chem. 2008;51:6642–6645. doi: 10.1021/jm800986w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Hsu TS, Chen C, Lee PT, et al. 7-Chloro-6-piperidin-1-yl-quinoline-5,8-dione (PT-262), a novel synthetic compound induces lung carcinoma cell death associated with inhibiting ERK and CDC2 phosphorylation via a p53-independent pathway. Cancer Chemother Pharmacol. 2008;62:799–808. doi: 10.1007/s00280-007-0667-5. [DOI] [PubMed] [Google Scholar]
- 218.Kast R, Schirok H, Figueroa-Perez S, et al. Cardiovascular effects of a novel potent and highly selective azaindole-based inhibitor of Rho-kinase. Br J Pharmacol. 2007;152:1070–1080. doi: 10.1038/sj.bjp.0707484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Castoreno AB, Smurnyy Y, Torres AD, et al. Small molecules discovered in a pathway screen target the Rho pathway in cytokinesis. Nat Chem Biol. 2010;6:457–463. doi: 10.1038/nchembio.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Patel RA, Liu Y, Wang B, et al. Identification of novel ROCK inhibitors with anti-migratory and anti-invasive activities. Oncogene. 2014;33:550–555. doi: 10.1038/onc.2012.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Maroney AC, Finn JP, Connors TJ, et al. Cep-1347 (KT7515), a semisynthetic inhibitor of the mixed lineage kinase family. J Biol Chem. 2001;276:25302–25308. doi: 10.1074/jbc.M011601200. [DOI] [PubMed] [Google Scholar]
- 222.Hudkins RL, Diebold JL, Tao M, et al. Mixed-lineage kinase 1 and mixed-lineage kinase 3 subtype-selective dihydronaphthyl[3,4-a]pyrrolo[3,4-c]carbazole-5-ones: optimization, mixed-lineage kinase 1 crystallography, and oral in vivo activity in 1-methyl-4-phenyltetrahydropyridine models. J Med Chem. 2008;51:5680–5689. doi: 10.1021/jm8005838. [DOI] [PubMed] [Google Scholar]
- 223.Tan I, Lai J, Yong J, et al. Chelerythrine perturbs lamellar actomyosin filaments by selective inhibition of myotonic dystrophy kinase-related Cdc42-binding kinase. FEBS Lett. 2011;585:1260–1268. doi: 10.1016/j.febslet.2011.03.054. [DOI] [PubMed] [Google Scholar]
- 224.Unbekandt M, Croft DR, Crighton D, et al. A novel small-molecule MRCK inhibitor blocks cancer cell invasion. Cell Commun Signal. 2014;12:54. doi: 10.1186/s12964-014-0054-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Lowe HI, Watson CT, Badal S, et al. Cycloartane-3,24,25-triol inhibits MRCKalpha kinase and demonstrates promising anti prostate cancer activity in vitro. Cancer Cell Int. 2012;12:46. doi: 10.1186/1475-2867-12-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Peterson JR, Lokey RS, Mitchison TJ, et al. A chemical inhibitor of N-WASP reveals a new mechanism for targeting protein interactions. Proc Natl Acad Sci U S A. 2001;98:10624–10629. doi: 10.1073/pnas.201393198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Peterson JR, Bickford LC, Morgan D, et al. Chemical inhibition of N-WASP by stabilization of a native autoinhibited conformation. Nat Struct Mol Biol. 2004;11:747–755. doi: 10.1038/nsmb796. [DOI] [PubMed] [Google Scholar]